Abstract
microRNAs (or miRNAs) are small noncoding RNAs (21 to 25 nucleotides) that are processed from longer hairpin RNA precursors and are believed to be involved in a wide range of developmental and cellular processes, by either repressing translation or triggering mRNA degradation (RNA interference). By using a computer-assisted approach, we have identified 46 potential miRNA genes located in the human imprinted 14q32 domain, 40 of which are organized as a large cluster. Although some of these clustered miRNA genes appear to be encoded by a single-copy DNA sequence, most of them are arranged in tandem arrays of closely related sequences. In the mouse, this miRNA gene cluster is conserved at the homologous distal 12 region. In vivo all the miRNAs that we have detected are expressed in the developing embryo (both in the head and in the trunk) and in the placenta, whereas in the adult their expression is mainly restricted to the brain. We also show that the miRNA genes are only expressed from the maternally inherited chromosome and that their imprinted expression is regulated by an intergenic germline-derived differentially methylated region (IG-DMR) located ∼200 kb upstream from the miRNA cluster. The functions of these miRNAs, which seem only conserved in mammals, are discussed both in terms of epigenetic control and gene regulation during development.
Over 200 distinct genes encoding 21- to 25-nucleotide (nt)-long noncoding RNAs, the microRNAs (miRNAs), have been identified through either computer-assisted approaches or cDNA cloning strategies in many organisms, including worm, plants, flies, mouse, and human (Lai 2003; Bartel 2004). They are encoded within irregular hairpins and generated by a two-step mechanism involving Drosha and Dicer, two distinct dsRNA-specific ribonucleases belonging to the RNAse III family. First, Drosha cleaves a larger RNA precursor (the pri-miRNA) in the nucleus to release an irregular ∼70-nt-long hairpin (the pre-miRNA; Lee et al. 2003) from which the miRNA is excised from one strand within the cytoplasm through the activity of Dicer (Bernstein et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). Then, the mature 21- to 23-nt-long miRNAs incorporate into the RISC/miRNP complexes containing proteins belonging to the Argonaute family (Schwarz and Zamore 2002) and base-pair with mRNAs to trigger either RNA degradation (RNA interference or RNAi) or translation repression (Hutvagner and Zamore 2002; Llave et al. 2002). Indeed, miRNAs perfectly complementary to the RNA target guide sequence-specific RNA cleavages (they function like small interfering RNAs or siRNAs), whereas miRNAs imperfectly complementary to the target repress its translation by an as-yet-unknown mechanism.
In plants, miRNAs exhibit perfect or near-perfect complementarities with their targets, and they can either guide mRNA degradation (Llave et al. 2002; Palatnik et al. 2003; Tang et al. 2003; Xie et al. 2003) or inhibit their translation (Aukerman and Sakai 2003; Chen 2004). Remarkably, most of predicted target mRNAs encode for transcription factors known to regulate leaf and flower development, embryonic patterning, and timing transition to flowering (Rhoades et al. 2002). By contrast, animal miRNAs seem to act predominantly as translational repressors, and they have been shown to play a key role in the establishment of temporal and spatial gene expression pattern (Lee et al. 1993; Reinhart et al. 2000; Johnston and Hobert 2003), in the regulation of tissue growth, apoptosis, and fat metabolism (Brennecke et al. 2003; Xu et al. 2003) and in the differentiation of hematopoietic lineages (Chen et al. 2004). However, although many computationally predicted mRNA targets have been recently reported in flies and in mammals (Enright et al. 2003; Lewis et al. 2003; Stark et al. 2003), only a very few miRNA targets have been experimentally tested in vivo, and other RNA-mediated gene silencing mechanisms can also be envisaged. In this regard, RNAi-related mechanisms are also required for the silencing of heterochromatin in Schizosaccharomyces pombe (Hall et al. 2002; Reinhart and Bartel 2002; Volpe et al. 2002; Schramke and Allshire 2003; Verdel et al. 2004), for the programmed chromosomal rearrangements in Tetrahymena (Yao et al. 2003) and for the transcriptional gene silencing in plants and in Drosophila (Mette et al. 2000; Hamilton et al. 2002; Pal-Bhadra et al. 2002, 2004; Zilberman et al. 2003). Thus, 21- to 25-nt-long RNAs are involved in a range of eukaryotic gene regulation phenomena, and their potential as modulators of gene expression is just beginning to be appreciated.
Genomic imprinting refers to an epigenetic phenomenon by which a subset of mammalian autosomal genes, the so-called imprinted genes, are expressed in a parent-of-origin specific manner (only one of the two parental alleles is expressed). Most of these imprinted genes are clustered within large chromosomal domains harboring both paternally and maternally expressed imprinted genes. Molecular mechanisms underlying genomic imprinting are not fully understood but include allele-specific DNA methylation and core histone modifications, cis-acting elements functioning as imprinting control centers, chromatin boundaries or insulators, and antisense noncoding RNAs (Verona et al. 2003). We have previously shown that the imprinted mouse distal 12 chromosome (human 14q32) encodes four miRNA genes—mir-134, mir-154, mir-136, and mir-127—of which the two latter are only expressed from the maternal allele and are transcribed in an antisense orientation to a retrotransposon-like gene (Rtl1; Fig. 1). Rtl1 is transcribed from the paternal chromosome only, raising the possibility that the RNAi machinery might negatively regulate the Rtl1 gene and/or contribute to its imprinting status (Lin et al. 2003; Seitz et al. 2003).
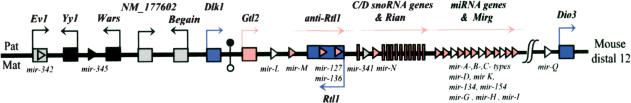
Figure 1.
Schematic representation of the imprinted mouse distal 12 domain (human 14q32). The position of several imprinted genes is indicated by squares (Gtl2 and protein-coding genes), vertical bars (snoRNA genes), or triangles (miRNA genes). Maternally expressed and paternally expressed genes are filled in pink and blue, respectively. Biallelically expressed genes and genes with undetermined imprinted status are colored in black and grey, respectively. Open triangles represent in silico-predicted miRNA gene that we failed to experimentally detect (i.e., pre-mir-L, pre-mir-H, pre-mir-G, pre-mir-I, pre-mir-Q). Two recently described miRNA genes—mir-342 and mir-345 (Kim et al. 2004)—map within an intron of Ev1 and between Yy1 and Wars genes, respectively. Imprinting status of mir-342 remains unsolved as we failed to detect it in embryos. An intergenic germ-line derived differentially methylated region (IG-DMR) located between Dlk1 and Gtl2 genes is represented by circles (filled indicates hypermethylated; open, hypomethylated). Mat indicates maternal chromosome; Pat, paternal chromosome. The figure is not drawn to scale.
In the present work, through a large and systematic computer-assisted approach within the ∼1-Mb human imprinted 14q32 locus/mouse distal 12 domain, we report the identification of 46 potential miRNA genes. Remarkably, most of these genes map within a ∼40-kb interval that represents, to our knowledge, the largest miRNA gene cluster described so far. All the miRNA genes tested exhibit a tissue-specific expression pattern, and they are only expressed from the maternally inherited allele with imprinted expression controlled by a differentially methylated region (DMR) located ∼200 kb away. Although their functions are still unknown, the unexpected characterization of these novel miRNA genes considerably improves our knowledge of gene organization at this imprinted domain by adding a novel layer of complexity in terms of noncoding RNA genes and their potential involvement in epigenetic controls.
RESULTS
Identification of miRNA Genes at Human 14q32 Domain
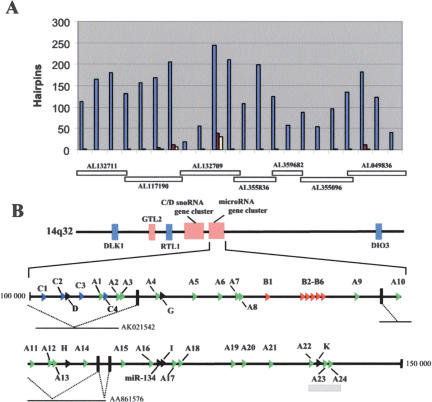
The tendency of miRNA genes to occur in clusters (Lagos-Quintana et al. 2001, 2003; Lau et al. 2001; Mourelatos et al. 2002; Reinhart et al. 2002; Dostie et al. 2003) prompted us to search systematically for additional miRNA genes encoded within the whole 1-Mb human imprinted 14q32 domain. This was achieved by identifying conserved sequences between mouse and human that have the potential to fold into imperfect hairpins of similar length to known miRNA genes. The sequences were filtered by using the MiRscan software recently developed by Lim and colleagues (2003), and a total of 35 conserved hairpins were eventually retained (Fig. 2A). Remarkably, most of them (29) are clustered within a 40-kb-long region that maps ∼25 kb downstream of the C/D snoRNA gene cluster (Cavaille et al. 2002). The remaining selected hairpins are encoded either around or within the Rtl1 gene (including the previously reported miR-127 and miR-136 [Lagos-Quintana et al. 2002] and three novel ones, miR-L, miR-M, miR-N) or upstream from the Dio3 gene (miR-Q; Figs. 1, 2A, Supplemental data S1).
Figure 2.
Identification of computationally predicted miRNA genes at the human imprinted 14q32 domain. (A) Distribution of the conserved hairpins between human and mouse along the 1-Mb-long imprinted 14q32 domain. Vertical bars indicate the number of in silico-predicted hairpins found within each 50-kb-long region. Blue bars indicate the number of human hairpins at the 14q32 domain; red bars, the number of conserved hairpins between mouse and human; and yellow bars, the number of conserved hairpins giving rise to a miRscan score >10 (Lim et al. 2003). The relative positions of the overlapping human BACs covering the analyzed domain are indicated below the histogram. (B) A large cluster of miRNA genes mapping downstream from the C/D snoRNA gene cluster. miRNA genes belonging to the A-, B-, and C-types are represented by green, red, and blue arrows, respectively, and miRNAs encoded by a single-copy gene (miR-D, G, H, I, K) are indicated as black arrows. Note that four miRNA genes—miR-A22, miR-K, miR-A23, and miRA24—are embedded within a conserved CpG island (indicated by a gray rectangle) from which the most conserved sequences lie within the pre-miRNA genes (data not shown). Dotted lines denote splicing events identified by ESTs analysis. Numbering indicates the relative position of the miRNA gene cluster (in nucleotides) within the BAC AL132709 sequence. The picture is drawn to scale.
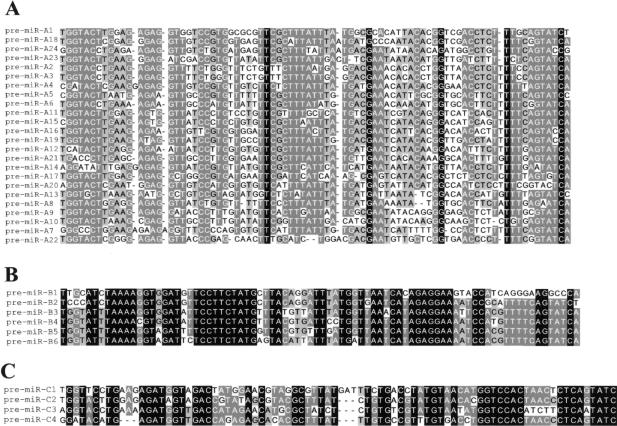
Multiple sequence alignments revealed that 15 predicted miRNA genes have sequences related to the previously described miR-154 gene precursor (Lagos-Quintana et al. 2002; Seitz et al. 2003), and a subsequent BLAST search with lower stringency conditions allowed us to identify eight additional human miR-154-related sequences (Figs. 2B, 3A), although six do not have clear mouse counterparts. These miR-154-related sequences have been grouped into the A-type family (A1 to A24 gene copies). Interestingly, within the same 40-kb interval other hairpins could also be ranked together into families of closely related sequences: the B-type family (six and three gene copies in human and mouse, respectively) and the C-type family (four gene copies both in human and in mouse; Figs. 2B, 3B,C). Thus, overall a total of 40 conserved in silico-predicted human miRNA genes, all of them mapping in the same relative orientation, have been isolated downstream of the C/D snoRNA gene cluster (Fig. 2B), and to our knowledge, they represent the largest cluster of miRNA genes described to date. A-, B-, and C-type pre-miRNA sequences appear to be related to each other (particularly at their terminal 5′ and 3′ ends; data not shown), and this might suggest that the three miRNA families have evolved through duplications from a common miRNA gene ancestor.
Figure 3.
Multiple sequence alignment of the three human miRNA families. The names of the pre-miRNA variants belonging to the A-, B-, and C-types are indicated at the left in A, B, and C, respectively. Conservation of each nucleotide position is denoted by shading according to GeneDoc software: black (100%), dark gray (80% to 99%), and clear gray (60% to 79%). Sequence alignment of the conserved murine pre-miRNAs belonging to the A-, B-, and C-types is shown in Supplemental data S2.
Experimental Detection of the In Silico-Predicted miRNAs
The expression of all the predicted miRNA species was first monitored in mouse by Northern blot analysis with oligonucleotide probes complementary to either the 5′ or the 3′ arm of the predicted fold-back structures. We failed to detect convincing signals for most of them, including miR-154 and miR-134, which were originally cloned from cDNA libraries (Lagos-Quintana et al. 2002; data not shown). Indeed, only miR-C3, miR-A24, and B-type miRNAs gave positive signals in total RNA isolated from whole embryo or adult brain (data not shown). Nevertheless, based on our current knowledge of the relative position of the mature miRNA within the pre-miRNA (Lim et al. 2003), we designed a novel specific set of primers for several miRNAs distributed along the cluster (Supplemental data S1) and performed a more sensitive primer extension analysis. By using this approach, we mapped the 5′ end of small RNA species encoded within pre-miR-A22 (3′ arm), pre-miR-A24 (3′ arm), pre-miR-B4 (3′ arm), pre-miR-B6 (3′ arm), pre-miR-C2 (3′ arm), pre-miR-C3 (3′ arm), pre-miR-154 gene (5′ arm), pre-miR-134 (5′ arm), pre-miR-K (3′ arm), pre-miR-M (3′ probe), and pre-miR-N (3′ probe; Table 1; data not shown). Because of potential cross-hybridization between related miRNA sequences, we have been unable to test unambiguously each miRNA gene candidate, hence the possibility that some of the in silico-predicted miRNA variants might not be expressed cannot be formally ruled out. However, three recently cloned miRNAs—miR-323, miR-329, and miR-300—are encoded within the 3′ strand of the A1, A2, and A9 pre-miRNA gene precursors (Houbaviy et al. 2003; Kim et al. 2004), respectively, validating our in silico approach and suggesting that others, if not all the A copies, might also give rise to mature miRNAs. Our failure to experimentally detect miRNAs within miR-D, miRG, miR-H, miR-I, miR-L, and miR-Q predicted hairpins might reflect expression of these miRNA genes at levels below the classical experimental detection. Consistent with this, miR-300 embedded within the in silico-predicted miR-D hairpin (5′ arm) has been cloned, but its transcripts have not been detected by Houbaviy et al. (2003).
Table 1.
List of Experimentally Detected or Cloned miRNAs at the Mouse Distal 12 Domain
| miRNAa | Sequence (5′ to 3′)b | Maternally expressed miRNAs | References |
|---|---|---|---|
| miR-342 | UCUCACACAGAAAUCGCACCCGUC | n.d. | Kim et al. 2004; this study |
| miR-345 | UGCUGACCCCUAGUCCAGUGC | — | Kim et al. 2004; this study |
| miR-127 | UCGGAUCCGUCUGAGCUUGGCU | + | Lagos-Quintana et al. 2002; Seitz et al. 2003 |
| miR-136 | ACUCCAUUUGUUUUGAUGAUGGA | + | Lagos-Quintana et al. 2002; Seitz et al. 2003 |
| miR-134 | UGUGACUGGUUGACCAGAGGG | + | Lagos-Quintana et al. 2002; this study |
| miR-154/miR-A19-5c | UAGGUUAUCCGUGUUGCCUUCG | + | Lagos-Quintana et al. 2002; this study |
| miR-323/miR-A1-3′ | GCACAUUACACGGUCGACCUCU | n.d. | Kim et al. 2004; this study |
| miR-329/miR-A2-3′ | AACACACCCAGCUAACCUUUUU | n.d. | Kim et al. 2004; this study |
| miR-300/miR-A9-3′ | UAUGCAAGGGCAAGCUCUCUUC | n.d. | Houbaviy et al. 2003; this study |
| miR-409/miR-A22-3′ | GAAUGUUGCUCGGUGAACCCCUU | n.d. | this study |
| miR-410/miR-A24-3′ | AAUAUAACACAGAUGGCCUGUU | + | this study |
| miR-376b/miR-B4 | AUCAUAGAGGAACAUCCACUUU | + | this study |
| miR-376/miR-B6 | AUCGUAGAGGAAAAUCCACGUU | + | this study |
| miR-411/miR-C2 | AACACGGUCCACUAACCCUCAGU | + | this study |
| miR-380-3p/miR-C3 | UAUGUAGUAUGGUCCACAUCUU | + | this study |
| miR-299/miR-D | UGGUUUACCGUCCCACAUACAU | n.d. | Houbaviy et al. 2003; this study |
| miR-412/miR-K | ACUUCACCUGGUCCACUAGCCGU | n.d. | this study |
| miR-337/miR-M | UCAGCUCCUAUAUGAUGCCUUUC | + | Kim et al. 2004; this study |
| miR-370/miR-Nd | GCCUGCUGGGGUGGAACCUGGUU | + | this study |
| miR-341e | UCGAUCGGUCGGUCGGUCAGU | n.d. | Kim et al. 2004 |
n.d. indicates not determined.
Novel miRNA sequences have been submitted to the miRNA registry (http://www.sanger.ac.uk/cgi-bin/Rfam/mirna/browse.pl.) and they have been given new names in accordance to the miRNA registry numbering (e.g. miR-A22-3′ is also called miR-409 in the miRNA registry).
The exact length of the miRNAs which have been detected solely by primer extension is not known. Thus, the two last nucleotides at the 3′-termini are in italics as they are only predicted based on a 23 nt theoretical long RNA species. Primer extension assay generates two cDNA products with a size differing by one nucleotide. Based on the miR-134 and miR-154 sequences obtained independently by cloning strategies (Lagos-Quintana et al. 2002), we have considered the shorter cDNA product as the correct 5′ termini while the longer one might correspond to in vivo processing heterogeneity and/or addition of an extra nucleotide in a matrix-independent manner by the AMV reverse transcriptase (Promega technical service, pers. comm.).
miR-154/A19 are processed from the 5′ side of A19 (Lagos-Quintana et al. 2002) while miR-323/A1, miR-329/A2, miR-A9, miR-A22 and miR-A24 are processed from the 3′ strand of the A1, A2, A9, A22 and A24 gene copies, respectively suggesting that both strands of A-type can potentially be converted to miRNAs.
Depending upon RNA samples, we could also detect a ladder-like pattern superimposed to the mature 5′ end of miR-N. Thus, this miRNA might not fulfill the stringent criteria described in (Ambros et al. 2003).
mir-341 was not found in our in silico search as it is not conserved at the human inprinted 14q32 interval.
Several miRNAs described here have been also detected experimentally in bovine and rat but not in chicken brain (data not shown). BLAST searches also failed to detect significant hits in Caenorhabditis elegans, Drosophila melanogaster, and Fugu rubripes genomes. Taken together, these observations strongly suggest that these miRNA sequences are specific to mammalian species. Based on conservation criteria between human and fish F. rubripes, a recent study argued that the number of miRNAs in human genome should not exceed ∼250, with ∼40 remaining to be determined (Lim et al. 2003). Our study clearly shows that for mammalian systems, this number may be an underestimate. Reinforcing this notion, poorly conserved embryonic stem-cell-specific miRNA genes, also organized in a tandem array, have been recently described and proposed to play a key role in the regulation of early mammalian development (Houbaviy et al. 2003).
Tissue-specific Expression Pattern of the Distal 12 miRNA Genes
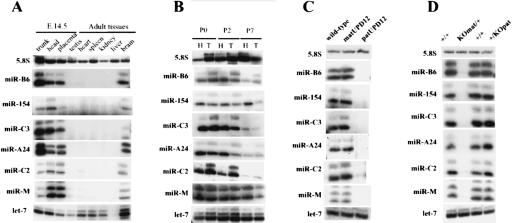
Many miRNA genes are expressed in a temporal and/or tissue-specific manner in line with their well-documented functions in development (Carrington and Ambros 2003). To gain insight into the miRNA gene expression pattern, primer extensions have been performed for several miRNAs by using total RNA extracted from several adult mouse tissues, embryos (head or trunk), and placenta. As shown in Figure 4A, in adults all tested miRNAs exhibit nearly the same tissue-specific expression pattern with signals detected only in the brain. This is consistent with miR-A1/miR-323 and miR-A2/miR-329 being originally isolated from rat neurones (Kim et al. 2004). Furthermore, within the developing embryos (E12.5, E14.5, and E17.5) all the tested miRNA genes are expressed at roughly the same levels not only in the head but also in the trunk and the placenta, indicating that other tissues distinct from the brain express them as well (Fig. 4A; data not shown). In neonates (P0, P2, P7), miRNAs are still detected in the trunk, even though we noticed a reproducible decrease in the expression level for most of them after birth (Fig. 4B). Curiously, although each primer extension product generally appears as a band doublet for a given miRNA, for samples extracted from the head or the trunk in neonates, several miRNAs exhibit a different intensity ratio between the two vicinal cDNA bands. The biological relevance of this difference, if any, remains unclear (see also Table 1). Further studies are needed to identify cell type-specific expression patterns as well as mechanisms of miRNA gene regulation.
Figure 4.
Expression of in silico-predicted miRNA genes. Tissue-specific (A) and developmental gene expression pattern (B) of the miRNAs analyzed by primer extension assay. H indicates head; T, trunk. 5.8S and let-7 probes have been used as gel loading controls (Northern blot analysis). (C) The miRNA genes are only expressed from the maternally inherited chromosome. Ten micrograms of total RNA extracted from wild-type embryos, embryos with maternal uniparental disomy for chromosome 12 (matUPD12), or embryos with paternal uniparental disomy for chromosome 12 (patUPD12) have been subjected to primer extension assay. (D) The miRNA imprinted gene expression is controlled by the intergenic germline-derived differentially methylated region (IG-DMR). Ten micrograms of total RNA extracted from embryo with a deletion of the IG-DMR from either the maternally or the paternally inherited chromosomes (as specified at the top of the picture) have been subjected to primer extension analysis.
Maternal Expression of the miRNA Gene Cluster Is Regulated by a Distal Intergenic, Germ-Line-Derived DMR (IG-DMR).
Because of their location within a previously characterized imprinted domain, we next examined whether the novel miRNA genes are subjected to genomic imprinting. As shown in Figure 4C, E15.5 embryos with a maternal uniparental disomy of chromosome 12 (matUPD12) express all the miRNAs that we have tested, whereas embryos having two paternal chromosome 12 copies (patUPD12) do not, showing that these miRNA genes are only expressed from the maternally inherited chromosome. We noticed that miRNA signals detected in the matUPD embryos are not twice as intense as those seen in the wild-type embryos. Although we can not exclude that these observations have a biological significance, we believe that our primer extension assay is not quantitative enough to reproducibly observe the expected twofold increase in the matUPD embryos. An IG-DMR located between the Dlk1 and Gtl2 genes has been recently shown to play a key role in controlling imprinted expression within the whole locus (Fig. 1). Deletion of the unmethylated IG-DMR from the maternally inherited chromosome results in the silencing of all the maternally expressed imprinted genes together with loss of imprinting (biallelic expression) of the maternally repressed genes. When inherited from the paternal allele, the deletion of the methylated IG-DMR does not affect imprinted gene expression (Lin et al. 2003). All the miRNAs that we have checked were absent from total RNA extracted from embryos having a maternal deletion of the IG-DMR, whereas those with the paternally inherited deletion had miRNA expression similar to that of the control unmanipulated littermates (Fig. 4D). We therefore conclude that, similar to the other maternally expressed noncoding RNA genes (anti-Rtl1, Gtl2, and the C/D snoRNA genes; Lin et al. 2003), the expression of the miRNA gene cluster is controlled by this distal cis-acting element.
The miRNAs Are Processed From Large Noncoding RNAs
All the miRNA genes described in this study share the same tissue-specific expression pattern with strongest expression in the adult brain. A similar gene expression pattern has been previously reported for the other neighboring maternally expressed noncoding RNA genes (Gtl2 and C/D RNA genes; Schmidt et al. 2000; Cavaille et al. 2002). Very little is known about miRNA transcription and its regulation in mammals (Bartel 2004). Although a few of these clustered miRNA genes are embedded within introns or exons of Mirg, a maternally expressed gene lacking a conserved protein-coding potential (Seitz et al. 2003), the genomic organization of most of them is still unclear (Figs. 2B, 5). Each pre-miRNA gene might be transcribed from its own tissue-specific promoter or alternatively it might be processed from (some) polycistronic-like RNA(s) as already reported for other clustered miRNAs (Lee et al. 2002). To address this second possibility, we have used RT-PCR to detect primary precursors encompassing several miRNAs relatively close to each other. As shown in Figure 5, transcripts encompassing several neighboring miRNAs are readily detected. Although we cannot rule out that some miRNA genes are transcribed from their own promoter, it seems more likely that most, if not all, pre-miRNA genes are generated through RNA processing either from polycistronic RNA and/or from intronic sequences rather than being individually expressed as ∼70-nt-long primary transcripts. Thus, the novel clustered miRNA genes share a number of common properties with the upstream C/D snoRNA genes (Cavaille et al. 2002). Both classes of small noncoding RNA genes are (1) relatively weakly conserved during evolution, (2) organized as repeated arrays and processed from larger noncoding RNA genes, (3) transcribed in the same direction from the maternal chromosome only, (4) regulated by the IG-DMR, and (5) expressed in a tissue-specific manner with strongest expression in the adult brain. Thus, it is tempting to speculate that C/D RNAs and miRNAs are processed from a single, large primary transcript that might initiate at the Gtl2 promoter. Future work is now needed to fully characterize this atypical and complex transcription unit.
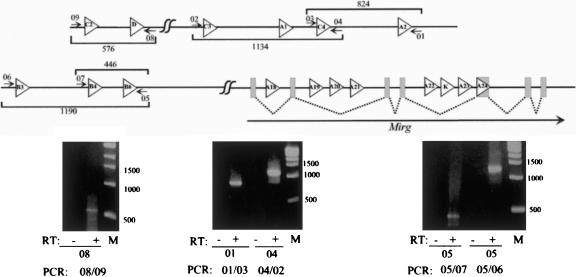
Figure 5.
Detection of large transcripts overlapping several miRNA genes. Ten micrograms of DNAse I-, proteinase K-treated total RNAs extracted from the head of mouse embryo (E17.5) was subjected to RT-PCR with appropriate primers (numbered arrows) mapping outside from the ∼70-nt pre-miRNA genes. The location of the primers and expected sizes of the amplification products are shown on the top part of the figure (not drawn to scale). Primer sequences are available on request. RT indicates reverse transcriptase (Superscript II, Invitrogen).
DISCUSSION
Most imprinted genes play important roles in embryonic and/or placental growth (Tycko and Morison 2002) and developmental abnormalities in mouse and human have been previously associated with the lack of expression of the maternally inherited genes at distal 12 and 14q32, respectively (Georgiades et al. 2000; Kurosawa et al. 2002). Given the known roles of miRNAs in development (Carrington and Ambros 2003), it is likely that these novel imprinted miRNAs mediate a function during mouse development. In contrast to the mir-127 and mir-136 genes encoded in the antisense strand of the Rtl1 gene (Seitz et al. 2003), none of these novel maternally expressed miRNAs exhibit full complementarity to a cellular mRNA, suggesting they probably inhibit mRNA translation rather than triggering mRNA degradation. Indeed, a role in translation control is supported by the association of miR-A1/miR-323 with the polyribosomes in rat primary cortical cultures (Kim et al. 2004). So far, despite the identification of >150 miRNAs from mouse and human, their biological functions remain highly elusive, mainly due to the difficulty in finding convincing RNA target(s). Through computer searches, cross genome comparison (i.e., pufferfish versus mammalian) are commonly used to predict conserved mRNA targets (Lim et al. 2003). Because the miRNAs described in this study seem to be restricted to mammalian species, assigning mRNA target(s) for these imprinted miRNAs might be a difficult task. Moreover, given that the sequences of most of clustered pre-miRNA genes are related to some extent, we could envision that (1) they regulate the same mRNA (or a common class of mRNAs) by interacting at various binding sites along the 3′ untranslated region (3′ UTR); or (2) conversely, the relative lack of strong selection pressure on several miRNAs (see Supplemental data S2) might also allow them to target multiple and distinct mRNA targets. In the sheep, deregulated gene expression at the Dlk1-Gtl2 domain (Charlier et al. 2001) is associated with the Callipyge (Clpg) phenotype, a muscular hypertrophy manifest only in heterozygous individuals who have inherited the Clpg mutation from their paternal chromosome, but not observed in Clpg/Clpg homozygotes. One mechanism postulated to explain this unexpected mode of inheritance is a trans interaction between the reciprocally imprinted genes (e.g., a trans-acting repressor expressed from the maternal chromosome could negatively regulate some paternally expressed genes; for a review, see Georges et al. 2003). The miRNA genes described in this study are obvious candidates for playing such a role.
RNA-mediated gene silencing mechanisms are thought to operate at the imprinted Igf2r-Air domain as well as in X-chromosome inactivation in female mammalian cells (Plath et al. 2002; Sleutels et al. 2002). Moreover RNA-mediated formation of heterochromatin has been observed in S. pombe (Hall et al. 2002; Volpe et al. 2002; Schramke and Allshire 2003; Verdel et al. 2004), and small RNAs play a role in the DNA methylation and silencing of target sequences in plants (Aufsatz et al. 2002). Thus, an emerging theme is the idea that noncoding RNAs have key functions in genome modification. Whether the clustered miRNAs can control gene expression at the transcriptional level is still an open and challenging question. We noticed that four miRNA genes—miR-A22, miR-A23, miR-A24, and miR-K—are embedded within a conserved CpG island located at the 3′ end of the miRNA gene cluster. The methylation status of this CpG-rich domain is under investigation.
Did this large cluster of miRNAs emerge within this imprinted domain before or after the onset of imprinting at this region? If the former, did it confer some aspect of imprinting control, or if the latter, is the imprinted expression of these small hairpin RNAs a mere consequence of more widespread expression of noncoding RNAs from the maternal chromosome? One thought-provoking idea pertaining to the evolution of the imprinting mechanism suggests that intragenomic parasitic sequences, including repeated sequences, retrotransposons, and other related elements, are heritably and variably silenced in order that they are rendered transcriptionally inactive, “hidden” by heterochromatinization, and/or immobilized (Barlow 1993; Bird 1997; Yoder et al. 1997; Martienssen 1998; Whitelaw and Martin 2001). Although this hypothesis was based on a function for DNA methylation, it could equally be applied to a small RNA-mediated mechanism. Thus, one can speculate that these repeated small RNA genes might have integrated into this chromosomal domain prior to the emergence of its imprinting, and their subsequent gene amplification may have been perceived by the host as “foreign DNA,” resulting in imposition of epigenetic regulation to the surrounding protein-coding genes. Conversely, the miRNA gene cluster might have been generated after the establishment of the imprinting, perhaps due to a specific chromatin architecture prone to gene duplication and/or to the different behavior of the two parental chromosomes, which is permissive for nondisruptive insertion. It remains to be determined whether the parental origin specific nature of the miRNA expression contributes to, or is a consequence of, genomic imprinting.
An exhaustive characterization of all the epigenetic marks along this large imprinted domain together with genetically engineered mouse models is now required to fully appreciate the potential involvement of these imprinted miRNA genes in mouse development and/or epigenetic control.
METHODS
Unless otherwise noted, all techniques for manipulating nucleic acids were performed according to standard protocols.
Oligodeoxynucleotides
They were all synthesised by Y. de Préval (LBME) on a PerSeptive Biosystems Expedite apparatus. Primer sequences are available on request
Searching for miRNA Genes at Human Imprinted 14q32 Domain
BAC sequences around DLK1/GTL2 positions were retrieved from the Human Genome Project Working Draft (http://genome.ucsc.edu/) and 100-nt-long stem-loop structures with a ΔG <–30 kcal/mole were systematically identified within the ∼1-Mb-long imprinted 14q32 region by using RNAfold software from http://www.tbi.univie.ac.at/~ivo/RNA/. We then retained the conserved hairpin structures between the murine and human orthologous imprinted loci (according to BLAT search at http://genome.ucsc.edu/) that give rise to a miRscan score >10 by using the software developed by D.P. Bartel and his colleagues (http://genes.mit.edu/mirscan/). Sequence alignment of the miRNA genes were obtained by MultiAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html), and conserved nucleotides were shaded by GeneDoc (http://www.Cris/com/~ketchup/genedoc.shtml).
Experimental Detection of the miRNAs
In silico-predicted miRNA genes were validated either by Northern blot analysis or by primer extension. Total RNA was isolated by the guanidium thiocyanate method from mouse tissues or from whole embryos freshly prepared according to French institutional and United Kingdom Home Office guidelines. For Northern blot analyses, 15 to 20 μg of DNAse I-, protease K-treated total RNA was separated on a denaturing 15% acrylamide/7M urea gel. Electrotransfer in 0.5× TBE buffer was performed onto nylon membranes (Hybond N+) followed by an UV light irradiation of the membrane. Hybridizations were carried out with 5′ (32P)-labeled oligodeoxynucleotide probes, through an overnight incubation in 5× SSPE, 1% SDS, 150 μg/mL yeast tRNA, and 5× Denhardt's. Membranes were washed twice with 2× SSPE and 0.1 % SDS at room temperature before autoradiography. For primer extension assay, after a heat denaturation step (5 min at 75°C), 10 μg of total RNA was annealed with a (32P)-labeled and gel-purified specific primer (see Supplemental data) and reverse transcribed during 2 h at 42°C by AMV reverse transcriptase (Promega), and the resulting cDNA products were analyzed on a 15% acrylamide /7 M urea gel.
NOTE ADDED IN PROOF
Human miRNAs derived from our computationally predicted pre-miR-B3 and pre-miR-A23 have been experimentally cloned by Suh et al. (2004). They have been named miR-368 and miR-369, respectively.
Acknowledgments
This work has been initiated in the Dr. Jean-Pierre Bachellerie's laboratory, and we thank him very much for his permanent support and for the critical reading of the manuscript. We are also grateful to D. Morello for her help with mouse manipulation. Work in the laboratory of J.C. is supported by the Ministère de l'éducation Nationale, de la Recherche et de la technologie (Action Concertée Incitative 2003: Biologie cellulaire moléculaire et structurale), and work in the laboratory of A.F-S. is funded by a CR-UK grant. S.-P.L. is funded by a graduate research scholarship from the Taiwanese Government; H.S., by a PhD fellowship Allocation de Moniteur Normalien (Ministère de l'Education Nationale, de la Recherche et de la technologie). The authors declare that they have no competing financial interests.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2743304. Article published online before print in August 2004.
[Supplemental material is available online at www.genome.org.]
References
- Aufsatz, W., Mette, M.F., van der Winden, J., Matzke, A.J., and Matzke, M. 2002. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. 99: 16499-16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, D.P. 1993. Methylation and imprinting: From host defense to gene regulation? Science 260: 309-310. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Bird, A. 1997. Does DNA methylation control transposition of selfish elements in the germline? Trends Genet. 13: 469-472. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25-36. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. 2003. Role of microRNAs in plant and animal development. Science 301: 336-338. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Seitz, H., Paulsen, M., Ferguson-Smith, A.C., and Bachellerie, J.P. 2002. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 11: 1527-1538. [DOI] [PubMed] [Google Scholar]
- Charlier, C., Segers, K., Karim, L., Shay, T., Gyapay, G., Cockett, N., and Georges, M. 2001. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat. Genet. 27: 367-369. [DOI] [PubMed] [Google Scholar]
- Chen, C.Z., Li, L., Lodish, H.F., and Bartel, D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83-86. [DOI] [PubMed] [Google Scholar]
- Chen, X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges, M., Charlier, C., and Cockett, N. 2003. The callipyge locus: Evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 19: 248-252. [DOI] [PubMed] [Google Scholar]
- Georgiades, P., Watkins, M., Surani, M.A., and Ferguson-Smith, A.C. 2000. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127: 4719-4728. [DOI] [PubMed] [Google Scholar]
- Hall, I.M., Shankaranarayana, G.D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232-2237. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell. 5: 351-358. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056-2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834-838. [DOI] [PubMed] [Google Scholar]
- Johnston, R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845-849. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Krichevsky, A.M., Grad, Y., Hayes, G.D., Kosik, K.S., and Church, G.M. 2004. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. 101: 360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa, K., Sasaki, H., Sato, Y., Yamanaka, M., Shimizu, M., Ito, T., Okuyama, T., Matsuo, M., Imaizumi, K., Kuroki, Y., et al. 2002. Paternal UPD14 is responsible for a distinctive malformation complex. Am. J. Med. Genet. 110: 268-272. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735-739. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. 2003. New microRNAs from mouse and human. RNA 9: 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C. 2003. microRNAs: Runts of the genome assert themselves. Curr. Biol. 13: R925-R936. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787-798. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lin, S.P., Youngson, N., Takada, S., Seitz, H., Reik, W., Paulsen, M., Cavaille, J., and Ferguson-Smith, A.C. 2003. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35: 97-102. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053-2056. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. 1998. Transposons, DNA methylation and gene control. Trends Genet. 14: 263-264. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9: 315-327. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669-672. [DOI] [PubMed] [Google Scholar]
- Plath, K., Mlynarczyk-Evans, S., Nusinow, D.A., and Panning, B. 2002. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 36: 233-278. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J. and Bartel, D.P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901-906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513-520. [DOI] [PubMed] [Google Scholar]
- Schmidt, J.V., Matteson, P.G., Jones, B.K., Guan, X.J., and Tilghman, S.M. 2000. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes & Dev. 14: 1997-2002. [PMC free article] [PubMed] [Google Scholar]
- Schramke, V. and Allshire, R. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069-1074. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S. and Zamore, P.D. 2002. Why do miRNAs live in the miRNP? Genes & Dev. 16: 1025-1031. [DOI] [PubMed] [Google Scholar]
- Seitz, H., Youngson, N., Lin, S.P., Dalbert, S., Paulsen, M., Bachellerie, J.P., Ferguson-Smith, A.C., and Cavaille, J. 2003. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34: 261-262. [DOI] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The noncoding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810-813. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila MicroRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M.R., Lee, Y., Kim, J.Y., Kim, S.K., Moon, S.H., Lee, J.Y., Cha, K.Y., Chung, H.M., Yoon, H.S., Moon, S.Y., et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270: 488-498. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko, B. and Morison, I.M. 2002. Physiological functions of imprinted genes. J. Cell. Physiol. 192: 245-258. [DOI] [PubMed] [Google Scholar]
- Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verona, R.I., Mann, M.R., and Bartolomei, M.S. 2003. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell. Dev. Biol. 19: 237-259. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Whitelaw, E. and Martin, D.I. 2001. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 27: 361-365. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784-789. [DOI] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790-795. [DOI] [PubMed] [Google Scholar]
- Yao, M.C., Fuller, P., and Xi, X. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300: 1581-1584. [DOI] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335-340. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu; Human Genome Project Working Draft.
- http://www.tbi.univie.ac.at~/ivo/RNA; RNA fold software.
- http://genes.mit.edu/mirscan; software developed by D.P. Bartel and colleagues.
- http://prodes.toulouse.inra.fr/multalin/multalin.html; MultiAlin.
- http://www.Cris.com~/ketchup/genedoc.shtml; GeneDoc.