Abstract
Human activities have resulted in the release and introduction into the environment of a plethora of aromatic chemicals. The interest in discovering how bacteria are dealing with hazardous environmental pollutants has driven a large research community and has resulted in important biochemical, genetic, and physiological knowledge about the degradation capacities of microorganisms and their application in bioremediation, green chemistry, or production of pharmacy synthons. In addition, regulation of catabolic pathway expression has attracted the interest of numerous different groups, and several catabolic pathway regulators have been exemplary for understanding transcription control mechanisms. More recently, information about regulatory systems has been used to construct whole-cell living bioreporters that are used to measure the quality of the aqueous, soil, and air environment. The topic of biodegradation is relatively coherent, and this review presents a coherent overview of the regulatory systems involved in the transcriptional control of catabolic pathways. This review summarizes the different regulatory systems involved in biodegradation pathways of aromatic compounds linking them to other known protein families. Specific attention has been paid to describing the genetic organization of the regulatory genes, promoters, and target operon(s) and to discussing present knowledge about signaling molecules, DNA binding properties, and operator characteristics, and evidence from regulatory mutants. For each regulator family, this information is combined with recently obtained protein structural information to arrive at a possible mechanism of transcription activation. This demonstrates the diversity of control mechanisms existing in catabolic pathways.
INTRODUCTION
Human activities have resulted in the release and introduction into the environment of a plethora of aromatic chemicals. Although the input of these synthetic chemicals may be smaller than the total amount of aromatic compounds released from decaying plant material, their novel structures or their quantities as single pure molecules induce major changes in microbial communities, which are the major recyclers of organic chemicals in nature (76). For example, large quantities of a single organic solvent offer a very specific metabolizable carbon source for specialized groups of microorganisms (161) and thus can act selectively for enrichment of those groups in the environment (87, 154). Compounds such as chlorinated solvents, herbicides, and pesticides often carry uncommon chemical structures, side chains, or functional groups, which can have toxic effects or provide new carbon sources for bacteria which have adapted their metabolism to degrade the compounds (191, 207, 261). The interest in discovering how bacteria are dealing with hazardous environmental pollutants has driven a large research community and has resulted in important biochemical, genetic, and physiological knowledge about the degradation capacities of microorganisms (40, 43, 76, 146, 191, 206, 207). A large variety of metabolic pathways have been discovered in very different microorganisms, fueling up-to-date online databases such as the Biocatalysis/Biodegradation Database (57). Knowledge about the biodegradation capacities of microorganisms is being applied directly in bioremediation practice (240), and individual biotransformation reactions are potentially very interesting for incorporation into chemical synthesis (230).
Less obviously associated with bioremediation applications, green chemistry, or production of pharmacy synthons have been studies of the regulatory mechanisms, which govern the expression of specific catabolic pathways (105). However, regulation of catabolic pathway expression has attracted the interest of numerous different groups, who have tried to unravel the molecular partners in the regulatory process, the signals triggering pathway expression, and the exact mechanisms of activation and repression. More recently, information about regulatory systems has attracted interest, with the potential of these systems being used as sensory mechanisms in whole-cell living bioreporters, genetically modified bacteria which can be used as sensors to measure the quality of aqueous, soil, and air environments (49, 96, 242). It soon was discovered that a large diversity of regulatory systems existed for mediating the expression of catabolic pathways. Furthermore, many related catabolic pathways did not carry the same regulatory system, which led to the hypothesis that regulatory systems and their target operons do not necessarily coevolve but seem to become associated independently (23, 39). As genomic, genetic, and biochemical data accumulated, regulatory proteins for catabolic gene expression were classified into other protein families, showing that they (as expected) were not unique to typical biodegradation pathways per se. In fact, many aromatic degradation pathways, typically those involved in the metabolism of aromatic amino acids, hydroxylated benzoates and phenylpropionic acids, ubiquinones, and aromatic amines, are very widespread among microbial species (43), leaving perhaps only the catabolic pathways for toxic and xenobiotic compounds to the groups of microorganisms usually considered for biodegradation (i.e., pseudomonads, sphingomonads, Rhodococcus spp., Ralstonia spp., and Burkholderia spp.). One would thus have to conclude that the capabilities of regulatory proteins to react specifically to substrates or intermediates of catabolic pathways have evolved many times independently; this is explained in more detail below.
Although excellent reviews exist on the diversity of catabolic pathways (76, 206), on evolutionary aspects (99, 262, 271), and on different details of specific regulator families (71, 139, 224, 233), no concise overview of the different regulatory systems involved in biodegradation pathways (or catabolic pathways, as we will refer to them often) exists in the literature, although a shorter treatise was published by Diaz and Priets (44). In our opinion, the topic of biodegradation is relatively coherent, and thus treatment of the different regulatory systems involved in catabolic pathways also deserves a coherent overview—although it is clear that there are no special “catabolic” regulatory proteins. In addition, several three-dimensional structures of regulatory proteins have recently been resolved, which allows a much more detailed prediction of how the actual activation and repression mechanisms of regulatory proteins take place. This might solve many questions which have arisen from more traditional genetic and biochemical approaches to transcription activation and repression. The current view of regulatory proteins is that of a set of sophisticated protein-DNA-RNA polymerase interaction machineries with sufficient diversity to promote or inhibit DNA transcription by RNA polymerase. What remains largely elusive, however, is how the machinery is turning.
The structure of this review is relatively straightforward. The various known regulatory families are treated individually, as far as possible with examples of catabolic pathways. Specific attention has been given to describing the genetic organization of the regulatory genes, promoters, and target operon(s); this is followed by a discussion integrating the knowledge of signaling molecules, DNA binding properties and operator characteristics, regulator-DNA structure, and evidence from regulatory mutants to arrive at a possible mechanism of transcription activation. What is not specifically treated in this review is how regulatory systems themselves are embedded in the host's physiology and controlled by other global regulatory systems. For information about these aspects, readers are referred to other specific overviews, such as references 23, 232, and 265. For sake of the overview, we have refrained from too many in-depth details of very specific individual transcription regulators.
LysR FAMILY OF TRANSCRIPTIONAL REGULATORS
Catabolic Operons Controlled by LysR-Type Regulators
LysR-type transcriptional regulators (LTTRs) comprise the largest family of prokaryotic regulatory proteins identified so far (84, 224). The family has expanded to over 100 members that have been identified in diverse bacterial genera. This diversity is also reflected in LTTRs associated with degradation pathways of aromatic compounds (Table 1) (44). A large group of LTTRs regulates a single target operon only, such as CatR controlling catBCA expression for catechol metabolism in Pseudomonas putida (215), ClcR controlling the clcABDE operon of plasmid pAC27 (33), TcbR controlling the tcbCDEF operon in plasmid pP51 of Pseudomonas sp. strain P51 (263), and CbnR controlling the cbnABCD operon for chlorocatechol metabolism in Ralstonia eutropha (163). TfdR (and/or its identical twin TfdS) also regulate an operon coding for chlorocatechol metabolism (i.e., the tfdDCEFB genes), but the same regulators also control the expression of two other operons, tfdA and tfdDIICIIEIIFIIBIIK, all of which are involved in degradation of 2,4-dichlorophenoxyacetate in R. eutropha JMP134 (and are located on plasmid pJP4 [Fig. 1A ]) (127). Similarly, CatR of P. putida can coregulate the expression of the pheBA operon for phenol degradation and of the catBCA genes when this operon is provided on an additional plasmid (104). Like TfdR and TfdS, the NahR protein is a master regulator for the regulon of naphthalene degradation and acts by controlling expression from both the nah operon, required for the metabolism of naphthalene to salicylate and pyruvate, and the sal operon, encoding the enzymes for salicylate conversion (225, 228, 274). In this case, both operons are present on the NAH7 plasmid. Two LTTRs have been implicated in the transcriptional control of benzoate degradation in Acinetobacter sp. strain ADP1. In this microbe, BenM regulates expression from the benABCDE and benPK operons, which encode the enzymes for conversion of benzoate to catechol. CatM also regulates expression from benPK but in addition regulates expression from the catBCIJFD operon, leading to further metabolism of cis,cis-muconate to the tricarboxylic acid cycle (Fig. 1B). The catA gene, encoding catechol 1,2-dioxygenase, which converts catechol to cis,cis-muconate, is not present in the two operons but is also regulated by both CatM and BenM (34, 212). CatM and BenM also interfere with expression of the pobA gene for 4-hydroxybenzoate metabolism, perhaps by preventing the expression of the pcaK uptake system (16). Although LTTRs are often associated with ortho-cleavage pathways of catechol, they are also involved in a large variety of other degradation pathways (Table 1). For example, NtdR controls the genes encoding nitroarene dioxygenase in Acidovorax sp. strain JS42 (125); the TsaR protein controls the expression of the tsaMBCD genes, which encode the first steps in the degradation of p-toluenesulfonate in Comamonas testosteroni T-2 (253); and BphR2 is involved in the regulation of a biphenyl catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707 (268). Preliminary data for regulation of the biphenyl pathway indicate that BphR2 is activating the expression of a first operon [bphA1A2(orf3)A3A4] whereas a second regulatory gene (bphR1, a regulator of the GntR family) is activating the expression of the second operon (bphX1X2X3D). LTTRs have also been identified in γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis (148), in aniline degradation in P. putida (67), and in catabolism of 3-phenylpropionic acid in Escherichia coli K-12 (42). A longer list of LTTRs involved in transcriptional control of aromatic compound degradation is presented in reference 44.
TABLE 1.
Regulatory proteins from the LysR family involved in expression control of pathways for degradation of aromatic compounds
| Regulatory protein | Bacterial genus | Regulated operon(s) | Pathway substrate | RBS | Position of:
|
Inducers | Accession no. (reference) | |
|---|---|---|---|---|---|---|---|---|
| RBS | ABS | |||||||
| CatR | Pseudomonas | catBCA (pheBA) | Catechol (phenol) | AGACC-N5-GGTAT | −79 to −54 | −47 to −34 | cis,cis-Muconate | A35118 |
| ClcR | Pseudomonas | clcABD | 3-Chlorocatechol | ATAC-N7-GTAT | −79 to −53 | −37 to −28 | 2-Chloro-cis,cis-muconate | A40641 |
| TcbR | Pseudomonas | tcbCDEF | 3,4-Dichlorocatechol | TTACG-N5-CGTAA | −71 to −56 | −34 to −24 | ND | A38861 |
| CbnR | Ralstonia | cbnABCD | 3-Chlorocatechol | TTACG-N5-CGTAA | −80 to −50 | −50 to −20 | 2-Chloro-cis,cis-muconate | BAA74529 |
| TfdR/S | Ralstonia | tfdDIICIIEIIFIIBIIK, tfdCDEFB, tfdA | 2,4-Dichlorophenol | ATAC-N7-GTAT (?) | −59 to 75 (?) | NDa | Chloro-cis,cis-muconate | P10086 |
| CatM | Acinetobacter | catBCIJFD, catA, benPK | Catechol | ATAC-N7-GTAT | −72 to 58 | ND | cis,cis-Muconate | P07774 |
| PcaQ | Agrobacterium | pcaDCHGB | Protocatechuate | TAA-N7-TTA (?) | ND | ND | β-Carboxy-cis,cis-muconate, γ-carboxymuconate | AAA91130 (179) |
| BenM | Acinetobacter | benABCDE (catA), benPK | Benzoate (catechol) | ATAC-N7-GTAT | −78 to −50 | −50 to −29 | Benzoate and cis,cis-muconate (synergistic activation) | AAC46441 |
| TfdT | Burkholderia | tfdC | 4-Chlorocatechol | ND | ND | ND | 3- or 4-Chlorocatechol, 2- or 3-chlorobenzoate | BAB56008 |
| NahR | Pseudomonas | nahABCFDE, salGHINL | Naphthalene, salicylate | TTCA-N6-TGAT | −80 to −47 | Near −3 | Salicylate | A31382 |
| NtdR | Acidovorax | ntdAaorf2AbAcAd | 2-Nitrotoluene | ND | ND | ND | 2,4- and 2,6-Dinitrotoluene, salicylate, anthranilate | AAP70492 |
| LinR | Sphingomonas | linE-linD | Chlorohydroquinone | ATTCA-N5-TGAAT (?) | ND | ND | 2,5- and 2,6-Dichloro- and chlorohydroquinone | BAA36280 |
ND, not determined.
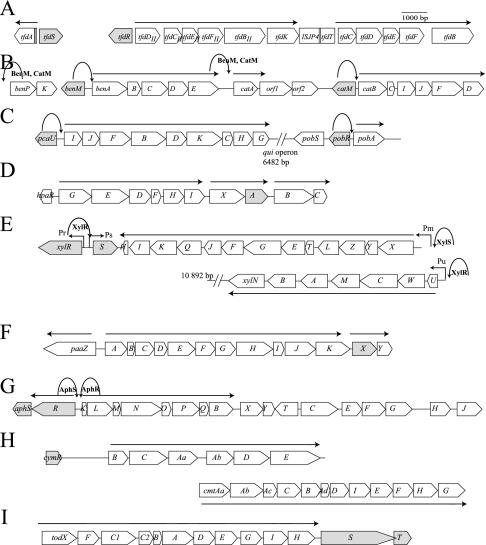
FIG. 1.
Typical examples of the genetic organization of pathways for degradation of aromatic compounds. The regulatory genes are indicated in grey. (A) The tfd operons of R. eutropha JMP134(pJP4). The LysR-type transcription regulator TfdR/S regulates the tfdA, tfdC, and tfdDII promoters. (B) The ben and cat operons of Acinetobacter sp. strain ADP1. benM and catM code for LysR-type transcription regulators, which act on four promoters: benP, benA, catA, and catB. (C) The pca and pob clusters of Acinetobacter sp. strain ADP1. The genes pcaU and pobR code for IclR-type regulators, acting on the pcaI and pobA promoters. (D) The hpa cluster of E. coli with HpaA as the XylS/AraC-type regulator. (E) The xyl operons for toluene degradation present on the TOL plasmid of P. putida mt2. XylR is the NtrC-type regulator acting on Pu and Ps, whereas XylS is the exemplar for the XylS/AraC family and acts on the Pm promoter. (F) The paa cluster in E. coli strain W, with the GntR-type regulator PaaX regulating the paaA promoter. (G) The aph cluster in C. testosteroni strain TA441, encoding the aphS (GntR-type) and aphR (XylR-type) regulators. (H) The cym and cmt clusters in P. putida F1 with the TetR-type regulator cymR. (I) The tod cluster of P. putida DOT-T1 with the two-component regulatory system todST. Straight arrows represent transcripts produced after specific regulatory action of the indicated regulator. Parabolic arrows point to the site of action of a specific regulator in cases when multiple regulators are involved. Hooked arrows indicate specific promoter names, where necessary.
In general, the gene for LTTRs lies upstream of its target-regulated gene cluster and is transcribed in the opposite direction (Fig. 1A and B), but exceptions to this rule exist. For example, the gene for the SalR regulator in Acinetobacter sp. strain ADP1 is present within the transcriptional unit salRA. The salA gene is coding for salicylate hydroxylase, which converts salicylate to catechol. Experimental evidence suggests that SalR is regulating the expression of itself and of salA (101). The phnS gene (coding for a further uncharacterized LTTR) is cotranscribed as the first gene of an operon including the phnFECDAL catabolic genes for naphthalene and phenanthrene degradation in Burkholderia sp. strain RP007 (119).
All identified LTTRs involved in aromatic degradation pathways act as transcriptional activators for their target metabolic operons in the presence of a chemical inducer, which is usually a pathway intermediate (33, 128, 212, 215, 227). In some cases, the effectors are (substituted) muconates which have lost their aromatic character, whereas in other systems aromatic compounds act as effectors (e.g., salicylate, catechol, and nitrotoluene [Table 1]). There is also experimental evidence for competition of several compounds on the regulatory protein. For example, fumarate reversibly inhibits the formation of the clcA transcript in in vitro transcription assays in the presence of purified ClcR and 2-chloro-cis,cis-muconate (138). The presence of cis,cis-muconate decreases the affinity of the BenM protein for benzoate (31). All LTTRs repress their own expression, and both autorepression and activation of the catabolic operon promoter are exerted from the same binding site, which is called the regulator or repressor binding site (RBS) (20, 216, 227). Autorepression was not influenced by the presence of an inducer in the case of BenM (20), but expression of the clcR and tcbR promoters was slightly enhanced in the presence of an inducer (33, 263). Relatively few data exist on autorepression mechanisms, since most studies on LTTRs have focused on the mechanisms of target gene activation.
Structure and Conformation
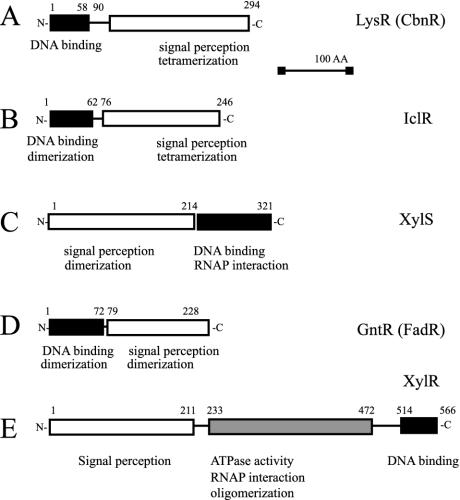
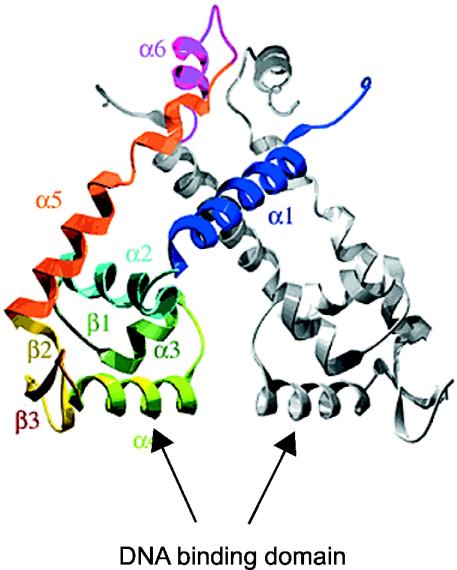
LTTRs involved in the degradation of aromatic compounds are composed of 394 to 403 amino acid residues with a molecular mass of between 32 and 37 kDa. All the evidence presented so far points to tetramers being the active form of LTTRs. ClcR and CatR have been identified as dimers in solution (32, 181), but two dimers are needed to bind DNA (141). CatM, BenM, and CbnR were found to form tetramers (20, 30, 155), and NahR probably also acts as a tetrameric form (226). The well-studied LTTR CysB, which is not involved in the activation of pathways for aromatic-compound degradation, also forms tetramers in solution (88, 145). Only TsaR remains as monomer in solution (253). LTTRs have a conserved domain organization, which has been determined from mutagenesis studies and sequence alignments (224). A DNA binding region with a predicted helix-turn-helix (HTH) motif is located in the 66 N-terminal amino acid residues of the protein, two regions located between residues 95 to 173 and residues 196 to 206 are involved in inducer recognition, and one region between residues 227 and 253 is supposed to be involved in multimerization (Fig. 2A).
FIG. 2.
Schematic domain organization of the different regulator families. The domain containing the HTH DNA binding motif is indicated in black. The domain bound by the chemical inducer is indicated in white.
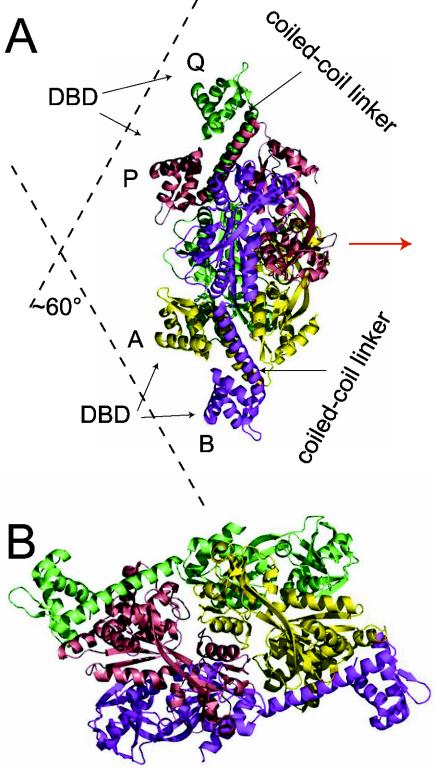
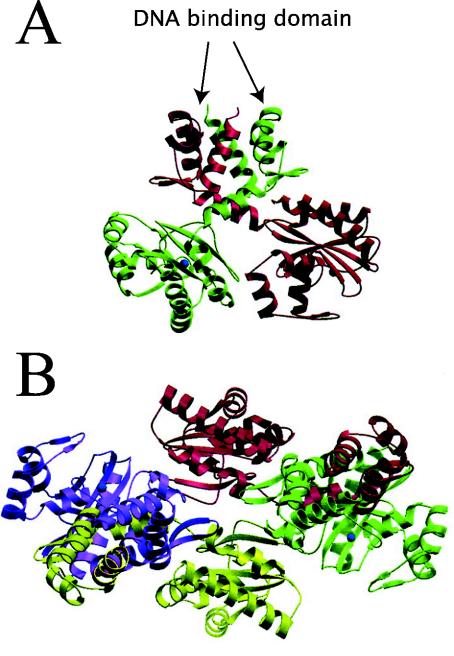
Recently, the first complete LTTR has been crystallized (CbnR) (155). Crystals of BenM and CatM devoid of their N-terminal DNA binding domain have also been obtained, but their structures have not been resolved yet (29). CbnR crystallized as a tetramer (Fig. 3) with two main parts, the four DNA binding domains (residues 1 to 58) and a central body (155). The four DNA binding domains have no interactions with each other, whereas the central body of the tetramer is composed of four intertwined regulatory domains (residues 88 to 294 of each subunit). The tetramer structure can be regarded as a dimer of a dimer, whereby each dimer is composed of two subunits in different configurations. The two subunits in each dimer are connected through the coiled-coil linker (residues 59 to 87), which at the same time separates the DNA binding domain and the central body (Fig. 3). Subunit A of the AB dimer interacts with the regulatory domains of both subunits Q and P of the other (PQ) dimer, whereas subunit B interacts only with P (Fig. 3). The structure seems to allow easy transmission of conformational changes, for example in the central body, the DNA binding domains. The DNA binding motifs in the tetramer are presented in such a way as to form a V-shaped structure, which matches exactly with the distance and configuration of the two DNA binding sites (Fig. 3). Although the mode of action and the location of the effector binding pockets still need better definition, this structural model beautifully fits and explains previous experimental evidence with the LTTRs.
FIG. 3.
Tetrameric structure of CbnR in side (A) and top (B) views (155). Subunits A, B, P, and Q are shown in yellow, magenta, cyan, and green, respectively. The symmetry axis in the molecule is shown as a red arrow. The location of the coiled-coil linkers (see the text) and DNA binding domains (DBD) are indicated, which would impose a 60°C bending angle on the binding site on the DNA. Reprinted from reference 155 with permission from the publisher and from the authors.
Mechanisms of Activation
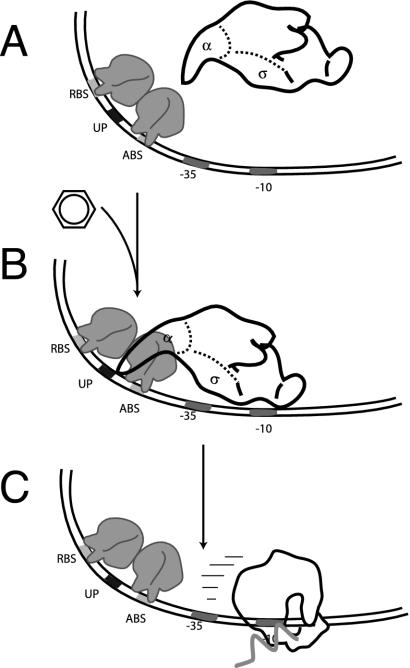
Due to their tetrameric form, LTTRs interact with several sites on the DNA of the promoter region. Classically, DNA interactions are shown by DNase I footprinting techniques and purified regulatory protein. Interestingly, DNase I cleavage patterns of the protein-bound nucleotide regions were similar for ClcR, CatR, TcbR, CbnR, and, to a lesser extent, NahR. The different interactions are explained in more detail for ClcR. ClcR binds the clcA promoter irrespective of the presence of inducer (32, 141, 182). However, in the absence of inducer, ClcR protects a 27-bp region (RBS) from −79 to −53 and a 10-bp region (activator binding site [ABS]) from −37 to −28 relative to the transcription start site. Each region is supposed to be bound by one of the two dimers in the tetramer (Fig. 4). The RBS contains an interrupted inverted repeat, ATAC-N7-GTAT, with the consensus LTTR binding motif T-N11-A (224). Two hypersensitive bands (−52/−51 and −42) also show up on ClcR-bound promoter DNaseI footprints. This may reflect the bending imposed on the DNA by the V-shaped configuration of the tetramer (Fig. 3). In the presence of inducer, the hypersensitive band at −42 disappeared and occupation of the ABS shifted from −37 to −41 (141), suggesting that the bending angle is relaxed on interaction with the effector. Conformational changes on effector binding could be detected for benzoate and cis,cis-muconate binding to BenM (31). Measurements of the bending angle of the clcA promoter in the absence (71°) and in the presence (55°) of effector corroborate this idea of bending relaxation (140). Similar bending angles were reported for CbnR at the cbnA promoter in the absence and presence of inducer, although for CbnR no changes in DNase I footprints were observed as for ClcR (163). The role of the RBS and ABS is not completely clarified. Although ClcR and CbnR contacted both the RBS and ABS in the absence of inducer, CatR did not (215, 216). It was concluded that the regulatory proteins have a higher binding affinity to the RBS than to the ABS, since a fragment with only the ABS is not bound by ClcR or CatR (139, 181). However, contacts to the ABS are supposed to be necessary for mediating interactions with RNA polymerase (RNAP), and fragments with only the RBS do not lead to in vitro transcript production in the presence of ClcR, RNAP, and inducer (139).
FIG. 4.
Schematic steps in transcription activation by LysR-type transcription regulators. (A) Two subunits of the regulator tetramer binds to the RBS and two other subunits bind to the ABS present on the −35 promoter region. (B and C) In the presence of an inducer, the regulatory protein shifts its interaction on the ABS to −42. By interacting with the α-CTD domain of the RNAP, it directs this to bind a so-called UP-DNA sequence motif, which is located between the RBS and ABS (B). This increases the binding affinity of RNAP for the promoter and initiates transcription (C). mRNA is drawn as a zig-zagged arrow.
Further contacts seem to occur between the regulatory protein complex and the C-terminal domain of the α-subunit (α-CTD) of RNAP. This was concluded from the lack of mRNA synthesis in in vitro transcription experiments with CatR (or ClcR), the catB (clcA) promoter, cis,cis-muconate, and RNAP devoid of the α-CTD (28, 139). A direct interaction of NahR with the α-CTD of RNAP has been demonstrated by using a yeast two-hybrid system (178), and this interaction was not influenced by the presence of salicylate. Chugani et al. (28) postulated that the regulatory complex was directing the α-CTD of RNAP to a region between the RBS and the ABS, which they called the UP-motif (Fig. 4). Interaction with the UP-motif would increase the affinity of RNAP to the promoter (28). However, exactly how this process would lead to transcription activation is not known. The degree of DNA supercoiling has also been implicated in controlling the level of transcription activation from LTTR-dependent promoters. This has been concluded from in vitro transcription experiments with CatR, in which supercoiled templates produced no mRNA transcript except in the presence of cis,cis-muconate whereas linearized templates did (28). Perhaps the regulatory protein can overcome the torsional constraint on the DNA in the presence of inducer, thereby facilitating open-complex formation by RNAP.
In some cases, a third DNA binding site further downstream of the transcriptional start site has been identified. For example, CatR binds with low affinity to a region between +162 and +196 of the catB gene (27). This site has been named the inhibitor binding site (IBS) and is supposed to prevent excessively high expression from the catBCA operon by titrating CatR protein. The affinity of binding of CatR to the IBS increased at higher inducer concentrations, and catB promoter fragments containing the IBS had a three- to fourfold lower expression than those without IBS (27). IBS regions have also been identified in the pheBA operon and clcABD operon (27).
BenM has the unique peculiarity (for the moment) that the inducers benzoate and cis,cis-muconate can have synergistic effects on the activation process compared to the effects of each inducer alone (20, 31, 34). In addition, BenM represses benA transcription in the absence of inducer. Benzoate and cis,cis-muconate alone had very different effects on the patterns observed in DNase I footprints of BenM on the benA promoter than did both inducers simultaneously. In the absence of inducer, BenM bound two areas with dyad symmetry, site 1 (ATAC-N7-GTAT) at positions −57 to −71 and site 3 (ATTC-N7-GTAT) at positions −5 to −19. Several hypersensitive sites were detected (at positions −50, −45, −39, −36, −34, −29, and −24), suggesting again a clear bending of the DNA imposed by the BenM regulatory complex. In the presence of cis,cis-muconate or benzoate, BenM still protected site 1 but no longer protected site 3. When both inducers were present simultaneously, the number of hypersensitive sites was reduced and instead a region called site 2 (which also contained a dyad symmetry motif, ATAC-N7-GTGT, located between −36 and −50) was protected from DNase I cleavage (20). The authors hypothesized that BenM binding to site 3 (which overlaps with the −10 region) causes the observed repressive effect in the absence of inducer. In the presence of inducer, is BenM released from site 3, enabling RNAP to access the promoter, whereas BenM binding to site 2 would lead to more productive contacts to RNAP and a higher transcriptional output.
IclR FAMILY OF TRANSCRIPTIONAL REGULATORS
Catabolic Operons Controlled by IclR Regulators
IclR-type regulators have a similar structure as the LysR-type regulators (224), but rather dissimilar amino acid sequences distinguish the two families. IclR-type regulators are generally transcriptional repressors (86, 132, 156, 245); however, those which control catabolic pathways have all been described as activators (Table 2). For example, PcaU of Acinetobacter sp. strain ADP1 (75), PcaR of P. putida (213), PcaR of Agrobacterium tumefaciens, and CatR and PcaR of Rhodococcus opacus 1CP (58, 59) are activators for the ortho-cleavage pathways which they regulate. However, in the absence of inducer they may still act as repressors, as was shown for PcaU (254). Further IclR members are MhpR, which activates the meta-cleavage pathway in 3-(3-hydroxyphenyl)propionic acid degradation by E. coli (61); PobR, which is the activator for the 4-hydroxybenzoate degradation pathway in Acinetobacter sp. strain ADP1 (45), and OhbR, controlling the genes for the oxygenolytic ortho dehalogenation of halobenzoates (256).
TABLE 2.
Major IclR family members regulating degradation pathways of aromatic compounds
| Regulatory protein | Bacterial genus | Regulated operon(s) | Pathway substrate | RBS | Positiona | Inducer | Accession no. |
|---|---|---|---|---|---|---|---|
| PcaU | Acinetobacter | pcaIJFBDKCHG | Protocatechuate | TGTTCGATaATCGAACCAA | −53 to −94 | Protocatechuate | AAC37157 |
| PobR | Acinetobacter | pobA | p-Hydroxybenzoate | TGTCCGATgATCGGACAAA | −53 to −94 | p-Hydroxybenzoate | A36893 |
| PcaR | Pseudomonas | pcaIJ, pcaBDC, pcaF, pcaK | Protocatechuate | GTTCGATaATCGCAC | Around −10 | β-Ketoadipate | Q52154 |
| MhpR | Escherichia | mhpABCDFE | 3-(3-Hydroxyphenyl)propionic acid | GGTGCACCtGGTGCACA | −50 to −66 | 3-(3-Hydroxyphenyl)propionic acid | P77569 |
Position relative to the transcription start site.
In general, the gene for the IclR-type regulator lies upstream of its target gene cluster and is transcribed in the opposite direction (Fig. 1C). An exception is the pcaR gene of P. putida (160). PcaR actually controls four distinct gene clusters required for the degradation of protocatechuate to tricarboxylic acid cycle intermediates and of the 4-hydroxybenzoate transporter PcaK (160). The pcaR gene lies upstream of pcaK and is transcribed in the same direction (90, 159, 213). PcaR from P. putida and PobR and PcaU from Acinetobacter sp. strain ADP1 repress their own expression (46, 81, 254). It is thought that the mechanism of autorepression is different among IclR-type regulators, since not all of them bind at the same position on the promoter region (75, 81, 213) and since for some of them the addition of effectors changes the expression of the regulatory gene itself. For example, expression of pcaR and pobR is independent of the usual effectors for PcaR and PobR (46, 81) but pcaU expression increases in the presence of effectors for PcaU (75).
Structure and Conformation
The size of IclR-type regulators is around 238 to 280 amino acid residues (25 to 30 kDa) (59, 61, 82, 112, 256). IclR-family members have an HTH DNA binding motif in the N-terminal domain (45, 61) and a C-terminal domain involved in subunit multimerization and effector binding (112). This was confirmed by the crystal structure of IclR from Thermotoga maritima 0065 (Fig. 5) (275), which showed that the amino acid residues previously identified to be involved in PobR inducer specificity (112) are present in IclR and comprise a pocket in the C-terminal domain (275). Although PcaU and PcaR from P. putida formed dimers in solution (81, 193), IclR crystallized as a dimer of a dimer with an asymmetric configuration (275). The two subunits within one IclR dimer interact solely at the interface of their DNA binding domains. As a consequence, the distance between the HTH motifs within one dimer is relatively short and results in a structure favorable for binding relatively short (12- to 14-bp) palindromic DNA sequences with specific contacts predominantly in the major groove of the DNA. The C-terminal domains do not contact each other in the dimer, but they do bridge with the C-terminal domains from the neighboring dimer, which is oriented in asymmetric fashion (Fig. 5). It is not really clear how the tetramer interacts with the DNA. Mass spectrometric data revealed that four IclR subunits are present per DNA containing one single palindrome (see below). The presumed ligand binding region is close to the region involved in tetramerization, suggesting that ligand binding and tetramerization may be linked. Moreover, the tetramer seems to be the active DNA binding form since, in mass spectrometric studies, four IclR subunits bound one synthetic DNA containing one single palindrome. The stoichiometry of two protein subunits for one DNA molecule was never detected (48).
FIG. 5.
IclR dimer and tetramer arrangements as derived from the crystal structure (275). (A) The dimer interface is formed exclusively between the two HTH DNA binding domains. Monomers are colored red or green. (B) The tetramer viewed from the top is composed of two asymmetric dimers shown in four different colors: red and green for one asymmetric unit, and yellow and magenta for the other. The tetramer interface is formed exclusively between signal binding domains. Reprinted from reference 275 with permission from the publisher and from the authors.
Mechanism of Transcription Activation
There is no clear consensus on the binding site for IclR members (Table 2). For example, the MhpR binding site is formed by one 15-bp palindrome that lies 50 to 66 nucleotides upstream of the mhpA transcription start site (252), but those of PcaU and PobR are three perfect 10-bp sequence repetitions that lie between 53 and 94 bp upstream of the pobA and pcaI transcription start sites, respectively (46, 75, 193). Two of the 10-bp sequence repetitions form one palindrome, and the third repeat is oriented opposite to the second one but separated by another 10 bp (193). In contrast to those, the PcaR binding site is formed by a series of 15 nucleotides present twice in the pcaI promoter and overlapping with the −10/−35 promoter region (81). One might thus conclude that when the regulators conform to their binding sites, they must be different between MhpR, PobR, PcaU, and PcaR.
IclR-type regulators bind their promoter DNA in the absence of effector, and adding effector molecules had no effect on the affinity of the protein-DNA interactions displayed by purified PobR, PcaU, PcaR, and MhpR (46, 75, 81, 252). When, however, the chemical effector was added to a mixture of regulator (in this case PcaR), purified σ70-RNAP, and a pcaI promoter fragment, the formation of a PcaR-RNAP-DNA complex was enhanced compared to the situation without effector (81). The authors suggested that the role of the regulatory protein might be to favor the recruitment of RNAP to the promoter, perhaps by optimizing the critical distance between the −35 and −10 elements in the pcaI promoter from 16 to 17 bp (81). The same effect of enhanced complex formation was also found for SoxR and MerR (6, 85). Also, in the mhpA promoter the −35 and −10 elements are separated by 16 bp (252). However, the pobA promoter (regulated by PobR) already has a spacing of 17 bp (75), and thus the generality of this promoter distance optimization as an activation mechanism for IclR-type regulators is debatable.
AraC/XylS FAMILY
Catabolic Operons Controlled by AraC/XylS-Type Regulators
For years the XylS protein was the only member of the AraC family which was involved in expression control of a catabolic operon, namely, the meta-cleavage pathway operon for degradation of meta-toluate located on the TOL plasmid in P. putida (92). Recent alignment studies have shown that more than 300 proteins, some of which may be involved in the control of catabolic pathways, contained a typical stretch in the C-terminal part of about 100 amino acids that would classify them as AraC/XylS-type regulators (251). The remaining part of the proteins can be very different, though (see below). AraC/XylS-type regulators for catabolic operons generally act as transcription activators in the presence of a chemical effector molecule (Table 3). Some of the more recently discovered catabolic gene regulators of the AraC/XylS family are PobC of P. putida WCS358 (13), PobR of Azotobacter chroococcum (198), and PobR of Pseudomonas sp. strain HR199 (173). PobC and PobR regulate the expression of a pobA gene for p-hydroxybenzoate hydroxylase. HpaA is a XylS-type protein found in E. coli and regulates the expression of the hpaCB genes for p-hydroxyphenylacetate hydroxylase (195). Other family members identified in degradation pathways are CbdS, controlling the 2-halobenzoate dioxygenase genes in Burkholderia sp. strain TH2 (246); BenR, controlling the benzoate 1,2-dioxygenase operon in P. putida (36); CadR, controlling the genes for 2,4-dichlorophenoxyacetic acid degradation in Bradyrhizobium sp. strain HW13 (109); and IpbR, which regulates the expression of isopropylbenzene degradation in P. putida RE204 (51).
TABLE 3.
Major AraC/XylS-type regulators controlling the expression of degradation pathways for aromatic compounds
| Regulator | Bacterial genus | Regulated operons | Pathway substrate | RBS | Positionsa | Inducer(s) | Accession no. |
|---|---|---|---|---|---|---|---|
| XylS | Pseudomonas | xylXYZLTE, GFKQKIH | m-Toluate | TGCA-N6-GGNTA | −35 to −49, −56 to −71 | Benzoate, 2- and 3-methyl-, 2,3-, 2,5-, and 3,4-dimethylbenzoate | AAA26029 |
| CbdS | Burkholderia | cbdABC | 2-Halobenzoate | TGCA-N7-GGATA (?) | −44 to −59(?), −67 to −81 | 2-Chloro-, 2-bromo-, and 2-iodobenzoate, o-toluate, benzoate | BAB21583 |
| PobC | Pseudomonas | pobA | p-Hydroxybenzoate | NDb | ND | p-Hydroxybenzoate, protocatechuate | CAB64665 |
| PobR | Azotobacter | pobA | p-Hydroxybenzoate | ND | ND | p-Hydroxybenzoate | AAF03756 |
| IpbR | Pseudomonas | ipbABCED | Isopropylbenzene | AAA(A/T)AACGGATA (?) | −35 to −46(?), −56 to −67 | Isopropylbenzene, naphthalene, trichloro- ethylene | AF006691 |
| HpaA | Escherichia | hpaBC | 4-Hydroxyphenylacetate | AAAAGT (*2) (?) | −41 to −68(?) | 3-, 4-Hydroxy- and phenylacetate | Z37980 |
| BenR | Pseudomonas | benABC | Benzoate | (G/T)GCA-N(5/6)-GGATA | −70 to −56(?), −49 to −35 | ND | AAF63447 |
| CadR | Bradyrhizobium | cadABKC | 2,4-Dichlorophenoxyacetic acid | ND (?) | ND | 2,4-dichloro- and 4-chlorophenoxyacetic acid | BAB78520 |
Relative to the transcription start site.
ND, not determined.
Most of the genes for XylS-type regulators lie upstream of their target operon, but, in contrast to lysR-type genes, they are transcribed in the same direction as the target genes. For example, cbdS, hpaA, cadR, benR, and phcT are all transcribed in the same direction as the genes they are regulating (36, 109, 196, 246). The pobR and pobC genes, on the other hand, are transcribed in the opposite direction (13, 173, 198). The xylS gene itself is also transcribed in the opposite direction to the meta-cleavage pathway operon (Fig. 1E) but is located downstream of it (93).
Expression of the genes for XylS-type regulators is not controlled just by themselves but in many cases by other activators or cascades. The best-studied example is XylS. Expression of xylS is strongly dependent on another regulatory protein XylR (see further below). XylR stimulates xylS-transcription from a σ54-dependent promoter (called Ps1) when cells are grown on xylenes (69, 134). In the absence of suitable aromatic inducers, the xylS gene is expressed at low constitutive levels from a σ70-dependent promoter called Ps2 (69). The hpaA regulatory gene has the unusual feature of being cotranscribed with the hpaX gene, which lies directly upstream of hpaA (194, 195). Expression of hpaA takes place from two promoters, one located directly upstream in the coding region of hpaX and the second located upstream of hpaX, in which case a bicistronic transcript is produced (195). Another example is provided by the phcT regulatory gene. PhcT is at the top of a regulatory cascade controlling phenol degradation (250). PhcT enhances transcriptional activation of the phc operon for phenol degradation by interacting with another regulator, PhcR, but at the same time it represses expression of the phcR gene itself. The dual action of PhcT is possible because its binding site is located in the intergenic region between phcR and the phc phenol operon, which themselves are oriented divergently (250).
Structure and Conformation
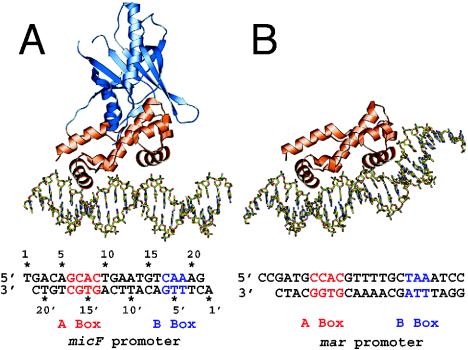
XylS/AraC-type regulators involved in degradation control of aromatic compounds are typically between 293 and 322 amino acids in size, with a molecular mass around 35 kDa. One exception is PhcT, which has only 257 amino acid residues with a predicted molecular mass of 28 kDa. As far as has been determined, most AraC members form dimers in solution (21, 24). The solution state of XylS has not been determined directly, since the protein is very difficult to purify. Ruiz et al. found that a peptide containing only the N-terminal end of XylS was able to dimerize (217). The C-terminal end of XylS/AraC-type regulators is the most highly conserved part among this protein family and contains two HTH motifs within a region of 100 or so amino acid residues (Fig. 2C) (71). In XylS the two HTH motifs comprise residues 231 to 252 and 282 to 305 (133), and a truncated XylS polypeptide with only the 112 C-terminal residues was capable of binding and activating transcription from the Pm promoter (103). Two AraC family members (i.e., Rob and MarA) have recently been crystallized (114, 210) (Fig. 6), but they do not match the most common AraC architecture, since MarA is a single-domain protein (with only a DNA binding domain) and Rob has the DNA binding domain located at the N terminus of the protein (52). MarA and Rob were crystallized as one monomer bound to a DNA fragment containing one binding site, CCAC-N7-TAA and GCAC-N7-CAA, respectively (Fig. 6). The two HTH motifs of MarA bind to adjacent segments of the major groove, with the helical axes of the recognition helices almost parallel to the DNA base pairs (Fig. 6). Because the recognition helices in the HTH motifs protrude from the same face of the protein, MarA binds to one face of the DNA. The two HTH motifs are connected by a rigid central linker helix that imposes their orientation and distance on the DNA binding site. Binding results in a bending of the DNA at an angle of 35°, because the two recognition helices are separated by 27Å whereas the pitch of the DNA B-form is 34 Å (210). The structure of the Rob DNA binding domain is quite similar to that of MarA, but two main differences were observed in the protein-DNA complex (Fig. 6). First, the DNA was not bent in the structure obtained with Rob, and second, only one protein helix was contacting the major groove of the DNA (114). In addition, nonspecifically attached Rob subunits were present in the crystallographic structure on the other side of the target DNA, which could be interpreted as an artifact (114). Martin and Rosner suggested that the presence of the second monomer prevented bending of the DNA and hence interaction of the second protein helix to the major groove of the DNA (137). On the other hand, it has been suggested that the Rob structure represents the state required for transcription activation, since conserved residues of one protein helix in the HTH-motif are not contacting the DNA but are available for interactions with the transcriptional machinery (114). Gallegos et al. had already previously suggested that conserved amino acids in this part of the HTH motif may contact the transcriptional machinery whereas the first helix would be more variable and involved in recognition of the target promoter (71).
FIG. 6.
Structures of the Rob-micF and the MarA-mar DNA complex (114). The structurally similar N-terminal DNA binding domains of Rob and MarA are colored orange, and the unique C-terminal domain of Rob is shaded blue. (A) The N-terminal HTH motif of Rob contacting bases of the binding site. (B) MarA and the induced bend on its DNA binding site. Both HTH modules are situated in adjacent major-groove surfaces on one side of the DNA. Reprinted from reference 114 with permission from the publisher and from the authors.
The N-terminal end (also called the regulatory domain) of XylS/AraC members is not well conserved. The N-terminal domain of AraC has been crystallized in the absence and in the presence of arabinose, but it has such a low percent identity to XylS (12.5%) and XylS-mediated transcription activation from Pm is so different from AraC and the PBAD promoter (52, 71) that extrapolation of the AraC structure to XylS is doubtful. Structural ideas were retrieved, however, from genetic evidence and different XylS mutants. For example, single point mutations in the N-terminal end of XylS resulted in proteins responsive to other aromatic inducers, which suggests a role for the N-terminal end in effector recognition (Fig. 2C) (144, 218). Furthermore, some mutations in this region impaired the in vitro formation of cross-linking products between XylS peptides containing only the N-terminal part fused to the maltose binding protein. This was interpreted such that the N-terminal part of XylS would be involved in dimerization as well (217, 239). Both C- and N-terminal parts can act independently (218, 219). For example, mutations in both the C- and N-terminal regions of XylS can yield semiconstitutive mutants (203) or suppressors (143). This suggests there is some form of “cross talking” between the two domains, but the mechanism of this remains to be resolved.
Mechanisms of Activation
An understanding of the activation mechanisms by the “catabolic” members of the XylS/AraC family may be biased by the relatively large body of knowledge about XylS compared to others. A first peculiarity of the XylS system is that activation can take place in two modes, i.e., at low xylS expression levels in the presence of an inducer (66, 204) and on overexpression in the absence of inducer (91, 94, 142, 202, 241), although the latter mechanism may have no physiological meaning for the xyl pathway. At least BenR seems to behave similarly to XylS in this respect (36).
XylS probably binds as two monomers or one dimer to two direct sequence repeats in the Pm promoter. This can be concluded from crystallographic data for the two AraC members Rob and MarA (114, 210). DNase I footprinting of the XylS interaction on the Pm promoter showed a protected region from nucleotides −78 to −28 upstream of the transcription start site (102, 103). The protein bound along one side of the DNA, covering four helices and thereby making specific contacts in four adjacent major grooves (102). By earlier site-directed mutagenesis studies, a consensus direct repeat (TGCA-N6-GGNTA), which is present twice in the Pm promoter between −35 and −49 and between −56 and −70, had been identified as the binding determinant (70, 78, 107). In the presence of m-toluate, the degree of protection by XylS on the Pm promoter increased (102, 103), suggesting that the affinity of XylS to its binding site had changed. Subsequent in vivo methylation studies showed an altered methylation protection pattern of the XylS-bound region in the presence of m-toluate, which indicates that conformational changes take place under inducing conditions (78).
Strangely, XylS-mediated transcription activation from the Pm promoter is independent of σ70-RNAP (136) but during exponential growth is dependent on the alternative sigma factor σ32, which normally is involved in the heat shock response (135). This has led to the suggestion that the presence of the chemical effector m-toluate is triggering a similar response for the cell to the heat shock response. In the late exponential and stationary phases, the sigma requirement for the Pm promoter changes to σ38 (135, 136). However, σ38-RNAP uses exactly the same promoter region in the Pm promoter as that used by σ32 (136). Mutants can be obtained with mutations in the N-terminal domain of XylS which change the requirement of XylS for σ38 to σ70 (203).
A large number of XylS/AraC members, including XylS itself, seem to require contact with α-CTD and with parts of the σ-factor in order to achieve full activation (15, 53, 116, 117, 131, 220). This was concluded mostly from in vitro transcription experiments and transcriptional reporter gene fusions with mutated RNAPs. A direct interaction between XylS and RNAP has not been demonstrated conclusively (251), but introducing 2 bp between the nucleotides at positions −36 and −37 of the Pm promoter abolished XylS-dependent transcription activation, probably because XylS and RNAP were offset and could no longer interact (77). In vivo dimethyl sulfate footprinting suggested that XylS can retain RNAP on the Pm promoter in the absence of inducer. In the presence of benzoate, RNAP was released, with concomitant transcription initiation (147).
What can be concluded about the role of XylS in the process of transcription activation? Apparently, XylS prevents open-complex formation in the absence of inducer but somehow recruits RNAP to bind to the Pm promoter. In the presence of inducer, XylS changes its own conformation and that of its binding site, which allows isomeratization of promoter-RNAP to form the open complex (251). Exactly how the effector mediates these changes and how this is transmitted to the DNA and RNAP remain elusive.
GntR-TYPE REGULATORS
Catabolic Operons Controlled by GntR-Type Regulators
Regulation by repression is rare for catabolic pathways. Ferrandez et al. were the first to report that the product of the paaX gene repressed the expression of two operons for phenylacetic acid (PA) degradation (paaABCDEFGHIJK and paaZ [Fig. 1E]) in E. coli strain W (63). These operons code for an aerobic catabolic pathway but have typical features of anaerobic degradation of aromatic compounds, because of activation of PA to phenylacetyl-coenzymeA (PA-CoA) by the action of a PA-CoA ligase (PaaK). By sequence similarity, the paaX gene product was classified as a GntR-type repressor. The paaX gene is located downstream of paaK and is transcribed in the same direction, although it is not part of the same transcript (63). Since then, a few other GntR members have been found for catabolic pathways (Table 4). The repressors PhcS and AphS regulate the expression of the phenol degradation pathway in C. testosteroni strains R5 and TA441 (7, 249). The phcS and aphS genes both belong to a small operon (phcRS and aphRS), of which the second gene is coding for an NtrC/XylR-type regulator (see below) (7). The aphRS and phcRS operons are located upstream of and transcribed in the opposite direction from their target catabolic operons (8, 249). The AphS protein acts as a factor for silencing the aph genes for phenol metabolism, whereas AphR is its transcription activator. Knockout mutations in the aphS gene were shown to enhance the transcription of the aph structural genes and of the aphR promoter as well (7). VanR, which represses the vanAB vanillate demethylase genes in Acinetobacter, is also a GntR-type regulator (149). The vanR gene is transcribed in the opposite direction to and overlaps with the vanB gene. Transcriptional regulation of biphenyl degradation in Ralstonia eutropha A5 (152), Pseudomonas sp. strain KKS102 (165), and P. pseudoalcaligenes KF707, is now also assumed to be mediated by GntR-type regulators (BphS) (269). The bphS gene in R. eutropha R5 is oriented in the opposite orientation to the first gene of the biphenyl operon (bphE) (152). In the strain KKS102, bphS is separated from its target operon by an insertion element (165). The third regulatory gene (orf0, now bphR2) present in a biphenyl catabolic regulon is also located upstream of the target operon but transcribed in the same direction (269). The Orf0 (now BphR2) protein is atypical in the GntR group since it is presumed to be an activator rather than a repressor.
TABLE 4.
GntR-type regulators controlling the degradation pathways of aromatic compounds
| Regulatory protein | Bacterial genus | Regulated operons | Pathway substrate | RBS | Positionsa | Effector compound(s) | Accession no. |
|---|---|---|---|---|---|---|---|
| BphS | Pseudomonas | bphEGF(orf4)A1A2A3BCD(orf1)A4R | (Chloro-)biphenyl | ATAATCTATTTTAT, AAATAACGTTATTT, AATGGCAACAATAT | −12 to +3, +6 to +23, +27 to +40, +44 to +58 | 2-Hydroxy-6-oxophenylhexa- 2,4-dienoic acid | BAB64312 |
| AAAAAAAGATTGTT | |||||||
| PaaX | Escherichia | paaABCDEFGHIJK | Phenylacetic acid | AATGTGATCGTGTT | +3 to +17 | Phenylacetyl-CoA | CAA66101 |
| Orf0 (BphR) | Pseudomonas | orf0, bphX0X1X2X3D | Biphenyl | ATATAAATATCGATTATC (?) | +5 to +22 | 2,3-Dihydroxybiphenyl and 2-hydroxy-6-oxo-6-phenylhexa- 2,4-dienoate | BAA12882 |
| AphS | Comamonas | aphKLMOPQB, aphR | Phenol | TATCGATA (?) | +44 to +50 (?) | NDb | BAA89295 |
| VanR | Acinetobacter | vanAB | Vanillate | ND | ND | Vanillate (?) | AAC27105 |
Relative to the transcription start site.
ND, not determined.
GntR family members that control the degradation of aromatic compounds (except Orf0) are transcriptional repressors in the absence of the pathway substrates. In the presence of the pathway substrate, repression is released by interaction of the regulator with the aromatic compounds or one of its metabolites (62, 149, 165, 249). This was interpreted as a loss of DNA binding affinity mediated by the chemical inducer (see below) (62, 165). The mechanism by which Orf0 activates transcription is not known. Strong evidence for its activator function comes from gel shift assays performed with the orf0 promoter region and purified Orf0. In these experiments, OrfO bound the promoter fragment more strongly in the presence of inducers (2,3-dihydroxybiphenyl and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate) than in their absence (269).
Structure and Conformation
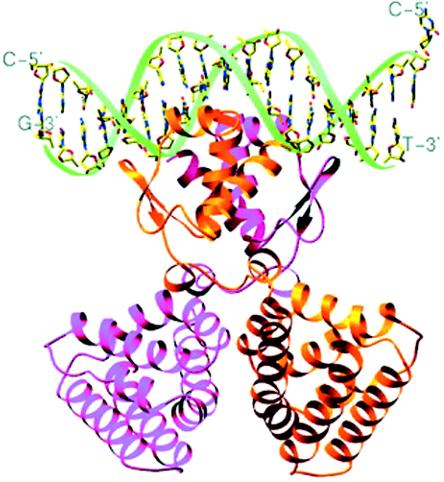
GntR-like regulators are between 239 and 254 amino acids long with a molecular mass of around 27 kDa. The PaaX protein is considerable bigger, with 316 amino acids and a molecular mass of 35 kDa. Structural information comes from GntR-type regulators that are not involved in pathways for degradation of aromatic compounds. Haydon and Guest first described bacterial regulators of the GntR family as having similar N-terminal DNA binding domains but displaying high heterogeneity among the various C-terminal effector binding and oligomerization domains (Fig. 2D) (83). This view has been largely confirmed by crystallographic structure determination and mutagenesis studies (199, 200, 259, 273). Exemplary for the GntR-type structure is FadR (211), whose structure has been resolved in the presence of its target DNA and its cognate inducer (Fig. 7). FadR exists as a homodimer in solution and binds DNA as a dimer (199, 259, 260, 273). The protein in the crystal structure showed an α/β N-terminal domain (residues 1 to 72) with the HTH motif and a C-terminal domain (residues 79 to 228) that is formed by seven α-helices with short connecting loops. The seven α-helices are packed together to form a bundle, and a large cavity within this bundle makes up the effector (myristoyl-CoA) binding pocket (259, 260). N- and C-terminal domains interact only via a short linker region comprising two short α-helices. The two monomers in the dimer are packed together in parallel fashion, resulting in reciprocal interactions of the domains and linker regions across the interface (273). Only the tip of the paired α-recognition helices projects orthogonally into the same major groove (259, 273). Each terminal portion of the α-recognition helix interacts in an identical fashion with the DNA. Part of the HTH motif also penetrates the minor groove of the DNA helix to make specific contacts with two bases (259). The DNA has a B-form conformation with a curvature of 20° toward the protein, and this results in a contraction of the central major groove and an expansion of the opposite minor groove (273).
FIG. 7.
Structure of the FadR-DNA complex (273). The paired recognition helices are in the major groove, and the two wings are within the flanking minor grooves. Note that the DNA is bent toward the FadR protein, leaving a slight contraction in the major groove as a result. Reprinted from reference 273 with permission from the publisher and from the authors.
Mechanisms of Regulation
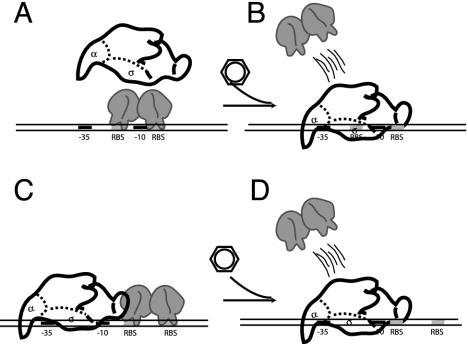
The repression mechanism of GntR-type regulators seems to be a simple hindrance for RNAP binding or open-complex formation (Fig. 8). GntR-type proteins bind either the promoter region itself or between the transcription and translation start sites. For example, four regions on the bphE promoter were bound by purified BphS of Pseudomonas sp. strain KKS102. The binding sites are located between positions −12 and +3 (called BSI), +6 and +23 (BSII), +27 and +40 (BSIII), and +44 and +58 (BSIV) compared to the transcription start site. BphS has more affinity to BSI and BSII than to BSIII and BSIV, a difference which was explained by the presence of inverted repeats in BSI and BSII (Table 4), and no such features in BSIII and BSIV. In fact, fragments with only BSI and BSII are sufficient for BphS to mediate repression, whereas transcription from an artificial promoter fragment containing only BSIII and BSIV was not repressed to a significant extent. Nevertheless, the strongest repression was obtained when all four binding sites were present (165). BphS loses its ability to bind to the promoter in the presence of the inducer 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (an intermediate in biphenyl metabolism).
FIG. 8.
Two modes of repression by GntR-type regulators. (A) The repressor bound to the −10 promoter region impairs RNA polymerase binding. (B) Binding of an inducer abolishes DNA binding upon which RNA polymerase can start transcription. (C) Repressor bound between the transcription and translation start site impairs open-complex formation. (D) Binding of the inducer to the repressor abolishes DNA binding activity. Regulatory proteins are shown as ellipsoids.
A 15-bp imperfect palindromic sequence was identified in the PaaX-dependent promoters paaZ and paaA. The palindrome was located between positions −30 and −16 in the paaZ promoter and between +3 and +17 in the paaA promoter. DNase I footprinting showed that the PaaX protein actually protects a larger region, which extends from −1 to +51 in the paaA promoter and from −30 to +17 in the paaZ promoter (62). Hypersensitive sites were spaced at approximately 10-nucleotide intervals, suggesting binding of PaaX to one side of the double helix. The fact that PaaX protected the −10 box in the paaZ promoter but not in the paaA promoter suggests a different mechanism of PaaX repression in these two promoters (as shown in the model in Fig. 8). In the presence of phenylacetate-CoA, binding of PaaX to the paaA and paaZ promoters was released (62). A sequence of events leading to the release of repression can be deduced from the FadR-DNA structure in the presence of myristoyl-CoA (the inducer for FadR) (259). Binding of the effector induces conformational changes in the sensor kink helix α8 (Fig. 7), which then forms two separate helices. As a result, some of the side chains on this helix are pushed toward the neighboring helix, α4 (linker region), which responds by shifting away from helix α8 toward the N-terminal DNA binding domain. This shift introduces additional contacts with the helix α1 in the N-terminal domain, which tilts away and causes a hinge-bending motion of both DNA binding domains of the dimer in opposite direction. This leads to a change in the distance between the two recognitions helices, which now lose their ability to interact with the DNA helix (259).
TetR-, MarR-, AND FNR-TYPE REGULATORS
TetR Family
Only one example of a TetR-type regulator involved in the regulation of an catabolic pathway for aromatic compounds is known. This protein, CymR, is a repressor for the expression of the genes for p-cymene (cym) and p-cumate (cmt) degradation in P. putida F1 (Fig. 1H; Table 5) (50). The cymR gene itself is positioned upstream of and transcribed in the same direction as the cym operon. Similar sequences have been identified in the cmt and cym promoter regions (Table 5) and may be the recognition sites for the CymR repressor protein (50, 164). CymR is supposed to impose its repressing effect by inhibiting RNAP access to the promoter. In the presence of the pathway effector, p-cumate, CymR is assumed to no longer bind to its operator site, in analogy to the TetR repressor in the presence of tetracycline (170). Strangely, p-cymene, the substrate for the cym-encoded pathway, is not an effector for CymR (50). The CymR protein is 203 amino acids long with a molecular mass of 23 kDa and is supposed to form a dimer in solution, like the TetR protein, for which the structure is known (170). Its N-terminal part contains an HTH motif (predicted for amino acids 19 to 64 in CymR), which is responsible for DNA binding, whereas the C-terminal domain binds tetracycline (170). The C-terminal domains interact in the dimer to form a core. The HTH motifs bind to the (palindromic) operator sequence, with the recognition helices aligned parallel to the major groove. The distance and the relative orientation of the HTH motifs distinguish between the induced and the noninduced status of TetR. Inducer binding triggers a sequence of α-helix displacements, which results in an increasing separation of the recognition helices. The shift of the recognition helices along the major groove disrupts the contacts between the DNA binding domains and the operator, causing dissociation of the TetR operator complex (170).
TABLE 5.
TetR, MarR, and FNR family members and two-component regulators in degradation pathways of aromatic compounds
| Family | Regulatory protein | Bacterial genus | Regulated operons | Pathway substrate(s) | RBS | Positiona | Inducer(s) or inducing condition(s) | Accession no. |
|---|---|---|---|---|---|---|---|---|
| TetR | CymR | Pseudomonas | cmt, cym | p-Cumate, p-cymene | ACAAACAGAC-N6-GTCTGTTTGT (?) | Around +1 (?) | p-Cumate | AAB62296 |
| MarR | BadR | Rhodopseudomonas | badDEFG | Benzoate | NDb | ND | Benzoyl-CoA (?) | AAC23923 |
| CbaR | Comamonas | cbaABC | Chlorobenzoate | GT(T/A)G-N(6/9)-(T/C)AAC | Around +1 and around +40 | Protocatechuate, 3-hy- droxybenzoate | AAG00065 | |
| NbzR | Pseudomonas | nbzJCaCbDGFEIH | Aminophenol | ? | ? | Nitrobenzene, amino- phenol, succinate | AF319593 | |
| HpcR (HpaR) | Escherichia | hpc(hpa)EGB-DGH | 3- and 4-hydroxy- and 3,4-dihydroxyphenyl- acetate | AATCATTAACATATTAATGATT (OPR1)- AATCATTAATATACAAACAGTT (OPR2) | OPR1 centered around +2, OPR2 around −210 | 3- and 4-hydroxy- and 3,4- dihydroxyphenylacetate | S56952, Z37980 | |
| HcaR | Acinetobacter | hcaABCDE | Hydroxycinnamate | ? | ? | Hydroxycinnamoyl-CoA | L05770 | |
| FNR | AadR | Rhodopseudomonas | hbaR, badDEFG | (4-Hydroxy)benzoate | TTGAT-N4-ATCAA (?) | −36 to −49 (?) | Anaerobiosis | B43334 |
| HbaR | Rhodopseudomonas | hbaA | 4-Hydroxybenzoate | ND | ND | 4-Hydroxybenzoate | AAF04013 | |
| CprK | Desulfitobacterium | cprBA | o-Chlorophenol | TTAAT-N4-ACTAA | −35 to −48 | 3-Chloro-4-hydroxyphenyl- acetic acid (?) | AAF67815 | |
| Two-component regulators | TodST | Pseudomonas | todXFC1C2ADEGIH | Toluene | ATAAAC-N4-GTTTAT | −113 to −98 | Toluene | CAB43735/C, AB43736 |
| StySR | Pseudomonas | styABCD | Styrene | ATAAAC-N4-GTTTAT | −52 to −37 | Styrene | AAC06272/A, AC06271 |
Relative to the transcription start site.
ND, not determined.
MarR-Type Regulators
The nbz operon for aminophenol degradation on plasmid pNB1 in P. putida HS12 is regulated by NbzR, a MarR-type repressor protein (176). The chemical inducer for the pathway has not been identified, and it was proposed that another trans-acting factor might be involved in regulating pathway expression. Other MarR-type regulators involved in catabolic pathways include HpcR (HpaR), the repressor of the hpc (hpa) operon for 3- and 4-hydroxy- and 3,4-dihydroxyphenylacetic acid (homoprotocatechuate) degradation in E. coli strains C (214) and W (68). In fact, current information from genome sequences shows that the pathways for homoprotocatechuate degradation are very common among Gamma- and Betaproteobacteria. CbaR, another MarR-type regulator, controls the cbaABC operon for 3-chlorobenzoate degradation present on plasmid pBRC60 in C. testosteroni BR60 (197). 3-Chlorobenzoate and protocatechuate are effectors for CbaR, leading to derepression (197). The genes for benzoyl-CoA reductase in Rhodopseudomonas palustris are also under the control of a MarR family member, named BadR. BadR activates rather than represses gene expression (55), although most other MarR family members are repressors of gene transcription (113, 171). Benzoate is assumed to be the effector for BadR-mediated activation (55). Additional pathway control is exerted by the AadR regulator, a FNR-type regulator, under anaerobic conditions (55). The regulatory genes nbzR, cbaR, and hpcR are located upstream of and are transcribed in the opposite direction to their target operons (176, 194, 197). badR is located upstream of and transcribed in the same direction as its target operons but is separated from them by one additional gene (55). HcaR, the repressor of the hydroxycinnimate (hca) genes in Acinetobacter sp. strain ADP1, is also a distant member to MarR (28% identical aligned residues), which reacts to a hydroxycinnamoyl-CoA thioester as an effector compound (180).
DNA binding studies have been performed with HpaR (68) and CbaR (197). CbaR contacts two sites (named BSI and BSII), which are located near the transcription start site. BSI is located approximately 40 nucleotides downstream of the transcription start site of cbaA, whereas BSII overlaps with it. An inverted repeat of 4 nucleotides separated by 6 and 9 nucleotides (GTTG-N6/9-CAAC) is present in the BSI region. Similar (but not perfect) inverted repeats are present in BSII (GTTG-N6-TAAC or GTAG-N9-TAAC), and it is presumed that CbaR interacts with the inverted-repeat sequences in some way. In DNase I footprinting experiments, CbaR bound to a BSI DNA fragment with higher affinity than to BSII, which led to the conclusion that independent CbaR proteins bind to each site. In the presence of 3-chlorobenzoate, benzoate, or protocatechuic acid, DNA binding by CbaR is disrupted, whereas 3-hydroxy- and 3-carboxybenzoate increased binding affinity by 60-fold. The physiological significance of this phenomenon is not understood (197). HpaR from E. coli is binding to two operator sites in the Pg promoter for the hpaGEDEFHI operon, one of which overlaps with the transcriptional start site (68). The operator around the transcriptional start site contains a perfect palindromic sequence (AATCATTAA-N4-TTAATGATT). Transcription from the Pg promoter is furthermore subject to strong catabolite repression by the cyclic AMP receptor protein.
Most MarR-type proteins carry a conserved HTH motif in the center of the protein, whose function would be to bind DNA (4). NbzR is atypical, since several conserved residues of the HTH motif are not present in NbzR. Instead, the protein possesses a putative leucine zipper motif (which is absent in other family members), which is thought to be responsible for the NbzR dimerization and DNA binding. Hence, Park and Kim suggested that the poor HTH motif of NbzR perhaps does not contribute to DNA binding (176). MarR-type regulators controlling catabolic operons are relatively small proteins. They contain between 148 and 196 amino acids and have a molecular mass of between 17 and 22 kDa. MarR has been crystallized (4, 130, 272). The structure revealed a MarR dimer with each subunit consisting of six helical regions. The N- and C-terminal regions encompassing residues 10 to 21 and 123 to 144, respectively, are closely juxtaposed and intertwine with the equivalent regions of the second subunit to form a domain that holds the dimer together (Fig. 9). These N- and C-terminal domains are linked to the remainder of the protein by two long antiparallel helices in each subunit. The helices lead to one globular domain, which includes residues 55 to 100 of each subunit and might be responsible for DNA binding. Although the globular DNA binding domains of the dimer are adjacent, they make minimal contact with each other. Four molecules of salicylate, which is an effector for MarR, were visible in the crystal structure: two at each subunit and two on either side of the proposed DNA binding helix (4).
FIG. 9.
The structure of the DNA binding domain of the MarR dimer (4). One subunit is in grey, and the second is coloured. The first six N-terminal residues of MarR are not included in the structure. The α1- and α5-helices are the linkers between the central domain and the N- and C-terminal part of the protein. The central domain contains the α4-helix, expected to be the recognition helix of the DNA binding motif. Reprinted from reference 4 with permission from the publisher and from the authors.
FNR-Type Regulators
Three members of the FNR family of regulators were found to be involved in controlling catabolic operon expression (Table 5). These include HbaR, which activates the expression of the gene for 4-hydroxybenzoate-CoA ligase in response to 4-hydroxybenzoate in R. palustris (56); AadR, also present in R. palustris (55); and CprK of Desulfitobacterium dehalogenans (238). AadR is the transcriptional activator for the hbaR gene itself. It is also, with BadR (see above), a coactivator for the badD benzoyl-CoA reductase gene in R. palustris (55, 56), functioning as an oxygen sensor and enhancing the expression of the anaerobic degradation pathway under oxygen exclusion. Upstream of the transcription start sites of hbaR and badD, interrupted inverted repeats (TTGAT-N4-ATCAA) are found, which were postulated to be AadR binding sites (55, 56). The cprK gene is part of a cluster for o-chlorophenol dehalogenase (238). One small transcript which encompasses the cprBA genes for the dehalogenase itself is induced under conditions of dehalorespiration; the remainder of the cpr genes are expressed constitutively. At several points within the cpr cluster, FNR-like binding motifs were detected. Within the cprB promoter, this motif (TTAAT-N4-ACTA) occurs at 41 nucleotides upstream of the transcription start site and is presumed to be a CprK binding site (238).
The proteins of the FNR family are generally between 233 and 242 amino acids in length with a calculated molecular mass of around 27 kDa. The best-characterized member of the family is FNR, a global regulator which represses the expression of genes involved in aerobic respiration and activates the expression of genes that permit the reduction of alternative electron acceptors under anoxic conditions. In its active DNA binding form, FNR is a homodimer containing one [4Fe-4S]2+ cluster per subunit (12). Integrity of the cluster is required for FNR dimerization, site-specific DNA binding, and transcription activation (108). The cluster is also essential for redox sensing by FNR. FNR is supposed to interact with the α-C-terminal domain and the sigma factor of the RNAP (121, 131). The amino acid sequence of AadR contains the essential conserved cysteine residues for iron-sulfur coordination and, as such, may be a redox sensor as well. In contrast, HbaR and CprK lack the characteristic N-terminal cysteines and are not specific sensors for anoxic conditions (56, 238).
TWO-COMPONENT REGULATORY SYSTEMS
Two-component signal transduction systems comprise the major mechanisms by which bacteria sense environmental signals and control global cellular processes. They typically consist of two individual proteins, a sensory histidine kinase and a response regulator. In the general model (reviewed in references 5, 54, 175, 209, and 243), signal perception by the N-terminal domain of the sensor kinase catalyzes an ATP-dependent phosphorylation of a conserved histidine in the central region of the protein. Once the histidine is phosphorylated, the phosphoryl group is transferred from the histidine to a conserved aspartate usually found at the N terminus of the cognate response regulator. Phosphorylation of the regulatory domain catalyzes the effector domain, which is the final mediator for gene expression control. Although this is a basic scheme, it is highly adaptable, and numerous variations have been found.
Involvement in Regulation of Catabolic Pathways
A number of two-component regulatory systems are known to control the expression of catabolic pathways. The first of these is the TodST system, which was identified in P. putida F1 and P. putida DOT-T1 (118, 151). TodST controls toluene degradation from the todX promoter, which lies upstream of the todXFC1C2BADEGIH operon. The todST genes are located downstream of but in the same direction as todH (Fig. 1I). The regulatory genes form their own transcriptional unit. Expression from the todX promoter is elicited by growth on toluene (Table 5) (118). In P. mendocina, the TmoST proteins regulate the expression of the tmo toluene-4-monooxygenase genes. The tmoST genes themselves are not located in the direct vicinity of the tmo operon (205). Another two-component system regulates the expression of benzylsuccinate synthase in Azoarcus sp. strain T and in Thauera aromatica strain K172. Benzylsuccinate synthase is the first enzyme in the anaerobic conversion of toluene and is under the control of a two-component system named TdiSR (3, 126). In T. aromatica strain T1, two potential two-component systems, tutC1B1 and tutCB, seem to be involved in the regulatory control of toluene degradation under aerobic and anaerobic conditions (35, 126). A further two-component system named BpdST was identified as the possible sensor/regulator of the biphenyl degradation pathway in Rhodococcus sp. strain M5 (115). As with the todST genes, bpdST is located downstream of the target operon. Transcription activation by BpdST takes place in the presence of biphenyl (115). Finally, a two-component system called StyRS has been described for the styABCD degradation pathway of styrene to phenylacetate (Table 5). StyRS-mediated expression from the styA promoter is inducible by styrene but repressed by phenylacetate (166). StyRS analogs have been found in Pseudomonas sp. strain Y2 (264), in P putida CA-3 (166), in Pseudomonas sp. strain VLB120 (174), and in P. fluorescens ST (222). In all these bacteria, the stySR genes are located upstream of and in the same direction as the styABCD operon.
Structure and Conformation of the Sensor Kinase Component
The sensor kinase proteins of the two-component systems involved in regulation of degradation pathways of aromatic compounds are structurally dissimilar. One example is the TodS protein, which is 978 amino acids long and has a molecular mass of 108 kDa. TodS has five predicted defined domains, of which the first contained a so-called bZIP motif. The bZIP motif consists of a stretch of several basic amino acid residues, which probably contact DNA directly, and an adjacent region with a heptad repeat of leucine residues (the leucine zipper) that mediates protein dimerization (74, 257). The presence of a bZIP domain in a sensory histidine kinase was very unusual. The TodS bZIP domain alone was shown to bind a DNA fragment upstream of the todS start codon. This region contained the sequence 5′-TGACTCA-3<29>′, which is identical to the recognition sequences of the eukaryotic proteins FOS and Jun, which also contain the bZIP motif (118). The possible physiological relevance of the TodS bZIP binding to the DNA is not known. The TodS protein also contains two identical histidine kinase domains, called Hk1 and Hk2, spanning amino acids 184 to 409 and 756 to 978, respectively. The histidines at positions 190 and 760 are the sites of autophosphorylation. The two domains also carry a peptide motif called the G1 and G2 blocks, which are involved in ATP binding. An intrinsic response regulator domain reminiscent of CheY (266) spans amino acids 443 to 570 and could be potentially phosphorylated by other signal-transducing proteins. Amino acids 34 to 153 contain a hypothetical chemical sensor domain, whereas the region between positions 592 and 735 is supposed to form the oxygen-sensing domain (26, 118). Oxygen-sensing domains are found in various redox and light sensor proteins (248, 276). The authors concluded that TodS might actually be a dual sensor that is capable of sensing both toluene as a primary signal and oxidative stress in the cytoplasm (118). Isolation of TodS effector specificity mutants proved that the aromatic compounds directly interact with the sensor kinase (26). From amino acid similarity comparisons, it was concluded that TutC, TmoS, and StyS are homologous to TodS (35, 205, 264).
In contrast to TodS, the TdiS protein of T. aromatica K172 is only 548 amino acids long and has a molecular mass of 63 kDa (126). TdiS has three distinct predicted domains. Two domains are found at the N terminus (residues 27 to 162 and 182 to 330); they are similar to each other and to other oxygen-sensing domains (see above). The C-terminal domain (residues 349 to 546) is a typical conserved histidine kinase domain. TdiS of Azoarcus sp. strain T and TutC1 of T. aromatica strain T1 are very similar in organization to TdiS. Leuthner and Heider suggested that the TdiS-type protein could be specific for anaerobic toluene metabolism whereas TodS (or StyS) would preferentially be involved in aerobic metabolism only (126). T. aromatica strain T1 can metabolize toluene under both aerobic and anaerobic conditions (60) and, interestingly, has two toluene sensor/response regulators, of which TutCB resembles TodST and TutC1B1 resembles TdiST. However, it is not known whether one of them really controls the aerobic pathway only and the other controls the anaerobic pathway.
The last type of sensory histidine kinase is found only in BpdS. BpdS is exceptionally long: 1576 amino acids with a molecular mass of 170 kDa (115). One region in the N terminus (residues 61 to 146) shows sequence similarity to a conserved domain of eukaryotic proteins, which use tyrosine or serine/threonine as acceptors of phosphoryl groups. Between residues 316 and 323, there is a Walker A motif that could bind ATP. The C terminus of BpdS contains the histidine kinase domain. In addition, seven potential hydrophobic transmembrane segments exist, which could mean that the C-terminal histidine kinase is located within the cytoplasm whereas the N-terminal part of the protein would stick in the periplasmic space. In this respect, BpdS resembles receptor or receptor-like kinase proteins, which carry out signal transduction processes in both plants and animals (115).
Structure and Conformation of the Response Regulators
Once the signals are perceived by the sensory kinases of the system, the kinases transmit the signal in form of phosphorylation to the response regulator. The response regulators (TodT, BpdT, etc.) are 206 to 227 amino acids long and have a molecular mass of between 23 and 25 kDa. The response regulators in the two-component systems controlling the aromatic compound degradation pathway have a very similar configuration. They contain an N-terminal regulatory domain (residues 10 to 115 in TodT) with an aspartate phosphorylation site and a C-terminal DNA binding domain (residues 144 to 187) with an HTH motif (118). TodT binds the todX promoter even in the absence of phosphorylation (118). The TodT binding site (the tod box) has been identified as a 12-bp inverted repeat with a 4-bp spacing (ATAAAC-N4-GTTTAT) centered at bp −105.5 from the todX transcriptional start site. The dyad symmetry of the binding sites suggests that TodT binds as a dimer (118).
The response regulator StyR of the StyRS system was shown to bind an identical sequence motif to TodT (i.e., ATAAAC-N4-GTTTAT); however, in this case the motif was centered at bp −44.5 from the styA transcription start site (Table 5) (222). The nonphosphorylated form of StyR was shown to be a monomer, but on phosphorylation the two monomers join to form a dimer (124). This is also known to occur in other two-component regulatory systems (37, 64, 89, 129, 243). StyR in dimeric form has a 10-fold-higher affinity for the styA promoter than does the monomer (124), which could mean that dimerization of the response regulator is needed for efficient recruitment of the protein on the promoter and subsequent transcription activation. Interactions of TodT or StyR with RNAP have not been experimentally probed, and it cannot be concluded that the transcription activation mechanisms should be the same.
XylR/NtrC-TYPE TRANSCRIPTIONAL REGULATORS INVOLVED IN CATABOLIC PATHWAYS
In contrast to the above-described regulatory proteins, which control transcription mediated by σ70-RNAP (except for XylS), XylR/NtrC-type regulators activate RNAP containing the alternative sigma factor σ54. The σ54-RNAP holoenzyme forms a stable complex with −12 and −24 promoters but is unable to start transcription without further activation (17, 192, 223). Isomerization to the open complex is strongly stimulated by specific regulatory proteins, generally known as prokaryotic enhancer binding proteins. The best-characterized member is NtrC, and often the family of prokaryotic enhancer binding proteins is referred to as the NtrC family. NtrC-type regulators bind to DNA regions called upstream activating sequences (UASs), which are usually located more than 100 bp upstream from the σ54-RNAP binding site (18, 19, 79, 150, 208). Interactions between σ54-RNAP bound to the −12/−24 region and the regulatory protein associated with the UASs are often facilitated by a bend in the intervening DNA, which can be the result of integration host factor binding (190).
Catabolic Operons Controlled by XylR/DmpR Subclass Regulators
Regulation of aromatic-compound degradation is very often mediated by NtrC-type regulators. The best-studied examples of those are the XylR and the DmpR proteins from P. putida (see below). For this reason, the specific subclass of NtrC-type regulatory proteins which act on promoters from catabolic pathways is also often referred to as the XylR/DmpR subclass, and we refer to them as such here as well. Briefly, XylR from P. putida mt-2 is one of the two main regulatory proteins for the xylene and toluene degradation pathway encoded on the TOL plasmid (the other being XylS [see above]) (201). XylR is responsive to chemical effectors such as m- and p-xylene, which are the substrates for what is called the upper pathway. XylR in combination with σ54-RNAP triggers expression from the Pu promoter (in front of the xylU gene) and from the Ps promoter, driving expression from xylS as well (2, 47, 91, 111, 202). The gene for XylR itself is located downstream of the meta-cleavage pathway operon and of xylS (Fig. 1E). In addition to being a transcriptional activator, XylR represses its own expression. Under inducing conditions, autorepression of xylR increases (14, 134). The reason for the autorepression seems to lie in the fact that the xylR promoter and the XylR binding sites in the intergenic regions for xylS and xylR overlap, which may prevent σ70-RNAP binding to the xylR promoter.
The other very extensively studied NtrC-type transcriptional regulator involved in the degradation of aromatic compounds is DmpR. DmpR regulates expression from the Po promoter, which drives transcription from one single large operon for phenol degradation (dmpKLMNOPQBCDEFGHI) that is present on the pVI150 plasmid in Pseudomonas sp. strain CF600 (234). The dmpR gene itself is located directly upstream of and is divergently oriented from dmpK. A large number of highly similar regulators to DmpR have been found, which all control phenol degradation operons (Table 6) (8, 153, 158, 177, 229, 249). A few other examples worth mentioning here are TbuT and TbmR, which control toluene monooxygenase gene expression in Burkholderia picketti PK01 (22) and Burkholderia sp. strain JS150 (98), respectively. Two other members are TouR, which controls the touABCDEF and dmp-like operons for toluene monooxygenase and phenol degradation in Pseudomonas stutzeri OX1 (10), and AreR, which is involved in the degradation pathway for aryl esters (areCBA) in Acinetobacter sp. strain ADP1 (100). The genes for TbuT and TouR are located downstream of the target operons tbuA1UBVA2C and touABCDEF, respectively, but are transcribed in the same direction (11, 22). Expression of tbuT is partly controlled by TbuT itself, since a transcript covering both the tbu structural genes and tbuT is synthesized in the presence of toluene, which starts at the TbuT-dependent promoter PtbuA1 (22). Also, areR is transcribed in the same direction as areCBA but is located upstream of it. Finally, HbpR is the regulatory protein which controls two small operons for 2-hydroxybiphenyl degradation (hbpCA and hbpD) in Pseudomonas azelaica HBP1 (97). The intergenic region between hbpR and hbpC looks as if it has been subject to recent duplication and deletion events, since a small 5′ part of a second hbpR copy is still present (96). Expression from hbpR itself is autoregulated by HbpR, which could be attributed to a secondary set of HbpR binding sites overlapping the hbpR promoter (96).
TABLE 6.
XylR/DmpR family members involved in degradation pathways of aromatic compounds
| Regulatory protein | Bacterial genus | Regulated operon(s) | Pathway substrate(s) | RBS | Positiona | Inducer(s) | Accession no. or reference |
|---|---|---|---|---|---|---|---|
| XylR | Pseudomonas | xylUWCMABN, xylS | Toluene, m- and p-xylene | TTGATCAAATCGACAG-N14-TTGATGATTTGCTCAA | −172 to −127 | Toluene, xylene, p-chlorobenzaldehyde, alkylbenzyl alcohols, m-amino- and o- and p- nitrotoluene | AAA26028 |
| DmpR | Pseudomonas | dmpKLMNOPQBCD EFGHI | Phenol | ATAAGCATTTGCTCAA-N14-TTGATCAAATGCTTAA | −171 to −126 | Phenol, o-, m-, and p-cresol, 3,4-dimethyl- phenol | A47074 |
| MopR | Acinetobacter | mopKLMNOP | Phenol | TTGATGAAATCAGCAT-N9-TTCATTATTTGATGAA (?) | −151 to −111 (?) | Chlorophenol, phenol, o-, and m-cresol | CAA62584 |
| AphR | Comamonas | aphKLMNOPQB, aphCEFGHJI | Phenol | GCTTGATCA-N2-TGATCATGC (?) | −181 to −162 (?) | NDb | BAA34177 |
| TouR | Pseudomonas | touABCDEF, dmp- like operon | Toluene, o-xylene, phenol | TTAATCATCTGGTGAA-N13-TTAACCAATTAATAA (?) | −149 to −106 (?) | o-, m-, and p-cresol, 2,3-, 2,4- 2,5-, and 3,4-dimethylphenol | CAB52211 |
| TbuT | Burkholderia | tbuA1UBVA2C | Toluene | TTACCAATTGATGAAATCAAC-N19-TGATCTGATCA AAAACTGA (?) | −207 to −145 (?) | Benzene, toluene, ethylbenzene, o-xylene, phenol, o- and m-cresol, trichloroethylene | AAC44567 |
| TbmR | Burkholderia | tbmABCDEF | Toluene/benzene | ACGATTTGATGAAAGCAGC-N4-GGCGGCTGCATCA AATCATCAA (?) | −201 to −156 (?) | Benzene, toluene, ethylbenzene, o-xylene, phenol, o- and m-cresol, trichloroethylene | 120 |
| ArcR | Acinetobacter | areCBA | Benzylalcohol esters | ATTGTACATTGTTCAGATTCTGGACAATGTACAAT | −139 to −105 | Benzaldehyde, benzylalcohol, acetate, propionate, butyrate, 2- and 4-hydroxy- benzylacetate | AF150928 |
| HbpR | Pseudomonas | hbpCA, hbpD | 2-Hydroxybiphenyl | TTAATAAATTTATGAA-N16-TTCATAATATGGTGA | −227 to −180 | 2-Hydroxy-, 2,2′-dihydroxy-, and 2-amino- biphenyl | AAA84988 |
Relative to the transcription start site.
ND, not determined.
The types of chemical effectors and cofactors required for XylR/DmpR-mediated activation have been very well studied (Table 6). One of the motivations for those studies has been the urge to modulate the effector spectrum of the regulators by mutagenesis (73, 237). XylR/DmpR-like activators require a chemical effector and ATP as the cofactor. The effector is usually the primary substrate of the target pathway or a compound related to this. Both XylR and DmpR hydrolyze ATP or dATP, which is essential for activation (188, 270). In addition, XylR can hydrolyze GTP, whereas DmpR only binds to but does not hydrolyze GTP, CTP, and UTP (270). Some aromatic compounds such as 2,4-dimethylphenol and 4-ethylphenol can bind DmpR and activate ATP hydrolysis without triggering transcription activation (unproductive effectors). This process is not well understood but may involve a slightly different conformational change required for ATP hydrolysis and for productive contacts with the transcription machinery exerted by productive and unproductive effectors (168, 169). Unproductive effectors can also act as competitive inhibitors for productive effectors (i.e., those which lead to transcription activation) (168). 3-Nitrotoluene can also bind XylR without mediating activation (73, 221). In several cases, inducing compounds which effect transcription activation are not direct substrates for the target pathway, such as 2-aminobiphenyl and HbpR (97).
Domain Organization
XylR/DmpR-type regulators are typically between 563 and 570 amino acids long (molecular mass of 62 to 68 kDa), with the exception of AreR, which is 600 amino acids long. Since XylR and DmpR have resisted full purification, there is no direct structural information on those proteins, except ideas which can be extrapolated from genetic experiments. We therefore use structural data from other NtrC-type regulators, which are not involved in the degradation of aromatic compounds, to illustrate possible structural motifs for XylR/DmpR subclass members. Members of the NtrC family in general display an ordered three-domain structure, although the domains themselves are not necessarily very similar among different regulators (150) (Fig. 2E). The C-terminal end (forming the so-called D-domain) contains the HTH motif responsible for DNA binding (162) and in some cases for dimerization (110, 184).
The central or C-domain is the most highly conserved region among NtrC family members, and seven highly conserved regions (C1 to C7) have been identified here (150, 172). Mutagenesis of the regions C1 (containing the Walker A motif), C4 (Walker B motif), and C7 in XylR and DmpR showed they are involved in ATP binding and hydrolysis (188, 270). Mutagenesis of and cross-linking experiments with DctD (an NtrC-type regulator not involved in catabolic pathways) showed the conserved C3 region to be interacting with RNAP through the β and σ54 subunits of the holoenzyme (106, 122, 267). In vitro interactions between σ54 and PspF (a noncatabolic activator) took place between the I-region of σ54 and the PspF C-domain, most probably involving the 6-amino-acid GAFTGA motif within the C3 region (25).
The so-called A-domain, which is located at the N-terminal end and is very specific for the different NtrC-type regulators, has been implicated in signal perception and hence determines the specificity of the protein (233). The A-domain of DmpR and XylR (211 amino acids long) was shown to interact directly with the inducing aromatic compounds, and various effector specificity mutations have been generated in this region of the protein (38, 73, 167, 168, 183, 221, 235, 236). One inducer binding site is present per monomer, which was demonstrated for DmpR (167) and which could be pinpointed to a subregion between amino acid residues 107 and 186 (237). A structural model of the XylR A-domain generated by a “fold recognition approach” (41) predicted eight α-helices and seven β-strands connected by 14 loops. Based on this model and the different XylR/DmpR effector specificity mutants, five different regions in the A-domain were postulated to form the aromatic compound binding site. These are loops 12 (position 174), 10 (position 142), 4 (position 65), 2 (positions 37), and 14 (position 203). The A-domain was also shown to modulate the ATPase activity of the C-domain. This was concluded from observations that regulatory proteins with their A-domain deleted displayed constitutive ATPase activity and activated transcription to the same extent as the complete protein in the presence of an inducer (9, 185, 229, 236). Successive deletion of the A-domain identified a subregion between residues 160 to 210 which was sufficient to repress XylR-dependent transcription activity in the absence of the inducer (187). Several other lines of evidence (167, 185, 229) have led to the current hypothesis that the A-domain acts as movable domain which, in its closed state, represses the ATPase activity of the C-domain but, in an open state that occurs under inducer binding, exposes the constitutive ATPase activity. The connecting amino acid sequence between the A- and C-domains is provided by a short region called the B-linker, which seems essential for the change from the closed to the open state. DNA binding data with purified HbpR suggested that the protein assembles to the oligomeric form irrespective of the presence of the effector (255).
Structure and Configurations
Based on sequence similarities, NtrC-family members were classified as members of the AAA+ superfamily (ATPases associated with various cellular activities) (157). Recent structural information has confirmed this sequence-predicted classification. The structure of the joined N-terminal regulatory and central ATPase domains of NtrC1 (NtrC1AC) from Aquifex aeolicus and the structure of the isolated central domain (NtrC1C) were recently determined in their ADP-bound form (Fig. 10) (123). The central ATPase domain in NtrC1AC is inactive, whereas that of NtrC1C is capable of oligomer assembly, ATP hydrolysis, and transcriptional activation. The resolution of these structures has been an important step in recognizing the structural differences between the active and inactive states of the central domain. NtrC1AC is a dimer in solution in which the regulatory domain of each monomer is adjacent to the central domain of its partner in the dimer (Fig. 10A). This might also be the state for XylR/DmpR proteins, since, for instance, DmpR without ATP, DNA, and inducer appeared as a dimer in solution as well (270). The main dimerization interface is formed by the B-linker and monomers are held in a front-to-front fashion in the dimer (Fig. 10A). The dimerization interface seems important for maintaining the off-state of the ATPase domain. In fact, mutations in the B-linker of XylR that disrupted the linker's secondary structure resulted in a much higher background expression of a single-copy chromosomal Pu::lacZ fusion (72), which seems in agreement with the presumed role of the B-linker.
FIG. 10.
Crystal structures of NtrC1AC and NtrC1C (123). (A) Dimer solution structure of NtrC1AC. The monomers are colored gray and gold. The GAFTGA and β-hairpin loops of the central ATPase domain are colored blue and green, respectively. ADP and the catalytic arginine residue (R293) are shown in a ball-and-stick representation. R293 cannot contact ADP in the dimer. (B) Heptameric structure of A-domain-deleted NtrCC. The top view (left) illustrates how the protomers (labeled A to G) pack to a heptamer. GAFTGA loops (blue) and β-hairpin (green) point toward the central pore in the heptamer. The side view is also shown (right). Reprinted from reference 123 with permission from the publisher and from the authors.
The central domain protein NtrC1C without the A-domain crystallized as a ring-like heptamer (Fig. 10B). The ultrastructure of purified PspF stabilized in oligomeric form with the ATP transition analogue ADP-aluminium fluoride was reconstructed as a trimer of dimers (or asymmetric hexamer) from cryoelectronmicrographs (231). The diameter and height of the NtrC1C heptamer rings were approximately 124 and 40 Å, respectively, with a front-to-back packing of the protomers. In this configuration, the ATPase domains contact two others simultaneously on each side. In the NtrCAC dimer, one side of the ATPase domain is contacting the other subunit whereas the second one (β-hairpin and GAFTGA in Fig. 10) is interacting with the B-linker. Because of this, an essential highly conserved Arg residue at position 293 is far away from the bound ADP molecule in the dimer whereas in the heptamer R293 comes close to the ADP of its neighboring protomer. The Arg-293 residue could thus actually be contacting the γ-phosphate of ATP and could catalyze ATP hydrolysis in the heptamer configuration. There is good evidence to assume that XylR/DmpR-like proteins also form heptamers or hexamers as part of their activation cycle. For example, addition of ATPγS and the inducer 2-methylphenol resulted in mainly hexameric forms of DmpR (270). Although XylR has resisted purification attempts, the effect of ATPγS and ATP on subunit oligomerization was detected with purified XylR-ΔA (186). Transmission electron microscopy shows that XylR-ΔA and a Pu DNA fragment were forming larger (XylR oligomers) and smaller (dimer) complexes, depending on the presence of ATP (72).
The GAFTGA-loop forms the motif that is assumed to be interacting with the RNA polymerase transcription machinery. In the NtrCC structure, all GAFTGA-loops pointed toward the middle central pore of the heptamer (Fig. 10B). This would create an extended binding surface for σ54-RNAP, although it is not clear where exactly the A-domains would go in the heptamer. For the PspF transcriptional regulator (which, however, lacks an A-domain), the interaction surface to σ54-RNAP was formed between the I-region of σ54-RNAP and the GAFTGA-loop of PspF (25). The hexameric state observed for PspF (231) and the heptameric NtrCC structure strongly suggest that XylR/DmpR-type regulators form similar oligomeric complexes. However, very different from PspF and NtrC1, which do not have a specific effector domain, must be the mechanism leading to effector-mediated transcription activation. Garmendia and de Lorenzo have proposed an activation cycle for XylR (72). In the activation cycle, XylR would change between dimer (unbound) and heptamer/hexamer (bound to the DNA), with the disassembly process being triggered by ATP hydrolysis (Fig. 11). For XylR/DmpR-like proteins, which carry an A-domain, there must thus be some sort of mechanical movement of the A-domain in the oligomeric form on induction, probably mediated by the B-linker, thereby perhaps exposing the GAFTGA-motifs in the central part. However, the typical signature stretches of the B-linker are not present in recognizably similar form in some other XylR/DmpR-like proteins members such as HbpR and TbuT (169), which may indicate that the mechanistic “movement” is not a requirement per se or can be achieved by other structures than the B-linker only.
FIG. 11.
Schematic mode of activation of XylR/DmpR-type regulators. (A) The inactive regulator dimer binds its inducer, which results in a protein conformation change. (B) Binding of ATP triggers multimerization to a hexamer (or heptamer). (C) ATP hydrolysis coupled to correct interaction with RNA polymerase triggers transcription activation. (D) Dissociation of the hexamer to a dimer on ATP hydrolysis and dissociation of the inducer. The stages during which the DNA is contacted are not shown.
DNA Binding during the Activation Process
The current activation model does not include a specific function for the DNA binding site occupied by the XylR/DmpR-subclass regulatory proteins. Two functions have been proposed: (i) to provide a “physical” support for the regulatory protein in the vicinity of σ54-RNAP and (ii) to aid in obtaining the active form of the regulatory complex. This was concluded from experiments in which the addition of the DNA fragment containing the binding site increased the ATPase activity of XylR-ΔA or TouR (and TouR-ΔA) (9, 188). However, it has to be said that similar experiments with DmpR showed a small or nonexistent stimulatory effect (270).
XylR/DmpR-like proteins bind to an approximately 60-bp sequence which is located far upstream of the transcription initiation site (the UAS) (Table 6). This was shown from DNase I footprinting analysis for purified HbpR, as well as for DmpR and XylR variants devoid of their N-terminal A-domains (189, 247, 255). Interestingly, the protection patterns were quite similar for the three proteins, and matching DNA sequences forming two imperfect palindromes were present in each of the promoters (189, 247, 255). For XylR, the UAS is localized between bp −127 and −172 from the Pu transcription start site and between bp −139 and −184 from Ps. The DmpR binding sequences lay between bp −126 and −171 from the Po promoter, whereas those for HbpR are located between bp −180 and −227 on the hbpC promoter and between bp −168 and −225 on the hbpD promoter (96, 255). A second set of HbpR-UAS is present in the intergenic region between hbpR and hbpC (bp −494 to −540), which is improperly positioned to act on the hbpC promoter but which was shown to be important for hbpR autoregulation (96). The two palindromic sequences in the promoter are approximately 16 bp long with a spacing of 29 to 42 bp between the centers of the palindromes.
XylR-ΔA bound one UAS on the Pu-DNA with higher affinity than it bound the other (189). At that point, the authors suggested that this could mean that two dimers bound the two binding sites independently. However, in the case of HbpR, both palindromic sequences were simultaneously needed for binding, since fragments containing only one were not maintained in a stable protein-DNA complex in gel mobility shift assays (255). Adding 2-hydroxybiphenyl and ATP resulted in a twofold change in binding affinity of HbpR to the promoter DNA (255). DNase I footprints of the Pu UAS fragment with purified XylR-ΔA and ATP showed an occupation and slightly expanded protected region of the UAS at lower protein concentrations compared to those in the assay without ATP (186). ATP hydrolysis itself is not required for occupation of the UASs by XylR, since the use of ATPγS, which in contrast to ATP, is not hydrolyzable, gave rise to identical footprinting patterns to those for ATP (186). This would be in agreement with the binding of a higher-order oligomeric form. In vivo footprinting of the Pu promoter showed that the presence of the inducer slightly altered the interaction between XylR and the DNA (1). More recently, Valls and de Lorenzo repeated the in vivo footprinting with a single-copy xylR gene and the Pu promoter (258). Their results showed that XylR interacts only very transiently with its binding site, with hardly any protection visible under both uninduced and induced conditions. In fact, the estimated number of XylR hexamers is around 5 to 30 per individual cell under stationary-phase conditions (65). This would equal a concentration of 10 to 50 nM, which is quite low compared to the 400 nM XylR-ΔA required to protect the DNA in in vitro DNase I footprints. However, it is clear that most static DNA binding studies fully neglect the kinetic aspects of the regulatory cycle in the microbes themselves.
PROSPECTS
As a result of many studies during the past decade, we now have a much better understanding of the behavior of different regulatory proteins in the process of mediating transcription. Although a few pivotal regulatory proteins in classical degradation pathways for aromatic compounds (i.e., XylS, XylR, and DmpR) have resisted purification, the solution state could be deduced for several others, sometimes in the presence of the DNA binding regions. In some cases it has been possible to deduce the structural changes induced in the protein or protein-DNA complex by the chemical effector. In contrast, little is currently known about the interactions between the regulatory protein and the effector compounds. The effector binding pockets and effector binding affinities are not well defined, and it is not clear how effector binding to the regulatory protein transmits to an “activation” signal for RNAP.
Although a relatively clear picture has emerged for the different components of the regulatory machineries, their interactions with the operator DNA and with RNA polymerase or with other auxiliary protein factors, and the types of signaling molecules, the picture is mostly static. Most models suggest a regulatory protein which is bound to its DNA binding site and ready to activate transcription (or disappear after derepression). Clear indications for XylR/DmpR-type regulators point at a cyclic model between the inactive regulator state and the active (multimeric) state, but with an equilibrium toward the inactive state and very little site occupancy by XylR. We might thus be very surprised to find our ideas about the activation (or derepression) processes changing when more kinetic or affinity equilibria data become available.
Our current perspective on regulatory systems is most probably somewhat biased by the choice of traditionally studied microorganisms with degradation capacities, such as Pseudomonas, Acinetobacter, Ralstonia, and Burkholderia, typical members of the Gamma- and Betaproteobacteria. It might very well be that new interesting systems are discovered, such as the two FNR-like and one MarR-like regulatory proteins in Rhodopseudomonas which control benzene degradation under anaerobic conditions. Also, regulatory proteins with possible membrane-spanning domains (such as BpdS) or with association with transport and chemotaxis (such as NahY of P. putida) (80) may provide very new insights in how bacteria accomplish recognition and expression regulation of aromatic degradation pathways. In the end and from an environmental perspective, it will again be necessary to refocus on the strategies used by bacteria to recognize and utilize aromatic compounds (or other polluting chemicals) at very low concentration ranges (micromolar or lower). Although in the laboratory we like to feed our pet microorganisms with much higher doses (millimolar), the organisms in their natural environments have to cope with very low carbon compound fluxes or (at best) fluctuating levels. Are regulatory proteins sufficiently sensitive to react and mediate specific catabolic gene expression under these conditions, or are the organisms using alternative strategies to metabolize low carbon fluxes? Gene expression data for the 2,4-dichlorophenoxyacetate degradative pathway in R. eutropha, for example, have suggested that only very slight bursts of transcription activation take place at 10 μM 2,4-dichlorophenoxyacetate (127). Similar experiments with S. paucimobilis have not shown any measurable gene induction of the lin pathway at 3 μM lindane (M. Suar, J. R. van der Meer, K. Lawlor, C. Holliger, and R. Lal, submitted for publication) but, rather, a low constitutive expression. Interestingly, a very clear difference exists between the apparent effector binding affinity of heavy-metal-responsive regulatory proteins (nano- to micromolar range) and those responsive to aromatic compounds (usually micromolar) (95). Obtaining a better view of the differences in the molecular binding affinities of the effector compounds to the regulator and the role of DNA binding in the affinity of effector binding may help to resolve this apparent mystery. This will also be an important challenge for the application of bacterial regulatory systems for the construction of environmental quality sensors, which should have sensitivities below micromolar concentrations.
Acknowledgments
We thank Victor de Lorenzo for critically reading the manuscript. We also gratefully acknowledge the many detailed suggestions by two unknown reviewers, which have helped to improve the manuscript.
Part of this work was financed under the EU 5th Framework Program, BIOCARTE contract QLK3-CT-2002-01923, and by the Swiss Federal Office for Education and Science.
REFERENCES
- 1.Abril, M. A., M. Buck, and J. L. Ramos. 1991. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J. Biol. Chem. 266:15832-15838. [PubMed] [Google Scholar]
- 2.Abril, M. A., C. Michan, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achong, G. R., A. M. Rodriguez, and A. M. Spormann. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 5.Alex, L. A., and M. I. Simon. 1994. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 10:133-138. [DOI] [PubMed] [Google Scholar]
- 6.Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. [DOI] [PubMed] [Google Scholar]
- 7.Arai, H., S. Akahira, T. Ohishi, and T. Kudo. 1999. Adaptation of Comamonas testosteroni TA441 to utilization of phenol by spontaneous mutation of the gene for a trans-acting factor. Mol. Microbiol. 33:1132-1140. [DOI] [PubMed] [Google Scholar]
- 8.Arai, H., S. Akahira, T. Ohishi, M. Maeda, and T. Kudo. 1998. Adaptation of Comamonas testosteroni TA441 to utilize phenol: organization and regulation of the genes involved in phenol degradation. Microbiology 144:2895-2903. [DOI] [PubMed] [Google Scholar]
- 9.Arenghi, F. L., P. Barbieri, G. Bertoni, and V. de Lorenzo. 2001. New insights into the activation of o-xylene biodegradation in Pseudomonas stutzeri OX1 by pathway substrates. EMBO Rep. 2:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arenghi, F. L., D. Berlanda, E. Galli, G. Sello, and P. Barbieri. 2001. Organization and regulation of meta cleavage pathway genes for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 67:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arenghi, F. L., M. Pinti, E. Galli, and P. Barbieri. 1999. Identification of the Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase regulatory gene (touR) and of its cognate promoter. Appl. Environ. Microbiol. 65:4057-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates, D. M., C. V. Popescu, N. Khoroshilova, K. Vogt, H. Beinert, E. Munck, and P. J. Kiley. 2000. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275:6234-6240. [DOI] [PubMed] [Google Scholar]
- 13.Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 147:1611-1620. [DOI] [PubMed] [Google Scholar]
- 14.Bertoni, G., S. Marqués, and V. de Lorenzo. 1998. Activation of the toluene-responsive regulator XylR causes a transcriptional switch between σ54 and σ70 promoters at the divergent Pr/Ps region of the TOL plasmid. Mol. Microbiol. 27:651-659. [DOI] [PubMed] [Google Scholar]
- 15.Bhende, P. M., and S. M. Egan. 2000. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with σ70. J. Bacteriol. 182:4959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzostowicz, P. C., A. B. Reams, T. J. Clark, and E. L. Neidle. 2003. Transcriptional cross-regulation of the catechol and protocatechuate branches of the β-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. Appl. Environ. Microbiol. 69:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck, M., and W. Cannon. 1992. Activator-independent formation of a closed complex between σ54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol. Microbiol. 6:1625-1630. [DOI] [PubMed] [Google Scholar]
- 18.Buck, M., and W. Cannon. 1992. Specific binding of the transcription factor σ54 to promoter DNA. Nature 358:422-424. [DOI] [PubMed] [Google Scholar]
- 19.Buck, M., W. Cannon, and J. Woodcock. 1987. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol. Microbiol. 1:243-249. [DOI] [PubMed] [Google Scholar]
- 20.Bundy, B. M., L. S. Collier, T. R. Hoover, and E. L. Neidle. 2002. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc. Natl. Acad. Sci. USA 99:7693-7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrne, A. M., and R. H. Olsen. 1996. Cascade regulation of the toluene-3- monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J. Bacteriol. 178:6327-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caswell, R., J. Williams, A. Lyddiatt, and S. Busby. 1992. Overexpression, purification and characterization of the Escherichia coli MelR transcription activator protein. Biochem. J. 287:493-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaney, M., R. Grande, S. R. Wigneshweraraj, W. Cannon, P. Casaz, M. T. Gallegos, J. Schumacher, S. Jones, S. Elderkin, A. E. Dago, E. Morett, and M. Buck. 2001. Binding of transcriptional activators to σ54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 15:2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi, E. N., M. C. Cho, Y. Kim, C. K. Kim, and K. Lee. 2003. Expansion of growth substrate range in Pseudomonas putida F1 by mutations in both cymR and todS, which recruit a ring-fission hydrolase CmtE and induce the tod catabolic operon, respectively. Microbiology 149:795-805. [DOI] [PubMed] [Google Scholar]
- 27.Chugani, S. A., M. R. Parsek, and A. M. Chakrabarty. 1998. Transcriptional repression mediated by LysR-type regulator CatR bound at multiple binding sites. J. Bacteriol. 180:2367-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chugani, S. A., M. R. Parsek, C. D. Hershberger, K. Murakami, A. Ishihama, and A. M. Chakrabarty. 1997. Activation of the catBCA promoter: probing the interaction of CatR and RNA polymerase through in vitro transcription. J. Bacteriol. 179:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark, T. J., S. Haddad, E. L. Neidle, and C. Momany. 2004. Crystallization of the effector-binding domains of BenM and CatM, LysR-type transcriptional regulators from Acinetobacter sp. ADP1. Acta Crystallogr. Ser. D 60:105-108. [DOI] [PubMed] [Google Scholar]
- 30.Clark, T. J., C. Momany, and E. L. Neidle. 2002. The benPK operon, proposed to play a role in transport, is part of a regulon for benzoate catabolism in Acinetobacter sp. strain ADP1. Microbiology 148:1213-1223. [DOI] [PubMed] [Google Scholar]
- 31.Clark, T. J., R. S. Phillips, B. M. Bundy, C. Momany, and E. L. Neidle. 2004. Benzoate decreases the binding of cis,cis-muconate to the BenM regulator despite the synergistic effect of both compounds on transcriptional activation. J. Bacteriol. 186:1200-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coco, W. M., M. R. Parsek, and A. M. Chakrabarty. 1994. Purification of the LysR family regulator, ClcR, and its interaction with the Pseudomonas putida clcABD chlorocatechol operon promoter. J. Bacteriol. 176:5530-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coco, W. M., R. K. Rothmel, S. Henikoff, and A. M. Chakrabarty. 1993. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J. Bacteriol. 175:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collier, L. S., G. L. Gaines, and E. L. Neidle. 1998. Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coschigano, P. W., and L. Y. Young. 1997. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl. Environ. Microbiol. 63:652-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowles, C. E., N. N. Nichols, and C. S. Harwood. 2000. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 182:6339-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Da Re, S., J. Schumacher, P. Rousseau, J. Fourment, C. Ebel, and D. Kahn. 1999. Phosphorylation-induced dimerization of the FixJ receiver domain. Mol. Microbiol. 34:504-511. [DOI] [PubMed] [Google Scholar]
- 38.Delgado, A., and J. L. Ramos. 1994. Genetic evidence for activation of the positive transcriptional regulator XylR, a member of the NtrC family of regulators, by effector binding. J. Biol. Chem. 269:8059-8062. [PubMed] [Google Scholar]
- 39.de Lorenzo, V., and J. Pérez-Martin. 1996. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol. 19:1177-1184. [DOI] [PubMed] [Google Scholar]
- 40.Deo, P. G., and N. G. Karanth. 1994. Biodegradation of hexachlorocyclohexane isomers in soil and food environment. Crit. Rev. Microbiol. 20:57-78. [DOI] [PubMed] [Google Scholar]
- 41.Devos, D., J. Garmendia, V. de Lorenzo, and A. Valencia. 2002. Deciphering the action of aromatic effectors on the prokaryotic enhancer-binding protein XylR: a structural model of its N-terminal domain. Environ. Microbiol. 4:29-41. [DOI] [PubMed] [Google Scholar]
- 42.Diaz, E., A. Ferrández, and J. L. García. 1998. Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. J. Bacteriol. 180:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Díaz, E., A. Ferrández, M. A. Prieto, and J. L. García. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Díaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 45.DiMarco, A. A., B. Averhoff, and L. N. Ornston. 1993. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J. Bacteriol. 175:4499-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiMarco, A. A., and L. N. Ornston. 1994. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J. Bacteriol. 176:4277-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixon, R. 1986. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol. Gen. Genet. 203:129-136. [DOI] [PubMed] [Google Scholar]
- 48.Donald, L. J., D. J. Hosfield, S. L. Cuvelier, W. Ens, K. G. Standing, and H. W. Duckworth. 2001. Mass spectrometric study of the Escherichia coli repressor proteins, IclR and GclR, and their complexes with DNA. Protein Sci. 10:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dua, M., A. Singh, N. Sethunathan, and A. K. Johri. 2002. Biotechnology and bioremediation: successes and limitations. Appl. Microbiol. Biotechnol. 59:143-152. [DOI] [PubMed] [Google Scholar]
- 50.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eaton, R. W., O. V. Selifonova, and R. M. Gedney. 1998. Isopropylbenzene catabolic pathway in Pseudomonas putida RE204: nucleotide sequence analysis of the ipb operon and neighboring DNA from pRE4. Biodegradation 9:119-132. [DOI] [PubMed] [Google Scholar]
- 52.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egan, S. M., A. J. Pease, J. Lang, X. Li, V. Rao, W. K. Gillette, R. Ruiz, J. L. Ramos, and R. E. Wolf, Jr. 2000. Transcription activation by a variety of AraC/XylS family activators does not depend on the class II-specific activation determinant in the N-terminal domain of the RNA polymerase alpha subunit. J. Bacteriol. 182:7075-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egger, L. A., H. Park, and M. Inouye. 1997. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 2:167-184. [DOI] [PubMed] [Google Scholar]
- 55.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an FNR family member. J. Bacteriol. 181:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egland, P. G., and C. S. Harwood. 2000. HbaR, a 4-hydroxybenzoate sensor and FNR-CRP superfamily member, regulates anaerobic 4-hydroxybenzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 182:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellis, L. B., B. K. Hou, W. Kang, and L. P. Wackett. 2003. The University of Minnesota Biocatalysis/Biodegradation Database: post-genomic data mining. Nucleic Acids Res. 31:262-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eulberg, D., and M. Schlömann. 1998. The putative regulator of catechol catabolism in Rhodococcus opacus 1CP-an IclR-type, not a LysR-type transcriptional regulator. Antonie Leeuwenhoek 74:71-82. [DOI] [PubMed] [Google Scholar]
- 60.Evans, P. J., D. T. Mang, K. S. Kim, and L. Y. Young. 1991. Anaerobic degradation of toluene by a denitrifying bacterium. Appl. Environ. Microbiol. 57:1139-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferrández, A., J. L. Garcia, and E. Díaz. 1997. Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 179:2573-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrández, A., J. L. Garcia, and E. Díaz. 2000. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J. Biol. Chem. 275:12214-12222. [DOI] [PubMed] [Google Scholar]
- 63.Ferrández, A., B. Miñambres, B. Garcia, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 64.Fiedler, U., and V. Weiss. 1995. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 14:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraile, S., F. Roncal, L. A. Fernández, and V. de Lorenzo. 2001. Monitoring intracellular levels of XylR in Pseudomonas putida with a single-chain antibody specific for aromatic-responsive enhancer-binding proteins. J. Bacteriol. 183:5571-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franklin, F. C., P. R. Lehrbach, R. Lurz, B. Rückert, M. Bagdasarian, and K. N. Timmis. 1983. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J. Bacteriol. 154:676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fukumori, F., and C. P. Saint. 1997. Nucleotide sequences and regulational analysis of genes involved in conversion of aniline to catechol in Pseudomonas putida UCC22(pTDN1). J. Bacteriol. 179:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galan, B., A. Kolb, J. M. Sanz, J. L. García, and M. A. Prieto. 2003. Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res. 31:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J. Bacteriol. 178:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. The TACAN4TGCA motif upstream from the −35 region in the σ70-σs-dependent Pm promoter of the TOL plasmid is the minimum DNA segment required for transcription stimulation by XylS regulators. J. Bacteriol. 178:6427-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garmendia, J., and V. de Lorenzo. 2000. The role of the interdomain B linker in the activation of the XylR protein of Pseudomonas putida. Mol. Microbiol. 38:401-410. [DOI] [PubMed] [Google Scholar]
- 73.Garmendia, J., D. Devos, A. Valencia, and V. de Lorenzo. 2001. A la carte transcriptional regulators: unlocking responses of the prokaryotic enhancer-binding protein XylR to non-natural effectors. Mol. Microbiol. 42:47-59. [DOI] [PubMed] [Google Scholar]
- 74.Gentz, R., F. J. Rauscher, C. Abate, and T. Curran. 1989. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science 243:1695-1699. [DOI] [PubMed] [Google Scholar]
- 75.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 180:1512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 77.Gonzalez-Pérez, M. M., S. Marqués, P. Dominguez-Cuevas, and J. L. Ramos. 2002. XylS activator and RNA polymerase binding sites at the Pm promoter overlap. FEBS Lett. 519:117-122. [DOI] [PubMed] [Google Scholar]
- 78.Gonzalez-Pérez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marqués. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286-2290. [DOI] [PubMed] [Google Scholar]
- 79.Gralla, J. D. 1996. Activation and repression of E. coli promoters. Curr. Opin. Genet. Dev. 6:526-530. [DOI] [PubMed] [Google Scholar]
- 80.Grimm, A. C., and C. S. Harwood. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo, Z., and J. E. Houghton. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the −35 and the −10 promoter elements. Mol. Microbiol. 32:253-263. [DOI] [PubMed] [Google Scholar]
- 82.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 83.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 84.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hidalgo, E., and B. Demple. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hindle, Z., and C. P. Smith. 1994. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 12:737-745. [DOI] [PubMed] [Google Scholar]
- 87.Hohnstock, A. M., K. G. Stuart-Keil, E. E. Kull, and E. L. Madsen. 2000. Naphthalene and donor cell density influence field conjugation of naphthalene catabolism plasmids. Appl. Environ. Microbiol. 66:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hryniewicz, M. M., and N. M. Kredich. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang, K. J., C. Y. Lan, and M. M. Igo. 1997. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. USA 94:2828-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hughes, E. J., M. K. Shapiro, J. E. Houghton, and L. N. Ornston. 1988. Cloning and expression of pca genes from Pseudomonas putida in Escherichia coli. J. Gen. Microbiol. 134:2877-2887. [DOI] [PubMed] [Google Scholar]
- 91.Inouye, S., A. Nakazawa, and T. Nakazawa. 1987. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc. Natl. Acad. Sci. USA 84:5182-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inouye, S., A. Nakazawa, and T. Nakazawa. 1981. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J. Bacteriol. 148:413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inouye, S., A. Nakazawa, and T. Nakazawa. 1986. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putida TOL plasmid and identification of the protein product. Gene 44:235-242. [DOI] [PubMed] [Google Scholar]
- 94.Inouye, S., A. Nakazawa, and T. Nakazawa. 1987. Overproduction of the xylS gene product and activation of the xylDLEGF operon on the TOL plasmid. J. Bacteriol. 169:3587-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jaspers, M. C. M. 2002. Using reporter bacteria to study the bioavailability of pollutants in aqueous environments. Ph.D. thesis. Swiss Federal Institute of Technology, Zürich, Switzerland.
- 96.Jaspers, M. C. M., C. Meier, A. J. B. Zehnder, H. Harms, and J. R. van der Meer. 2001. Measuring mass transfer processes of octane with the help of an alkSalkB::gfp-tagged Escherichia coli. Environ. Microbiol. 3:512-524. [DOI] [PubMed] [Google Scholar]
- 97.Jaspers, M. C. M., W. A. Suske, A. Schmid, D. A. Goslings, H.-P. E. Kohler, and J. R. van der Meer. 2000. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol. 182:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson, G. R., and R. H. Olsen. 1997. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 63:4047-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson, G. R., and J. C. Spain. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110-123. [DOI] [PubMed] [Google Scholar]
- 100.Jones, R. M., L. S. Collier, E. L. Neidle, and P. A. Williams. 1999. areABC genes determine the catabolism of aryl esters in Acinetobacter sp. strain ADP1. J. Bacteriol. 181:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones, R. M., V. Pagmantidis, and P. A. Williams. 2000. sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J. Bacteriol. 182:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaldalu, N., T. Mandel, and M. Ustav. 1996. TOL plasmid transcription factor XylS binds specifically to the Pm operator sequence. Mol. Microbiol. 20:569-579. [DOI] [PubMed] [Google Scholar]
- 103.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 182:1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kasak, L., R. Horak, A. Nurk, K. Talvik, and M. Kivisaar. 1993. Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J. Bacteriol. 175:8038-8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Keasling, J. D. 1999. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 17:452-460. [DOI] [PubMed] [Google Scholar]
- 106.Kelly, M. T., and T. R. Hoover. 1999. Mutant forms of Salmonella typhimurium σ54 defective in transcription initiation but not promoter binding activity. J. Bacteriol. 181:3351-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1993. Identification of a cis-acting sequence within the Pm promoter of the TOL plasmid which confers XylS-mediated responsiveness to substituted benzoates. J. Mol. Biol. 230:699-703. [DOI] [PubMed] [Google Scholar]
- 108.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181-185. [DOI] [PubMed] [Google Scholar]
- 109.Kitagawa, W., S. Takami, K. Miyauchi, E. Masai, Y. Kamagata, J. M. Tiedje, and M. Fukuda. 2002. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Klose, K. E., A. K. North, K. M. Stedman, and S. Kustu. 1994. The major dimerization determinants of the nitrogen regulatory protein NtrC from enteric bacteria lie in its carboxy-terminal domain. J. Mol. Biol. 241:233-245. [DOI] [PubMed] [Google Scholar]
- 111.Kohler, T., S. Harayama, J. L. Ramos, and K. N. Timmis. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J. Bacteriol. 171:4326-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1998. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J. Bacteriol. 180:5058-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Komeda, H., M. Kobayashi, and S. Shimizu. 1996. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc. Natl. Acad. Sci. USA 93:4267-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424-430. [DOI] [PubMed] [Google Scholar]
- 115.Labbe, D., J. Garnon, and P. C. Lau. 1997. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Landini, P., J. A. Bown, M. R. Volkert, and S. J. Busby. 1998. Ada protein-RNA polymerase sigma subunit interaction and alpha subunit-promoter DNA interaction are necessary at different steps in transcription initiation at the Escherichia coli Ada and aidB promoters. J. Biol. Chem. 273:13307-13312. [DOI] [PubMed] [Google Scholar]
- 117.Landini, P., and S. J. Busby. 1999. The Escherichia coli Ada protein can interact with two distinct determinants in the σ70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J. Bacteriol. 181:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lau, P. C., Y. Wang, A. Patel, D. Labbe, H. Bergeron, R. Brousseau, Y. Konishi, and M. Rawlings. 1997. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc. Natl. Acad. Sci. USA 94:1453-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leahy, J. G., G. R. Johnson, and R. H. Olsen. 1997. Cross-regulation of toluene monooxygenases by the transcriptional activators TbmR and TbuT. Appl. Environ. Microbiol. 63:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee, D. J., H. J. Wing, N. J. Savery, and S. J. Busby. 2000. Analysis of interactions between Activating Region 1 of Escherichia coli FNR protein and the C-terminal domain of the RNA polymerase alpha subunit: use of alanine scanning and suppression genetics. Mol. Microbiol. 37:1032-1040. [DOI] [PubMed] [Google Scholar]
- 122.Lee, J. H., and T. R. Hoover. 1995. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a σ45-dependent transcriptional activator, interacts with σ45 and the beta subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 92:9702-9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee, S. Y., A. de La Torre, D. Yan, S. Kustu, B. T. Nixon, and D. E. Wemmer. 2003. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 17:2552-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leoni, L., P. Ascenzi, A. Bocedi, G. Rampioni, L. Castellini, and E. Zennaro. 2003. Styrene-catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem. Biophys. Res. Commun. 303:926-931. [DOI] [PubMed] [Google Scholar]
- 125.Lessner, D. J., R. E. Parales, S. Narayan, and D. T. Gibson. 2003. Expression of the nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol. 185:3895-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leuthner, B., and J. Heider. 1998. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol. Lett. 166:35-41. [DOI] [PubMed] [Google Scholar]
- 127.Leveau, J. H. J., F. König, H.-P. Füchslin, C. Werlen, and J. R. van der Meer. 1999. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol. Microbiol. 33:396-406. [DOI] [PubMed] [Google Scholar]
- 128.Leveau, J. H. J., and J. R. van der Meer. 1996. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 178:6824-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lewis, R. J., D. J. Scott, J. A. Brannigan, J. C. Ladds, M. A. Cervin, G. B. Spiegelman, J. G. Hoggett, I. Barak, and A. J. Wilkinson. 2002. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J. Mol. Biol. 316:235-245. [DOI] [PubMed] [Google Scholar]
- 130.Lim, D., K. Poole, and N. C. Strynadka. 2002. Crystal structure of the MexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 131.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 132.Maloy, S. R., and W. D. Nunn. 1982. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J. Bacteriol. 149:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Manzanera, M., S. Marqués, and J. L. Ramos. 2000. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 476:312-317. [DOI] [PubMed] [Google Scholar]
- 134.Marqués, S., M. T. Gallégos, M. Manzanera, A. Holtel, K. N. Timmis, and J. L. Ramos. 1998. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 180:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marqués, S., M. T. Gallégos, and J. L. Ramos. 1995. Role of σs in transcription from the positively controlled Pm promoter of the TOL plasmid of Pseudomonas putida. Mol. Microbiol. 18:851-857. [DOI] [PubMed] [Google Scholar]
- 136.Marqués, S., M. Manzanera, M. M. Gonzalez-Pérez, M. T. Gallégos, and J. L. Ramos. 1999. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with σ32 or σ38 depending on the growth phase. Mol. Microbiol. 31:1105-1113. [DOI] [PubMed] [Google Scholar]
- 137.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 138.McFall, S. M., B. Abraham, C. G. Narsolis, and A. M. Chakrabarty. 1997. A tricarboxylic acid cycle intermediate regulating transcription of a chloroaromatic biodegradative pathway: fumarate-mediated repression of the clcABD operon. J. Bacteriol. 179:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McFall, S. M., S. A. Chugani, and A. M. Chakrabarty. 1998. Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene 223:257-267. [DOI] [PubMed] [Google Scholar]
- 140.McFall, S. M., T. J. Klem, N. Fujita, A. Ishihama, and A. M. Chakrabarty. 1997. DNase I footprinting, DNA bending and in vitro transcription analyses of ClcR and CatR interactions with the clcABD promoter: evidence of a conserved transcriptional activation mechanism. Mol. Microbiol. 24:965-976. [DOI] [PubMed] [Google Scholar]
- 141.McFall, S. M., M. R. Parsek, and A. M. Chakrabarty. 1997. 2-Chloromuconate and ClcR-mediated activation of the clcABD operon: in vitro transcriptional and DNase I footprint analyses. J. Bacteriol. 179:3655-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mermod, N., J. L. Ramos, A. Bairoch, and K. N. Timmis. 1987. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol. Gen. Genet. 207:349-354. [DOI] [PubMed] [Google Scholar]
- 143.Michán, C., B. Kessler, V. de Lorenzo, K. N. Timmis, and J. L. Ramos. 1992. XylS domain interactions can be deduced from intraallelic dominance in double mutants of Pseudomonas putida. Mol. Gen. Genet. 235:406-412. [DOI] [PubMed] [Google Scholar]
- 144.Michán, C., L. Zhou, M. T. Gallégos, K. N. Timmis, and J. L. Ramos. 1992. Identification of critical amino-terminal regions of XylS. The positive regulator encoded by the TOL plasmid. J. Biol. Chem. 267:22897-22901. [PubMed] [Google Scholar]
- 145.Miller, B. E., and N. M. Kredich. 1987. Purification of the cysB protein from Salmonella typhimurium. J. Biol. Chem. 262:6006-6009. [PubMed] [Google Scholar]
- 146.Mishra, V., R. Lal, and Srinivasan. 2001. Enzymes and operons mediating xenobiotic degradation in bacteria. Crit. Rev. Microbiol. 27:133-166. [DOI] [PubMed] [Google Scholar]
- 147.Miura, K., S. Inouye, and A. Nakazawa. 1998. Protein binding in vivo to OP2 promoter of the Pseudomonas putida TOL plasmid. Biochem. Mol. Biol. Int. 46:933-941. [DOI] [PubMed] [Google Scholar]
- 148.Miyauchi, K., H. S. Lee, M. Fukuda, M. Takagi, and Y. Nagata. 2002. Cloning and characterization of linR, involved in regulation of the downstream pathway for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 68:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morawski, B., A. Segura, and L. N. Ornston. 2000. Repression of Acinetobacter vanillate demethylase synthesis by VanR, a member of the GntR family of transcriptional regulators. FEMS Microbiol. Lett. 187:65-68. [DOI] [PubMed] [Google Scholar]
- 150.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mosquéda, G., M. I. Ramos-Gonzalez, and J. L. Ramos. 1999. Toluene metabolism by the solvent-tolerant Pseudomonas putida DOT-T1 strain, and its role in solvent impermeabilization. Gene 232:69-76. [DOI] [PubMed] [Google Scholar]
- 152.Mouz, S., C. Merlin, D. Springael, and A. Toussaint. 1999. A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol. Gen. Genet. 262:790-799. [DOI] [PubMed] [Google Scholar]
- 153.Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann. 1996. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 178:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Müller, T. A., C. Werlen, J. Spain, and J. R. van der Meer. 2003. Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol. 5:163-173. [DOI] [PubMed] [Google Scholar]
- 155.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 156.Nasser, W., S. Reverchon, and J. Robert-Baudouy. 1992. Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol. Microbiol. 6:257-265. [DOI] [PubMed] [Google Scholar]
- 157.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 158.Ng, L. C., C. L. Poh, and V. Shingler. 1995. Aromatic effector activation of the NtrC-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J. Bacteriol. 177:1485-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nichols, N. N., and C. S. Harwood. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida β-ketoadipate pathway. J. Bacteriol. 177:7033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nishino, S. F., J. C. Spain, L. A. Belcher, and C. D. Litchfield. 1992. Chlorobenzene degradation by bacteria isolated from contaminated groundwater. Appl. Environ. Microbiol. 58:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.North, A. K., K. E. Klose, K. M. Stedman, and S. Kustu. 1993. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J. Bacteriol. 175:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ogawa, N., S. M. McFall, T. J. Klem, K. Miyashita, and A. M. Chakrabarty. 1999. Transcriptional activation of the chlorocatechol degradative genes of Ralstonia eutropha NH9. J. Bacteriol. 181:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ohta, Y., M. Maeda, and T. Kudo. 2001. Pseudomonas putida CE2010 can degrade biphenyl by a mosaic pathway encoded by the tod operon and cmtE, which are identical to those of P. putida F1 except for a single base difference in the operator-promoter region of the cmt operon. Microbiology 147:31-41. [DOI] [PubMed] [Google Scholar]
- 165.Ohtsubo, Y., M. Delawary, K. Kimbara, M. Takagi, A. Ohta, and Y. Nagata. 2001. BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276:36146-36154. [DOI] [PubMed] [Google Scholar]
- 166.O'Leary, N. D., W. A. Duetz, A. D. Dobson, and K. E. O'Connor. 2002. Induction and repression of the sty operon in Pseudomonas putida CA-3 during growth on phenylacetic acid under organic and inorganic nutrient-limiting continuous culture conditions. FEMS Microbiol. Lett. 208:263-268. [DOI] [PubMed] [Google Scholar]
- 167.O'Neill, E., L. C. Ng, C. C. Sze, and V. Shingler. 1998. Aromatic ligand binding and intramolecular signalling of the phenol-responsive σ54-dependent regulator DmpR. Mol. Microbiol. 28:131-141. [DOI] [PubMed] [Google Scholar]
- 168.O'Neill, E., C. C. Sze, and V. Shingler. 1999. Novel effector control through modulation of a preexisting binding site of the aromatic-responsive σ54-dependent regulator DmpR. J. Biol. Chem. 274:32425-32432. [DOI] [PubMed] [Google Scholar]
- 169.O'Neill, E., P. Wikström, and V. Shingler. 2001. An active role for a structured B-linker in effector control of the σ54-dependent regulator DmpR. EMBO J. 20:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 171.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 172.Osuna, J., X. Soberon, and E. Morett. 1997. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 6:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Overhage, J., A. U. Kresse, H. Priefert, H. Sommer, G. Krammer, J. Rabenhorst, and A. Steinbüchel. 1999. Molecular characterization of the genes pcaG and pcaH, encoding protocatechuate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Panke, S., B. Witholt, A. Schmid, and M. G. Wubbolts. 1998. Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl. Environ. Microbiol. 64:2032-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pao, G. M., and M. H. Saier, Jr. 1995. Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J. Mol. Evol. 40:136-154. [DOI] [PubMed] [Google Scholar]
- 176.Park, H. S., and H. S. Kim. 2001. Genetic and structural organization of the aminophenol catabolic operon and its implication for evolutionary process. J. Bacteriol. 183:5074-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Park, S. M., H. H. Park, W. K. Lim, and H. J. Shin. 2003. A new variant activator involved in the degradation of phenolic compounds from a strain of Pseudomonas putida. J. Biotechnol. 103:227-236. [DOI] [PubMed] [Google Scholar]
- 178.Park, W., C. O. Jeon, and E. L. Madsen. 2002. Interaction of NahR, a LysR-type transcriptional regulator, with the α subunit of RNA polymerase in the naphthalene degrading bacterium, Pseudomonas putida NCIB 9816-4. FEMS Microbiol. Lett. 213:159-165. [DOI] [PubMed] [Google Scholar]
- 179.Parke, D. 1996. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J. Bacteriol. 178:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Parke, D., and L. N. Ornston. 2003. Hydroxycinnamate (hca) catabolic genes from Acinetobacter sp. strain ADP1 are repressed by HcaR and are induced by hydroxycinnamoyl-coenzyme A thioesters. Appl. Environ. Microbiol. 69:5398-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Parsek, M. R., D. L. Shinabarger, R. K. Rothmel, and A. M. Chakrabarty. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174:7798-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parsek, M. R., R. W. Ye, P. Pun, and A. M. Chakrabarty. 1994. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region. catBC promoter activation by CatR. J. Biol. Chem. 269:11279-11284. [PubMed] [Google Scholar]
- 183.Pavel, H., M. Forsman, and V. Shingler. 1994. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J. Bacteriol. 176:7550-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pelton, J. G., S. Kustu, and D. E. Wemmer. 1999. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292:1095-1110. [DOI] [PubMed] [Google Scholar]
- 185.Pérez-Martín, J., and V. de Lorenzo. 1995. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc. Natl. Acad. Sci. USA 92:9392-9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pérez-Martín, J., and V. de Lorenzo. 1996. ATP binding to the σ54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell 86:331-339. [DOI] [PubMed] [Google Scholar]
- 187.Pérez-Martín, J., and V. de Lorenzo. 1996. Identification of the repressor subdomain within the signal reception module of the prokaryotic enhancer-binding protein XylR of Pseudomonas putida. J. Biol. Chem. 271:7899-7902. [DOI] [PubMed] [Google Scholar]
- 188.Pérez-Martín, J., and V. de Lorenzo. 1996. In vitro activities of an N-terminal truncated form of XylR, a σ54-dependent transcriptional activator of Pseudomonas putida. J. Mol. Biol. 258:575-587. [DOI] [PubMed] [Google Scholar]
- 189.Pérez-Martín, J., and V. de Lorenzo. 1996. Physical and functional analysis of the prokaryotic enhancer of the σ54-promoters of the TOL plasmid of Pseudomonas putida. J. Mol. Biol. 258:562-574. [DOI] [PubMed] [Google Scholar]
- 190.Pérez-Martín, J., K. N. Timmis, and V. de Lorenzo. 1994. Co-regulation by bent DNA. Functional substitutions of the integration host factor site at σ54-dependent promoter Pu of the upper-TOL operon by intrinsically curved sequences. J. Biol. Chem. 269:22657-22662. [PubMed] [Google Scholar]
- 191.Pieper, D. H., and W. Reineke. 2001. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11:262-270. [DOI] [PubMed] [Google Scholar]
- 192.Popham, D. L., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629-635. [DOI] [PubMed] [Google Scholar]
- 193.Popp, R., T. Kohl, P. Patz, G. Trautwein, and U. Gerischer. 2002. Differential DNA binding of transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 184:1988-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Prieto, M. A., E. Díaz, and J. L. García. 1996. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J. Bacteriol. 178:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Prieto, M. A., and J. L. García. 1997. Identification of a novel positive regulator of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Biochem. Biophys. Res. Commun. 232:759-765. [DOI] [PubMed] [Google Scholar]
- 196.Prieto, M. A., and J. L. García. 1994. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component enzyme. J. Biol. Chem. 269:22823-22829. [PubMed] [Google Scholar]
- 197.Providenti, M. A., and R. C. Wyndham. 2001. Identification and functional characterization of CbaR, a MarR-like modulator of the cbaABC-encoded chlorobenzoate catabolism pathway. Appl. Environ. Microbiol. 67:3530-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Quinn, J. A., D. B. McKay, and B. Entsch. 2001. Analysis of the pobA and pobR genes controlling expression of p-hydroxybenzoate hydroxylase in Azotobacter chroococcum. Gene 264:77-85. [DOI] [PubMed] [Google Scholar]
- 199.Raman, N., P. N. Black, and C. C. DiRusso. 1997. Characterization of the fatty acid- responsive transcriptional factor FadR. Biochemical and genetic analyses of the native conformation and functional domains. J. Biol. Chem. 272:30645-30650. [DOI] [PubMed] [Google Scholar]
- 200.Raman, N., and C. C. DiRusso. 1995. Analysis of acyl coenzyme A binding to the transcription factor FadR and identification of amino acid residues in the carboxyl terminus required for ligand binding. J. Biol. Chem. 270:1092-1097. [DOI] [PubMed] [Google Scholar]
- 201.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 202.Ramos, J. L., N. Mermod, and K. N. Timmis. 1987. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol. Microbiol. 1:293-300. [DOI] [PubMed] [Google Scholar]
- 203.Ramos, J. L., C. Michán, F. Rojo, D. Dwyer, and K. Timmis. 1990. Signal- regulator interactions. Genetic analysis of the effector binding site of xylS, the benzoate-activated positive regulator of Pseudomonas TOL plasmid meta-cleavage pathway operon. J. Mol. Biol. 211:373-382. [DOI] [PubMed] [Google Scholar]
- 204.Ramos, J. L., A. Stolz, W. Reineke, and K. N. Timmis. 1986. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc. Natl. Acad. Sci. USA 83:8467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ramos-González, M. I., M. Olson, A. A. Gatenby, G. Mosquéda, M. Manzañera, M. J. Campos, S. Vichez, and J. L. Ramos. 2002. Cross-regulation between a novel two-component signal transduction system for catabolism of toluene in Pseudomonas mendocina and the TodST system from Pseudomonas putida. J. Bacteriol. 184:7062-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 207.Reineke, W., and H.-J. Knackmuss. 1988. Microbial degradation of haloaromatics. Annu. Rev. Microbiol. 42:263-287. [DOI] [PubMed] [Google Scholar]
- 208.Reitzer, L. J., and B. Magasanik. 1986. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 209.Reizer, J., and M. H. Saier, Jr. 1997. Modular multidomain phosphoryl transfer proteins of bacteria. Curr. Opin. Struct. Biol. 7:407-415. [DOI] [PubMed] [Google Scholar]
- 210.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA- binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 212.Roméro-Arroyo, C. E., M. A. Schell, G. L. Gaines, and E. L. Neidle. 1995. catM encodes a LysR-type transcriptional activator regulating catechol degradation in Acinetobacter calcoaceticus. J. Bacteriol. 177:5891-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Roper, D. I., T. Fawcett, and R. A. Cooper. 1993. The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol. Gen. Genet. 237:241-250. [DOI] [PubMed] [Google Scholar]
- 215.Rothmel, R. K., T. L. Aldrich, J. E. Houghton, W. M. Coco, L. N. Ornston, and A. M. Chakrabarty. 1990. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J. Bacteriol. 172:922-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rothmel, R. K., D. L. Shinabarger, M. R. Parsek, T. L. Aldrich, and A. M. Chakrabarty. 1991. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxyl-radical footprinting. J. Bacteriol. 173:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ruíz, R., S. Marqués, and J. L. Ramos. 2003. Leucines 193 and 194 at the N- terminal domain of the XylS protein, the positive transcriptional regulator of the TOL meta-cleavage pathway, are involved in dimerization. J. Bacteriol. 185:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ruíz, R., and J. L. Ramos. 2002. Residues 137 and 153 at the N terminus of the XylS protein influence the effector profile of this transcriptional regulator and the σ factor used by RNA polymerase to stimulate transcription from its cognate promoter. J. Biol. Chem. 277:7282-7286. [DOI] [PubMed] [Google Scholar]
- 219.Ruíz, R., and J. L. Ramos. 2001. Residues 137 and 153 of XylS influence contacts with the C-terminal domain of the RNA polymerase α subunit. Biochem. Biophys. Res. Commun. 287:519-521. [DOI] [PubMed] [Google Scholar]
- 220.Ruíz, R., J. L. Ramos, and S. M. Egan. 2001. Interactions of the XylS regulators with the C-terminal domain of the RNA polymerase α subunit influence the expression level from the cognate Pm promoter. FEBS Lett. 491:207-211. [DOI] [PubMed] [Google Scholar]
- 221.Salto, R., A. Delgado, C. Michán, S. Marqués, and J. L. Ramos. 1998. Modulation of the function of the signal receptor domain of XylR, a member of a family of prokaryotic enhancer-like positive regulators. J. Bacteriol. 180:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Santos, P. M., L. Leoni, I. Di Bartolo, and E. Zennaro. 2002. Integration host factor is essential for the optimal expression of the styABCD operon in Pseudomonas fluorescens ST. Res. Microbiol. 153:527-536. [DOI] [PubMed] [Google Scholar]
- 223.Sasse-Dwight, S., and J. D. Gralla. 1988. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Natl. Acad. Sci. USA 85:8934-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 225.Schell, M. A. 1985. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene 36:301-309. [DOI] [PubMed] [Google Scholar]
- 226.Schell, M. A., P. H. Brown, and S. Raju. 1990. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J. Biol. Chem. 265:3844-3850. [PubMed] [Google Scholar]
- 227.Schell, M. A., and E. F. Poser. 1989. Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J. Bacteriol. 171:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schell, M. A., and P. E. Wender. 1986. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J. Bacteriol. 166:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schirmer, F., S. Ehrt, and W. Hillen. 1997. Expression, inducer spectrum, domain structure, and function of MopR, the regulator of phenol degradation in Acinetobacter calcoaceticus NCIB8250. J. Bacteriol. 179:1329-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Schoemaker, H. E., D. Mink, and M. G. Wubbolts. 2003. Dispelling the myths—biocatalysis in industrial synthesis. Science 299:1694-1697. [DOI] [PubMed] [Google Scholar]
- 231.Schumacher, J., X. Zhang, S. Jones, P. Bordes, and M. Buck. 2004. ATP-dependent transcriptional activation by bacterial PspF AAA+ protein. J. Mol. Biol. 338:863-875. [DOI] [PubMed] [Google Scholar]
- 232.Shingler, V. 2003. Integrated regulation in response to aromatic compounds: from signal sensing to attractive behaviour. Environ. Microbiol. 5:1226-1241. [DOI] [PubMed] [Google Scholar]
- 233.Shingler, V. 1996. Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19:409-416. [DOI] [PubMed] [Google Scholar]
- 234.Shingler, V., M. Bartilson, and T. Moore. 1993. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J. Bacteriol. 175:1596-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shingler, V., and T. Moore. 1994. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J. Bacteriol. 176:1555-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shingler, V., and H. Pavel. 1995. Direct regulation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol. Microbiol. 17:505-513. [DOI] [PubMed] [Google Scholar]
- 237.Skarfstad, E., E. O'Neill, J. Garmendia, and V. Shingler. 2000. Identification of an effector specificity subregion within the aromatic-responsive regulators DmpR and XylR by DNA shuffling. J. Bacteriol. 182:3008-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 240.Spain, J. 1997. Synthetic chemicals with potential for natural attenuation. Bioremed. J. 1:1-9. [Google Scholar]
- 241.Spooner, R. A., M. Bagdasarian, and F. C. Franklin. 1987. Activation of the xylDLEGF promoter of the TOL toluene-xylene degradation pathway by overproduction of the xylS regulatory gene product. J. Bacteriol. 169:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sticher, P., M. C. M. Jaspers, K. Stemmler, H. Harms, A. J. B. Zehnder, and J. R. van der Meer. 1997. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol. 63:4053-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 244.Reference deleted.
- 245.Sunnarborg, A., D. Klumpp, T. Chung, and D. C. LaPorte. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Suzuki, K., N. Ogawa, and K. Miyashita. 2001. Expression of 2-halobenzoate dioxygenase genes (cbdSABC) involved in the degradation of benzoate and 2-halobenzoate in Burkholderia sp. TH2. Gene 262:137-145. [DOI] [PubMed] [Google Scholar]
- 247.Sze, C. C., A. D. Laurie, and V. Shingler. 2001. In vivo and in vitro effects of integration host factor at the DmpR-regulated σ54-dependent Po promoter. J. Bacteriol. 183:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Teramoto, M., S. Harayama, and K. Watanabe. 2001. PhcS represses gratuitous expression of phenol-metabolizing enzymes in Comamonas testosteroni R5. J. Bacteriol. 183:4227-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Teramoto, M., K. Ohnishi, S. Harayama, and K. Watanabe. 2002. An AraC/XylS family member at a high level in a hierarchy of regulators for phenol-metabolizing enzymes in Comamonas testosteroni R5. J. Bacteriol. 184:3941-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Torres, B., G. Porras, J. L. García, and E. Díaz. 2003. Regulation of the mhp cluster responsible for 3-(3-hydroxyphenyl)propionic acid degradation in Escherichia coli. J. Biol. Chem. 278:27575-27585. [DOI] [PubMed] [Google Scholar]
- 253.Tralau, T., J. Mampel, A. M. Cook, and J. Ruff. 2003. Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 69:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Trautwein, G., and U. Gerischer. 2001. Effects exerted by transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:873-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tropel, D., and J. R. van der Meer. 2002. Identification and physical characterization of the HbpR binding sites of the hbpC and hbpD promoters. J. Bacteriol. 184:2914-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Turner, R., and R. Tjian. 1989. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science 243:1689-1694. [DOI] [PubMed] [Google Scholar]
- 258.Valls, M., and V. de Lorenzo. 2003. Transient XylR binding to the UAS of the Pseudomonas putida σ54 promoter Pu revealed with high intensity UV footprinting in vivo. Nucleic Acids Res. 31:6926-6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.van Aalten, D. M., C. C. DiRusso, and J. Knudsen. 2001. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 20:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.van Aalten, D. M., C. C. DiRusso, J. Knudsen, and R. K. Wierenga. 2000. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 19:5167-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 262.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.van der Meer, J. R., A. C. Frijters, J. H. J. Leveau, R. I. L. Eggen, A. J. B. Zehnder, and W. M. de Vos. 1991. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEF chlorocatechol oxidative operon, and analysis of the regulatory region. J. Bacteriol. 173:3700-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Velasco, A., S. Alonso, J. L. García, J. Perera, and E. Díaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Vicente, M., K. F. Chater, and V. De Lorenzo. 1999. Bacterial transcription factors involved in global regulation. Mol. Microbiol. 33:8-17. [DOI] [PubMed] [Google Scholar]
- 266.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 267.Wang, Y. K., J. H. Lee, J. M. Brewer, and T. R. Hoover. 1997. A conserved region in the σ54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol. Microbiol. 26:373-386. [DOI] [PubMed] [Google Scholar]
- 268.Watanabe, T., H. Fujihara, and K. Furukawa. 2003. Characterization of the second LysR-type regulator in the biphenyl-catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 185:3575-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Watanabe, T., R. Inoue, N. Kimura, and K. Furukawa. 2000. Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 275:31016-31023. [DOI] [PubMed] [Google Scholar]
- 270.Wikström, P., E. O'Neill, L. C. Ng, and V. Shingler. 2001. The regulatory N- terminal region of the aromatic-responsive transcriptional activator DmpR constrains nucleotide-triggered multimerisation. J. Mol. Biol. 314:971-984. [DOI] [PubMed] [Google Scholar]
- 271.Williams, P. A., and J. R. Sayers. 1994. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195-217. [DOI] [PubMed] [Google Scholar]
- 272.Wu, R. Y., R. G. Zhang, O. Zagnitko, I. Dementieva, N. Maltzev, J. D. Watson, R. Laskowski, P. Gornicki, and A. Joachimiak. 2003. Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor. J. Biol. Chem. 278:20240-20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Xu, Y., R. J. Heath, Z. Li, C. O. Rock, and S. W. White. 2001. The FadR-DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 276:17373-17379. [DOI] [PubMed] [Google Scholar]
- 274.Yen, K. M., and I. C. Gunsalus. 1985. Regulation of naphthalene catabolic genes of plasmid NAH7. J. Bacteriol. 162:1008-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhang, R. G., Y. Kim, T. Skarina, S. Beasley, R. Laskowski, C. Arrowsmith, A. Edwards, A. Joachimiak, and A. Savchenko. 2002. Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family. J. Biol. Chem. 277:19183-19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]