Abstract
Neomorphic mutations in isocitrate dehydrogenase 1 (IDH1) are driver mutations in acute myeloid leukemia (AML) and other cancers. We report the development of new allosteric inhibitors of mutant IDH1. Crystallographic and biochemical results demonstrated that compounds of this chemical series bind to an allosteric site and lock the enzyme in a catalytically inactive conformation, thereby enabling inhibition of different clinically relevant IDH1 mutants. Treatment of IDH1 mutant primary AML cells uniformly led to a decrease in intracellular 2-HG, abrogation of the myeloid differentiation block and induction of granulocytic differentiation at the level of leukemic blasts and more immature stem-like cells, in vitro and in vivo. Molecularly, treatment with the inhibitors led to a reversal of the DNA cytosine hypermethylation patterns caused by mutant IDH1 in AML patients’ cells. Our study provides proof-of-concept for the molecular and biological activity of novel allosteric inhibitors for targeting different mutant forms of IDH1 in leukemia.
Graphical Abstract
Introduction
Isocitrate dehydrogenase (IDH) catalyzes the conversion of isocitrate to α-ketoglutarate in the Krebs (citric acid) cycle. This enzymatic pathway is associated with various molecular processes regulating the cellular epigenetic state, including histone and DNA modifications. Point mutations affecting IDH1(R)-132 and IDH2(R)-172 or (R)-140 are observed in a variety of cancers including low-grade gliomas, secondary glioblastomas, melanoma, angioimmunoblastic T-cell lymphomas, myeloproliferative neoplasms, myelodysplastic syndrome (MDS), and acute myeloid leukemia (AML) 1–5. Mutations in IDH1 or IDH2 are mutually exclusive in AML, and lead to the loss of the enzyme’s normal catalytic activity with concomitant gain of a neomorphic function that promotes the overproduction of the R-enantiomer of 2-hydroxyglutarate (2-HG) and reduction of α-ketoglutarate (α-KG) 4,6. It has been shown that IDH1 mutations result in global DNA and histone hypermethylation partly through disruption of ten eleven translocation 2 (TET2) function as well as the accumulation of 2-HG and by this mechanism promote a block in cellular differentiation and progression to leukemia 7–12. Recent studies have revealed that the mutant IDH1 protein and its oncometabolite R-2-HG lead to cytokine independent growth and a block in erythropoietin-induced differentiation of the TF-1 erythroleukemia cell line 13,14.
Patient outcomes in AML remain poor, especially for patients above 60 years of age who represent the vast majority 15,16. Cure rates in these patients, who typically do not tolerate high-dose chemotherapy and stem cell transplantation, are below 20% and have not been improved significantly in the past 4 decades. Hence novel, targeted therapies are urgently needed. Approximately 10–40% of normal karyotype AML patients have mutations in IDH1 or IDH2, making these proteins obvious therapeutic targets in AML 17,18. Recently, inhibition of mutant IDH1 was shown to have preclinical efficacy in IDH1 mutant glioma cells 19, and inhibitors of mutant IDH2 have been developed for differentiation therapy of IDH2 mutant AML and have shown promising preclinical and early phase clinical activity 20,21. First generation inhibitors of mutant IDH1 are in preclinical and clinical testing, however, the specific molecular and functional effects of IDH1 inhibitors in AML, including in primary patients’ cells, have not been reported yet.
In the current study, we report novel, allosteric inhibitors of mutant IDH1 and examine the biophysical, biochemical, cell biological, and molecular mechanisms by which they exert anti-leukemic effects in IDH1 mutant AML patient’s cells both ex vivo and in vivo.
Results
Identification, biochemical, structural characterization
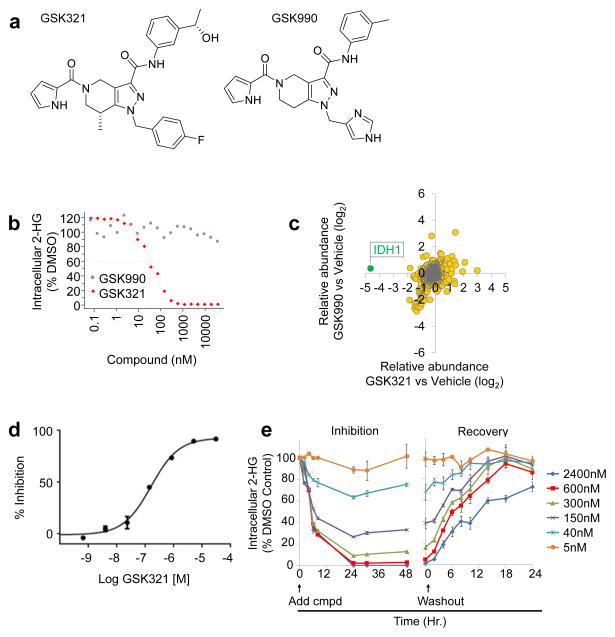
A high-throughput biochemical screen (Online methods) targeting an IDH1 heterodimer composed of R132H and wild-type (WT) IDH1 led to the identification of a tetrahydropyrazolopyridine (THPP) series of inhibitors. Biochemical potency optimization revealed preferences for meta-substitution on the aniline ring and substitution of the THPP core with groups such as an (R)-7-methyl (Supplementary Results, Supplementary Table 1 and 2). Additional optimization of the aniline moiety led to the identification of GSK321 (Fig. 1a, Supplementary Notes), a highly potent inhibitor of mutant IDH1 enzymes, with IC50 values of 4.6 nM against R132H, 3.8 nM against R132C, and 2.9 nM against R132G. GSK321 showed modest activity against WT IDH1 (46 nM) and exhibited minimal to no inhibitory activity against IDH2 proteins in biochemical and functional assays (Fig. 1b). Biochemical potencies were comparable whether measured by 2-HG production or NADPH oxidation (Supplementary Fig. 1a). A structurally related cell-permeable analog (as determined by artificial membrane and Caco-2 permeability assays (Online methods and Supplementary Table 3), GSK990 was utilized as an IDH1 inactive inhibitor (Fig. 1a and Table 1).
Figure 1. Identification of novel allosteric inhibitors of mutant IDH1.
(a) Chemical structures of GSK321, an active IDH1 mutant inhibitor, and GSK990, a structurally-related inactive inhibitor. (b) Potent inhibition of IDH1 mutant enzyme leads to decreased production of intracellular R-2 hydroxyglutarate (2-HG) in IDH1 R132C mutant HT-1080 cells after treatment with IDH1 GSK321 for 24 hours. Absence of inhibition of IDH1 mutant enzymatic activity using GSK990. Half-maximal effective concentration (EC50) values for decreased intracellular 2-HG levels are indicated. Graphs are representative of 2 to 8 independent experiments. (c) GSK321 selectively interacts with IDH1. A GSK321 derivative was immobilized to sepharose beads and used to pull-down cellular target proteins from HT-1080 cell lysate. Mass spectrometry was utilized to identify peptides from proteins bound by the IDH1 inhibitor matrix. A comparison of peptide enrichment observed in the presence of vehicle or competing compound demonstrated that GSK321 uniquely competes IDH1 from the matrix. (d) Dose response curve showing GSK321 binding affinity for IDH1 in HT-1080 cell lysate. Binding of IDH1 to the IDH1 inhibitor matrix was assessed in the presence of increasing concentrations of free GSK321 to determine the percent inhibition. Error bars indicate mean ± S.D. for 2 experimental replicates. (e) GSK321 inhibits 2-HG production within 24 hours and is rapidly reversible upon compound washout. Data presented as mean ± standard error (N = 4).
Table 1.
Biochemical and cellular IC50 data in nM for GSK321 and GSK990.
| GSK321 (Active inhibitor) | ||||
| IDH1 IC50 (nM) | ||||
| R132H | R132C | R132G | WT | HT1080 cell 2-HG |
| 4.6a | 3.8a | 2.9a | 46 | 85 |
| IDH2 IC50 (nM) | ||||
| R140Q | R172S | WT | R172 U87MG | SW1353 cell 2-HG |
| 1358 | 1034 | 496 | 7940 | 4689 |
| GSK990 (Inactive inhibitor) | ||||
| IDH1 IC50 (nM) | ||||
| R132H | R132C | R132G | WT | HT1080 cell 2-HG |
| 10915 | 4495 | 23255 | >100000 | >36000 |
| IDH2 IC50 (nM) | ||||
| R140Q | R172S | WT | R172 U87MG | SW1353 cell 2-HG |
| >100000 | >100000 | >100000 | ND | ND |
Determined using tight binding equation as described in methods
ND, Not determined due to lack of biochemical inhibition
Mechanistic cellular activity of GSK321 and GSK990 was evaluated in HT1080 fibrosarcoma cells which harbor an R132C IDH1 mutation and have markedly elevated levels of 2-hydroxyglutarate (2-HG). After 24 h compound treatment, GSK321 potently inhibited intracellular 2-HG production in HT1080 cells with a half maximal effective concentration (EC50) of 85 nM by LC-MS/MS analysis (Fig. 1b). No changes in intracellular 2-HG were observed with GSK990. Furthermore, GSK321 was evaluated for selectivity using a chemoproteomic approach capable of identifying proteins that interact with a compound in the context of a cellular lysate, which demonstrated high selectivity for IDH1 (Fig. 1c,d and Supplementary Data Set 1). In addition, examination of the temporal kinetics of 2-HG inhibition demonstrated that 2-HG levels declined rapidly following addition of compound and maximal inhibition was achieved by 24 hours (Fig. 1e). The effect of the compound was found to be reversible as 2-HG levels recovered to within 20% of the pre-treatment levels only 14 hours after compound washout (Fig. 1e).
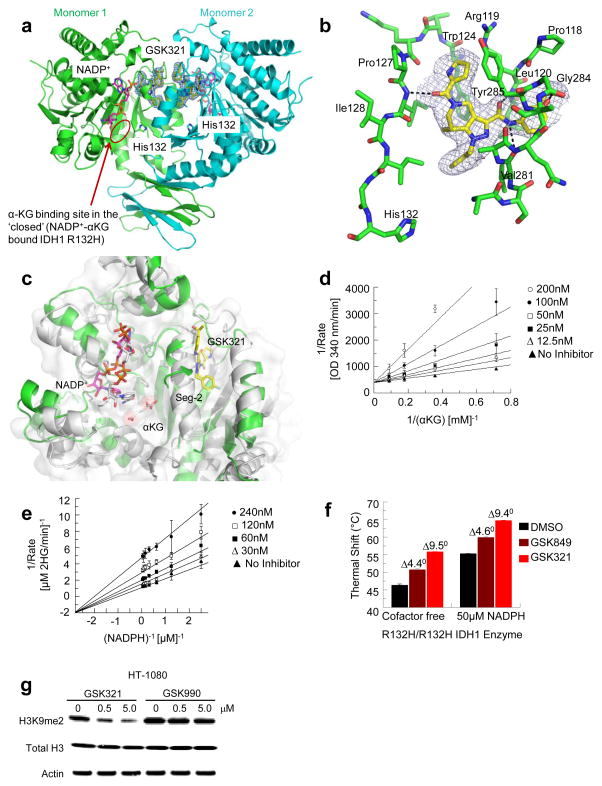
Next, a crystal structure of GSK321 bound to R132H IDH1 homodimer in the presence of NADP+ was obtained at 2.25 Å, with density found for both inhibitor and cofactor binding to both subunits (Fig. 2a and Supplementary Table 4). GSK321 binds to an allosteric pocket formed when the human IDH1 enzyme is in an open, cofactor-bound form 22,23. The inhibitor’s 4-fluorophenyl group resides favorably in a lipophilic pocket (Fig. 2a). In contrast, GSK990 contains a polar imidazole ring which may clash with the lipophilic residues that line this pocket and contribute to its poor IDH1 inhibitor potency. In the GSK321-IDH1 complex, the inhibitor does not make contact with NADP+ or with the mutated residue at position 132 (His132). The ability of GSK321 to potently inhibit several different, clinically relevant R132 mutants is consistent with this allosteric binding mode (Fig. 2a and 1b). One molecule of GSK321 binds to each of the IDH1 R132H monomers in a pocket lined on three sides by Ile128, Pro127, Trp124, Arg119 and Leu120 (Fig. 2b). The remaining side is formed by residues (e.g. Val281, Gly284 and Tyr285) in a dynamic segment of the polypeptide chain referred to as Seg-2. Seg-2 acquires a helical conformation in the closed IDH1-NADP+-αKG ternary complex but is mostly disordered in the open IDH1-NADP+ binary complex suggesting that it undergoes a loop to helix transition during the catalytic cycle. Though Seg-2 is disordered in the binary complex 24, it acquires a partial helical structure in the ternary complex upon interacting with GSK321 (Fig. 2c).
Figure 2. Structural and biochemical characterization.
(a) Crystal structure of GSK321 bound to the R132H IDH1 homodimer. GSK321 (yellow) is bound in an allosteric pocket in both monomers of the IDH1 R132H dimer. NADP+ (magenta) and His132 are also shown. (b) Detailed view of the allosteric binding pocket for GSK321. GSK321 (yellow) is bound mainly through H-bonds to the backbone of IDH1 (green). (c) Overlay of one monomer of the IDH1 R132H-NADP+ binary complex (open form, green) bound to GSK321 (yellow) and the IDH1 R132H-NADP+-Ca2+/αKG ternary complex (closed form, grey). The inhibitor is wedged next to Seg-2 preventing its full organization into the active enzyme conformation. (d) and (e) Biochemical MOI of GSK849 Competitive inhibition is observed between GSK849 and αKG (Vmax = 20 ± 1 min−1; KαKG = 2.2 ± 0.4 mM; GSK849 Ki = 31 ± 5.2 nM) while mixed/noncompetitive inhibition is observed betweenGSK849) and NADPH (Vmax = 62 ± 1.5 min−1; KNADPH = 1.0 ± 0.10 μM; GSK849 Kis = 205 ± 102 nM and GSK Kii = 70 ± 5.0 nM). (f) Thermal stabilization data for GSK849 and GSK321. Binding was observed with either cofactor free or NADPH saturated enzyme (mean and S.D. are shown for a total of N = 6 replicates). (g) GSK321, but not GSK990, leads to reduction of histone H3K9 dimethylation (H3K9me2). Representative gel depicted of N = 6 total replicates. R132C IDH1 expressing HT-1080 cells were treated for 48 hours with either GSK321 or GSK990. Total H3 and Actin are shown as loading controls (see full gel images in Supplementary Fig. 1b).
To determine the mechanism of inhibition (MOI) of this inhibitor scaffold, we utilized a slightly weaker analog of the same chemical series, GSK849, to avoid complications that exist when attempting to determine MOI for the inhibitors with Ki values below the enzyme concentration of the assay (Supplementary Table 2). Kinetically, GSK849 displays a competitive mode of inhibition versus αKG despite not binding in the same pocket as the substrate (Fig. 2d). This can be attributed to the interaction of the inhibitor with Seg-2, which precludes the loop-to-helix transition required for turnover. GSK849 displays a mixed/non-competitive mode of inhibition versus NADPH (Fig. 2e). Previous studies revealed that mutant IDH1 employs an ordered kinetic mechanism, with NADPH binding preceding that of alpha-ketoglurate (αKG) 25. While orthosteric inhibitors, such as N-oxalyl glycine, have been shown to display an uncompetitive pattern of inhibition versus NADPH due to the obligatory binding order, the mixed/non-competitive pattern we observed for GSK849 is consistent with its allosteric nature where multiple MOIs are possible 26. This MOI was further confirmed by thermal shift analysis of cofactor depleted R132H as we have previously described 25. A lower Tm was observed for the NADPH-free form of recombinant human IDH1 R132H compared to the protein incubated with excess, saturating NADPH (50 μM). However, a similar positive thermal shift (ΔTm) was observed for binding of THPP compounds GSK321 and GSK849 to IDH1 R132H both in the absence and presence of NADPH, which demonstrated that both inhibitors can bind to both cofactor free and NADPH saturated enzyme (Fig. 2f). Finally, since it is known that elevated 2-HG levels can inhibit αKG dependent enzymes such as Jmj histone demethylases, we evaluated the effect of GSK321 and GSK990 on histone H3K9me2 in R132C IDH1 mutant expressing HT1080 fibrosarcoma cells. As expected, within 48 hours of treatment, GSK321 induced markedly decreased H3K9me2 levels (Fig. 2g and Supplementary Fig. 1b). Together these studies demonstrated that GSK321, but not GSK990, interacted uniquely with IDH1. Hence GSK321 was selected for further studies based on its potency and selectivity, to elucidate its biochemical mechanism of action and biological consequences in primary IDH1 mutant cells from patients with AML.
Cellbiologic effects of GSK321 in primary IDH1 mutant AML
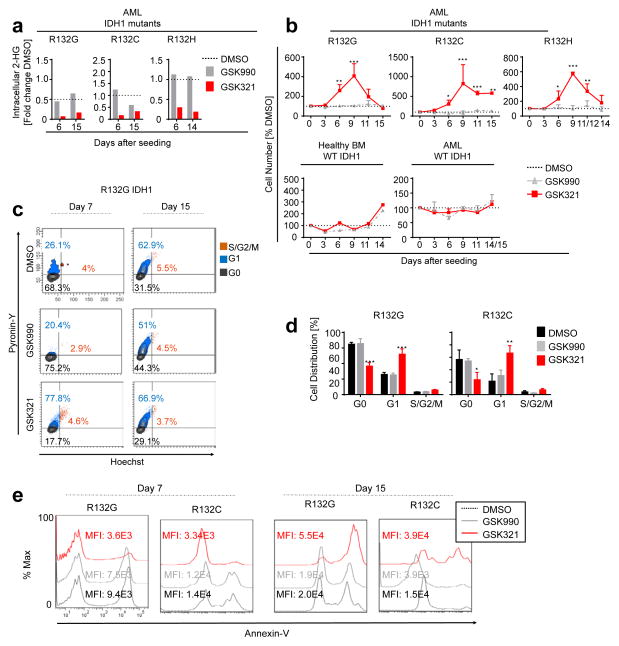
We treated R132G IDH1 AML cells ex vivo with increasing concentrations of GSK321 IDH1 mutant inhibitor, GSK990 inactive inhibitor, or 0.3% DMSO as a vehicle control (Supplementary Fig. 1d). We observed a concentration-dependent decrease in intracellular 2-HG levels with 78% inhibition at a concentration of 1.7 μM GSK321. GSK990 showed only modest inhibitory activity at concentrations greater than 5.1 μM. Based on these observations, we treated IDH1 wild type (WT), R132G, R132C and R132H IDH1 mutant AML and bone marrow (BM) cells from healthy donors with 3 μM GSK321 or GSK990. Following 6 days of treatment in suspension culture, we observed a significant decrease in intracellular 2-HG with GSK321 (R132G, 0.13-fold ± 0.1-fold; R132C, 0.15-fold ± 0.2-fold; R132H, 0.29-fold), in contrast to cells treated with either DMSO or GSK990 (Fig. 3a). Stable inhibition of intracellular 2-HG was maintained after 14 to 15 days (Fig. 3a), and up to 22 days after treatment in suspension cultures (Supplementary Fig. 1e).
Figure 3. GSK321 decreases intracellular 2-HG and affects proliferation of primary IDH1 mutant AML cells.
(a to e) PB or BM cells from IDH1 mutant or WT AML patients were cultured in the presence of 0.3% DMSO or 3μM GSK990 or GSK321 for indicated time points. (a) Reduction of intracellular 2-HG following the treatment of IDH1 mutant AML cells with GSK321 or GSK990 compared to DMSO treated cells. Shown are mean values for 2 biological replicates. 2-HG levels are normalized to total cell numbers and expressed as fold change relative to the DMSO control. P-values were determined by unpaired student’s t-test. (b) Transient increase in absolute cell numbers after treatment of IDH1 mutant AML cells with GSK321. Cell numbers are displayed relative to DMSO treated cells (dotted lines). Error bars depict mean ± S.E.M. (N = 3 independent biological samples each for R132G, R132C and R132H, and 2 WT IDH1 biological AML samples in technical duplicates (total N=4); N = 2 technical replicates for healthy BM WT IDH1. P-values were determined using two-way ANOVA. (c) Representative FACS plots of cell cycle analysis of R132G IDH1 AML cells after treatment in suspension culture for 7 and 15 days respectively. (d) Quantification of cell cycle distribution. Hoechst and Pyronin-Y staining were performed. Total N=3 for 2 independent biological samples each for R132G and R132C. P-values were calculated using two-way ANOVA. (e) Evaluation of cell death by Annexin-V staining of IDH1 mutant AML patients’ cells treated for indicated time points. Representative FACS plots are depicted for each mutant. [*p<0.05; **p<0.01; ***p<0.001]
Interestingly, we observed a significant, initial increase in cell numbers (2-fold to 15-fold) in IDH1 mutant AML cells in the presence of GSK321, whereas no such effect could be observed with either GSK990 or DMSO treated cells, or following treatment of healthy donor or AML cells with WT IDH1 (Fig. 3b). This effect was only transient, peaking at day 9 after initiation of treatment, and reverting to control levels after 14 or 15 days. After 7 days of treatment with GSK321, we observed a reproducible and significant decrease in quiescent (G0)-phase cells in R132G IDH1 (36.7 ± 4.4% GSK321 vs. 65.7 ± 6.5% GSK990 vs. 64.9 ± 2.4% DMSO) and R132C IDH1 (24.0 ± 11.8% GSK321 vs. 54.1 ± 3.8% GSK990 vs. 56.6 ± 15.3% DMSO) AML cells (Fig. 3c,d). Additionally, there was a significant increase in cells in G1-phase in R132G IDH1 (57.9 ± 6.9% GSK321 vs. 25.3 ± 2.5% GSK990 vs. 27.0 ± 2.6% DMSO) and R132C IDH1 (66.8 ± 12.0% GSK321 vs. 30.6 ± 8.9% GSK990 vs. 21.6 ± 11.8% DMSO). Similar cell cycle changes were observed in R132H IDH1 mutant AML cells after treatment for 6 days in suspension culture (Supplementary Fig. 2a). By the 15th day of treatment, no notable differences in cell cycling were observed (Fig. 3c). Consistent with these results, an initial increase of viable (Annexin-V−) cells in R132G and R132C IDH1 mutant AML cells after 7 days of treatment with GSK321 was followed by decreased viability and increased cell death after 15 days (Fig. 3e).
Differentiation of IDH1 mutant blasts and stem-like cells
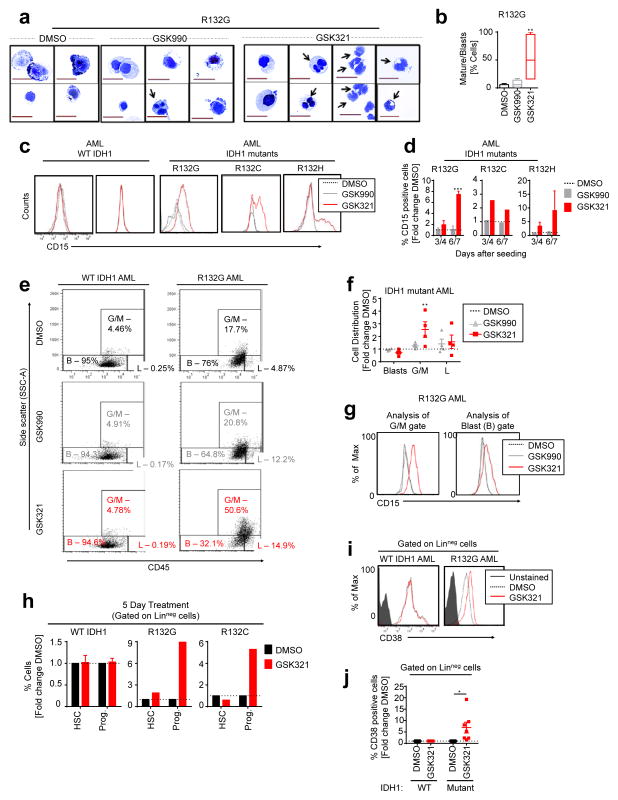
AML cells are hallmarked by a block in differentiation and a lack of responsiveness to cytokines, resulting in reduced suspension culture growth under differentiation-inducing conditions in vitro. We therefore determined the effects of treatment with GSK321 or controls (DMSO or GSK990) on cellular differentiation. In GSK321-treated IDH1 mutant AML cells, we observed morphological changes consistent with granulocytic differentiation, with an increase in cells with indented and segmented nuclei (Fig. 4a). Quantification of cells with signs of maturation in comparison to immature blasts revealed a highly significant increase in differentiating cells upon GSK321 treatment in comparison to either control (Fig. 4b). No morphological changes suggestive of granulocytic differentiation were observed in WT IDH1 AML cells treated with DMSO, GSK990, or GSK321 (Supplementary Fig. 2b).
Figure 4. Induction of differentiation in primary IDH1 mutant AML blasts and immature stem-like cells.
(a to l) Treatment of primary IDH1 mutant or IDH1 WT AML BM or PB mononuclear (MNC) cells in differentiating media in the presence of 0.3% DMSO or 3μM GSK990 or GSK321 after indicated times. (a) Cytospins showing signs of granulocytic maturation (black arrows) after 9 days of treatment. 63X magnification, scale bars 20 μm. (b) Quantification of myeloid differentiation after 9 days of treatment of R132G IDH1 mutant AML (Box = 25th–75th percentiles; whiskers = min and max values; p-value determined by one-way ANOVA Friedman Test). (c) FACS plots of CD15 expression after 7 days of treatment. (d) Increase in the number of CD15+ cells in IDH1 mutant AML cells with GSK321 treatment compared to DMSO or GSK990 controls; expressed as fold change of DMSO after 3–4 and 6–7 days, respectively. (N=3 R132G, N=2 R132C, and N=3 R132H independent biological samples). (e) Representative CD45/SSC FACS dot plots with 3 gates (G/M [Granulocytes/monocytes – SSChigh CD45+], L [Lymphocytes -SSClow CD45high] and B [Blasts -SSClow CD45low/+]) after 7 days of treatment. (f) Quantification of cell distribution in blast, G/M and L gates as displayed in (e). Each dot represents an independent biological sample (N = 4) (p-value determined using 2-way ANOVA). (g) Histogram plots of CD15 expression on cells within the blast and G/M gates in (e). (h) FACS analysis of immature hematopoietic stem cell (HSC) and progenitor (Prog.) cell compartments. Error bars depict mean ± S.D. (N = 3 independent WT IDH1 biological samples). (i) Representative FACS plots of CD38 expression within lineage-negative (Linneg) cells, after 5 days of treatment. (j) Percentages of CD38 expressing cells within the Linneg gate are quantified as fold change relative to DMSO (unpaired Student’s t-test). Each dot represents an independent biological sample (N = 3 WT AML; and 7 IDH1 mutant AML). [*p<0.05; **p<0.01; ***p<0.001]
Induction of granulocytic differentiation was confirmed by flow cytometry for expression of the granulocytic marker CD15 27 (Fig. 4c,d). We observed a consistent increase of CD15-positive cells (Fig. 4c) in R132G, R132C and R132G IDH1 mutants after 7 days of treatment with GSK321. Quantification of the percentage of CD15 expressing differentiating cells identified a significant increase following 6 to 7 days of treatment with GSK321 (7.4 ± 0.4-fold R132G, 1.9 ± 0.6-fold R132C, 9.0 ± 7.2-fold R132H) compared to GSK990 treated cells (1.1 ± 0.4-fold R132G, 0.92 ± 0.4-fold R132C, 1.2 ± 0.3-fold R132H) (Fig. 4d). To further distinguish the residual bulk leukemic cells from differentiated cells within the culture, we quantified leukemic blast cells (B; CD45low/+ SSClow cells) by flow cytometric analysis as previously described 28. After 7 days of treatment of R132G IDH1 AML cells with GSK321, in comparison to DMSO treated cells, we observed a decrease in the fraction of leukemic blast cells (0.7 ± 0.1-fold GSK321 vs. 1.0 ± 0.04-fold GSK990) and a significant increase in the granulocytic/monocytic (G/M; SSChigh CD45+) cell fraction (2.5 ± 0.6-fold GSK321 vs. 1.3 ± 0.1-fold GSK990) (Figure 4e,f). Of note, we found upregulation of CD15 in both blast and G/M cell fractions upon treatment of IDH1 mutant AML cells with GSK321 (Fig. 4g). In contrast, no changes in either blast or G/M fractions were observed in DMSO, GSK990 or GSK321 treated WT IDH1 AML cells (Fig. 4e). Additional analysis including HLA-DR and c-Kit markers showed a significant decrease in the leukemic blasts following treatment with GSK321 (0.4 ± 0.2-fold) vs. GSK990 (1.5 ± 0.4-fold) (Supplementary Fig. 2d) 29. Similarly, evaluation of the G/M and blast (B) cell fractions for expression of an alternative myeloid differentiation marker, CD33, revealed specific increase in the percentage of CD33 expressing cells within the G/M gate for GSK321 treated cells (Supplementary Fig. 2e).
Next, we examined the effect of GSK321 at the level of more immature cells including phenotypic hematopoietic stem cells (HSC; Linneg CD34+ CD38−) and progenitor cells (Prog.; Linneg CD34+ CD38+) (Fig. 4h). Analysis of WT IDH1 cells revealed no significant changes in either the HSC or progenitor cell compartments after 5 days of treatment. Interestingly, we observed a consistent and highly significant increase in the percentage of phenotypic progenitor cells upon treatment of R132G and R132C IDH1 mutant AML cells with GSK321 (Fig. 4h). Next, we specifically evaluated linneg IDH1 mutant versus WT AML cells for surface expression of CD38 protein, a marker of cell cycling and differentiation at the stem/progenitor cell level, and observed a significant increase in CD38+ cells (Fig. 4i,j). Of note, analysis of CD38 positive cells after 6 and 18 days of treatment in suspension culture showed a significant decrease in the CD38 positive cell fraction at later time points (Supplementary Fig. 2f).
Finally, we plated primary bone marrow (BM) cells in methylcellulose containing human recombinant cytokines, followed by treatment with DMSO, GSK990, or GSK321. Treatment of IDH1 mutant AML cells with GSK321 in semi-solid media for 10–12 days led to decreased intracellular 2-HG (Supplementary Fig. 2g) and increase in more mature G/M colonies (Supplementary Fig. 2h). To distinguish the contribution of IDH1-mutant versus residual normal cells to colony initiation, we picked individual colonies and performed Sanger sequencing. DNA sequencing revealed a significant enrichment for IDH1 mutant allele-positive clones upon treatment with GSK321 (Supplementary Fig. 2i), indicating that inhibition of mutant IDH1 restores cytokine responsiveness and colony-forming capacity of IDH1 mutant AML cells. Similar observations were made when we plated FACS-sorted LinnegCD34+CD38− IDH1 mutant AML cells, again demonstrating that GSK321 is effective in immature stem-like cells (Supplementary Fig. 2j). Additionally, Sanger sequencing of unfractionated cells from IDH1 mutant AML patients grown in suspension culture revealed that IDH1 mutant AML cells regained their cytokine responsiveness and competitive growth in the presence of co-existing wild type cells in ex vivo cultures upon GSK321 treatment, while DMSO or GSK990 treatment led to more rapid disappearance of IDH1-mutant positive cells from the culture by day 14 (Supplementary Fig. 2k). Taken together, these findings show that inhibition of mutant IDH1 by GSK321 overcomes the pathognomonic differentiation block of AML cells, and induces myeloid differentiation of IDH1 mutant cells at the level of leukemic blasts and more stem-like cells.
GSK IDH1 inhibitor reduces leukemic blasts in vivo
In order to explore the effects of IDH1 inhibition in vivo, a compound structurally related to GSK321 and referred to as GSK864 was developed with improved pharmacokinetic properties (Supplementary Fig. 3a and Supplementary Table 2). Similar to GSK321, GSK864 inhibited 2-HG production in R132C IDH1 mutant HT1080 fibrosarcoma cells with an EC50 of 320 nM by LCMS/MS analysis (Supplementary Fig. 3b,c). Following intraperitoneal (IP) administration in CD-1 mice, significant concentrations of GSK864 were maintained in peripheral blood samples of mice for up to 24 hours (Supplementary Fig. 3d,e).
We utilized GSK864 to assess the effects of IDH1 inhibition on primary IDH1-mutant human AML cells in an in vivo xenografting model. IDH1 WT or mutant AML patient BM cells were transplanted into sublethally irradiated NSG mice (Supplementary Fig. 4a), and comparable levels of AML engraftment were observed in BM aspirates 3 weeks after transplantation, and prior to treatment with either vehicle or 150 mg/kg GSK864. The percentage of total human (CD45+) engrafted cells was not significantly different pre-and post-treatment in the GSK864 versus the vehicle group (Supplementary Fig. 4b). Following treatment we observed a decrease in 2HG in IDH1-mutant AML cells of GSK321 treated mice (Supplementary Fig. 3f). After treatment with GSK864, we observed a significant decrease in the percentage of blast cells (SSClow CD45low/+) and a relative increase in mature lymphoid and granulocytic/monocytic cells (Supplementary Fig. 4c). Analysis of BM cells for expression of markers of early differentiation revealed slightly increased numbers of huCD45+ CD38+ cells in R132C or R132H IDH1 mutant engrafted mice treated with GSK864 (Supplementary Fig. 4d). Cytomorphology analysis of sorted, viable mCD45.1− huCD45+ cells from bone marrow identified cells with signs of myeloid differentiation, including increased cytoplasm-to-nuclear ratio and nuclear indentation, in mice treated with GSK864 (Supplementary Fig. 4e). While GSK864 represents an early tool compound requiring further optimization, these data provide proof-of-concept for reduction of leukemic blasts and stimulation of myeloid differentiation of primary IDH1 mutant AML cells upon treatment with allosteric IDH1 mutant inhibitors in vivo.
Molecular effects of GSK321 in IDH1 mutant AML cells
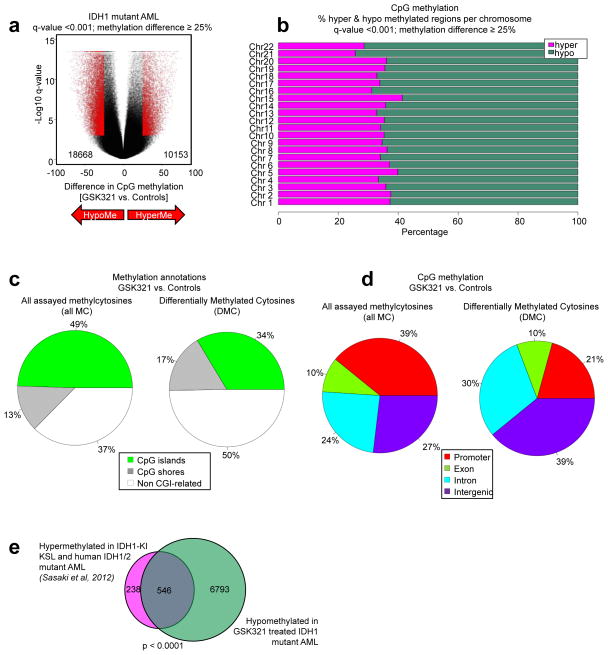
Performing enhanced reduced representation bisulfite sequencing (ERRBS) analysis of primary IDH1 mutant AML cells treated with either 3 μM of GSK321 or controls (DMSO or GSK990) for 6 days, we found overall hypomethylation, with 18668 hypomethylated and 10153 hypermethylated CpG loci (q-value < 0.001; methylation difference ≥ 25%) following treatment of R132G IDH1 mutant AML with GSK321 in comparison to controls (Fig. 5a). We observed a higher number of hypomethylated CpGs out of all covered CpGs, within all somatic chromosomes (q-value < 0.001; methylation difference ≥ 25%; Fig. 5b and Supplementary Fig. 5). Interestingly, within all assayed methylcytosines, we observed that the most striking differences in methylation did not occur within CpG islands (CGIs), CGI shores or proximal promoter regions, but within non CGI-related regions and in intergenic and intronic regions (Fig. 5c,d). The majority of differentially methylated cytosines (DMCs) were located at <5 to <500 kilobases (kb) from the nearest transcription start site (TSS) (Supplementary Fig. 6a).
Figure 5. GSK321 leads to genome-wide DNA cytosine hypomethylation in IDH1-mutant AML cells.
IDH1 R132G AML cells were treated in suspension culture with controls (DMSO or 3 μM GSK990) or 3 μM GSK321 for 6 days. (a) Volcano plot depicting differentially methylated CpGs. (b) Stacking barplots showing the percentage of hyper-and hypomethylated CpGs out of all covered CpGs for each somatic chromosome for GSK321 inhibitor treatment compared to controls. (c) Pie chart illustrating all assayed methylation sites and the proportion of differentially methylated cytosines (DMCs) annotated to (epi)genomic features upon treatment with GSK321. Green denotes CpG islands, grey denotes CpG shores and white denotes non CGI-related regions. (d) Pie chart illustrating all assayed methylation sites and the proportion of differentially methylated cytosines (DMCs) upon GSK321 treatment annotated to promoter regions (red), exons (green), introns (blue) and intergenic regions (purple) throughout the genome. (e) Venn diagram representing the overlap between hypomethylated annotated CpGs (7339) in GSK321 treated IDH1 mutant AML cells and hypermethylated CpGs (784) in IDH1-knock in (KI) LSK and human IDH1/2 mutant AML7. Out of these 784 genes hypermethylated in IDH-mutant AML cells, 546 were hypomethylated upon treatment with the active IDH1 inhibitor. p-value < 0.0001.
Specific evaluation of all annotated hypomethylated genes in GSK321 versus control treated IDH1 mutant AML cells demonstrated significant enrichment of genes and biological processes associated with the regulation of cell cycle, myelomonocytic differentiation, and stem cell fate and differentiation (Supplementary Fig. 6b, Supplementary Table 5–7). Interestingly, one of the hypomethylated genes was CD38, which we had found upregulated in vitro (Fig. 4h–j and Supplementary Fig. 2f), in vivo (Supplementary Fig. 4d) and by microarray analysis (Supplementary Fig. 7 and Supplementary Data Set 2) following treatment with either GSK321 or GSK864.
Analysis of differentially methylated regions (DMRs) identified 2075 hypomethylated and 1157 hypermethylated regions (1kb tiles; q-value < 0.001; methylation difference ≥ 25%) (Supplementary Fig. 6c). Similar to the DMC analysis, we identified a large number of changes occurring within non CGI-related and intergenic regions (Supplementary Fig. 6d,e). The majority of DMRs were located at >5 to <500 kilobases (kb) from the nearest transcription start site (TSS) (Supplementary Fig. 6f). Consistently, these annotated hypomethylated genes showed enrichment of genes associated with “proliferation and differentiation of stem cells”, “colony formation of cells”, “differentiation of hematopoietic progenitor cells”, and “cell death/apoptosis” (Supplementary Fig. 6g).
We compared our data upon treatment with allosteric IDH1 inhibitor with a methylation signature from a previous study which had identified 784 hypermethylated genes in IDH1/2 mutant AML cells 7. Strikingly, treatment of IDH1 mutant AML cells with GSK321 led to hypomethylation of approximately 70% (546 out of 784) of CpGs associated with genes of this signature (Fig. 5e and Supplementary Table 8), and thus indicates a significant molecular reversal of the hypermethylation phenotype induced by mutant IDH1. Similarly, comparison of the hypomethylated genes as annotated from DMRs upon GSK321 treatment to the hypermethylated genes determined by Sasaki et al also showed a significant overlap (Supplementary Fig. 6h).
Finally, we measured gene expression profiles of DMSO, GSK990, or GSK321 treated R132G IDH1 AML cells (Supplementary Fig. 7a,b and Supplementary Data Set 2). Ingenuity pathway analysis and gene set enrichment analysis of genes differentially expressed between GSK321 vs. control (DMSO or GSK990) treated cells identified enrichment of genes/pathways associated with several cell growth, differentiation, and leukemia-relevant processes (Supplementary Fig. 7c,d).
Discussion
Blocked differentiation is a hallmark of AML, and targeting this differentiation block rather than cell cycling of AML cells is a strategy that holds much promise for the development of potentially curative treatments for this devastating disease 30–32. ‘Differentiation therapy’ of APL (acute promyelocytic leukemia), a molecularly defined subtype of AML, by all trans retinoic acid (ATRA) has dramatically increased cure rates in patients carrying the PML-RARA gene fusion. In recent years, several new disease alleles have been discovered that play important roles in mediating the differentiation block in AML, including mutations of the IDH family of genes. Besides AML, recurrent IDH1 and IDH2 mutations have been reported in other cancer types 33,34. Inhibitors of mutant IDH2 have recently been reported and have shown very encouraging characteristics in preclinical and clinical phase I/II studies. In this study, we developed and evaluated allosteric inhibitors of mutant IDH1 for their anti-leukemic effects on primary IDH1 mutant cells from AML patients. The IDH1 inhibitor GSK321 stably decreased 2-hydroxyglutarate (2-HG) production in several different IDH1 mutant AML cells within a 2–3 week time frame ex vivo. Based on both structure and kinetic evaluations, GSK321 binds in an allosteric site on IDH1. Binding of GSK321 is proposed to inhibit turnover by locking the enzyme into an open, catalytically inactive conformation. Because of the allosteric nature of the interaction, GSK321 is able to inhibit multiple mutant forms of IDH1, which is consistent with our cell biological observations. An additional advantage of this type of mechanism (non-competitive versus NADPH) is that GSK321 retains excellent potency in cells, since it does not compete with the tightly bound cofactor 25.
During the preparation of this manuscript the first crystal structure of an inhibitor (VVS in PDB code 4UMX) bound to the IDH1 allosteric site was published 23. However, both the binding mode and MOI of VVS is distinct from GSK321 despite occupying spatially overlapping portions of the binding pocket. Whereas the THPPs bind completely within each subunit, VVS contains an imidazole group which protrudes into the dimer interface. Due to this difference, our structure contains two THPP inhibitors bound per enzyme dimer, whereas VVS binds to only one subunit of IDH1. One result of these different binding modes is that VVS is proposed to inhibit one subunit of IDH1 via direct interaction with the metal binding amino acid Asp279, and through disruption of catalytically required structural elements in the other subunit in an asymmetric manner. However, in our structure GSK321 does not interact with Asp279, as it lies within an intrinsically disordered region known as Seg2. The binding of VVS induces more of a conformational change in this segment than does GSK321. A second result of these different binding modes is that the MOI of VVS and GSK321 are distinct. While GSK321 is competitive versus αKG, VVS was shown to be non-competitive versus this substrate. This may reflect the different conformational states induced by the two classes of inhibitors. Consistent with this idea, VVS is reported to display time-dependent inhibition of mutant IDH1, whereas GSK321 does not. Thus, the discrepancy in MOI may also be due to the time-dependent nature of VVS.
In addition to these reports on the allosteric IDH1 inhibitor binding site, two different ligand binding sites have previously been described for the mutant IDH enzymes 21,35. The first is a distinct allosteric site on IDH2, which lies along the dimer interface and displays non-time dependent, noncompetitive inhibition versus αKG 20. The second is an orthosteric site, referred to as isocitrate site 1 35. Additionally, several other IDH1 inhibitors have been disclosed 19,36,37, however no ligand bound structures were reported and the MOI was found to be uncompetitive versus NADPH, contrasting with GSK849 and VVS.
In our study, we demonstrate a transient and specific increase in cell numbers upon treatment with GSK321, which was attributable to the induction of cellular differentiation of leukemic cells. These findings are highly reminiscent of the responses that have been observed following the treatment of APL cells with ATRA 38,39, and are similar to the effects observed following IDH2 inhibition 20,40. Indeed, GSK321 treatment of primary IDH1 mutant AML cells, but not of cells from healthy donors or from wild-type (WT) IDH1 AML patient samples, led to a significant decrease in leukemic blasts. Of note, the differentiation-inducing effect of GSK321 was also observed in more immature stem-like cells. Additionally, although with lower potency, we found that treatment of xenotransplanted NSG mice with the bioavailable GSK864, resulted in decreased levels of leukemic blasts, increased CD38 surface expression and morphological changes consistent with cell differentiation. These results suggest the potential use of (further optimized) derivatives of GSK321 or GSK864 for “differentiation therapy” of IDH1-mutant AML.
Previous studies have shown that IDH1 mutations result in global DNA and histone hypermethylation 7–11,41. We found that treatment with GSK321 led to reduced levels of the H3K9me2 mark in IDH1 R132C expressing HT-1080 cells. Furthermore, we found that GSK321 induced a widespread reversal of aberrant DNA hypermethylation driven by the IDH1 mutation in primary AML patients’ cells. This reversal was particularly pronounced in intergenic and intronic regions, which suggests a role of distal gene regulatory sites in the pathogenesis of IDH1 mutant AML. Annotation of loci within these regions corresponded to enrichment of genes and pathways consistent with our cell biological observations. Interestingly, in a previous report by Rohle et al 19 studying the inhibition of mutant IDH1 in a glioma cell line, no changes in DNA methylation were detected, suggesting that functional effects of mutant IDH1 and its pharmacological inhibition are mediated through different mechanisms in AML and glioma cells.
In summary, we have developed a novel series of inhibitors of mutant IDH1 and demonstrate that pharmacological, allosteric inhibition of mutant IDH1 is feasible and effective in primary AML patients’ cells bearing heterozygous IDH1 mutations. Additional preclinical studies are warranted to explore the combined use of IDH1 mutant inhibitors and other traditional chemotherapeutic agents. Our study provides a rationale for further development and investigation of allosteric inhibitors of mutant IDH1 and the testing for their potential use in the treatment of patients with IDH1 mutant AML and other cancers.
Online Methods
Compound synthesis
Compounds
Synthesis and analysis of compounds is described in detail in the Supplementary Notes file. GSK990 and GSK321 were prepared as 20 mM stocks in DMSO and diluted in DMSO for subsequent studies. Both compounds are stable in media for at least 14 days. For use in vivo, GSK864 was prepared to a final concentration of 16.6 mg/mL in propylene glycol (PG), DMSO, PEG-400 and water in the ratio 16.7:3.3:40:40.
Compound analysis
A selective and sensitive UPLC/MS/MS method was developed to quantify GSK864 in mouse blood (diluted 1:1 v/v with water). Samples were analyzed for GSK864 using protein precipitation with acetonitrile containing an internal standard, a structural analog of GSK864, followed by UPLC/MS/MS analysis. The UPLC system employed was a Waters Acquity and was coupled to an ABSciex API 5000 triple quadrupole mass spectrometer utilizing positive-ion Turbo Spray ionization. Separation was achieved on a Waters Acquity UPLC BEH C18 (2.1 × 50 mm, 1.7 μm) column using a quick (<1 min) linear gradient elution of 0.05% formic acid in water and 0.05% formic acid in acetonitrile. The flow rate was set to 1.0 mL/min and was introduced into the source of the mass spectrometer without any post-column split.
Standards (10–12 concentrations in duplicate) were prepared fresh daily by spiking a known amount of GSK864 into the appropriate matrix and performing the analysis of standards in parallel with authentic samples. The precursor and product ion transitions selected for the analyte and internal standard were specific to the corresponding compound and no interference from endogenous peaks was observed. Individual standard concentration values were generally within 20% of the interpolated concentration and the signal to noise ratio at the lower limit of quantification was >8:1. The lower limit of quantification was 1 ng/mL and the calibration curve was linear over a 10000-fold concentration range.
High Throughput Screen
50 nL of 1 mM test compound stocks (in 100% DMSO) were pre-stamped into 1536 well black plates. 2 μL of enzyme solution (187 nM R132H/wt IDH1) in base buffer (100 mM Tris, pH=8.0, 10 mM MgCl2, 1 mM CHAPS, 0.1 mg/mL BSA, 0.2 mM DTT) was added to the plate using a Multidrop Combi, excepting control wells in columns 35 and 36 where buffer alone was dispensed. Immediately following, 2 μL of substrate solution (5 mM 2-HG, 7.5 μM NADP+, 25 μM resazurin, 2.5 U/mL diaphorase) in base buffer was added to the entire plate using a second Multidrop Combi. The plates were centrifuged for 1 minute at 500 rpm to ensure mixing and then incubated at room temperature for 2 hours. Reactions were quenched with the addition of 2 μL of stop solution (250 mM EDTA in 100 mM Tris buffer, pH=8.0) added with a third Multidrop Combi, plates were centrifuged a second time as before, and finally read on a Viewlux imager for fluorescence intensity (ex=525nm/em=589nm/561nm dichroic filter).
Primary compound responses were corrected via plate pattern detection (iterative smoothing) to reduce systematic plate effects. The 2.14 million GSK collection was tested in single shot, and compounds with a corrected inhibition response exceeding 3 robust SDs (>22% inhibition) above robust mean of the test population were marked as primary hits (2.2% hit rate). Property-biased hit marking was employed to boost identification of ligand efficient hits, yielding a total hit pool of 55k compounds (2.6%). Elimination of weak hits with high molecular size (MW>450), high lipophilicity (cLogP>4), or low efficiency (PEI<1) yielded a final list of 26k compounds (1.2%) for confirmation testing. Replicate re-testing in single shot and interference testing were utilized to identify 3910 compounds for dose response testing. 1167 of these compounds exhibited IC50 values ≤ 10 μM or better inhibition of IDH1 in at least one dose response replicate. Compounds which interfered with the coupling system were removed at both the single shot and dose response testing stages. Hits were further qualified through direct binding to mutant R132H IDH1 by thermal shift analysis, confirmation of enzyme inhibition in the forward direction of α-KG to 2-HG directly by Mass Spectrometry (see Biochemical IC50s methods), chemical resynthesis, and measurement of 2-HG reduction in cellular assays.
Crystallization, structure determination, and refinement of hIDH1 R132H
For structure, the IDH1 R132H homodimer was concentrated to 15 mg/mL in storage buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 1 mM EDTA) for crystallization after removal of the His6 purification tag, and aliquots were frozen at −80 °C until used. The crystals of the IDH1 R132H + NADP + (open form) used for soaking in the inhibitor grew readily by vapor diffusion in sitting drops at 4 °C from 18%–23% PEG3350, 200 mM ammonium sulfate, 100 mM Bis-Tris pH 7.0, 10 mM NADP+ with seeding. The sitting drops were prepared by mixing 1.5 μL well solution, 0.5 μL of the seed dilution, and 2 μL of protein. The plates were sealed with clear tape and incubated at 4 °C in the dark. Crystals started to appear in 24–48 hrs and grew to full size by the second week. Fresh or frozen seed crystals were added to the crystallization drop at a 1/10,000 dilution. Seed stocks were prepared by crushing of crystals from a previous crystallization experiment by combining 3 drops, crushing, and diluting the seeds with 1 mL of stabilizing solution (24% PEG 3350, 200 mM ammonium sulfate, 100 mM Bis-Tris pH 7.0, 5 mM NADP+). A 100 mM stock of NADP+ (Sigma #N8160-15VL, 10 mg/vial) was prepared by dissolving the contents of the vial in 120 μL 100 mM Tris pH 9 and adjusting the pH to ~7.5 with NaOH. The stock solution was stored frozen at −20 °C. For soaking experiments, the soaking solutions containing 2–20 mM inhibitor (from 200–500 mM stocks in DMSO), 1%–5% DMSO, 28% PEG 3350, 200 mM ammonium sulfate, 100 mM Bis-Tris pH 7.0, 5 mM NADP+. The soaking solution containing the inhibitor was gently mixed stepwise with the crystallization drop. The reservoir solution was replaced with 300 μL of stabilizing solution (without NADP+) and the plate resealed. Crystals were soaked between 16 hrs and up to 6 days in the cold room. To prepare the crystals for data collection, the majority of the soaking solution was slowly exchanged with the stabilizing solution containing inhibitor, NADP+, and 20% ethylene glycol. The crystals were then frozen by immersing them in liquid nitrogen. The diffraction data were collected and processed as previously described.
Biochemical IC50 determinations
Compound potencies with mutant IDH1 and IDH2 were determined by measuring the formation of 2HG with the RapidFire-MS/MS system or through the oxidation of NADPH by absorbance. Compound potencies with wild type IDH1 and IDH2 were determined by the formation of NADPH with the diaphorase/resazurin coupled assay 25.
RapidFire-MS/MS measurements of mutant IDH1 and IDH2 reactions were conducted at room temperature in 384-well Greiner polypropylene microtiter plates in a total volume of 20 μL of assay buffer. Final compound concentrations were typically varied from 0.5 to 100,000 nM, though in the case of tight-binding inhibition a range of 0.4 nM to 2 μM was used with a 1.5-fold dilution scheme. The αKG concentration was fixed at 500 μM, the NADPH concentration was fixed at 50 μM, and mutant IDH1 and IDH2 were fixed at 15 nM. Reactions were conducted in duplicate and quenched with final concentrations of 100 mM EDTA. After each addition plates were centrifuged for 60 s to ensure complete mixing of reagents. Endpoint data was obtained after 2 hours for R132C IDH1, R132G IDH1, R140Q IDH2 and R172S IDH2; and after 4 hours for R132H IDH1. For the RapidFire-MS/MS analysis, a 0.5 μL aliquot of each well was diluted 200-fold into a separate analysis plate containing 100 μL of a freshly prepared methanol/acetonitrile mixture (1:1) with 0.075 μg/mL (~0.49 μM) d4-2HG as an internal standard.
Absorbance measurements of the mutant IDH1 and IDH2 reactions were conducted at room temperature in 96-well Corning half-area microtiter plates in a total volume of 100 μL of assay buffer. Final compound concentrations were varied as in the RapidFire-MS/MS experiments. The αKG concentration was fixed at 1.5 mM, the NADPH concentration was fixed at 50 μM, and mutant IDH1 was fixed at either 100 nM for R132H and R132C or 75 nM for R132G. Mutant IDH2 was fixed at either 50 nM for R140Q or 40 nM for R172S. Reactions were initiated by addition of enzyme and NADPH, and absorbance (340 nm) was monitored continuously for the first 10 minutes to obtain linear reaction rates.
For WT IDH1 and IDH2, reactions were conducted at room temperature in 384-well Greiner black microtiter plates in a total volume of 10 μL of assay buffer. Final compound concentrations were typically varied from 5 to 100,000 nM; the isocitrate concentration was fixed at 10 μM, the NADP+ concentration was fixed at 5 μM, and WT IDH1 and IDH2 were fixed at 0.1 nM. Reactions were conducted in duplicate and run kinetically. After each addition, plates were centrifuged for 60 s to ensure complete mixing of reagents.
Data was fit to following equations to determine IC50
In the case of IC50s ≫ [Enzyme] a simple 2 parameter fit was used, with data normalized to controls:
Where y is the % of normalized enzyme activity, x is the concentration of inhibitor, and s is the Hill slope factor.
In the case of IC50s = or < [Enzyme] a tight binding equation was used to globally fit the data, more accurately determine IC50 values, and account for the significant fraction of bound inhibitor (Copeland RA, 2005):
where vi/v0 is the fractional activity, I is the concentration of inhibitor, E is the concentration of enzyme, and appKi is the IC50.
Biochemical mechanism of inhibition analysis
In the direction of 2HG formation we determined the MOI of IDH1 inhibitors using the NADPH oxidation absorbance decrease assay when αKG was varied and the RapidFire-MS/MS assay when NADPH was varied as described previously 25 with the following modifications. Final concentrations of αKG were 1.4 to 11.2 mM in 2-fold increments and GSK849 were 0 and 12.5 nM to 200 nM (200 nM ○, 100 nM ●, 50 nM □, 25 nM ■, 12.5 nM □, or no inhibitor ▲) in 2-fold increments at 50 μM NADPH (~50KM) and 40 nM homodimeric R132H active sites. Data in duplicate at each inhibitor concentration were fitted to a competitive inhibition model described by eq 3 in 25. Final concentrations of NADPH were 0.4 μM to 25.6 μM in 2-fold increments and GSK849 were 0 and 30 nM to 240 nM (240 nM ●, 120 nM □, 60 nmM ■, 30 nM □, or no inhibitor ▲) in 2-fold increments at 2 mM aKG (~KM) at 15 nM homodimeric R132H active sites. Data in duplicate at each inhibitor concentration were fitted to a mixed/noncompetitive inhibition model described by eq 4 in 25.
Artificial membrane permeability (AMP) assay
Permeability was determined using an artificial membrane permeability assay: a 1.8% lipid (phosphatidylcholine, egg) in 1% cholesterol decane solution was applied to a Millicell 96-well, 0.4 μm, PCF culture plate. 250 μL and 100 μL of 50 mM phosphate buffer pH 7.4 with 0.5% encapsin were applied to the donor and receiver compartments, respectively. 2.5 μL of a 10 mM stock solution of compound in DMSO was added to the donor compartment. The plate was incubated at room temperature for 3 hours. Samples from both donor and receiver compartments were analyzed by HPLC with UV detection (215 nm) and permeability was calculated. Data are reported as the mean of at least 2 individual experiments. GSK990 AMP = 59 nm/sec (moderate); GSK321 AMP = 324 nm/sec (high).
Caco-2 cell permeability assay
Caco-2 cells (clone C2BBe1) were obtained from American Type Culture Collection (Manassas, VA). Cell monolayers were grown to confluence on collagen-coated, microporous, polycarbonate membranes in 12-well Costar Transwell plates. The permeability assay buffer was Hanks’ balanced salt solution containing 10 mM HEPES and 15 mM glucose at a pH of 7.4. The buffer in the receiver chamber also contained 1% bovine serum albumin. Cell monolayers were dosed on the apical side (A-to-B) or basolateral side (B-to-A) and incubated at 37 °C with 5% CO2 in a humidified incubator. Samples were taken from the donor and receiver chambers at 120 minutes. Each determination was performed in duplicate. The flux of co-dosed lucifer yellow was also measured for each monolayer to ensure no damage was inflicted to the cell monolayers during the flux period. All samples were assayed by LC-MS/MS using electrospray ionization.
Proteomics based inhibitor profiling
In order to generate a probe matrix, GSK838 compound, an amine-functionalized derivative of GSK321, was immobilized on NHS-activated Sepharose 4 Fast Flow beads (Amersham Biosciences) at a ligand density of 1 mM. Derivatized beads were incubated over night at room temperature in darkness on an end-over-end shaker and non-reacted NHS-groups were blocked by incubation with aminoethanol at room temperature on the end-over-end shaker, overnight. Beads were washed with 10 mL of DMSO and were stored in isopropanol at −20 °C. Prior to use, beads were washed three times with 5 – 10 volumes of DP buffer (50 mM TRIS pH 7.4, 5% Glycerol, 150 mM NaCl, 25 mM NaF, 1.5 mM MgCl2, 0.4% Igepal CA-630), collected by centrifugation for 1 min at 311 x g in a Heraeus centrifuge and finally re-suspended in DP buffer to prepare a 5% beads slurry. Affinity profiling assays were carried out as described previously 42,43 with minor modifications. HT-1080 lysate was diluted with DP buffer to a protein concentration of 5 mg/mL and cleared by centrifugation at 145,000 x g. Aliquots of cell extracts (1 mL) were incubated with test compounds GSK321 or GSK990 or vehicle for 45 min, then 35 μL derivatized sepharose beads were added per sample and incubated on an end-over-end shaker for 1 h at 4 °C. Beads were transferred to disposable columns (MoBiTec), washed with DP buffer containing 0.2% Igepal CA-630 and eluted with 50 μL 2X SDS sample buffer. Proteins were alkylated with 200 mg/ml iodoacetamide for 30 min, partially separated on 4–12% NuPAGE (Invitrogen), and stained with colloidal Coomassie. Gels were cut into three slices and subjected to in-gel digestion 42. Peptide extracts were labeled with TMT reagents in 90 mM triethylammoniumbicarbonate, pH 8.53. After quenching of the reaction with glycine, labeled extracts were combined. Extracts from vehicle-treated sample were labeled with TMT reagent 131, and combined with extracts from compound-treated samples labeled with TMT reagents 126 & 128, fractionated with reversed-phase chromatography at pH 12 (1 mm Xbridge column, Waters) into 9 fractions as previously described (Kruse U et al, 2011). Samples were dried in vacuo and resuspended in 0.05% TFA in water. 50% of the sample was injected into an Ultimate3000 nanoRLSC (Dionex) coupled to a Q Exactive (Thermo Scientific™). Peptides were separated on custom-made 50 cm × 100 μM (ID) reversed-phase columns (Reprosil) at 40 °C. Gradient elution was performed from 2% acetonitrile to 40% acetonitrile in 0.1% formic acid over 2 hours. Samples were online injected into a Q-Exactive mass spectrometer operating with a data-dependent top ten method. MS spectra were acquired using 70,000 resolution and an ion target of 3E6. HCD scans were performed with 35% NCE at 35,000 resolution (at m/z 200) and an ion target setting of 2E5. The instrument was operated with Tune 2.2 and Xcalibur 2.7.
Production of recombinant proteins for structure and kinetics
IDH1 wild type and mutant enzymes were produced as previously described 44. The cDNA for the human IDH2 gene (UniProtKB/Swiss-Prot Accession No. P48735) was PCR amplified from human kidney Marathon ready cDNA and then subcloned into a Baculovirus expression vector. The gene was modified with the removal of the N-term mitochondria localization signal sequence and the addition of a C-term Flag tag, resulting in IDH2(40-452)Flag. The R140Q and R172S IDH2 mutations were introduced using a QuikChange site directed mutagenesis kit (Agilent Technologies). Both P1 virus and baculovirus-infected insect cells (BIICs) of Wild-type, R140Q and R172S were generated using the Bac-to-Bac baculovirus expression system (Invitrogen 10359-016). Protein was expressed by infection with 10,000-fold diluted BIIC stock at 2×106 cells/ml in Spodoptera frugiperda (Sf9) cells. The cell paste was harvested by centrifugation at 66 hours after infection.
Sf9 cells which over-expressed the target IDH2 proteins by Baculovirus infection were lysed in lysis buffer (50 mM Tris, pH7.5, 300 mM NaCl, 10% glycerol, 1% Triton) by Avestin emulsiflex C50. The supernatants were mixed with anti-Flag resin at 4°C overnight and then eluted from the resin by the addition of 100 μg/ml Flag peptide in lysis buffer (without Triton). The eluate was concentrated and loaded onto a Superdex 200 size exclusion column and eluted with sizing buffer (25 mM Tris, 100 mM NaCl, 1 mM DTT, pH 7.5). Fractions containing dimer were pooled and concentrated for use in kinetics. Specific mutations were confirmed by LC/MS and protein peptide mapping analysis.
Peptide and protein identification and quantification
Mascot 2.2 (Matrix Science) was used for protein identification using a 10 ppm mass tolerance for peptide precursors and 20 mDa (HCD) mass tolerance for fragment ions. Carbamidomethylation of cysteine residues and TMT modification of lysine residues were set as fixed modifications and methionine oxidation, N-terminal acetylation of proteins and TMT modification of peptide N-termini were set as variable modifications. The search database consisted of a customized version of the IPI protein sequence database combined with a decoy version of this database created using a script supplied by Matrix Science. Unless stated otherwise, we accepted protein identifications as follows: i) For single spectrum to sequence assignments, we required this assignment to be the best match and a minimum Mascot score of 31 and a 10x difference of this assignment over the next best assignment. Based on these criteria, the decoy search results indicated <1% false discovery rate (FDR); ii) for multiple spectrum to sequence assignments and using the same parameters, the decoy search results indicate <0.1% false discovery rate. Only proteins identified by at least two uniquely matching peptide were quantified. Reporter ion intensities were read from raw data using in-house software 43 and multiplied with ion accumulation times (the unit is milliseconds) to yield a measure proportional to the number of ions, this measure is referred to as ion area. Spectra matching to peptides were filtered according to the following criteria: mascot ion score >15, signal-to-background of the precursor ion >4 and signal-to-interference >0.5 45,46. Fold-changes were corrected for isotope purity as described and adjusted for interference caused by co-eluting nearly isobaric peaks as estimated by the signal-to-interference measure 47. Protein quantification was derived from individual spectra matching to unique peptides using a sum-based bootstrap algorithm 43.
The calculation of significance of differences in relative protein abundances (log2 scale) was performed for each treatment to DMSO comparison using all the relative protein abundances to estimate the underlying normal distribution and from that distribution the p-values for each comparison were inferred. Subsequently a correction for multiple hypothesis testing was performed using Benjamini-Hochberg analysis 48.
Pharmacokinetic analysis
Concentration-time data was analyzed by non-compartmental methods using the computer program Phoenix WinNonlin v6.1 (Pharsight Corporation). Area under the blood concentration-time curve was calculated using the linear-log trapezoidal rule. The terminal elimination rate constant (λz) was derived from the log-linear disposition phase of the concentration-time curve using least-squares regression analysis. AUC(0-inf) was estimated as the sum of AUC(0-t) and C(t)/λz, where C(t) is the predicted concentration from the log-linear regression analysis at the last measurable time point. The elimination half-life (t½) was calculated as ln (2)/λz. The mean residence time (MRT), the apparent blood clearance (CLb) and the volume of distribution at steady-state (Vss) were also estimated. CLb was calculated by dividing the dose by AUC(0-inf). MRT was estimated as [AUMC(0-inf)/AUC(0-inf)] and Vss was calculated using the following equation: Vss = CL*MRT.
Pharmacokinetic analysis in mouse
All studies were conducted after review by the Institutional Animal Care and Use Committee at GSK and in accordance with the GSK Policy on the Care, Welfare and Treatment of Laboratory Animals. Six mice (30–40 g) had cannulae surgically implanted in the femoral artery by Laboratory Animal Sciences (LAS), GlaxoSmithKline (Collegeville, PA). The mice were allowed to recover from surgery for at least 48 h. Mice received GSK864 (213 mg/kg, 10 mL/kg) via intraperitoneal (ip) administration. IP formulation was PG:DMSO:PEG400:H2O (16.7:3.3:40:40) and was prepared by the Pharmaceutical Development group (GSK). Serial blood samples (~0.030 mL) were collected via the femoral artery catheter at 15, 30, 45, 60, 120, 180 min following administration. A terminal 1440 min sample was collected by cardiac puncture following anesthetization in a CO2 chamber. Blood aliquots (25 μL) were collected and transferred to tubes containing 25 μL of purified water. The tubes were lightly vortex mixed, placed on dry ice, and then stored at about −30°C prior to analysis.
Quantification of intracellular 2-HG in HT-1080 cells
R132C IDH1 HT-1080 chondrosaroma cells (ATCC) in log phase growth were treated with a range of compound concentrations for 24 hours prior to extracting R-2-hydroxyglutarate (R-2-HG) with 70% (1:1) acetonitrile/methanol diluted in ddH2O (−20 °C). R-2-HG was then quantified by LC-MS using an API 4000 triple-quadrupole tandem mass spectrometer (Applied Biosystems). LC was performed on a Phenomenex Luna C18 analytical column. R-2-HG was monitored by the transition of 146.7 m/z > 128.9 m/z using a DP −35.0, collision energy −16.0, and CXP −9.0, and the internal standard (4D) 2-hydroxyglutarate was monitored by the transition of 151.0 m/z > 131.9 m/z using a DP −35.0, collision energy −16.0, and CXP −9.0. Calibration curves were set up using R-2-HG standards dissolved in extraction buffer. Data were acquired and processed with Analyst for Windows (version 1.4.2). The peak areas for R-2-HG were normalized using the peak area of the internal standard. R-2-HG concentrations were normalized to the DMSO control treated cells and expressed as a percent of control activity. EC50 values were calculated by fitting data to a four parameter, non-linear, (variable slope) concentration response curve with XLfit (IDBS). Compounds were evaluated for their ability to inhibit R-2-HG production in R132C IDH1 mutant HT-1080 cells. EC50 values for GSK321 and GSK864 were 0.085 ± 0.074 μM and 0.320 ± 0.257 μM, respectively. The structurally-related inactive analog GSK990 did not inhibit R-2-HG up to 36 μM.
Chemical and reagents
Cavitron (hydroxypropyl β-cyclodextrin) was purchased from Cargill, Inc. HPLC-grade acetonitrile was purchased from Sigma Aldrich and formic acid was purchased from EMD Millipore Corp. All other chemicals and reagents were of standard laboratory reagent grade or better.
Cells
Cryopreserved bone marrow (BM) or peripheral blood (PB) mononuclear cells (MNCs) of healthy individuals and leukemia patients were utilized in this study. Informed consent was obtained from all subjects. All experiments were approved by the IRB of the Albert Einstein College of Medicine (#2008-942). HT-1080 cells obtained from ATCC were confirmed to be authentic via STR profiling and were maintained in EMEM media supplemented with 10% heat-inactivated fetal bovine serum.
Genotyping
Genomic DNA was extracted by suspending cells in 50 mM NaOH incubated for 5 min at 95°C. Following cooling, samples were resuspended in 1M Tris HCl. PCR amplification of IDH1 and IDH2 exon 4 using TaqPro Red complete mix (Denville) was performed using 2 sets of primers detailed in (Supplementary Table 10). Mutations in patient samples are listed in Supplementary table 11. PCR consisted of an initial denaturation step of 95°C for 2 min, followed by 35 cycles of a denaturation step for 95°C for 30 s, primer specific annealing temperature for 30 s and an extension step for 72°C for 1 min. PCR products were then purified using the Qiaquick PCR purification kit (Qiagen) and submitted for Sanger sequencing. IDH1 or IDH2 nested PCR was performed by first amplifying exon 4 using FW1/RV1 primer pairs, followed by DNA purification and amplification of PCR product using FW2/RV2 primers.
2-Hydroxyglutarate (2-HG) measurement
Intracellular R-2-hydroxyglutarate (R-2-HG) in IDH1 mutant or wild-type AML cells treated with DMSO, GSK990, or GSK321 was extracted using (1:1) acetonitrile/methanol diluted in ddH2O. R-2-HG was then quantified by LC-MS using an API 4000 triple-quadrupole tandem mass spectrometer (Applied Biosystems). LC was performed on a Phenomenex Luna C18 analytical column. R-2-HG was monitored by the transition of 146.7 m/z > 128.9 m/z using a DP −35.0, collision energy −16.0, and CXP −9.0, and the internal standard (4D) 2-hydroxyglutarate was monitored by the transition of 151.0 m/z > 131.9 m/z using a DP −35.0, collision energy −16.0, and CXP −9.0. Calibration curves were set up using R-2-HG standards dissolved in extraction buffer. Data were acquired and processed with Analyst for Windows (version 1.4.2). The peak areas for R-2-HG were normalized using the peak area of the internal standard. R-2-HG concentrations were normalized to the DMSO control treated cells and expressed as a percent of control activity.
Western blot analysis
R132C expressing HT-1080 cells were seeded into plates and allowed to equilibrate overnight at 37°C and 5% CO2. Cells were then treated with varying concentrations of GSK321 or GSK990 for 48 hours. Cell lysates were prepared and global histone modification and specific control protein levels were determined by western blot analysis using antibodies specific for beta-Actin (Sigma, #A2228), total histone H3 (Cell Signaling, #4499P) and H3K9me2 (Cell Signaling, #4658P) 49.
Cell culture and proliferation assay
Human BM or PB MNCs from healthy and AML samples were thawed and incubated in serum free media containing recombinant human cytokines 10 ng/mL IL-3, 25 ng/mL IL-6, 50 ng/mL SCF, 100 ng/mL TPO, 50 ng/mL FLT-3L, 1:500 primocin (50 mg/mL; InvivoGen) and 40 μg/mL Low Density Lipoprotein (Sigma) for 12–16 h. All cytokines were purchased from Preprotech, NJ, USA. Following pre-incubation, viable cells were seeded at 1.5–3 × 105 cells per 500 μL media in a 48-well plate and treated with 0.3% DMSO and 3 μM GSK990 or GSK321 inhibitors and were monitored for up to 22 days. The numbers of viable cells were determined by hemocytometer using trypan blue exclusion at the indicated time points.
HT-1080 cell culture and lysate preparation
HT-1080 cells were grown in 15 cm diameter plates to subconfluency in MEM medium (Invitrogen,) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 10% FCS (HI) + 1 mM sodium pyruvate + 0.1 mM non-essential amino acids. Cells were harvested by scraping and washed once in 1 x PBS; pellets were frozen in liquid nitrogen and stored at −80 °C. HT-1080 lysate preparation was performed at 4 °C. Frozen HT-1080 cell pellets were homogenized in lysis buffer (50 mM Tris-HCl, 0.8% Igepal-CA630, 5% glycerol, 150 mM NaCl, 1.5 mM MgCl2, 25 mM NaF, 1 mM sodium vanadate, 1 mM DTT, pH 7.5). One complete EDTA-free protease inhibitor tablet (Roche) per 25 ml was added. The sample was dispersed using a Dounce homogenizer, kept rotating for 30 min and spun for 10 min at 20,000g. The supernatant was spun again for 1 h at 145,000 x g. The protein concentration was determined by Bradford assay (BioRad), and aliquots were snap frozen in liquid nitrogen and stored at −80 °C.
Colony formation assay
1.5×105 to 5×105 IDH1 mutant AML cells per mL were plated in HSC003 methylcellulose media containing IL-3, SCF, GM-CSF, and EPO (R&D Systems), and Primocin in the presence of DMSO (0.3%), 3 μM GSK321 or GSK990 IDH1 inhibitors. Colonies were scored after 10–12 days at 37 °C and 5% CO2 and individual clones were isolated for genomic DNA extraction using an inverted microscope (Fisher Scientific, USA).
Apoptosis analysis
Apoptotic cells were assessed by staining PBS washed cells with phycoerythrin (PE)-conjugated Annexin-V (BD Pharmingen) for 10–15 min in incubation buffer (Annexin-V-FLUOS Staining Kit, Roche). Fluorescence was recorded by flow cytometry using the FACSAria (BD Biosciences).
Cell cycle analysis
Cell cycle activity was measured using Hoechst 33342 (Invitrogen) and Pyronin-Y (Sigma) incorporation as previously described 50. Cell cycle distribution was measured by BD FACSAria and analyzed by FlowJo software.
Mice
Male CD-1 mice were obtained from Charles River Laboratories. Non-obese diabetic severe combined immunodeficiency (NOD-SCID) -Interleukin 2 receptor gamma null (IL-2Rγnull) (NSG) mice were housed in the Albert Einstein College of Medicine animal facility. Experimental procedures were approved by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (IACUC; protocol #20131202). Mice were included in the study without any further preselection or formal randomization and comprised balanced numbers form both genders. Investigators were not blinded to group allocations.
Xenotransplantation and bone marrow aspiration
For xenotransplantation, sublethally irradiated (2Gy), 4–8 week old NSG mice were intravenously injected with up to 3×106 viable human primary IDH1 mutant AML cells. Engraftment of leukemic cells was confirmed by BM aspirations from femurs of anesthetized mice at 3 weeks after transplantation and at indicated time after treatment. Mice with detectable initial engraftment were included in the study and randomly subjected to compound or vehicle treatment. Investigators were not blinded to treatment group allocations. BM aspirates were then analyzed in downstream assays. Viability was assessed by trypan blue exclusion. Animal numbers were chosen based on previous experiences with drug evaluation studies in AML xenograft models 51,52. P-values are indicated.
Flow cytometric analysis of BM engraftment
Femoral aspirates were washed in PBS and red blood cells were lysed using ACK lysis buffer. The following antibodies were used: PE-CY5-conjugated anti-mouse CD45.1 (A20; eBioscience), PE-conjugated anti-human CD45 (HI30; BD Biosciences), PE-CY7-conjugated anti-human CD38 (HIT2; eBioscience), APC-conjugated anti-human CD34 (581; BD Pharmingen), PE-conjugated anti-human CD33 (WM53; eBioscience), and FITC-conjugated anti-human CD15. 4′,6-diamidino-2-phenylindole (DAPI) evaluated on the ultraviolet (UV) channel was utilized as a viability control. Data were collected on a FACSAria II instrument and analyzed with BD FACS Diva software.
Cell differentiation and blasts evaluation
Cytospins were prepared at indicated time points using the Cytofuge (Statspin) at 600x g for 6 min. Morphology was examined after DiffQuik (IMEB) staining, followed by evaluation and documentation using an Axiovert 200M microscope with an AxioCam color camera (Zeiss). Flow cytometric assessment of the differentiation state of the cells was performed with PE-CY7-conjugated anti-human CD38 (HIT2; eBioscience), PE-conjugated anti-human CD33 (WM53; eBioscience), and FITC-or Pacific blue (PB)-conjugated mouse anti-human CD15 antibody (VIMC6; eBioscience or Invitrogen).
Using PE-conjugated mouse anti-human CD45 antibody (BD Pharmingen) as previously described 28, we set up 3 gates on the CD45/SSC dot plot: gate L for lymphocytes (SSClow CD45high), G/M for granulocytes and monocytes (SSChigh CD45+) and B for blast cells (SSClow CD45low/+). Stained cells were analyzed by Flow cytometry using a FACS Aria II (BD Biosciences). Flow cytometry data were analyzed with BD FACS Diva and FlowJo softwares.
Additional flow cytometric markers for HLA-DR (G46-6; BD Pharmingen) and c-Kit/CD117 (104D2; Biolegend) were utilized for additional leukemic blast analysis 29.
The distribution of leukemic hematopoietic stem and progenitor cell compartment was evaluated by flow cytometry after treatment for the specified times. In brief; cells were stained for 30 minutes in the dark on ice with antibodies against lineage antigens: PE-Cy5-conjugated CD2 (RPA.2.10; eBioscience), CD3 (UCHT1; eBioscience), CD4 (S3.5; Invitrogen), CD7 (CD7-6B7; Invitrogen), CD8 (3B5; Invitrogen), CD11b (VIM12; Invitrogen), CD14 (3G8; Invitrogen), CD19 (H1B19; eBioscience), CD20 (2H7; eBioscience), CD56 (MEM-188; invitrogen), Glycophorin A (GA-R2), and CD-10 (eBioCB-CALLA; eBioscience). Following this first staining procedure, the cells were washed with PBS and stained for 30 minutes in the dark on ice with hematopoietic stem and progenitor markers: APC conjugated CD34 (581; BD Pharmingen) and PE-Cy7 conjugated CD38 (HIT2; eBioscience). After staining protocols, washed cells were evaluated by FACSAria II Special Order System (BD Biosciences) as previously described53,54. For colony formation assays, lineage-negative CD34-positive CD38-negative (Lin−CD34+CD38−) IDH1 mutant AML cells were isolated after staining as described above. Sorted cells were grown in semi-solid media as described in “Colony formation assay”.
Preparation of nucleic acids
Fractionated or unfractionated cells were treated for 6 days with 0.3% DMSO, 3 μM GSK990 or GSK321, washed with 1X phosphate buffered serum (PBS) and then resuspended in RLT buffer Plus (Qiagen) for DNA extraction and stored at −80°C. High molecular weight genomic DNA was extracted from homogenized cells using the AllPrep RNeasy kit (Qiagen) according to the manufacturer’s instructions.
Enhanced Reduced Representation Bisulfite Sequencing
High molecular weight genomic DNA was extracted using standard techniques and quality was assessed by gel electrophoresis for all samples used. DNA methylation profiling was performed by enhanced reduced representation bisulfite sequencing (ERRBS) as previously described 55,56. Briefly, 10ng of DNA was digested with MspI and subjected to end pair and ligation of paired-end Illumina sequencing adaptors. This was followed by size selection (150–400 base pairs) by gel extraction (Qiagen) and bisulfite treatment with the EZ DNA methylation kit (Zymo research). PCR amplification with Illumina PCR PE1.0 and 2.0 primers was followed by purification of library products with Agencort AMPure XP beads (Beckman Coulter). Assessment for quantity and quality was performed using a Qubit dsDNA High Sensitivity Assay (Life Technologies) and a high sensitivity DNA kit on a Bioanalyzer 2100 (Agilent Technologies). Libraries were sequenced using a 50-base pair single end approach on a HiSeq2000 (Illumina). Image capture, analysis and base calling were performed using CASAVA 1.8.2 (Illumina). Data alignment to hg19 was performed by the Weill Cornell Medical College Epigenomics Core as previously described 55,56.
ERRBS analysis
Methylation “call” files were analyzed with a software package for DNA-methylation analysis, methylKit (version 0.5.3)56, and annotation from high-throughput bisulfite sequencing in the statistical programming language R/Bioconductor (version 2.15.2) with non-overlapping DMCs 1 kb in length (≥10X coverage; ≥25% difference in methylation; q-value < 0.001). Differences in methylation were assessed in GSK321 treated IDH1 mutant AML samples versus controls (DMSO and GSK990 treated). DMCs were annotated for CpG islands, CpG shores (2 kilobases (kb) upstream and downstream of CpG islands), promoters (1 kilobase upstream and downstream of transcription start sites (TSS)), regions farther than 1 kilobase upstream or downstream of Refseq genes were considered intergenic. Differentially Methylated Regions (DMRs) were annotated for 1kb tiles, ≥ 25% difference in methylation and q-value < 0.001. Using the Genomic Regions Enrichment of Annotations Tool (GREAT) 57 differentially methylated cytosines (hypermethylated or hypomethylated) were annotated by assigning each differentially methylated locus to the single nearest gene within 1000kb of the gene transcription start site, in both directions. Pathway analysis of annotated differentially methylated cytosines was performed using Ingenuity Pathway Analysis software (Ingenuity Systems) and the Ingenuity knowledge database as reference. Circos plots of differentially expressed regions (DMRs; ≥25% difference in methylation; q-value < 0.00001) were performed as described on the developer’s website, circos.ca.
Gene expression microarray analysis
Total RNA extracted was extracted using RNeasy Micro kit (Qiagen, Valencia, CA). RNA was amplified from Control (DMSO or 3μM GSK990) or 3μM GSK321 treated IDH1 AML cells, using the NuGEN Ovation Pico WTA System V2 (PN 3302) which generates sense strand cDNA from total RNA follows with fragmentation and labeling using the Encore® Biotin Module (PN 4200) for use with mouse Affymetrix GeneChip Sense Target (ST) Array. Briefly, Total RNA is reverse transcribed using chimeric primer mix and reverse transcriptase priming method that primes mRNA and converts into double stranded cDNA using first and second strand enzyme from the kit. The product is synthesized using SPIA amplification technology from the double stranded SPIA tag to yield sscDNA. The sscDNA is chemically fragmented and biotin-labeled made into a hybridization cocktail using hybridization kit (PN 901524) according to the Affymetrix Genechip protocol, which was then hybridized to Human 2.0 ST GeneChip probe array (PN 902113). The array image is generated by a high-resolution GeneArray Scanner 3000 7G (Affymetrix, Santa Clara, CA,). The data were analyzed with GeneChip® Command Console® Software (AGCC) using Affymetrix default analysis settings. Following RMA normalization, differentially expressed genes (fold change 1.5 and pvalue < 0.1) were evaluated using the transcriptome analysis console (TAC) software (Affymetrix). Ingenuity pathway analysis (IPA™; Ingenuity® Systems www.ingenuity.com) and gene set enrichment analysis (GSEA; Broad Institute) softwares were used to further evaluate the differentially expressed genes and/or disrupted pathways.
Statistical analysis
Data statistics were performed using the Chi-square analysis, Student’s t-test, one-way ANOVA or two-way ANOVA with Dunnett’s multiple comparisons test. Statistical significance was set at p-value < 0.05. Statistical analyses were performed with Graphpad Prism 6 (Graphpad Software) or Excel (Microsoft). To assess statistical similarities between hypomethylated genes in GSK321 treated IDH1 mutant cells and the 784 hypermethylated genes in IDH1 mutant Lys-M Knock In and human IDH1 mutant AML cells, we performed two-tailed Chi-square with Yates’ Correction. Most experiments reported in this paper were carried out with primary samples from patients with IDH1 mutations and were designed to provide proof-of-principle for effects of the tested IDH1-mutant inhibitors. Sample sizes chosen are indicated in the individual figure legends and were not based on formal power calculations to detect pre-specified effect sizes. Animal numbers were chosen based on previous experiences with similar types of studies. Structural data were deposited in the Protein Data Bank (PDB) under PDB code 5DE1. Profiling data have been deposited in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI), accession numbers GSE72152, and GSE72253.
Supplementary Material
Chemical probe table
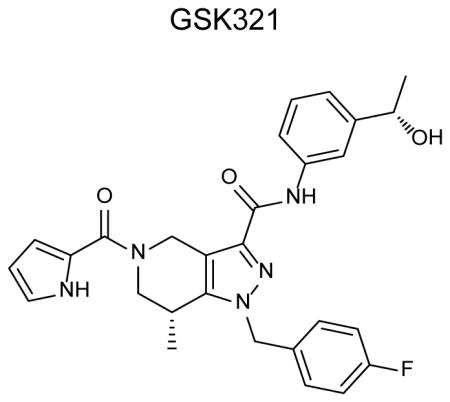
| ||
|---|---|---|
| Category | Parameter | Description |
| Compound | Additional names | (7R)-1-[(4-fluorophenyl)methyl]-N-{3-[(1S)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrol-2-ylcarbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3c]pyridine-3-carboxamide |
| Citation | ||
| Chemical descriptors | ||
| Chemical compound page | ||
| Entries in chemical databases | ||
| Availability | Online methods | |
| Additional comments | ||
|
| ||
| In vitro profiling | Target | Human isocitrate dehydrogenase isoform 1 (O75874 - IDHC_HUMAN) |
| Potency | 4.6nM vs R132H IDH1; 3.8nM vs R132C IDH1, 2.9nM vs R132G IDH1; see Figure 1C | |
| Selectivity | 46nM vs wild type IDH1; 1358nM vs R140Q IDH2, 1034nM vs R172S IDH2, 496nM vs wild type IDH2; see Figure 1C | |
| Potential reactivities | GSK321 has no obvious reactive groups such as thiol modifying moieties/Michael acceptors, primary alkyl halides, acid halides, esters, aldehydes, azos, azides, etc. | |
| SAR | Limited SAR available in supplementary table 1 | |
| Mechanism of action | Inhibitor, MoA competitive vs αKG and mixed vs NADPH, by analogy with close analog | |
| Structure of target-probe complex | Structure of GSK321 with R132H | |
| Additional comments | ||
|
| ||
| Cellular profiling | Validation of cellular target | 85nM against R132C IDH1 in HT1080 cells, which naturally harbor this mutation. Inhibition was measured by reduction in 2-HG by LC-MS/MS, Figure 1D |
| Validation of cellular specificity | Chemoproteomics was carried out using an amine-functionalized analog of GSK321 captured on bead. Only IDH1 was identified from an HT1080 lysate pulldown experiment as binding to the compound; see Figure 1e,f and supplementary table 2 | |
| Additional comments | ||
Acknowledgments
We thank Daqian Sun of the Einstein Human Stem Cell FACS and Xenotransplantation facility (Supported by NSTEM grant C024172), Jiahao Chen, Caroline K. Sheridan and Francine Garrett-Bakelman of the Weill Cornell Medical College Epigenomics Core, and the Albert Einstein College of Medicine Epigenomics Core for expert technical assistance and advice. Funding: This work was supported by the Albert Einstein Cancer Center Core Support grant (P30CA013330), NYSTEM grant #24306, and GlaxoSmithKline (GSK). E.P. is supported by NIH U24CA114737 and U10CA180827. U.C.O. is supported by NIH F31CA162770 and the MSTP grant T32 GM007288. U.S. is a Research Scholar of the Leukemia & Lymphoma Society, and the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
Footnotes
Author contributions
UCO, BB, BW, MTM, NA, AA, BS and US designed the study and experiments; UCO, EG, BP, AR, CR, CQ, AS, KJW, AJR, SJS, HQ, HZ, GJ, MFS, MB, GD, CD, PB, AJ, JC, IAD, KM, LC, MTM, NDA, PB, BS, HRW, HM, YK, and CR performed research and analyzed data; BB performed bioinformatic analysis; UCO, IAD, KM and SN conducted in vivo studies; UCO, AV, BB, BW, MC, EP, HM, NC, NA, BS, MTM, JM, AV, US interpreted experiments and directed the study; and UCO, NA, BS, MTM, and US wrote the manuscript; All authors discussed results and contributed to the writing and editing of the manuscript.
Competing interests: This work was supported by GlaxoSmithKline (GSK). EG, BP, ARR, CR, CQ, AS, KW, AR, SS, HQ, HZ, CD, GD, PB, AG, GJ, MFS, MB, GD, NC, NDA, BS, and MTM are employees of GlaxoSmithKline.
References
- 1.Rakheja D, Konoplev S, Medeiros LJ, Chen W. IDH mutations in acute myeloid leukemia. Hum Pathol. 2012;43:1541–51. doi: 10.1016/j.humpath.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R, et al. Expanding the spectrum of IDH1 mutations in gliomas. Mod Pathol. 2013;26:619–25. doi: 10.1038/modpathol.2012.210. [DOI] [PubMed] [Google Scholar]
- 3.Luchman HA, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14:184–91. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol. 2011;178:1395–402. doi: 10.1016/j.ajpath.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–73. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 9.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 10.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye D, Ma S, Xiong Y, Guan KL. R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis. Cancer Cell. 2013;23:274–6. doi: 10.1016/j.ccr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuendgen A, Germing U. Emerging treatment strategies for acute myeloid leukemia (AML) in the elderly. Cancer Treat Rev. 2009;35:97–120. doi: 10.1016/j.ctrv.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Burnett AK. Treatment of acute myeloid leukemia: are we making progress? Hematology Am Soc Hematol Educ Program. 2012;2012:1–6. doi: 10.1182/asheducation-2012.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Abdel-Wahab O, Patel J, Levine RL. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am. 2011;25:1119–33. doi: 10.1016/j.hoc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Molenaar RJ, Radivoyevitch T, Maciejewski JP, van Noorden CJ, Bleeker FE. The driver and passenger effects of isocitrate dehydrogenase 1 and 2 mutations in oncogenesis and survival prolongation. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbcan.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 21.Yen K, et al. AG-221 Offers a Survival Advantage In a Primary Human IDH2 Mutant AML Xenograft Model. Blood. American Society of Hematology Annual Meeting and Exposition; 2013. [Google Scholar]
- 22.Xu X, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–57. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 23.Deng G, et al. Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (IDH1) via Disruption of a Metal Binding Network by an Allosteric Small Molecule. J Biol Chem. 2014 doi: 10.1074/jbc.M114.608497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B, Zhong C, Peng Y, Lai Z, Ding J. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010;20:1188–200. doi: 10.1038/cr.2010.145. [DOI] [PubMed] [Google Scholar]
- 25.Rendina AR, et al. Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism. Biochemistry. 2013;52:4563–77. doi: 10.1021/bi400514k. [DOI] [PubMed] [Google Scholar]
- 26.van Westen GJ, Gaulton A, Overington JP. Chemical, target, and bioactive properties of allosteric modulation. PLoS Comput Biol. 2014;10:e1003559. doi: 10.1371/journal.pcbi.1003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy NC, Albrechtsen MT, Kerr MA. Characterization of a human granulocyte differentiation antigen (CDw15) commonly recognized by monoclonal antibodies. Biosci Rep. 1985;5:933–41. doi: 10.1007/BF01119906. [DOI] [PubMed] [Google Scholar]
- 28.Lacombe F, et al. Flow cytometry CD45 gating for immunophenotyping of acute myeloid leukemia. Leukemia. 1997;11:1878–86. doi: 10.1038/sj.leu.2400847. [DOI] [PubMed] [Google Scholar]
- 29.Sandes AF, et al. Combined flow cytometric assessment of CD45, HLA-DR, CD34, and CD117 expression is a useful approach for reliable quantification of blast cells in myelodysplastic syndromes. Cytometry B Clin Cytom. 2013;84:157–66. doi: 10.1002/cyto.b.21087. [DOI] [PubMed] [Google Scholar]
- 30.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 31.Pandolfi A, Barreyro L, Steidl U. Concise review: preleukemic stem cells: molecular biology and clinical implications of the precursors to leukemia stem cells. Stem Cells Transl Med. 2013;2:143–50. doi: 10.5966/sctm.2012-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Will B, et al. Minimal PU.1 Reduction Induces a Preleukemic State and Promotes Development of Acute Myeloid Leukemia. Nat Med. 2015 doi: 10.1038/nm.3936. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13:345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27:1986–98. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng B, et al. Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase. ACS Med Chem Lett. 2013;4:542–546. doi: 10.1021/ml400036z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis MI, et al. Biochemical, Cellular and Biophysical Characterization of a Potent Inhibitor of Mutant Isocitrate Dehydrogenase IDH1. J Biol Chem. 2014 doi: 10.1074/jbc.M113.511030. [DOI] [PMC free article] [PubMed]
- 37.Brooks E, et al. Identification and Characterization of Small-Molecule Inhibitors of the R132H/R132H Mutant Isocitrate Dehydrogenase 1 Homodimer and R132H/Wild-Type Heterodimer. J Biomol Screen. 2014;19:1193–1200. doi: 10.1177/1087057114541148. [DOI] [PubMed] [Google Scholar]
- 38.Chen SJ, et al. Rearrangements in the second intron of the RARA gene are present in a large majority of patients with acute promyelocytic leukemia and are used as molecular marker for retinoic acid-induced leukemic cell differentiation. Blood. 1991;78:2696–701. [PubMed] [Google Scholar]
- 39.Tallman MS, et al. All-trans-retinoic acid in acute promyelocytic leukemia. N Engl J Med. 1997;337:1021–8. doi: 10.1056/NEJM199710093371501. [DOI] [PubMed] [Google Scholar]
- 40.Kats LM, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–41. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan CG, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22:2339–55. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–44. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 43.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–65. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 44.Pietrak B, et al. A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry. 2011;50:4804–12. doi: 10.1021/bi200499m. [DOI] [PubMed] [Google Scholar]
- 45.Savitski MM, et al. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J Am Soc Mass Spectrom. 2010;21:1668–79. doi: 10.1016/j.jasms.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Savitski MM, et al. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal Chem. 2011;83:8959–67. doi: 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
- 47.Savitski MM, et al. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J Proteome Res. 2013;12:3586–98. doi: 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 49.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–94. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth M, et al. Eltrombopag inhibits the proliferation of leukemia cells via reduction of intracellular iron and induction of differentiation. Blood. 2012;120:386–94. doi: 10.1182/blood-2011-12-399667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Will B, et al. Effect of the nonpeptide thrombopoietin receptor agonist Eltrombopag on bone marrow cells from patients with acute myeloid leukemia and myelodysplastic syndrome. Blood. 2009;114:3899–908. doi: 10.1182/blood-2009-04-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barreyro L, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120:1290–8. doi: 10.1182/blood-2012-01-404699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Will B, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120:2076–86. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akalin A, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.