Abstract
Functionally related genes often cluster into a genome region under coordinated regulation, forming a local regulome. To understand regulation of the CHRNA5/CHRNA3/CHRNB4 nicotinic receptor gene cluster, we integrate large-scale RNA expression data (brain and peripheral) from GTEx (Genotype Tissue Expression), clinical associations (GRASP) and linkage disequilibrium data (1,000 Genomes) to find candidate SNPs representing independent regulatory variants. CHRNA3, CHRNA5, CHRNB4 mRNAs, and a well-expressed CHRNA5 antisense RNA (RP11-650L12.2) are co-expressed in many human tissues, suggesting common regulatory elements. The CHRNA5 enhancer haplotype tagged by rs880395 not only increases CHRNA5 mRNA expression in all tissues, but also enhances RP11-650L12.2 and CHRNA3 expression, suggesting DNA looping to multiple promoters. However, in nucleus accumbens and putamen, but not other brain regions, CHRNA3 expression associates uniquely with a haplotype tagged by rs1948 (located in the CHRNB4 3′UTR). Haplotype/diplotype analysis of rs880395 and rs1948 plus rs16969968 (a nonsynonymous CHRNA5 risk variant) in GWAS (COGEND, UW-TTURC, SAGE) yields a nicotine dependence risk profile only partially captured by rs16969968 alone. An example of local gene clusters, this nicotinic regulome is controlled by complex genetic variation, with broad implications for interpreting GWAS.
Keywords: human, smoking, gene expression, functional genomics, regulome, nicotinic receptor subunit, genetic association studies, genetic variation, eQTL
Introduction
Nicotinic acetylcholine receptors are ligand-gated ion channels activated by acetylcholine and nicotine, mediating synaptic transmission in brain and diverse functions in the periphery. Composed of five α and/or β subunits encoded by nicotinic receptor genes, nicotinic receptors display multiple compositions. Main mediators of nicotine’s actions in the brain, α4 and β2 subtypes are prevalent in dopaminergic neurons in the ventral tegmental area that project to nucleus accumbens or prefrontal cortex (the reward circuit), while α3 and β4 subtypes are well expressed in the habenulo-interpeduncular midbrain pathway (the aversion pathway). Both receptors can incorporate CHRNA5 (α5) leading to altered channel properties further influenced by genetic and post-translational factors (Consortium et al. 2015). This study focuses on the genomic region encompassing the adjacent CHRNA5 (MIM# 118505)-CHRNA3 (MIM# 118503)-CHRNB4 (MIM# 118509) genes, encoding α5, α3 and β4 subunits (Figure 1), implicated in clinical phenotypes including nicotine dependence (Thorgeirsson et al. 2008; Tobacco and Genetics Consortium 2010; Thorgeirsson and Stefansson 2010; Liu et al. 2010; Saccone et al. 2007; Saccone et al. 2010) and smoking related disorders (Amos et al. 2008; Hung et al. 2008;Thorgeirsson et al. 2008; Weiss et al. 2008; Doyle et al. 2011; Tyndale et al. 2015; Minicã et al. 2016). Among genetic variants implicated in nicotine dependence, the nonsynonymous rs16969968 is associated with the strongest risk (Saccone et al. 2010).
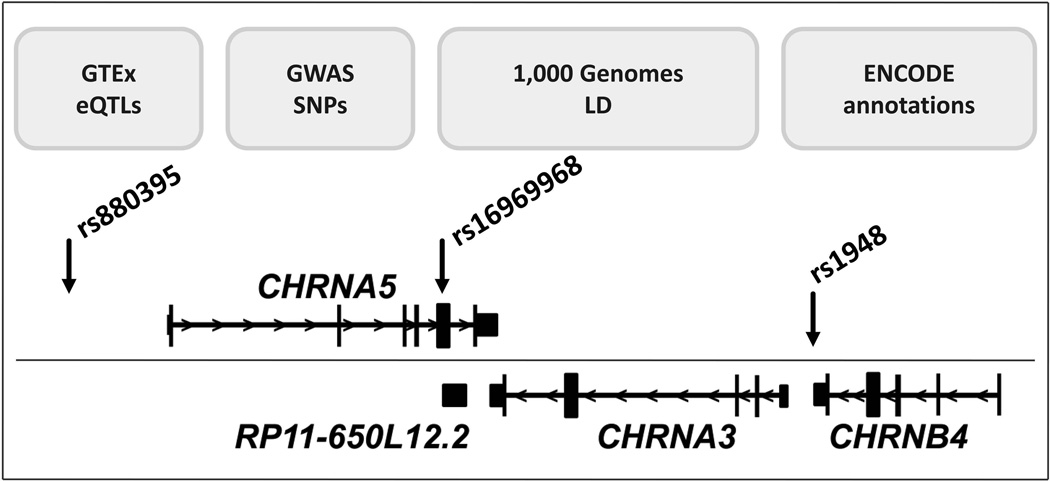
Figure 1.
CHRNA5/CHRNA3/CHRNB4 nicotinic receptor locus and analysis approach. A 110 kb region (UCSC hg38 chr15: 78,827,000-78,937,000) encompassing protein coding and noncoding RNAs: PSMA4, CHRNA5, RP11-650L12.2, CHRNA3 and CHRNB4. > indicates 5′ to 3′. Relevant SNPs are indicated with arrows.
These nicotinic receptor genes and several other adjacent genes such as PSMA4 appear to be in part co-expressed with one another (Flora et al. 2000), suggesting coordinated expression over a broad region. The locus is characterized by long regions of frequent SNPs in high linkage disequilibrium (LD) with each other, with four main haplotypes identified across CHRNA5-CHRNA3-CHRNB4 (Weiss et al. 2008). Here we test the hypothesis that this genomic region is subject to regulatory processes that extend beyond single genes, forming a regulome characterized by multiple enhancer and promoter elements that interact with each other via long range DNA looping. Understanding coordinated regulation is essential for interpreting phenotype associations attributed to specific variants in the cluster.
Variants in the CHRNA5/CHRNA3/CHRNB4 cluster have also been associated with other diseases, both peripheral (cardiac arrest, lung cancer) and central (addiction to other agents, schizophrenia) (GWAS catalog). In this study we focus on identifying haplotypes affecting regulation in the brain, in comparison to regulatory processes in peripheral tissues. Owing to the complex LD structure in this locus (Supp. Figure S1), it has proven difficult to identify causative variants and target genes, with some exceptions. A known nonsynonymous CHRNA5 SNP, rs16969968, and an enhancer haplotype (tagged by rs880395) on an opposite allele, influence α5 function and association with nicotine addiction (Bierut et al. 2008; Weiss et al. 2008; Wang et al. 2009; Liu et al. 2010; Saccone et al. 2010; Doyle et al. 2011; Frahm et al. 2011; Smith et al. 2011). The effect of enhancer rs880395 was found significant when considered in the context of rs16969968 (Smith et al. 2011). In this study we use genomics databases to search for regulatory variants across the CHRNA5/CHRNA3/CHRNB4 cluster and ask how these variants interact with each other, defining a local regulome (Figure 1). We then test for clinical associations to explore a new paradigm of integrated genetic influence of a gene cluster rather than single variants.
Materials and Methods
Datasets
GWAS and mRNA expression data were downloaded from dbGaP. This project is compliant with the regulations of the Ohio State University (OSU) Institutional Review Board and operates under a protocol reviewed and approved by a duly constituted ethics committee.
Genotype and Tissue Expression Project (GTEx): tissue specific expression, eQTLs, and allelic ratios. We acquired tissue specific RNA sequencing data from GTEx via dbGaP Project #5358 through the dbGaP repository, accession number phs000424.v5.p1 in April 2014 and the GTEx Portal in January–May 2016. Tissue specific eQTLs were calculated by GTEx using normalized RPKM (Reads Per Kilobase of transcript per Million mapped reads) and imputed genotypes with PEER factors and gender as covariates. eQTLs were defined by GTEx using a gene-specific p-value threshold based on permutation testing; see (Lonsdale et al. 2013) and (http://www.gtexportal.org/home/documentationPage). We focus in part on skeletal muscle (n = 429) and nucleus accumbens (n = 113) RNA profiles.
Clinical associations were derived from NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) and Genome-Wide Repository of Associations Between SNPs and Phenotypes (GRASP) (http://grasp.nhlbi.nih.gov/Overview.aspx). The GWAS Catalog provides SNP associations with a p value <10−5 in published GWAS, while GRASP applies p < 0.05.
Nicotine dependence GWAS: The Study of Addiction: Genetics and Environment (SAGE) (phs000092.v1.p1) includes COGA (Collaborative Study on Genetics of Alcoholism), FSCD (Family Study on Cocaine Dependence) and COGEND (Collaborative Genetic Study of nicotine dependence). The Genetic Architecture of Smoking and Smoking Cessation (phs000404.v1.p1) includes additional COGEND and UW-TTURC subjects (University of Wisconsin Transdisciplinary Tobacco Use Research Center). The gender imbalance in the study population precludes analysis of the impact of gender, but gender was included as a covariate.
Linkage disequilibrium (LD)
LD plots for the 500 kb region were generated with Haploview v 4.2 HapMap Download Version 3, Release R2. R2 and D′ were calculated using the ‘ld’ function in the ‘snpStats’ package implemented in R. LD was calculated based on imputed genotypes reported by GTEx in Caucasians only (n = 377) for plots of R2 versus p value.
Co-expression of mRNAs
Co-expression was defined as significant (Bonferroni corrected p value < 0.05) correlation between RPKM values in a given tissue using the cor.test function in R.
Testing clinical significance of candidate variants
We used FTND score ≥4 (Fagerström Test for Nicotine Dependence) to define nicotine dependence. The analysis was restricted to Caucasians who smoked at least 100 lifetime cigarettes. The strict definition for nicotine independence (having significant smoking exposure without dependence) limited the number of controls (3:1 ratio for cases (N=2756) and controls (N=845)) (Supp. Table S1). Haplotype and diplotype probabilities were generated with SVS Golden Helix SNP & Variation Suite v 8.3.4 using the Expectation Maximization (EM) algorithm; estimations deviated from predicted values 0, 0.5, or 1 by a maximum of 0.04. Significant haplotypes and diplotypes were tested as predictors of nicotine dependence with age and gender as covariates. Odds ratios and confidence intervals were calculated from the estimated effect size and standard error in the linear model. Results were compared to those obtained with ‘BayHap’ (Bayesian analysis of haplotype association using Markov Chain Monte Carlo) package in R. Additive logistic models were compared using ANOVA with a likelihood ratio test (LRT) implemented in R. GLM p values are reported for the standard Wald’s test and are reported in ANOVA for LRT (unless otherwise stated). Effect sizes reported are beta coefficients derived from GLMs.
Results
RNA expression across tissues in the extended gene cluster
A 500 kb window around the CHRNA5/CHRNA3/CHRNB4 cluster (GRCh38: 15:78428191-78906250) includes 20 genes (8 protein coding and 12 non-coding) (Supp. Figure S1). We considered RNA expression profiles reported in GTEx, for those genes adjacent to CHRNA5/CHRNA3/CHRNB4 that overlap with long haplotype structures (Supp. Figure S1), excluding poorly expressed genes (more than 50% of samples with 0 RPKM) or not reported in GTEx (Supp. Table S2). While CHRNA5 and CHRNA3 are expressed in multiple brain regions, CHRNB4 mRNA is detectable only in peripheral tissues; therefore, our subsequent analyses of CHRNB4 are restricted to peripheral tissues.
Coordinated expression between transcripts
Analysis of pairwise co-expression shows that CHRNA5/CHRNA3/CHRNB4 mRNAs and the CHRNA5 antisense RNA RP11-650L12.2 are co-expressed in most tissues (Supp. Figure S2). Most adjacent protein coding genes are not co-expressed, except for PSMA4 and MORF4L1 (Supp. Figure S3), and are not further considered, other than PSMA4 because it shares eQTLs with the nicotinic genes (Supp. Figure S4A) and is part of this LD block (Supp. Figure S1).
In further analysis, we focus on two tissues: skeletal muscle representing the periphery and nucleus accumbens, a brain region relevant to addiction, because of available sample size (n = 429 and n = 113, respectively) and abundant eQTLs (n = 785 and n = 238) (Supp. Table S3). In both tissues CHRNA3, CHRNA5, and RP11-650L12.2 are significantly co-expressed (Supp. Figure S2).
Expression quantitative loci (eQTLs) for nicotinic receptor and adjacent genes
To identify genetic variants modulating expression patterns, we considered SNPs identified as eQTLs in GTEx. While some eQTLs associate with expression of only a single gene in a single tissue, others associate with multiple genes in multiple tissues. To determine which genes appear to be under shared genetic control, we searched for common eQTLs (Supp. Figure S4B). In nucleus accumbens, three genes have identified eQTLs: RP11-650L12.2, CHRNA3, and CHRNA5. Whereas most SNPs are similarly associated with expression of all three genes in skeletal muscle, eQTLs are distinct for CHRNA3 in nucleus accumbens. Common eQTLs can either result from LD with a single causative eQTL or suggest the presence of more than one regulatory variant. With hundreds of eQTLs in the region, we first identify the most significant SNP for each gene in each tissue, followed by LD analysis to determine whether a single or multiple haplotypes account for the observed eQTLs.
CHRNA5-enhancer haplotype as ‘master regulator’ of the nicotinic cluster
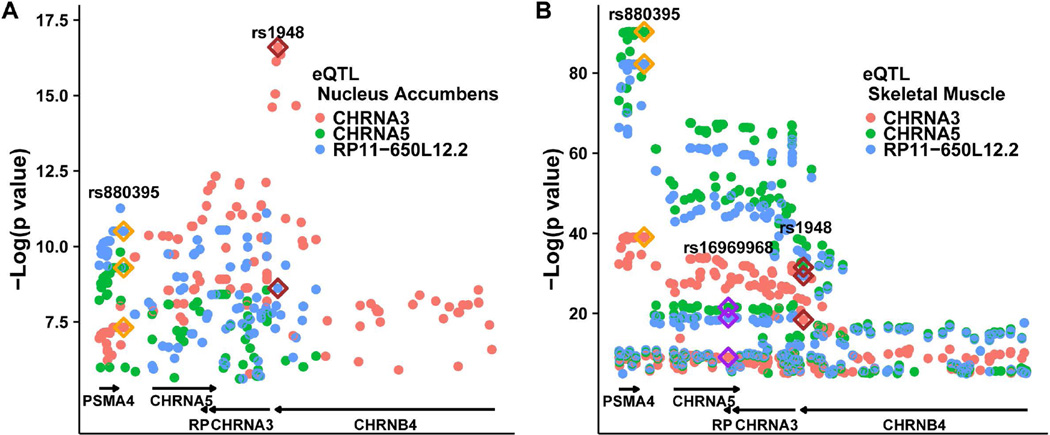
Figure 2 displays all significant eQTLs for CHRNA5, CHRNA3, and antisense RP11-650L12.2, in nucleus accumbens and skeletal muscle. The SNP with the most significant p value varied across tissues; however, top scoring SNPs for CHRNA5 mRNA, spread across a considerable genomic distance, were in high LD (D′>0.97, R2>0.91 in Caucasians) (Supp. Table S4). In GTEx, these CHRNA5 enhancer SNPs are the most significant eQTL for CHRNA5 in skeletal muscle (lowest p value 2.8e-91, effect size = −1.1). The tight LD structure makes it difficult to pinpoint the causative variant(s); therefore, we select for further analysis the haplotype-tagging SNP rs880395, previously shown to be associated with a near fourfold increase in CHRNA5 mRNA expression in brain (Smith et al. 2011). The same CHRNA5-enhancer SNP was also the strongest signal for RP11-650L12.2 expression across several tissues, and for CHRNA3 in skeletal muscle but not in nucleus accumbens (Supp. Table S4). Similarly distributed eQTL profiles for CHRNA5 and RP11-650L12.2 in nucleus accumbens, and for CHRNA5, CHRNA3, and antisense RP11-650L12.2 in skeletal muscle (Figure 2) indicate similar genetic regulation patterns (Supp. Figure S2). The CHRNA5-enhancer variant increases the nicotinic genes, but decreases PSMA4 expression in heart, artery, skin and skeletal muscle (Supp. Table S4). The CHRNA3 eQTL profile differs from that of CHRNA5 and RP11-650L12.2 in the nucleus accumbens (Figure 2A), suggesting distinct regulation of CHRNA3 in this brain region.
Figure 2.
eQTLs for the nicotinic receptor locus by genomic location. Y-axis:-log (p value) in nucleus accumbens (A) and skeletal muscle (B) for eQTL SNPs associated with expression of CHRNA3 (red), CHRNA5 (green), and RP11-650L12.2 (antisense to CHRNA5, blue) (GTEx data). eQTLs of interest are highlighted with a diamond: rs880395 (orange), rs1948 (red), and rs16969968 (purple; nonsynonymous SNP). In nucleus accumbens, rs1948 is the top eQTL for CHRNA3 but it is not significant for CHRNA5.
Identifying key regulatory elements
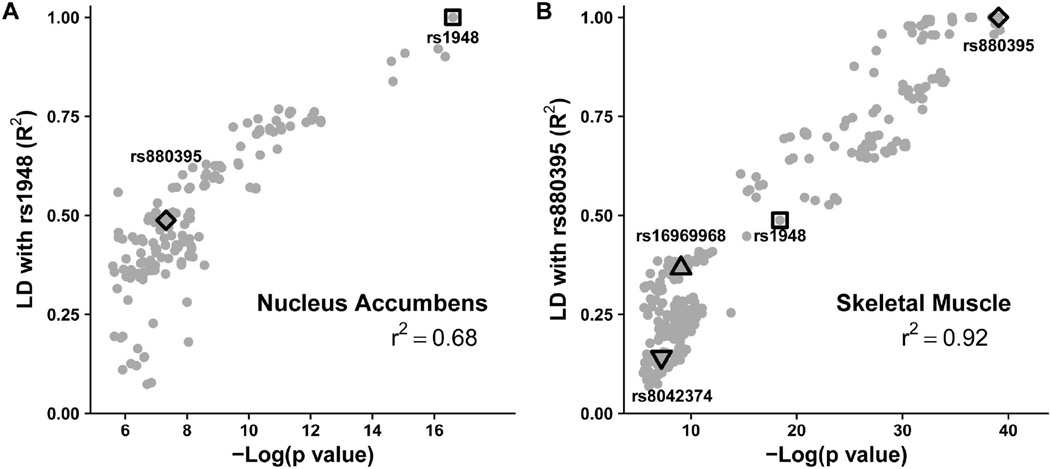
eQTLs in the CHRNA5/CHRNA3/CHRNB4 cluster distribute across a large region over 200 kb with high LD. Assuming one causative variant for a given gene, all other eQTL p values should correlate with LD (R2) to the causative variant marked by the highest scoring SNP. Plotting eQTL p values against LD for CHRNA5 and CHRNA3 in skeletal muscle and nucleus accumbens (Figure 3 and Supp. Figure S5) reveals strong correlations (r2 0.68–0.92). The CHRNA5-enhancer variant (marked by rs880395) predominantly affects CHRNA3, CHRNA5, and RP11-650L12.2 RNA expression in muscle and other tissues, while in the nucleus accumbens, the CHRNA3-enhancer haplotypes (rs1948) affects CHRNA3 (Figure 3A) and RP11-650L12.2 RNA.
Figure 3.
Correlation between eQTL p values and LD (R2) between all eQTLs and the top eQTL in nucleus accumbens (rs1948, correlation r2=0.68, A) and skeletal muscle (rs880395, correlation r2=0.92, B) for CHRNA3. Diamonds: CHRNA5-enhancer rs880395, triangle: nonsynonymous CHRNA5 rs16969968, square: CHRNA3-enhancer (brain) rs1948, and a potential additional regulatory haplotype for CHRNA3 tagged by rs8042374 (inverted triangle, not studied further). While the nonsynonymous rs16969968 SNP is listed as an eQTL in muscle, this result is attributable to LD with rs880395.
Only three brain regions show significant eQTLs for CHRNA3 mRNA, all within the basal ganglia (caudate, nucleus accumbens and putamen). While the rank order of the top SNP varies somewhat between different regions in the basal ganglia, all are in high LD with the eQTL signal marked by rs1948 (potential CHRNA3-enhancer SNPs). Located in the 3′UTR of CHRNB4, associations of rs1948 may have been falsely attributed to CHRNB4 (Flora et al. 2013; Gallego et al. 2013). rs1948 is associated with higher CHRNA3 expression in the nucleus accumbens (p=2.5e-17, effect size =0.99), which is further supported by considering allelic mRNA ratios (Supp. Figure S6).
The novel association between rs1948 and CHRNA3 expression in brain was tested in a separate database, Braineac (The Brain eQTL Almanac), reporting eQTLs for 10 brain regions. Of basal ganglia tissues, only putamen is represented in Braineac. As in GTEx, rs1948 is associated with CHRNA3 expression (Supp. Figure S7) in putamen but in none of the other 9 brain regions measured, indicating brain region selective regulation.
Overlapping eQTLs, significant GWAS SNPs, and LD between them
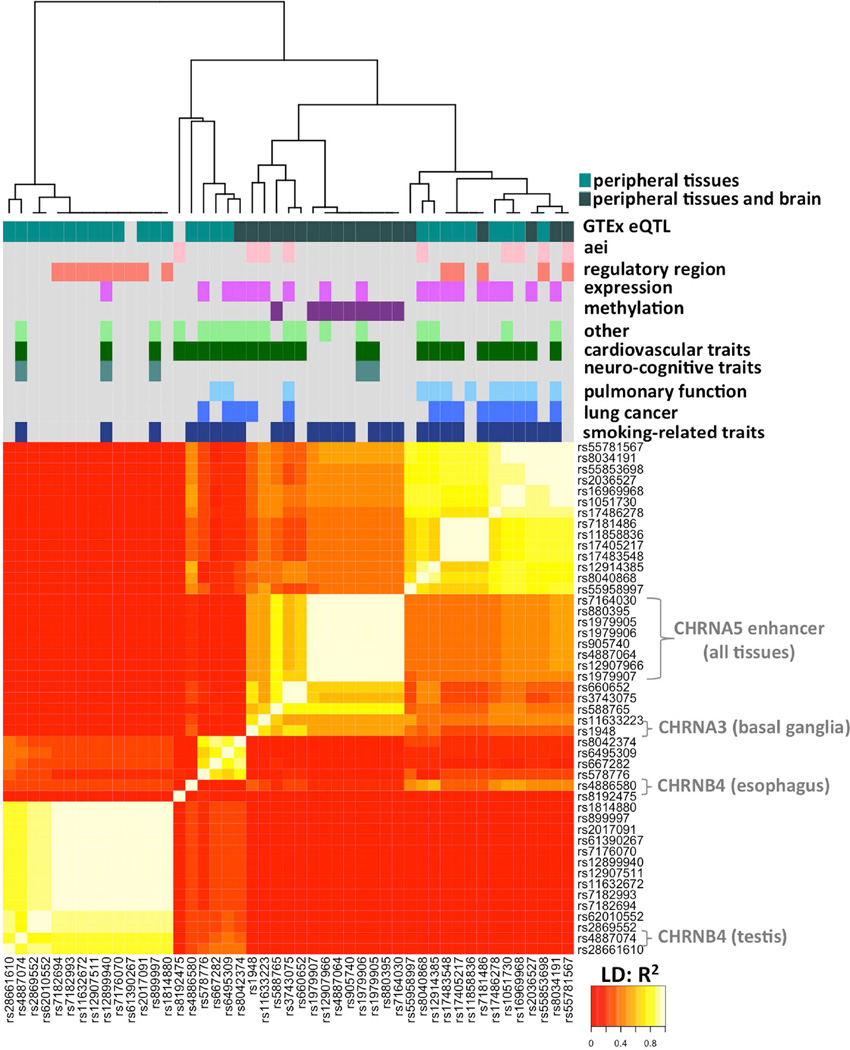
To test relationships between eQTLs and GWAS hits, as a function of LD, we overlaid data from dbGaP, GTEx, and the 1,000 Genomes Project, and visualized connections in a heatmap representing LD between candidate variants (Figure 4). SNPs are not ordered by genomic location but clustered by R2. Candidate SNPs were selected as follows: top eQTLs for all three nicotinic receptor genes and RP11-650L12.2, overlapping eQTLs and GWAS hits, placed in regions with genomic marks of regulatory potential (ENCODE), associated with allelic RNA ratios in GTEx or imbalanced allelic DNA ratios in DNAse hypersensitivity sites (Maurano et al. 2015), and published phenotype associations (Supp. Table S5; including guidelines for SNP selection).
Figure 4.
Heat map of LD values (R2) between candidate SNPs (47) with expression and regulatory annotations, and clinical associations. SNPs are grouped by their LD in CEU population, independent of genomic location. Colored blocks arranged in rows (top) indicate functional annotations for the selected SNPs. Dendrogram on the top depicts relatedness of groups and SNP sources listed in Supp. Table S5.
This approach identified four main groups of SNPs with high LD in Caucasians (Figure 4), each marked by a SNP discussed earlier. The nonsynonymous CHRNA5 (rs16969968) and CHRNA5-enhancer (rs880395) reside in different groups (reflective of their negative LD), while the CHRNA3-enhancer (rs1948) shows moderate LD with the CHRNA5-enhancer (indicated in orange). The CHRNB4 eQTLs also reside in distinct blocks, indicating distinct genetic effects. Thus, additional groups of SNPs in LD may be relevant to clinical associations; here we will focus on the CHRNA3- and CHRNA5-enhancer variants and their interactions with the nonsynonymous CHRNA5 rs16969968 SNP.
Association of nonsynonymous CHRNA5 SNP, CHRNA5-enhancer, and CHRNA3-enhancer variants with nicotine dependence
Our results reveal the presence of two main regulatory variants, marked by rs880395 (labeled CHRNA5-enhancer) and rs1948 (CHRNA3-enhancer), active only in the basal ganglia. Both rs880395 and rs1948 are in partial LD with each other and in negative LD with the nonsynonymous CHRNA5 SNP rs16969968. Here we address the combined effects of these variants on nicotine dependence in 3,601 Caucasians from SAGE (Study of Addiction: Genetics and Environment) and the Genetic Architecture of Smoking and Smoking Cessation. Nicotine dependence was defined by smoking history and the Fagerstrom Test for Nicotine Dependence (FTND) score (range 0–10, ND ≥4). The CHRNA5-enhancer rs880395 was not directly genotyped, but is well marked by rs8053 (R2 =0.97; D′=1); to simplify, we refer to this SNP as rs880395.
Testing each SNP using ANOVA confirmed that rs16969968 is a risk factor for nicotine dependence (Bierut et al. 2008; Smith et al. 2011). Although rs880395 and rs1948 are not individually significant, both are significant in the context of a second variant – rs1948 is significant when considered with rs16969968, and rs880395 when considered with rs1948 (Table 1). The estimated effect of rs16969968 is greatest in a recessive model (dominant model: estimate 0.26, p=0.001; recessive model: estimate 0.5, p=0.0002), possibly because in heterozygous carriers, the frequent CHRNA5-enhancer variant (increased expression) can occur on the main rs16969968 allele, which could dilute the effect of the minor rs16969968 allele. Considering only the most significant models from the ANOVA, the minor allele (A) of rs16969968 is associated with risk of nicotine dependence, the minor allele (A) of rs880395 is protective (negative effect size), and the minor allele (A) of rs1948 is associated with risk (Table 2).
Table 1.
Analysis of variance (ANOVA) comparing ability of models with different SNP combinations to account for nicotine dependence (recessive model).
| Variable of interest |
ANOVA p value |
Model 1 | Model 2 |
|---|---|---|---|
| rs16969968 | 6.0e-06 | ND ~ sex + age | ND ~ sex + age + rs16969968 |
| rs880395 | 0.16 | ND ~ sex + age | ND ~ sex + age + rs880395 |
| rs1948 | 0.92 | ND ~ sex + age | ND ~ rs1948 + sex + age |
| rs880395 in context of rs16969968 |
0.11 | ND ~ rs16969968 + sex + age |
ND ~ rs16969968 + rs880395 + sex + age |
| rs880395 in context of rs1948 |
0.04 | ND ~ rs1948 + sex + age | ND ~ rs1948 + rs880395 + sex + age |
| rs1948 in context of rs880395 |
0.12 | ND ~ sex + age_int + rs880395 |
ND ~ sex + age_int + rs880395 + rs1948 |
| rs1948 in context of rs16969968 |
0.007 | ND ~ rs16969968 + sex + age |
ND ~ rs16969968 + rs1948 + sex + age |
| rs16969968 in the context of rs880395 |
4.3e-06 | ND ~ sex + age_int + rs880395 |
ND ~ sex + age_int + rs16969968 + rs880395 |
| rs16969968 in the context of rs1948 |
1.4e-07 | ND ~ sex + age_int + rs1948 |
ND ~ sex + age_int + rs16969968 + rs1948 |
Table 2.
Linear models show protective effect for rs880395 and risk for rs1948 and rs16969968 for nicotine dependence (ND).
| rs16969968 | rs880395 | rs1948 | |
|---|---|---|---|
| ND ~ rs16969968 + sex + age | 0.27a (7.5e-06)b |
||
| ND ~ rs16969968 + rs1948 + sex + age | 0.36 a (1.6e-07) b |
0.19 a (0.007) b |
|
| ND ~ rs880395 + rs1948 + sex + age | −0.17 a (0.04) b |
0.13 a (0.12) b |
effect size;
p value
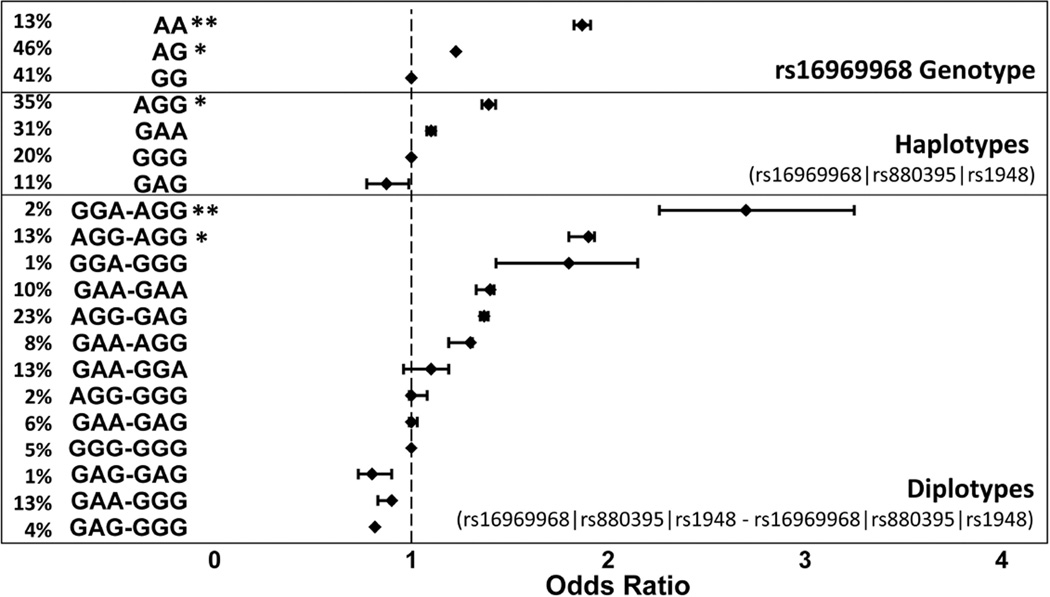
To evaluate how these SNPs in combination contribute to nicotine dependence, we consider haplotypes. Because of the LD relationships between rs16969968 (G/A), rs880395 (G/A) and rs1948 (G/A) (alleles always presented in this order), haplotypes can be assigned in a majority of subjects with high confidence (Figure 5). The GGG ‘wild-type’ haplotype represents only 20% of subjects while AGG is most prevalent in Caucasians (35%). Hence, the minor A allele of rs16969968 is largely restricted to occur together with the main G alleles of the two enhancer SNPs. A simple allele test reveals significant differences in risk effect between haplotypes, with AGG conveying risk (OR 1.4; p=0.03).
Figure 5.
Association of nicotine dependence with rs16969968 genotype (G>A), haplotypes, and diplotypes comprised of: rs16969968 (G>A), rs880395 (G>A), and rs1948 (G>A), reported as odds ratio with 95% confidence intervals. Significance levels denoted by p * < 0.05, ** <0.01. Frequency of each group is indicated on far left (% rounded to nearest integer).
Because of the LD structure, the number of prevalent diplotypes is also reduced, enabling assessment of diplotype risk factors. Thirteen diplotypes with frequencies of 1–23%, most assigned unambiguously, account for >99% of the subjects. The GGG-GGG diplotype is represented in only 5% of subjects with an assigned odds ratio of 1; odds ratios of the other diplotypes range from 0.8 to 2.7 (Figure 5). Consistent with the haplotype analysis, the AGG-AGG diplotype (13%) conveys risk with an odds ratio of 1.9 (p = 0.004). The odds ratio for the nonsynonymous variant on the background of the CHRNA5-enhancer, the AGG-GAG diplotype, is not significant (p = 0.76), as carriers of the A allele of rs8803965 tend to cluster in the lower risk groups. On the other hand, carriers of the A allele of rs1948 tend to experience higher risk, with the highest odds ratio of 2.7 (p = 0.02) for AGG-GGA. The results support a protective role for the rs880395 A allele with higher expression of CHRNA5 and suggest that the rs1948 A allele, similarly causing higher expression of CHRNA3 mRNA, contributes to risk (Figure 5). The results show that each of the three variants exerts robust effects in the context of haplotypes/diplotypes, generating a gradient of risk not captured by rs16969968 alone (Figure 5).
Discussion
Our goal was to assess the influence of genetic variation of the CHRNA5/CHRNA3/CHRNB4 cluster as an interactive regulome on nicotine dependence. High LD between countless SNPs spread across a large genomic region suggest the influence of genetic selection pressures that shape the overall genomic architecture, rather than selecting single SNPs. Analyses of large-scale genomics databases identifies at least 4 high LD blocks carrying regulatory variants, in addition to the nonsynonymous SNP rs16969968, the predominant risk factor for nicotine dependence in the region. Frequent eQTLs for CHRNA5, CHRNA3, and RP11-650L12.2, some tissue selective, reside on long overlapping haplotypes, while eQTLs for CHRNB4 are separate and were not detectable in brain regions because of low mRNA levels (GTEx). This study focuses on two main eQTL haplotypes regulating expression of CHRNA5, CHRNA3, and RP11-650L12.2, and their influence, combined with rs16969968, on nicotine dependence.
CHRNA5-enhancer and CHRNA3-enhancer variants
We show here that a previously reported CHRNA5-enhancer haplotype tagged by rs880395 not only associates with a robust increase in CHRNA5 mRNA expression in brain (Smith et al. 2011) and peripheral tissues, but also enhances expression of CHRNA3 and RP11-650L12.2 RNA while reducing PSMA4 mRNA expression in most tissues. The role of the RP-11 antisense RNA remains to be clarified. Our result indicates that the CHRNA5-enhancer serves as a master regulator, likely affecting the expression of multiple genes via DNA looping processes (Li et al. 2012; Sanyal et al. 2012; Zhang et al. 2013; Heidari et al. 2014; D. Wang et al. 2014; Whalen, Truty, and Pollard 2016).
In the basal ganglia of the brain, a distinct regulatory variant tagged by rs1948 increases CHRNA3 mRNA expression (the CHRNA3-enhancer), demonstrating distinct regulation in brain regions germane to nicotine dependence. The CHRNA5-enhancer and CHRNA3-enhancer variants are in partial LD with each other, despite separation by a large genomic distance, a critical feature that needs to be reflected in GWAS analyses. In contrast, the nonsynonymous SNP rs16969968 minor A allele resides nearly exclusively on an allele opposite to that of the CHRNA5-enhancer variant rs880395 (Supp. Table S6). As a result, the effect of the minor allele of rs16969968 is diluted in heterozygous carriers if the main allele carries the CHRNA5-enhancer haplotype with high expression. Similarly, the rs16969968 A allele resides predominantly on a haplotype together with the main G allele of the CHRNA3-enhancer variant rs1948, expressing less CHRNA3 mRNA than the minor A allele in the basal ganglia.
Because of high LD between numerous CHRNA5-enhancer and CHRNA3-enhancer SNPs, we cannot identify the causative variants from these results, except to infer that they are likely well marked by the top enhancer SNPs identified here. Moreover, we cannot exclude the possibility that multiple causative variants in high LD exist at the CHRNA3- and CHRNA5-enhancer sites.
Genetic effects of rs16969968 and the CHRNA3-enhancer variant
Previous studies have demonstrated that the nonsynonymous CHRNA5 SNP rs16969968 (D398N) reduces response to nicotine (Bierut et al. 2008), causing desensitization (Kuryatov, Berrettini, and Lindstrom 2011). rs16969968 does not appear to affect gene expression per se, even though it is assigned an eQTL in GTEx, a result of the LD structure of the locus.
The CHRNA5-enhancer region has been analyzed previously (Smith et al. 2011). The CHRNA3-enhancer haplotype active in the basal ganglia tagged by rs1948, while residing in the 3′UTR of CHRNB4, regulates CHRNA3 mRNA expression in this brain region. Previous studies have reported associations between rs1948 alcohol and tobacco use (Stephens et al. 2013; Schlaepfer, Hoft, and Ehringer 2008) and blood pressure (Kaakinen et al. 2012). While Flora et al. (2013) found no effect of rs1948 on TF binding in vitro using EMSA, rs1948 did alter transcriptional activity in a luciferase assay in a lung cancer cell line, but not in neuronal cells. The causative variant remains to be identified unambiguously.
Interplay between the CHRNA5-enhancer and CHRNA3-enhancer variants with rs16969968 affecting nicotine dependence
In contrast to rs16969968, neither of the CHRNA5- and CHRNA3-enhancers, represented by rs880395 and rs1948, respectively, have significant effects on nicotine dependence when tested in isolation. Rather, GWAS of nicotine dependence reveals that the effect of each variant depends on the others. Both the CHRNA5- and CHRNA3-enhancers may affect nicotine dependence by altering relative abundance of receptor subunits and receptor composition, specifically in the basal ganglia important for addiction. The CHRNA3-enhancer tends to convey risk, while the CHRNA5-enhancer is protective.
Because of the extensive LD structure, only a limited number of haplotypes are extant in the population (Caucasians in this study), enabling the assessment of haplotype (in order rs16969968 (G>A) – rs880395 (G>A) – rs1948 (G>A)) in our GWAS cohort (Caucasians). As expected, AGG carries the greatest risk. We also assign diplotypes to 13 groups accounting for >99% of the GWAS cohort, which convey risk in a gradual fashion from lowest (GGG-GAG) to highest (AGG-AGG and AGG-GGA). The minor allele of rs16969968 is uncommon (MAF 1–5%) in Asian and African populations, precluding analysis in these populations. In Caucasians, risk assignments with use of diplotypes yield substantially different results, compared to rs16969968 genotype alone, including high risk predictions for GGG-GGA and AGG-GGA, classified by rs16969968 genotype alone (GG) as low risk. The influence of the CHRNA3/5 enhancers in other ethnic groups requires additional studies. Since addictive behaviors are influenced by multiple gene loci that are typically added into a composite risk score, even small errors in risk assessment at a given gene locus can lead to substantial errors in a multigenic overall score.
In conclusion, our results show that a genomic region with functionally related genes, such as the CHRNA5/CHRNA3/CHRNB4 cluster, is under coordinated regulatory control. As a result, clinical association studies employing individual independent genetic variants in a complex gene locus are bound to miss a critical portion of the overall genetic variability conveyed by this type of gene cluster. Our approach suggests re-interpretation of genetic effects of the CHRNA5/CHRNA3/CHRNB4 cluster and serves as a roadmap for studying the ‘regulome’ of gene clusters relevant to complex disorders.
Supplementary Material
Acknowledgments
Funding statement
The funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources (NCRR) or the National Institutes of Health (NIH) or other funders. This work was supported in part by NIH grant U01 GM092655 (W.S.), TL1 TR001069 (KH), computing time provided by the Ohio Supercomputer Center: Grant #: PAS0885-2 Project: Collaboration Environment for Pharmacogenomics and the CCTS (Center for Clinical and Translational Science) at OSU under Award Number UL1RR025755 from the NCRR. Funding for outside databases provided in the supplement.
References
- Amos Christopher I, Wu Xifeng, Broderick Peter, Gorlov Ivan P, Gu Jian, Eisen Timothy, Dong Qiong, et al. Nature Genetics. 5. Vol. 40. Nature Publishing Group; 2008. Genome-Wide Association Scan of Tag SNPs Identifies a Susceptibility Locus for Lung Cancer at 15q25.1; pp. 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut Laura Jean, Stitzel Jerry A, Wang Jen C, Hinrichs Anthony L, Grucza Richard A, Xuei Xiaoling, Saccone Nancy L, et al. Variants in Nicotinic Receptors and Risk for Nicotine Dependence. The American Journal of Psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle Glenn A, Wang Min-Jung, Chou Andrew D, Oleynick John U, Arnold Steven E, Buono Russell J, Ferraro Thomas N, Berrettini Wade H. In Vitro and Ex Vivo Analysis of CHRNA3 and CHRNA5 Haplotype Expression. PloS One. 2011;6(8):e23373. doi: 10.1371/journal.pone.0023373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora A, Schulz R, Benfante R, Battaglioli E, Terzano S, Clementi F, Fornasari D. Transcriptional Regulation of the Human alpha5 Nicotinic Receptor Subunit Gene in Neuronal and Non-Neuronal Tissues. European Journal of Pharmacology. 2000;393(1–3):85–95. doi: 10.1016/s0014-2999(00)00040-6. http://www.ncbi.nlm.nih.gov/pubmed/10771001. [DOI] [PubMed] [Google Scholar]
- Flora Amber V, Zambrano Cristian A, Gallego Xavier, Miyamoto Jill H, Johnson Krista A, Cowan Katelyn A, Stitzel Jerry A, Ehringer Marissa A. Functional Characterization of SNPs in CHRNA3/B4 Intergenic Region Associated with Drug Behaviors. Brain Research. 2013 Sep;1529:1–15. doi: 10.1016/j.brainres.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm Silke, Ślimak Marta A, Ferrarese Leiron, Santos-Torres Julio, Antolin-Fontes Beatriz, Auer Sebastian, Filkin Sergey, et al. Neuron. 3. Vol. 70. Elsevier; 2011. Aversion to Nicotine Is Regulated by the Balanced Activity of β4 and α5 Nicotinic Receptor Subunits in the Medial Habenula; pp. 522–535. [DOI] [PubMed] [Google Scholar]
- Gallego Xavier, Cox Ryan J, Laughlin James R, Stitzel Jerry A, Ehringer Marissa A. Alternative CHRNB4 3’-UTRs Mediate the Allelic Effects of SNP rs1948 on Gene Expression. PloS One. 2013;8(5):e63699. doi: 10.1371/journal.pone.0063699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley Paul, Kadakkuzha Beena M, Faghihi Mohammad Ali, Magistri Marco, Zeier Zane, Khorkova Olga, Coito Carlos, Hsiao Jane, Lawrence Matthew, Wahlestedt Claes. Regulation of the Apolipoprotein Gene Cluster by a Long Noncoding RNA. Cell Reports. 2014;6(1):222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari Nastaran, Phanstiel Douglas H, He Chao, Grubert Fabian, Jahanbani Fereshteh, Kasowski Maya, Zhang Michael Q, Snyder Michael P. Genome-Wide Map of Regulatory Interactions in the Human Genome. Genome Research. 2014;24(12):1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Rayjean J, McKay James D, Gaborieau Valerie, Boffetta Paolo, Hashibe Mia, Zaridze David, Mukeria Anush, et al. Nature. 7187. Vol. 452. Nature Publishing Group; 2008. A Susceptibility Locus for Lung Cancer Maps to Nicotinic Acetylcholine Receptor Subunit Genes on 15q25; pp. 633–637. [DOI] [PubMed] [Google Scholar]
- Kaakinen Marika, Ducci Francesca, Sillanpää Mikko J, Läärä Esa, Järvelin Marjo-Riitta. Associations between Variation in CHRNA5-CHRNA3-CHRNB4, Body Mass Index and Blood Pressure in the Northern Finland Birth Cohort 1966. PloS One. 2012;7(9):e46557. doi: 10.1371/journal.pone.0046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov Alexander, Berrettini Wade, Lindstrom Jon. Acetylcholine Receptor (AChR) α5 Subunit Variant Associated with Risk for Nicotine Dependence and Lung Cancer Reduces (α4β2)2 α5 AChR Function. Molecular Pharmacology. 2011;79(1):119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Jae Yeon, Yoo Seung Soo, Kang Hyo-Gyoung, Jin Guang, Bae Eun Young, Choi Yi Young, Choi Jin Eun, et al. A Functional Polymorphism in the CHRNA3 Gene and Risk of Chronic Obstructive Pulmonary Disease in a Korean Population. Journal of Korean Medical Science. 2012;27(12):1536–1540. doi: 10.3346/jkms.2012.27.12.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Guoliang, Ruan Xiaoan, Auerbach Raymond K, Sandhu Kuljeet Singh, Zheng Meizhen, Wang Ping, Poh Huay Mei, et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell. 2012;148(1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Jason Z, Tozzi Federica, Waterworth Dawn M, Pillai Sreekumar G, Muglia Pierandrea, Middleton Lefkos, Berrettini Wade, et al. Meta-Analysis and Imputation Refines the Association of 15q25 with Smoking Quantity. Nature Genetics. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale John, Thomas Jeffrey, Salvatore Mike, Phillips Rebecca, Lo Edmund, Shad Saboor, Hasz Richard, et al. The Genotype-Tissue Expression (GTEx) Project. Nature Genetics. 2013;45(6):580–585. doi: 10.1038/ng.2653. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano Matthew T, Haugen Eric, Sandstrom Richard, Vierstra Jeff, Shafer Anthony, Kaul Rajinder, Stamatoyannopoulos John A. Large-Scale Identification of Sequence Variants Influencing Human Transcription Factor Occupancy in Vivo. Nature Genetics. 2015;47(12):1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minicã CC, Mbarek H, Pool R, Dolan CV, Boomsma DI, Vink JM. Pathways to Smoking Behaviours: Biological Insights from the Tobacco and Genetics Consortium Meta-Analysis. Molecular Psychiatry. 2016 Mar; doi: 10.1038/mp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay Jessica E, Rhodes C. Harker, Thirtamara-Rajamani Keerthi, Smith Ryan M. Genetic Influences on Nicotinic α5 Receptor (CHRNA5) CpG Methylation and mRNA Expression in Brain and Adipose Tissue. Genes and Environment. 2015;37(1):14. doi: 10.1186/s41021-015-0020-x. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone Nancy L, Culverhouse Robert C, Schwantes-An Tae-Hwi, Cannon Dale S, Chen Xiangning, Cichon Sven, Giegling Ina, et al. Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: A Meta-Analysis and Comparison with Lung Cancer and COPD. In: Gibson Greg., editor. PLoS Genetics. 8. Vol. 6. Public Library of Science; 2010. p. e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone Scott F, Hinrichs Anthony L, Saccone Nancy L, Chase Gary A, Konvicka Karel, Madden Pamela A F, Breslau Naomi, et al. Cholinergic Nicotinic Receptor Genes Implicated in a Nicotine Dependence Association Study Targeting 348 Candidate Genes with 3713 SNPs. Human Molecular Genetics. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal Amartya, Lajoie Bryan R, Jain Gaurav, Dekker Job. The Long-Range Interaction Landscape of Gene Promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer Isabel R, Hoft Nicole R, Ehringer Marissa A. The Genetic Components of Alcohol and Nicotine Co-Addiction: From Genes to Behavior. Current Drug Abuse Reviews. 2008;1(2):124–134. doi: 10.2174/1874473710801020124. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2600802&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Ryan M, Alachkar Houda, Papp Audrey C, Wang Danxin, Mash Deborah C, Wang Jen-Chyong, Bierut Laura J, Sadee Wolfgang. Nicotinic α5 Receptor Subunit mRNA Expression Is Associated with Distant 5’ Upstream Polymorphisms. European Journal of Human Genetics?: EJHG. 2011;19(1):76–83. doi: 10.1038/ejhg.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens Sarah H, Hartz Sarah M, Hoft Nicole R, Saccone Nancy L, Corley Robin C, Hewitt John K, Hopfer Christian J, et al. Distinct Loci in the CHRNA5/CHRNA3/CHRNB4 Gene Cluster Are Associated with Onset of Regular Smoking. Genetic Epidemiology. 2013;37(8):846–859. doi: 10.1002/gepi.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Stefansson K. International Journal of Epidemiology. 2. Vol. 39. Oxford University Press; 2010. Commentary: Gene-Environment Interactions and Smoking-Related Cancers; pp. 577–579. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson Thorgeir E, Geller Frank, Sulem Patrick, Rafnar Thorunn, Wiste Anna, Magnusson Kristinn P, Manolescu Andrei, et al. Nature. 7187. Vol. 452. Nature Publishing Group; 2008. A Variant Associated with Nicotine Dependence, Lung Cancer and Peripheral Arterial Disease; pp. 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium, The Tobacco and Genetics. Genome-Wide Meta-Analyses Identify Multiple Loci Associated with Smoking Behavior. Nature Genetics. 2010;42(5):441–447. doi: 10.1038/ng.571. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndale Rachel F, Zhu Andy Z X, George Tony P, Cinciripini Paul, Hawk Larry W, Schnoll Robert A, Swan Gary E, Benowitz Neal L, Heitjan Daniel F, Lerman Caryn. Lack of Associations of CHRNA5-A3-B4 Genetic Variants with Smoking Cessation Treatment Outcomes in Caucasian Smokers despite Associations with Baseline Smoking. PloS One. 2015;10(5):e0128109. doi: 10.1371/journal.pone.0128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Danxin, Poi Ming J, Sun Xiaochun, Gaedigk Andrea, Leeder J Steven, Sadee Wolfgang. Common CYP2D6 Polymorphisms Affecting Alternative Splicing and Transcription: Long-Range Haplotypes with Two Regulatory Variants Modulate CYP2D6 Activity. Human Molecular Genetics. 2014;23(1):268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, Duan W, et al. Human Molecular Genetics. 16. Vol. 18. Oxford University Press; 2009. Risk for Nicotine Dependence and Lung Cancer Is Conferred by mRNA Expression Levels and Amino Acid Change in CHRNA5; pp. 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, et al. Molecular Psychiatry. 5. Vol. 14. Nature Publishing Group; 2009. Genetic Variation in the CHRNA5 Gene Affects mRNA Levels and Is Associated with Risk for Alcohol Dependence; pp. 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss Robert B, Baker Timothy B, Cannon Dale S, Niederhausern Andrew von, Dunn Diane M, Matsunami Nori, Singh Nanda A, et al. A Candidate Gene Approach Identifies the CHRNA5-A3-B4 Region as a Risk Factor for Age-Dependent Nicotine Addiction. PLoS Genetics. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen Sean, Truty Rebecca M, Pollard Katherine S. Enhancer-Promoter Interactions Are Encoded by Complex Genomic Signatures on Looping Chromatin. Nature Genetics. 2016;48(5):488–496. doi: 10.1038/ng.3539. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Chen, Hu Zhibin, Yu Dianke, Huang Liming, Jin Guangfu, Liang Jie, Guo Huan, et al. Genetic Variants on Chromosome 15q25 Associated with Lung Cancer Risk in Chinese Populations. Cancer Research. 2009;69(12):5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- Zhang Yubo, Wong Chee-Hong, Birnbaum Ramon Y, Li Guoliang, Favaro Rebecca, Ngan Chew Yee, Lim Joanne, et al. Chromatin Connectivity Maps Reveal Dynamic Promoter-Enhancer Long-Range Associations. Nature. 2013;504(7479):306–310. doi: 10.1038/nature12716. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.