ABI5 is a prominent regulator of late seed maturation in legumes, coupling seed vigor with raffinose family oligosaccharide and late embryogenesis abundant protein accumulation and degreening.
Abstract
The preservation of our genetic resources and production of high-quality seeds depends on their ability to remain viable and vigorous during storage. In a quantitative trait locus analysis on seed longevity in Medicago truncatula, we identified the bZIP transcription factor ABSCISIC ACID INSENSITIVE5 (ABI5). Characterization of Mt-abi5 insertion mutant seeds revealed that both the acquisition of longevity and dormancy were severely impaired. Using transcriptomes of developing Mt-abi5 seeds, we created a gene coexpression network and revealed ABI5 as a regulator of gene modules with functions related to raffinose family oligosaccharide (RFO) metabolism, late embryogenesis abundant (LEA) proteins, and photosynthesis-associated nuclear genes (PhANGs). Lower RFO contents in Mt-abi5 seeds were linked to the regulation of SEED IMBIBITION PROTEIN1. Proteomic analysis confirmed that a set of LEA polypeptides was reduced in mature Mt-abi5 seeds, whereas the absence of repression of PhANG in mature Mt-abi5 seeds was accompanied by chlorophyll and carotenoid retention. This resulted in a stress response in Mt-abi5 seeds, evident from an increase in α-tocopherol and upregulation of genes related to programmed cell death and protein folding. Characterization of abi5 mutants in a second legume species, pea (Pisum sativum), confirmed a role for ABI5 in the regulation of longevity, seed degreening, and RFO accumulation, identifying ABI5 as a prominent regulator of late seed maturation in legumes.
INTRODUCTION
Seed longevity, i.e., the maintenance of viability during storage, is a crucial factor for the conservation of genetic resources and to ensure proper seedling establishment and crop yield. Longevity is progressively acquired during maturation, after seed filling, until the seed reaches a dry, quiescent state (Probert et al., 2007; Verdier et al., 2013; Righetti et al., 2015). During this period of seed development, late embryogenesis abundant (LEA) protein and raffinose family oligosaccharides (RFOs) accumulate, and chlorophyll is degraded (Rosnoblet et al., 2007; Chatelain et al., 2012; Verdier et al., 2013, Righetti et al., 2015). Seed dispersion or harvest in the case of crops can occur any time during late seed maturation (Probert et al., 2007). Therefore, the content of LEA, RFO, and/or chlorophyll in the seeds is indicative of seed maturity and vigor (Jalink et al., 1998; Sinniah et al., 1998; Vandecasteele et al., 2011; de Souza Vidigal et al., 2016). Also, during seed maturation, the progressive loss of water induces the formation of a so-called cytoplasmic glass, an amorphous matrix resembling a solid-like state where the mobility and relaxation rates of molecules are severely slowed down, thereby imposing a state of quiescence and conferring longevity (reviewed in Buitink and Leprince, 2004).
The role of LEA proteins and small heat shock proteins in longevity has been documented (Hundertmark et al., 2011; Chatelain et al., 2012; Prieto-Dapena et al., 2006). Additional protective factors conferring longevity include a set of antioxidants against oxidation occurring during storage, such as glutathione (Kranner et al., 2006), tocochromanols (Mène-Saffrané et al., 2010; Vom Dorp et al., 2015), and seed coat components, including flavonoids and cutin (Debeaujon et al., 2000; De Giorgi et al., 2015). Seed longevity has also been shown to be associated with DNA and protein repair systems, such as DNA ligase (Waterworth et al., 2010), protein l-isoaspartyl methyltransferase (Ogé et al., 2008), and methionine sulfoxide reductases (Châtelain et al., 2013). The role of RFO in seed longevity remains circumstantial. It is based on observations that RFO contents increase concomitantly with the acquisition of longevity (Sinniah et al., 1998; Verdier et al., 2013). In Arabidopsis thaliana, seeds of alpha-galactosidase2 (agal2) mutants exhibited reduced longevity (Righetti et al., 2015), and galactinol was found to be a marker for seed longevity in several species (de Souza Vidigal et al., 2016). However, there is no direct evidence for a causal relationship between the protective properties of RFO via a role in the stability of the cytoplasmic glass and longevity (Buitink et al., 2000). Alternative roles for RFOs during establishment of seed longevity include a possible function in carbon storage to provide energy that can easily be mobilized during germination after ageing, a function in mitigating environmental stress and protection against oxidative damage (Nishizawa et al., 2008).
In most plant species, chlorophyll is degraded during the late stages of seed maturation, before the seed reaches the dry state (Nakajima et al., 2012; Delmas et al., 2013; Teixeira et al., 2016). Chlorophyll retention in dry oily seeds appears to be detrimental to seed longevity. Seeds of the Arabidopsis green-seeded (grs) mutant contained twice as much chlorophyll as the wild type and exhibited reduced storage stability (Clerkx et al., 2003). Likewise, seeds of the non-yellow coloring1 (nyc1) nyc1-like (nol) double mutant affected in the conversion of chlorophyll b to chlorophyll a, the first step of chlorophyll degradation, contained 10-fold more chlorophyll than the wild type and had a strongly reduced longevity (Nakajima et al., 2012). However, the cause-effect relationship between seed longevity and chlorophyll content remains elusive.
Whereas several upstream regulators of desiccation tolerance have been identified, information is scarce on the regulatory network leading to the acquisition of longevity. Arabidopsis loss-of-function mutants of the master regulators ABSCISIC ACID INSENSITIVE3 (ABI3) and LEAFY COTYLEDON1 (LEC1) produce seeds that lose their viability during desiccation or during the first few weeks after harvest (Ooms et al., 1993; Nambara et al., 1994; Sugliani et al., 2009; Delahaie et al., 2013). Overexpression of the ABI3-regulated heat shock transcription factor A-9 from sunflower (Helianthus annuus; HaHSFA9) led to increased stability during accelerated aging in tobacco (Nicotiana tabacum) seeds (Prieto-Dapena et al., 2006; Personat et al., 2014). The A-9 transcription factor also interacts with the sunflower drought-responsive factor HaDREB2 (DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN B2) in a seed-specific manner to enhance stability during accelerated aging (Almoguera et al., 2009). Using a trait-based gene significance measure and gene network analysis, a coexpression module related to longevity was found that led to the identification of 11 transcription factors, such as WRKY3 and NUCLEAR FACTOR, X-BOX LIKE1 (NFXL1), whose mutations in Arabidopsis led to reduced longevity (Righetti et al., 2015). Another important regulator of seed maturation is DELAY OF GERMINATION1 (DOG1), originally identified for its role in dormancy, but also shown to play a role in seed storage stability and regulation of many seed maturation genes in interaction with ABI3 (Bentsink et al., 2006; Dekkers et al., 2016).
With the aim of identifying further loci involved in longevity of legume seeds, we performed a quantitative trait loci (QTL) study on a recombinant inbred line (RIL) population of Medicago truncatula that we previously used to identify loci for traits related to seed and seedling vigor and nonreducing soluble sugar content (Vandecasteele et al., 2011). Here, we identified M. truncatula ABI5 as a candidate gene involved in longevity through its colocation with a QTL of longevity and further characterized its function during seed maturation in legume seeds. ABI5 is a member of the basic leucine zipper (bZIP) family referred to as abscisic acid (ABA) response element binding (AREB) or ABA-responsive promoter element (ABRE) binding factors (ABFs), which mediate cellular responses to ABA in seeds and vegetative tissues (Finkelstein and Lynch, 2000; Jakoby et al., 2002; Fujita et al., 2005; Yoshida et al., 2010; Wang et al., 2015). Acting downstream and/or synergistically with ABI3, ABI5 functions as homo- or heterodimers that bind to ABRE elements found in the regulatory regions of many stress-related genes (Carles et al., 2002; Lopez-Molina et al., 2001; Nakabayashi et al., 2005; De Giorgi et al., 2015) as well as light-signaling genes (Lee et al., 2012; Sakuraba et al., 2014).
In Arabidopsis, ABI5 regulates seed germination, dormancy, and seedling growth. After germination, ABI5 is necessary for ABA-induced growth arrest under unfavorable conditions (Lopez-Molina et al., 2001) and the reinduction of desiccation tolerance in germinated seeds of Arabidopsis (Maia et al., 2014) and M. truncatula (Terrasson et al., 2013). ABI5 also integrates light and ABA signals during germination and seedling growth (Lee et al., 2012; Tang et al., 2013). However, little is known about a possible role of ABI5 during seed maturation. In M. truncatula, Mt-ABI5 was found to be a hub in the gene network regulating seed survival in the dry state (Verdier et al., 2013), but a role in seed longevity was not investigated.
Using mutants of two legume species, M. truncatula and pea (Pisum sativum), here, we demonstrate a role for Mt-ABI5 in the acquisition of seed longevity during seed maturation. Further transcriptomic and biochemical analysis demonstrate a role for Mt-ABI5 during seed maturation, regulating the accumulation of LEA proteins, RFO, and expression of photosynthetic genes and dismantling of the photosynthetic apparatus. Our results reveal that Mt-ABI5 is an important regulator of late seed maturation in legumes, connecting degreening with the acquisition of seed longevity.
RESULTS
The bZIP Transcription Factor Gene ABI5 Colocates with QTL for Longevity, RFO Content, and Germination Rate of M. truncatula Seeds
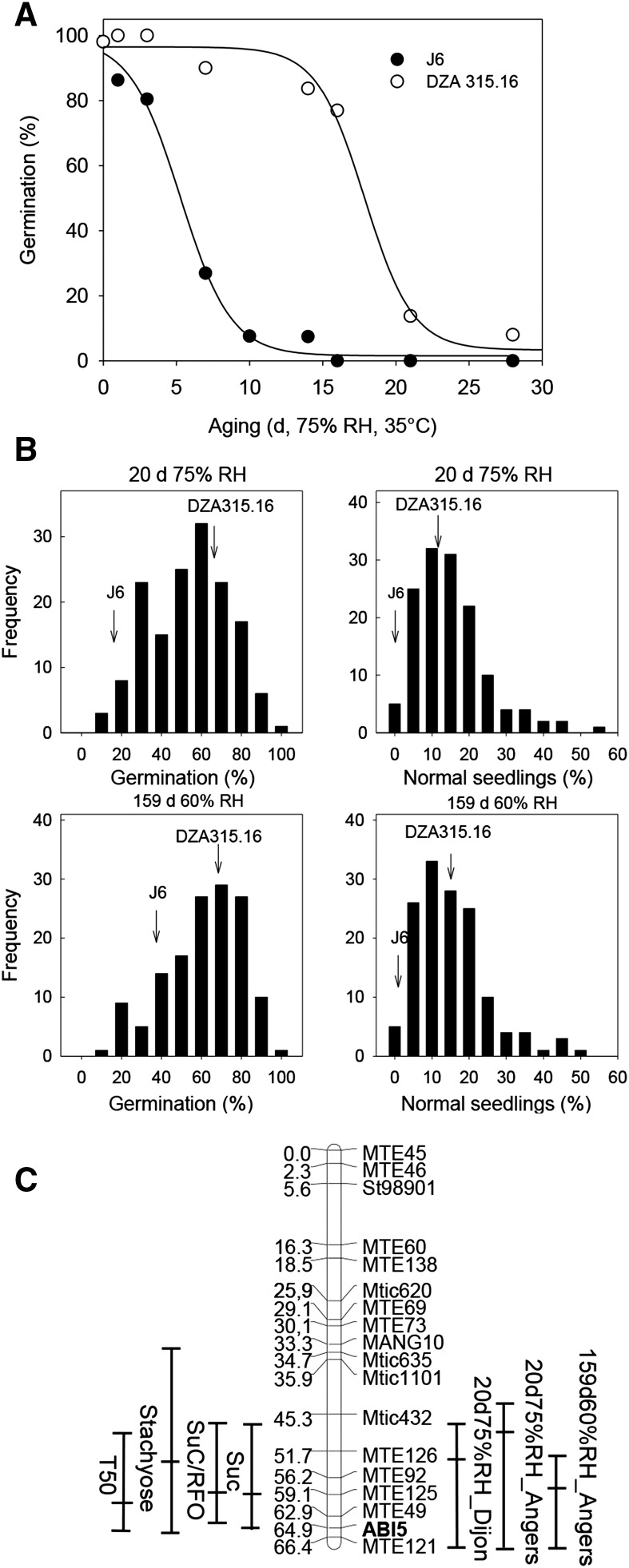
To identify M. truncatula populations with genetic variability in seed longevity, we screened the parents of different sets of RIL populations. Thus, a strong genotypic difference in longevity was detected between the M. truncatula genotypes Jemalong-6 (A17) and DZA315.16, with seed longevity being 4-fold higher in DZA315.16 (Figure 1A). Based on these parental differences, the variability in longevity was assessed in 171 RILs derived from the cross between J6 and DZA315.16 (i.e., the LR-4 population). Two storage conditions were chosen to assess seed life span: (1) 75% relative humidity (RH) at 35°C, known to induce a fast deterioration (Verdier et al., 2013); and (2) 60% RH at 35°C, corresponding to moderate aging. The percentage of final germination and normal seedling morphology was determined after 20 and 159 d of storage under the two conditions, respectively. A large variation was found in these parameters, with germination percentages varying between 10 and 100%, and normal seedling percentages between 0 and 50% (Figure 1B). Ten QTL were detected for seed longevity during accelerated aging, and five during moderate aging, corresponding to six chromosomal regions (Table 1). Three regions showed a colocation between QTL for accelerated aging and moderated aging, explaining between 10 and 21% of the variation (Table 1).
Figure 1.
QTL Analysis of Seed Longevity.
(A) Loss of viability during storage at 75% RH, 35°C of both parents (J6 and DZA 315.16) using triplicates of 30 to 50 seeds obtained from three to five plants. Data (means ± se) were fitted with sigmoidal curves.
(B) Frequency distribution of percentages of germination and normal seedlings after storage obtained from the LR4 RILs. Time and conditions of storage are indicated together with the value obtained from the respective parents.
(C) Genetic map of linkage group 7 showing the QTL mapping for time to 50% germination (T50), sugar content, and viability after indicated storage times and conditions from seeds produced in Angers and Dijon (France). The lines indicate the confidence intervals and the peak positions.
Table 1. Summary of Detected QTL for Aging Traits in the M. truncatula LR4 Population Originating from a Cross between J6 and DZA315.16.
| Trait | LG | Peak Position (cM) | Confidence Interval (cM) | LOD | R2 (%) | Additive Effect J6 (%) |
|---|---|---|---|---|---|---|
| 20 d aging 75% RH on %FG | 1 | 67.8 | 62.8–81.5 | 5.2 | 13.2 | 6.9 |
| 159 d aging 60% RH on %NS | 1 | 86.3 | 45–68.3 | 3.1 | 9.7 | 2.8 |
| 20 d aging 75% RH on %FG | 3 | 59.3 | 53.6–62.2 | 5.4 | 13.7 | −9.4 |
| 20 d aging 75% RH on %NS | 3 | 59.3 | 51.2–62.2 | 3.7 | 10.2 | −9.4 |
| 159 d aging 60% RH on %FG | 3 | 63.5 | 61.2–66.4 | 6.2 | 17.4 | −9.8 |
| 14 d aging 75% RH on %FG | 3 | 65.5 | 55.5–66.4 | 3.6 | 12.8 | −9.1 |
| 159 d aging 60% RH on %NS | 3 | 65.5 | 41.9–66.4 | 3.5 | 10.3 | −4.1 |
| 159 d aging 60% RH on %FG | 6 | 15.1 | 0–35.6 | 4.0 | 12.1 | 6.6 |
| 20 d aging 75% RH on %NS | 7 | 47.3 | 39.0–66.4 | 4.0 | 10.7 | −2.1 |
| 20 d aging 75% RH on %NS Dijon | 7 | 54.2 | 47.4–66.6 | 5.3 | 13.2 | −8.9 |
| 159 d aging 60% RH on %NS | 7 | 58.2 | 52.2–66.4 | 4.8 | 14.1 | −3.8 |
| 20 d aging 75% RH on %NS | 8 | 48.3 | 40.8–54.6 | 6.0 | 15.7 | −8.3 |
| 14 d aging 75% RH on %FG | 8 | 49.7 | 43.1–56.1 | 6.6 | 21.1 | −9.2 |
| 14 d aging 75% RH on %NS | 8 | 49.7 | 41.7–74.7 | 3.3 | 11.8 | −6.1 |
| 20 d aging 75% RH on %FG | 8 | 50.3 | 41.2–66 | 5.1 | 13.0 | −6.7 |
Aging assays were performed at two RHs (60% and 75%) and 35°C. Percentages of final germination (%FG) and of normal seedlings (%NS) were determined at the indicated days of storage. Position in centimorgans (cM) from the LR4 map are described by Vandecasteele et al. (2011). LG, linkage group; LOD, logarithm of the odds; R2, percentage of phenotypic variance explained.
To identify candidate genes that might be responsible for these QTL, genes encoding transcription factors that were previously identified as being correlated with survival in the dry state (Verdier et al., 2013) were localized on the chromosomes. The Mt-ABI5 gene, a bZIP transcription factor and the homolog of Arabidopsis ABI5 (Wang et al., 2015), was located at position 64.9 on LG 7, within the confidence interval of three QTL for longevity both for moderate and accelerated aging with seeds obtained from two different growth locations (Angers and Dijon) (Figure 1C, Table 1). This chromosomal region also contained QTL for the composition of soluble sugars Suc and stachyose (Sta), for the ratio Suc/RFO, and a QTL for germination rate, i.e., T50 (time to reach 50% germination; Figure 1C) that were previously identified by our group (Vandecasteele et al., 2011). The Mt-ABI5 gene was amplified by PCR from both parental lines. Several polymorphisms were detected in the upstream promoter region and intron regions and one single nucleotide polymorphism was found in an exon, resulting in a missense variant R177G (Supplemental Table 1). Mapping of the gene confirmed its localization on linkage group 7 (Supplemental Table 1; Figure 1C).
Mt-ABI5 Regulates the Acquisition of Seed Longevity and Dormancy during Seed Maturation in M. truncatula
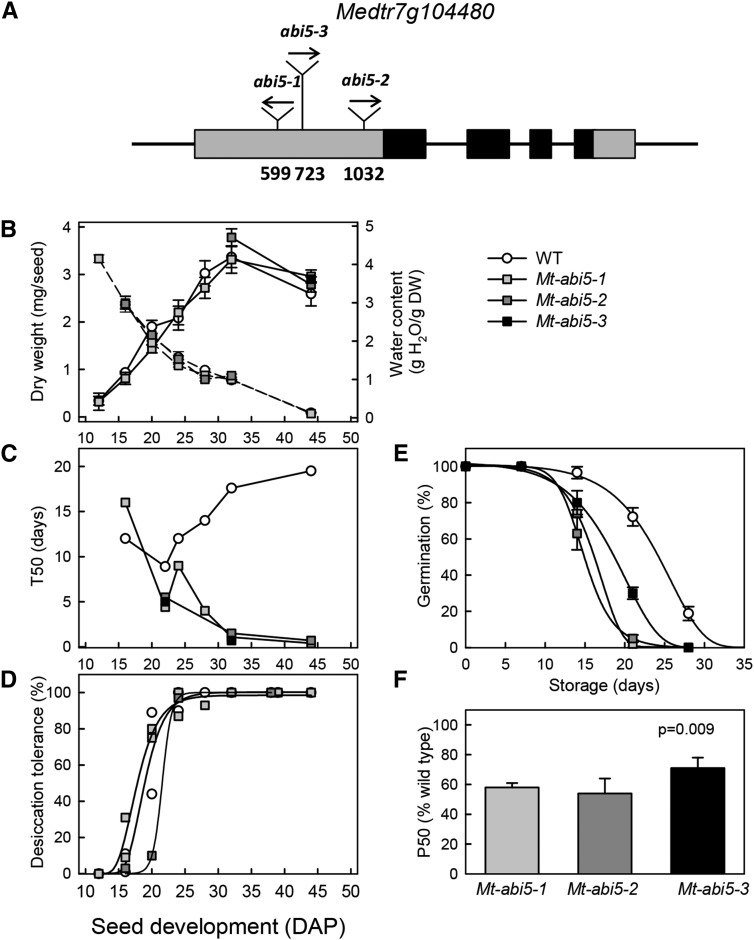
The role of Mt-ABI5 in seed longevity in M. truncatula was investigated using Tnt1 insertion mutants. Previously, we reported ABA insensitivity during germination for two Mt-abi5 alleles (Terrasson et al., 2013), corresponding to Mt-abi5-1 and Mt-abi5-2. Here, we identified a third insertion mutant allele, Mt-abi5-3, for which ABA insensitivity during germination was also confirmed (Supplemental Figure 1). The insertions of the Tnt1 transposons were all located 5′ to the bZIP domain (Figure 2A). To assess the impact of the Mt-abi5 mutation on seed development, physiological indicators of maturation were monitored from seed filling onwards. No differences in the accumulation of dry weight or the loss of water content during maturation were observed between wild-type and mutant seeds (Figure 2B). Total time of maturation until abscission of the pod was also comparable for all lines, being around 44 d after pollination (DAP). All genotypes acquired their capacity to germinate at the same time at around 15 d. However, in wild-type seeds, time to germination (as assessed by T50) increased during maturation from 10 to 18 d, indicative of the establishment of dormancy (Bolingue et al., 2010; Figure 2C). By contrast, time to germination of Mt-abi5 seeds steadily decreased during maturation, with a T50 of only 12 h for fully mature seeds (Figure 2C; Supplemental Figure 1). This value was considerably lower than that of completely after-ripened wild-type seeds (T50 = 23 h; Bolingue et al., 2010), indicating that not only dormancy but also germination rate is affected in the Mt-abi5 mutants.
Figure 2.
Mt-ABI5 Affects Seed Physiological Quality during Seed Development.
(A) Gene structure of Medtr7g104480 showing the Tnt1 insertions in the three Mt-abi5 alleles. Boxes and lines indicate exons and introns, respectively. The gray boxes indicate the open reading frame and the black boxes indicate the bZIP domain.
(B) Changes in seed dry weight and water content during maturation. Data are the means (±se) of four replicates of five seeds for immature seeds and 60 seeds for mature seeds.
(C) Changes in dormancy evaluated as the time necessary to obtain 50% germination. Data from two independent cultures are shown and were obtained from 50 seeds for each time point.
(D) Acquisition of desiccation tolerance evaluated after fast drying and imbibition using 30 to 50 seeds from a pool of 20 plants.
(E) Representative loss of viability during storage at 75% RH, 35°C using triplicates of 30 to 50 seeds from a pool of 20 plants. Data (means ± se) were fitted with sigmoidal curves to estimate the P50 (time necessary to obtain a loss of viability of 50% during storage).
(F) Longevity of Mt-abi5 seeds as assessed by calculating P50 during storage of seeds obtained from two to four independent cultures and storage experiments. Data (means ± se) are expressed as percentage of wild-type seeds from plants grown concomitantly with mutants. The level of significance (P) between mutants and the wild type using an ANOVA test (see Supplemental Data Set 3) is indicated.
Seeds of Mt-abi5 acquired desiccation tolerance at the same developmental stage as wild-type seeds (Figure 2D). Longevity was determined by the construction of survival curves, showing the decrease in germination at the indicated time after storage at 75% RH, 35°C (Figure 2E). Viability of the Mt-abi5 seeds was lost earlier during storage than that of the wild type. The P50, the time of storage needed to obtain a loss of 50% of germination, was consistently reduced in the Mt-abi5 seeds to 60 to 70% of the wild-type values, for different harvests (Figure 2F).
A Gene Network-Based Analysis of the Mt-abi5 Transcriptome Reveals Distinct Gene Modules with Specific Biological Functions during Seed Maturation
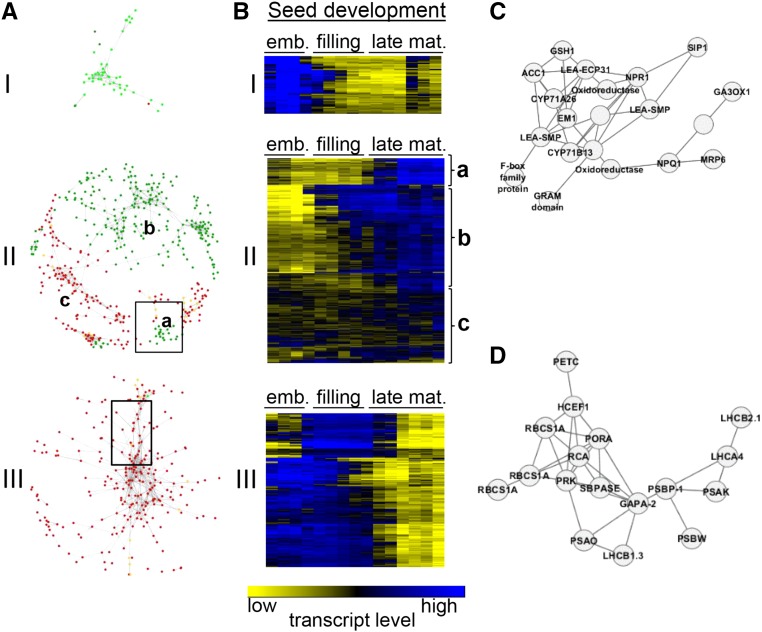
The reduction of dormancy and longevity in the Mt-abi5 mutant lines indicates that the late seed maturation is deregulated. To understand the basis for the deregulation, the transcriptomes of wild-type and Mt-abi5 seeds were analyzed at three stages during seed maturation: 16 DAP (beginning of seed filling); 24 DAP (mid seed filling, when desiccation tolerance is acquired); and pod abscission, when longevity has been fully established. A total of 2042 and 869 probes were up- and downregulated respectively, in Mt-abi5 mutants compared with wild-type seeds (Supplemental Data Set 1). Venn diagrams showed that this differential expression occurred mostly at abscission, with 1868 (91%) and 730 (84%) transcripts exhibiting increased and decreased levels, respectively (Supplemental Data Set 1). Only a few probes differed between Mt-abi5 and the wild type at 16 DAP (17 up- and 66 downregulated) and 24 DAP (28 up- and 57 downregulated) (see Venn diagram in Supplemental Data Set 1). Using the seed maturation transcriptome that we previously generated (Righetti et al., 2015; see http://bar.utoronto.ca/efpmedicago/cgi-bin/efpWeb.cgi?dataSource=medicago_seed), we noticed that differentially expressed probes in developing Mt-abi5 seeds can be grouped in different sets of expression profiles compared with the wild type. To capture the temporal regulation of these probes during seed development, a coexpression network was constructed using the intensities (transcript levels) of the wild-type and Mt-abi5 mutant seeds together with those obtained at 15 time points during wild-type seed maturation (Figure 3).
Figure 3.
Gene Coexpression Network of Deregulated Transcripts in Mt-abi5 Mutant Seeds during Development.
(A) Gene coexpression modules (I to III) during development of Mt-abi5 seeds. Temporal analysis of nodes is depicted by coloring genes according to their specific expression in comparison with wild-type seeds. Light-green nodes represent downregulated transcripts in Mt-abi5 seeds at 16 DAP. Dark-yellow nodes represent upregulated transcripts in Mt-abi5 seeds at 24 DAP. Dark-green and dark-red nodes correspond to down- and upregulated transcripts, respectively, in Mt-abi5 at 40 DAP (mature seeds). Gene interactions were visualized using Cytoscape using an edge-weighted spring-embedded layout.
(B) Expression profiles of the nodes shown in (A) during seed development in wild-type seeds. Data are derived from Righetti et al. (2015). The color scale corresponds to transcript level (low = yellow; high = blue). Emb, embryogenesis; late mat., late maturation.
(C) Focus on the regulation of genes involved in protection against desiccation (box shown in module IIa).
(D) Focus on the regulation of genes involved in photosynthesis (box shown in module III).
Three main gene modules were detected that regrouped 1147 nodes (probes) with 2409 edges and that are hereafter referred to as modules I, II, and III (Figure 3A). To understand the topology of the network, nodes were colored based on their expression profile. Expression of most of the probes was deregulated (repressed or increased) at final maturation (Figures 3A and 3B). Module I contained 51 transcripts that were differentially downregulated at 16 DAP in the Mt-abi5 seeds compared with wild-type seeds (Figure 3A). This module was significantly enriched with Gene Ontology (GO) terms associated with cell wall organization and pectin esterase activity, with gene products predicted to be localized in the cell wall or apoplast (Table 2). Expression profiles of probes in this module in wild-type seeds showed elevated transcripts during late embryogenesis and early seed filling, after which transcript levels decreased (Figure 3B, top).
Table 2. GO Enrichment Analysis of Maturation Modules Obtained from Gene Coexpression Analysis.
| GO Term | Ontology | Description | No. in Input List | #BG/Ref | FDR |
|---|---|---|---|---|---|
| Module I | |||||
| GO:0042545 | P | Cell wall modification | 4 | 386 | 0.0081 |
| GO:0030599 | F | Pectin esterase activity | 4 | 148 | 1.60E-11 |
| GO:0005618 | C | Cell wall | 10 | 618 | 7.90E-20 |
| GO:0048046 | C | Apoplast | 5 | 406 | 1.70E-06 |
| Module II, cluster a | |||||
| GO:0009793 | P | Embryonic development ending in seed dormancy | 6 | 465 | 0.00014 |
| GO:0048316 | P | Seed development | 6 | 530 | 0.00046 |
| GO:0006952 | P | Defense response | 7 | 766 | 0.00082 |
| GO:0009725 | P | Response to hormone stimulus | 8 | 982 | 0.00091 |
| GO:0009628 | P | Response to abiotic stimulus | 10 | 1471 | 0.0012 |
| GO:0009266 | P | Response to temperature stimulus | 5 | 485 | 0.0036 |
| GO:0019748 | P | Secondary metabolic process | 5 | 489 | 0.0036 |
| GO:0048608 | P | Reproductive structure development | 7 | 978 | 0.0076 |
| Module II, cluster b | |||||
| GO:0015979 | P | Photosynthesis | 7 | 162 | 5.00E-19 |
| GO:0006091 | P | Generation of precursor metabolites and energy | 5 | 285 | 0.00028 |
| GO:0006952 | P | Defense response | 7 | 766 | 0.023 |
| Module II, cluster c | |||||
| GO:0006915 | P | Apoptosis | 4 | 159 | 2.80E-07 |
| GO:0012501 | P | Programmed cell death | 4 | 244 | 0.00035 |
| GO:0006457 | P | Protein folding | 4 | 275 | 0.00083 |
| GO:0009408 | P | Response to heat | 3 | 161 | 0.0011 |
| GO:0006952 | P | Defense response | 6 | 766 | 0.0088 |
| GO:0009743 | P | Response to carbohydrate stimulus | 3 | 240 | 0.036 |
| Module III | |||||
| GO:0015979 | P | Photosynthesis | 13 | 162 | 5.30E-26 |
| GO:0010218 | P | Response to far red light | 5 | 57 | 3.70E-09 |
| GO:0009637 | P | Response to blue light | 5 | 69 | 2.40E-07 |
| GO:0006066 | P | Alcohol metabolic process | 9 | 270 | 2.10E-05 |
| GO:0006629 | P | Lipid metabolic process | 16 | 841 | 0.00043 |
| GO:0009416 | P | Response to light stimulus | 12 | 596 | 0.0024 |
| GO:0006461 | P | Protein complex assembly | 5 | 134 | 0.0031 |
| GO:0009314 | P | Response to radiation | 12 | 613 | 0.0031 |
| GO:0009409 | P | Response to cold | 8 | 328 | 0.0047 |
| GO:0009699 | P | Phenylpropanoid biosynthetic process | 5 | 141 | 0.0047 |
| GO:0006952 | P | Defense response | 12 | 766 | 0.04 |
Analysis was performed using AgriGO applying a χ2 test with the Yekutieli multitest adjustment method. #BG/ref, number of genes annotated in GO term in background; FDR, false discovery rate; P, biological process; F, molecular function, C, cellular component.
In module II, three subclusters were identified and analyzed separately. Cluster IIa contained genes with both up- and downregulated transcripts in the Mt-abi5 seeds compared with wild-type seeds, in which their level increased during the final stages of seed maturation (Figure 3B). Cluster IIb was characterized by overrepresented biological functions related to “embryonic development ending in seed dormancy,” “defense response,” “response to hormone stimulus and temperature,” as well as “secondary processes” and “reproductive structure development” (Table 2). A zoom on part of this cluster containing probes with downregulated transcripts in the Mt-abi5 seeds compared with the wild type revealed several transcripts encoding LEA proteins including EM1 and several members of the seed maturation protein family (SMP, also referred to as D-34 family; Figure 3C). This cluster also contained homologs of γ-GLUTAMYLCYSTEINE SYNTHETASE (GSH1), which catalyzes the first step of glutathione biosynthesis; NON-PHOTOCHEMICAL QUENCHING1 (NPQ1) involved in the xanthophyll cycle; and SEED IMBIBITION PROTEIN1 (SIP1), a raffinose synthase involved in RFO synthesis. Cluster IIb contained probes with decreased transcript levels in the Mt-abi5 seeds, whereas they increased from mid seed filling onwards during wild-type seed maturation (Figure 3B). This cluster was overrepresented with GO categories associated with photosynthesis, including nodes representing 14 probes encoding chloroplastic Arabidopsis homologs (Table 2). Cluster IIc contained probes with increased transcript levels in the Mt-abi5 mutants during late maturation compared with wild-type seeds. Most of these probes did not show differential regulation during wild-type seed development and transcript levels were low (Figure 3B). This cluster was overrepresented by genes related to programmed cell death and protein folding, including many small heat shock proteins and chaperonins (Table 2), indicating a stress response in developing mutant seeds.
Module III contained 253 probes with transcripts that were higher in the Mt-abi5 seeds at pod abscission compared with wild-type seeds (Figure 3A). In wild-type seeds, these genes are highly expressed during embryogenesis and seed filling, while expression decreases during late seed maturation (Figure 3B), indicating that this downregulation is impaired in the Mt-abi5 mutants. GO enrichment analysis showed a strong overrepresentation in categories associated with photosynthesis and light response (Table 2). Figure 3D shows a detail of the module enriched in these genes: many encode subunits or core units of both PSI and PSII: PHOTOSYSTEM I LIGHT HARVESTING COMPLEX GENE1 (LHCA1), PHOTOSYSTEM I SUBUNIT D-2 (PsaD-2), PHOTOSYSTEM I SUBUNIT F (PsaF), PsaK, PsaG, and PsaH-2 for PSI and LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5 (Lhcb5), PHOTOSYSTEM II SUBUNIT P (PsbP), PsbQ, and PHOTOSYSTEM II REACTION CENTER W (PsbW) for PSII. Likewise, a number of genes encoding proteins involved in the Calvin cycle are part of this cluster, as well as a PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A (PORA), involved in the reduction of protochlorophyllide to chlorophyllide (Figure 3D, Table 2).
Mt-ABI5 Regulates Nonreducing Sugar Metabolism and Accumulation of Longevity-Associated LEA Proteins during Late Seed Maturation
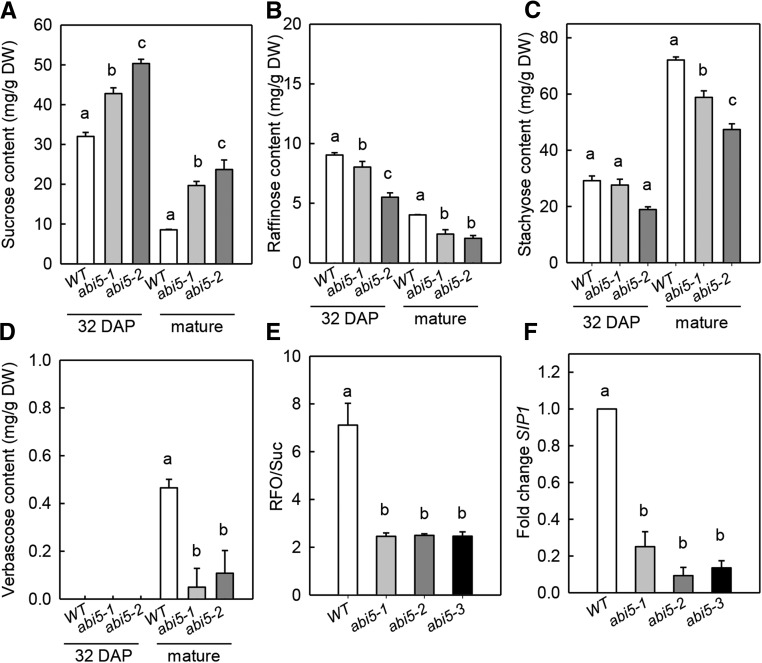
The presence of an Arabidopsis homolog of the raffinose synthase SIP1 in gene module IIb together with the colocation of RFO and longevity QTL suggests a role of Mt-ABI5 in regulating nonreducing sugar metabolism during seed maturation. To test this hypothesis, soluble sugars were determined in Mt-abi5 and wild-type seeds at two time points during seed development: 32 DAP, corresponding to the end of seed filling, beginning of maturation drying, and chlorophyll loss; and maturity, being around 44 DAP. In wild-type seeds, Suc content decreased between 32 DAP and mature seeds, whereas Stac, the major RFO in M. truncatula and verbascose (Ver) contents increased (Figures 4A to 4D). At 32 DAP, Mt-abi5 seeds contained already significantly more Suc (ANOVA, P value < 0.001; Supplemental Data Set 3), and this difference remained detectable in mature seeds (Figure 4A). By contrast, no significant difference was observed in Sta and Ver contents at 32 DAP between Mt-abi5 and wild-type seeds, whereas a slight difference was detectable for Raf. During further maturation, Sta and Ver accumulation also were compromised in the Mt-abi5 seeds (Figures 4C and 4D). Consequently, the ratio of RFO/Suc was lower for seeds of the three Mt-abi5 mutant alleles, suggesting that the conversion of Suc into RFO is in part under regulation of Mt-ABI5 during maturation drying (Figure 4E). A possible role for this conversion could be through the transcriptional regulation of SIP1 from gene module IIb (Figure 3C). RT-qPCR analysis confirmed that the transcript level of this gene was strongly reduced in mature Mt-abi5 mutant seeds (Figure 4F). In silico analysis of the upstream promoter sequence of SIP1 revealed the presence of an ABRE, suggesting a possible binding site for Mt-ABI5. Yeast one-hybrid assays confirmed that a 56-bp promoter fragment of Mt-SIP1 containing the ABRE interacted with the Mt-ABI5 protein, whereas this interaction was abolished when the ABRE was mutated (Supplemental Figure 2).
Figure 4.
Sugar Metabolism Is Affected in Mt-abi5 Seeds.
(A) to (D) Contents in sucrose, raffinose, stachyose, and verbascose in wild-type and Mt-abi5 seeds harvested at 32 DAP and at pod abscission (mature).
(E) Ratio of RFO/Suc. Data are the average (±se) of four replicates of 10 seeds each from a pool of 20 plants.
(F) Expression of Mt-SIP1 (SEED IMBIBITION PROTEIN1/RAFFINOSE SYNTHASE) in mature wild-type and Mt-abi5 seeds. Expression was normalized using Mt-MSC27 and Mt-ACTIN11 as reference genes. Data are the means of three independent biological replicates. Significantly different values (assessed by ANOVA, P < 0.05; Supplemental Data Set 3) were ranked into groups as indicated by the respective letter using the Student-Newman-Keuls test.
In addition, module IIa contained several transcripts encoding LEA proteins (Figure 3C). For a number of these genes, we previously reported that their corresponding polypeptide abundance increased in parallel with the acquisition of longevity in M. truncatula seeds (Chatelain et al., 2012). Considering the posttranscriptional regulation of these LEA proteins (Verdier et al., 2013), we investigated whether the decrease in LEA transcripts in Mt-abi5 seeds also resulted in decreased LEA protein accumulation. A protein fraction enriched in LEA polypeptides was isolated, taking advantage of their hydrophilic and unstructured character, which allows them to remain in solution after heating the crude protein extract at 95°C (Boudet et al., 2006). The polypeptides present in this so-called “heat-stable” fraction were previously identified to establish a reference map for subsequent characterization of the LEA proteome in M. truncatula (Chatelain et al., 2012 for A17; Delahaie et al., 2013 for R-108) (Supplemental Figure 3A).
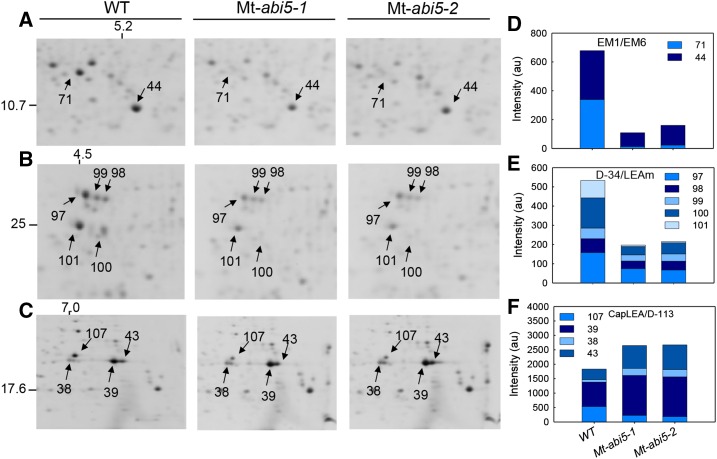
The abundance of seventeen polypeptides was at least twofold reduced in the Mt-abi5 seeds compared with the wild type (Supplemental Data Set 2 and Supplemental Figures 3B to 3D). The polypeptide showing the strongest change (18-fold) was the EM1 protein, whereas its family member, EM6, was reduced 4-fold (Figure 5A). Both polypeptides together represented 8% of the total spot intensities of the 2D gel (Supplemental Data Set 2). Almost all the polypeptides corresponding to SEED BIOTINYLATED PROTEIN65 (SBP65) were reduced in abundance (Supplemental Data Set 2). Also, the abundance of all the spots corresponding to D-34 proteins or a mix of D-34 with LeaM (spot 100), a mitochondrial LEA protein, was strongly reduced in the Mt-abi5 seeds (Figure 5B). All of the transcripts of these LEA proteins were downregulated in the Mt-abi5 mutants, except for LeaM. This suggests that the reduction of intensity of spot 100 is due to a reduction in D-34 rather than LeaM.
Figure 5.
Mt-ABI5 Affects LEA Polypeptide Accumulation.
(A) to (C) Focus on specific areas of a representative 2D gel showing differentially expressed polypeptides between mature wild-type and Mt-abi5 seeds. Spot numbers indicate the following polypeptides. (A): 44, EM6; 71, EM1. (B): 97, D34-I; 98, D-34-I/LEAm; 99, LEAm; 100, PM25/D35-II; 101, D34-II/PM25. (C): 39, 38, 107, Cap LEA 1; 43, Protein D-113.II.
(D) to (F) Polypeptide abundance of the indicated LEA polypeptides. Data are the average of spot intensities obtained from five to six gels.
Four highly abundant LEA polypeptides, comprising 13% of the total spot intensity, were over 2-fold more abundant in the heat-stable proteome of the Mt-abi5 seeds compared with the wild type. They corresponded to PM10, a dehydrin (DHN), a member of the D113 family D-113 II (spot 43), and a CapLEA polypeptide (spot 38) (Figure 5C). These results corroborated the transcriptomic analysis in which a significant upregulation was found for D-113 II and CapLEA (Supplemental Data Set 1). Next to LEA polypeptides, three polypeptides that showed the highest increase in abundance in the Mt-abi5 seeds compared with the wild type were identified as GLYCINE-RICH PROTEIN2, PROTEIN DISULFIDE ISOMERASE and CHLOROPLAST CHAPERONIN10 (CPN10), all of which were involved in protein folding upon the stress response (Supplemental Data Set 2).
Chloroplast Pigment Composition and Photosystem-Associated Gene Expression Are Altered in Mature Mt-abi5 Seeds
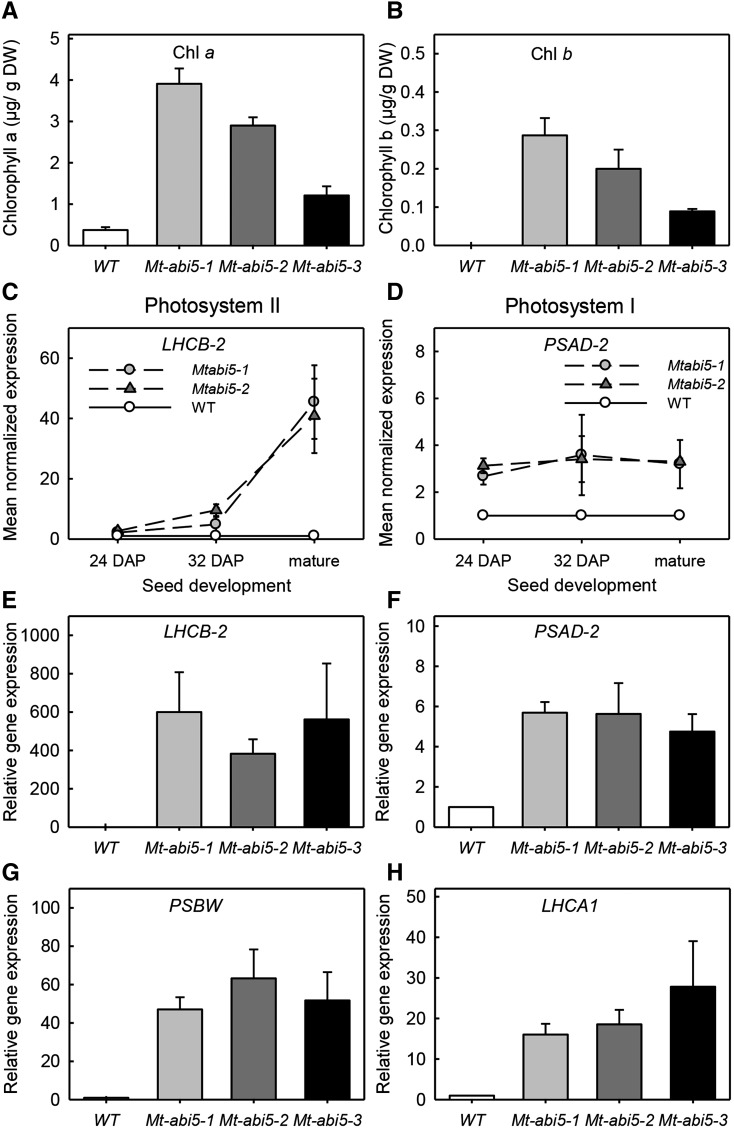
Considering that coexpression modules affected by Mt-abi5 were enriched with categories associated with photosynthesis genes (Figure 3, Table 2), we investigated whether chlorophyll degradation was also affected (Figures 6A and 6B). In mature wild-type seeds, only a residual amount of chlorophyll a was detected, whereas by contrast, Mt-abi5 seeds contained 3- to 10-fold more chlorophyll a than the wild type (Figure 6A). The Mt-abi5 seeds also contained small amounts of chlorophyll b, whereas it had completely disappeared from wild-type seeds (Figure 6B). During seed development, a difference in chlorophyll degradation was already evident at 24 and 32 DAP, although the differences were not significant due to high variation between the biological replicates (Supplemental Figure 4).
Figure 6.
Chlorophyll and Photosystem-Associated Gene Expression Are Affected in Mt-abi5 Seeds.
(A) and (B) Chlorophyll a and chlorophyll b contents in mature seeds. Data are the means (±se) of three replicates of 100 seeds from a pool of 20 plants.
(C) and (D) Level of expression of Mt-LCHB-2 and Mt-PSAD-2during seed maturation at 24 and 32 DAP and in mature seeds. Data were normalized using expression values found for wild-type seeds at each specific stage.
(E) to (H) Level of expression of PSII genes (Mt-LCHB-2 and Mt-PSBW) and PSI genes (Mt-LCHA1 and Mt-PSAD-2) in mature wild-type and Mt-abi5 seeds. Data were normalized using expression values of wild-type seeds. Mt-MSC27 and Mt-ACTIN11 were used as reference genes. Data are the means (±se) of three independent biological replicates. All values for Mt-abi5 seeds were significantly different from the wild type, except for Mt-LCHB-2 gene expression at 24 DAP.
Chlorophyll b must be converted to chlorophyll a by NYC1 or NOL as a first step toward chlorophyll degradation (Christ and Hörtensteiner, 2014). Therefore, we assessed whether the expression of genes involved in the chlorophyll breakdown pathway was affected in Mt-abi5 seeds (Supplemental Figure 5). No significant difference in Mt-NYC1 expression between wild-type and mutant seeds was detected by RT-qPCR (Supplemental Figure 5B). A closer inspection of the transcriptome data showed that the levels of transcripts encoding enzymes involved in the degradation of chlorophyll were either not significantly different or higher in Mt-abi5 compared with mature wild-type seeds (NYC1, NYE1, and PHEOPHORBIDE A OXYGENASE [PAO]) (Supplemental Figure 5C; Supplemental Data Set 1).
ABI5 has been shown to be involved in the delay of dark-induced senescence in Arabidopsis seedlings (Sakuraba et al., 2014). In contrast to Arabidopsis, no difference could be observed in dark-induced senescence in leaves from the M. truncatula Mt-abi5 mutant compared with the wild type. Whereas chlorophyll content decreased during senescence, no significant difference was detected between the Mt-abi5 mutant plants and wild-type lines (Supplemental Figure 6). Therefore, none of these findings support a direct role for Mt-ABI5 in the chlorophyll degradation at the final steps of maturation.
The transcriptome analysis identified numerous transcripts from genes encoding different subunits of PSI and PSII that remained high in the mature Mt-abi5 seeds, whereas transcripts were completely degraded in wild-type seeds (Figure 3; Supplemental Figure 7A). We confirmed these observations by RT-qPCR analysis on a new harvest of Mt-abi5 and wild-type seeds (Figures 6C to 6H; Supplemental Figure 7B). Transcript levels of Mt-LHCB-2 and Mt-PSAD-2, belonging to PSII and PSI, respectively, were already 2-fold higher at 24 DAP in Mt-abi5 seeds compared with the wild type (Figures 6C and 6D). During seed maturation, this difference increased progressively for Mt-LHCB-2, resulting in relative amounts of transcripts that were 50-fold higher in mature Mt-abi5 seeds compared with the wild type (Figure 6C), whereas the difference remained stable for Mt-PSAD-2 (Figure 6D). Transcript levels for several genes increased 6- to 600-fold in mature Mt-abi5 seeds (Figures 6E to 6H), with the strongest changes for transcripts of PSII (Figures 6E and 6G).
Carotenoids are present in the reaction center and, like chlorophyll, are known to decrease during seed maturation (Gonzalez-Jorge et al., 2013; Monma et al., 1994). Therefore, to support the hypothesis that chloroplasts are not entirely degraded in Mt-abi5 mutants, we assessed whether the steady state level of carotenoids was affected in Mt-abi5 seeds during maturation by HPLC (Table 3). The decrease in the carotenoids β-carotene and lutein started between 24 and 32 DAP in both wild-type and Mt-abi5 plants, but no significant differences were found between the genotypes at these stages (Supplemental Figure 7C). However, in mature Mt-abi5 seeds, β-carotene and lutein contents were 2- and 5- to 6-fold higher, respectively (Table 3; Supplemental Figure 7C). We also detected two other carotenoids that were significantly more abundant in mutant seeds, tentatively identified as zeaxanthin and antheraxanthin, based on their retention time and absorbance spectra of the peaks (Thayer and Björkman, 1990; Table 3).
Table 3. Contents of Carotenoids and Tocochromanols of Mature Seeds of M. truncatula.
| Compound | Genotype |
|||
|---|---|---|---|---|
| Wild Type | Mt-abi5-1 | Mt-abi5-2 | Mt-abi5-3 | |
| Carotenoids (µg g DW−1) | ||||
| Lutein | 1.2 ± 0.1 (a) | 9.6 ± 0.6 (b) | 6.9 ± 1.2 (b) | 7.1 ± 0.7 (b,c) |
| β-Carotene | 1.2 ± 0.9 (a) | 5.0 ± 0.8 (b) | 3.8 ± 0.3 (c) | 2.3 ± 0.7 (d) |
| Antheraxanthin | 0.2 ± 0.2 (a) | 3.6 ± 0.2 (b) | 2.8 ± 0.1 (c) | 1.8 ± 0.2 (d) |
| Zeaxanthin | 1.8 ± 0.3 (a) | 10.2 ± 1.0 (b) | 10.0 ± 0.1 (b) | 9.0 ± 0.6 |
| Tocochromanol (nmol g DW−1) | ||||
| α-Tocopherol | 752 ± 17 (a) | 931 ± 25 (b) | 934 ± 17 (b) | ND |
| β-Tocopherol | 8.8 ± 1.7 | 10.5 ± 0.7 | 9.5 ± 0.5 | ND |
| λ-Tocopherol | 34.0 ± 0.8 | 35.3 ± 2.9 | 39.9 ± 2.9 | ND |
| PC-8 | 3.0 ± 1.9 (a,b) | 0.8 ± 0.5 (a) | 5.8 ± 1.4(b) | ND |
For tocopherols, data are the means (±se) of five replicates using 20 seeds from a pool of 20 plants. For carotenoids, data are the means (±se) of three replicates of 100 seeds from a pool of 20 plants. Values for antheraxanthin and zeaxanthins are expressed in β-carotene equivalents. When significantly different (assessed by ANOVA; Supplemental Data Set 3), values were ranked into groups as indicated by the respective letter using a Newmans-Keuls test. ND, not determined; PC-8, plastochromanol-8; DW, dry weight.
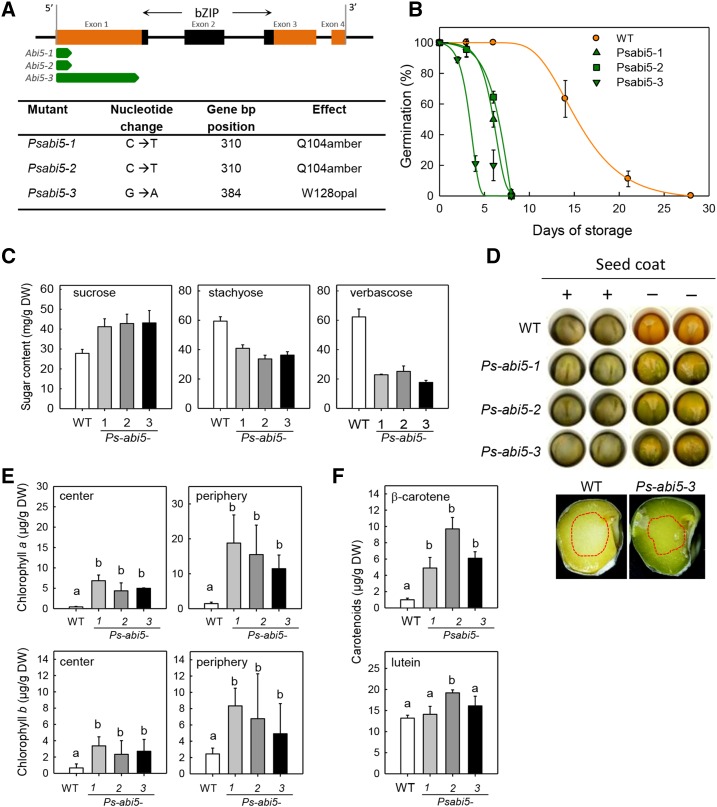
Analysis of Ps-abi5 Mutant Seeds Confirms a Role of ABI5 in Longevity and Chlorophyll Degradation in Legume Seeds
Longevity is affected in the Mt-abi5 mutant seeds, whereas, to the best of our knowledge, this phenotype has not been reported for Arabidopsis abi5 mutant seeds. Therefore, we characterized abi5-1 seeds of Arabidopsis (Finkelstein, 1994). We confirmed the ABA insensitivity and a strong reduction of the LEA EM1 polypeptide in mature seeds of the Arabidopsis abi5-1 mutant by immunoblotting (Supplemental Figures 8A and 8B). Soluble sugar contents were affected, with Suc levels accumulating to a lesser extent in abi5-1 mature seeds, whereas total RFO was slightly higher, shifting the RFO ratio from 4.6 in the wild-type to 3.8 in abi5-1 (Supplemental Figures 8C and 8D). Yet, whereas the Raf content was higher, the amounts of longer oligosaccharides Stach and Ver were lower. These data suggest that the conversion from Raf to Stach and Verb is impaired. However, the other phenotypes related to late maturation that are prominent in Mt-abi5 mutants were less evident in the abi5-1 seeds of Arabidopsis (Supplemental Table 2). A slight defect in dormancy was observed for abi5-1 seeds that were harvested at 19 DAP since 93% of these immature seeds germinated whereas 66% of germination was observed for wild-type seeds (Supplemental Figure 8E). Depending on the growth conditions and the maturity stage, longevity was not affected or was slightly increased in the abi5-1 mutant (Supplemental Figure 8E). In addition, no significant difference was found in chlorophyll content, although a tendency for higher chlorophyll contents was observed for mutant compared with wild-type seeds (Supplemental Figure 8F).
Considering these contrasting results in longevity between M. truncatula and Arabidopsis, we investigated whether defects in ABI5 would also affect seed maturation and longevity in pea. Pea EMS mutant alleles of Ps-abi5 were obtained using a TILLING approach (Dalmais et al., 2008), which allowed the identification of three independent nonsense mutations that were located 5′ to the bZIP domain (Figure 7A). Mutant plants were backcrossed to the wild-type Cameor line and then cultivated under the same growth conditions before seeds were harvested at maturity. No differences in seed weight or water content were observed between wild-type and mutant seeds. To test their longevity, seeds were stored for increasing periods of time at 45°C and 75% RH and germination was assessed (Figure 7B). Ps-abi5 seeds exhibited a severe reduction in life span, with a decrease in P50 values of more than 3-fold compared with the wild type (Figure 7B). As in M. truncatula, nonreducing sugar contents were also strongly affected in mature Ps-abi5 seeds, both in the axis (Figure 7C) and cotyledons (Supplemental Table 2). Suc levels increased by 50% in Ps-abi5 mutant seeds, whereas the major RFO (Stach and Ver) decreased by 1.5- and 2.8-fold, respectively.
Figure 7.
Characterization of abi5 Mutant Seeds of Pea.
(A) Gene structure of Ps-ABI5 and confirmed Ps-abi5 EMS mutants detected by TILLING screening. The position of the DNA region encoding the bZIP domain is indicated in black. The open reading frame of the wild-type ABI5 gene is indicated in orange. The green arrows depict the open reading frames for the mutant lines that carry premature stop codons.
(B) Survival curves (percentage of germination) of mature seeds during storage at 45°C, 75% RH. Data are the means (±se) of three replicates of 30 seeds from a pool of 30 plants.
(C) Sugar contents in the axis of mature uncoated pea seeds. Data are the means (se) of four replicates of 10 seeds from a pool of 30 plants.
(D) Representative photographs of dry mature seeds before (+) and after (−) removing the seed coat and after splitting the seed open to show the inside of cotyledons.
(E) Chlorophyll contents of mature dry cotyledons. Tissues were harvested in the center and periphery of the cotyledons as outlined in (D) (red dashed lines).
(F) Carotenoid contents of mature dry seeds. Data from (E) and (F) are the means of four replicates of 10 seeds from a pool of 30 plants. When significantly different (assessed by ANOVA, P < 0.05; Supplemental Data Set 3), values were ranked into groups as indicated by the respective letter using the Student-Newman-Keuls test.
Ps-abi5 and wild-type seeds appeared green when the seed coat was present (Figure 7D). However, a strong difference in the maturation-induced degreening was found after removing the seed coat. Indeed, mutant seeds were much greener than wild-type seeds, particularly in the periphery of the cotyledons (Figure 7D). HPLC analysis of the seed coats revealed no difference in chlorophyll a and b between the wild type and Ps-abi5 (Supplemental Figure 9). Biochemical analysis of uncoated seeds showed that chlorophyll a and b were more abundant in the mutant seeds, both in the periphery and in the center of the cotyledons, with the strongest difference in the periphery (Figure 7E). β-Carotene contents were 2- to 5-fold higher in total seed extracts of mutant seeds compared with the wild type (Figure 7F). Similar differences were found for the tentatively identified carotenoids zeaxanthin and antheraxanthin, whereas lutein content was not affected in Ps-abi5 seeds. Overall, the phenotype was comparable between Ps-abi5 and Mt-abi5 mutants, and even stronger in pea seeds, suggesting that in legumes, ABI5 regulates late seed maturation traits.
Tocopherol Content Is Affected in Mature Mt-abi5 Seeds
During chlorophyll breakdown, the phytol released via hydrolysis is the main source of prenyl moieties for tocopherol biosynthesis (Vom Dorp et al., 2015). Seeds accumulate large amounts of tocopherols and other tocochromonanols. When the synthesis of these lipophilic antioxidants is impaired during seed development, longevity is strongly affected, due to an increased lipid peroxidation during storage (Mène-Saffrané et al., 2010; Vom Dorp et al., 2015). Therefore, we tested whether the contents of these lipophilic antioxidants were affected in mature seeds of Mt-abi5 mutants as a result of chlorophyll retention (Table 3). α-Tocopherol represented more than 90% of the total tocopherol content in seeds of M. truncatula. Unexpectedly, α-tocopherol contents were 20% higher in Mt-abi5 seeds, whereas other tocochromanols were not significantly affected. Consistent with these observations, M. truncatula PHEOPHYTIN PHEOPHORBIDE HYDROLASE (Mt-PPH) together with VITAMIN E DEFECTIVE3 (Mt-VTE3) and Mt-VTE4 transcript levels, involved in chlorophyll hydrolysis, the synthesis of λ-tocopherol, and its conversion to α-tocopherol, respectively, were higher in mature Mt-abi5 seeds, but not at 32 DAP (Supplemental Figure 5C and Supplemental Data Set 1). This increase in α-tocopherol contents might be indicative of an oxidative stress response within the chloroplast in order to protect thylakoid membrane lipids from oxidation (Havaux et al., 2005).
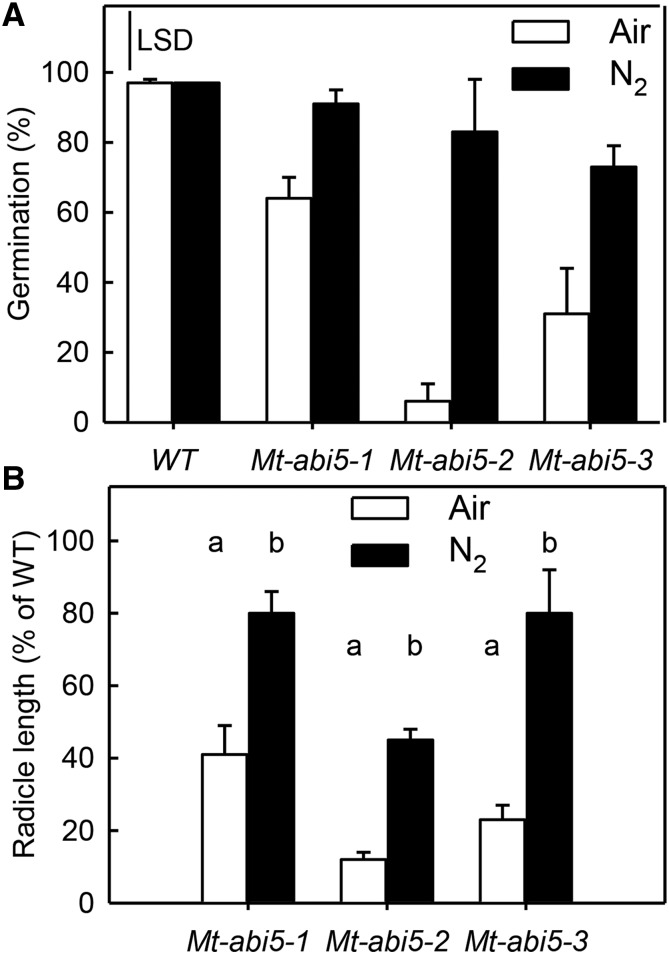
Mt-abi5 Seeds Exhibit Enhanced Sensitivity to Oxygen
To further investigate whether Mt-abi5 seeds are more sensitive to oxidative damage, we performed an aging experiment by storing seeds under N2 to test whether longevity could be restored in the Mt-abi5 mutants in the absence of oxygen. Seeds were first equilibrated at 75% RH and then stored hermetically for 14 d at 35°C. When stored in air, Mt-abi5 seeds showed a significant decrease in viability, whereas the percentage of germination of wild-type seeds was 97% (Figure 8A). When stored under a N2 atmosphere, the germination percentage of Mt-abi5 seeds increased sharply and was no longer significantly different compared with wild-type seeds. For those seeds that germinated after storage, we also measured the radicle length after 48 h of imbibition (Figure 8B). Seeds of the three mutant lines did not fully regain their vigor because radicle length remained shorter compared with wild-type seeds, when stored under N2, suggesting that additional oxygen-independent mechanisms participated in the deterioration of Mt-abi5 seeds during storage. We also checked whether high light intensity would impair storage stability and germination of Mt-abi5 seeds. When stored at 75% RH and 580 µmol photons m−2 s−2 for 14 d, the viability of the Mt-abi5 mutant seeds was not significantly different from that of the wild type. Furthermore, germination in high light intensity did not affect germination, cotyledon greening, or seedling emergence compared with the wild type. Therefore, Mt-abi5 seeds do not appear to be more susceptible to photooxidation than do wild-type seeds.
Figure 8.
Mature Mt-abi5 Seeds Are More Sensitive Than the Wild Type to Oxygen.
(A) Percentage of viability after 14 d of hermetic storage at 35°C under ambient air or nitrogen atmosphere after a 4 d equilibration at 75% RH. Data are the means of triplicates of 50 seeds.
(B) Radicle length of aged Mt-abi5 survivors obtained after 14 d of storage. Data are the means of 10 to 12 radicles (±se) measured after 48 h of imbibition and are expressed as percentage of radicle length of aged wild-type seeds. When significantly different (ANOVA, P < 0.05; Supplemental Data Set 3), values were ranked into groups as indicated by the respective letter using a Student-Newman-Keuls test.
DISCUSSION
Using a QTL approach in M. truncatula by screening seeds under accelerated or moderate aging conditions to identify loci of seed longevity, we revealed Mt-abi5 as a putative regulator of longevity and RFO content. Until now, ABI5 was known to regulate developmental processes during imbibition and seedling establishment, including germination, dormancy, and reestablishment of desiccation tolerance in germinated seedlings (Lopez-Molina et al., 2001; Terrasson et al., 2013; Maia et al., 2014; Supplemental Table 3). Here, the characterization of M. truncatula and pea abi5 mutants revealed that ABI5 governs an array of late maturation-specific events in seeds before reaching the dry state. While seed weight and desiccation tolerance were not affected, both longevity and dormancy, normally acquired during maturation, were severely reduced in Mt-abi5 and Ps-abi5 mutants. A network-based approach on seed transcriptomes collected from Mt-abi5 mutants during maturation revealed a deregulation of several developmentally regulated gene modules with functions related to RFO metabolism, LEA protein accumulation, and regulation of photosynthesis-associated nuclear genes (Figure 9). Mature abi5 seeds of M. truncatula and pea retained chlorophyll and carotenoids (Figures 6A, 6B, 7E, and 7F, Table 3). An additional gene module contained stress response genes whose transcripts were increased during late stages of Mt-abi5 seed development. The gene modules that show different developmental profiles of expression indicate that ABI5 is involved in an intricate network with additional transcription factors that regulate pathways pivotal to the seed dispersal characteristics and seed fitness, but independent from seed filling. It is noteworthy that the total maturation time was not altered between wild-type and mutant seeds. Therefore, abi5 defects cannot simply be explained by a shorter maturation phase. Transcriptome comparison based on DAP between mutants and the wild type might introduce potential biases due to delayed development. Especially genes that are differential only at one stage based on seed age might be prone to this bias. In our study, only a few probes were differentially expressed between Mt-abi5 and the wild type at 16 or 24 DAP (see Venn diagram in Supplemental Data Set 1). For the majority of these probes, differences were found either only at pod abscission or maintained until abscission. In our study, pod abscission corresponds to a physiological stage and is not dependent on the age of the seed, thereby removing this bias.
Figure 9.
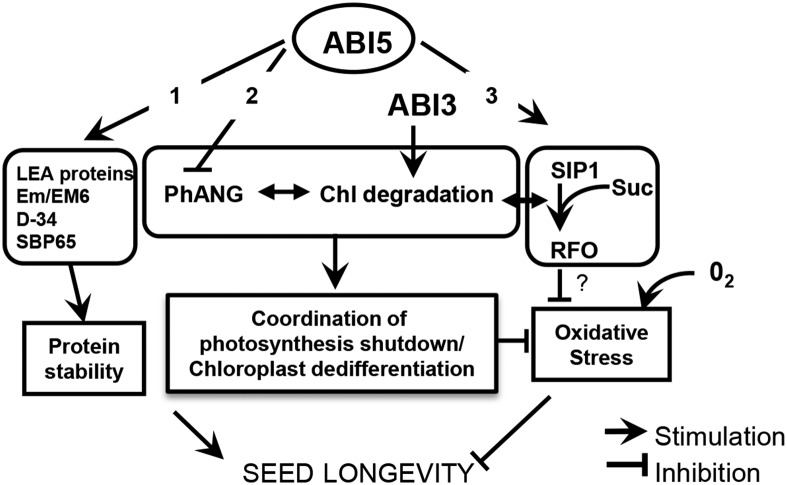
Model Summarizing the Different Actions of ABI5 in the Regulation of Late Seed Maturation in Legumes.
(1) Production of LEA proteins Em/Em6, D-34, and SBP65 that protect proteins from aggregation during desiccation/rehydration. (2) Downregulation of expression of photosynthesis associated nuclear genes, which is accompanied by chlorophyll and carotenoid degradation (although not directly regulated by ABI5) and leads to chloroplast dedifferentiation, which altogether prevents oxidative stress in the mature seed. (3) Induced expression of SIP1, thus producing RFO, which might inhibit oxidative stress both inside and outside the chloroplast.
The chlorophyll retention observed in the mature pea and M. truncatula abi5 seeds could suggest that chlorophyll degradation is under the regulation of ABI5. In Arabidopsis and Brassica napus (oilseed rape), chlorophyll retention in mature seeds has been attributed to defects in the first steps of chlorophyll breakdown (Christ and Hörtensteiner 2014), including the conversion of chlorophyll b to chlorophyll a by NYC1 and NOL (Nakajima et al., 2012) and the oxidation of pheophorbide a by PAO (Chung et al. 2006) and STAY-GREEN (SGR), whose function remains to be secured (Christ and Hörtensteiner 2014). In Arabidopsis seeds, chlorophyll degradation is under the control of ABI3, NAC016, and ABF4 (Nakajima et al., 2012; Delmas et al., 2013), whereas in senescent leaves, it is under the control of ABI5, since ABI5 binds directly to the promoter of NYC1, NOL, and SGR genes (Sakuraba et al., 2014). However, despite the green phenotypes of pea and M. truncatula abi5 seeds, our RT-qPCR and transcriptome data showed that genes encoding enzymes of the PAO pathway were either not affected or even upregulated during maturation in the legume abi5 mutants (Supplemental Figure 5). In addition, chlorophyll breakdown during dark-induced senescence was not affected in Mt-abi5 seedlings compared with the wild type (Supplemental Figure 6). Thus, chlorophyll retention is likely to be an indirect consequence of a defect in ABI5 that acts on other regulatory mechanisms still awaiting characterization.
Developing Mt-abi5 seeds also showed a strong deregulation of a large number of photosynthesis-related nuclear and chloroplastic genes (Figure 6; Supplemental Data Set 1). The concomitant regulation of chloroplast and nuclear genes encoding photosynthesis proteins is crucial for the photosynthetic competence of chloroplasts and disassembly during senescence. One of the functions of ABA is to coordinate the expression of these genes in response to developmental and environmental cues (Cutler et al., 2010; Fujita et al., 2011; Yamburenko et al., 2013). In Mt-abi5 seeds, this deregulation occurred before the onset of chlorophyll loss since transcript levels of both Mt-LHCB2 and Mt-PSAD-2, encoding subunits of PSII and PSI, respectively, were already 2-fold higher in mutant seeds at 24 DAP (Figures 6C and 6D). During seed maturation, this difference increased progressively for Mt-LHCB-2, resulting in relative amounts of transcripts that were 50-fold higher in mature Mt-abi5 seeds compared with the wild type (Figure 6C). Consistent with this observation, the Arabidopsis LHCB2 and FERREDOXIN C2 (FDC2; encoding a chloroplastic ferredoxin capable of alternative electron partitioning in PSII) were identified as genes bound by ABI5 in vivo in imbibed seeds, suggesting a direct regulation of PSII genes by ABI5 (De Giorgi et al., 2015). Further inspection of the common transcripts upregulated in mature seeds of both Mt-abi5 and Arabidopsis abi5-7 (Nakabayashi et al., 2005) revealed 135 differentially expressed genes, out of which five genes were related to PSI, PSII, and light-harvesting complex.
In oily non-chlorophyllous seeds, the presence of chlorophyll in mature seeds is an undesirable trait leading to poor nutritional quality, reduced longevity and economic losses (Jalink et al., 1998; Clerkx et al., 2003; Chung et al., 2006; Nakajima et al., 2012; Delmas et al., 2013; Teixeira et al., 2016). Although the link between chlorophyll retention and poor seed quality has been suggested for a long time, a direct cause-effect relationship with longevity has been established only recently. Arabidopsis seeds of the nyc1 nol double mutant affected in the first step of chlorophyll degradation contained 10-fold more chlorophyll than the wild type and had a strongly reduced longevity (Nakajima et al., 2012). In pea and M. truncatula abi5 mutant seeds, chlorophyll degradation and longevity were both affected. More chlorophyll was retained in mature dry Ps-abi5 seeds compared with Mt-abi5 seeds, and longevity was also much more affected in the Ps-abi5 seeds (Figures 2E, 2F, and 7B), thus underscoring the detrimental effect of chlorophyll retention in dry seeds. The reasons behind the impaired storage stability observed here but also in seeds of Arabidopsis and oilseed rape remain elusive. There are several plausible explanations for this cause-effect relationship that are not mutually exclusive.
The first explanation is that the impaired chlorophyll degradation leads to accumulation of phototoxic products and chlorophyll catabolites. To avoid photooxidative damage and cell death during chlorophyll degradation in leaves and fruits, detoxification of photoactive chlorophyll and chlorophyll catabolites must occur (Christ and Hörtensteiner, 2014). Whether phototoxic products resulting from chlorophyll degradation accumulate in seeds during desiccation is unknown. However, in soybean (Glycine max) seeds of the stay-green mutants cytG and Gd1d2 that retain chlorophyll, the catabolites chlorophyllides and phaeophytins were barely detectable (Canfield et al., 1995). In M. truncatula, dry, mature abi5 mutant seeds were not particularly sensitive to high light in comparison to the wild type. Likewise, germination of green seeds from the grs and nyc nol mutants was not more sensitive to light than darkness (Clerkx et al., 2003; Nakajima et al., 2012). So far, there is no evidence suggesting that phototoxic intermediates accumulate to such an extent that longevity would be impaired.
The second explanation for the link between chlorophyll retention and longevity could be an incomplete regulation of the chloroplast dedifferentiation that leads to oxidative damage and reduced seed fitness. Several lines of evidence from this work show that developing Mt-abi5 seeds had to cope with oxidative stress within the chloroplasts during late maturation. To scavenge reactive oxygen species in the chloroplasts, tocopherols and nonphotochemical quenching through the xanthophyll cycle act redundantly, thereby protecting thylakoid membrane lipids from photooxidation (Havaux et al., 2005). In this study, α-tocopherol contents were increased by 20% in dry mutant seeds and this was accompanied by increased transcript levels of its biosynthetic genes. Furthermore, the operation of the xanthophyll cycle appeared to be disrupted in dry abi5 mutant seeds from M. truncatula and pea: The xanthophyll carotenoids zeaxanthin and antheraxanthin exhibited higher amounts, suggesting an increase in deepoxidation in response to maturation drying (Table 3). The partial restoration of the longevity phenotype when Mt-abi5 seeds were stored under N2 also points to a deficiency in the oxidative stress response. When developing Arabidopsis seeds were treated with DCMU, which disrupts the electron flow at PSII, mature seeds deteriorated faster during accelerated aging (Allorent et al., 2015). In light of these observations, we speculate that ABI5 might participate in the progressive shutdown of photosynthesis by repressing photosynthesis nuclear gene expression prior to reaching the quiescent dry state. In this way, ABI5 in legumes might be implicated in the coordination of shutting down photosynthesis during seed maturation to avoid oxidative stress before reaching the quiescent state. Our transcriptome and proteome study provides further evidence suggesting that Mt-abi5 seeds responded to stress during late maturation (Figure 3, cluster IIc). A set of LEA proteins and transcripts of HSP were specifically upregulated, including the chloroplast co-chaperonin 20 (CPN20) as well as other stress-responsive bZIP AREB and ABF transcription factors, such as homologs of AREB3/DPBF2 and AREB1/ABF2, a key regulator enhancing drought tolerance and LEA gene expression (Fujita et al., 2005).
In addition to its role in chloroplast dedifferentiation during maturation drying, ABI5 was found to regulate a set of genes involved in the synthesis of protective compounds (Figures 3 to 5). Previously, it was demonstrated that ABI5 binds directly to the promoters of the EM1 and EM6 genes, and Arabidopsis abi5 mutants exhibited reduced accumulation of the corresponding proteins during maturation (Finkelstein and Lynch, 2000; Carles et al., 2002). Our transcriptome and proteome studies extend these observations as the abundance of several LEA proteins was lower in mutant seeds. In particular, the EM/EM6 proteins, SBP65, and members of the D-34 family were severely reduced. Their role in seed longevity remains to be established, but in vitro, PM25, a member of the D-34 family, and Em6 were found to increase the density of sugar glasses (Tunnacliffe et al., 2010) and act against protein aggregation during rehydration (Boucher et al., 2010). Cytoplasmic glasses severely slow down molecular motion and deleterious reactions, thereby contributing to the stability of the cytoplasm during long-term storage (reviewed in Buitink and Leprince, 2004). Interestingly, the proteome analysis also revealed that several other LEA proteins increased in abundance in the mutants, possibly in an attempt to counteract the absence of the abovementioned LEA proteins. However, this is apparently not sufficient to compensate for the decreased longevity in the Mt-abi5 mutants.
QTL analysis revealed that three out of five longevity QTL colocated with QTL of RFO composition of seeds from the same harvest (Table 1; Vandecasteele et al., 2011). The discovery that RFO accumulation was affected in the Mt-abi5 mutants, concomitant with the decrease in longevity, reinforces the genetic link between these two traits. Evidence for a direct link between RFO metabolism and longevity was also recently secured in Arabidopsis by Righetti et al. (2015), who showed that the agal2 mutant exhibited reduced longevity together with increased Suc and decreased RFO content, as observed in the abi5 seeds of pea and M. truncatula. Consistent with this, the transcript of Mt-SIP1, a Raf synthase, was deregulated in the Mt-abi5 mutant seeds. A direct interaction was demonstrated between Mt-ABI5 and the ABRE in the Mt-SIP1 promoter (Supplemental Figure 2), suggesting that ABI5 transactivates Mt-SIP1 expression to regulate the synthesis of RFO. Here, we showed that the Raf to Stach/Ver conversion is also impaired in Arabidopsis abi5-1 mutants (Supplemental Figure 8), likely via the deregulation of SIP1. ChIP-seq analysis identified SIP1 as a direct target of ABI5 in Arabidopsis (De Giorgi et al., 2015). The role of RFO in seed longevity remains elusive. Our earlier work (Buitink et al., 2000) ruled out a role in the biophysical properties of the cytoplasmic glass state conferring long-term stability to dry seeds. There is evidence that RFO could act as scavenger of reactive oxygen species, in particular protecting photosynthesis from oxidative stress (Nishizawa et al., 2008). Indeed, Raf, being synthesized in the cytosol, can be imported into the chloroplast by an active membrane bound transporter (Schneider and Keller, 2009). In support of this hypothesis, longevity could be partially restored in Mt-abi5 seeds by storing them under nitrogen. That RFO metabolism might be coupled to chloroplast dedifferentiation and photosynthesis shutdown during maturation is also evident from the observation that DCMU treatment leading to decreased longevity in Arabidopsis (Allorent et al., 2015) also resulted in a 3-fold decrease in galactinol content, the precursor of RFO. Alternatively, the conversion from Suc to RFO might be related to a sugar signaling pathway as supported by the overrepresentation of Suc-responsive elements in the promoter of genes upregulated during the acquisition of longevity (Righetti et al., 2015).
Whereas in the two legume species, ABI5 played a major role in late maturation processes, related phenotypes in Arabidopsis were less pronounced (Supplemental Table 3). Dormancy, LEA protein accumulation, and RFO metabolism were affected in a similar way, but longevity and chlorophyll retention were not significantly different from the wild-type seeds (Supplemental Table 3). This might be due to redundancy and/or compensation mechanisms among the bZIP family factors involved in ABA signaling in Arabidopsis. ABI5, ABF3, and ABF1 were found to have overlapping effects, and the loss of either factor was compensated for by increased expression of other family members (Finkelstein et al., 2005; Yoshida et al., 2010). In the double mutant dog1-1 abi3-1, exhibiting reduced longevity, redundancy was suggested with ABF4 (Dekkers et al., 2016), which also binds to the NYC1 promoter (Nakajima et al., 2012). The known ability of bZIP transcription factors, such as AREB/ABF for heterodimerization resulting in alternative DNA binding repertoires associated with ABA signaling (Bensmihen et al., 2002; Finkelstein et al., 2005; Yoshida et al., 2010), would therefore provide a combinatorial control of the different seed maturation processes. Such combinations could vary during the course of evolution. In legumes, similar components are present (Wang et al., 2015), but their molecular wiring appears different from Brassicaceae.
To summarize, Figure 9 unveils ABI5 as a master regulator of late seed maturation in legumes by coordinating the acquisition of seed longevity and dormancy via three modes of action: (1) production of LEA proteins Em/Em6, D-34, and SBP65 that protect proteins from aggregation during desiccation/rehydration; (2) downregulation of expression of photosynthesis-associated nuclear genes, which is accompanied by chlorophyll and carotenoid degradation (although probably not directly regulated by ABI5) and leads to chloroplast dedifferentiation, which altogether prevents oxidative stress in the mature seed; and (3) induced expression of SIP1, thus producing RFO, which might inhibit oxidative stress both inside and outside the chloroplast. Thus, ABI5 appears to be a prominent regulator of late maturation in legumes, and this finding sheds light on the relationship between seed vigor and RFO accumulation (Vandecasteele et al., 2011) and chlorophyll retention (Jalink et al., 1998; Nakajima et al., 2012; Delmas et al., 2013; Teixeira et al., 2016). Together with previous studies showing a correlation between longevity and chlorophyll retention in Arabidopsis (Clerkx et al., 2003; Nakajima et al., 2012), our data suggest that degreening cannot be uncoupled from long-term survival in the dry state, an important aspect of seed vigor and seedling establishment. This crosstalk offers new opportunities to tackle the green seed problem as well as antinutritional characteristics of RFO in legumes.
METHODS
Plant Material
Medicago truncatula ssp tricycla (R-108) plants were cultivated according to Chatelain et al. (2012). Seeds were harvested upon pod abscission or at the indicated stages of development and stored at −196°C in liquid nitrogen. Three independent Tnt1 insertion lines in the Mt-ABI5 gene were used: NF4383, referred to as Mt-abi5-1; NF3376, referred to as Mt-abi5-2, both previously characterized by Terrasson et al. (2013); and NF7680, referred to as Mt-abi5-3, that was characterized in this study. Tnt1 insertions were verified by PCR using forward and reverse primers listed in Supplemental Table 4. The Mt-abi5-1 line was backcrossed twice with R-108. Pea (Pisum sativum) mutants and wild-type (var Cameor) were isolated by TILLING according to Dalmais et al. (2008) and grown under greenhouse conditions at 18°C/22°C and a 16-h photoperiod (200 µmol m−2 s−1) in a mixture of vermiculite and soil. Mutants were backcrossed twice and mutations were verified by dCAPS markers (Supplemental Table 4). Seeds were harvested at shedding.
Arabidopsis thaliana ecotype Wassilewskija and abi5-1 mutants (N8105) (Finkelstein, 1994) were obtained from the NASC. The abi5-1 mutation was confirmed by DNA sequencing using primers indicated in Supplemental Table 4). Arabidopsis seeds were multiplied by growing plants in a growth chamber (23°C, 16 h light, 100 µmol m−2 s−1). Seeds were harvested at 19 DAP (Arabidopsis) or at shedding. Seeds from Arabidopsis and M. truncatula were dried after harvest for 2 d at 44% RH, 20°C, and pea seeds were dried at 20°C in air (67% RH) for 2 weeks.
Phenotyping and QTL Analysis
The LR4 population consisted of 171 RILs and was derived from a cross between A17 and DZA315.16 (Vandecasteele et al., 2011). Longevity was determined on seed samples pooled from three plants. Before measurements, seeds were stored at 20°C at 60% RH for 5 months to allow for dormancy release, after which they were stored for an additional 7 months at 10°C prior to the longevity experiments. Four repetitions of 40 seeds were scarified, equilibrated for 7 d at 60% RH (moderate aging) or at 75% RH (accelerated aging), sealed in aluminum bags (Barrierfoil; Moore and Buckle) and stored for 159 d and 20 d, respectively, at 35°C. After storage, seeds were imbibed on filter paper and the percentages of final germination (radicle protrusion) and of morphologically normal seedlings were scored after 10 d at 20°C in the dark. Petri dishes were randomized following a complete-block design with four blocks, each containing one biological replicate. All statistical analyses were performed with SAS 8.1 (SAS Institute, 2000) as described by Vandecasteele et al. (2011).
Genetic maps and molecular data for the LR4 population are described by Vandecasteele et al. (2011). QTL mapping was performed using the MCQTL software (Jourjon et al., 2005) as described by Vandecasteele et al. (2011). For each QTL identified, additive genetic effect, percentage of phenotypic variance explained (R2), F-test value, as well as the confidence interval were determined and equivalent LOD scores calculated (Table 1). The map of linkage group 7 was drawn using MapChart software (Voorrips, 2002). Primers used for the mapping of Mt-ABI5 are indicated in Supplemental Table 4.
Physiological Characterization of abi5 Mutants
For germination assays, three replicates of 30 to 50 scarified seeds of M. truncatula or 100 to 200 Arabidopsis seeds were imbibed on filter paper with 4 and 1 mL, respectively, of distilled water at 20°C in the dark. For pea, three replicates of 30 seeds were imbibed in 100 mL of distilled water with folded filter paper. Seeds were considered as germinated when the radicle protruded from the seed coat. For ABA sensitivity assays, three replicates of 30 to 50 scarified M. truncatula mature seeds were imbibed in the presence or absence of 10 µM of ABA (Sigma-Aldrich). For Arabidopsis, 100 to 200 mature seeds were imbibed on two filter papers in the presence or absence of 1 µM ABA and incubated for 3 d at 4°C in dark followed by 8 d at 20°C with a 16-h photoperiod.
To assess longevity, mature seeds (scarified for M. truncatula) were stored in the dark or in the light (580 µmol m−2 s−1) at 75% RH using hermetically closed containers containing a saturated NaCl solution. Storage temperature was 35°C for M. truncatula and Arabidopsis and 45°C for pea. At different storage time intervals, seeds were imbibed and final germination percentage was counted after 10 d and 8 d, respectively. For the storage experiment in N2, replicates of 30 seeds were first equilibrated at 75% RH, 20°C for 4 d, then transferred into an aluminum bag and sealed. Bags were pierced with a needle and N2 was flushed for 20 s, after which the bag was resealed and placed at 35°C. Controls were seeds that received the same preequilibration treatment and were subsequently stored at 75% RH and 35°C for 14 d. Thereafter, germination was counted and radicle measured after 48 h of imbibition.
For the seed coat permeability assay, four replicates of 5 to 10 scarified mature seeds were incubated at 20°C and 100% RH and weighed from 0 to 8 h. Rates of imbibition were calculated by averaging the rate of water uptake during step increments of 2 h. The leaf senescence experiment was adapted from Sakuraba et al. (2014). Leaves of 4-week-old M. truncatula plants were detached and incubated in water at 21°C in the dark. After 0, 4, or 8 d of incubation, photographs were taken and leaves were frozen in liquid nitrogen. Between 2 and 10 mg of lyophilized powder was weighed and chlorophyll was extracted and spectrophotometrically measured as described below.
Tocopherol Measurement
Five replicates of 20 mature seeds were frozen in liquid nitrogen and ground to a fine powder (Precellys homogenizer). Tocopherols were isolated from the ground tissue using diethylether and 0.2 M H3PO4/1 M KCl. The upper organic phase was dried under air flow and the lipids were dissolved in n-hexane for HPLC analysis (Zbierzak et al., 2009).
Chlorophyll and Carotenoid Assays
For spectrometry analysis, replicates of 25 M. truncatula seeds or 5 to 7 mg of Arabidopsis seeds, or leaf material, were lyophilized overnight, weighed, and ground in 300 μL or 600 μL of N,N-dimethylformamide (Sigma-Aldrich), respectively. After overnight incubation at 4°C, samples were vortexed for 30 s and centrifuged for 5 min at 10,000g. Chlorophyll concentrations were measured according to Moran (1982) on a FluoStar Omega spectrophotometer (BMG Labtech). For HPLC analysis, pigment extraction and quantification were performed as described by Perrin et al. (2016). Briefly, the carotenoid measurements were performed with a Shimadzu HPLC equipped with a thermostated autosampler (SIL-10ADVP) and a diode array detector (SPD-M10AVP). Carotenoids were identified using standards of lutein and β-carotene.
Sugar Determination
Soluble sugar contents were assessed by HPLC (Dionex) according to Rosnoblet et al. (2007). Analysis was performed on four replicates of 10 mature seeds of M. truncatula, four replicates of five mature pea seeds (cotyledons or embryonic axes), and 15 mg of mature Arabidopsis seeds.
RNA Extraction, Microarray Analysis, and Gene Coexpression Network
RNA extraction was performed on three replicates of 50 seeds at 16 and 24 DAP and at abscission, for genotypes R-108, Mt-abi5-1, and Mt-abi5-2. Seeds were ground in liquid nitrogen. The extraction was performed with the Nucleospin Extract II, Total RNA Purification Kit (Macherey Nagel). RNA amplification, labeling, and hybridization of Nimblegen Medtr_v1.0 12x135K arrays were performed according to Terrasson et al. (2013). Three biological replicates were analyzed per comparison using the dye-switch method, and statistical analysis on the gene expression data was performed according to Verdier et al. (2013). Genes were considered differentially expressed when the log2 ratio between Mt-abi5 and the wild type was >1 or ≤1 at a given time point for each of the two mutants, with a P value < 0.01. Nimblegen microarray data were deposited in the NCBI Gene Expression Omnibus database (accession no. GSE87313).
The gene coexpression network of the 2911 differentially expressed genes (Supplemental Data Set 1) of the Mt-abi5 mutants was constructed using the normalized intensities of the wild-type and Mt-abi5 mutants together with those obtained at 15 time points during seed maturation, derived from Righetti et al. (2015). The network was constructed using Biolayout software (Goldovsky et al., 2005) with a correction value of 0.92, giving rise to a network with 1147 nodes and 2409 edges. Gene interactions were visualized using the open source software Cytoscape (version 3.3.0) with an edge-weighted spring-embedded layout.
RT-qPCR Analysis
For RT-qPCR, 1 μg total RNA was reverse-transcribed using the Quantitect reverse transcription kit (Qiagen), and qPCR was performed on a CFX96 real-time detection system (Bio-Rad Laboratories) according to the manufacturer’s instructions with SsoFast Eva Green Supermix (Bio-Rad Laboratories). Reference genes were MSC27 (Bolingue et al., 2010) and Mt-ACTIN11. Forward and reverse primers used for the different genes in this study are listed in Supplemental Table 4. Relative expression levels were calculated using the comparative 2∆(Ct) method (Schmittgen and Livak, 2008). Each data point represents the mean of three biological replicates of 30 seeds.
Yeast One-Hybrid Assay
Yeast one-hybrid assays were performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech) according to the manufacturer’s instructions. Primers used for the construction of the prey and bait are indicated in Supplemental Table 4. Fifty-six base pair oligonucleotides of the promoter region of Mt-SIP1 (−1348 to 1298) containing the ABRE consensus sequence or a mutated version were cloned into the pAbAi vector, carrying the AUR1-C gene. The plasmid was linearized and integrated into the genome of yeast (Y1H Gold). Yeast cells were then transformed with pGADT7-AD plasmid containing the coding region of Mt-ABI5. DNA-protein interaction was determined based on the ability of the cotransformants to grow on SD/-Leu medium in the presence of 200 to 500 ng mL−1 of aureobasidin A.
Protein Extraction, 2D-PAGE, and Immunoblotting
Soluble protein extraction, preparation of heat-soluble fractions, and analysis using 2D-PAGE were performed according to Chatelain et al. (2012). The experiments were set up in a randomized block design where six gels corresponding to three independent protein extractions of 50 seeds were run in parallel. Polypeptides were identified using the reference gel previously obtained by Chatelain et al. (2012) and Delahaie et al. (2013). Immunoblotting analysis was performed according to Boudet et al. (2006). Briefly, 10 μg of heat-stable protein extract was separated by SDS-PAGE on a 12% (w/v) polyacrylamide gel and transferred to a nitrocellulose membrane (Schleicher and Schuell). Immunodetection was performed by chemiluminescence using a rabbit polyclonal antibody raised against the recombinant Mt-EM6 protein (dilution 1:10,000; Davids Biotechnologie), an anti-rabbit IgG-peroxidase conjugate (dilution 1:50,000; Sigma-Aldrich A0545 Lot 32K4882), and the Amersham ECL Plus Western Blotting Detection reagents (GE Healthcare Life Sciences), following the manufacturer’s instructions. Chemiluminescence was monitored by a Chemidoc TM XRS molecular imager (Bio-Rad Laboratories).
Accession Numbers
Sequence data in this study can be found in the GenBank/EMBL data libraries under the following accession numbers: Mt-ABI5, Medtr7g104480; Mt-ACTIN11, Medtr2g008050; Mt-MSC27, TC85211; Mt-SIP1, Medtr3g077280; and Ps-ABI5, PsCam017634. Transcriptomic data obtained in this study have been deposited in the Gene Expression Omnibus database under accession number GSE87313.
Supplemental Data
Supplemental Figure 1. Germination of Mt-abi5 seeds is insensitive to ABA during imbibition in the dark.
Supplemental Figure 2. Yeast one-hybrid binding assays of Mt-ABI5 to the ABRE in the promoter region of Mt-SIP1.
Supplemental Figure 3. Two-dimensional electrophoresis gels of the heat stable proteome of M. truncatula seeds.
Supplemental Figure 4. Chlorophyll contents in developing wild-type and Mt-abi5 seeds.
Supplemental Figure 5. Expression of genes involved in the PAO degradation pathway in mature seeds of Mt-abi5.
Supplemental Figure 6. Dark-induced senescence in young seedlings of the wild type and Mt-abi5.
Supplemental Figure 7. Photosynthesis-associated gene expression and carotenoid content during seed development of wild-type and Mt-abi5 seeds.
Supplemental Figure 8. Longevity of wild-type and abi5-1 seeds of Arabidopsis together with sugar content and EM protein levels in dry mature seeds.
Supplemental Figure 9. Chlorophyll contents in seed coats of mature wild-type and Psabi5 mature seeds.
Supplemental Table 1. Detected polymorphisms between Jemalong J-6 and DZA315.16 in the region of Mt-ABI5 (Medtr7g104480) on chromosome 7.
Supplemental Table 2. Contents in soluble sugars of mature pea cotyledons.
Supplemental Table 3. Comparative analysis of affected phenotypes in seeds of the abi5 mutants of Medicago truncatula, Pisum sativum, and Arabidopsis thaliana.
Supplemental Table 4. List of primer combinations used for mutant selection and gene expression validation by qRT-PCR.
Supplemental Data Set 1. Differentially expressed genes between Mt-abi5-1 and Mt-abi5-2 and wild-type (R-108) seeds during maturation.
Supplemental Data Set 2. Identification and quantification of polypeptide abundance in mature seeds of Mt-abi5-1, Mt-abi5-2, and the wild type (genotype R-108).
Supplemental Data Set 3. Statistical test results showing the level of significance of data shown in the figures.
Supplementary Material
Acknowledgments
This work was funded in part by Génoplante 2010 QualityLegSeed Project GPLA06036G and by a grant from the Region des Pays de la Loire, France (QUALISEM 2009-2013). We thank Sandra Pelletier and the ANAN platform of the SFR Quasav, Angers, France for the assistance with the microarray analysis and access to hybridization facilities and INEM for the greenhouse facilities (IRHS, Angers, France). We thank Séverine Gagné for help with the HPLC analysis of carotenoids. The TILLING platform is funded by Initiative d’Excellence Paris-Saclay (Lidex-3P, ANR-11-IDEX-0003-02) and the European Research Council (ERC-SEXYPARTH).
AUTHOR CONTRIBUTIONS
O.L. and J.B. conceived the project and research plans, supervised the experiments, and wrote the article with contributions from all authors. J.Z., D.L., E.T., and E.C. performed most of the experiments and analyzed the data with contributions from K. Gutbrod, P.D., C.V., E.G., and K. Gallardo. B.L.V., M.D., and C.D.-L. provided technical assistance. C.L.S., K. Gallardo, M.D., and A.B. performed the genetic screenings of mutant pea and provided the Ps-ABI5 mutant lines.
Glossary
- LEA
late embryogenesis abundant
- RFO
raffinose family oligosaccharide
- QTL
quantitative trait loci
- ABA
abscisic acid
- AREB
ABA response element binding
- ABRE
ABA-responsive promoter element
- RIL
recombinant inbred line
- DAP
days after pollination
- GO
Gene Ontology
References
- Allorent G., Osorio S., Vu J.L., Falconet D., Jouhet J., Kuntz M., Fernie A.R., Lerbs-Mache S., Macherel D., Courtois F., Finazzi G. (2015). Adjustments of embryonic photosynthetic activity modulate seed fitness in Arabidopsis thaliana. New Phytol. 205: 707–719. [DOI] [PubMed] [Google Scholar]
- Almoguera C., Prieto-Dapena P., Díaz-Martín J., Espinosa J.M., Carranco R., Jordano J. (2009). The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolingue W., Ly Vu B., Leprince O., Buitink J. (2010). Characterization of dormancy behaviour in seeds of the model legume Medicago truncatula. Seed Sci. Res. 20: 97–107. [Google Scholar]
- Boucher V., Buitink J., Lin X., Boudet J., Hoekstra F.A., Hundertmark M., Renard D., Leprince O. (2010). MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 33: 418–430. [DOI] [PubMed] [Google Scholar]
- Boudet J., Buitink J., Hoekstra F.A., Rogniaux H., Larré C., Satour P., Leprince O. (2006). Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol. 140: 1418–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J., Hemminga M.A., Hoekstra F.A. (2000). Is there a role for oligosaccharides in seed longevity? An assessment of intracellular glass stability. Plant Physiol. 122: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J., Leprince O. (2004). Glass formation in plant anhydrobiotes: survival in the dry state. Cryobiology 48: 215–228. [DOI] [PubMed] [Google Scholar]
- Canfield M.R., Guiamét J.J., Noodén L.D. (1995). Alteration of soybean seedling development in darkness and light by the stay-green mutation cytG and Gd1d2. Ann. Bot. (Lond.) 75: 143–150. [Google Scholar]
- Carles C., Bies-Etheve N., Aspart L., Léon-Kloosterziel K.M., Koornneef M., Echeverria M., Delseny M. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30: 373–383. [DOI] [PubMed] [Google Scholar]
- Chatelain E., Hundertmark M., Leprince O., Le Gall S., Satour P., Deligny-Penninck S., Rogniaux H., Buitink J. (2012). Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 35: 1440–1455. [DOI] [PubMed] [Google Scholar]
- Châtelain E., Satour P., Laugier E., Ly Vu B., Payet N., Rey P., Montrichard F. (2013). Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc. Natl. Acad. Sci. USA 110: 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Hörtensteiner S. (2014). Mechanism and significance of chlorophyll breakdown. J. Plant Growth Regul. 33: 4–20. [Google Scholar]
- Chung D.W., Pruzinská A., Hörtensteiner S., Ort D.R. (2006). The role of pheophorbide a oxygenase expression and activity in the canola green seed problem. Plant Physiol. 142: 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx E.J.M., Vries H.B., Ruys G.J., Groot S.P.C., Koornneef M. (2003). Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiol. 132: 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dalmais M., Schmidt J., Le Signor C., Moussy F., Burstin J., Savois V., Aubert G., Brunaud V., de Oliveira Y., Guichard C., Thompson R., Bendahmane A. (2008). UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 9: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgi J., Piskurewicz U., Loubery S., Utz-Pugin A., Bailly C., Mène-Saffrané L., Lopez-Molina L. (2015). An endosperm-associated cuticle is required for Arabidopsis seed viability, dormancy and early control of germination. PLoS Genet. 11: e1005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Léon-Kloosterziel K.M., Koornneef M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122: 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B.J.W., He H., Hanson J., Willems L.A.J., Jamar D.C.L., Cueff G., Rajjou L., Hilhorst H.W.M., Bentsink L. (2016). The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 85: 451–465. [DOI] [PubMed] [Google Scholar]
- Delahaie J., Hundertmark M., Bove J., Leprince O., Rogniaux H., Buitink J. (2013). LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J. Exp. Bot. 64: 4559–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas F., Sankaranarayanan S., Deb S., Widdup E., Bournonville C., Bollier N., Northey J.G.B., McCourt P., Samuel M.A. (2013). ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 110: E3888–E3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Vidigal D., Willems L., van Arkel J., Dekkers B.J., Hilhorst H.W., Bentsink L. (2016). Galactinol as marker for seed longevity. Plant Sci. 246: 112–118. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S.L., Lynch T.J., Thomas T.L., Rock C.D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59: 253–267. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R. (1994). Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol. 105: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124: 509–525. [DOI] [PubMed] [Google Scholar]
- Goldovsky L., Cases I., Enright A.J., Ouzounis C.A. (2005). BioLayout(Java): versatile network visualisation of structural and functional relationships. Appl. Bioinformatics 4: 71–74. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Jorge S., et al. (2013). Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Eymery F., Porfirova S., Rey P., Dörmann P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M., Buitink J., Leprince O., Hincha D.K. (2011). The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Sci. Res. 21: 165–173. [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111. [DOI] [PubMed] [Google Scholar]
- Jalink H., van der Schoor R., Frandas A., van Pijlen J.G., Bino R.J. (1998). Chlorophyll fluorescence of Brassica oleracea seeds as a non-destructive marker for seed maturity and seed performance. Seed Sci. Res. 8: 437–443. [Google Scholar]
- Jourjon M.-F., Jasson S., Marcel J., Ngom B., Mangin B. (2005). MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21: 128–130. [DOI] [PubMed] [Google Scholar]
- Kranner I., Birtić S., Anderson K.M., Pritchard H.W. (2006). Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death? Free Radic. Biol. Med. 40: 2155–2165. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26: 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.-H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia J., Dekkers B.J.W., Dolle M.J., Ligterink W., Hilhorst H.W.M. (2014). Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytol. 203: 81–93. [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L., Jones A.D., DellaPenna D. (2010). Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 17815–17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monma M., Ito M., Saito M., Chikuni K. (1994). Carotenoid components in soybean seeds varying with seed color and maturation stage. Biosci. Biotechnol. Biochem. 58: 926–930. [Google Scholar]
- Moran R. (1982). Formulae for determination of chlorophyllous pigments extracted with n,n-dimethylformamide. Plant Physiol. 69: 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41: 697–709. [DOI] [PubMed] [Google Scholar]
- Nakajima S., Ito H., Tanaka R., Tanaka A. (2012). Chlorophyll b reductase plays an essential role in maturation and storability of Arabidopsis seeds. Plant Physiol. 160: 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Keith K., McCourt P., Naito S. (1994). Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol. 35: 509–513. [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Shigeoka S. (2008). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147: 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogé L., Bourdais G., Bove J., Collet B., Godin B., Granier F., Boutin J.-P., Job D., Jullien M., Grappin P. (2008). Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20: 3022–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms J., Leon-Kloosterziel K.M., Bartels D., Koornneef M., Karssen C.M. (1993). Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (a comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 102: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin F., Brahem M., Dubois-Laurent C., Huet S., Jourdan M., Geoffriau E., Peltier D., Gagné S. (2016). Differential pigment accumulation in carrot leaves and roots during two growing periods. J. Agric. Food Chem. 64: 906–912. [DOI] [PubMed] [Google Scholar]
- Personat J.-M., Tejedor-Cano J., Prieto-Dapena P., Almoguera C., Jordano J. (2014). Co-overexpression of two Heat Shock Factors results in enhanced seed longevity and in synergistic effects on seedling tolerance to severe dehydration and oxidative stress. BMC Plant Biol. 14: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P., Castaño R., Almoguera C., Jordano J. (2006). Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol. 142: 1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert R., Adams J., Coneybeer J., Crawford A., Hay F. (2007). Seed quality for conservation is critically affected by pre-storage factors. Aust. J. Bot. 55: 326–335. [Google Scholar]
- Righetti K., Vu J.L., Pelletier S., Vu B.L., Glaab E., Lalanne D., Pasha A., Patel R.V., Provart N.J., Verdier J., Leprince O., Buitink J. (2015). Inference of longevity-related genes from a robust co-expression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell 27: 2692–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnoblet C., Aubry C., Leprince O., Vu B.L., Rogniaux H., Buitink J. (2007). The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 51: 47–59. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.-Y., Kim J., Paek N.-C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5: 4636. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schneider T., Keller F. (2009). Raffinose in chloroplasts is synthesized in the cytosol and transported across the chloroplast envelope. Plant Cell Physiol. 50: 2174–2182. [DOI] [PubMed] [Google Scholar]
- Sinniah U.R., Ellis R.H., John P. (1998). Irrigation and seed quality development in rapid-cycling Brassica: soluble carbohydrates and heat-stable proteins. Ann. Bot. (Lond.) 82: 647–655. [Google Scholar]
- Sugliani M., Rajjou L., Clerkx E.J.M., Koornneef M., Soppe W.J.J. (2009). Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol. 184: 898–908. [DOI] [PubMed] [Google Scholar]
- Tang W., Ji Q., Huang Y., Jiang Z., Bao M., Wang H., Lin R. (2013). FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 163: 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira R.N., Ligterink W., França-Neto J.B., Hilhorst H.W., da Silva E.A. (2016). Gene expression profiling of the green seed problem in soybean. BMC Plant Biol. 16: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrasson E., Buitink J., Righetti K., Ly Vu B., Pelletier S., Zinsmeister J., Lalanne D., Leprince O. (2013). An emerging picture of the seed desiccome: confirmed regulators and newcomers identified using transcriptome comparison. Front. Plant Sci. 4: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer S.S., Björkman O. (1990). Leaf xanthophyll content and composition in sun and shade determined by HPLC. Photosynth. Res. 23: 331–343. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A., Hincha D.K., Leprince O., Macherel D. (2010). LEA proteins: versatility of form and function. In Dormancy and Resistance in Harsh Environments: Topics in Current Genetics 21, E. Lubzens, J. Cerda, and M. Clark, eds (Berlin, Heidelberg: Springer-Verlag; ), pp. 91–108. [Google Scholar]
- Vandecasteele C., et al. (2011). Quantitative trait loci analysis reveals a correlation between the ratio of sucrose/raffinose family oligosaccharides and seed vigour in Medicago truncatula. Plant Cell Environ. 34: 1473–1487. [DOI] [PubMed] [Google Scholar]
- Verdier J., et al. (2013). A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 163: 757–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Dorp K., Hölzl G., Plohmann C., Eisenhut M., Abraham M., Weber A.P.M., Hanson A.D., Dörmann P. (2015). Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis. Plant Cell 27: 2846–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R.E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cheng K., Wan L., Yan L., Jiang H., Liu S., Lei Y., Liao B. (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16: 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth W.M., Masnavi G., Bhardwaj R.M., Jiang Q., Bray C.M., West C.E. (2010). A plant DNA ligase is an important determinant of seed longevity. Plant J. 63: 848–860. [DOI] [PubMed] [Google Scholar]
- Yamburenko M.V., Zubo Y.O., Vanková R., Kusnetsov V.V., Kulaeva O.N., Börner T. (2013). Abscisic acid represses the transcription of chloroplast genes. J. Exp. Bot. 64: 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685. [DOI] [PubMed] [Google Scholar]
- Zbierzak A.M., Kanwischer M., Wille C., Vidi P.-A., Giavalisco P., Lohmann A., Briesen I., Porfirova S., Bréhélin C., Kessler F., Dörmann P. (2009). Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 425: 389–399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.