Abstract
Mesenchymal stem cells (MSCs) are characterized by their self-renewing capacity and differentiation potential into multiple tissues. Thus, management of the differentiation capacities of MSCs is important for MSC-based regenerative medicine, such as craniofacial bone regeneration, and in new treatments for metabolic bone diseases, such as osteoporosis. In recent years, histone modification has been a growing topic in the field of MSC lineage specification, in which the Su(var)3–9, enhancer-of-zeste, trithorax (SET) domain-containing family and the Jumonji C (JmjC) domain-containing family represent the major histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs), respectively. In this review, we summarize the current understanding of the epigenetic mechanisms by which SET domain-containing KMTs and JmjC domain-containing KDMs balance the osteogenic and adipogenic differentiation of MSCs.
Keywords: adipogenesis, histone methylation, histone lysine methyltransferase, histone lysine demethylase, mesenchymal stem cells, osteogenesis
Introduction
Mesenchymal stem cells (MSCs) are multipotent adult stem cells that can self-renew and differentiate into cells of the mesodermal lineages both in vitro and in vivo, such as osteocytes, adipocytes, and chondrocytes.1,2,3 Although human MSCs were originally isolated from bone marrow, they have since been found in many other adult tissues.4,5 In addition to their differentiation capability and easy access, MSCs can be recruited to sites of tissue damage and be activated to express immunosuppressive molecules and growth factors, thereby regulating immune homeostasis and facilitating tissue repair.6,7 Hence, MSCs are considered to be of great promise for regenerative medicine.
The osteogenic and adipogenic differentiation of MSCs has been widely studied.8 A balance between these two pathways is critical to maintain homeostasis under physiological conditions, and disruption of this balance leads to bone-related metabolic diseases, such as osteoporosis,9,10 which features excessive accumulation of bone marrow adipocytes and a decrease in bone mass.11 Thus, bone repair with MSCs can be facilitated by enhancing osteogenic differentiation and inhibiting adipogenic differentiation. Therefore, understanding of MSC differentiation is critical in the development of new treatments for metabolic bone diseases.
The differentiation of MSCs is a complex process that involves multiple mechanisms. Various signaling pathways, such as Wnt, Notch, transforming growth factor (TGF)-β, and nuclear factor-κB (NFκB), have been demonstrated in the regulation of MSC differentiation.12,13,14 Among these, intricate cross-talk has been identified.15,16 In addition, several transcription factors are considered to be essential modulators of MSC differentiation, such as runt-related transcription factor 2 (RUNX2) in osteogenesis and peroxisome proliferator-activated receptor-gamma (PPAR-γ) in adipogenesis.17,18 In the last decade, growing evidence has shown that epigenetic mechanisms play an important role in regulating MSCs differentiation.19,20,21
The term “epigenetic mechanism” refers to inheritable changes in gene expression without altering the DNA sequence and includes DNA methylation, histone modification, and small non-coding RNA-associated regulation.22 In eukaryotic cells, chromatin structure is highly dynamic, regulated in large part by histone modifications, such as histone acylation and methylation.23 During histone methylation, methyl groups are transferred to histone proteins of chromosomes, generally at lysine residues. Specifically, these lysines can be unmodified (me0), mono- (me1), di- (me2), or tri-methylated (me3). The resulting histone methylation states, which contribute to chromatin structural changes, are important in regulating transcription factors' access to the related gene promoters.24
Recent studies have highlighted histone methylation state as an important modulator in stem cell differentiation.24 Regulation of these methylation states is tightly controlled by the opposing activities of histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs),25 which are recruited to specific histone lysine residues and are responsible for the establishment and regulation of histone methylation and demethylation, respectively. The Su(var)3–9, enhancer-of-zeste, trithorax (SET) domain-containing family and the Jumonji C (JmjC) domain-containing family represent the major KMTs and KDMs.24,25
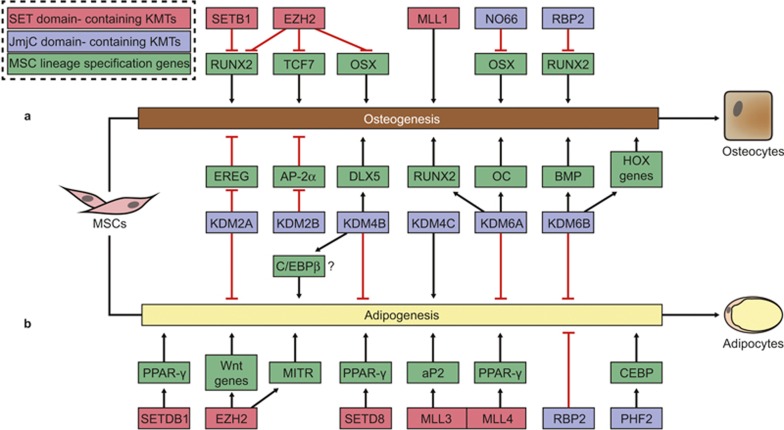
In this review, we summarize the current understanding of the epigenetic effects of histone methylation on balancing osteogenic and adipogenic differentiation of MSCs, specifically focusing on the roles of SET domain-containing KMTs and JmjC domain-containing KDMs (Figure 1).
Figure 1.
The role of SET domain-containing KMTs and JmjC domain-containing KDMs in osteogenesis and adipogenesis of MSCs. (a) The differentiation of MSC involves a lot of lineage specification genes, including transcription factors and other genes. SET domain-containing KMTs and JmjC domain-containing KDMs regulate the expression levels or transcription activity of the genes to determine the cell-fate. (a) The role of SET domain-containing KMTs and JmjC domain-containing KDMs in osteogenesis of MSCs. (b) The role of SET domain-containing KMTs and JmjC domain-containing KDMs in adipogenesis of MSCs. The related KMTs and KDMs are shown in red and blue boxes, respectively. The red question marker indicates the contradictory reported role of KDM4B in adipogenesis of MSCs. aP2, adipocyte protein 2; AP-2α, activating enhancer-binding protein 2α BMP, bone morphogenetic protein; C/EBPβ, CCAAT/enhancer-binding protein beta; DLX5, distal-less homeobox 5; EREG, epiregulin; EZH2, enhancer of zeste homologue 2; HOX, homeobox; KDM2A/JHM1A, JmjC domain-containing histone demethylase JmjC, Jumonji C; KDM, histone lysine demethylase; MSC, mesenchymal stem cell; MITR, myocyte enhancer factor-2 interacting transcriptional repressor; MLL, myeloid/lymphoid or mixed-lineage leukemia; RBP, retinol-binding protein; OC, osteocalcin; OSX, osterix; PHF2, plant homeodomain finger 2; PPAR-γ, peroxisome proliferator-activated receptor-gamma; RUNX2, runt-related transcription SET, Su(var)3–9, enhancer-of-zeste, trithorax; SETD8, SET domain-containing 8; SETDB1, SET domain bifurcated 1; TCF, transcription factor.
SET domain-containing histone methyltransferases
The evolutionarily conserved SET domain functions as the catalytic domain of KMTs by transferring a methyl group from s-adenosylmethionine to the ε-amine on the side chain of histone lysine residues.25 Aside from DOT1L and the WRAD complex,26,27 it is predicted that most KMTs are of a SET domain. These SET domain-containing KMTs seem to be particularly sensitive to posttranslational methylation at target lysine sites (Table 1). Although it has been five decades since histone methylation's discovery,28 its correlation with gene regulation was only established recently.29 SET domain bifurcated 1 (STDB1) and enhancer of zeste homologue 2 (EZH2) are the most widely studied SET domain-containing KMTs associated with MSC differentiation.
Table 1. The role of SET domain-containing KMTs during osteogenesis and adipogenesis (shaded) of MSCs.
| KMTs | Specificity | Target | Finding | Reference |
|---|---|---|---|---|
| SETDB1 | H3K9 | RUNX2 target genes | SETDB1 inhibits osteogenesis by reducing transcriptional activity of RUNX2 | 32,33 |
| PPAR-γ target genes | SETDB1 inhibits adipogenesis by transcriptional activity of PPAR-γ | 35 | ||
| EZH2 | H3K27 | RUNX2, TCF7, OC | EZH2 inhibits osteogenesis by catalyzing tri-methylation of H3K27 in the promoters of RUNX2, TCF7, and OC | 42,43 |
| Wnt genes, MITR | EZH2 promotes adipogenesis by catalyzing tri-methylation of H3K27 in the promoters of Wnt genes and MITR | 45,46 | ||
| SETD8 | H4K20 | PPAR-γ and PPAR-γ target genes | SETD8 promotes adipogenesis by catalyzing mono-methylation of H4K20 in the promoters of PPAR-γ and PPAR-γ target genes | 50,51 |
| MLL1 | H3K4 | Not known | Mice with SET domain mutated MLL1 exhibit skeletal defects | 52,53 |
| MLL3, MLL4 | H3K4 | PPAR-γ, aP2 | MLL3 and MLL4 promote adipogenesis by catalyzing methylation of H3K4 in the promoters of PPAR-γ and aP2 | 54,57 |
aP2, adipocyte protein 2; EZH, Enhancer of zeste homologue; KMT, histone lysine methyltransferase; MLL, myeloid/lymphoid or mixed-lineage leukaemia; MITR, myocyte enhancer factor-2 interacting transcriptional repressor; MSC, mesenchymal stem cells; OC, osteocalcin; PPAR, peroxisome proliferator-activated receptor; RUNX2, runt-related transcription factor 2; SET, Su(var)3–9, Enhancer-of-zeste, Trithorax; SETD8, SET domain-containing 8; STDB1, SET domain bifurcated 1; TCF, transcription factor.
SETDB1
SETDB1, also known as KMT1E/ESET, catalyzes the tri-methylation of histone 3 at lysine 9 (H3K9).30 The SETDB1 protein contains two major functional domains: the C-terminal SET domain catalyzes methylation, while the N-terminal domain is responsible for interaction with other chromatin-modifying enzymes.31 Previous studies have shown that SETDB1 is essential for early embryonic development. Indeed, the ubiquitous deletion of the SETDB1 leads to lethality between E3.5 and E5.5, due to defective growth of inner cell mass.30 Recently, growing evidence shows that SETDB1 is critical in the regulation of MSC osteoblastic and adipogenic differentiation.14
Specifically, SETDB1 is reported to inhibit osteogenic differentiation of MSCs.32 After generating SETDB1-deficient mice driven by Prx1-Cre, Lawson et al. found that mesenchymal deletion of SETDB1 led to long bone defects and significant decrease in trabecular bone in both embryos and postnatal mice.32,33 These mice also exhibited decreased osteoblasts in long bones, but normal osteoclast formation. These data indicate that SETDB1 is required for differentiation of MSCs into osteoblasts. Moreover, Lawson et al. also observed impaired osteoblastic activity in SETDB1-deficient MSCs from bone marrow, with hyperactivity of RUNX2. Consistent with these results, knockdown of SETDB1also enhances RUNX2-mediated gene transcription in vitro, which may be caused by the decrease of H3K9me3 in the promoters of RUNX2 target genes.33 Although RUNX2 is necessary for osteoblast differentiation, when transgenically over-expressed, it can inhibit osteoblast maturation.34 Collectively, SETDB1 inhibits osteogenic differentiation of MSCs, but it is required for normal skeletal formation due to its ability to repress hyperactive RUNX2-mediated transcription.
SETDB1 also plays a role in regulating adipogenic differentiation of MSCs. SETDB1 is down-regulated by PPAR-γ in rodent models of obesity, such as high-fat feeding mice and the genetically predisposed obese ob/ob mice. A similar down-regulation of SETDB1 is observed in adipogenic differentiation of MSCs in vitro, while siRNA-mediated knockdown of SETDB1 promotes adipogenesis with upregulated expression of PPAR-γ, CCAAT/enhancer-binding protein α (C/EBPα), and other adipogenesis-related genes.35
EZH2
EZH2, also known as KMT6A, catalyzes the addition of methyl groups to H3K27.36 This methyltransferase functions in the polycomb repressive complex 2 (PRC2) with suppressor of zeste 12 (SUZ12) and embryonic ectoderm development (EED).37 Notably, SUZ12 and EED are necessary for the catalytic activity of EZH2. Ubiquitous deletion of any of these components causes embryonic death during the post-implantation period.38,39,40 For decades, EZH2 has been known to be important for stem cell maintenance and differentiation.36,41
In terms of MSCs differentiation, EZH2 inhibits osteogenesis and promotes adipogenesis. To demonstrate this, suppression of EZH2 by cyclin-dependent kinase 1 (CDK1) results in MSC differentiation to osteoblasts.42 Likewise, chemical enzymatic inhibition and siRNA knockdown studies also show that EZH2 is a negative regulator of osteogenesis of MSCs.41 A genome-wide screen of polycomb target genes demonstrates that EZH2 is present on osteogenesis-related genes in non-differentiated MSCs, such as RUNX2 and transcription factor 7 (TCF7).36,42 Mechanistically, EZH2 expression is down-regulated under osteogenic inductive conditions and disassociates from the promoter RUNX2.43 Conversely, a recent study on neural crest cells (NCCs) reported that EZH2 is required for neural crest-derived bone formation. As a result of conditional ablation of EZH2 in NCCs, the formation of multiple skeletal elements was inhibited and severe craniofacial defects were caused.44 Therefore, it is likely that EZH2 has different tissue-specific functions in osteogenesis.
EZH2 methyltransferase activity is required for adipogenesis. Deletion of EZH2 in brown preadipocytes leads to decrease of H3K27me3 on the promoters of Wnt genes, resulting in severe defects in adipogenesis. These differentiation defects could be rescued by ectopic EZH2 and inhibitors of Wnt/β-catenin signaling. This suggests that EZH2 promotes adipogenesis by repression of Wnt genes.45 In addition to Wnt/β-catenin signaling, myocyte enhancer factor-2 interacting transcriptional repressor (MITR) also prevents adipogenesis by inhibiting the transcriptional activity of PPAR-γ. Acting to promote adipogenesis, EZH2 is present at the MITR promoter, where it inhibits MITR expression in adipocytes. Dissociation of EZH2 from the MITR promoter increases MITR expression and inhibits adipogenesis, while simultaneously enhancing the osteogenic differentiation of MSCs.46
Other SET family member genes
SET domain-containing 8 (SETD8), also known as KMT5A, catalyzes mono-methylation of H4K20.47 It has been reported to be essential for development and cell cycle progression.48 Deletion of SETD8 is lethal in flies49 and causes arrested embryonic development during the cleavage stage in mice.47 Over-expression of SETD8 by retrovirus enhances adipogenesis, while the siRNA-mediated knockdown of SETD8 impairs adipogenesis.50 As a requisite for committed preadipocyte differentiation, SETD8 promotes expression of PPAR-γ and PPAR-γ target genes through H4K20 mono-methylation.51 Like SETDB1, expression of SETD8 is regulated by PPAR-γ during adipogenesis. Following adipogenic stimuli, PPAR-γ promotes SETD8 expression, which catalyzes H4K20 mono-methylation in the promoters of PPAR-γ and of PPAR-γ target genes to enhance adipogenesis.50
Myeloid/lymphoid or mixed-lineage leukaemia 1 (MLL1), also known as KMT2A, catalyzes methylation at H3K4.27 The structure of MLL is complex, including three AT-hook motifs, a DNA methyltransferase homology domain in the amino-terminal half of the protein, plant homeodomain (PHD) region, and a SET domain at carboxyl-terminal.27 Due to the multiple functions of MLL1, deletion of MLL1 in mice has been reported to be lethal, and MLL1 heterozygous mice show retarded growth, hematopoietic abnormalities, and skeletal malformations.52 To investigate the specific function of SET domain in MLL1, Terranova et al. generated mutated mice, which expressed a SET domain-truncated form of MLL1.53 The mutated mice exhibit bidirectional homeotic transformations of the axial skeleton, and altered transcription of selected target HOX genes during development.53 The skeletal defects and bone malformations indicate the regulation of osteogenesis by MLL1.
MLL3 and MLL4, the paralogues of MLL1, catalyze methylation at H3K4.54,55 Deletion of the catalytic region of MLL3 leads to partial embryonic lethality, growth retardation, and female infertility.55,56 Notably, MLL3-null mice have little white adipose tissue, but normal brown adipose tissue (BAT). Compared to wild-type mouse embryonic fibroblasts (MEFs), MEFs isolated from MLL3-null mice show striking defects in adipogenesis. Upon adipogenic induction, MLL3 and MLL4 are recruited to the promoter of adipocyte protein 2 (aP2), an adipogenic marker gene, in a time-dependent manner.54 In addition, MLL3 and MLL4 can form a complex with Pax transactivation domain-interacting protein (PTIP). PTIP-deficient MEFs, white preadipocytes, and brown preadipocytes all exhibit impaired adipogenic potential, with decreased enrichment of PTIP and MLL4 on the PPAR-γ promoter.57 Collectively, MLL3 and MLL4 facilitate adipogenesis through their KMT activity to promote PPAR-γ and aP2 expression.
In summary, there is increasing evidence showing that SET domain-containing KMTs are essential for MSC differentiation. Major methylation sites mediated by SET domain-containing KMTs are located in the tail histones H3 and H4, such as H3K4, H3K9, H3K27, H3K36, and H4K20.58 In general, methylation at H3K4 and H3K36 is associated with transcriptional activation, while methylation at H3K9 and H3K27 is related to transcriptional repression. The role of mono-methylation at H4K20 could be either transcriptional activation or repression.25,58 Here, we summarize several SET domain-containing KMTs that integrate upstream stimulation and regulate osteogenesis and adipogenesis of MSCs by methylating the promoter of lineage-specific genes (Table 1). Although additional KMTs have been identified in association with osteogenesis and adipogenesis,46,59,60 further investigation to explore their underlying mechanism is necessary.
JmjC domain-containing KDMs
Unlike KMTs, KDMs remove methyl groups from histone lysine residues. This reversibility of KMTs was first discovered in 2004.61 To date, more than 30 histone demethylases have been identified, and they are classified into two classes: the lysine-specific demethylase family and the JmjC family. JmjC domain-containing proteins represent the largest class of potential histone demethylases and act by catalyzing the removal of mono-, di-, and tri-methyl residues via a 2OG-Fe(II)-dependent dioxygenase reaction.62,63 On the basis of emerging findings, JmjC domain-containing KDMs (Table 2) have been recognized and shown to play crucial roles in transcription regulation, stem cell differentiation, and animal development.62,64,65
Table 2. The role of JmjC domain-containing KDMs during osteogenesis and adipogenesis (shaded) of MSCs.
| KMTs | Specificity | Target | Finding | Reference |
|---|---|---|---|---|
| KDM2A | H3K4me3, H3K36me1/2 | EREG | KDM2A inhibits osteogenesis by catalyzing demethylation of H3K4me3 and H3K36me2 in the promoter of EGER through interaction with BCOR complex | 72 |
| Not known | KDM2A inhibits adipogenesis, while depletion of KDM2A in SCAPs enhances adipogenic differentiation | 68 | ||
| KDM2B | H3K4me3, H3K36me1/2 | AP-2α | KDM2B inhibits osteogenesis by catalyzing demethylation of H3K4me3 and H3K36me2 in the promoter of AP-2α through interaction with BCOR complex | 71 |
| KDM4B | H3K9me2/3, H3K36me2/3 | DLX5 | KDM4B inhibits osteogenesis by catalyzing demethylation of H3K9me3 in the promoter of DLX5 | 80 |
| Not known | KDM4B inhibits adipocyte lineage specification from human bone marrow-derived MSCs | 80 | ||
| C/EBPβ target genes | KDM4B promotes adipogenesis by catalyzing demethylation of H3K9me3 in the promoter of C/EBPβ target genes in preadipocytes | 81 | ||
| KDM4C | H3K9me2/3, H3K36me2/3 | Not known | KDM4C is required for adipogenesis from preadipocytes, which can be blocked by IDH mutation | 82 |
| KDM6A | H3K27me2/3 | RUNX2, OC | KDM6A promotes osteogenesis by catalyzing demethylation of H3K27me3 at transcription start sites of RUNX2 and OC | 43 |
| Not known | KDM6A inhibits adipogenesis, while the depletion of KDM6A in human bone marrow-derived MSCs enhances adipogenic differentiation | 43 | ||
| KDM6B | H3K27me2/3 | BMP and HOX genes | KDM6B promotes osteogenesis by catalyzing demethylation of H3K27me3 in the promoters of BMP and HOX genes | 80 |
| Not known | KDM6B inhibits adipogenesis, while depletion of KDM6B in human bone marrow-derived MSCs enhances adipogenic differentiation | 80 | ||
| NO66 | H3K4me1/2/3, H3K36me2/3 | OSX target genes | NO66 inhibits osteogenesis by catalyzing demethylation of H3K4me3 and H3K36me3 in the promoters of OSX target genes, such as BSP | 89-91 |
| RBP2 | H3K4me1/2/3 | RUNX2 target genes | RBP2 inhibits osteogenesis by catalyzing demethylation of H3K4me3 in the promoters of RUNX2 target genes, such as OSX and OC | 59 |
| Not known | RBP2 inhibits adipogenesis from preadipocytes, and depletion of RBP2 in preadipocytes enhances adipogenic differentiation potential | 94 | ||
| PHF2 | H3K9me2 | C/EBPα | PHF2 with C/EBPα promotes adipogenesis by catalyzing demethylation the H3K9me2 in the promoters of adipose genes | 98 |
AP-2α, activating enhancer binding protein 2α BMP, bone morphogenetic protein; C/EBP, CCAAT/enhancer binding protein; DLX5, distal-less homeobox 5; EREG, epiregulin; HOX, homeobox; JmjC, Jumonji C; KDM, histone lysine demethylase; KMT, histone lysine methyltransferase; MCS, mesenchymal stem cell; OC, osteocalcin; OSX, osterix; RBP, retinol binding protein; RUNX2, runt-related transcription factor 2.
KDM2 cluster
The KDM2 cluster, also known as JmjC domain-containing histone demethylase 1 (JHDM1) subfamily, catalyzes the removal of tri-methyl marks at H3K4, as well as mono- and di-methyl marks at H3K36.66,67,68 The KDM2 subfamily is comprised of two member genes: KDM2A (JHDM1A/FBXL11) and KDM2B (JHDM1B/FBXL10). KDM2A has shown to be important for proliferation of MSCs, and KDM2B has also been demonstrated to regulate cell proliferation and senescence.69 KDM2B-deficient mice show increased apoptosis in the neuroepithelium and mesenchyme at E9.5 and exhibit a failure of cranial neural tube closure.70 More recently, KDM2A and KDM2B were reported to regulate MSC differentiation by associating with the BCOR (BCL-6 co-repressor) complex.68,71,72
While BCOR mutation is lethal in males, BCOR mutation in females causes oculofaciocardiodental (OFCD) syndrome, which is characterized by canine teeth with extremely long roots, congenital cataracts, craniofacial defects, and congenital heart disease.71 KDM2B together with the BCOR complex was found to be recruited to the promoter of activating enhancer-binding protein 2α (AP-2α), which is required for osteogenesis. Consequently, KDM2B represses AP-2α expression by removing the H3K4me3 and H3K36me2 marks in the promoter.71 This mechanism serves as an explanation for the extremely long roots of canine teeth found in OFCD patients. Like KDM2B, KDM2A is also found to interact with BCOR and inhibit odontogenic differentiation of human stem cells from apical papilla (SCAPs), a type of dental MSCs.68 In addition, KDM2A and the BCOR complex are reported to inhibit osteogenesis by increasing histone H3K4/36 methylation in the epiregulin (EREG) promoter, thereby repressing EREG transcription. EREG is required for the expression of osteogenic-related genes, such as osterix (OSX) and distal-less homeobox 5 (DLX5).72
Although Dong et al. reported that the depletion of KDM2A in SCAPs enhances adipogenic differentiation in vitro,68 the effect of KDM2A on adipogenic differentiation is still not fully understood. Additional efforts, especially in vivo studies, are needed to explore the role of KDM2 member genes in adipogenesis.
KDM4 cluster
The KDM4 cluster, also known as the Jumonji domain-containing 2 subfamily, catalyzes the removal of di- and tri-methyl marks at H3K9 and H3K36.73,74 Five functional KDM4 member genes (KDM4A-E) have been identified in the human genome, of which KDM4A-C are broadly expressed in human tissues, while KDM4D and KDM4E are specifically enriched in the human testes.73,75 Partly because they have redundant roles, KDM4C or KDM4D knockout mice are viable without gross abnormalities.75,76 However, conditional deletion of KDM4B was recently reported to delay mammary gland development in mice,77 while KDM4A is required for skeletal muscle differentiation and neural crest specification.78,79 These findings indicate the roles of KDM4 member genes on transcription regulation, stem cell differentiation, and embryonic development.
More recently, KDM4B was reported to enhance osteogenic differentiation of human MSCs.19 In the bone marrow of aged mice, KDM4B expression is reduced, accompanying decreased osteogenesis. Knockdown of KDM4B by shRNA inhibits osteogenic differentiation in human bone marrow-derived MSCs, while over-expression of KDM4B enhances osteogenesis. After subcutaneous transplantation of MSCs with an HA scaffold, KDM4B is also required for MSC-mediated bone formation in vivo. Mechanistically, KDM4B was found to remove the H3K9me3 marks in the promoter of DLX5, activating its expression during osteogenesis. The DLX5 gene plays a critical role in osteoblast differentiation through controlling OSX expression.80
The role of KDM4 member genes in adipogenesis is still controversial. Ye et al. reported that the knockdown of KDM4B by shRNA enhanced adipogenic differentiation of MSCs and increased the expression of PPAR-γ and CD36 (also known as fatty acid translocase).80 These results indicate that KDM4B inhibits adipocyte lineage specification from MSCs. However, KDM4B was also found to act as a co-factor of C/EBPβ to promote differentiation of preadipocytes.81 After KDM4B interacts with C/EBPβ, they are recruited to the promoters of the target genes of C/EBPβ, such as cell division cycle 45 homolog, mini-chromosome maintenance complex component 3, GINS complex subunit 1, and cell division cycle 25 homolog c. By removing the H3K9me3 marks, KDM4B facilitates C/EBPβ target gene expression.81 It appears that the conflicting effects are caused by opposing functions of KDM4B at different stages of adipogenesis.
In addition, KDM4C is reported to be required for adipogenesis. Specifically, the inhibition of KDM4C induced by isocitrate dehydrogenase (IDH) mutation or 2-hydroxyglutarate (the product of mutated IDH) is sufficient to block differentiation of preadipocytes.82
KDM6 cluster
The KDM6 cluster catalyzes the removal of di- and tri-methyl marks at H3K2764,83 and contains three member genes: KDM6A (UTX), KDM6B (JMJD3), and UTY. KDM6A and KDM6B were the first to be identified as H3K26me2/me3 demethylases, followed by UTY, which has only recently been found to have overlapping redundancy with KDM6A in embryonic development.84 While ubiquitous deletion of KDM6B is lethal,85 approximately 25% KDM6A knockout mice survive to adulthood, partly due to the redundancy of UTY.84 Based on mice and cell culture models, KDM6A and KDM4B have been demonstrated to be critical in the regulation of stem cells, including M2 macrophage differentiation, muscle differentiation, and neuronal stem cell differentiation.85,86,87 Recent studies also found that KDM6A and KDM6B play a role in MSC lineage specification.
Both KDM6A and KDM6B promote osteogenic differentiation of MSCs. Hemming et al. reported that transcription levels of KDM6A are down-regulated during the process of osteogenesis of human bone marrow MSCs.43 Additionally, siRNA-mediated knockdown of KDM6A inhibits osteogenesis. Conversely, retroviral-mediated over-expression of KDM6A enhances the potential to differentiate into mineral forming osteoblasts and ectopic bone formation in vivo.43 This over-expression of KDM6A results in a decrease in H3K27me3 at both RUNX2 and osteocalcin (OC) transcription start sites, which subsequently increases the expression of RUNX2 and OC.43
In addition, KDM6B was shown to be required for osteogenic differentiation of MSCs.80,88 Upon osteogenic induction, KDM6B is recruited to the promoters of bone morphogenetic protein (BMP) and homeobox (HOX) genes, each of which promote the osteogenic differentiation of MSCs. When MSCs are depleted of KDM6B, they demonstrate impaired osteogenic differentiation and increased adipogenic differentiation in vitro, as well as decreased bone formation after subcutaneous transplantation with a scaffold.80 More recently, KDM6B was found to facilitate odontogenic differentiation of dental MSCs by removing the H3K27me3 mark from the BMP promoter.88
KDM6A and KDM6B are both reported to inhibit adipogenic differentiation of MSCs. Under adipogenic differentiation conditions, enforced over-expression of KDM6A results in decreased mRNA level of PPAR-γ, C/EBPα, and adiposin. Supporting these findings, functional studies show that the over-expression of KDM6A in MSCs decreases potential to form lipid in vitro. Conversely, knockdown of KDM6A shows an increase in lipid formation43 and depletion of KDM6B facilitates adipogenesis and promotes PPAR-γ and CD36 expression. In aged mice, bone marrow MSCs exhibit excessive adipogenic differentiation, which supports evidence of elevated H3K27me3 and decreased KDM6B.80 These findings indicate a negative role of KDM6A and KDM6B in adipogenesis. However, the mechanism for KDM6A and KDM6B regulation of adipogenic differentiation is still not clear.
Other related JmjC family member genes
NO66, another JmjC domain-containing protein, catalyzes the removal of mono-, di-, and tri-methyl marks at H3K4, as well as di- and tri-methyl marks at H3K36.89 NO66 has been found in all the developing bones, such as the E15.5 vertebrae and mandible and E18.5 femur, tibia, and fibula, indicating that NO66 plays a role in osteogenesis.89 Depletion of NO66 in preosteoblasts accelerates their differentiation and maturation, and knockdown of NO66 significantly increases the expression levels of osterix (OSX)-dependent matrix-forming genes.89 Mechanistically, NO66 interacts with OSX under osteogenic inductive conditions to inhibit OSX transcriptional activity.89,90 Both NO66 and OSX can be found present in the promoter of bone sialoprotein (BSP), resulting in demethylation of H3K4me3 and H3K36me3 and repression of BSP expression. A structural study revealed that the hinge domain-dependent oligomerization of NO66 is essential for the interaction with OSX.90 However, in OSX-deficient preosteoblasts, NO66 occupancy is increased in the chromatin of the BSP gene.91 Hence, it is clear that while NO66 may be recruited to the promoters by different mechanisms, NO66 contributes to the repression of OSX target genes and inhibits osteogenesis.
Retinol-binding protein 2 (RBP2), also known as KDM5A, catalyzes the removal of mono-, di-, and tri-methyl marks at H3K4.92,93 Deletion of the JmjC domain of RBR-2 (Caenorhabditis elegans RBP2 homolog) in C. elegans demonstrates that it is important for vulva development.92 During mouse embryonic stem cell differentiation, RBP2 is also found to regulate HOX gene expression levels, which are related to development and osteogenesis. However, mice with RBP2 deficiency are viable and grossly normal, except for behavioral abnormalities when held upside down.93 RBP2 was originally found to repress osteogenesis in Soas-2 cells (a human osteogenic sarcoma cell line),94 and has subsequently been reported to inhibit osteogenesis in adipose-derived MSCs by repression of RUNX2-mediated transcriptional activity.59 Depletion of RBP2 results in increased expression levels of osteogenic associated genes, promotes osteogenesis in vitro, and increases bone formation after subcutaneous transplantation in vivo. Moreover, the increased osteogenesis can be abolished by the knockdown of RUNX2.95 In addition, RBP2 is shown to inhibit adipogenesis from preadipocytes, and depletion of RBP2 in preadipocytes enhances lipid formation in vitro.94 Other studies show that RBP2 seems to exert inhibitory effects during both osteogenic and adipogenic differentiation.95,96
Plant homeodomain finger 2 (PHF2), also known as JHDM1E, catalyzes the removal of di-methyl marks at H3K9.20 However, the enzymatic activity of PHF2 depends on phosphorylation mediated by protein kinase A (PKA).97 About 70% of mice with PHF2 deficiency die within 3 days after birth, while the rest of the mice show a decrease in body weight, indicating that adipogenesis is impaired in these mice. Also, deletion of PHF2 in adipose tissue results in a 50% reduction in WAT, while BAT appears normal. Mechanistically, PHF2 was found to be a co-activator of C/EBPα and recruited to the promoter of C/EBP responsive elements, such as C/EBPα, PPAR-γ, and fatty acid-binding protein 4 (FABP4). Depletion of PHF2 in preadipocytes inhibits adipose gene expression and lipid formation.98 Together, PHF2 with C/EBPα promotes adipogenesis through demethylation of the H3K9me2 mark in the promoters of adipose genes.
In summary, these findings have revealed an important role of JmjC family genes in the regulation of the balance between osteogenic differentiation and adipogenic differentiation of MSCs. Unlike KMTs, JmjC family genes control transcriptional activity by removing the methylation marks at the specific histone lysine residues. A summary of the current opinions on regulation of osteogenesis and adipogenesis by various JmjC domain-containing KDMs can be found in Table 2. While some JmjC family genes take part in MSC differentiation at an early stage, such as KDM4B and KDM6B in osteogenesis,80 others play roles at a later stage, such as NO66 in osteogenesis91 and KDM4C in adipogenesis.82 However, the mechanism of JmjC family genes in adipogenesis is still not well understood. To further investigate, results based on cell culture models need to be verified by in vivo studies.
Outlook
Eukaryotic chromatin is organized in both euchromatin (active) and heterochromatin (inactive) forms. Histone methylation marks are key to define these functional states, particularly in promoter regions, as they affect the physical proximity of lysine residues to each other. However, during MSC differentiation, the cross-talk among the histone methylation marks is still elusive to us, and should be taken into account in future studies. For instance, the euchromatic mark H3K4me3 prevents tri-methylation of H3K9 by SETDB1,99 but the heterochromatin mark H3K9me3 prevents mono-methylation of H3K4 by SET7.100 Notably, the cross-talk among histone methylation marks is not restricted to methylation at lysine residues. More information can be found in previous reviews.58,101,102,103
Another important question concerns the role of opposing functions of KMTs and KDMs in establishing histone methylation states. It is plausible that both methylation and demethylation occur in the promoters of different lineage-specific genes. For instance, EZH2 and KDM6A, which both target H3K27, act as an epigenetic switch to regulate MSC lineage specification.43 However, the methyltransferase activity of SET domain-containing KMTs seems to be specific to one site, while JmjC domain-containing KDMs exhibit more redundancy and tissue specificity.
Interestingly, KMTs and KDMs may interact with each other. For instance, JMJ was shown to be required for efficient binding of the PRC2 complex.104 A study of mouse embryonic stem cells revealed that KDM4C could interact with the components of PRC2 and assist Ezh2 to fully repress target genes.104 In addition, KDM5C, a H3K4 demethylase, may interact with H3K9 methylases, which in turn may couple H3K9 methylation to H3K4 demethylation.64 These interactions may also be critical in MSC differentiation, especially in its initial stages.
Conclusion
MSCs hold great promise for the treatment of difficult bone defects. However, to employ MSCs for clinical use, we must first understand the mechanisms of MSC differentiation. Histone methylation has recently been identified as a key modulator of MSC lineage specification. Furthermore, epigenetic regulation of MSC differentiation is also critical for bone metabolism. In this review, we summarized the epigenetic mechanisms of histone methylation in relation to osteogenesis and adipogenesis of MSCs (Figure 1). Further investigation is still required to gain a more complete understanding, which will in turn enable the use of MSCs in regenerative medicine of large bone defects and metabolic bone diseases.
Acknowledgments
This study was supported by the National Institute of Dental and Craniofacial Research grants, K08DE024603-02, DE019412, and DE01651, and by a grant from 111 Project of MOE, China. Publication of this manuscript is supported by Open Fund of State Key Laboratory of Oral Diseases, Sichuan University.
References
- Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 2007; 213(2): 341–347. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006; 8(4): 315–317. [DOI] [PubMed] [Google Scholar]
- Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell 2008; 2(4): 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguell JJ, Erices A, Conget P. Mesenchymal stem cells. Exp Biol Med (Maywood) 2001; 226(6): 507–520. [DOI] [PubMed] [Google Scholar]
- Romanov YA, Darevskaya AN, Merzlikina NV et al. Mesenchymal stem cells from human bone marrow and adipose tissue: isolation, characterization, and differentiation potentialities. Bull Exp Biol Med 2005; 140(1): 138–143. [DOI] [PubMed] [Google Scholar]
- Ma S, Xie N, Li W et al. Immunobiology of mesenchymal stem cells. Cell Death Differ 2014; 21(2): 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Su J, Zhang L et al. Species variation in the mechanisms of mesenchymal stem cell-mediated immunosuppression. Stem Cells 2009; 27(8): 1954–1962. [DOI] [PubMed] [Google Scholar]
- James AW. Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica (Cairo) 2013; 2013: 684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragojevič J, Logar DB, Komadina R et al. Osteoblastogenesis and adipogenesis are higher in osteoarthritic than in osteoporotic bone tissue. Arch Med Res 2011; 42(5): 392–397. [DOI] [PubMed] [Google Scholar]
- Hoshiba T, Kawazoe N, Chen G. The balance of osteogenic and adipogenic differentiation in human mesenchymal stem cells by matrices that mimic stepwise tissue development. Biomaterials 2012; 33(7): 2025–2031. [DOI] [PubMed] [Google Scholar]
- Johnston CCJr, Bjarnason NH, Cohen FJ et al. Long-term effects of raloxifene on bone mineral density, bone turnover, and serum lipid levels in early postmenopausal women: three-year data from 2 double-blind, randomized, placebo-controlled trials. Arch Intern Med 2000; 160(22): 3444–3450. [DOI] [PubMed] [Google Scholar]
- Chang J, Wang Z, Tang E et al. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med 2009; 15(6): 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman EJ, Teng K, Lipschitz DA et al. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-γ2 transcription factor and TGF-β/BMP signaling pathways. Aging Cell 2004; 3(6): 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada I, Kouzmenko AP, Kato S. Wnt and PPARγ signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol 2009; 5(8): 442–447. [DOI] [PubMed] [Google Scholar]
- Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J Cell Biochem 2011; 112(12): 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Liu F, Lee M et al. NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proc Natl Acad Sci U S A 2013; 110(23): 9469–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KE, Park NR, Che X et al. Core binding factor β of osteoblasts maintains cortical bone mass via stabilization of Runx2 in mice. J Bone Miner Res 2015; 30(10): 1943. [DOI] [PubMed] [Google Scholar]
- Chawla A, Schwarz EJ, Dimaculangan DD et al. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 1994; 135(2): 798–800. [DOI] [PubMed] [Google Scholar]
- Johansson C, Tumber A, Che K et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics 2014; 6(1): 89–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol 2010; 20(11): 662–671. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Agger K et al. Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb Symp Quant Biol 2008; 73: 253–263. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun YE. Epigenetic regulation of stem cell differentiation. Pediatr Res 2006; 59(4 Pt 2): 21R–215R. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293(5532): 1074–1080. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet 2008; 17(R1): R28–R36. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 2007; 8(4): 307–318. [DOI] [PubMed] [Google Scholar]
- et al. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 2003; 112(5): 711–723. [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V et al. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem 2011; 286(5): 3359–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A 1964; 51: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG et al. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A 1999; 96(26): 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H et al. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 2004; 24(6): 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci 2003; 28(2): 59–62. [DOI] [PubMed] [Google Scholar]
- Yang L, Lawson KA, Teteak CJ et al. ESET histone methyltransferase is essential to hypertrophic differentiation of growth plate chondrocytes and formation of epiphyseal plates. Dev Biol 2013; 380(1): 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Teteak CJ, Gao J et al. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett 2013; 587(24): 3961–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Toyosawa S, Furuichi T et al. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol 2001; 155(1): 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Okada H, Shirai K et al. Comparison of two pediatric flail chest cases. Scand J Trauma Resusc Emerg Med 2015; 23(1): 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez G, González M, Isabal S et al. Method to quantify live and dead cells in multi-species oral biofilm by real-time PCR with propidium monoazide. AMB Express 2013; 3(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 2008; 20(3): 266–273. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23(20): 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 2001; 21(13): 4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B et al. The eed mutation disrupts anterior mesoderm production in mice. Development 1995; 121(2): 273–285. [DOI] [PubMed] [Google Scholar]
- Tonini T, D'Andrilli G, Fucito A et al. Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements. J Cell Physiol 2008; 214(2): 295–300. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen YH, Li LY et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011; 13(1): 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming S, Cakouros D, Isenmann S et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014; 32(3): 802–815. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Varum S, Zemke M et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development 2014; 141(4): 867–877. [DOI] [PubMed] [Google Scholar]
- Wang L, Jin Q, Lee JE et al. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A 2010; 107(16): 7317–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Yeh FL, Yeh SP et al. Myocyte enhancer factor-2 interacting transcriptional repressor (MITR) is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferator-activated receptor gamma-2. J Biol Chem 2011; 286(12): 10671–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 2009; 29(8): 2278–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Blanpain C. Mechanisms regulating epidermal stem cells. EMBO J 2012; 31(9): 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D et al. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 2005; 19(4): 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Okamura M, Tsutsumi S et al. The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 2009; 29(13): 3544–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 2011; 12(11): 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995; 378(6556): 505–508. [DOI] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA 2006; 103(17): 6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Saha PK, Yang QH et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA 2008; 105(49): 19229–19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA 2006; 103(42): 15392–15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Hahn M, Schotta G. Epigenetic regulation of development by histone lysine methylation. Heredity 2010; 105(1): 24–37. [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong S, Jin Q et al. Histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis. Cell Metab 2009; 10(1): 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O. On your histone mark, SET, methylate! Epigenetics 2013; 8(5): 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Shi L, Zhou Y et al. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells 2011; 29(7): 1112–1125. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008; 14(5): 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119(7): 941–953. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6(11): 838–849. [DOI] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X et al. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 2008; 50(7): 886–896. [DOI] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol 2008; 20(3): 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 2007; 8(11): 829–833. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439(7078): 811–816. [DOI] [PubMed] [Google Scholar]
- Gao R, Dong R, Du J et al. Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15INK4B and p27Kip1. Mol Cell Biochem 2013; 379(1/2): 115–122. [DOI] [PubMed] [Google Scholar]
- Dong R, Yao R, Du J et al. Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp Cell Res 2013; 319(18): 2874–2882. [DOI] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y et al. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15Ink4b. Nat Struct Mol Biol 2008; 15(11): 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Tokunaga A, Sakamoto R et al. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci 2011; 46(3): 614–624. [DOI] [PubMed] [Google Scholar]
- Fan Z, Yamaza T, Lee JS et al. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol 2009; 11(8): 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Ma Y, Ma P et al. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells 2013; 31(1): 126–136. [DOI] [PubMed] [Google Scholar]
- Labbé RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am J Transl Res 2013; 6(1): 1–15. [PMC free article] [PubMed] [Google Scholar]
- Lorbeck MT, Singh N, Zervos A et al. The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene 2010; 450(1–2): 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamori N, Zhao M, Meistrich ML et al. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod 2011; 84(6): 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MT, Agger K, Laugesen A et al. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol Cell Biol 2014; 34(6): 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M, Saso K, Tong KI et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 2011; 6(3): e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier L, Escaffit F, Chailleux C et al. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet 2011; 7(6): e1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell 2010; 19(3): 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Fan Z, Yu B et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012; 11(1): 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Li X, Huang JX et al. Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ 2012; 19(12): 1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483(7390): 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme J, Lettier G, Sidoli S et al. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet 2012; 8(5): e1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S et al. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 2012; 8(9): e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010; 11(10): 936–944. [DOI] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 2010; 29(8): 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One 2008; 3(8): e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yu B, Hong C et al. KDM6B epigenetically regulates odontogenic differentiation of dental mesenchymal stem cells. Int J Oral Sci 2013; 5(4): 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Coombes MM et al. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 2010; 29(1): 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Wu M, Zhou X et al. Structural insights into histone demethylase NO66 in interaction with osteoblast-specific transcription factor osterix and gene repression. J Biol Chem 2013; 288(23): 16430–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Zhou X et al. Osterix and NO66 histone demethylase control the chromatin of Osterix target genes during osteoblast differentiation. J Bone Miner Res 2014; 29(4): 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 2007; 128(6): 1063–1076. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 2007; 128(5): 889–900. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P et al. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005; 18(6): 623–635. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC et al. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 2008; 31(4): 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev 2008; 22(10): 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba A, Ohtake F, Okuno Y et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol 2011; 13(6): 668–675. [DOI] [PubMed] [Google Scholar]
- Okuno Y, Ohtake F, Igarashi K et al. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes 2013; 62(5): 1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, LeRoy G, Bua DJ et al. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 2010; 5(8): 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 2001; 8(6): 1207–1217. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol 2003; 15(2): 172–183. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21(3): 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci 2009; 66(8): 1419–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009; 139(7): 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]