Abstract
BACKGROUND AND PURPOSE:
The presence of the apolipoprotein E ε4 allele is the strongest sporadic Alzheimer disease genetic risk factor. We hypothesized that apolipoprotein E ε4 carriers and noncarriers may already differ in imaging patterns in midlife. We therefore sought to identify the effect of apolipoprotein E genotype on brain atrophy across almost the entire adult age span by using advanced MR imaging–based pattern analysis.
MATERIALS AND METHODS:
We analyzed MR imaging scans of 1472 participants from the Study of Health in Pomerania (22–90 years of age). We studied the association among age, apolipoprotein E ε4 carrier status, and brain atrophy, which was quantified by using 2 MR imaging–based indices: Spatial Pattern of Atrophy for Recognition of Brain Aging (summarizing age-related brain atrophy) and Spatial Pattern of Abnormality for Recognition of Early Alzheimer Disease (summarizing Alzheimer disease-like brain atrophy patterns), as well as the gray matter volumes in several Alzheimer disease- and apolipoprotein E–related ROIs (lateral frontal, lateral temporal, medial frontal, and hippocampus).
RESULTS:
No significant association was found between apolipoprotein E ε4 carrier status and the studied ROIs or the MR imaging–based indices in linear regression models adjusted for age, sex, and education, including an interaction term between apolipoprotein E and age.
CONCLUSIONS:
Our study indicates that measurable apolipoprotein E–related brain atrophy does not occur in early adulthood and midlife and suggests that such atrophy may only occur more proximal to the onset of clinical symptoms of dementia.
The presence of the apolipoprotein E gene (APOE) ε4 allele is the strongest genetic sporadic Alzheimer disease (AD) risk factor.1,2 The APOE gene has 2 additional codominant alleles (APOE ε2 and ε3). These alleles code for the 3 corresponding apolipoprotein isoforms (ApoE ε2, ε3, and ε4). Compared with individuals with the ε3 allele,2 which is the most common in the general population, those with the APOE ε4 allele have a higher risk of AD, whereas the APOE ε2 allele is associated with a lower risk of AD. The APOE genotype modifies the age at which cognitively healthy subjects present biomarker changes that define AD preclinical stages.3,4 There are several mechanisms that have been associated with the influence of APOE ε4 on AD risk: decreased clearance of amyloid from the brain, impaired synaptic plasticity, altered blood-brain barrier permeability, and immune response and impaired membrane repair following neuronal injury.5–7 In line with these results, neuropathologic studies have shown that plaques and tangles mediate APOE-related clinical changes.8 ApoE is present in plasma and CSF, but there is a weak correlation between ApoE concentrations within these 2 compartments.9,10 In APOE genotype–adjusted models, CSF, but not plasma, ApoE levels are associated with cognitive changes and brain atrophy.9 Nevertheless, a large longitudinal study reported associations with plasma ApoE levels that dropped to borderline significance when adjusted for the APOE genotype.11
Most APOE-related findings regarding brain structure have been reported in elderly subjects. However, due to the long AD preclinical stage, it could be expected that subtle neuroimaging changes may appear at younger ages and become more pronounced with advancing age. In line with this hypothesis, there is limited information regarding associations related to brain structural changes and APOE genotypes in younger subjects. Previous studies have reported lower hippocampal, orbitofrontal, and entorhinal volumes in subjects with APOE ε4 in the first decades of life.12,13 Other studies have shown brain hypometabolism in AD-related areas14 and increased activation of the default mode network15 in young APOE ε4 carriers but no differences in gray matter volume. The aforementioned changes reported in these studies would be taking place during the 3 decades before the ages that show at least 1% AD prevalence. These findings suggest that AD could have a neurodevelopmental component. However, the relation between regional brain atrophy and the APOE genotype needs to be evaluated across the adult life span.
We hypothesized that APOE ε4 carriers and noncarriers could present different imaging patterns that start differing in midlife, much earlier than clinical diagnosis. To investigate this hypothesis, we sought to model structural MR imaging brain changes associated with the APOE genotype in a large population-based study spanning the third-to-ninth decades of life by using sensitive pattern-based aging and AD summary indices.
Materials and Methods
Participants from the Study of Health in Pomerania
The Study of Health in Pomerania (SHIP) is a prospective cohort whose subjects are recruited from the general population of the German State of Mecklenburg–Western Pomerania. It is led by the Institute for Community Medicine at the Medical Faculty of the University of Greifswald.16 SHIP started at baseline with SHIP-0 between 1997 and 2001. After about 5 years, all participants were re-invited for a follow-up visit (ie, SHIP-1). From 2008 to 2012, the second follow-up examination, SHIP-2, was performed. Concurrent with SHIP-2, a new population sample from the same area was drawn, and similar examinations were undertaken between 2008 and 2012 (SHIP-TREND). SHIP-2 and SHIP-TREND included whole-body MR imaging scans,17 which were not present in SHIP-0 and SHIP-1. Trained certified radiologists, each with >5 years of MR imaging interpretation experience, visually inspected head MR imaging scans for artifacts and clinical findings. In our study, we included 1472 subjects of the total 3256 subjects with T1-weighted brain scans available in SHIP-2 and SHIP-TREND, 22–90 years of age at enrollment. We excluded 1784 subjects from this analysis on the basis of the following: 1) the presence of stroke, multiple sclerosis, epilepsy, cerebral tumor, intracranial cyst, or hydrocephalus (n = 150); 2) a high level of motion artifacts (n = 98); 3) failed quality control of the automatically skull-stripped data (n = 121); 4) lack of genomewide association study data (n = 1008); 5) an unidentifiable APOE genotyping (n = 35); and 6) lack of clinical data (cognitive scores, n = 190, or covariates, n = 182).
Clinical data, including sociodemographic factors and medical history, were collected in a standardized computer-assisted face-to-face interview. Years of education were recorded and grouped into 3 categories: <8 years, 8–10 years, and >10 years. In SHIP, two cognitive tests were obtained: the Verbal Learning and Memory Test (the German version of the California Verbal Learning and Memory Test18) for SHIP-2 and the Nuremberg Age Inventory for SHIP-TREND. The Nuremberg Age Inventory is a German test developed to measure the cognitive abilities during brain aging.19 It consists of subtests including, but not limited to, speed (eg, numbers) and memory (eg, words, numbers, and images). A description of the final SHIP sample included in our analysis (n = 1472) is shown in Table 1. The Ethics Committee of the Medical Faculty of the University of Greifswald approved SHIP.
Table 1:
Description of the SHIP sample included in this study (n = 1472)
| Characteristic | SHIP-2 | SHIP-TREND | SHIP-2 and SHIP-TREND (Analysis Sample) |
|---|---|---|---|
| Age (median) (SD) (yr) | 56.3 (12.2) | 51.1 (13.4)a | 53.3 (13.0) |
| Education (No.) (%) | |||
| <8 yr | 137 (18.4) | 77 (10.6)a | 214 (14.5) |
| 8–10 yr | 430 (57.8) | 415 (57.0)a | 845 (57.4) |
| >10 yr | 177 (23.8) | 236 (32.4)a | 413 (28.1) |
| APOE genotype: at least 1 ϵ4 allele (No.) (% positive) | 145 (19.5) | 177 (24.3)a | 322 (21.9) |
| Female sex (No.) (%) | 399 (53.6) | 421 (57.8) | 820 (55.7) |
| Verbal Learning and Memory Test (mean) (SD) | 8.6 (3.0) | ||
| Nuremberg Age Inventory (mean) (SD) | 11.2 (2.5) |
Significant difference at P <.05.
Image Acquisition
In SHIP-2 and SHIP-TREND, a comprehensive whole-body MR imaging protocol was used. A full description of the image-acquisition parameters for SHIP can be found in Hegenscheid et al.17 The neurocranium unit of SHIP included, among others, T1-weighted and fluid-attenuated inversion recovery sequences. Briefly, all images were obtained by using a 1.5T MR imaging scanner (Magnetom Avanto; Siemens, Erlangen, Germany). In this study, we used only the T1-weighted axial MPRAGE images for measuring regional patterns of AD-related brain atrophy. The T1-weighted image was acquired with the following parameters: 1 × 1 mm in-plane spatial resolution, section thickness = 1.0 mm, flip angle = 15°, TE = 3.4 ms, TR = 1900 ms.
Image Processing
An automated multiatlas segmentation method was applied on the T1-weighted image of each subject to calculate a brain mask, by removing extracranial material on the T1-weighted image.20 Total intracranial volume was estimated by calculating the volume of a subject's brain mask, which included the volumes of gray matter, white matter, ventricles, and the CSF that were contained within the outer brain boundary. Each brain mask was visually inspected for quality, by M.H., and all low-quality brain masks (including either under- or oversegmented brain) were excluded.
A multiatlas label fusion–based segmentation method21 was applied for segmentation of the brain into a set of anatomic ROIs. The volumes of ROIs were calculated and were normalized by total intracranial volume. In this analysis, we selected a set of gray matter ROIs on the basis of areas that were associated with AD-related atrophy and previously reported APOE-related findings. Further details are given in the On-line Appendix (On-line Method 1).
MR Imaging Pattern Classification
A pattern-classification method was previously proposed to derive Spatial Pattern of Abnormality for Recognition of Early AD (SPARE-AD),22 an index summarizing the high-dimensional imaging data with a single value that quantifies the atrophy patterns in AD-related regions. SPARE-AD has been shown to discriminate between normal cognition and mild cognitive impairment (MCI)23 and conversion from MCI to AD.24 We calculated, in this study, the SPARE-AD index for the SHIP population by using a model based on a linear support vector machine,25 which was trained on the external training dataset described in Da et al24 and had been validated earlier.22 More positive SPARE-AD implies a more Alzheimer disease–like brain structure, while more negative values reflect more normal brain structure.
Similar to the calculation of the SPARE-AD index, a linear support vector machine–based model was designed to predict an individual's age from the MR image and was used for quantifying Brain Aging (BA), summarized by the SPARE-BA index. The classification model was trained for optimally discriminating young and old subject groups and has been recently described.26,27 The SPARE-BA index for an individual implied fewer brain aging patterns for higher (positive) values and the presence of more aging-related atrophy patterns for lower (negative) values. While the SPARE-AD index captured more localized atrophy patterns in AD-related regions, such as in the hippocampus and temporal lobe, the SPARE-BA index captured more global aging patterns, distributed in the cortex, but particularly in the frontal lobe. Further details are given in the On-line Appendix (On-line Method 2).
APOE Determination in SHIP
The APOE genotypes were determined on the basis of rs429358(C;C) and rs7412(T;T) from the resulting imputation of the SHIP genotyping. More details can be found in the On-line Appendix (On-line Method 3).
Statistical Analysis
We studied the association between age, the APOE ε4 genotype, and AD-related gray matter volume (lateral frontal, lateral temporal, medial frontal, and hippocampus), as well as SPARE-AD and SPARE-BA, for the 1472 SHIP participants included in this study. In this study, subjects with at least 1 ε4 allele were considered to have the APOE ε4 genotype.
We applied linear regression models, which included total intracranial volume normalized ROI volumes as outcomes, and age square (if significant), age, APOE ε4 carrier status, and sex as predictors adjusting for education level and study cohort. We also investigated the significance of interaction terms between the APOE ε4 status and the variables: age2, age, education, and sex, after adding those interaction terms one at a time to the regression models as predictors. Results were considered statistically significant if the 2-sided P value was <.05. No multiple comparison adjustment was applied because analyses were performed on a small number of predictors that were selected on the basis of previously published findings rather than determined on the basis of the data derived therein. To identify potentially cognitively impaired subjects on the basis of the residuals of linear regressions between age and the available cognitive scores, we labeled those subjects with a z-score of less than−1.5 SDs as possibly cognitively impaired (n = 98) (On-line Fig 1). Spearman rank correlation coefficients were used to assess correlations between variables. The Student t test was used to compare continuous variables, and the Pearson χ2 test, to compare categoric variables. Analyses were performed by using the R statistical and computing software, Version 3.1 (http://www.r-project.org/).28
Results
Subjects Included in the Study
A total of 1472 participants, from which 744 belonged to SHIP-2 and 728 belonged to SHIP-TREND, with ages ranging from 22 to 90 years (median, 53.3 years), were included in the analyses. Cohorts did not differ in sex, but significant differences were present with respect to education level (P < .001), age (P < .0001), and the number of APOE ε4 carriers (P = .03). No differences were seen between subjects included in the study and those who were excluded based on the inclusion criteria described in the “Materials and Methods” section, except for sex (On-line Table 1).
Association between the MR Imaging–Based Indices and Age
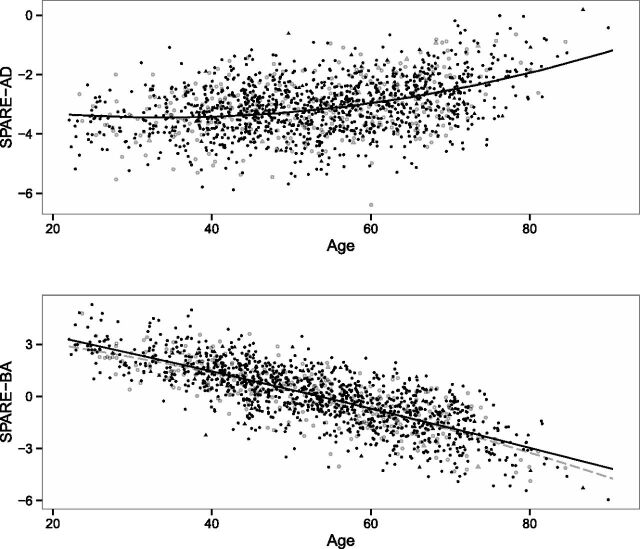
The Spearman rank correlation coefficient was r = 0.36 between age and SPARE-AD, and r = −0.77 between age and SPARE-BA (P < .001). The Spearman rank correlation coefficient between SPARE-AD and SPARE-BA was r = −0.44 (P < .001).
Association between APOE and MR Imaging Findings
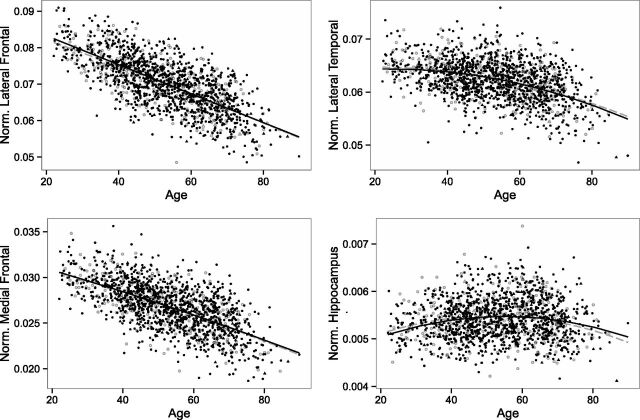
Figure 1 shows the relationship between the AD-related gray matter regions and age in APOE ε4 carrier and noncarrier individuals. Similarly, Fig 2 shows the relationship between the MR imaging–based indices and age. In the age- and sex-adjusted models that included the interaction term between APOE and age, no significant association was found between APOE ε4 carrier status and the studied MR imaging measurements in the regression models that included the ROI volumes or the SPARE indices as outcomes (Table 2). Similarly, there was no significant interaction between APOE ε4 carrier status and age (P > .1). In all models, older age was associated with greater brain atrophy, and a nonlinear square age term was significant for the models that included the lateral temporal regions, hippocampus, or SPARE-AD scores as outcomes (Table 2). Besides age, male participants showed smaller regional volumes, higher SPARE-AD, and lower SPARE-BA values over the studied life span. In additional analyses limiting the sample to younger (22–40 years) or older (60–90 years) participants, we did not find any APOE genotype–related differences (On-line Tables 2 and 3).
Fig 1.
Relationship between AD-related gray matter regions (lateral frontal, lateral temporal, medial frontal, and hippocampus; all regions were normalized by total intracranial volume) and age within APOE ε4 carrier (open objects, gray) and noncarrier (filled objects, black) SHIP individuals. Circles indicate cognitively healthy individuals, and triangles indicate cognitively impaired ones.
Fig 2.
Relationship between age and SPARE-AD and between age and SPARE-BA in APOE ε4 carrier (open objects, gray) and noncarrier (filled objects, black) individuals. Circles indicate cognitively healthy individuals, and triangles indicate cognitively impaired ones.
Table 2:
Linear regression models between age and ROI volumes (normalized by total intracranial volume) and SPARE-AD and SPARE-BAa
| Outcome | Age | Age2 | Female | APOE ϵ4 Carriers |
|---|---|---|---|---|
| Lateral frontal volume | −0.0003900 (<.001)b | – | 0.0021720 (<.001)b | −0.0002459 (.433) |
| Lateral temporal volume | 0.0001020 (.040)b | −0.0000021 (<.001)b | 0.0003695 (.040)b | 0.0001112 (.605) |
| Medial frontal volume | −0.0001291 (<.001)b | – | 0.0010690 (<.001)b | −0.0000055 (.964) |
| Hippocampal volume | 0.0000415 (<.001)b | −0.0000005 (<.001)b | 0.0001919 (<.001)b | 0.0000118 (.649) |
| SPARE-AD | −0.0438960 (<.001)b | 0.0006549 (<.001)b | −0.1775256 (<.001)b | 0.0293579 (.582) |
| SPARE-BA | −0.1062550 (<.001)b | – | 0.6416590 (<.001)b | −0.0514570 (.452) |
If age2 was not significant in a model, the coefficients were recalculated after excluding. Data are coefficient (P value).
Significant at P < .05. Models are adjusted for education and study cohort effects. Including interaction terms between APOE status and age2, age, sex, and education one at a time to those regression models showed no significant interaction (P > .1).
Association between APOE and Cognitive Scores
No significant association was detected among the Verbal Learning and Memory Test, Nuremberg Age Inventory, and APOE ε4 carrier status in a linear regression model after adjusting for age, sex, and education (On-line Tables 4 and 5).
Discussion
Although the APOE ε4 allele is the strongest genetic sporadic AD risk factor accounting for most of AD genetic-related risk and is associated with an earlier clinical onset of dementia and preclinical biomarker changes, we did not find brain volume differences between APOE ε4 carriers and noncarriers.
Here we studied the association between the APOE genotype and brain structure in a large cohort of subjects with a wide age range recruited from the general population whose MR imaging scans were obtained by using a standardized sequence in all subjects. We quantified structural brain changes by using 2 approaches: 1) an ROI approach, guided by previous findings; and 2) a pattern-based analysis by using 2 machine-learning-based summary indices. The imaging indices, the SPARE-AD and the SPARE-BA, were developed to quantify AD- and age-related brain changes, respectively. SPARE-AD has been shown to predict early AD-related changes23 and MCI stages.24 The SPARE-AD index outperformed ROI-defined volumes in detecting the earliest changes associated with initial cognitive symptoms and showed that 1 single brain volume measure alone is unlikely to adequately reflect the complexity of neurodegeneration in AD, as previously demonstrated in Fan et al.29 Therefore, it is unlikely that a lack of sensitivity of our approaches can explain the absence of associations between APOE and the structural MR imaging measures.
The difference in SPARE-AD scores of APOE ε4 carriers and noncarriers was statistically insignificant. Our results indicate that APOE-related MR imaging atrophy is not prominent at younger ages and suggest that atrophy may only be observable more proximal to the onset of clinical symptoms. Whereas older subjects with more severe degrees of cognitive impairment might be underrepresented in our population-based sample and thus lead to an underrepresentation of APOE ε4 carriers in the elderly (21.9% APOE ε4 carriers among the individuals older than 60 years of age), this underrepresentation would have not been the case for the younger subjects (27.0% APOE ε4 carriers among the individuals younger than 40 years of age). Therefore, we would not expect any recruitment bias to explain the lack of observed associations in the younger individuals included in our study.
Our findings agree with studies following cognitively healthy subjects with autosomal dominant mutations with an expected age at onset of disease, which report structural MR imaging changes taking place within ≤1 decade from the diagnosis of dementia.30,31 In addition, recent CSF and postmortem biomarker studies have shown that amyloid deposition starts in the fifth decade,3,32 usually without the individual having scans positive for amyloid. On the basis of the current AD biomarker33 model that predicts occurrence of amyloid β deposition before brain atrophy, a finding of brain atrophy in the third decade of life would be unlikely.
Previous studies have reported lower hippocampal, orbitofrontal, and entorhinal volumes in subjects with APOE ε4 in the early decades of life.12,13 The possible reasons for conflicting results could be differences in selection of the sample, the sample size, or processing pipelines of the MR imaging scans. Another potential explanation is that negative results are less likely to be reported than positive ones. Our study included a similar, or even larger, sample size than that in previous studies and applied sensitive methods to detect even subtle MR imaging changes associated with early AD-related changes and aging. However, it is possible that differences in functional changes associated with the APOE genotype at a younger age, as reported previously, precede structural changes14,15 and potentially even widespread amyloid β deposition, though this hypothesis still requires larger samples to validate initial reports based on small-sized cohorts.
In recent work, AD pathology from preclinical-to-clinical stages was associated with a decrease in the metabolism of the posterior cingulate cortex, even before any sign of AD.34 The appearance of hypometabolism before clinical change could vary depending on cognitive reserve. The results on cognitive reserve and APOE are controversial in the literature. However, APOE does not seem to modulate the effect of cognitive reserve on cognitive function.35 Due to the lack of functional imaging measures in our cohort, we could not evaluate the presence of these changes in the framework of SHIP.
While we did not find associations between APOE and brain volume measures, several studies reported the effects of the APOE genotype on white matter microstructure, which we did not assess in the current study. APOE was associated with white matter microstructure in 2 of 14 tracts in elderly persons free of dementia and preclinical dementia.36 Furthermore, APOE ε4 was associated with a decline in fractional anisotropy, a marker of white matter integrity, compared with noncarriers.37 On the other hand, APOE ε2 carriers had more robust white matter integrity.38 The mechanisms underlying the white matter microstructural changes are still not well-understood,39 but it is possible that detection of more subtle associations between APOE and brain structure in preclinical dementia requires microstructural assessment.
Limitations of our study include the use of imputed genotype data from single nucleotide polymorphism (SNP) arrays and not a specific assay for APOE40 to determine the APOE genotype, the limited clinical testing data available for the subjects, and the lack of longitudinal MR imaging scans.
Conclusions
Our study indicates that measurable APOE-related brain atrophy does not occur during early adulthood and midlife and suggests that APOE-related MR imaging atrophy may only be present later in life, more proximal to clinical disease onset.
Supplementary Material
Acknowledgments
SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg–West Pomerania. Genomewide data in SHIP and MR imaging scans in SHIP and SHIP-TREND have been supported by a joint grant from Siemens, Erlangen, Germany, and the Federal State of Mecklenburg–West Pomerania. Genomewide genotyping in SHIP-TREND-0 was supported by the Federal Ministry of Education and Research (grant No. 03ZIK012). M.H. was supported by the Alfried Krupp von Bohlen und Halbach Foundation. J.B.T. is supported by P01 AG032953, PO1 AG017586, P30 AG010124. and P50 NS053488. S.M.R. is supported by the Intramural Research Program, National Institute on Aging, National Institutes of Health.
ABBREVIATIONS:
- AD
Alzheimer disease
- APOE
apolipoprotein E
- MCI
mild cognitive impairment
- SHIP
Study of Health in Pomerania
- SNP
single nucleotide polymorphism
- SPARE-AD
Spatial Pattern of Abnormality for Recognition of Early Alzheimer Disease
- SPARE-BA
Spatial Pattern of Atrophy for Recognition of Brain Aging
Footnotes
Disclosures: Mohamad Habes—RELATED: Other: Supported by the Alfried Krupp von Bohlen und Halbach Foundation.* Jon B. Toledo—UNRELATED: Grants/Grants Pending: National Institute of Health grants.* Sandra Van der Auwera—RELATED: Grant: German Federal Ministry of Education and Research (grant No. 01ZX1314E).* Hans J. Grabe—UNRELATED: Grants/Grants Pending: German Research Foundation,* Grants from the German Ministry of Education and Research.* *Money paid to the institution.
The corresponding authors (M.H. and J.B.T.) confirm that each author has contributed to all of the following: drafting the article or revising it critically for important intellectual content, final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. In addition, each author's contribution was the following: M.H., J.B.T.: conception and design, acquisition of data, analysis and interpretation of data; S.M.R.: analysis and interpretation of data; J.D.: analysis and interpretation of data; S.V.d.A.: acquisition of data; G.E.: analysis and interpretation of data; D.J.: acquisition of data; K.H.: acquisition of data; G.H.: acquisition of data; H.V.: acquisition of data, analysis and interpretation of data; W.H.: acquisition of data, analysis and interpretation of data; and H.J.G. and C.D.: conception and design, acquisition of data, and analysis and interpretation of data.
References
- 1. Corder E, Saunders A, Strittmatter W, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993;261:921–23 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 2. Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 1993;90:1977–81 10.1073/pnas.90.5.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toledo JB, Zetterberg H, van Harten AC, et al. ; Alzheimer's Disease Neuroimaging Initiative. Alzheimer's disease cerebrospinal fluid biomarker in cognitively normal subjects. Brain J Neurol 2015;138:2701–15 10.1093/brain/awv199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jack CR, Wiste HJ, Weigand SD, et al. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: a cross-sectional study. Lancet Neurol 2014;13:997–1005 10.1016/S1474-4422(14)70194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keene CD, Cudaback E, Li X, et al. Apolipoprotein E isoforms and regulation of the innate immune response in brain of patients with Alzheimer's disease. Neurobiol Dis 2011;21:920–28 10.1016/j.conb.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zlokovic BV. Cerebrovascular effects of apolipoprotein E: implications for Alzheimer disease. JAMA Neurol 2013;70:440–44 10.1001/jamaneurol.2013.2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verghese PB, Castellano JM, Holtzman DM. Apolipoprotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol 2011;10:241–52 10.1016/S1474-4422(10)70325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mungas D, Tractenberg R, Schneider JA, et al. A 2-process model for neuropathology of Alzheimer's disease. Neurobiol Aging 2014;35:301–08 10.1016/j.neurobiolaging.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toledo J, Da X, Weiner M, et al. ; Alzheimer's Disease Neuroimaging Initiative. CSF Apo-E levels associate with cognitive decline and MRI changes. Acta Neuropathol (Berl) 2014;127:621–32 10.1007/s00401-013-1236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martínez-Morillo E, Hansson O, Atagi Y, et al. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer's disease patients and controls. Acta Neuropathol (Berl) 2014;127:633–43 10.1007/s00401-014-1266-2 [DOI] [PubMed] [Google Scholar]
- 11. Rasmussen KL, Tybjærg-Hansen A, Nordestgaard BG, et al. Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann Neurol 2015;77:301–11 10.1002/ana.24326 [DOI] [PubMed] [Google Scholar]
- 12. Shaw P, Lerch JP, Pruessner JC, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol 2007;6:494–500 10.1016/S1474-4422(07)70106-0 [DOI] [PubMed] [Google Scholar]
- 13. O'Dwyer L, Lamberton F, Matura S, et al. Reduced hippocampal volume in healthy young ApoE4 carriers: an MRI study. PLoS One 2012;7:e48895 10.1371/journal.pone.0048895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reiman EM, Chen K, Alexander GE, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A 2004;101:284–89 10.1073/pnas.2635903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filippini N, MacIntosh BJ, Hough MG, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A 2009;106:7209–14 10.1073/pnas.0811879106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Völzke H, Alte D, Schmidt CO, et al. Cohort profile: the Study of Health in Pomerania. Int J Epidemiol 2011;40:294–307 10.1093/ije/dyp394 [DOI] [PubMed] [Google Scholar]
- 17. Hegenscheid K, Kühn JP, Völzke H, et al. Whole-body magnetic resonance imaging of healthy volunteers: pilot study results from the population-based SHIP study. Rofo 2009;181:748–59 10.1055/s-0028-1109510 [DOI] [PubMed] [Google Scholar]
- 18. Woods SP, Delis DC, Scott JC, et al. The California Verbal Learning Test–second edition: test-retest reliability, practice effects, and reliable change indices for the standard and alternate forms. Arch Clin Neuropsychol 2006;21:413–20 10.1016/j.acn.2006.06.002 [DOI] [PubMed] [Google Scholar]
- 19. Fleischmann UM, Oswald WD. Nürnberger-Alters-Inventar (NAI); NAI-Testmanual und -Textband. Göttingen: Hogrefe, Verlag für Psychologie; 1999 [Google Scholar]
- 20. Doshi J, Erus G, Ou Y, et al. Multi-atlas skull-stripping. Acad Radiol 2013;20:1566–76 10.1016/j.acra.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doshi J, Erus G, Ou Y, et al. ; Alzheimer's Neuroimaging Initiative. MUSE: multi-atlas region segmentation utilizing ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 2016;127:186–95 10.1016/j.neuroimage.2015.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan Y, Shen D, Gur RC, et al. COMPARE: Classification of Morphological Patterns Using Adaptive Regional Elements. Med Imaging IEEE Trans 2007;26:93–105 10.1109/TMI.2006.886812 [DOI] [PubMed] [Google Scholar]
- 23. Toledo J, Weiner M, Wolk D, et al. ; Alzheimer's Disease Neuroimaging Initiative. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta Neuropathol Commun 2014;2:26 10.1186/2051-5960-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Da X, Toledo JB, Zee J, et al. ; Alzheimer's Neuroimaging Initiative. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin 2014;4:164–73 10.1016/j.nicl.2013.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vapnik VN. The Nature of Statistical Learning Theory. New York: Springer-Verlag; 2000 [Google Scholar]
- 26. Habes M, Erus G, Toledo JB, et al. White matter hyperintensities and imaging patterns of brain ageing in the general population. Brain 2016;139(pt 4):1164–79 10.1093/brain/aww008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Habes M, Janowitz D, Erus G, et al. Advanced brain aging: relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Transl Psychiatry 2016;6:e775 10.1038/tp.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R Core Team R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2014 [Google Scholar]
- 29. Fan Y, Batmanghelich N, Clark CM, et al. ; Alzheimer's Disease Neuroimaging Initiative. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage 2007;39:1731–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bateman RJ, Xiong C, Benzinger TLS, et al.; Dominantly Inherited Alzheimer Network. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med 2012;367:795–804 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villemagne VL, Burnham S, Bourgeat P, et al. ; Australian Imaging Biomarkers and Lifestyle (AIBL) Research Group. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol 2013;12:357–67 10.1016/S1474-4422(13)70044-9 [DOI] [PubMed] [Google Scholar]
- 32. Pletnikova O, Rudow GL, Hyde TM, et al. Alzheimer lesions in the autopsied brains of people 30 to 50 years of age. Cogn Behav Neurol 2015;28:144–52 10.1097/WNN.0000000000000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jack CR Jr, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013;12:207–16 10.1016/S1474-4422(12)70291-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teipel S, Grothe MJ; Alzheimer's Disease Neuroimaging Initiative. Does posterior cingulate hypometabolism result from disconnection or local pathology across preclinical and clinical stages of Alzheimer's disease? Eur J Nucl Med Mol Imaging 2016;43:526–36 10.1007/s00259-015-3222-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward DD, Summers MJ, Saunders NL, et al. The BDNF Val66Met polymorphism moderates the relationship between cognitive reserve and executive function. Transl Psychiatry 2015;5:e590 10.1038/tp.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laukka EJ, Lövdén M, Kalpouzos G, et al. Microstructural white matter properties mediate the association between APOE and perceptual speed in very old persons without dementia. PLoS One 2015;10:e0134766 10.1371/journal.pone.0134766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Persson J, Lind J, Larsson A, et al. Altered brain white matter integrity in healthy carriers of the APOE epsilon4 allele: a risk for AD? Neurology 2006;66:1029–33 10.1212/01.wnl.0000204180.25361.48 [DOI] [PubMed] [Google Scholar]
- 38. Chiang GC, Zhan W, Schuff N, et al. White matter alterations in cognitively normal apoE ϵ2 carriers: insight into Alzheimer resistance? AJNR Am J Neuroradiol 2012;33:1392–7 10.3174/ajnr.A2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gold BT, Johnson NF, Powell DK, et al. White matter integrity and vulnerability to Alzheimer's disease: preliminary findings and future directions. Biochim Biophys Acta 2012;1822:416–22 10.1016/j.bbadis.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saykin AJ, Shen L, Foroud TM, et al. ; Alzheimer's Disease Neuroimaging Initiative. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement 2010;6:265–73 10.1016/j.jalz.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.