Abstract
The emergence of artemisinin resistance among Plasmodium falciparum in the Greater Mekong subregion threatens malaria control interventions and is associated with multiple unique mutations in K13 (PF3D7_1343700). The aim of this study was to survey Cambodian Plasmodium vivax for mutations in the K13 ortholog (K12, PVX_083080) that might similarly confer artemisinin resistance. Extracted DNA from Cambodian isolates collected between 2009 and 2012 was pooled by province and year and submitted for next-generation sequencing. Single-nucleotide polymorphisms (SNPs) were identified using a pile-up approach that detected minority SNPs. Among the 14 pools, we found six unique SNPs, including three nonsynonymous SNPs, across six codons in K12. However, none of the SNPs were orthologous to artemisinin resistance–conferring mutations in PF3D7_1343700, and nonsynonymous changes did not persist through time within populations. These results suggest a lack of selection in the P. vivax population in Cambodia due to artemisinin drug pressure.
Multiple reports of artemisinin resistance in Plasmodium falciparum have come from Cambodia and the Thai–Burmese border.1–5 A molecular correlate of this resistance was recently discovered, and resistance was found to be associated with specific mutations in the BTB/POZ and propeller domains of the P. falciparum kelch protein, K13 (PF3D7_1343700).6–10 K13 mutations are now being used as molecular markers for surveillance of artemisinin resistance.9,10
Artemisinin resistance has not been reported in Plasmodium vivax.3,11 However, in Cambodia, antimalarial drugs are often dispensed without species-level diagnosis, and coinfections are common, likely resulting in artemisinin exposure to P. vivax. Moreover, dihydroartemisinin–piperaquine replaced chloroquine for treatment of P. vivax malaria in Cambodia in 2012 due to concerns of chloroquine resistance. Resistance-conferring mutations to antimalarial drugs in both P. falciparum and P. vivax have previously been described for several orthologous genes.12–14 This historical pattern suggests that surveillance of mutations in the orthologous gene to K13 among P. vivax (K12, PVX_083080) in Cambodia is warranted. In the case of falciparum malaria, mutations in K13 existed in the population before widespread artemisinin-based combination therapies (ACTs) use. Thus, as ACTs are increasingly used to treat P. vivax, a baseline evaluation of K12 mutations and a means of rapid surveillance for potential resistance mutations is needed.5,6,11,12
The aim of this study was to develop a rapid method to survey K12 for mutations that might confer artemisinin resistance and conduct a baseline evaluation of the reservoir of genetic diversity in K12. We modified our previously described pooled next-generation sequencing (NGS) approach and validated it using controls and simulations.15 NGS can reliably detect single-nucleotide polymorphisms (SNPs) in minority strains from complex infections that can otherwise be overlooked by traditional molecular–epidemiological methods, which often rely on Sanger sequencing.16 As such, previous surveillance of the P. vivax K12 gene may have overlooked minority strain polymorphisms and missed emerging resistance at low frequency in the population.17 In addition, using an NGS with pooled clinical isolates has proven to be accurate, sensitive, and cost-effective as compared with Sanger sequencing.15,16
Whole blood from 304 patients presenting with uncomplicated P. vivax malaria to seven clinical sites throughout western and northern Cambodia were collected from 2009 to 2012 by the Armed Forces Research Institute of Medical Science (Study Protocol WR1576). The study was approved by the Walter Reed Army Institute of Research Institutional Review Board (IRB), the Cambodian National Ethics Committee for Health Research and the Biomedical IRB at University of North Carolina. All participants provided written informed consent before sample collection.
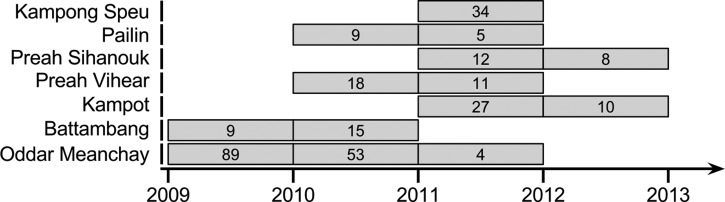
DNA was extracted from individual samples and pooled in an isovolumetric manner by province and year to create 14 pools (Figure 1 and Supplemental Figure 1). Two additional samples containing DNA extracted from a monoclonal monkey-adapted P. vivax infection were sequenced as controls. Polymerase chain reaction (PCR) used the FastStart High Fidelity PCR System (Roche, Mannheim, Germany). A 2,139-bp fragment encompassing the entire K12 gene was amplified using primers Pv.kelch-447706-G-Fwd (GAAAGCTGCCAAGGGAAGTA) and Pv.kelch-449945-G-Rev (CTCCCCATCTGTTCCATGTC). In 50 μL PCR reactions, 0.5 μL of Roche FastStart Hi-Fidelity Taq, 400 nM of forward and reverse primers, 200 nM of deoxynucleoside triphosphates, 2.25 mM of MgCl2, 5 μL of 10X reaction buffer, and 4 μL of template were used. Amplification cycling conditions included 95°C for 3 minutes, 16 cycles at 95°C for 20 seconds, a 20-second annealing step ranging from 70°C to 63°C with a ΔT of 1°C per two cycles, and an extension of 72°C for 3 minutes. This was followed by 24 cycles of 95°C for 20 seconds, 63°C for 20 seconds, and an extension of 72°C for 3 minutes, and a final extension of 72°C for 5 minute. PCR products were purified using the Invitrogen PureLink® PCR Purification Kit (ThermoFisher Scientific, Chicago, IL) and sheared on a Covaris S2 instrument (Woburn, MA) to an average fragment size of 350 bps. Sheared amplicons were prepared with the Ion Plus Fragment Library Kit and Ion Xpress Barcode Adaptors (Life Technologies, ThermoFisher Scientific, Chicago, IL) and submitted to the Ion Torrent Personal Genome Machine® Ion 318™ Chip with 400-base chemistry (ThermoFisher Scientific, Chicago, IL).
Figure 1.
Pooling strategy of clinical isolates by province and year included in the study. Each pool is represented by a box with the number of isolates pooled noted within the box.
Sequencing reads were aligned using Bowtie2 with default settings to the P. vivax Sal-1 reference strain (v3; http://www.plasmodb.org, accessed March 28, 2016), and the minor allele frequency (MAF) at all positions across the amplicon was determined using a revised version of our previously described Minor Allele Catcher (MAC).15,18 The revised MAC (rMAC) was rewritten in C++ and modified to account for homopolymer errors common with Ion Torrent, which could result in reduced detection of alleles or result in false-positive alleles at low frequency. Bases and reads were excluded from analysis if they had a mapping quality less than or equal to 10, a read length less than or equal to 200 bp, a base quality of less than or equal to 20, a base depth of less than or equal to 5,000, if SNPs occurred in the first or last 25 bases of a read, or if 30% or fewer of the reads were on the forward or reverse strand. Plots were generated with the R statistical packages dplyr, tidyr, ggplot2, ggmap, easyGgplot2, rgdal, sp, and maptools.19
From the 14 P. vivax pools (Figure 1), we obtained a total of 3,650,121 reads, of which 2,376,860 passed quality filters and were included in the analysis. In our two control samples, we generated 448,139 reads of which 329,624 reads passed quality filters and were included in the analysis. Overall, we achieved at least 5,000-fold coverage on ≥ 91% of the nucleotide positions analyzed (Supplemental Figure 2). A conservative MAF cutoff of 1.0% for SNP discovery was chosen based on our previous work and the two control samples, which would predict < 1 false-positive SNP across all of the samples.15
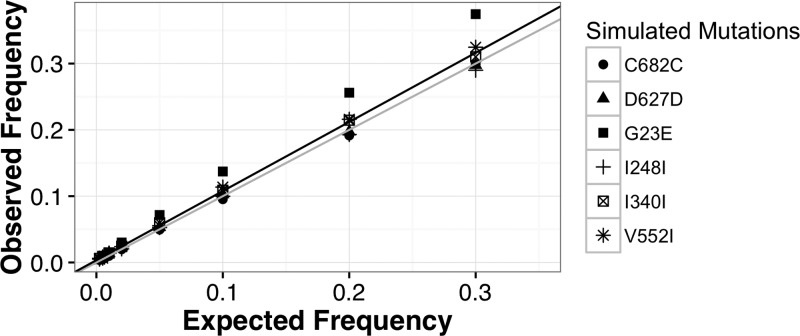
To validate the rMAC, we used a combination of simulations and monoclonal control samples, as few clonal lines were available for creating mixtures to empirically justify our 1% cutoff. The rMAC detected no SNPs in the two control samples. We then simulated read libraries for each of our unique mutations (three private alleles; three shared alleles) with MAF varying at 0.25%, 0.5%, 1%, 2%, 5%, 10%, 20%, and 30%, respectively. Reads were simulated to include common PCR and sequencing errors.20 These reads were then subjected to the same analysis as the clinical and control samples. The rMAC was able to identify simulated SNPs as low as 0.25% in 12/13 simulated pools, failing to detect G23E in the 0.25% simulation. Across different simulated allele frequencies, we saw a high degree of correlation between expected and observed values (r2 = 0.98) (Figure 2 ), supporting our conservative cutoff of a 1% MAF.
Figure 2.
Detection of simulated minor allele frequency (MAF) using the revised Minor Allele Catcher (rMAC) pipeline. Reads were simulated with mutations that matched our observed single-nucleotide polymorphisms (Table 1) at frequencies of 30%, 20%, 10%, 5%, 2%, 1%, 0.5%, and 0.25%, respectively. Polymerase chain reaction (PCR) error was simulated using an in-house script that incorporated the effects of starting template amount and polymerase error rates. It propagates polymerase errors such that early round errors appear at a higher frequency than errors in latter rounds. PCR-simulated sequences were then fragmented with read length distribution similar to patient samples and processed with 454Sim to incorporate sequencing errors similar to Ion Torrent sequencing. The scatter plot shows a distribution of the observed simulated frequencies calculated by the rMAC (observed frequency) vs. the eight expected MAFs ranging from 0.25% to 30% (expected frequency). A line of ideal fit (y = 1x + 0; colored gray) and a linear regressed line of best fit (y = 1.04x + 0.003, r2 = 0.98; colored black) are overlain on the plot. Simulations were run with 11,745–191,130 starting template copies, depending on the pooled sample and a polymerase error rate of 3.5 × 10−6.
Among the 14 pools, we found 12 SNPs across six codons. Of these, three were nonsynonymous mutations (Table 1). Three mutations were identified as private alleles, while the others were shared between two or more pools. Two synonymous mutations persisted through time: I248I in the KP population and I340I in the PL population. The V552I mutation shared between the OM2011 and KS2011 pools has been previously reported in Ratanakiri Province among samples collected in 2013.17 The highest MAF detected in this study was 8.80% from the OM region in 2010. Most mutations were confined to the BTB/POZ domain or the fifth blade of the kelch propeller domain of the K12 gene product and were found at low frequencies (1–4%). None of the observed mutations were in orthologous locations to artemisinin resistance–conferring mutations in PF3D7_1343700.9
Table 1.
Observed K12 polymorphisms in pools of Cambodian Plasmodium vivax isolates
| Province | Year | Nucleotide position | Ref base | Minor base | MAF (%) | AA locus* | AA change | Domain |
|---|---|---|---|---|---|---|---|---|
| BB | 2009 | 447,795 | G | A | 2.19 | G23E | NonSyn | Upstream |
| 2010 | 448,747 | A | T | 1.16 | I340I | Syn | BTB/POZ | |
| KP | 2011 | 448,471 | A | T | 3.66 | I248I | Syn | BTB/POZ |
| 2012 | 448,471 | A | T | 3.57 | I248I | Syn | BTB/POZ | |
| KS | 2011 | 449,381 | G | A | 2.99 | V552I | NonSyn | Blade 3 |
| OM | 2010 | 448,747 | A | T | 4.09 | I340I | Syn | BTB/POZ |
| 2010 | 449,381 | G | A | 5.47 | V552I | NonSyn | Blade 3 | |
| 2010 | 449,608 | C | T | 8.82 | D627D | Syn | Blade 5 | |
| PL | 2010 | 448,747 | A | T | 1.66 | I340I | Syn | BTB/POZ |
| 2011 | 448,747 | A | T | 2.22 | I340I | Syn | BTB/POZ | |
| PN | 2011 | 449,773 | T | C | 4.14 | C682C | Syn | Blade 6 |
| PV | 2010 | 448,471 | A | T | 1.26 | I248I | Syn | BTB/POZ |
AA = amino acid; BB = Battambang; KP = Kampot; KS = Kampong Speu; MAF = minor allele frequency; NonSyn = nonsynonymous; OM = Oddor Mean Chey; PL = Pailin; PN = Preah Sihanouk; PV = Preah Vihear; Syn = synonymous. Domain correspondence was determined by the homologous amino acid from the Plasmodium falciparum orthologue (PF3D7_1343700). Nucleotide positions are 0 based.
Polymorphisms shared between populations are in bold.
These results suggest that P. vivax contains a repertoire of standing variation within the K12 gene, but no evidence of resistance-conferring mutations before 2013. We would expect K12 mutations that confer resistance would be under directional selection and increase in frequency over time given enough ACT drug use. Previous studies have found a similarly low number of mutations in the K12 gene among Cambodian P. vivax (2/284 clinical isolates).15,17 This standing variation in K12 is similar to the current understanding of K13 mutations in African falciparum populations.9,15
We have developed and validated a rapid pooled NGS approach for surveillance of K12 mutations. Although we found only a small number of ephemeral nonsynonymous mutations, our results show that pooled NGS surveillance for allele frequency assessment may provide a more complete picture of gene diversity than traditional Sanger sequencing methods.17 In addition, this study provides a baseline to measure future changes to the P. vivax K12. Cambodia's recent shift to dihydroartemisinin–piperaquine as first-line therapy for vivax malaria in 2012 will result in strong selective pressure on the parasite and may eventually result in selection of mutations in this gene.11,12 Future surveillance of the K12 gene with NGS techniques is warranted to capture any potential homologous artemisinin resistance–conferring mutations among P. vivax.
Supplementary Material
ACKNOWLEDGMENTS
We want to thank the participants of the study. Nicholas F. Brazeau was funded by the IDSA Medical Scholars Program. This research was supported by a University of North Carolina-Chapel Hill Junior Faculty Development Award, the Armed Forces Health Surveillance Center/Global Emerging Infections Surveillance and Response System and the Military Infectious Disease Research Program.
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of the Army, the Department of Defense, or the U.S. Government.
Footnotes
Authors' addresses: Nicholas F. Brazeau, University of North Carolina School of Medicine, Chapel Hill, NC, E-mail: nbrazeau@med.unc.edu. Nicholas Hathaway, School of Medicine, University of Massachusetts Medical School, Worcester, MA, E-mail: nickjhathaway@gmail.com. Christian M. Parobek, Department of Genetics, University of North Carolina School of Medicine, Chapel Hill, NC, E-mail: christian_parobek@med.unc.edu. Jessica T. Lin, Division of Infectious Diseases, University of North Carolina School of Medicine, Chapel Hill, NC, E-mail: jessica_lin@med.unc.edu. Jeffrey A. Bailey, Division of Transfusion Medicine, Department of Medicine, University of Massachusetts Medical School, Worcester, MA, and Program in Bioinformatics and Integrative Biology, University of Massachusetts, Worcester, MA, E-mail: jeffrey.bailey@umassmed.edu. Chanthap Lon and David L. Saunders, Department of Immunology and Medicine, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand, E-mails: chanthapl@afrims.org and david.l.saunders.mil@mail.mil. Jonathan J. Juliano, Division of Infectious Diseases, Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, E-mail: jonathan_juliano@med.unc.edu.
References
- 1.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lon C, Manning JE, Vanachayangkul P, So M, Sea D, Se Y, Gosi P, Lanteri C, Chaorattanakawee S, Sriwichai S, Chann S, Kuntawunginn W, Buathong N, Nou S, Walsh DS, Tyner SD, Juliano JJ, Lin J, Spring M, Bethell D, Kaewkungwal J, Tang D, Chuor CM, Satharath P, Saunders D. Efficacy of two versus three-day regimens of dihydroartemisinin-piperaquine for uncomplicated malaria in military personnel in northern Cambodia: an open-label randomized trial. PLoS One. 2014;9:e93138. doi: 10.1371/journal.pone.0093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NPJ, White NJ, Anderson TJC, Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NTT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer H-P, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in southeast Asia. J Infect Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, Imwong M, Woodrow C, Manske M, Stalker J, Drury E, Campino S, Amenga-Etego L, Thanh T-NN, Tran HT, Ringwald P, Bethell D, Nosten F, Phyo AP, Pukrittayakamee S, Chotivanich K, Chuor CM, Nguon C, Suon S, Sreng S, Newton PN, Mayxay M, Khanthavong M, Hongvanthong B, Htut Y, Han KT, Kyaw MP, Faiz MA, Fanello CI, Onyamboko M, Mokuolu OA, Jacob CG, Takala-Harrison S, Plowe CV, Day NP, Dondorp AM, Spencer CCA, McVean G, Fairhurst RM, White NJ, Kwiatkowski DP. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MalariaGEN Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. eLife. 2016;5:e08714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leang R, Barrette A, Bouth DM, Menard D, Abdur R, Duong S, Ringwald P. Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob Agents Chemother. 2013;57:818–826. doi: 10.1128/AAC.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JT, Patel JC, Kharabora O, Sattabongkot J, Muth S, Ubalee R, Schuster AL, Rogers WO, Wongsrichanalai C, Juliano JJ. Plasmodium vivax isolates from Cambodia and Thailand show high genetic complexity and distinct patterns of P. vivax multidrug resistance gene 1 (pvmdr1) polymorphisms. Am J Trop Med Hyg. 2013;88:1116–1123. doi: 10.4269/ajtmh.12-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suwanarusk R, Chavchich M, Russell B, Jaidee A, Chalfein F, Barends M, Prasetyorini B, Kenangalem E, Piera KA, Lek-Uthai U, Anstey NM, Tjitra E, Nosten F, Cheng Q, Price RN. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis. 2008;198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eldin de Pécoulas P, Basco LK, Tahar R, Ouatas T, Mazabraud A. Analysis of the Plasmodium vivax dihydrofolate reductase–thymidylate synthase gene sequence. Gene. 1998;211:177–185. doi: 10.1016/s0378-1119(98)00118-8. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Mårtensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SM, Parobek CM, Aragam N, Ngasala BE, Mårtensson A, Meshnick SR, Juliano JJ. Pooled deep sequencing of Plasmodium falciparum isolates: an efficient and scalable tool to quantify prevailing malaria drug-resistance genotypes. J Infect Dis. 2013;208:1998–2006. doi: 10.1093/infdis/jit392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popovici J, Kao S, Eal L, Bin S, Kim S, Ménard D. Reduced polymorphism in the Kelch propeller domain in Plasmodium vivax isolates from Cambodia. Antimicrob Agents Chemother. 2015;59:730–733. doi: 10.1128/AAC.03908-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team R: A Language and Environment for Statistical Computing. 2016. https://www.R-project.org/ Available at. Accessed March 28, 2016.
- 20.Lysholm F, Andersson B, Persson B. An efficient simulator of 454 data using configurable statistical models. BMC Res Notes. 2011;4:449. doi: 10.1186/1756-0500-4-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.