Abstract
Since double-stranded RNA (dsRNA) has not until recently generally been thought to be deliberately expressed in cells, it has commonly been assumed that the major source of cellular dsRNA is viral infections. In this view, the cellular responses to dsRNA would be natural and perhaps ancient antiviral responses. While the cell may certainly react to some dsRNAs as an antiviral response, this does not represent the only response or even, perhaps, the major one. A number of recent observations have pointed to the possibility that dsRNA molecules are not seen only as evidence of viral infection or recognized for degradation because they cannot be translated. In some instances they may also play important roles in normal cell growth and function. The purpose of this review is to outline our current understanding of the fate of dsRNA in cells, with a focus on the apparent fact that their fates and functions appear to depend critically not only on where in the cell dsRNA molecules are found, but also on how long they are and perhaps on how abundant they are.
INTRODUCTION
The flow of genetic information generally proceeds from DNA to mRNA to protein. While variations on this theme have been noted over the course of the past few decades, this notion remains generally true. But what happens in the cell when mistakes are made along this pathway of gene expression? If DNA synthesis results in mistakes, elaborate response and repair pathways are accessed. If proteins are misfolded, they are often degraded. Likewise, mistakes in RNA synthesis and processing are dealt with by surprisingly sophisticated cellular machineries, some of which have been discussed or reviewed recently (136, 159, 188, 213, 224, 229, 249, 250, 259, 407).
Since double-stranded RNA (dsRNA) has not until recently generally been thought to be deliberately expressed in cells, it has commonly been assumed that the major source of cellular dsRNA is viral infections. In this view, the cellular responses to dsRNA would be natural and perhaps ancient antiviral responses. While the cell may certainly react to some dsRNAs as an antiviral response, this does not represent the only response or even perhaps the major one. A number of recent observations have pointed to the possibility that dsRNA molecules are not only seen as evidence of viral infection or recognized for degradation because they cannot be translated. In some instances they may also play important roles in normal cell growth and function. The purpose of this review is to outline our current understanding of the fate of dsRNA in cells, with a focus on the apparent fact that their fates and functions appear to depend critically on not only where in the cell dsRNA molecules are found, but also on how long they are and perhaps on how abundant they are.
ORIGINS AND PREVALENCE OF ENDOGENOUS dsRNA
While dsRNA may occur in cells as a result of viral infections, there appears to be an abundance of it expressed from within. The first report of endogenous dsRNA came from studies of heterogeneous nuclear ribonucleoprotein particles, where dsRNA was found to exist in a small fraction of them (154). Later, two groups confirmed this observation and reported that 2 to 5% dsRNA is present in native heterogeneous nuclear ribonucleoproteins isolated from HeLa cells (33, 87). This work showed that about 3% of isolated heterogeneous nuclear RNA from HeLa cells is resistant to RNase T1 but sensitive to RNase III, and such fractions can hybridize to cellular DNA following denaturation.
In recent years this picture has become clearer, and important advances have been made in our understanding not only of the extent and nature of antisense RNA expression within cells, but also of the cellular fates of dsRNAs. Until several years ago, while there were a number of reports of naturally occurring antisense RNA in cells of higher eukaryotes, the number of documented cases was relatively small (188). Unexpectedly, however, more recent results have shown that endogenous antisense RNA is rather common. Several groups have suggested that about 1% of all human genes might be transcribed from both strands (86, 207, 312, 344). Most recently, Yelin et al. (423) and Rosok and Sioud (315) used novel computational tools and expressed sequence tag datasets, along with experimental validation studies, to show that the true number may in fact be far higher; at least 5 to 10% of them are impacted by antisense. This antisense frequently lies in the 5′ or 3′ untranslated region of mRNAs.
Antisense expression to 3′ untranslated regions of mRNAs may indeed turn out to be important for the regulation of some gene expression. Lipman (215) observed that the 3′ untranslated regions of about 30% of vertebrate mRNAs are conserved. Why is this so? The purpose of such sequence conservation seems unlikely to reflect a need for numerous highly specific protein-RNA interactions. Rather, an attractive interpretation was that long stretches of conserved 3′ untranslated region sequences may function in RNA-RNA interactions (215). Taken together with the recent findings of common antisense transcription, this may further point to a heretofore unappreciated and pervasive role for antisense regulation within the cell.
Importantly, the estimate that about 8% of genes express natural antisense RNA in cells may still be too low. In the work reported by Yelin et al. (423), only polyadenylated RNAs were examined. Some antisense is almost certainly not polyadenylated. Also, not all transcribed sequences were available for analysis by the methods employed, trans-encoded antisense transcripts were not examined, and antisense RNAs that do not span introns were not included. This work also did not address what may be a large number of small noncoding antisense RNAs (169) or dsRNA resulting from bidirectional transcription from repetitive and transposable elements, which constitute almost half of the entire human genome (59). For example, LINEs (long interspersed nuclear elements) are non-long terminal repeat retrotransposons that are present in about 105 copies in mammalian genomes, constituting about 17% of genomic DNA (198). Most of these (>99.8%) are defective owing to rearrangements, truncations, and mutations (141, 198). A large amount of RNA in cells is related to LINE elements, and a significant fraction of this RNA is of the antisense orientation (27, 232). One might thus reasonably expect there to be significant amounts of cellular dsRNA related to many or most other repetitive and transposable elements.
The message from all of this is that dsRNA is frequently formed in cells. But what happens to it? Its fate turns out to depend critically on at least two parameters: how long it is, and where it is. In the cytoplasm, dsRNAs longer than 30 bp (called long dsRNAs throughout this article) activate the potent interferon and protein kinase R PKR antiviral pathways, resulting in non-sequence-specific effects that can include apoptosis (188). On the other hand, exciting recent work has shown that RNA duplexes of only 21 to 25 bp (short dsRNAs) in the cytoplasm can enter the sequence-specific RNA interference (RNAi) pathway, where they mediate the destruction of targeted mRNAs (123). In the nucleus, many or most long dsRNAs are edited by adenosine deaminases that act on double-stranded RNA (ADARs). Finally, several lines of evidence suggest that nuclear dsRNAs can also lead to gene silencing and heterochromatin formation in an epigenetic, sequence-independent fashion.
FATE OF dsRNA IN THE CYTOPLASM
In higher eukaryotic cells, there is generally thought to be little or no cytoplasmic dsRNA under normal growth conditions. However, it has been known for many years that introduction into the cytoplasm of synthetic dsRNAs [poly(I)-poly(C) or poly(A)-poly(U) duplexes] or naturally occurring dsRNAs can have enhancing effects on immune responses in animals, and can produce resistance to viral infection (56, 89, 197). It is now clear that the cytoplasm possesses not one but several potent response pathways to dsRNA. The best studied of these appear to have evolved as cellular responses to viral infection and are triggered by long dsRNAs.
Long Cytoplasmic dsRNAs Trigger Nonspecific and Global Effects
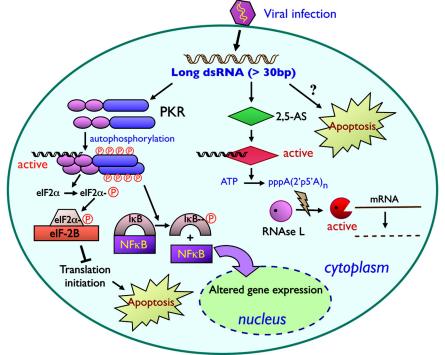
In mammalian cells, long cytoplasmic dsRNAs can trigger the interferon signaling pathway and lead to nonspecific inhibition of gene expression. Since dsRNAs are formed in almost all viral infections, the dsRNA-triggered interferon response has been thought to be primarily an antiviral defense system. In addition to the interferon system, three complementary and independent systems have been implicated in cytoplasmic dsRNA activity: the dsRNA-activated protein kinase R (PKR), the 2-5A system (2′-5′-oligoadenylate synthetase and RNase L), and a newly described Toll-like receptor 3 (TLR3)-mediated dsRNA response. These systems have been studied in detail in recent years, and there are a number of recent reviews of them (108, 129, 188, 235, 321, 409). The basic aspects of the PKR, 2-5A, and RNase L response pathways are summarized in Fig. 1.
FIG. 1.
Signaling pathways of long dsRNAs in the cytoplasm. The major known pathways of signaling by long cytoplasmic dsRNAs are shown and are discussed in detail in the text. Most long cytoplasmic dsRNAs result from virus infections. Such dsRNAs can bind to and activate PKR and 2′,5′-AS as well as activating the interferon pathway. Activated PKR phosphorylates a number of targets, including the translation factor eIF2α and the IκB protein, leading to translation inhibition and effects on gene expression in the nucleus. Activated 2′,5′-AS leads to activation of RNase L, which degrades mRNAs. As discussed in the text, long dsRNAs can also lead to apoptosis.
Interferons.
Interferons are members of a multigene family of inducible cytokines that modulate host immunological functions and can inhibit tumor cell growth and virus multiplication (reviewed in references 108, 321, and 357). Many viral infections or the introduction of long dsRNAs (more than 30 bp) into the cytoplasm induce alpha and beta interferons (61, 260). After induction, interferons are secreted into the extracellular compartment, and can function on neighboring cells as paracrine cytokines, where they initiate interferon signaling pathways, activate transcription factors, and lead to the upregulation of interferon-induced proteins (287), including PKR, 2-5-oligoadenylate synthetase, RNase L, and a cytoplasmic form of the RNA-editing enzyme ADAR1-L (see below) (285). As the interferon pathway is a potent antiviral response of cells, many viruses have evolved countermeasures to allow more efficient replication within host cells. There are a number of reports of viral proteins that inhibit interferon signaling, some of which were reviewed before (188) as well as more recently (17, 64, 94-97, 130, 261, 318, 341, 366).
dsRNA-activated protein kinase PKR.
PKR is a central player in the cytoplasmic response to dsRNA. PKR is an interferon-inducible, dsRNA-activated Ser/Thr protein kinase (49, 299). This enzyme is normally present only at low levels in cells and exists in an unphosphorylated, inactive form (138, 320). In interferon-treated cells, PKR is found predominantly in the cytoplasm and associated with ribosomes (288). However, a fraction of PKR is also found in the nucleus, primarily in the nucleoli (153, 157, 188), which suggests that PKR has multiple functions in cells, some of which are yet to be identified. In fact, it has been reported that a structured RNA element in the 3′ untranslated region of the tumor necrosis factor alpha mRNA binds PKR in the nucleus and that this interaction regulates the splicing of this message (273).
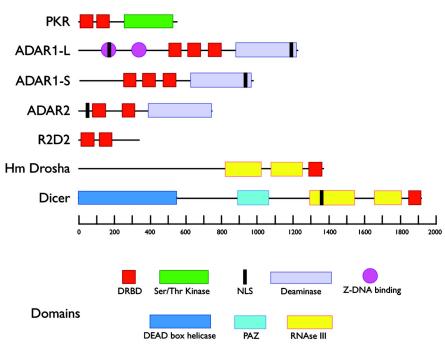
PKR contains two dsRNA binding domains and a kinase domain (246). Figure 2 shows the essential domain structure of this protein, along with that of some other important cellular dsRNA binding proteins that will be discussed here. In vitro, PKR has been shown to be activated by binding to RNAs containing extensive duplex secondary structures (228). When dsRNAs enter cells or are produced in the cytoplasm, they may bind to PKR and induce dimerization, which results in a conformational change, the unmasking of its catalytic domain, and autophosphorylation (Fig. 1). The activation of PKR is independent of the sequence of dsRNAs but depends on both their concentration and on their length. PKR is activated by low concentrations of dsRNAs but inhibited by higher concentrations (322). Manche et al. (228) showed that while a duplex sequence of 11 bp could bind to PKR, 33 bp was the minimum length for activation, and maximal activation was achieved with 80 bp. Furthermore, simultaneous binding of PKR to both dsRNA-binding domains is required for its activation (356). This conclusion, however, needs to be tempered by the observation that at least some short dsRNAs can also induce the PKR pathway in a concentration-dependent manner (see below, in the RNAi section).
FIG. 2.
Domain structures of some key proteins involved in the response of cells to dsRNA. The proteins are discussed in detail in the text, and the conserved domains are shown in colored boxes and identified at the bottom. The numbers refer to amino acid chain lengths.
After PKR is activated, it can phosphorylate a number of substrates, including eukaryotic initiation factor 2 (eIF-2α) (324), the transcription factor inhibitor IκB (186), the human immunodeficiency virus Tat protein (243), nuclear factor 90 of activated T cells, NFAT-90 (199), and the M-phase-specific dsRNA-binding phosphoprotein MPP4 (283). Phosphorylation of eIF2α has important consequences for cellular translation. Phosphorylated eIF2α binds to eIF2B very strongly, which impairs the eIF2B-catalyzed guanine nucleotide exchange reaction, resulting in inhibition of protein synthesis (Fig. 1) (50, 322).
Activated PKR can also mediate signal transduction in response to dsRNAs (299). It can phosphorylate IκB, releasing it from the transcription factor NF-κB, which can now be translocated to the nucleus, where it activates the expression of genes having NF-κB binding sites. These genes include beta interferon (369), Fas (70), p53 (55), Bax (103), and others (186, 187, 409). PKR has also been shown to influence the activity of the transcription factors STAT1 and IRF-1 (187, 408, 412), but the mechanism of this activation is still unclear.
Finally, there is evidence that dsRNAs can trigger apoptosis through PKR (103, 108, 208). PKR can activate apoptotic gene expression and induce apoptosis by activation of the Fas-associated death domain/caspase 8 pathway (12) or of caspase 9 (104). Vorburger et al. (395) reported that PKR also plays a role in E2F-1-mediated apoptosis. These authors further showed that PKR-null mouse embryo fibroblasts demonstrated significant resistance to E2F-1-induced apoptosis. Intriguingly, mice expressing PKR without its dsRNA-binding domains were sensitive to virus-induced apoptosis, while mice expressing PKR lacking its catalytic domain were not (13). Recent evidence suggests that activation of the c-Jun NH2-terminal kinases (JNK) family of mitogen-activated protein kinases and RNase L are also part of this apoptotic pathway (208). These results all serve to illustrate the complexity and diversity of effects that dsRNAs, through interaction with PKR, can exert on cells.
2′,5′-AS/RNase L.
In addition to the PKR pathway, the 2′,5′-oligoadenylate synthetase (2′,5′-AS)/RNase L pathway responds to dsRNA (Fig. 1). 2′,5′-AS is upregulated and activated in interferon-treated and virus-infected cells (173). 2′,5′-AS is activated upon binding to dsRNA, and RNA duplexes of at least 70 bp appear to be required for this activation (188, 248). The recent crystal structure of the porcine 2′,5′-AS reveals two noncontiguous domains which assemble to form an interface for dsRNA binding. After dsRNA binding, a conformational change leads to enzyme activation (124). Activated 2′,5′-AS is capable of polymerizing ATP and other nucleotides into products having novel 2′-5′ linkages.
RNase L, a widely expressed cytoplasmic endoribonuclease, dimerizes and is activated by 2′,5′AS (69). Activated RNase L catalyzes the degradation of viral and cellular RNAs (211), including 18S and 28S rRNAs and mRNAs, thus inhibiting protein synthesis (126, 143). RNase L negatively regulates PKR expression and activity and might cleave PKR mRNA. It has been shown that the absence of RNase L leads to selective stabilization of PKR mRNA, extensive eIF2α phosphorylation, and inhibition of viral protein synthesis (177). Activated RNase L can also induce apoptosis (39, 208). However, Martinand et al. showed that dsRNA can also induce an RNase L inhibitor, which inhibits 2′,5′-AS binding to RNase L (233). These observations suggest that a level of regulation of this cellular response to dsRNAs remains to be clarified.
TLR3-mediated dsRNA response.
Toll-like receptors (TLRs) are a family of innate immune recognition cell surface receptors that recognize a variety of microbial nucleic acid derivatives and metabolites to induce antimicrobial immune responses. Ten human family members have been identified so far. Each TLR family member recognizes distinct pathogen-derived ligands (272, 380). One TLR family member, TLR3, recognizes dsRNA and can induce an antiviral response by the activation of NF-κB and the induction of beta interferon (2, 236). Human TLR3 is expressed ubiquitously in most human tissues, including dendritic cells and intestinal epithelial cells (235). Although TLR3 is a type I transmembrane protein, some (perhaps a different isoform) might exist within the cytoplasm. Although TLR3 lacks an apparent dsRNA-binding domain, it recognizes very specific structural features in dsRNA, because neither dsDNA, poly(dI:dC), single-stranded RNA poly(rU), nor poly(rC) can induce the TLR3-mediated signaling pathway (236). Intracellular TLR3 can recognize some mRNAs, likely through secondary structure features (167). After TLR3 binds to dsRNA, tyrosine phosphorylation occurs in the intracellular Toll interleukin 1 resistance domain, which is essential for downstream signaling (326) via interaction with a number of adaptor proteins (235, 247).
Other response pathways to long dsRNA
It remains possible that other pathways exist by which long cytoplasmic dsRNAs exert effects on cells, but these pathways have not been characterized in molecular detail. For example, dsRNAs appear to be able to directly bind to and inactivate eIF-2, but this effect requires rather high intracellular concentrations of dsRNA (162, 188). In addition, dsRNAs can activate the p38 mitogen-activated protein kinase and JNK pathways, perhaps independently of the PKR or 2′,5′-AS/RNase L system (143).
Long Cytoplasmic dsRNAs Might Have Sequence-Specific Effects
There might exist in the cytoplasm of many mammalian cells an enzyme that can bind to long dsRNAs and deaminate many adenosine residues to inosines. This enzyme is the 150-kDa species of ADAR1, a member of a family of adenosine deaminases that act on RNA (18), which are primarily nuclear and will be described in more detail below. ADAR1 acts on both cellular and viral dsRNAs and catalyzes the hydrolytic deamination of adenosines to inosines (296). This editing can occur promiscuously on long dsRNAs and at site-specific positions, such as in the glutamate GluR-B receptor subunit (339), the serotonin 2C receptor (31), and hepatitis delta virus (152, 295, 413, 415). In vertebrates but not in invertebrates (18, 280), ADAR1 is expressed as two forms, a long form (p150, or ADAR1-L) and short form (p110, or ADAR1-S) (284). ADAR1-L is produced from an interferon-inducible promoter, while ADAR1-S is constitutively expressed (101, 102).
Both immunofluorescence studies and cellular fractionation showed that ADAR1-L is localized in both the cytoplasm and the nucleus, while constitutively expressed ADAR1-S is predominantly present in the nucleus (284). Owing to its cytoplasmic location and its ability to recognize and deaminate dsRNAs, it has been suggested that the cytoplasmic ADAR1-L may play a role in antiviral defense against viruses that replicate in the cytoplasm (18). While no direct evidence for such an activity has yet been reported, it has recently been shown that ADAR1-L activity is especially high in the cytoplasm (414). Thus, a potential important role of ADAR1-L in the cytoplasmic response to long dsRNA cannot be ruled out.
Short Cytoplasmic dsRNAs Elicit Specific Gene Silencing
In the preceding section, we discussed nonspecific inhibition of gene expression induced by long dsRNA. However, there is no evidence that these effects exist in lower organisms, such as plants, yeasts, Caenorhabditis elegans, trypanosomes, and Drosophila melanogaster. All of these organisms appear to lack (both genetically and biochemically) the interferon and PKR response systems, but in these organisms there is an alternative pathway, which is highly sequence specific, known as RNA interference (RNAi). This pathway was only uncovered relatively recently in higher eukaryotes, at least partly because it responds to short (21 to 23 nucleotide) dsRNAs, and its effects are generally masked by the overwhelmingly strong PKR response when dsRNAs longer than 25 bp are present in the cytoplasm (419). The RNAi pathway has been described in detail in a number of excellent recent reviews (60, 74, 77, 91, 121, 123, 142, 238, 242, 262, 292, 332, 342, 343, 345, 372, 426).
RNAi was first discovered, almost accidentally, in C. elegans by Fire and Mello, who observed that introducing a mixture of sense and antisense RNAs into adult nematodes led to substantially more effective gene silencing than introduction of either strand alone (92, 361). Related phenomena known as cosuppression in many species of plants (264), quelling in Neurospora crassa fungi (314), and posttranscriptional gene silencing in plants (185, 210, 239, 292, 383, 385, 386, 405) have been described. dsRNA also causes specific gene silencing in Trypanosome brucei (265), the hydra (220), zebrafish (402), frogs (270), and the slime mold Dictyostelium discoideum (230). Importantly, RNAi has also been observed in Drosophila melanogaster, cultured mammalian cells, in mouse embryos and even adult mice and rats (26, 32, 34, 42, 81, 119, 127, 171, 240, 416, 425). This broad conservation suggests that RNAi is an ancient and general mechanism for gene regulation which might have evolved to have both developmental and antiviral roles.
With the exception of recent work suggesting a role in nuclear gene silencing via heterochromatin induction (see below), essentially all available data are consistent with RNAi's acting primarily or exclusively in the cytoplasm (for example, see reference 429); however, possible RNAi effects in the nucleus have been reported (25, 212). Most of the RNAi machinery in the cell is located primarily in the cytoplasm, and many laboratories have observed that RNAi-mediated gene silencing is more successful when targeting open reading frames or sequences within spliced mRNAs rather than intronic sequences or transcriptional promoter elements.
Molecular mechanism of RNAi.
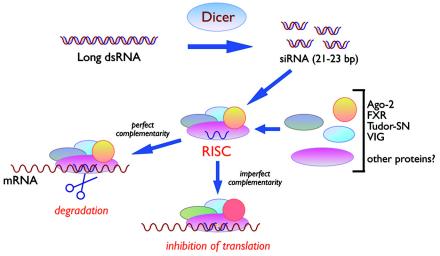
Although intensive biochemical and genetic studies have been carried out on RNAi during the past several years, its detailed mechanism of action has remained elusive. Figure 3 summarizes the key steps of the RNAi pathway as they are now understood, and Table 1 lists some of the key RNAi components that have been described in a variety of model organisms.
FIG. 3.
Mechanism of RNAi. In mammalian cells, long dsRNAs are trimmed in the nucleus by Dicer to siRNAs of 21 to 23 bp, which are then assembled into RISCs. During or after the assembly process, the two siRNA strands are unwound, and only one remains in active RISCs. These recognize their cytoplasmic mRNA targets by complementarity base pairing and direct mRNA degradation. In the case of micro-RNAs, nuclear precursors are first trimmed by the Drosha enzyme into highly structured RNAs of about 70 nucleotides in length. In the cytoplasm, these are further processed by Dicer to yield mature micro-RNAs that assemble into complexes related to RISCs. In this case, target sequences in mRNA 3′ untranslated regions are recognized by imperfect base pairing (indicated as a yellow bar in the siRNA-mRNA hybrid at the bottom) and lead to translation inhibition by a still-unclear mechanism. See the text for details.
TABLE 1.
RNAi-related and RISC proteins from various organismsa
| Gene or protein | Homo sapiens | Drosophila melanogaster | Caenorhabditis elegans | Schizosaccharomyces pombe | Arabidopsis thaliana |
|---|---|---|---|---|---|
| RNase III family | Dicer (21, 67) | Dcr1, Dcr2 (21) | DCR-1 (21, 183) | Dcr1 (21, 115, 116) | DCL-1 (CAF/SIN1/SUS1) (21, 90, 331) |
| Dicer-associated protein | R2D2 (216) | RDE-4 (216, 363) | HYL1 (389) | ||
| Argonaute family | eIF2Cs/Grep95 (67) | Ago1, Ago2, Aubergine (120) | RDE-1 (85, 362, 374) | Ago1 (115, 116) | AGO1/SGS4 (85, 253) |
| Fragile X family | FMRP (41, 144) | dFXR/dFMR1 (41, 144) | |||
| Vasa intronic gene | PAI-RBP-1 (41) | VIG (41) | F56d12.5/VIG-1 (41) | ||
| Nuclease | P100 (40) | Tudor-SN (40) | F10G7.2/TSN-1 (40) | ||
| RNase D | MUT-7 (175) | ||||
| DExH box helicase | DRH-1, DRH-2 (363) | ||||
| DEAD box helicase | Spindle E (172) | MUT-14 (176, 371) | |||
| RNA helicase | Armitage (54, 376) | SMG2 (68) | SDE3 (57) | ||
| Putative RdRp | EGO-1, RRF-1/RDE-9, RRF-3 (350, 354) | Rdp1 (115, 116) | SGS2/SDE1, SGS3 (57, 258) | ||
| Transmembrane protein | SID-1 (88) |
Primary references are shown in parentheses.
RNAi is carried out in two distinct steps. In the first step, long dsRNAs are processed into short 21- to 23-nucleotide-long effector dsRNAs called small interfering RNAs (siRNAs, 21 to 25 nucleotides long in plants) (82, 117, 118, 428). These short siRNAs have been isolated from lysed Drosophila cells after addition of dsRNAs (119) and from C. elegans (282). In the second step, the siRNAs are assembled into RNA-induced silencing complexes (RISCs), which direct the specific cleavage of target mRNAs. In these complexes, the short dsRNA duplex is unwound, generating active RISCs containing single siRNA strands (269). In principle, either of the two siRNA strands can be incorporated into RISCs. In practice, however, there appear to be rules that govern the selection or stability, resulting in a bias of one strand chosen over the other (178, 336).
Dicer cleaves long dsRNAs into siRNAs.
The initial cleavage reaction that produces siRNAs (but not the subsequent cleavage of mRNA targets) is mediated by a multidomain RNase III family enzyme, Dicer (21, 269, 362; reviewed in references 36 and 196). Dicer is a large protein of 220 kDa and contains an N-terminal heDExH/DEAH RNA helicase motif/ATPase domain, a PAZ protein-protein interaction domain, two RNase III-like catalytic domains, and a C-terminal dsRNA-binding domain (300) (Fig. 2). This enzyme is relatively well conserved in eukaryotes and has been found both in the nucleus and in the cytoplasm (23). So far, it has been identified in Arabidopsis, Dictyostelium, the fission yeast Schizosaccharomyces pombe, C. elegans, Drosophila melanogaster, and mammalian cells. It has not, however, been found in the budding yeast Saccharomyces cerevisiae. While most organisms have a single identifiable Dicer gene, D. melanogaster has two (dcr-1 and dcr-2), and Arabidopsis has four (331). So far, most biochemical characterizations have been carried out with the Drosophila enzymes, which might have distinct but complementary activities (145, 205, 375). dcr-1 is essential for mature micro-RNA (which is another class of small noncoding RNA, see below) processing, while dcr-2 is essential for the production of functional siRNAs. Consistent with its homology to RNase III class enzymes, Dicer cleaves dsRNAs into 21- to 23-bp double-stranded siRNAs which process two nucleotide 3′ extensions and phosphates at the 5′ ends.
Dicer function appears to be important for normal development. Mutations in the plant homologue of Dicer caused developmental abnormalities and infertility (107, 142, 149). Similarly, deletion of Dicer in C. elegans led to sterility (174, 183). Drosophila dcr-2 null mutants are viable, fertile, and morphologically normal but have a severe RNAi defect, while dcr-1 mutants displayed no apparent RNAi defect but exhibited morphological defects. This is consistent with a model in which dcr-1 is essential for the micro-RNA pathway while dcr-2 is essential for the RNAi pathway (205). Recently, genetic deletion of the Dicer gene in fission yeast caused defects in chromosome segregation (115). These results further pointed to a nuclear function for this enzyme, as would also be inferred from its proposed activity in processing micro-RNAs, which are generated from precursors transcribed in the nucleus (see below). Recent reports indicate that Dicer is also essential for mouse (22) and zebrafish (406) development.
RISC complex.
The composition of RISCs has still not been completely determined and remains somewhat controversial. Biochemical analyses in a number of systems have identified numerous components (37, 40, 41, 67, 85, 120, 144, 216, 253, 269, 291, 363, 410), and recent work has suggested that there are common mechanisms of RNAi between plants and animals (364).
Size estimates of RISC have ranged from less than 200 kDa for a human complex (41), to 360 kDa (269) and 500 kDa (120) for the Drosophila complex, to even larger complexes (216). Possible biochemical components are listed in Table 1 and include Dicer proteins (67, 145, 205, 291, 375), the dsRNA-binding protein R2D2 and its homologues (216, 363), members of the Argonaute gene family in S. pombe (115), plants (AGO1/SGS4) (85), Neurospora crassa (201), Trypanosoma brucei (76), C. elegans (362, 374), D. melanogaster (37, 85, 120, 410), and mammals (67, 370; reviewed in reference 37), RNA helicases (54, 376), components having yet unknown functions (for example, the Drosophila argonaute-2 protein along with the Drosophila homolog of the fragile X syndrome proteins FMRP and FXR1) (41, 144), and putative nucleases that mediate the cleavage of target mRNAs by RISC. The identification of the nuclease that is responsible for mRNA cleavage has been controversial. Recently, the first RISC subunit containing a recognizable nuclease domain (tudor staphylococcal nuclease) was reported (40). However, recent research from another group argued against this possibility (114, 334).
RNAi may be intimately connected to translation.
A number of lines of evidence point to a mechanistic connection between the RNAi response and translation, though a direct biochemical connection has not yet been described. First, the cytoplasmic RNAi machinery is commonly found to colocalize with polysomes. For example, in D. melanogaster, components of the RNAi machinery clearly interact with the translation machinery (144). Second, the RNAi response is almost certainly mechanistically related to the micro-RNA pathway, which is thought to regulate translation efficiency (shared components, and micro-RNAs can be mutated to act as siRNAs) (see below). Third, recent work has suggested that RNAs that are not translated are refractory to siRNA inhibition, while those being actively translated are effective targets. Thus, untranslated RNA virus “negative” strands appear to be resistant to RNAi cleavage, while their complementary “positive” strands are sensitive (24, 172). An alternative interpretation is that “resistant” RNAs are simply packaged in such a way as to be refractory to siRNA interactions. In human immunodeficiency virus studies, some researchers found nontranslated, infecting human immunodeficiency virus type 1 RNAs to be resistant to RNAi (139); however, others saw the opposite result (150). Finally, it is possible that RNAi has a connection to the phenomenon of translation-associated nonsense-mediated mRNA decay, since SMG2−/− mutants of C. elegans, which are deficient in nonsense-mediated mRNA decay, have also been shown to be deficient in RNAi (51, 68).
Regulation of RNAi.
There are a number of ways in which the RNAi response might be regulated. First, individual proteins important for RNAi might be directly regulated in their expression or activity. This aspect of regulation has not yet been systematically evaluated, largely because the list of components of the RNAi machinery is not yet complete. Second, the siRNAs or mRNA targets may contain features that impact recognition by the RNAi machinery. This has been an active area of investigation in many groups and depends not only on which siRNA strand is incorporated into the RISC complex (178, 336) but also on kinetic aspects of target cleavage. Most recently it has been reported that different regions of siRNA have different contributions to cleavage: the 5′ ends of siRNAs contribute more to the binding to target mRNA, while the central regions and 3′ end of siRNAs contribute to providing a helical geometry which is required for cleavage (114).
Finally, cellular or viral proteins that interfere with the RNAi response pathway might be expressed. Recent work in C. elegans identified a protein, Eri-1, that contains a nucleic acid binding domain and an exonuclease domain which can inhibit RNAi activity. Since Eri-1 is conserved evolutionally, this negative regulation might be general in other species (170). In plants, RNAi has an important antiviral activity. However, a tombaviral protein, p19, has been reported to suppress RNAi in infected hosts (44, 195, 384, 421; reviewed in references 146 and 427).
Persistence and spreading of the RNAi response.
In some but not all cells, the RNAi response can persist for multiple cell generations and can even spread from cell to cell (92, 112). This effect is most evident in C. elegans (92, 112) and in plants (277, 387). One way in which the RNAi effect could be maintained for long periods of time would be via the activity of an RNA-dependent RNA polymerase (RdRp) activity, which uses siRNAs as primers to convert RNA into dsRNAs that are degraded to produce new siRNAs, called secondary siRNAs, thereby amplifying the gene-silencing effect in the cells. Another outcome of such an activity would be the spreading of the RNAi effect to sequences upstream or downstream of the originally targeted sequence.
An RdRp enzyme is required for RNA silencing in a number of organisms (53, 214, 230, 347, 354) (Table 1). However, RdRp is probably not required for RNAi in D. melanogaster or mammals. Schwartz et al. (337) showed that RNAs lacking a 3′ hydroxyl group cannot be extended by RdRp but can nevertheless generate a robust RNAi response in D. melanogaster. Another group drew the same conclusion based on data obtained in mammalian oocytes (358). Further, RNAi is exon specific in D. melanogaster (42), also supporting the conclusion that RdRp is not required. Most recently, Chi et al. (46, 65) showed, using microarrays and human 293T cells, that siRNA-induced gene silencing is highly gene specific and that secondary siRNA is not detectable.
Both in worms and in plants, the RNAi effect can spread systemically, from cell to cell (92, 112, 277, 387). This is likely due to the presence of specific cell surface receptors for siRNAs. It was recently reported that the C. elegans Sid-1 protein mediates siRNA uptake and is essential for systemic RNAi (88). When expressed in D. melanogaster, this protein also allowed the insect cells to take up dsRNA. A recent genetic screen for the RNAi spreading defect in C. elegans isolated genes rsd2, rsd3, rsd4, and rsd6 (rsd stands for RNAi spreading defective), which are required for systemic gene silencing (373).
Micro-RNAs Are Related to siRNAs and May Use a Similar Pathway
Micro-RNAs represent an abundant and important class of small, ≈22-nucleotide noncoding RNA species in cells. These RNAs are involved in many processes, including regulation of gene expression during development and defense against viruses. While they are not generated from perfect duplex RNA precursors and do not act by perfectly matching their targets through complementary base pairing, micro-RNAs nevertheless must be included in any discussion of RNAi, as they appear to function through the same underlying cellular machinery. There are a number of excellent recent reviews of micro-RNA processing and function (4-6, 14-16, 38, 91, 113, 140, 192, 338, 392).
In the past year there has been enormous progress in the biochemical and computational identification of novel micro-RNA species (109, 190, 191, 193, 311). Hundreds of these small RNAs are now known, but the functions of the vast majority of them are still unclear. Recently it has been shown in the mouse that miR-181 is expressed in hematopoietic cells and controls hematopoiesis (45), while the C. elegans lys-6 micro-RNA controls neuronal left-right asymmetry (160). Intriguingly, even viruses may encode micro-RNAs to regulate host or viral gene expression (290).
Pre-micro-RNAs are transcribed as longer precursors which are processed in two steps. In the first step, the primary transcripts (pri-micro-RNA) are cleaved to shorter precursors of ≈70 nucleotides (pre-micro-RNA) by an RNase III family member related to Dicer, called Drosha, in the nucleus (204) (Fig. 2). These precursors exist as highly structured, imperfect hairpin RNAs that are further processed by Dicer in the cytoplasm to mature ≈21-nucleotide micro-RNAs (204). Recently, a Ran-dependent importin-β-related receptor, exportin-5, has been shown to mediate efficient nuclear export of pre-micro-RNAs (181, 223, 424).
Unlike many or most siRNAs, however, only one of the two strands produced by Dicer cleavage is generally assembled into a RISC-like structure. The structural components of micro-RNPs may differ somewhat from those in RISCs (257); however, the two complexes have many common components (72, 158, 257, 388).
While most micro-RNAs have unknown functions, a general picture based on detailed mechanistic studies of several individual species is becoming clear (4, 38). The best-characterized micro-RNAs are lin-4 and let-7 of C. elegans. lin-4 and let-7 regulate endogenous genes involved in developmental timing in C. elegans. let-7 and lin-4 mutant worms show abnormal development (203, 310). lin-4 is antisense to sequences in the 3′untranslated region of mRNAs lin-14 and lin-28, while let-7 is complementary to the 3′ untranslated region of lin-41. These micro-RNAs act by inhibiting protein synthesis through an unknown translational repression pathway. Evidence from lin-4 studies suggests that mRNA stability, polyadenylation level, and translational initiation are not affected (310).
This type of regulation likely exists as well in mammalian cells because overexpression of miR-30 and miR-21 can repress gene expression without changing mRNA stability (430, 431). It is reasonable to predict that some novel micro-RNAs, if not all, will turn out to regulate the expression of genes such as lin-4 and let-7. However, we cannot exclude the possibility that some micro-RNAs may target mRNA regions other than the 3′ untranslated region, and it remains possible that some micro-RNAs may interact functionally with proteins rather than RNAs.
Although siRNAs and micro-RNAs are very similar in how they are produced and assembled into macromolecular complexes, their effects on gene expression are distinct and in at least some instances appear to be related more to how they interact with their targets than with how they are produced. Thus, mutation of a micro-RNA to be perfectly complementary to a target mRNA can lead to RNA degradation, while mutation of an siRNA to be imperfectly complementary to a target can lead to translational inhibition rather than mRNA degradation (66, 432). Also, in plants, micro-RNAs with imperfect complementarity to their targets can nevertheless mediate mRNA cleavage (276). Finally, micro-RNA can direct the cleavage of HOXB8 mRNA in mammalian cells (422), suggesting that micro-RNAs and siRNAs might in some instances regulate gene silencing in an overlapping way.
Applications of RNAi and siRNA Technology
Owing to the lack of an interferon/PKR response pathway in C. elegans and D. melanogaster, long dsRNA successfully induces an RNAi response in these organisms. However, as discussed above, introduction of long dsRNAs into mammalian cells will activate the interferon, PKR, and RNase L pathways and result in nonspecific inhibition of gene expression. This fact severely limits the applications of dsRNA in mammalian cells. However, the Tuschl group first showed that this limitation could be overcome by introduction by transfection of synthetic short double-stranded siRNAs (21 to 23 bp) into cultured mammalian cells (81). Specific inhibition of expression of the target gene was achieved without activation of the nonspecific pathway. This work paved the way to apply RNAi to specifically knock down endogenous genes, transgenes, or viral genes in a wide variety of eukaryotic cells, including human (245).
RNAi has now become a powerful tool for reverse genetics studies and antiviral studies in the laboratory. Researchers are increasingly using RNAi combined with traditional molecular or genetic methods to characterize the functions of proteins in cell growth and development (42, 164, 222, 227, 396). However, caution is warranted, as it has been reported that high doses of siRNAs can in fact lead to activation of the interferon and PKR pathways (29, 351).
The past few years have witnessed the development of a large number of useful approaches that take advantage of RNAi as a tool to study basic biological processes or in the production of novel therapeutics and antiviral agents. There are a number of useful recent reviews on this subject, for uses in both plants and animals (1, 8, 35, 105, 106, 128, 139, 150, 156, 161, 165, 200, 202, 234, 297, 302, 305, 346, 349, 367, 368, 382, 383, 398, 401, 405, 411, 417).
Particular success has already been achieved in the application of RNAi-based technologies to the inhibition of the replication of a number of viruses, including human immunodeficiency virus-1 (52, 150, 202, 268), other retroviruses (139), Hepatitis B virus (241) and recently, influenza A virus (100).
In this rapidly developing area, a number of powerful new methods have been developed for the introduction of siRNAs into cells or animals or for their production within cells. One can readily purchase synthetic RNAs for use in transfection experiments, or one can produce them in the laboratory by in vitro transcription or by digestion of dsRNAs with recombinant Dicer or RNase III (263, 418). There are a number of reports of the development of expression vectors that produce intracellular hairpin structures that can be processed by Dicer into functional siRNAs (30, 274, 275, 425). Both DNA-based (360) and lentiviral RNA virus-based (7, 316) delivery vectors have been described. Recently, a CRE-lox-based strategy has been developed for temporal or tissue-specific knockdown in animals (422).
Finally, the RNAi pathway may have evolved to be a major antiviral pathway in plants. This is evidenced by the evolution of plant virus genes that target the RNAi machinery as a way of evading the host's innate defenses. For example, Flock house virus infects both plant and D. melanogaster cells, and this virus might also be able to infect mammalian cells. In insect cells, infection by this nodavirus leads to accumulation of siRNAs specific for the viral genome. However, the Flock house virus B2 protein functions to block RNAi silencing in both plant and insect cells (209). These results also serve to point out the extent of conservation of the RNAi pathway between the plant and animal kingdoms.
Specificity of siRNA
Although studies have shown that a single mismatch between an siRNA and target mRNA can abolish silencing (83), recently more and more investigations on the specificity of RNAi have arrived at more cautious conclusions. A number of groups have now reported that there is sometimes unexpected off-target gene silencing by the RNAi or micro-RNA machinery (147, 328). siRNAs can activate the interferon response, PKR, 2′,5′-AS, TLR3, or other dsRNA response pathways in a concentration-dependent manner (166, 289, 351). Such activation of nonspecific dsRNA response pathways may depend on how the siRNAs are made.
There are three common ways to make 21- to 23-nucleotide siRNAs: chemical synthesis, in vitro transcription by bacteriophage polymerases, and RNA polymerase III promoter-driven vector-based short heterochromatic RNA. Independent studies showed that siRNAs and short heterochromatic RNAs made by bacteriophage polymerase or RNA polymerase III promoter-driven vectors can induce the interferon and dsRNA response pathways, while chemically synthesized siRNAs are less able to do so (29, 179). However, the interferon response can be alleviated by adjusting polymerase III-driven vector sequences or by eliminating the 5′ triphosphate of bacteriophage polymerase-produced transcripts, which appears to play a role in initiating an interferon response (286, 323). Therefore, caution must be exerted in the design and use of siRNAs.
FATE OF dsRNA IN THE NUCLEUS
While dsRNA in the cytoplasm is generally thought to arise after viral infection, most or all naturally occurring sense-antisense dsRNAs are made within the nucleus, and there is no direct evidence that the resulting duplex RNAs ever end up in the cytoplasm under normal growth conditions. Additionally, there is no evidence that nuclear dsRNAs trigger the PKR, interferon, or 2′,5′-AS pathway. Nuclear dsRNA must therefore be treated differently than that in the cytoplasm. We will discuss two nuclear fates, ADAR editing and gene silencing by formation of heterochromatin.
Adenosine Deaminases That Act on RNA
There may be two distinct fates for dsRNA in the nucleus, and the link between these fates is still obscure. The first and clearest fate of nuclear dsRNAs is to be A-to-I edited by members of the ADAR enzyme family. ADARs are ubiquitously expressed in the nuclei of higher eukaryotes, including C. elegans, D. melanogaster, and mammalian cells. There are a number of good recent reviews of the biochemistry and biological functions of these enzymes (18, 225, 226, 330, 340).
In eukaryotes, a dsRNA unwinding or modifying activity was first discovered in Xenopus laevis (19, 266, 306). The enzyme, ADAR1, was subsequently found to be a member of a small gene family whose members catalyze the conversion of adenosines to inosines within dsRNA (20, 266, 397) by hydrolytic deamination (296). Mammals express two active enzymes, ADAR1 and ADAR2; C. elegans likewise expresses two forms, adr-1 and adr-2; but D. melanogaster has but a single ADAR gene, dADAR. The human ADAR1 and ADAR2 enzymes have slightly distinct but overlapping substrate specificities (206).
Long nuclear dsRNAs are promiscuously edited by ADAR.
ADAR editing is highly sensitive to the length of the duplex. Perfect RNA duplexes of less than 15 bp are modified only inefficiently in vitro and perhaps not at all in vivo (266). Optimal activity is generally seen with dsRNAs of at least 25 to 30 bp and preferably greater than 100 bp in length (20, 266). Thus, short RNA stem-loop structures and duplexes are generally refractory to editing, while more extensively base-paired molecules are favored editing substrates. In long perfect duplexes, about 50% of the A's on each strand can be edited in an almost random pattern, with the exception of a clear 5′ neighbor preference for A or U (294). The resulting RNAs contain I-U base pairs which make the RNA duplex unstable and may lead to partial or complete unwinding (20). In fact, extensive unwinding of edited duplexes might be considered a likely fate, as RNA helicase activity appears to be closely connected to editing in vivo (28, 308).
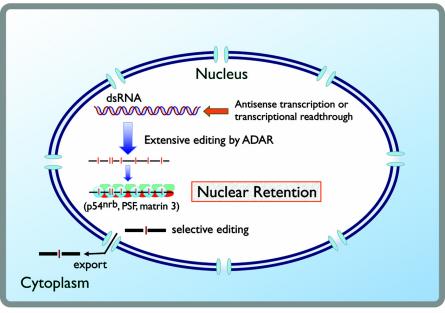
In the mouse polyoma virus model system, it was found that at late times in infection, large amounts of long dsRNA are produced in the nucleus (189, 219). These molecules are promiscuously edited by ADAR, but the resulting inosine-containing RNAs are not exported to the cytoplasm (189). Further biochemical analysis revealed that many hyperedited RNAs appeared to bind tightly and specifically to a protein complex that resulted in their retention in the nucleus (433). The highly conserved and abundant nuclear protein p54nrb binds hyperedited RNAs with striking specificity. This protein exists in a complex with the splicing factor PSF and the inner nuclear matrix structural protein matrin 3, which confers highly cooperative binding to inosine-containing RNA and leads to nuclear retention, most likely via attachment to the nuclear matrix (433).
p54nrb is the first identified nuclear RNA-binding protein that requires inosine for high-affinity binding to RNA. This protein has also recently been reported to exist in novel nuclear compartments called paraspeckles (93). These data led to the important conclusion that nuclear antisense RNA leads to hyperediting and subsequent nuclear retention of target transcripts. Messages with only one or a few inosines (resulting from editing of short duplex regions in pre-mRNAs) escape and can be delivered to the cytoplasm. This discrimination between selectively edited RNAs and promiscuously edited RNAs provides the cell a useful way in which antisense RNA can regulate gene expression and is diagrammed in Fig. 4.
FIG. 4.
One fate of long dsRNAs in the nucleus. Duplex RNAs produced by antisense transcription or transcriptional readthrough are promiscuously edited by ADAR enzymes, generating RNAs with extensive adenosine-to-inosine modifications. These molecules are bound tightly and cooperatively to a nuclear matrix-associated complex of p54nrb, PTB-associated splicing factor, and matrin 3, which prevents their export to the cytoplasm (433). RNAs with only one or a few inosines are not tightly bound and are allowed to leave the nucleus.
ADARs can also edit RNAs in a site-selective manner.
ADARs, however, can also edit RNAs in a highly site-selective manner, but this is not dependent on perfect cRNA duplexes. Selective editing of only one or a few A's in an mRNA molecule can lead to the expression of specifically altered proteins and, in the case of the hepatitis delta virus antigenomic RNA, can regulate gene expression. Selective editing leads to altered proteins because inosines base-pair preferentially with cytosines and are recognized as G's by the translation machinery. A-to-I editing thus can lead to altered coding potential in mRNAs but cannot generate stop codons. Selective editing has been reported primarily in mRNAs that are important for the function of the nervous system, which led to speculation that editing has an ancient role in the evolution of nervous system function and behavior (307).
Among the best-studied examples is the mRNA encoding the mammalian glutamate receptor subunits (135, 339, 355). Interestingly, selective editing in this as well as other genes results from double-stranded secondary structures formed by base pairing between exons and downstream intron elements (79, 131, 221). Transcripts encoding the 2C subtype of the neurotransmitter serotonin receptor also undergo RNA editing events in which genomically encoded adenosine residues are converted to inosines (31, 267). In this system, as for the glutamate receptor, editing requires the interaction of exon sequences with downstream intron sequences. Editing in these systems is not at all promiscuous but rather is directed to specific adenosines which are imbedded in favorable secondary structures, often involving an unpaired adenosine (163, 182, 271, 327, 415). Thus, while 15-bp perfect duplexes cannot be edited by ADAR in vitro, a minimal natural selective editing substrate consisting of a 15-bp dsRNA stem with a single-base mismatch was sufficient for editing, though longer substrates were certainly more optimal (133). Very recent studies combining comparative genomics and experimental approaches allowed the exciting discovery of a large number of new ADAR substrates (137). Interestingly, these new substrates follow the general pattern of intron-exon interactions.
The observation that selective editing involves intron-exon interactions leads to several important conclusions that might be important as well for hyperediting. First, editing must be fast, and ADAR must be present in or near the elongating RNA polymerase II enzyme complex. This is consistent both with studies on the general localization of ADAR enzymes throughout the nucleus and to sites of transcription (though they may sometimes concentrate somewhat in the nucleolus) (78, 359) and with the observation that ADAR exists in large ribonucleoprotein particles containing splicing factors (303). Second, there must exist regulation and coordination of the editing versus splicing events in these genes, since rapid splicing would preclude editing, and failure to resolve RNA secondary structures might interfere with the splicing process. Indeed, the involvement of a specific RNA helicase enzyme, the D. melanogaster maleless protein, and its mammalian homologue, RNA helicase A, in the coordination of editing and splicing has been observed (28, 308).
Editing within noncoding regions might also be important for the regulation of gene expression. Morse et al. (255, 256) developed a method to identify inosine-containing RNAs. Using this method, they found ADAR substrates within 3′ untranslated regions, introns, and noncoding RNAs in C. elegans and in human brain. Since repetitive elements such as LINE and Alu sequences were found to contain edited bases, and since these elements are commonly present in the intronic or untranslated regions, this finding also raised the possibility that ADAR might be involved in the regulation of repetitive elements or transposon expression or functions in mammalian genome (254).
ADAR structure and function.
The ADARs from all organisms contain variable numbers of dsRNA-binding domains and a highly conserved C-terminal catalytic domain. Figure 2 illustrates the domain structure of the human ADAR1 long and short isoforms. The longer form, ADAR1-L, is found in both the nucleus and the cytoplasm, while the short form, ADAR1-S, is mostly nuclear.
The nucleocytoplasmic distribution of ADAR1-L is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain (359). The catalytic domain is sufficient for deaminase activity on some selectively edited minimal substrates but not on long dsRNAs (133). Three dsRNA-binding motifs are important for the catalytic activity of ADAR1, but the contributions of the three dsRNA-binding domains are different, with dsRNA-binding domains III being fundamentally important for deaminase activity (217, 218). ADAR1-L (Fig. 2) contains two N-terminal Z-DNA binding domains (335), three double-stranded RNA binding motifs (dsRBD I, II, and III) (180), and a C-terminal deaminase catalytic domain (194, 218). The Z-DNA binding domain is not required for catalytic activity, but it has been suggested that this binding domain might target ADAR1-L to sites of active transcription in the nucleus (132). Recently it has also been shown that ADAR1 is dynamically associated with the nucleolus, from which it can be recruited to sites of editing I the nucleoplasm (63, 325).
The individual ADAR1 dsRNA-binding domains have distinct in vivo localization capabilities, which may be important for chromosomal targeting, substrate recognition, and editing specificity (73). Recent studies also showed that ADAR1 is a nucleocytoplasmic shuttling protein with a nuclear localization signal and nuclear export signals (298). In their active state, ADARs appear to exist as homodimers (47, 98, 151).
Biological importance of ADARs.
The importance of ADAR activity for viability and development has been revealed through the construction and analysis of knockout mutant organisms. The first animal in which ADAR was knocked out was D. melanogaster (281). While mutant flies were viable, they exhibited striking defects in adult nervous system function and integrity. C. elegans has two ADAR genes, adr-1 and adr-2, with adr-1 being expressed in most cells of the nervous system and developing vulva (378). Genetic knockouts have shown that, while not essential for viability, both ADARs are important for normal behavior, with mutants showing defects in chemotaxis (378).
More recently, gene knockout studies in transgenic mice have provided very interesting insights into ADAR function in mammals. ADAR1 appears to be more important for development and viability than ADAR2. ADAR2 has only a single essential in vivo target, a CAG codon in the GluR-B gene (134). ADAR2−/− mice die of neurological disorders but appear normal if the GluR-B gene is replaced with one in which a single codon is replaced with an edited version (168). An earlier study indicated that ADAR1−/− homozygous mice die as embryos, while ADAR1+/− mice have defects in erythropoiesis in the liver (399). More recent studies have shown that ADAR1−/− mice die by embryonic day 11.5 with widespread apparent apoptosis. ADAR1−/− fibroblasts are prone to apoptosis induced by serum deprivation (400). Also, Hartner et al. (125) found, studying knockout mice in vivo, that ADAR1 selectively edits two of the five known edited adenosines in the serotonin 5-HT2C receptor pre-mRNA. Further, homozygous knockout of ADAR1 leads to embryo-lethal defects in liver structure and hematopoiesis. Thus, ADAR1 is clearly important in the development of nonneuronal tissues.
Possible connections between ADARs, PKR, and RNAi.
Finally, some highly structured RNAs point to complex and subtle effects that relate to the interplay between the different dsRNA response pathways of PKR, RNAi, and ADAR editing. The hepatitis delta virus genome is largely but not completely duplex RNA. While it can be edited in a site-selective manner by ADAR and can activate PKR (48, 313), it cannot be cleaved by Dicer (43). On the other hand, fragile X syndrome transcripts encode trinucleotide repeats that can form RNA hairpins that cannot activate PKR but are efficiently cut by Dicer (122). Also, as mentioned above, ADAR knockouts of C. elegans show chemotaxis defects. However, these defects could be rescued specifically by crossing an adar−/− strain to RNAi-defective strains carrying rde-1 or rde-4 (377).
RNAi Machinery in the Nucleus
Role of the RNAi machinery in the establishment of heterochromatin.
Based on the considerations raised so far in this review, it would seem logical to conclude that, since most of the RNAi machinery is localized in the cytoplasm, this compartment is where most of the important RNAi-related effects are manifested. However, recent work from elegant genetic studies in the fission yeast S. pombe may force a reevaluation of this paradigm. In the past several years, it has become more evident that nuclear dsRNA can lead to gene silencing and heterochromatin formation in a sequence-independent, epigenetic fashion (3, 58, 62, 84, 110, 111, 115, 116, 155, 231, 244, 278, 309, 393, 394, 403). Unexpectedly, this gene silencing appears to be mediated not by the ADAR editing machinery but by the RNAi machinery (Fig. 5). This surprising insight has come both from work on RNA-mediated transcriptional gene silencing in plants (see reference 351 for a recent review) and from elegant genetic studies carried out in the fission yeast S. pombe (3, 11, 116, 301, 393, 394), and the suspicion is that similar phenomena will be observed in higher eukaryotes.
FIG. 5.
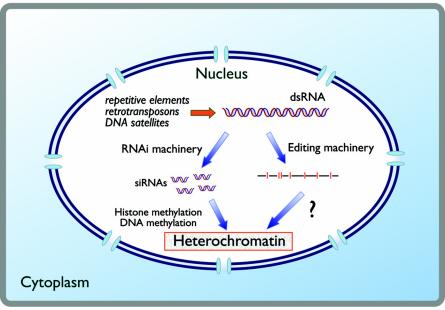
Nuclear dsRNAs might also induce heterochromatin formation. Transcription from repetitive elements, retrotransposons, and DNA satellite sequences, such as in centromeres and telomeres, might generate dsRNAs. In one model, developed primarily from work with S. pombe (see the text for details), this RNA is processed by the RNAi machinery to generate siRNAs that, by an unknown mechanism, lead to histone methylation, DNA methylation, and ultimately heterochromatin. On the right, and discussed in the text, is suggested a hypothetical pathway by which the editing machinery could also influence the formation of heterochromatin, either by an independent mechanism or by modulating the RNAi-based pathway.
One of the first demonstrations that RNA could affect gene expression in the nucleus came from studies in plants showing RNA-directed DNA methylation (404). This requires the processing of long dsRNAs to smaller 21- to 24-nucleotide species, which are linked to gene silencing through DNA methyltransferases as well as histone modifications (9, 148, 434; see reference 237 for a recent review). In S. pombe, genetic studies revealed that RNAi proteins, including Dicer (Dcr1), Argonaute (Ago1), and RdRp (Rdp1), are important for the formation of heterochromatin and centromere silencing by promoting H3 lysine 9 methylation (393, 394). Methylation of H3 lysine 9 can recruit the yeast Swi6 protein, which is the homologue of the higher eukaryote heterochromatin protein 1 (HP1), which can then direct heterochromatin formation and the spreading of gene silencing to the surrounding sequences (3, 115, 116, 393, 394). Thus, some transcription must occur in the region of centromeric repeats. This transcription could either be bidirectional, resulting in dsRNA, or lead to dsRNA by the action of RdRp, which could itself be primed by siRNAs. In this model, dsRNAs are processed by the enzyme Dicer into RNAi-related siRNAs or short heterochromatic RNAs (116, 251, 309, 381, 394). These RNAs then lead to the recruitment of factors involved in heterochromatization to the genomic region from which they derive.
Recent work has provided direct evidence to connect nuclear siRNAs to heterochromatin assembly. Verdel et al. isolated an RNA-induced initiation of transcriptional gene silencing complex (RITS), which is required for heterochromatin assembly in S. pombe (390). This complex contains Ago1, Chp1, Tas3, and Dicer-cleaved siRNAs. In this complex, Ago1 is known to be a RISC component that binds to siRNAs, while Chp1 has been shown to bind centromeres. Moreover, the siRNAs in the complex were found to be homologous to centromeric repeats. Therefore, there appears to be a direct connection between siRNAs and centromeres. This work suggested a mechanism of epigenetic gene silencing at specific chromosomal loci by siRNAs in S. pombe (80, 111, 390). RITS-mediated epigenetic gene silencing might also be conserved in other systems. For example, in D. melanogaster, Argonaute proteins and polycomb proteins have been shown to be required for repeat-induced transcriptional gene silencing (278, 279). While the molecular mechanism by which nuclear siRNAs can recruit factors necessary for heterochromatin formation remains fairly unclear, interesting genetic screens in C. elegans for mutants defective in RNAi revealed that a large fraction of these mutants encode gene products that are chromatin associated (75).
In S. pombe, small centromeric siRNAs have been observed (309), and Volpe et al. showed that RdRp is bound to the centromeric DNA repeats by chromatin immunoprecipitation (393). These results suggested that RdRp may transcribe the second strand from nascent centromeric transcripts and generate dsRNA. The resulting long dsRNA would then be cleaved by Dicer into RNAi-related siRNAs. The role of RdRp in such gene silencing might be to amplify the RNA signals, leading to maintenance of the repressed state. Martienssen (231) presents a nice model explaining how single-copy elements cannot sustain silencing by this pathway but tandem arrays can. Consistent with this model, silencing at the mating type locus of S. pombe also depends on a sequence similar to centromeric repeats (10) but is not maintained well because there are no tandem repeats. Finally, Schramke et al. (333) recently showed in S. pombe that the introduction of hairpin RNA structures into this organism can induce RNA-mediated, chromatin-based epigenetic gene silencing.
How do the above RNAi-related gene-silencing mechanisms of plants and fission yeast apply to higher eukaryotes? While there is currently no direct biochemical evidence for RNAi-mediated chromatin silencing in higher eukaryotes, there are tantalizing clues that a connection will soon be made. Heterochromatin is commonly associated with chromosomal regions that are rich in repetitive sequences. Transcription from such regions could conceivably generate dsRNAs, which could enter a nuclear RNAi pathway. For a number of years it has been observed that increasing the number of copies of transgenes in mammals often leads to lower rather than higher expression (99) and can lead to regions of locally high concentrations of dsRNA (317). This might result from spurious bidirectional transcription, which in turn leads to gene silencing. Thus, more is not always better when introducing transgenes into cells. Furthermore, it has been reported that RNAi defects relieve the silencing of tandem transgene arrays in Neurospora crassa, Arabidopsis thaliana, C. elegans (53, 176, 258), and D. melanogaster (279).
In the C. elegans germ line, transposons are silenced, but they are mobile in somatic cells (175, 362). Some mutants that cannot silence RNA are also defective in RNAi (348). In D. melanogaster, some mutants carrying mutations in RNAi components lost heterochromatic silencing (279). In one interesting study, antisense RNA from a human gene locus was shown to lead to gene silencing and methylation of CpG islands, suggesting that in mammalian cells nuclear dsRNA can induce transcriptional gene silencing associated with DNA methylation within promoter regions, as has been seen in plants (379). Finally, RNA synthesis has now been found in regions of the genome that were once thought to be transcriptionally silent. For example, it has recently been shown that a human centromere is transcriptionally competent (319).
Role of dsRNA in imprinting and X chromosome inactivation.
Finally, there is a possibility that RNAi-related gene silencing mechanisms may even extend to genomic imprinting and X chromosome inactivation in the nucleus. While the mechanisms of genomic imprinting and X chromosome inactivation have not been completely uncovered, in both cases long nuclear dsRNAs have been suggested to play a critical role. Genomic imprinting affects many mammalian genes and results in the expression of those genes from only one of the two parental chromosomes (for reviews, see references 304 and 352). So far, about 20% of known imprinted genes are associated with antisense transcripts, most of which are noncoding RNA and may have regulatory functions. Recent work showed that the Air antisense RNA, which overlaps the maternally expressed Igf2r gene, has an active role and is required for genomic imprinting (353). X chromosome inactivation is the transcriptional silencing of one X chromosome in female mammalian cells (see reference 169 for a recent review). It requires a region of the X chromosome known as the X inactivation center. Within the X inactivation center, there are two noncoding transcripts, Xist and Tsix. Tsix and Xist have the potential to form dsRNAs, and together they regulate the choice of X chromosome inactivation (reviewed in references 293 and 391).
Connection between RNAi and RNA Editing
Is there a connection between RNAi and RNA editing? While there has been understandable excitement in the past few years concerning a potential role of the RNAi machinery in nuclear gene silencing, we caution that, especially in higher eukaryotic cells, this model needs to be examined critically and biochemical pathways need to be more clearly identified. Tandem transgenes are silenced in D. melanogaster (71), but in this organism no RdRp is evident. Vertebrate genomes also lack RdRp, suggesting that perhaps a mode of silencing other than RNAi exists. We suggest that in higher eukaryotes, the ADAR editing machinery may be intimately linked with dsRNA-mediated nuclear gene silencing and may either cooperate with, replace, or compete with the RNAi machinery (Fig. 5).
As we have seen, the dsRNA-induced RNA editing machinery appears to be active throughout the nucleus. In higher eukaryotes, therefore, dsRNAs derived from centromeric DNA repeats or tandem transgene arrays are quite likely be efficient substrates for ADARs. In vitro data showed that RNAi is inhibited if the dsRNA is edited by ADAR2 (329). Edited dsRNAs contain mismatched I-U base pairs, which may lead to partial or complete unwinding of duplexes. Such molecules are poor substrates for Dicer cleavage. Also, when C. elegans mutants lacking ADAR activity were examined, it was found that normally expressed transgenes were now silenced by the RNAi machinery, indicating that ADAR activity can modulate the RNAi response in the nucleus (184).
Thus, in order for the RNAi machinery to play a key role in heterochromatin formation in higher eukaryotes, it would have to function in such a way as to overcome the editing machinery or to synergize with it. It is possible that RNA editing does in fact occur in heterochromatic regions; however, some dsRNA sequences might remain relatively refractory to editing owing to the inherent base preferences of the ADAR enzymes, and this subset of dsRNAs would then be targeted for Dicer cleavage.
DNA Elimination in Tetrahymena thermophila
Finally, there have been very interesting reports of the involvement of the RNAi machinery in gene regulation in T. thermophila. In T. thermophila, RNAi proteins have been linked to the development of the somatic macronucleus from the germ micronucleus (reviewed in reference 252). In this development, there are chromosomal rearrangements followed by DNA elimination of repetitive elements and transposons. This elimination requires an Argonaute protein homolog, TWI1, and two chromodomain proteins, PDD1 and PDD3. Histone H3 lysine 9 methylation is also involved, and small RNAs are seen (251, 365). Furthermore, injection of dsRNA into Tetrahymena cells at specific developmental stages triggers deletion of the targeted genomic regions (420).
CONCLUSIONS
As we have seen, there are a rich diversity of cellular responses to dsRNA, with the potential for subtle and unexpected cross talk between them. dsRNA has a variety of profound effects on cells, but these effects differ depending both on the intrinsic nature of the dsRNA trigger and on the cellular localization of the duplexes. In the cytoplasm, long and short dsRNAs have strikingly different effects. In the cytoplasm, long dsRNAs trigger nonspecific, global effects, while short dsRNAs enter the sequence-specific RNAi or micro-RNA pathway. In the nucleus, the situation is reversed: long dsRNAs appear to be edited and retained in a sequence-specific manner, while short dsRNAs may have a role in epigenetic gene silencing. At this time, the cytoplasmic response systems are understood in greater molecular detail than the nuclear systems, but it is the nucleus where the bulk of cellar dsRNA is produced and processed. Future work needs to focus more on the nuclear fate of dsRNA and promises to not only continue to provide surprising new insights into the central role that dsRNA plays in the regulation of gene expression, but also to point to new ways in which RNA can be used as a tool to inhibit target gene expression.
Acknowledgments
We thank J. DeCerbo, R. Gu, J. Podoloff, and Z. Zhang for helpful suggestions throughout this work.
This work was supported by grants CA45382 and GM066816 from the National Institutes of Health.
REFERENCES
- 1.Adelman, Z. N., I. Sanchez-Vargas, E. A. Travanty, J. O. Carlson, B. J. Beaty, C. D. Blair, and K. E. Olson. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 76:12925-12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappa B by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Allshire, R. 2002. Molecular biology. RNAi and heterochromatin-a hushed-up affair. Science 297:1818-1819. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673-676. [DOI] [PubMed] [Google Scholar]
- 5.Ambros, V. 2001. MicroRNAs: tiny regulators with great potential. Cell 107:823-826. [DOI] [PubMed] [Google Scholar]
- 6.Ambros, V., R. C. Lee, A. Lavanway, P. T. Williams, and D. Jewell. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13:807-818. [DOI] [PubMed] [Google Scholar]
- 7.An, D. S., Y. Xie, S. H. Mao, K. Morizono, S. K. Kung, and I. S. Chen. 2003. Efficient lentiviral vectors for short hairpin RNA delivery into human cells. Hum. Gene Ther. 14:1207-1212. [DOI] [PubMed] [Google Scholar]
- 8.Arenz, C., and U. Schepers. 2003. RNA interference: from an ancient mechanism to a state of the art therapeutic application? Naturwissenschaften 90:345-359. [DOI] [PubMed] [Google Scholar]
- 9.Aufsatz, W., M. F. Mette, J. Van Der Winden, M. Matzke, and A. J. Matzke. 2002. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21:6832-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayoub, N., I. Goldshmidt, R. Lyakhovetsky, and A. Cohen. 2000. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailis, J. M., and S. L. Forsburg. 2002. RNAi hushes heterochromatin. Genome Biol. 3:1035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balachandran, S., C. N. Kim, W. C. Yeh, T. W. Mak, K. Bhalla, and G. N. Barber. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 17:6888-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltzis, D., S. Li, and A. E. Koromilas. 2002. Functional characterization of PKR gene products expressed in cells from mice with a targeted deletion of the N terminus or C terminus domain of PKR. J. Biol. Chem. 277:38364-38372. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee, D., and F. Slack. 2002. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays 24:119-129. [DOI] [PubMed] [Google Scholar]
- 15.Bartel, B., and D. P. Bartel. 2003. MicroRNAs: At the Root of Plant Development? Plant Physiol. 132:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 17.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 18.Bass, B. L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bass, B. L., and H. Weintraub. 1987. A developmentally regulated activity that unwinds RNA duplexes. Cell 48:607-613. [DOI] [PubMed] [Google Scholar]
- 20.Bass, B. L., and H. Weintraub. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55:1089-1098. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein, E., S. Y. Kim, M. A. Carmell, E. P. Murchison, H. Alcorn, M. Z. Li, A. A. Mills, S. J. Elledge, K. V. Anderson, and G. J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 5:5. [DOI] [PubMed] [Google Scholar]
- 23.Billy, E., V. Brondani, H. Zhang, U. Muller, and W. Filipowicz. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 98:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosher, J. M., P. Dufourcq, S. Sookhareea, and M. Labouesse. 1999. RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics 153:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutla, A., C. Delidakis, I. Livadaras, M. Tsagris, and M. Tabler. 2001. Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol. 11:1776-1780. [DOI] [PubMed] [Google Scholar]
- 27.Branciforte, D., and S. L. Martin. 1994. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 14:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bratt, E., and M. Ohman. 2003. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA 9:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 8:8. [DOI] [PubMed] [Google Scholar]
- 30.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 31.Burns, C. M., H. Chu, S. M. Rueter, L. K. Hutchinson, H. Canton, E. Sanders-Bush, and R. B. Emeson. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303-308. [DOI] [PubMed] [Google Scholar]
- 32.Calegari, F., W. Haubensak, D. Yang, W. B. Huttner, and F. Buchholz. 2002. Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl. Acad. Sci. USA 99:14236-14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvet, J. P., and T. Pederson. 1977. Secondary structure of heterogeneous nuclear RNA: two classes of double-stranded RNA in native ribonucleoprotein. Proc. Natl. Acad. Sci. USA 74:3705-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caplen, N. J., J. Fleenor, A. Fire, and R. A. Morgan. 2000. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252:95-105. [DOI] [PubMed] [Google Scholar]
- 35.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 36.Carmell, M. A., and G. J. Hannon. 2004. RNAse III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11:214-218. [DOI] [PubMed] [Google Scholar]
- 37.Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733-2742. [DOI] [PubMed] [Google Scholar]
- 38.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 39.Castelli, J. C., B. A. Hassel, A. Maran, J. Paranjape, J. A. Hewitt, X. L. Li, Y. T. Hsu, R. H. Silverman, and R. J. Youle. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 5:313-320. [DOI] [PubMed] [Google Scholar]
- 40.Caudy, A. A., R. F. Ketting, S. M. Hammond, A. M. Denli, A. M. Bathoorn, B. B. Tops, J. M. Silva, M. M. Myers, G. J. Hannon, and R. H. Plasterk. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425:411-414. [DOI] [PubMed] [Google Scholar]
- 41.Caudy, A. A., M. Myers, G. J. Hannon, and S. M. Hammond. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celotto, A. M., and B. R. Graveley. 2002. Exon-specific RNAi: a tool for dissecting the functional relevance of alternative splicing. RNA 8:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang, J., P. Provost, and J. M. Taylor. 2003. Resistance of human hepatitis delta virus RNAs to dicer activity. J. Virol. 77:11910-11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, C., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83-86. [DOI] [PubMed] [Google Scholar]
- 46.Chi, J. T., H. Y. Chang, N. N. Wang, D. S. Chang, N. Dunphy, and P. O. Brown. 2003. Genomewide view of gene silencing by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:6343-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho, D. S., W. Yang, J. T. Lee, R. Shiekhattar, J. M. Murray, and K. Nishikura. 2003. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278:17093-17102. [DOI] [PubMed] [Google Scholar]
- 48.Circle, D. A., O. D. Neel, H. D. Robertson, P. A. Clarke, and M. B. Mathews. 1997. Surprising specificity of PKR binding to delta agent genomic RNA. RNA 3:438-448. [PMC free article] [PubMed] [Google Scholar]
- 49.Clemens, M. J. 1997. PKR—a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol. 29:945-949. [DOI] [PubMed] [Google Scholar]
- 50.Clemens, M. J. 1996. Protein kinases that phosphorylate eIF-2 and eIF-2B, and their role in eukaryotic cell translational control, p. 139-172. In M. J. Clemens (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Clissold, P. M., and C. P. Ponting. 2000. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10:R888-R890. [DOI] [PubMed] [Google Scholar]
- 52.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cogoni, C., and G. Macino. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166-169. [DOI] [PubMed] [Google Scholar]
- 54.Cook, H. A., B. S. Koppetsch, J. Wu, and W. E. Theurkauf. 2004. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116:817-829. [DOI] [PubMed] [Google Scholar]
- 55.Cuddihy, A. R., S. Li, N. W. Tam, A. H. Wong, Y. Taya, N. Abraham, J. C. Bell, and A. E. Koromilas. 1999. Double-stranded-RNA-activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol. Cell. Biol. 19:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cunnington, P. G., and J. D. Naysmith. 1975. Naturally occurring double-stranded RNA and immune responses. Effects on plaque-forming cells and antibody formation. Immunology 28:451-468. [PMC free article] [PubMed] [Google Scholar]
- 57.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543-553. [DOI] [PubMed] [Google Scholar]
- 58.Dawe, R. K. 2003. RNA interference, transposons and the centromere. Plant Cell. 15:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deininger, P. L., and M. A. Batzer. 2002. Mammalian retroelements. Genome Res. 12:1455-1465. [DOI] [PubMed] [Google Scholar]
- 60.Denli, A. M., and G. J. Hannon. 2003. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28:196-201. [DOI] [PubMed] [Google Scholar]
- 61.Deonarain, R., A. Alcami, M. Alexiou, M. J. Dallman, D. R. Gewert, and A. C. Porter. 2000. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 74:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dernburg, A. F., and G. H. Karpen. 2002. A chromosome RNAissance. Cell 111:159-162. [DOI] [PubMed] [Google Scholar]
- 63.Desterro, J. M., L. P. Keegan, M. Lafarga, M. T. Berciano, M. O'Connell, and M. Carmo-Fonseca. 2003. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 116:1805-1818. [DOI] [PubMed] [Google Scholar]
- 64.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dillin, A. 2003. The specifics of small interfering RNA specificity. Proc. Natl. Acad. Sci. USA 100:6289-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13:41-46. [DOI] [PubMed] [Google Scholar]
- 68.Domeier, M. E., D. P. Morse, S. W. Knight, M. Portereiko, B. L. Bass, and S. E. Mango. 2000. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289:1928-1931. [DOI] [PubMed] [Google Scholar]
- 69.Dong, B., and R. H. Silverman. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133-4137. [DOI] [PubMed] [Google Scholar]
- 70.Donze, O., J. Dostie, and N. Sonenberg. 1999. Regulatable expression of the interferon-induced double-stranded RNA dependent protein kinase PKR induces apoptosis and fas receptor expression. Virology 256:322-329. [DOI] [PubMed] [Google Scholar]
- 71.Dorer, D. R., and S. Henikoff. 1994. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77:993-1002. [DOI] [PubMed] [Google Scholar]
- 72.Dostie, J., Z. Mourelatos, M. Yang, A. Sharma, and G. Dreyfuss. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9:180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doyle, M., and M. F. Jantsch. 2003. Distinct in vivo roles for double-stranded RNA-binding domains of the Xenopus RNA-editing enzyme ADAR1 in chromosomal targeting. J. Cell Biol. 161:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dudley, N. R., and B. Goldstein. 2003. RNA interference: Silencing in the cytoplasm and nucleus. Curr. Opin. Mol. Therapeut. 5:113-117. [PubMed] [Google Scholar]
- 75.Dudley, N. R., J. C. Labbe, and B. Goldstein. 2002. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA 99:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durand-Dubief, M., and P. Bastin. 2003. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 78.Eckmann, C. R., and M. F. Jantsch. 1999. The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J. Cell Biol. 144:603-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egebjerg, J., V. Kukekov, and S. F. Heinemann. 1994. Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc. Natl. Acad. Sci. USA 91:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ekwall, K. 2004. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell 13:304-305. [DOI] [PubMed] [Google Scholar]
- 81.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 82.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elgin, S. C., and S. I. Grewal. 2003. Heterochromatin: silence is golden. Curr. Biol. 13:R895-R898. [DOI] [PubMed] [Google Scholar]
- 85.Fagard, M., S. Boutet, J. B. Morel, C. Bellini, and H. Vaucheret. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97:11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fahey, M. E., T. F. Moore, and D. G. Higgins. 2002. Overlapping antisense transcription in the human genome. Comp. Funct. Genom. 3:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fedoroff, N., P. K. Wellauer, and R. Wall. 1977. Intermolecular duplexes in heterogeneous nuclear RNA from HeLa cells. Cell 10:597-610. [DOI] [PubMed] [Google Scholar]
- 88.Feinberg, E. H., and C. P. Hunter. 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301:1545-1547. [DOI] [PubMed] [Google Scholar]
- 89.Field, A. K., C. W. Young, I. H. Krakoff, A. A. Tytell, G. P. Lampson, M. M. Nemes, and M. R. Hilleman. 1971. Induction of interferon in human subjects by poly I:C. Proc. Soc. Exp. Biol. Med. 136:1180-1186. [DOI] [PubMed] [Google Scholar]
- 90.Finnegan, E. J., R. Margis, and P. M. Waterhouse. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13:236-240. [DOI] [PubMed] [Google Scholar]
- 91.Finnegan, E. J., and M. A. Matzke. 2003. The small RNA world. J. Cell Sci. 116:4689-4693. [DOI] [PubMed] [Google Scholar]
- 92.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 93.Fox, A. H., Y. W. Lam, A. K. Leung, C. E. Lyon, J. Andersen, M. Mann, and A. I. Lamond. 2002. Paraspeckles. A novel nuclear domain. Curr. Biol. 12:13-25. [DOI] [PubMed] [Google Scholar]
- 94.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-8114. [DOI] [PubMed] [Google Scholar]
- 95.Francois, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 98.Gallo, A., L. P. Keegan, G. M. Ring, and M. A. O'Connell. 2003. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22:3421-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garrick, D., S. Fiering, D. I. Martin, and E. Whitelaw. 1998. Repeat-induced gene silencing in mammals. Nat. Genet. 18:56-59. [DOI] [PubMed] [Google Scholar]
- 100.Ge, Q., L. Filip, A. Bai, T. Nguyen, H. N. Eisen, and J. Chen. 2004. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA 101:8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.George, C. X., and C. E. Samuel. 1999. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229:203-213. [DOI] [PubMed] [Google Scholar]
- 102.George, C. X., and C. E. Samuel. 1999. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. USA 96:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gil, J., and M. Esteban. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5:107-114. [DOI] [PubMed] [Google Scholar]
- 104.Gil, J., M. A. Garcia, and M. Esteban. 2002. Caspase 9 activation by the dsRNA-dependent protein kinase, PKR: molecular mechanism and relevance. FEBS Lett. 529:249-255. [DOI] [PubMed] [Google Scholar]
- 105.Gitlin, L., and R. Andino. 2003. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 77:7159-7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 107.Golden, T. A., S. E. Schauer, J. D. Lang, S. Pien, A. R. Mushegian, U. Grossniklaus, D. W. Meinke, and A. Ray. 2002. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130:808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 109.Grad, Y., J. Aach, G. D. Hayes, B. J. Reinhart, G. M. Church, G. Ruvkun, and J. Kim. 2003. Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11:1253-1263. [DOI] [PubMed] [Google Scholar]
- 110.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 111.Grewal, S. I., and J. C. Rice. 2004. Regulation of heterochromatin by histone methylation and small RNA. Curr. Opin. Cell Biol. 16:230-238. [DOI] [PubMed] [Google Scholar]
- 112.Grishok, A., H. Tabara, and C. C. Mello. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287:2494-2497. [DOI] [PubMed] [Google Scholar]
- 113.Grosshans, H., and F. J. Slack. 2002. Micro-RNAs: small is plentiful. J. Cell Biol. 156:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haley, B., and P. D. Zamore. 2004. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11:599-606. [DOI] [PubMed] [Google Scholar]
- 115.Hall, I. M., K. Noma, and S. I. Grewal. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 117.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 119.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 120.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 121.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 122.Handa, V., T. Saha, and K. Usdin. 2003. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 31:6243-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 124.Hartmann, R., J. Justesen, S. N. Sarkar, G. C. Sen, and V. C. Yee. 2003. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell 12:1173-1185. [DOI] [PubMed] [Google Scholar]
- 125.Hartner, J. C., C. Schmittwolf, A. Kispert, A. M. Mueller, M. Higuchi, and P. H. Seeburg. 2003. Liver disintegration in the mouse embryo by deficiency in RNA editing enzyme ADAR1. J. Biol. Chem. 279:4894-4902. [DOI] [PubMed] [Google Scholar]
- 126.Hassel, B. A., A. Zhou, C. Sotomayor, A. Maran, and R. H. Silverman. 1993. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 12:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hasuwa, H., K. Kaseda, T. Einarsdottir, and M. Okabe. 2002. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 532:227-230. [DOI] [PubMed] [Google Scholar]
- 128.Havelda, Z., C. Hornyik, A. Crescenzi, and J. Burgyan. 2003. In situ characterization of cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77:6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heim, M. H. 2000. Intracellular signalling and antiviral effects of interferons. Dig. Liver Dis. 32:257-263. [DOI] [PubMed] [Google Scholar]
- 130.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Herb, A., M. Higuchi, R. Sprengel, and P. H. Seeburg. 1996. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc. Natl. Acad. Sci. USA 93:1875-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herbert, A., J. Alfken, Y. G. Kim, I. S. Mian, K. Nishikura, and A. Rich. 1997. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. USA 94:8421-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Herbert, A., and A. Rich. 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. USA 98:12132-12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Higuchi, M., S. Maas, F. N. Single, J. Hartner, A. Rozov, N. Burnashev, D. Feldmeyer, R. Sprengel, and P. H. Seeburg. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406:78-81. [DOI] [PubMed] [Google Scholar]
- 135.Higuchi, M., F. N. Single, M. Köhler, B. Sommer, R. Sprengel, and P. H. Seeburg. 1993. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75:1361-1370. [DOI] [PubMed] [Google Scholar]
- 136.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 137.Hoopengardner, B., T. Bhalla, C. Staber, and R. Reenan. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301:832-836. [DOI] [PubMed] [Google Scholar]
- 138.Hovanessian, A. G. 1989. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J. Interferon Res. 9:641-647. [DOI] [PubMed] [Google Scholar]
- 139.Hu, W., C. Myers, J. Kilzer, S. Pfaff, and F. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301. [DOI] [PubMed] [Google Scholar]
- 140.Hunter, C., and R. S. Poethig. 2003. miSSING LINKS: miRNAs and plant development. Curr. Opin. Genet. Dev. 13:372-378. [DOI] [PubMed] [Google Scholar]
- 141.Hutchison III, C. A., S. C. Hardies, D. D. Loeb, W. R. Shehee, and M. H. Edgell. 1989. LINEs and related retroposons: long interspersed repeated sequences in the eukaryotic genome, p. 593-617. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 142.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 143.Iordanov, M. S., J. M. Paranjape, A. Zhou, J. Wong, B. R. Williams, E. F. Meurs, R. H. Silverman, and B. E. Magun. 2000. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol. Cell. Biol. 20:617-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ishizuka, A., M. C. Siomi, and H. Siomi. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16:2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jabri, E. 2004. RISCy business. Nat. Struct. Mol. Biol. 11:300. [DOI] [PubMed] [Google Scholar]
- 146.Jabri, E. 2004. Sizing up small RNAs. Nat. Struct. Mol. Biol. 11:112. [DOI] [PubMed] [Google Scholar]
- 147.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 148.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the kryptonite histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 149.Jacobsen, S. E., M. P. Running, and E. M. Meyerowitz. 1999. Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126:5231-5352. [DOI] [PubMed] [Google Scholar]
- 150.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jaikaran, D. C., C. H. Collins, and A. M. MacMillan. 2002. Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem. 277:37624-37629. [DOI] [PubMed] [Google Scholar]
- 152.Jayan, G. C., and J. L. Casey. 2002. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J. Virol. 76:12399-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jeffrey, I. W., S. Kadereit, E. F. Meurs, T. Metzger, M. Bachmann, M. Schwemmle, A. G. Hovanessian, and M. J. Clemens. 1995. Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp. Cell Res. 218:17-27. [DOI] [PubMed] [Google Scholar]
- 154.Jelinek, W., and J. E. Darnell. 1972. Double-stranded regions in heterogeneous nuclear RNA from HeLa cells. Proc. Natl. Acad. Sci. USA 69:2537-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jenuwein, T. 2002. An RNA-guided pathway for the epigenome. Science 297:2215-2218. [DOI] [PubMed] [Google Scholar]
- 156.Jia, Q., and R. Sun. 2003. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 77:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jimenez-Garcia, L. F., S. R. Green, M. B. Mathews, and D. L. Spector. 1993. Organization of the double-stranded RNA-activated protein kinase DAI and virus-associated VA RNAI in adenovirus-2-infected HeLa cells. J. Cell Sci. 106:11-22. [DOI] [PubMed] [Google Scholar]
- 158.Jin, P., D. C. Zarnescu, S. Ceman, M. Nakamoto, J. Mowrey, T. A. Jongens, D. L. Nelson, K. Moses, and S. T. Warren. 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7:113-117. [DOI] [PubMed] [Google Scholar]
- 159.Johnson, C., D. Primorac, M. McKinstry, J. McNeil, D. Rowe, and J. B. Lawrence. 2000. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150:417-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Johnston, R. J., and O. Hobert. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426:845-849. [DOI] [PubMed] [Google Scholar]
- 161.Joost Haasnoot, P. C., D. Cupac, and B. Berkhout. 2003. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 162.Kaempfer, R., and J. Kaufman. 1973. Inhibition of cellular protein synthesis by double-stranded RNA: inactivation of an initiation factor. Proc. Natl. Acad. Sci. USA 70:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kallman, A. M., M. Sahlin, and M. Ohman. 2003. ADAR2 A-I editing: site selectivity and editing efficiency are separate events. Nucleic Acids Res. 31:4874-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 165.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kariko, K., P. Bhuyan, J. Capodici, and D. Weissman. 2004. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J. Immunol. 172:6545-6549. [DOI] [PubMed] [Google Scholar]
- 167.Kariko, K., H. Ni, J. Capodici, M. Lamphier, and D. Weissman. 2004. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 279:12542-12550. [DOI] [PubMed] [Google Scholar]
- 168.Kask, K., D. Zamanillo, A. Rozov, N. Burnashev, R. Sprengel, and P. H. Seeburg. 1998. The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc. Natl. Acad. Sci. USA 95:13777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kelley, R. L., and M. I. Kuroda. 2000. Noncoding RNA genes in dosage compensation and imprinting. Cell 103:9-12. [DOI] [PubMed] [Google Scholar]
- 170.Kennedy, S., D. Wang, and G. Ruvkun. 2004. A conserved siRNA-degrading RNAse negatively regulates RNA interference in C. elegans. Nature 427:645-649. [DOI] [PubMed] [Google Scholar]
- 171.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 172.Kennerdell, J. R., S. Yamaguchi, and R. W. Carthew. 2002. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 16:1884-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNase D. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 176.Ketting, R. F., and R. H. Plasterk. 2000. A genetic link between co-suppression and RNA interference in C. elegans. Nature 404:296-298. [DOI] [PubMed] [Google Scholar]
- 177.Khabar, K. S., Y. M. Siddiqui, F. Al-Zoghaibi, L. Al-Haj, M. Dhalla, A. Zhou, B. Dong, M. Whitmore, J. Paranjape, M. Al-Ahdal, F. Al-Mohanna, B. R. Williams, and R. H. Silverman. 2003. RNase L mediates transient control of interferon response through modulation of the double-stranded RNA dependent protein kinase PKR. J. Biol. Chem 278:20124-20132. [DOI] [PubMed] [Google Scholar]
- 178.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209-216. [DOI] [PubMed] [Google Scholar]
- 179.Kim, D. H., M. Longo, Y. Han, P. Lundberg, E. Cantin, and J. J. Rossi. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22:321-325. [DOI] [PubMed] [Google Scholar]
- 180.Kim, U., Y. Wang, T. Sanford, Y. Zeng, and K. Nishikura. 1994. Molecular cloning of a cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. USA 91:11457-11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim, V. N. 2004. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 14:156-159. [DOI] [PubMed] [Google Scholar]
- 182.Klaue, Y., A. M. Kallman, M. Bonin, W. Nellen, and M. Ohman. 2003. Biochemical analysis and scanning force microscopy reveal productive and nonproductive ADAR2 binding to RNA substrates. RNA 9:839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293:2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Knight, S. W., and B. L. Bass. 2002. The role of RNA editing by ADARs in RNAi. Mol. Cell 10:809-817. [DOI] [PubMed] [Google Scholar]
- 185.Kooter, J. M., M. A. Matzke, and P. Meyer. 1999. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4:340-347. [DOI] [PubMed] [Google Scholar]
- 186.Kumar, A., J. Haque, J. Lacoste, J. Hiscott, and B. R. Williams. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. USA 91:6288-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappa B. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kumar, M., and G. G. Carmichael. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. USA 94:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lagos-Quintana, M., R. Rauhut, J. Meyer, A. Borkhardt, and T. Tuschl. 2003. New microRNAs from mouse and human. RNA 9:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735-739. [DOI] [PubMed] [Google Scholar]
- 192.Lai, E. C. 2003. MicroRNAs: runts of the genome assert themselves. Curr. Biol. 13:R925-936. [DOI] [PubMed] [Google Scholar]
- 193.Lai, E. C., P. Tomancak, R. W. Williams, and G. M. Rubin. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lai, F., R. Drakas, and K. Nishikura. 1995. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 270:17098-17105. [DOI] [PubMed] [Google Scholar]
- 195.Lakatos, L., G. Szittys, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lamontagne, B., S. Larose, J. Boulanger, and S. A. Elela. 2001. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 3:71-78. [PubMed] [Google Scholar]
- 197.Lampson, G. P., A. A. Tytell, A. K. Field, M. M. Nemes, and M. R. Hilleman. 1967. Inducers of interferon and host resistance. I. Double-stranded RNA from extracts of Penicillium funiculosum. Proc. Natl. Acad. Sci. USA 58:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 199.Langland, J. O., P. N. Kao, and B. L. Jacobs. 1999. Nuclear factor-90 of activated T-cells: A double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38:6361-6368. [DOI] [PubMed] [Google Scholar]
- 200.Lawrence, D. 2002. RNAi could hold promise in the treatment of HIV. Lancet 359:2007. [DOI] [PubMed] [Google Scholar]
- 201.Lee, D. W., R. J. Pratt, M. McLaughlin, and R. Aramayo. 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 203.Lee, R. C., R. L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 204.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 205.Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He, E. J. Sontheimer, and R. W. Carthew. 2004. Distinct roles for Drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell 117:69-81. [DOI] [PubMed] [Google Scholar]
- 206.Lehmann, K. A., and B. L. Bass. 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39:12875-12884. [DOI] [PubMed] [Google Scholar]
- 207.Lehner, B., G. Williams, R. D. Campbell, and C. M. Sanderson. 2002. Antisense transcripts in the human genome. Trends Genet. 18:63-65. [DOI] [PubMed] [Google Scholar]
- 208.Li, G., Y. Xiang, K. Sabapathy, and R. H. Silverman. 2003. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J. Biol. Chem. 21:21. [DOI] [PubMed] [Google Scholar]
- 209.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 210.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 211.Li, X. L., J. A. Blackford, and B. A. Hassel. 1998. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 72:2752-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liang, X. H., Q. Liu, and S. Michaeli. 2003. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl. Acad. Sci. USA 100:7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lipardi, C., Q. Wei, and B. M. Paterson. 2001. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107:297-307. [DOI] [PubMed] [Google Scholar]
- 215.Lipman, D. J. 1997. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 25:3580-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Liu, Q., T. A. Rand, S. Kalidas, F. Du, H. E. Kim, D. P. Smith, and X. Wang. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921-1925. [DOI] [PubMed] [Google Scholar]
- 217.Liu, Y., C. X. George, J. B. Patterson, and C. E. Samuel. 1997. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 272:4419-4428. [DOI] [PubMed] [Google Scholar]
- 218.Liu, Y., and C. E. Samuel. 1996. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 70:1961-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu, Z., D. B. Batt, and G. G. Carmichael. 1994. Targeted nuclear antisense RNA mimics natural antisense-induced degradation of polyoma virus early RNA. Proc. Natl. Acad. Sci. USA 91:4258-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lohmann, J. U., I. Endl, and T. C. Bosch. 1999. Silencing of developmental genes in Hydra. Dev. Biol. 214:211-214. [DOI] [PubMed] [Google Scholar]
- 221.Lomeli, H., J. Mosbacher, T. Melcher, T. Hoger, J. R. Geiger, T. Kuner, H. Monyer, M. Higuchi, A. Bach, and P. H. Seeburg. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266:1709-1713. [DOI] [PubMed] [Google Scholar]
- 222.Lum, L., S. Yao, B. Mozer, A. Rovescalli, D. Von Kessler, M. Nirenberg, and P. A. Beachy. 2003. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299:2039-2045. [DOI] [PubMed] [Google Scholar]
- 223.Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 303:95-98. [DOI] [PubMed] [Google Scholar]
- 224.Lykke-Andersen, J. 2001. mRNA quality control: Marking the message for life or death. Curr. Biol. 11:R88-91. [DOI] [PubMed] [Google Scholar]
- 225.Maas, S., and A. Rich. 2000. Changing genetic information through RNA editing. Bioessays 22:790-802. [DOI] [PubMed] [Google Scholar]
- 226.Maas, S., A. Rich, and K. Nishikura. 2003. A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 278:1391-1394. [DOI] [PubMed] [Google Scholar]
- 227.Maine, E. M. 2001. RNAi As a tool for understanding germline development in Caenorhabditis elegans: uses and cautions. Dev. Biol. 239:177-189. [DOI] [PubMed] [Google Scholar]
- 228.Manche, L., S. R. Green, C. Schmedt, and M. B. Mathews. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12:5238-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104:173-176. [DOI] [PubMed] [Google Scholar]
- 230.Martens, H., J. Novotny, J. Oberstrass, T. L. Steck, P. Postlethwait, and W. Nellen. 2002. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Martienssen, R. A. 2003. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 35:213-214. [DOI] [PubMed] [Google Scholar]
- 232.Martin, S. L., and D. Branciforte. 1993. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell. Biol. 13:5383-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Martinand, C., T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1998. The RNase L inhibitor (RLI) is induced by double-stranded RNA. J. Interferon Cytokine Res. 18:1031-1038. [DOI] [PubMed] [Google Scholar]
- 234.Martinez, M. A., B. Clotet, and J. A. Este. 2002. RNA interference of HIV replication. Trends Immunol. 23:559-561. [DOI] [PubMed] [Google Scholar]
- 235.Matsumoto, M., K. Funami, H. Oshiumi, and T. Seya. 2004. Toll-like receptor 3: a link between toll-like receptor, interferon and viruses. Microbiol. Immunol. 48:147-154. [DOI] [PubMed] [Google Scholar]
- 236.Matsumoto, M., S. Kikkawa, M. Kohase, K. Miyake, and T. Seya. 2002. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem. Biophys. Res. Commun. 293:1364-1369. [DOI] [PubMed] [Google Scholar]
- 237.Matzke, M., W. Aufsatz, T. Kanno, L. Daxinger, I. Papp, M. F. Mette, and A. J. Matzke. 2004. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta 1677:129-141. [DOI] [PubMed] [Google Scholar]
- 238.Matzke, M. A., W. Aufsatz, T. Kanno, M. F. Mette, and A. J. Matzke. 2002. Homology-dependent gene silencing and host defense in plants. Adv. Genet. 46:235-275. [DOI] [PubMed] [Google Scholar]
- 239.Matzke, M. A., A. J. Matzke, G. J. Pruss, and V. B. Vance. 2001. RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11:221-227. [DOI] [PubMed] [Google Scholar]
- 240.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 241.McCaffrey, A. P., H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion, and M. A. Kay. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21:639-644. [DOI] [PubMed] [Google Scholar]
- 242.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 243.McMillan, N. A., R. F. Chun, D. P. Siderovski, J. Galabru, W. M. Toone, C. E. Samuel, T. W. Mak, A. G. Hovanessian, K. T. Jeang, and B. R. Williams. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413-424. [DOI] [PubMed] [Google Scholar]
- 244.Mette, M. F., W. Aufsatz, J. van Der Winden, M. A. Matzke, and A. J. Matzke. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19:5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Metzlaff, M. 2002. RNA-mediated RNA degradation in transgene- and virus-induced gene silencing. Biol. Chem. 383:1483-1489. [DOI] [PubMed] [Google Scholar]
- 246.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 247.Meylan, E., K. Burns, K. Hofmann, V. Blanchetau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503-507. [DOI] [PubMed] [Google Scholar]
- 248.Minks, M. A., D. K. West, S. Benvin, J. J. Greene, P. O. Ts'o, and C. Baglioni. 1980. Activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2′-O-methylated poly (inosinic acid) poly(cytidylic acid): correlations with interferon-inducing activity. J. Biol. Chem. 255:6403-6407. [PubMed] [Google Scholar]
- 249.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 250.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10:193-198. [DOI] [PubMed] [Google Scholar]
- 251.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 252.Mochizuki, K., and M. A. Gorovsky. 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr. Opin. Genet. Dev. 14:181-187. [DOI] [PubMed] [Google Scholar]
- 253.Morel, J. B., C. Godon, P. Mourrain, C. Beclin, S. Boutet, F. Feuerbach, F. Proux, and H. Vaucheret. 2002. Fertile hypomorphic Argonaute (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 14:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Morse, D. P., P. J. Aruscavage, and B. L. Bass. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. USA 99:7906-7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Morse, D. P., and B. L. Bass. 1997. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 36:8429-8434. [DOI] [PubMed] [Google Scholar]
- 256.Morse, D. P., and B. L. Bass. 1999. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. USA 96:6048-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mourrain, P., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, D. Jouette, A. M. Lacombe, S. Nikic, N. Picault, K. Remoue, M. Sanial, T. A. Vo, and H. Vaucheret. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533-542. [DOI] [PubMed] [Google Scholar]
- 259.Muhlemann, O., C. S. Mock-Casagrande, J. Wang, S. Li, N. Custodio, M. Carmo-Fonseca, M. F. Wilkinson, and M. J. Moore. 2001. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell 8:33-43. [DOI] [PubMed] [Google Scholar]
- 260.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 261.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Murchison, E. P., and G. J. Nannon. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16:223-229. [DOI] [PubMed] [Google Scholar]
- 263.Myers, J. W., J. T. Jones, T. Meyer, and J. E. Ferrell. 2003. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 21:324-328. [DOI] [PubMed] [Google Scholar]
- 264.Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Nishikura, K. 1992. Modulation of double-stranded RNAs in vivo by RNA duplex unwindase. Ann. N. Y. Acad. Sci. 660:240-250. [DOI] [PubMed] [Google Scholar]
- 267.Niswender, C. M., E. Sanders-Bush, and R. B. Emeson. 1998. Identification and characterization of RNA editing events within the 5-HT2C receptor. Ann. N. Y. Acad. Sci. 861:38-48. [DOI] [PubMed] [Google Scholar]
- 268.Novina, C. D., M. F. Murray, P. J. B. D.M. Dykxhoorn, J. Riess, S.-K. Lee, R. G. Collman, J. Liberman, P. Shankar, and P. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:1-6. [DOI] [PubMed] [Google Scholar]
- 269.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 270.Oelgeschlager, M., J. Larrain, D. Geissert, and E. M. De Robertis. 2000. The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature 405:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ohman, M., A. M. Kallman, and B. L. Bass. 2000. In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6:687-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.O'Neill, L. A., and C. A. Dinarello. 2000. The IL-1 receptor/toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol. Today 21:206-209. [DOI] [PubMed] [Google Scholar]
- 273.Osman, F., N. Jarrous, Y. Ben-Asouli, and R. Kaempfer. 1999. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13:3280-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Paddison, P. J., A. A. Caudy, and G. J. Hannon. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Palatnik, J. F., E. Allen, X. Wu, C. Schommer, R. Schwab, J. C. Carrington, and D. Weigel. 2003. Control of leaf morphogenesis by microRNAs. Nature 425:257-263. [DOI] [PubMed] [Google Scholar]
- 277.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pal-Bhadra, M., U. Bhadra, and J. A. Birchler. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9:315-327. [DOI] [PubMed] [Google Scholar]
- 279.Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra, J. A. Birchler, and S. C. Elgin. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669-672. [DOI] [PubMed] [Google Scholar]
- 280.Palladino, M. J., L. P. Keegan, M. A. O'Connell, and R. A. Reenan. 2000. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6:1004-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Palladino, M. J., L. P. Keegan, M. A. O'Connell, and R. A. Reenan. 2000. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102:437-449. [DOI] [PubMed] [Google Scholar]
- 282.Parrish, S., J. Fleenor, S. Xu, C. Mello, and A. Fire. 2000. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell. 6:1077-1087. [DOI] [PubMed] [Google Scholar]
- 283.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 274:20432-20437. [DOI] [PubMed] [Google Scholar]
- 284.Patterson, J. B., and C. E. Samuel. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15:5376-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Patterson, J. B., D. C. Thomis, S. L. Hans, and C. E. Samuel. 1995. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210:508-511. [DOI] [PubMed] [Google Scholar]
- 286.Pebernard, S., and R. D. Iggo. 2004. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation 72:103-111. [DOI] [PubMed] [Google Scholar]
- 287.Pellegrini, S., and C. Schindler. 1993. Early events in signalling by interferons. Trends Biochem. Sci. 18:338-342. [DOI] [PubMed] [Google Scholar]
- 288.Persaud, S. J., and P. M. Jones. 1994. Antisense oligonucleotide inhibition of gene expression- application to endocrine systems. J. Mol. Endocrinol. 12:127-130. [DOI] [PubMed] [Google Scholar]
- 289.Persengiev, S. P., X. Zhu, and M. R. Green. 2004. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 10:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 291.Pham, J. W., J. L. Pellino, Y. S. Lee, R. W. Carthew, and E. J. Sontheimer. 2004. A dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117:83-94. [DOI] [PubMed] [Google Scholar]
- 292.Plasterk, R. H. 2002. RNA silencing: the genome's immune system. Science 296:1263-1265. [DOI] [PubMed] [Google Scholar]
- 293.Plath, K., S. Mlynarczyk-Evans, D. A. Nusinow, and B. Panning. 2002. Xist RNA and the mechanism of x chromosome inactivation. Annu. Rev. Genet. 36:233-278. [DOI] [PubMed] [Google Scholar]
- 294.Polson, A. G., and B. L. Bass. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Polson, A. G., B. L. Bass, and J. L. Casey. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380:454-456. [DOI] [PubMed] [Google Scholar]
- 296.Polson, A. G., P. F. Crain, S. C. Pomerantz, J. A. McCloskey, and B. L. Bass. 1991. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry 30:11507-11514. [DOI] [PubMed] [Google Scholar]
- 297.Pooggin, M., P. V. Shivaprasad, K. Veluthambi, and T. Hohn. 2003. RNAi targeting of DNA virus in plants. Nat. Biotechnol. 21:131-132. [DOI] [PubMed] [Google Scholar]
- 298.Poulsen, H., J. Nilsson, C. K. Damgaard, J. Egebjerg, and J. Kjems. 2001. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 21:7862-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Proud, C. G. 1995. PKR: a new name and new roles. Trends Biochem. Sci. 20:241-246. [DOI] [PubMed] [Google Scholar]
- 300.Provost, P., D. Dishart, J. Doucet, D. Frendewey, B. Samuelsson, and O. Radmark. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21:5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Provost, P., R. A. Silverstein, D. Dishart, J. Walfridsson, I. Djupedal, B. Kniola, A. Wright, B. Samuelsson, O. Radmark, and K. Ekwall. 2002. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA 99:16648-16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Raitskin, O., D. S. Cho, J. Sperling, K. Nishikura, and R. Sperling. 2001. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. USA 98:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Rand, E., and H. Cedar. 2003. Regulation of imprinting: a multi-tiered process. J. Cell. Biochem. 88:400-407. [DOI] [PubMed] [Google Scholar]
- 305.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Rebagliati, M. R., and D. A. Melton. 1987. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell 48:599-605. [DOI] [PubMed] [Google Scholar]
- 307.Reenan, R. A. 2001. The RNA world meets behavior: A-I pre-mRNA editing in animals. Trends Genet. 17:53-56. [DOI] [PubMed] [Google Scholar]
- 308.Reenan, R. A., C. J. Hanrahan, and G. Barry. 2000. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron 25:139-149. [DOI] [PubMed] [Google Scholar]
- 309.Reinhart, B. J., and D. P. Bartel. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 310.Reinhart, B. J., F. J. Slack, M. Basson, A. E. Pasquinelli, J. C. Bettinger, A. E. Rougvie, H. R. Horvitz, and G. Ruvkun. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901-906. [DOI] [PubMed] [Google Scholar]
- 311.Rhoades, M. W., B. J. Reinhart, L. P. Lim, C. B. Burge, B. Bartel, and D. P. Bartel. 2002. Prediction of plant microRNA targets. Cell 110:513-520. [DOI] [PubMed] [Google Scholar]
- 312.Rinn, J. L., G. Euskirchen, P. Bertone, R. Martone, N. M. Luscombe, S. Hartman, P. M. Harrison, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. The transcriptional activity of human chromosome 22. Genes Dev. 17:529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Robertson, H. D., and M. B. Mathews. 1996. The regulation of the protein kinase PKR by RNA. Biochimie 78:909-914. [DOI] [PubMed] [Google Scholar]
- 314.Romano, N., and G. Macino. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6:3343-3353. [DOI] [PubMed] [Google Scholar]
- 315.Rosok, O., and M. Sioud. 2004. Syst. identification of sense-antisense transcripts in mammalian cells. Nat. Biotechnol. 22:104-108. [DOI] [PubMed] [Google Scholar]
- 316.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 317.Rudert, F., S. Bronner, J. M. Garnier, and P. Dolle. 1995. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome 6:76-83. [DOI] [PubMed] [Google Scholar]
- 318.Ruggli, N., J. D. Tratschin, M. Schweizer, K. C. McCullough, M. A. Hofmann, and A. Summerfield. 2003. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of Npro. J. Virol. 77:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Saffery, R., H. Sumer, S. Hassan, L. H. Wong, J. M. Craig, K. Todokoro, M. Anderson, A. Stafford, and K. H. Choo. 2003. Transcription within a functional human centromere. Mol. Cell 12:509-516. [DOI] [PubMed] [Google Scholar]
- 320.Samuel, C. E. 1991. Antiviral actions of interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology 183:1-11. [DOI] [PubMed] [Google Scholar]
- 321.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Samuel, C. E. 1993. The eIF-2-alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603-7606. [PubMed] [Google Scholar]
- 323.Samuel, C. E. 2004. Knockdown by RNAi-proceed with caution. Nat. Biotechnol. 22:280-282. [DOI] [PubMed] [Google Scholar]
- 324.Samuel, C. E. 1979. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc. Natl. Acad. Sci. USA 76:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sansam, C. L., K. S. Wells, and R. B. Emeson. 2003. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 100:14018-14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Sarkar, S. N., H. L. Smith, T. M. Rowe, and G. C. Sen. 2003. Double-stranded RNA signaling by Toll-like receptor 3 requires specific tyrosine residues in its cytoplasmic domain. J. Biol. Chem. 278:4393-4396. [DOI] [PubMed] [Google Scholar]
- 327.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Scacheri, P. C., O. Rozenblatt-Rosen, N. J. Caplen, T. G. Wolfsberg, L. Umayam, J. C. Lee, C. M. Hughes, K. S. Shanmugam, A. Bhattacharjee, M. Meyerson, and F. S. Collins. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 101:1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Scadden, A. D., and C. W. Smith. 2001. RNAi is antagonized by A-to-I hyper-editing. EMBO Rep. 2:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Schaub, M., and W. Keller. 2002. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie 84:791-803. [DOI] [PubMed] [Google Scholar]
- 331.Schauer, S. E., S. E. Jacobsen, D. W. Meinke, and A. Ray. 2002. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7:487-491. [DOI] [PubMed] [Google Scholar]
- 332.Scherr, M., M. A. Morgan, and M. Eder. 2003. Gene silencing mediated by small interfering RNAs in mammalian cells. Curr. Med. Chem. 10:245-256. [DOI] [PubMed] [Google Scholar]
- 333.Schramke, V., and R. Allshire. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301:1069-1074. [DOI] [PubMed] [Google Scholar]
- 334.Schwartz, D. S., Y. Tomari, and P. D. Zamore. 2004. The RNA-induced silencing complex is a Mg(2+) dependent endonuclease. Curr. Biol. 14:787-791. [DOI] [PubMed] [Google Scholar]
- 335.Schwartz, T., K. Lowenhaupt, Y. G. Kim, L. Li, B. A. Brown 2nd, A. Herbert, and A. Rich. 1999. Proteolytic dissection of Zab, the Z-DNA-binding domain of human ADAR1. J. Biol. Chem. 274:2899-2906. [DOI] [PubMed] [Google Scholar]
- 336.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199-208. [DOI] [PubMed] [Google Scholar]
- 337.Schwarz, D. S., G. Hutvagner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 338.Schwarz, D. S., and P. D. Zamore. 2002. Why do miRNAs live in the miRNP? Genes Dev. 16:1025-1031. [DOI] [PubMed] [Google Scholar]
- 339.Seeburg, P. H. 1996. The role of RNA editing in controlling glutamate receptor channel properties. J. Neurochem. 66:1-5. [DOI] [PubMed] [Google Scholar]
- 340.Seeburg, P. H. 2002. A-to-I editing: new and old sites, functions and speculations. Neuron 35:17-20. [DOI] [PubMed] [Google Scholar]
- 341.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 342.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 343.Sharp, P. A., and P. D. Zamore. 2000. Molecular biology. RNA interference. Science 287:2431-2433. [DOI] [PubMed] [Google Scholar]
- 344.Shendure, J., and G. M. Church. 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biology 3:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Shi, Y. 2003. Mammalian RNAi for the masses. Trends Genet. 19:9-12. [DOI] [PubMed] [Google Scholar]
- 346.Shlomai, A., and Y. Shaul. 2003. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology 37:764-770. [DOI] [PubMed] [Google Scholar]
- 347.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465-476. [DOI] [PubMed] [Google Scholar]
- 348.Sijen, T., and R. H. A. Plasterk. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426:310-314. [DOI] [PubMed] [Google Scholar]
- 349.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Simmer, F., M. Tijsterman, S. Parrish, S. Koushika, M. Nonet, A. Fire, J. Ahringer, and R. Plasterk. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12:1317. [DOI] [PubMed] [Google Scholar]
- 351.Sledz, C. A., M. Holko, M. J. De Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 24:24. [DOI] [PubMed] [Google Scholar]
- 352.Sleutels, F., D. P. Barlow, and R. Lyle. 2000. The uniqueness of the imprinting mechanism. Curr. Opin. Genet. Dev. 10:229-233. [DOI] [PubMed] [Google Scholar]
- 353.Sleutels, F., R. Zwart, and D. P. Barlow. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810-813. [DOI] [PubMed] [Google Scholar]
- 354.Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin, and E. M. Maine. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10:169-178. [DOI] [PubMed] [Google Scholar]
- 355.Sommer, B., M. Kohler, R. Sprengel, and P. H. Seeburg. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67:11-19. [DOI] [PubMed] [Google Scholar]
- 356.Spanggord, R. J., M. Vuyisich, and P. A. Beal. 2002. Identification of binding sites for both dsRBMs of PKR on kinase-activating and kinase-inhibiting RNA ligands. Biochemistry 41:4511-4520. [DOI] [PubMed] [Google Scholar]
- 357.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 358.Stein, P., P. Svoboda, M. Anger, and R. M. Schultz. 2003. RNAi: Mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Strehblow, A., M. Hallegger, and M. F. Jantsch. 2002. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol. Biol. Cell 13:3822-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Tabara, H., A. Grishok, and C. C. Mello. 1998. RNAi in C. elegans: soaking in the genome sequence. Science 282:430-431. [DOI] [PubMed] [Google Scholar]
- 362.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 363.Tabara, H., E. Yigit, H. Siomi, and C. C. Mello. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109:861-871. [DOI] [PubMed] [Google Scholar]
- 364.Tang, G., B. J. Reinhart, D. P. Bartel, and P. D. Zamore. 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 366.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 367.Tenllado, F., D. Barajas, M. Vargas, F. A. Atencio, P. Gonzalez-Jara, and J. R. Diaz-Ruiz. 2003. Transient expression of homologous hairpin RNA causes interference with plant virus infection and is overcome by a virus encoded suppressor of gene silencing. Mol. Plant-Microbe Interact. 16:149-158. [DOI] [PubMed] [Google Scholar]
- 368.Tenllado, F., B. Martinez-Garcia, M. Vargas, and J. R. Diaz-Ruiz. 2003. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 3:3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Thanos, D., and T. Maniatis. 1995. NF-kappa B: a lesson in family values. Cell 80:529-532. [DOI] [PubMed] [Google Scholar]
- 370.Thonberg, H., C. Scheele, C. Dahlgren, and C. Wahlestedt. 2004. Characterization of RNA interference in rat PC12 cells: requirement of GREp95. Biochem. Biophys. Res. Commun. 318:927-934. [DOI] [PubMed] [Google Scholar]
- 371.Tijsterman, M., R. F. Ketting, K. L. Okihara, T. Sijen, and R. H. Plasterk. 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295:694-697. [DOI] [PubMed] [Google Scholar]
- 372.Tijsterman, M., R. F. Ketting, and R. H. Plasterk. 2002. The genetics of RNA silencing. Annu. Rev. Genet. 36:489-519. [DOI] [PubMed] [Google Scholar]
- 373.Tijsterman, M., R. C. May, F. Simmer, K. L. Okihara, and R. H. Plasterk. 2004. Genes required for systemic RNAi interference in Caenorhabditis elegans. Curr. Biol. 14:645-649. [DOI] [PubMed] [Google Scholar]
- 374.Tijsterman, M., K. Okihara, K. Thijssen, and R. Plasterk. 2002. PPW-1, a PAZ/PIWI protein required for efficient germline rNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 12:1535. [DOI] [PubMed] [Google Scholar]
- 375.Tijsterman, M., and R. H. A. Plasterk. 2004. Dicers at RISC: the mechanism of RNAi. Cell 117:1-4. [DOI] [PubMed] [Google Scholar]
- 376.Tomari, Y., T. Du, B. Haley, D. S. Schwartz, R. Bennett, C. H. A., and B. S. Koppetsch. 2004. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116:831-841. [DOI] [PubMed] [Google Scholar]
- 377.Tonkin, L. A., and B. L. Bass. 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Tonkin, L. A., L. Saccomanno, D. P. Morse, T. Brodigan, M. Krause, and B. L. Bass. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21:6025-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tufarelli, C., J. A. Stanley, D. Garrick, J. A. Sharpe, H. Ayyub, W. G. Wood, and D. R. Higgs. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34:157-165. [DOI] [PubMed] [Google Scholar]
- 380.Underhill, D. M., and A. Ozinsky. 2002. Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol. 14:103-110. [DOI] [PubMed] [Google Scholar]
- 381.Vaistij, F. E., L. Jones, and D. C. Baulcombe. 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14:857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Valdes, V. J., A. Sampieri, J. Sepulveda, and L. Vaca. 2003. Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J. Biol. Chem. 278:19317-19324. [DOI] [PubMed] [Google Scholar]
- 383.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 384.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. T. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 385.Vaucheret, H., C. Beclin, and M. Fagard. 2001. Post-transcriptional gene silencing in plants. J. Cell Sci. 114:3083-3091. [DOI] [PubMed] [Google Scholar]
- 386.Vaucheret, H., and M. Fagard. 2001. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17:29-35. [DOI] [PubMed] [Google Scholar]
- 387.Vaucheret, H., L. Nussaume, J. C. Palauqui, I. Quillere, and T. Elmayan. 1997. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell 9:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Vaucheret, H., F. Vazquez, P. Crata, and D. P. Bartel. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18:1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Vazquez, F., V. Gasciolli, P. Crete, and H. Vaucheret. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 390.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Verona, R. I., M. R. W. Mann, and M. S. Bartolomei. 2003. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Biol. 19:237-259. [DOI] [PubMed] [Google Scholar]
- 392.Voinnet, O. 2003. RNA silencing bridging the gaps in wheat extracts. Trends Plant Sci. 8:307-309. [DOI] [PubMed] [Google Scholar]
- 393.Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng, R. A. Martienssen, and R. C. Allshire. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11:137-146. [DOI] [PubMed] [Google Scholar]
- 394.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 395.Vorburger, S. A., A. Pataer, K. Yoshida, G. N. Barber, W. Xia, P. Chiao, L. M. Ellis, M. C. Hung, S. G. Swisher, and K. K. Hunt. 2002. Role for the double-stranded RNA activated protein kinase PKR in E2F-1-induced apoptosis. Oncogene 21:6278-6288. [DOI] [PubMed] [Google Scholar]
- 396.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 397.Wagner, R. W., J. E. Smith, B. S. Cooperman, and K. Nishikura. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA 86:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wang, M. B., D. Abbott, and P. M. Waterhouse. 2000. A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol. Plant Pathol. 1:347-356. [DOI] [PubMed] [Google Scholar]
- 399.Wang, Q., J. Khillan, P. Gadue, and K. Nishikura. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290:1765-1768. [DOI] [PubMed] [Google Scholar]
- 400.Wang, Q., M. Miyakoda, W. Yang, J. Khillan, D. L. Stachura, M. J. Weiss, and K. Nishikura. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279. [DOI] [PubMed]
- 401.Wang, Q. C., Q. H. Nie, and Z. H. Feng. 2003. RNA interference: antiviral weapon and beyond. World J. Gastroenterol. 9:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wargelius, A., S. Ellingsen, and A. Fjose. 1999. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 263:156-161. [DOI] [PubMed] [Google Scholar]
- 403.Wassenegger, M. 2000. RNA-directed DNA methylation. Plant Mol. Biol. 43:203-220. [DOI] [PubMed] [Google Scholar]
- 404.Wassenegger, M., S. Heimes, L. Riedel, and H. L. Sanger. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76:567-576. [DOI] [PubMed] [Google Scholar]
- 405.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 406.Wienholds, E., M. J. Koudijs, F. J. van Eeden, E. Cuppen, and R. H. Plasterk. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35:217-218. [DOI] [PubMed] [Google Scholar]
- 407.Wilkinson, M. F., and A. B. Shyu. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23:775-787. [DOI] [PubMed] [Google Scholar]
- 408.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]
- 409.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed]
- 410.Williams, R. W., and G. M. Rubin. 2002. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. USA 99:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Wojtkowiak, A., A. Siek, M. Alejska, A. Jarmolowski, Z. Szweykowska-Kulinska, and M. Figlerowicz. 2002. RNAi and viral vectors as useful tools in the functional genomics of plants. Construction of BMV-based vectors for RNA delivery into plant cells. Cell Mol. Biol. Lett. 7:511-522. [PubMed] [Google Scholar]
- 412.Wong, A. H., N. W. Tam, Y. L. Yang, A. R. Cuddihy, S. Li, S. Kirchhoff, H. Hauser, T. Decker, and A. E. Koromilas. 1997. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 16:1291-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Wong, S. K., and D. W. Lazinski. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 99:15118-15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Wong, S. K., S. Sato, and D. W. Lazinski. 2003. Elevated activity of the large form of ADAR1 in vivo: very efficient RNA editing occurs in the cytoplasm. RNA 9:586-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Wong, S. K., S. Sato, and D. W. Lazinski. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7:846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Xia, H., Q. Mao, H. L. Paulson, and B. L. Davidson. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20:1006-1010. [DOI] [PubMed] [Google Scholar]
- 417.Yamamoto, T., S. Omoto, M. Mizuguchi, H. Mizukami, H. Okuyama, N. Okada, N. K. Saksena, E. A. Brisibe, K. Otake, and Y. R. Fuji. 2002. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol. Immunol. 46:809-817. [DOI] [PubMed] [Google Scholar]
- 418.Yang, D., F. Buchholz, Z. Huang, A. Goga, C. Y. Chen, F. M. Brodsky, and J. M. Bishop. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Yang, S., S. Tutton, E. Pierce, and K. Yoon. 2001. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 21:7807-7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Yao, M. C., P. Fuller, and X. Xi. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300:1581-1584. [DOI] [PubMed] [Google Scholar]
- 421.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Yekta, S., I. H. Shih, and D. P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594-596. [DOI] [PubMed] [Google Scholar]
- 423.Yelin, R., D. Dahary, R. Sorek, E. Y. Levanon, O. Goldstein, A. Shoshan, A. Diber, S. Biton, Y. Tamir, R. Khosravi, S. Nemser, E. Pinner, S. Walach, J. Bernstein, K. Savitsky, and G. Rotman. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21:379-386. [DOI] [PubMed] [Google Scholar]
- 424.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-miRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Yu, J.-Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Zamore, P. D. 2002. Ancient pathways programmed by small RNAs. Science 296:1265-1269. [DOI] [PubMed] [Google Scholar]
- 427.Zamore, P. D. 2004. Plant RNAi: how a viral silencing suppressor inactivates siRNA. Curr. Biol. R198-200. [DOI] [PubMed]
- 428.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 429.Zeng, Y., and B. R. Cullen. 2002. RNA interference in human cells is restricted to the cytoplasm. RNA 8:855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Zeng, Y., and B. R. Cullen. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]
- 432.Zeng, Y., R. Yi, and B. R. Cullen. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 100:9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Zhang, Z., and G. G. Carmichael. 2001. The fate of dsRNA in the nucleus. A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465-475. [DOI] [PubMed] [Google Scholar]
- 434.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]