AUG-stops in the 5′ untranslated regions of Arabidopsis genes, including NIP5;1, which encodes a boron influx transporter, regulate B-dependent mRNA accumulation through ribosome stalling.
Abstract
Upstream open reading frames (uORFs) are often translated ahead of the main ORF of a gene and regulate gene expression, sometimes in a condition-dependent manner, but such a role for the minimum uORF (hereafter referred to as AUG-stop) in living organisms is currently unclear. Here, we show that AUG-stop plays an important role in the boron (B)-dependent regulation of NIP5;1, encoding a boric acid channel required for normal growth under low B conditions in Arabidopsis thaliana. High B enhanced ribosome stalling at AUG-stop, which was accompanied by the suppression of translation and mRNA degradation. This mRNA degradation was promoted by an upstream conserved sequence present near the 5′-edge of the stalled ribosome. Once ribosomes translate a uORF, reinitiation of translation must take place in order for the downstream ORF to be translated. Our results suggest that reinitiation of translation at the downstream NIP5;1 ORF is enhanced under low B conditions. A genome-wide analysis identified two additional B-responsive genes, SKU5 and the transcription factor gene ABS/NGAL1, which were regulated by B-dependent ribosome stalling through AUG-stop. This regulation was reproduced in both plant and animal transient expression and cell-free translation systems. These findings suggest that B-dependent AUG-stop-mediated regulation is common in eukaryotes.
INTRODUCTION
In eukaryotes, the scanning model predicts that ribosomes scan mRNAs from the 5′-end and identify the first AUG codon to start translation (Jackson et al., 2010). However, the first open reading frame (ORF) is not necessarily the main ORF of a gene. Short ORFs present in the 5′-untranslated region (UTR), known as upstream ORFs (uORFs), are often translated ahead of the main ORF. uORFs affect the expression of the main ORF because translation of the main ORF occurs either by leaky scanning and/or reinitiation of translation. During leaky scanning, the AUG codon of the uORF is not recognized and the 43S preinitiation complex (i.e., a complex of the 40S subunit, methionyl-tRMAIMet, and a few initiation factors) continues scanning for a downstream AUG (Supplemental Figure 1A). During reinitiation of translation, the 40S subunit does not dissociate from the mRNA after translation termination of the uORF and reinitiates translation at a downstream AUG (Supplemental Figure 1B) (Hellens et al., 2016).
Among the translated uORFs, some simply downregulate the expression of the main ORF, while others regulate the expression of the main ORF in a condition-dependent manner (Morris and Geballe, 2000; Hinnebusch, 2005; Hood et al., 2009; Ebina et al., 2015). For example, in Arabidopsis thaliana, S-adenosylmethionine decarboxylase 1 (AdoMetDC1) is required for producing spermidine and spermine. The ribosome stalls at the termination codon of a uORF of AdoMetDC1 in response to polyamine, leading to the downregulation of AdoMetDC1 (Uchiyama-Kadokura et al., 2014). In another example, in yeast (Saccharomyces cerevisiae), the ribosome stalls at the termination codon of a uORF of carbamyl phosphate synthetase 1 (CPA1) in an arginine-dependent manner, leading to the repression of CPA1 production required for arginine biosynthesis (Gaba et al., 2005). Ribosome stalling at the uORF adversely affects translation of the main ORF (Supplemental Figure 1C).
Minimum ORFs, in which the start codon is directly followed by a stop codon, referred to as “AUG-stops” in this article, are widely found in the 5′-UTRs of eukaryotic genes. According to our calculations, 23% of mRNAs in Arabidopsis carry uORFs (excluding those that overlap with the main ORF), of which 14% (i.e., 4% of Arabidopsis mRNA) have at least one AUG-stop in their 5′-UTR. AUG-stop affects the expression of a downstream ORF in human immunodeficiency virus type I (Krummheuer et al., 2007), but the role of AUG-stops in condition-dependent gene expression remains unclear.
Arabidopsis NIP5;1 (Takano et al., 2006) is among the genes that carry AUGUAA in their 5′-UTRs. NIP5;1 is a diffusion facilitator of boron (B) that is required for the efficient uptake of B by the roots. B is an essential nutrient for plants (Warington, 1923), algae (Smyth and Dugger, 1981), and cyanobacteria (Bonilla et al., 1990) that is also required for some animals (Fort et al., 1998; Rowe and Eckhert, 1999; Lanoue et al., 2000) but is toxic when present in excess (Blevins and Lukaszewski, 1998; Kabu and Akosman, 2013). B in neutral solution exits mostly as boric acid (H3BO3), which is a very weak Lewis acid with a pKa of 9.24 (Woods, 1996). B is directly or indirectly involved in crucial biological processes in various organisms, including both plants and animals (Brown et al., 2002; Chen et al., 2002; Dembitsky et al., 2002). In plants, B cross-links rhamnogalacturonan II (RG-II), a component of the pectic polysaccharides in the cell wall. The cis-diol groups of apiose in RG-II form an ester with boric acid, creating a borate-dimeric RG-II complex, which is important for normal growth in Arabidopsis (O’Neill et al., 2001, 2004). B is essential for cell elongation, and a number of physiological changes have been reported for plants under low B conditions (Blevins and Lukaszewski, 1998). Due to its essential nature and toxicity at high concentrations, it is important for plants to maintain B homeostasis for proper growth, and the regulation of the B transport process plays a crucial role in B homeostasis.
NIP5;1 is essential for normal growth of Arabidopsis under a limited B supply, and NIP5;1 mRNA accumulates to high levels under low B conditions but not under high B conditions (Takano et al., 2006). NIP5;1 mRNA is destabilized under a high supply of B, resulting in reduced B uptake (Tanaka et al., 2011). A 19-nucleotide region, −131 to −113 nucleotides (relative to the translation start site of NIP5;1) in the 5′-UTR is required for this regulation (Supplemental Figures 2A and 2B), which plays an important role in normal growth under excess B supply in Arabidopsis (Tanaka et al., 2011). NIP5;1 has four uORFs, two of which are AUGUAA (i.e., the start codon is immediately followed by a stop codon), located at −122 and −312 nucleotides in its 5′-UTR (herein referred to as −122AUG-stop and −312AUG-stop, respectively) (Supplemental Figures 2A and 2B). The region required for the B-dependent NIP5;1 mRNA degradation includes −122AUG-stop (Tanaka et al., 2011).
In this study, we investigated the mechanisms of the AUG-stop-mediated regulation of NIP5;1 expression in Arabidopsis. We found that AUG-stop plays crucial roles in B-dependent ribosome stalling and the regulation of transcript accumulation, as well as in the translation of the main ORF of this gene.
RESULTS
B-Dependent Downregulation of NIP5;1 through AUG-Stop in Vivo
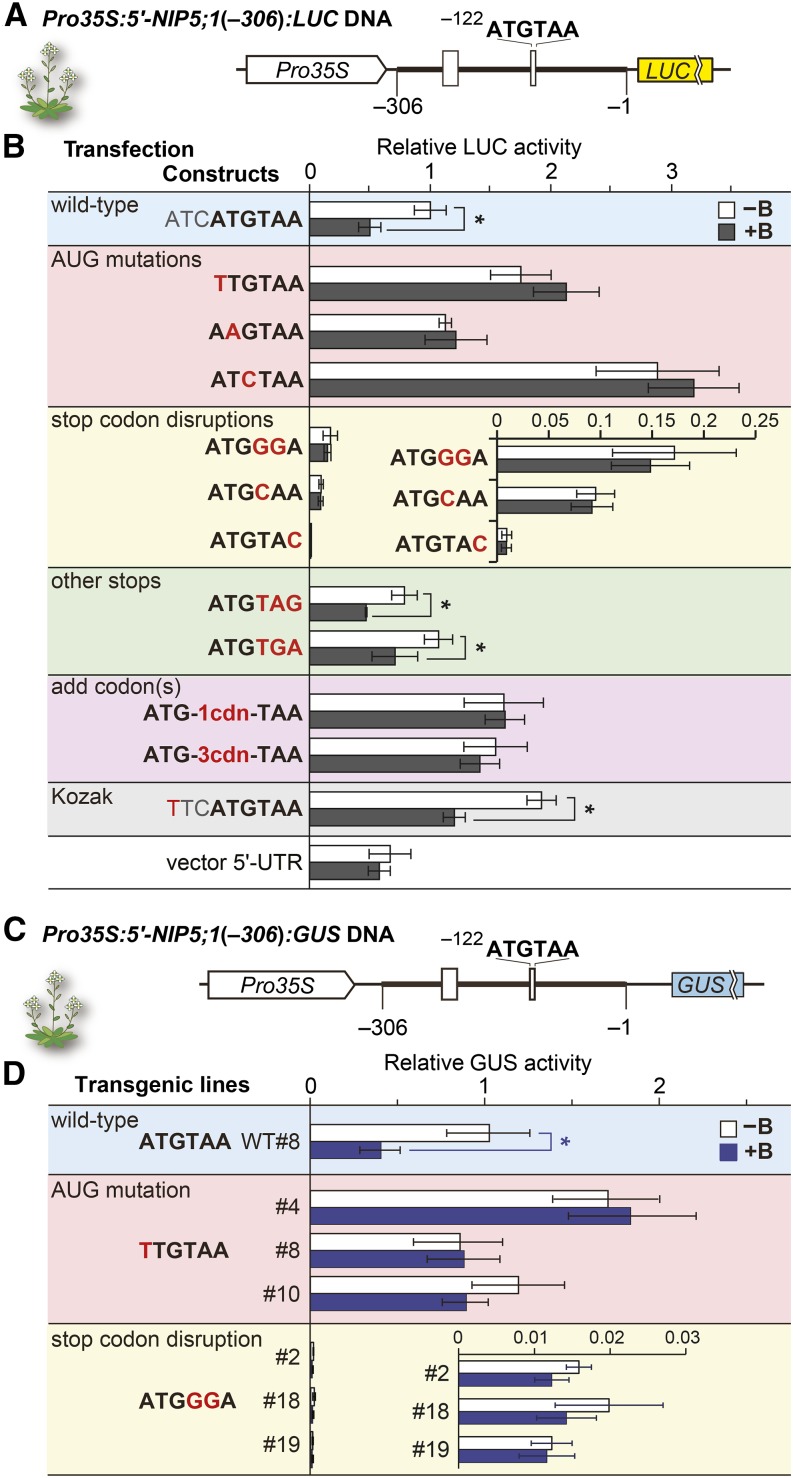
The −131- to −113-nucleotide region in the NIP5;1 5′-UTR, which is responsible for its B-dependent mRNA degradation (Supplemental Figures 2A and 2B) (Tanaka et al., 2011), includes −122AUG-stop. We first examined the possible involvement of −122AUG-stop in the B-dependent regulation of NIP5;1. The −306 to −1 nucleotide region of the NIP5;1 5′-UTR sequence was fused to the luciferase (LUC) reporter gene and placed downstream of a constitutive 35S RNA promoter (referred to as Pro35S) of cauliflower mosaic virus [Pro35S:5′-NIP5;1(−306):LUC] (Figure 1A). The DNA was used to transfect Arabidopsis MM2d suspension cells (Menges and Murray, 2002). A series of mutations were introduced into −122AUG-stop, and their effects on reporter activities were examined under high and low B conditions (Figure 1B).
Figure 1.
B-Dependent Downregulation of NIP5;1.
(A) Schematic representation of Pro35S:5′-NIP5;1(−306):LUC DNA. Open boxes represent uORFs. The thick line represents the sequence corresponding to the 5′-UTR of NIP5;1. Nucleotide numbers are relative to the translation start site (+1). The thin lines represent 18- and 15-nucleotide linker sequences at the 5′ and 3′ ends, respectively, of the 5′-UTR.
(B) Transfection experiments using cultured Arabidopsis cells. Constructs represent the sequences of, and around, the AUGUAA in each construct with the altered nucleotides shown in red. Transfected protoplasts were incubated under 500 μM B (+B) or 1 μM B (−B) conditions. The LUC activity of each transfected cell extract was normalized with RLUC activity from the cotransfected internal control plasmid and shown as the relative LUC activity. Means ± sd of relative LUC activities are shown (n = 3). Asterisks indicate a significant reduction in +B compared with −B conditions (P < 0.05). The “vector 5′-UTR” negative control carries only the vector’s 5′-UTR sequence. The nucleotide and corresponding amino acid sequences of ATG-1cdn-TAA and ATG-3cdn-TAA are shown in Supplemental Figure 2C.
(C) Schematic representation of Pro35S:5′-NIP5;1(−306):GUS DNA. The thin lines represent 37- and 59-nucleotide linker sequences at the 5′ and 3′ ends, respectively, of the 5′-UTR.
(D) Effect of B on GUS activities in transgenic plants. Transgenic plants were grown under 100 µM B (+B) or 0.3 µM B (−B) conditions. Relative GUS activities in roots were determined. Transgenic line WT#8 carries a wild-type −122AUG-stop (Tanaka et al., 2011). Three each of independent transgenic lines were used for the mutant constructs. Means ± sd of GUS activities relative to that of WT#8 under −B are shown (n = 4 to 5). Asterisks indicate significant reductions under +B conditions (P < 0.05).
In the construct carrying the wild-type −122AUG-stop, reporter activity was reduced to approximately half under high B conditions compared with low B conditions (Figure 1B, shaded in blue). Introduction of mutations into ATG abolished the response to high B conditions, and the reporter activities became generally higher in both low and high B conditions. Disruption of the stop codon (shaded in yellow) abolished the response to high B conditions and, in this case, the reporter activities were much reduced in both low and high B conditions, whereas changing TAA to other stop codons, i.e., TAG or TGA (shaded in green), retained the B-dependent suppression of the reporter activity. Placing extra codon(s) between ATG and TAA (shaded in purple) also abolished the response. These results indicate that the −122AUG-stop sequence is required for B-dependent downregulation and that the AUG codon needs to be directly followed by a stop codon. In the construct carrying a mutation in the Kozak sequence (a consensus sequence for efficient translation initiation) (Kozak, 1986; Joshi et al., 1997) of −122AUG-stop, the response to B was maintained, although the reporter activities increased under both B conditions (Figure 1B, shaded in gray).
The general upregulation of reporter activities observed in the AUG mutations can be explained by the lack of ribosome assembly at uORF. The general upregulation in the construct carrying a mutation in the Kozak sequence, which is likely to reduce recognition of AUG in AUGUAA, can be explained by the increased leaky scanning. In the constructs in which stop codons were disrupted, the ORF that starts from the −122AUG is extended into the LUC reporter gene in a different reading frame (Supplemental Figure 2D). Therefore, the downregulation of LUC activity in the stop codon disruption mutants suggests that most of the ribosomes start translation at −122AUG and that leaky scanning does not contribute much to the reporter activities observed in the wild-type construct (Supplemental Figures 1A and 1B).
To test whether −122AUG-stop is necessary for the B response in vivo, we generated transgenic Arabidopsis plants carrying the NIP5;1 5′-UTR (−306 to −1 nucleotides) fused to a GUS reporter gene driven by Pro35S [Pro35S:5′-NIP5;1(−306):GUS] (Figure 1C) and exposed the plants to high and low B conditions (Figure 1D). As a positive control, we used one of the transgenic lines that we described previously (P35SUTR+7-GUS #8) (Tanaka et al., 2011). In transgenic plants carrying the wild-type −122AUG-stop, the reporter activity was reduced under high B conditions compared with low B conditions, corroborating our previous report (Tanaka et al., 2011), while introduction of a mutation into either ATG or TAA of the −122AUG-stop abolished the response. Thus, AUG-stop is required for the B-dependent downregulation of NIP5;1 in vivo.
Detection of B-Dependent NIP5;1 mRNA Degradation Intermediates in Arabidopsis Roots
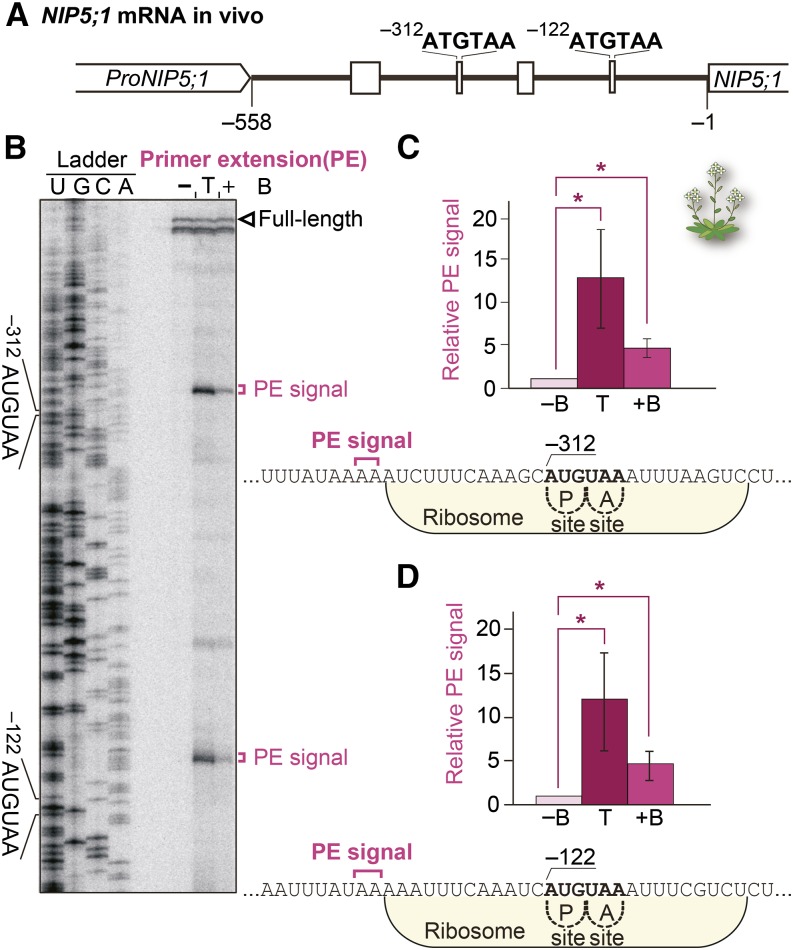
It is possible that the degradation of NIP5;1 mRNA goes through a specific degradation intermediate(s). To examine the positions of the 5′-ends of degradation intermediates, we conducted primer extension analysis using mRNA extracted from the roots of Col-0 wild-type Arabidopsis (Figures 2A and 2B). The Arabidopsis Information Resource database (TAIR9/10) (Lamesch et al., 2012) lists the transcription start site of NIP5;1 at the −312 nucleotide, whereas our primer extension analysis positioned it at −558 (Figure 2B), which corroborated well with that of the Plant Promoter Database (Hieno et al., 2014). Therefore, we used the −558 nucleotide in the NIP5;1 5′-UTR as the position of the transcription start site in this study.
Figure 2.
B-Dependent NIP5;1 mRNA Degradation Intermediates in Vivo
(A) Schematic representation of the NIP5;1 5′-UTR.
(B) Primer extension analysis. Col-0 plants were grown for 28 d with 100 µM B (+B) or 0.3 µM B (−B). For the sample in lane T, Col-0 plants grown for 28 d with 0.3 µM B were transferred to 100 µM B for 10 min. Total RNA extracted from the roots was used for the primer extension reaction. Loading was adjusted to ensure full-length signal intensities were at similar levels. A sequence ladder was generated using the same primer. The open arrowhead marks the 5′-end of the full-length mRNA. Primer extension signals corresponding to the 5′-ends of RNA degradation intermediates upstream of −312AUG-stop and −122AUG-stop are marked with magenta brackets.
(C) and (D) Means ± sd (n = 3) of relative primer extension signal intensities are shown for −122AUG-stop (C) and −312AUG-stop (D). Asterisks indicate significant differences (P < 0.05). Nucleotide sequences around the AUG-stops, with primer extension signal positions, are shown. Ribosomes occupying the RNA having the AUG codon positioned at the P-site are shown.
Our primer extension analysis detected two major signals representing the 5′-end points of mRNA degradation intermediates from plants grown under high B conditions (Figures 2B, lane +B, 2C, and 2D). Even stronger signals were detected in plants 10 min after being transferred from low B to high B conditions (Figures 2B, lane T, 2C, and 2D), implying that mRNA degradation is temporarily activated in response to the increased B concentration. These signals were found in a region 13 to 14 nucleotides upstream of both −312AUG-stop and −122AUG-stop (counting from the A of AUG), respectively (Figures 2C and 2D). No mRNA degradation intermediate was detected under low B conditions (Figure 2B, lane B).
The mRNA degradation intermediates were detected in experiments independent from ours. Degradome data sets available at the Parallel Analysis of RNA Ends (PARE) database (German et al., 2008) provided us with information about the 5′-ends of mRNA decay intermediates in plants grown in soil (note that normal soil culture conditions correspond to a mild-high B condition in this study). Prominent 5′-ends of 5′-truncated NIP5;1 mRNA are present at −326 and −134 nucleotides of the 5′-UTR, and the signals are enhanced in the exoribonuclease 4 (xrn4) mutant, which lacks 5′-3′ exoribonuclease activity (German et al., 2008) (Supplemental Figures 3A and 3B). These positions correspond to 14 and 12 nucleotides upstream of −312AUG-stop and −122AUG-stop, respectively. These positions correspond well with the 5′-ends of degradation intermediates detected in our primer extension analysis (Figures 2C and 2D).
OsNIP3;1 and ZmNIP3;1, rice (Oryza sativa) and maize (Zea mays) orthologs, respectively, of Arabidopsis NIP5;1, also carry AUGUAA sequences. The PARE database shows mRNA decay intermediates at 12 and 13 nucleotides upstream of the AUG-stops in these genes (Li et al., 2010; Liu et al., 2014) (Supplemental Figures 3C and 3D), suggesting that the same mechanism is exploited in these plant species as in Arabidopsis NIP5;1.
B-Dependent NIP5;1 mRNA Degradation Is Independent of the Nonsense-Mediated mRNA Decay Pathway
uORFs sometimes induce the destabilization of mRNA through the nonsense-mediated mRNA decay (NMD) mechanism because the uORF’s stop codon may be recognized as a premature termination codon (Amrani et al., 2006). UPF1 and UPF3 play critical roles in the induction of mRNA degradation by NMD (Hori and Watanabe, 2005; Yoine et al., 2006b). We therefore determined the accumulation levels of NIP5;1 mRNA under high and low B conditions in the upf1-1 missense mutant (Yoine et al., 2006a), upf3-1 knockout mutant (Hori and Watanabe, 2005), and upf3-3 knockout mutant (Arciga-Reyes et al., 2006) in vivo (Supplemental Figure 4A). The upf3-1 knockout mutant was also used to measure NIP5;1 mRNA stability (Supplemental Figures 4B and 4C). Neither NIP5;1 mRNA accumulation levels nor NIP5;1 mRNA half-lives were affected by the mutations tested, indicating that NMD is not responsible for the B-dependent regulation of NIP5;1 mRNA decay.
B-Dependent NIP5;1 mRNA Degradation in Vitro
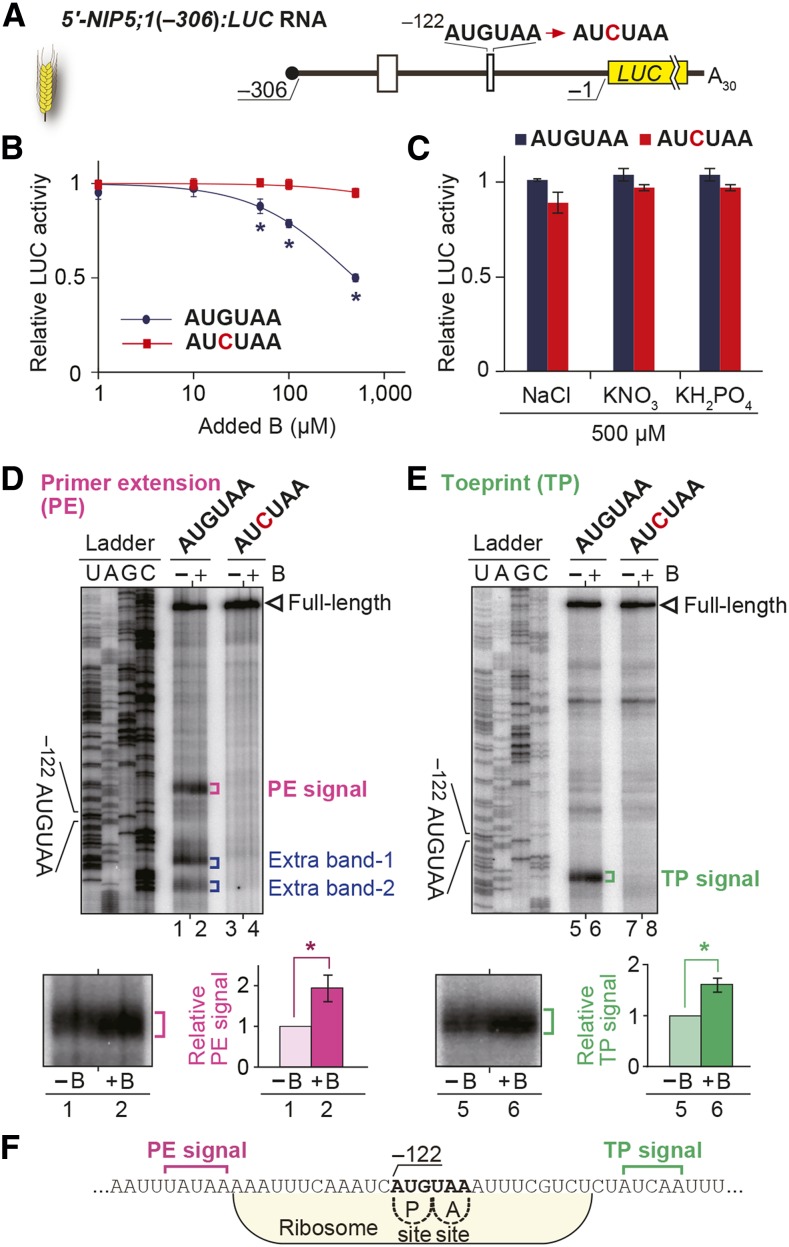
To establish an in vitro system to analyze B-dependent regulation through AUG-stop, we used a wheat germ extract (WGE) in vitro translation system. In vitro-transcribed RNA carrying −306 to −1 nucleotides of the NIP5;1 5′-UTR fused to the LUC reporter gene [5′-NIP5;1(−306):LUC] was translated in the presence of various concentrations of B (Figures 3A and 3B). When a transcript containing the wild-type AUG-stop sequence was used, the reporter activity was reduced as the B concentration increased, while no such change was observed when −122AUG-stop was mutated (Figure 3B). These results establish that the in vitro translation system is capable of reproducing the response to supplemental B.
Figure 3.
AUG-Stop Affects B-Dependent Downregulation of NIP5;1.
(A) Schematic representation of 5′-NIP5;1(−306):LUC RNA. The thick line represents the mRNA sequence and open boxes represent uORFs.
(B) Effect of B on translation in vitro. RNA carrying 5′-NIP5;1(−306):LUC with or without a mutation in AUGUAA was translated in WGE. Means ± sd of relative LUC activities under various B conditions are shown (n = 3). Asterisks indicate significant reductions in RNA carrying −122AUGUAA (P < 0.05).
(C) Effects of NaCl, KNO3, and KH2PO4 at 500 μM on reporter expression in vitro. RNA carrying 5′-NIP5;1(−306):LUC with or without a mutation in AUGUAA was translated in WGE. Means ± sd of relative LUC activities are shown (n = 3)
(D) and (E) Primer extension (D) and toeprint (E) analyses in WGE with 300 µM B (+B) or without B supplementation (−B). Signals are enlarged under the main image of the AUGUAA lane, and their means ± sd (n = 3) of relative intensities are shown. Asterisks indicate significant differences (P < 0.05).
(F) Nucleotide sequence around −122AUG-stop. Positions of PE (magenta) and TP signals (green) are marked. Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
We also investigated whether AUG-stop-mediated regulation is observed for other elements. We tested the effects of major nutrients, nitrogen (KNO3), phosphorous (KH2PO4), and potassium (KNO3 and KH2PO4), together with NaCl. Addition of these salts to WGE at a concentration of 500 µM, a concentration in which boric acid reduced reporter expression by half (Figure 3B), showed no differential effect on reporter activity (Figure 3C). These results demonstrate that the response to boric acid is specific among the salts and ions examined.
We then examined mRNA degradation in WGE. B-dependent signals of mRNA degradation intermediates were detected by primer extension analysis using RNA extracted from the in vitro translation mixture after 60 min of translation. B-dependent primer extension signals were detected at 13 to 17 nucleotides upstream of −122AUG-stop (Figure 3D, lanes 1 and 2), which overlapped with the end points detected in vivo (Figures 2C and 2D). No such signal was detected when −122AUG-stop was mutated (Figure 3D, lanes 3 and 4). Extra bands were also detected downstream of −122AUG-stop, but these bands do not represent a part of the B-dependent downregulation mechanisms because the signal intensities were not dependent on B conditions (Supplemental Figure 5). These results indicate that the B-dependent downregulation via the −122AUG-stop observed in vivo is recapitulated in the WGE in vitro translation system.
Either of the Two AUG-Stops in the 5′-UTR of NIP5;1 mRNA Is Sufficient for B-Dependent Regulation
Endogenous NIP5;1 mRNA carries two AUG-stops in its 5′-UTR, and two signals from mRNA degradation intermediates were observed in vivo (Figure 2B), corresponding to the two major mRNA 5′-ends listed in the PARE database (Supplemental Figure 3B). It is likely that both −312AUG-stop and −122AUG-stop are functional.
To obtain experimental evidence for the function of −312AUG-stop in B-dependent downregulation of the expression and degradation of mRNA, we conducted various experiments using an in vitro-transcribed RNA carrying the full-length NIP5;1 5′-UTR (−558 to −1 nucleotides) (Supplemental Figure 6A). B-dependent downregulation of reporter activity was observed when either or both of the AUG-stops were present, but not when both AUG-stops were deleted (Supplemental Figure 6B). Furthermore, primer extension signals were detected at 13 to 17 nucleotides upstream of the −312AUG-stop (Supplemental Figures 6C, lanes 3 and 4, and 6E), indicating that −312AUG-stop is also functional in the B-dependent downregulation of the expression and degradation of mRNA in vitro.
Ribosomes Stall at AUG-Stop in the 5′-UTR of NIP5;1 mRNA in Vitro in a B-Dependent Manner
The 5′-end points of the mRNA degradation intermediates (Figure 3D; Supplemental Figure 6C, marked with magenta brackets) suggest that stalled ribosomes are involved in mRNA degradation because the 5′-ends of these mRNA degradation intermediates are located near the 5′-edge of a ribosome when the ribosome’s P-site is positioned at AUG (Sachs et al., 2002; Yamashita et al., 2014) (Figure 3F; Supplemental Figure 6E). To obtain direct evidence for ribosome stalling, we performed primer extension inhibition (toeprint) analysis using RNA from the in vitro translation mixture after 30 min of translation in WGE. In a standard toeprint reaction, cycloheximide (CHX) was added after the translation reaction to stabilize the ribosome (Sachs et al., 2002). B-dependent toeprint signals were detected at 17 to 21 nucleotides downstream of the −122AUG-stop (counting from the A of AUG) (Figures 3E, lanes 5 and 6, and 3F). Consistent results were observed with RNA carrying −312AUG-stop. B-dependent toeprint signals were detected at 14 to 16 nucleotides downstream of the −312AUG-stop (Supplemental Figures 6D, lanes 7 and 8, and 6E). Thus, ribosomes stall at both AUG-stops in a B-dependent manner, representing an intriguing regulatory role for AUG-stops through ribosome stalling.
We also performed a toeprint analysis without CHX (Supplemental Figure 7). Toeprint signals were detected at a similar position to that detected in the presence of CHX, and the signals were stronger under B supplementation, although the overall signal intensities were reduced (Supplemental Figures 7B and 7C). These results indicate that ribosome stalling is dependent on B treatment and that CHX enhanced the toeprint signals.
Possible Involvement of Translation Reinitiation in B-Dependent Expression of the Main ORF of NIP5;1
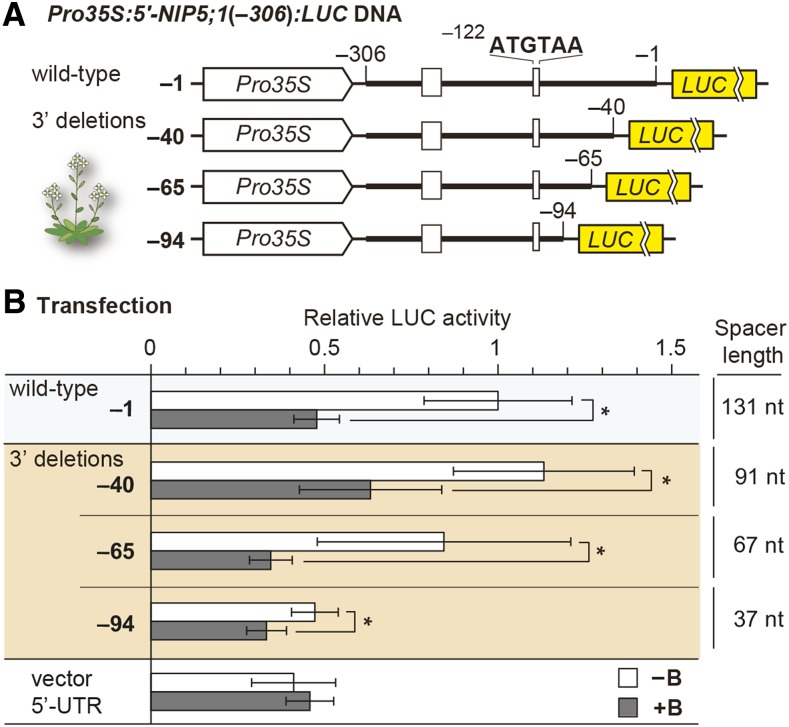
Translation of the main ORF in uORF-containing mRNA occurs via leaky scanning and/or reinitiation of translation (Supplemental Figures 1A and 1B) (Hellens et al., 2016). As mentioned above, the overall reduction in reporter activity in stop codon disruption mutant constructs (Figure 1B) suggests that leaky scanning makes only a minor contribution to the reporter activity observed in the wild-type construct and that leaky scanning is not affected by B conditions. This finding, in turn, suggests that reinitiation plays a major role in the translation of the main ORF and that the efficiency of reinitiation is affected by B conditions. Thus, we tested whether reinitiation is affected by B conditions. The involvement of reinitiation can be assessed by modulating the length between the site of ribosome stalling and the main ORF, referred to as the spacer (Kozak, 1987, 2001; Rajkowitsch et al., 2004). The efficiency of reinitiation is reduced if the spacer is less than 50 nucleotides long (Child et al., 1999; Zhou et al., 2010).
Arabidopsis MM2d cells were transfected with plasmids containing a series of 3′-deletions of the 5′-UTR (Figure 4A). The B response was retained even when the spacer was only 37 nucleotides long, but LUC activity under low B conditions was reduced when the spacer was 37 nucleotides long compared with 67 nucleotides or longer (Figure 4B). The extent of the B response was reduced when a construct with a 37 nucleotide spacer, which is expected to reduce reinitiation efficiency, was used. These results corroborate the hypothesis that reinitiation is reduced under high B conditions, although other possibilities remain (e.g., mRNA stability is affected by changes in spacer length).
Figure 4.
B-Dependent Reinitiation of NIP5;1.
(A) Schematic representation of 3′-deletion series of Pro35S:5′-NIP5;1(−306):LUC DNA. The thick line represents the sequence corresponding to NIP5;1 mRNA. The thin lines represent the 18- and 15-nucleotide linker sequences at the 5′ and 3′ ends, respectively, of the 5′-UTR.
(B) Effects of spacer deletions on the B response in transfection experiments. Transfected protoplasts were incubated under 500 μM B (+B) or 1 μM B (−B) conditions, and means ± sd of relative LUC activities (n = 8) are shown. Asterisks indicate significant reductions under +B compared with −B conditions (P < 0.05). The “vector 5′-UTR” negative control carries only the vector’s 5′-UTR sequence.
Insertion of AUG-Stop into the 5′-UTR of a Non-B-Responsive Gene Confers B-Dependent Regulation
To examine whether AUG-stop is capable of inducing B-dependent regulation, the AUGUAA sequence was introduced into the 5′-UTR of the non-B-responsive gene MOLYBDATE TRANSPORTER2 (MOT2) (Gasber et al., 2011) and Dof zinc finger DNA binding protein gene Dof1.8 (Yanagisawa, 2002) (Figure 5A; Supplemental Figure 8A), both of which lack uORFs, including AUG-stops, in their 5′-UTRs and whose 5′-UTRs are roughly the same size (333 and 296 nucleotides, for MOT2 and Dof1.8, respectively) as that of our experimental NIP5;1 constructs used to analyze the functions of −122AUGUAA (306 nucleotide). Since spacer length is important for B-dependent regulation (Figure 4B), we inserted AUGUAA at −122 nucleotides of their 5′-UTRs (Figure 5B; Supplemental Figure 8B), a position identical to that of −122AUG-stop in NIP5;1.
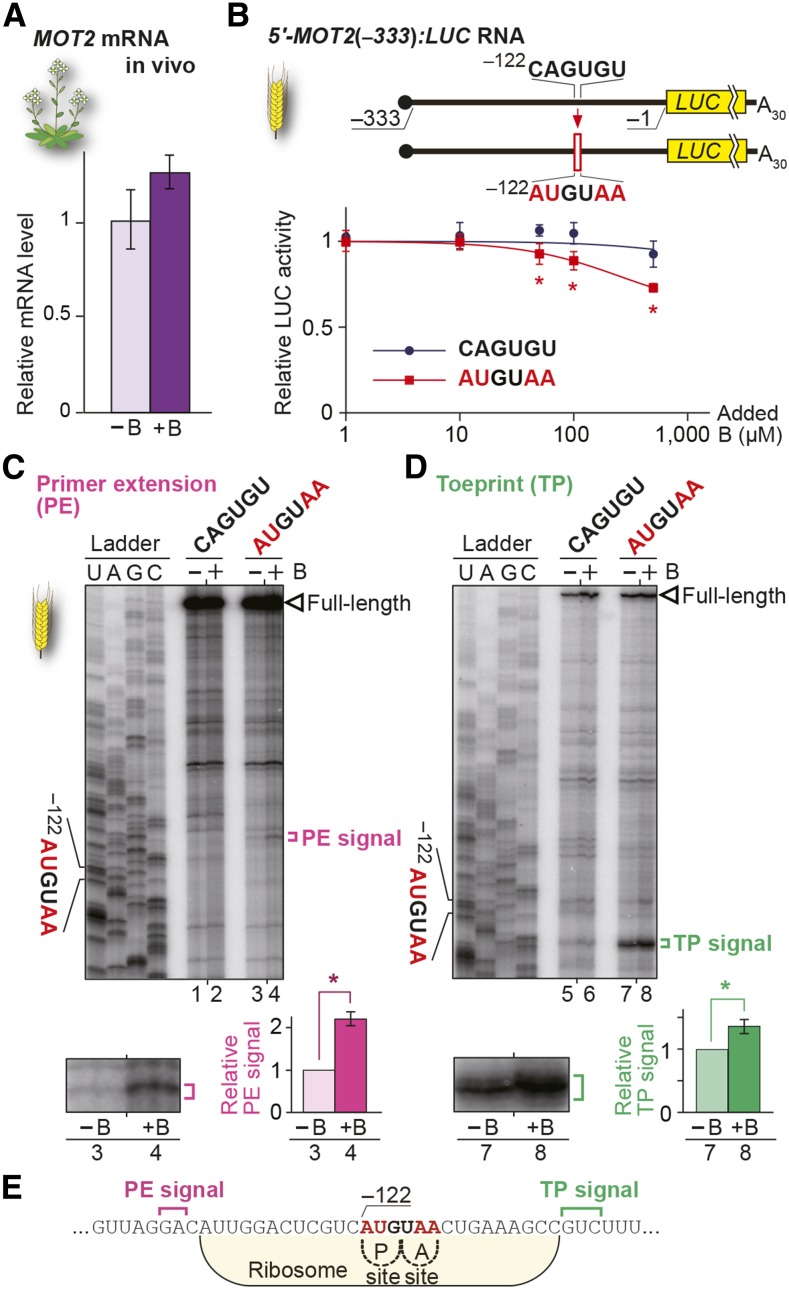
Figure 5.
B-Dependent Downregulation Conferred by Introduction of AUG-Stop into MOT2 5′-UTR.
(A) mRNA accumulation in Col-0 roots under 100 µM B (+B) and 0.3 µM B (−B) conditions. Means ± sd of relative mRNA accumulation (n = 3) are shown.
(B) Schematic representation of 5′-MOT2(−333):LUC RNA and the effect of AUGUAA introduction on the B response. RNA carrying 5′-MOT2(−333):LUC with or without AUGUAA was translated in WGE in the presence of various B concentrations. Means ± sd of relative LUC activities (n = 3) are shown. Asterisks indicate significant reduction of reporter activity in RNA having an AUGUAA compared with that in RNA having the original MOT2 5′-UTR sequence (P < 0.05).
(C) and (D) Primer extension (C) and toeprint (D) analyses in WGE with 300 µM B (+B) or without B supplementation (−B). Signals are enlarged under the main image of the AUGUAA lane, and means ± sd of relative intensities are shown (n = 3). Asterisks indicate significant differences (P < 0.05).
(E) Nucleotide sequence around AUG-stop and the positions of PE (magenta) and TP signals (green). Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
Reporter assays in WGE revealed that introducing the AUGUAA sequence led to B-dependent downregulation of the expression in both genes (Figure 5B; Supplemental Figure 8B). To further examine the effect of AUG-stop insertion, primer extension and toeprint analyses were performed using a construct carrying the MOT2 5′-UTR. Primer extension and toeprint signals were detected 14 to 15 nucleotides upstream (Figure 5C, lanes 3 and 4) and 15 to 17 nucleotides downstream (Figure 5D, lanes 7 and 8), respectively, of the introduced AUGUAA sequence, and the signal intensities increased under high B conditions, suggesting that ribosome stalling and mRNA degradation similar to those in NIP5;1 occurred at the introduced AUGUAA (Figure 5E). These gain-of-function experiments suggest that AUG-stop is capable of inducing B-dependent ribosome stalling and the mRNA degradation of non-B-responsive genes.
Genome-Wide Analysis of Ribosome Stalling at AUG-Stop in Arabidopsis
To examine genome-wide ribosome stalling at AUG-stops, we analyzed the Arabidopsis ribosome profiling data set (Juntawong et al., 2014). This data set was obtained from Arabidopsis plants grown under “normal” conditions, which correspond to mild-high B conditions. The extent of ribosome stalling at each combination of the two codons, which is expected to be located in the P-site (codon 1) and A-site (codon 2), was estimated by counting the number of reads. The counts were divided by the average read count of the corresponding mRNA for normalization (Supplemental Figure 9). The presence of AUG at the first codon and the stop codon at the second codon tend to cause ribosome stalling (Supplemental Figure 9A). It is likely that ribosomes generally stall on AUG-stops with high frequency compared with other two-codon combinations (Supplemental Figure 9B).
Identification of Other Genes that Are Regulated in a B-Dependent Manner through AUG-Stop in Arabidopsis
To assess the generality of B-dependent mRNA degradation through AUG-stops, we looked for Arabidopsis genes that are regulated in a similar manner to that of NIP5;1. Microarray analysis was performed under high and low B conditions in Arabidopsis seedlings. Five genes, NIP5;1, CONSERVED PEPTIDE UPSTREAM OPEN READING FRAME47 (CPuORF47), transcription factor gene ABS2/NGAL1, At4g19370, and cupredoxin superfamily protein gene SKU5, that showed the strongest downregulation among those carrying AUG-stops in their 5′-UTRs (Figure 6A; Supplemental Figure 10A and Supplemental Table 1) were used for the analysis. The level of downregulation was judged by the FC+B/−B values, in which the expression levels under high B conditions were divided by those under low B conditions.
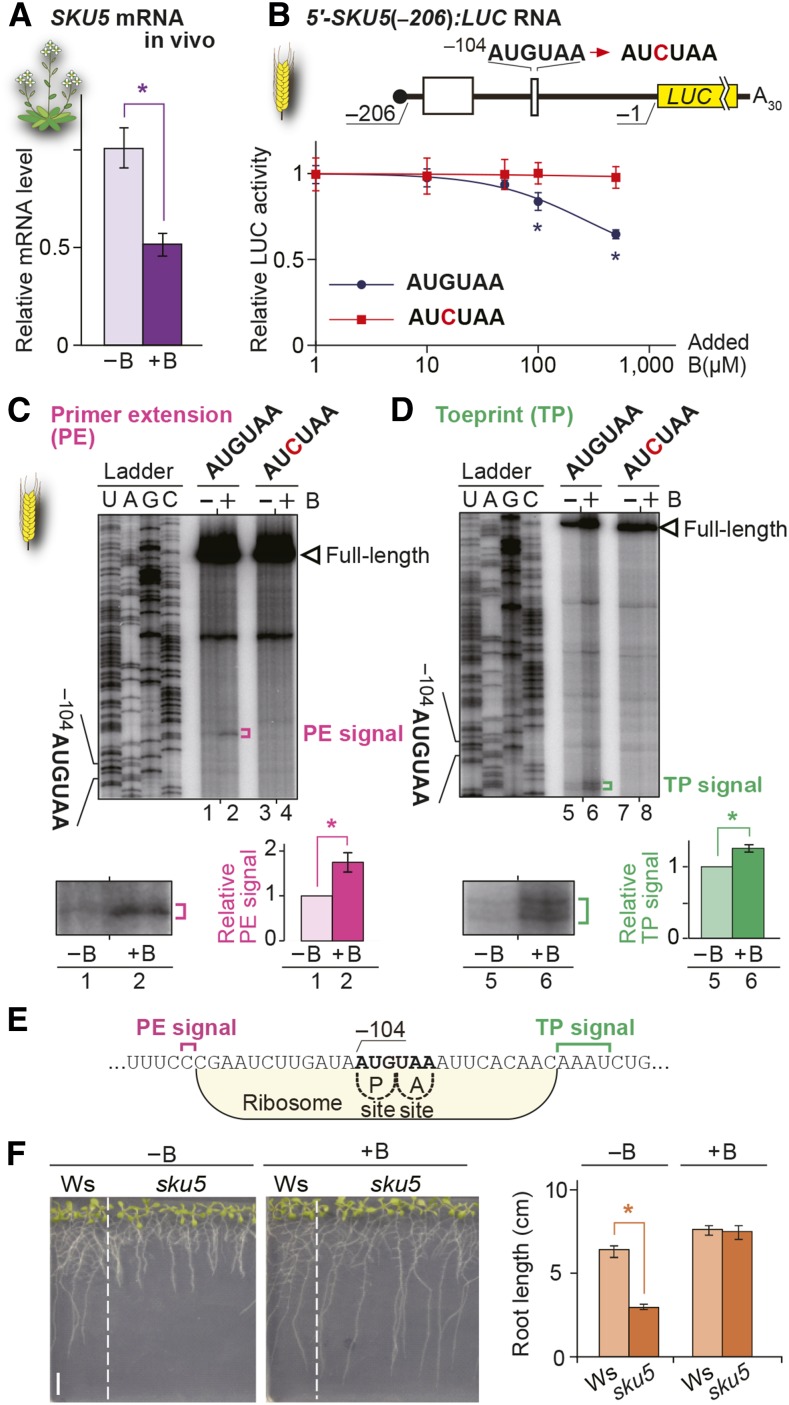
Figure 6.
Role of AUG-Stop in B-Dependent Downregulation of SKU5.
(A) mRNA accumulation in Col-0 roots under 100 µM B (+B) and 0.3 µM B (−B). Means ± sd of relative mRNA accumulation (n = 3) are shown. Asterisk indicates significant reduction under +B condition (P < 0.05).
(B) Schematic representation of the 5′-SKU5(−206):LUC RNA. Open boxes represent uORFs. 5′-SKU5(−206):LUC RNA was translated in WGE, and means ± sd of relative LUC activities (n = 3) are shown. Asterisks indicate significant reductions of relative reporter activity in RNA carrying AUGUAA (P < 0.05).
(C) and (D) Primer extension (C) and toeprint (D) analyses in WGE with 300 µM B (+B) or without B supplementation (−B). Signals are enlarged below the main image of the AUGUAA lane, and means ± sd of relative signal intensities are shown (n = 3). Asterisks indicate significant differences (P < 0.05).
(E) Nucleotide sequence around AUG-stop and the positions of primer extension (magenta) and toeprint signals (green) are marked. Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
(F) Arabidopsis Wassilewskija (Ws) and sku5 mutant plants were grown under 0.3 µM B (−B) or 100 µM B (+B) conditions for 10 d, and root length was measured. Means ± sd of root length (n = 5 to 10) are shown. Asterisks indicate significant reductions in root lengths of sku5 mutants compared with wild-type Wassilewskija (P < 0.05). Bar = 10 mm.
The five genes were tested to determine whether they are regulated through AUG-stop. All genes other than At4g19370 exhibited B-dependent suppression of translation (Figure 6B; Supplemental Figure 10B and Supplemental Table 1). Introduction of a mutation in the AUG-stop eliminated the B-dependent responses in ABS2/NGAL1 and SKU5 (Figure 6B; Supplemental Figure 10B), but not in CPuORF47 (Supplemental Table 1), in the reporter assay and in vitro translation system. These results indicate that B-dependent ribosome stalling is not specific to NIP5;1, while the results for CPuORF47 suggest the presence of a B-dependent regulatory mechanism(s) other than AUG-stop-mediated regulation.
SKU5 and ABS2/NGAL1 were analyzed in more detail. In both genes, toeprint and primer extension signals were detected, and the distance from the AUG-stop of SKU5 and ABS2/NGAL1 to the toeprint signals (15 to 18 and 16 to 17 nucleotides, respectively) and to the 5′-ends of the mRNA degradation intermediates (13 and 14 nucleotides, respectively) corroborated well with those in NIP5;1. The signal intensities were higher under high B conditions than under low B conditions (Figures 6C to 6E; Supplemental Figures 10C to 10E).
SKU5, an AUG-Stop-Mediated B-Regulated Gene, Functions in Low-B Conditions in Arabidopsis
SKU5 encodes a plant-specific transcription factor involved in directional root growth (Sedbrook et al., 2002). Since cell expansion requires B, SKU5 expression may be regulated in a B-dependent manner through AUG-stop. To assess the importance of SKU5 in B responses in Arabidopsis, we examined the growth of a sku5 knockout mutant (Sedbrook et al., 2002) under low B conditions. The mutant seedlings exhibited growth defects under low B, but not under high B conditions (Figure 6F), indicating that SKU5 functions under low B conditions as does NIP5;1 (Takano et al., 2006). The expression of SKU5, as well as NIP5;1, could be effectively suppressed under high B conditions via regulation through AUG-stop.
A Sequence Upstream of AUG-Stop Is Required for Promoting mRNA Degradation
Only a subset of genes containing AUGUAA in their 5′-UTRs are under the control of the AUG-stop-mediated downregulation of mRNA accumulation, implying that sequences other than AUGUAA are also required for this regulation. An alignment of NIP5;1, ABS2/NGAL1, and SKU5 and the rice and maize orthologs of Arabidopsis NIP5;1 sequences around AUGUAA (Supplemental Table 2) showed that the region 12 to 19 nucleotides upstream of AUGUAA is well conserved (UUU[C/A]AA[A/C]A), and U is present 9 nucleotides downstream of AUGUAA, while no such occurrence was observed in AUGUAA-containing non-B-responding genes (Figure 7). The upstream conserved region covers the positions of the 5′-ends of mRNA degradation intermediates (Figure 3F), and we hypothesized that the upstream conserved sequence is necessary for promoting mRNA degradation. To investigate this hypothesis, we conducted an in vitro translation study (Figure 8A). Disruption of the upstream conserved sequence strongly decreased the mRNA degradation intermediate signals under both high and low B conditions (Figures 8B, lanes 5 and 6, and 8D), although the effect was less prominent than when AUG-stop was mutated, when virtually no signal was detected irrespective of B conditions (Figure 8B, lanes 3 and 4). It should also be noted that disruption of the upstream conserved sequence had little or no effect on B-dependent toeprint signals (Figures 8C, lanes 7, 8, 11, and 12, and 8D). These results suggest that the upstream conserved sequence acts in enhancing mRNA degradation, but not in ribosome stalling.
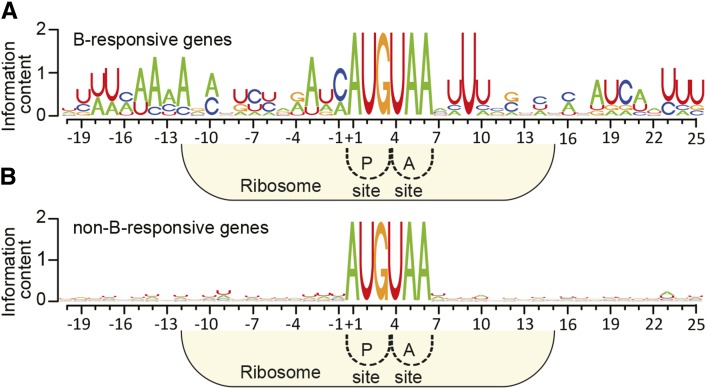
Figure 7.
Sequence Consensus around AUG-Stop in B-Responsive Genes.
Sequence logos around AUGUAA. NIP5;1, ABS2/NGAL1, SKU5, and the rice and maize orthologs of Arabidopsis NIP5;1 subjected to B-dependent AUG-stop-mediated mRNA degradation (Supplemental Table 2) (A) and non-B-responsive genes represented by 100 genes whose FC+B/−B values were closest to 1.0 based on microarray analysis (B) were used to construct the sequence logos. Positions are shown with the A of AUG as +1. Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
Figure 8.
The Upstream Conserved Sequence Affects B-Dependent mRNA Degradation.
(A) Schematic representation of 5′-NIP5;1(−306):LUC RNA. Open boxes represent uORFs.
(B) and (C) RNA carrying 5′-NIP5;1(−306):LUC with or without a mutation in AUGUAA or with a mutation in the region 17 to 14 nucleotides upstream of −122AUG-stop was translated in WGE. Primer extension (B) and toeprint (C) analyses in WGE with 300 µM B (+B) or without B supplementation (−B). Signals are enlarged under the main image of the AUGUAA lane, and their relative intensities are shown. Means ± sd are shown (n = 3). Asterisks indicate significant differences (P < 0.05).
(D) Nucleotide sequence around −122AUG-stop. Positions of primer extension (magenta) and toeprint signals (green) are marked. Ribosome occupation of RNA having the AUG codon positioned at the P-site is shown.
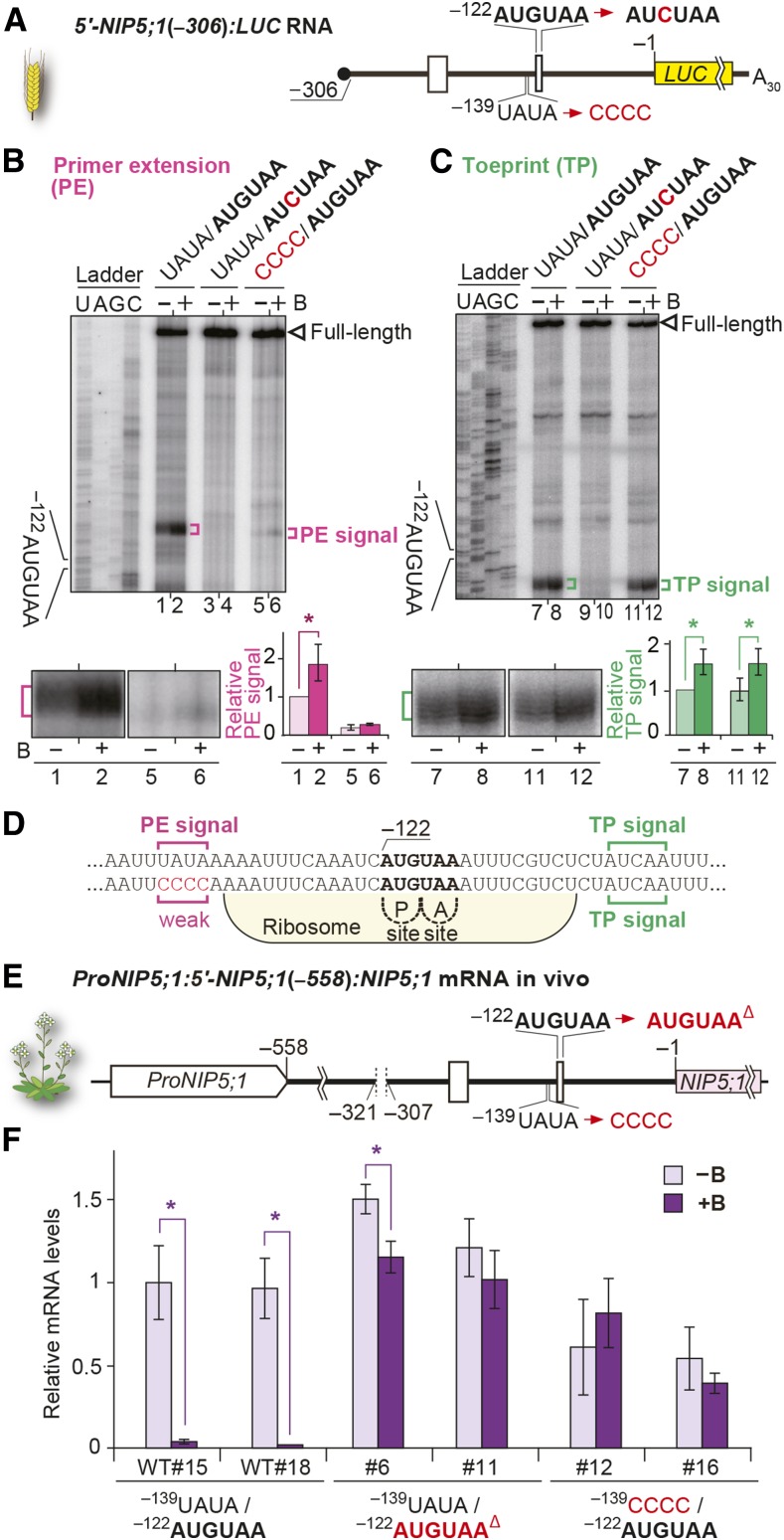
(E) Schematic representation of ProNIP5;1:5′-NIP5;1(−558):NIP5;1. Deletion of −312AUGUAA, a mutation in the upstream conserved sequence, and the deletion of −122AUGUAA are indicated.
(F) mRNA accumulation in transgenic plant roots grown under 100 µM B (+B) and 0.3 µM B (−B) conditions. Means ± sd of relative mRNA accumulation (n = 3) are shown. Asterisks indicate significant reductions under +B condition (P < 0.05). Two independently transformed lines were used for each construct.
To further examine whether the upstream conserved sequence is capable of promoting B-dependent mRNA degradation in a context different from that of NIP5;1, the upstream conserved sequence was introduced into the 5′-UTR of MOT2 (Supplemental Figure 11A). The insertion of the upstream conserved sequence increased mRNA degradation intermediate signals under both high and low B conditions (Supplemental Figures 11B, lanes 3 to 6, and 11D), while little or no effect on B-dependent toeprint signals was observed (Supplemental Figures 11C, lanes 9 to 12, and 11D), suggesting that the upstream conserved sequence is necessary for enhancing mRNA degradation, but not for ribosome stalling in the context of MOT2 5′-UTR, corroborating the observations with NIP5;1 (Figures 8B and 8C).
We also investigated whether the conserved U at the 9 nucleotide downstream of AUGUAA is involved in B-dependent regulation. Replacement of U by C at this position did not affect the B response (Supplemental Figure 12), suggesting that the conserved U at 9 nucleotides downstream of AUGUAA is not involved in B-dependent downregulation.
To further confirm the importance of the upstream conserved sequence in the B-dependent regulation of NIP5;1 mRNA accumulation in vivo, a construct carrying a NIP5;1 genome sequence (−1669 to +3587) [ProNIP5;1:5′-NIP5;1(−558):NIP5;1] (Figure 8E) was introduced into nip5;1-1 mutant plants, in which the NIP5;1 mRNA level was reduced to ∼1% that of wild-type Col-0 plants (Takano et al., 2006). Since one of the AUG-stops is sufficient for B-dependent downregulation (Supplemental Figure 6), −312AUG-stop was deleted in all of the constructs (−321 to −307). Mutations were then introduced into −122AUG-stop or the upstream conserved sequence (14 to 17 nucleotides upstream of −122AUG-stop), and their effects on mRNA accumulation were examined (Figure 8F). In transgenic plants carrying wild-type −122AUG-stop, NIP5;1 mRNA accumulation was significantly reduced under high B conditions compared with low B conditions, whereas B-dependent NIP5;1 mRNA accumulation was abolished or very much weakened in transgenic plants carrying mutations either in −122AUG-stop or in the upstream conserved sequence. These results demonstrate that the upstream conserved sequence plays a role in B-dependent NIP5;1 mRNA accumulation in vivo.
Physiological Importance of the AUG-Stop-Mediated Regulation of NIP5;1
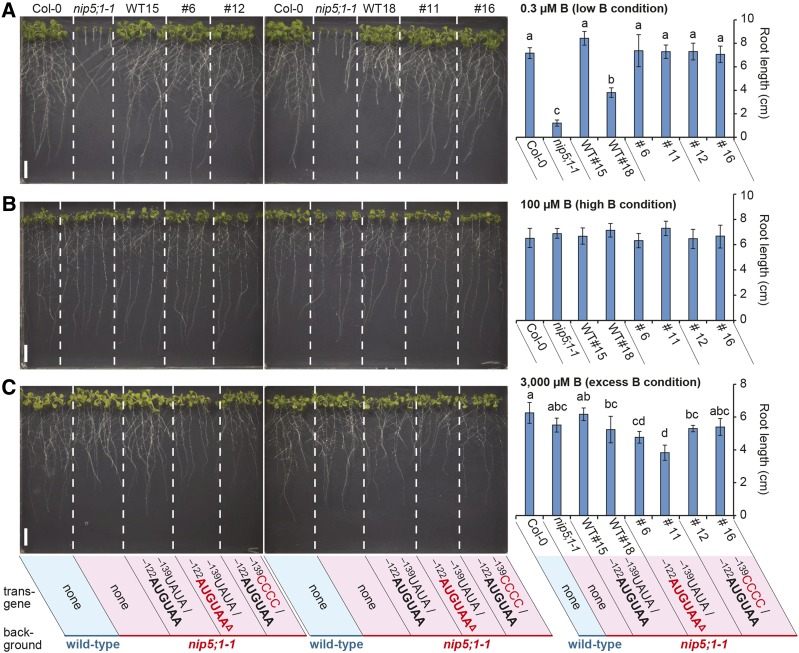
We examined the roles of AUG-stop and the upstream conserved sequence in the growth of Arabidopsis under different B conditions. Transgenic plants used in Figure 8E were grown on plates containing B at 0.3 μM (low B), 100 μM (high B), and 3000 μM (excess B), and their root growth was examined (Figure 9). Under the 0.3 µM B conditions, all of the transgenic plants we tested showed growth recovery compared with nip5;1-1 mutant plants (Figure 9A), suggesting that mutations in −122AUG-stop and the upstream conserved sequence do not affect NIP5;1 (the trans-gene) functions under low B conditions, in which B-dependent downregulation through NIP5;1 5′-UTR does not occur. The growths of all plants was indistinguishable under 100 µM B conditions, in which the nip5;1-1 mutant grew normally (Figure 9B). In contrast, under 3000 μM B conditions, in which wild-type growth was reduced by the toxic effect of B, the root growth of transgenic plants carrying the deletion of −122AUG-stop was poorer than that of Col-0 (Figure 9C), suggesting that impairment of AUG-stop-mediated regulation is important for avoiding the toxic effect of B. This pattern is identical to that observed with transgenic plants carrying a larger deletion of NIP5;1 5′-UTR that we reported previously (Tanaka et al., 2011). One of the two independent lines carrying the construct with a mutation in the upstream conserved sequence (line #12) exhibited poorer growth than wild-type Col-0, while the other transgenic line (#16) did not show significant differences from the wild type (Figure 9C). Together, these results suggest that the effect of the upstream conserved sequence may have weaker physiological importance than the presence of AUG-stop, corroborating the results of in vitro experiments examining the regulation of B-dependent ribosome stalling and mRNA degradation (Figures 8B and 8C).
Figure 9.
AUG-Stop Affects Root Growth under Excess B conditions.
Col-0, nip5;1-1, and the transgenic lines used in Figure 8F were grown on plates containing 0.3 μM B for 13 d (A), 100 μM B for 10 d (B), and 3000 μM B for 14 d (C), and root lengths were measured. Photographs were taken after the roots of Col-0 plants reached more than 6 cm. Means ± sd are shown (n = 4 to 10). Different letters indicate significant differences (P < 0.05) by Tukey test. Bars = 10 mm.
B-Dependent Regulation via AUG-Stop in Animal Systems
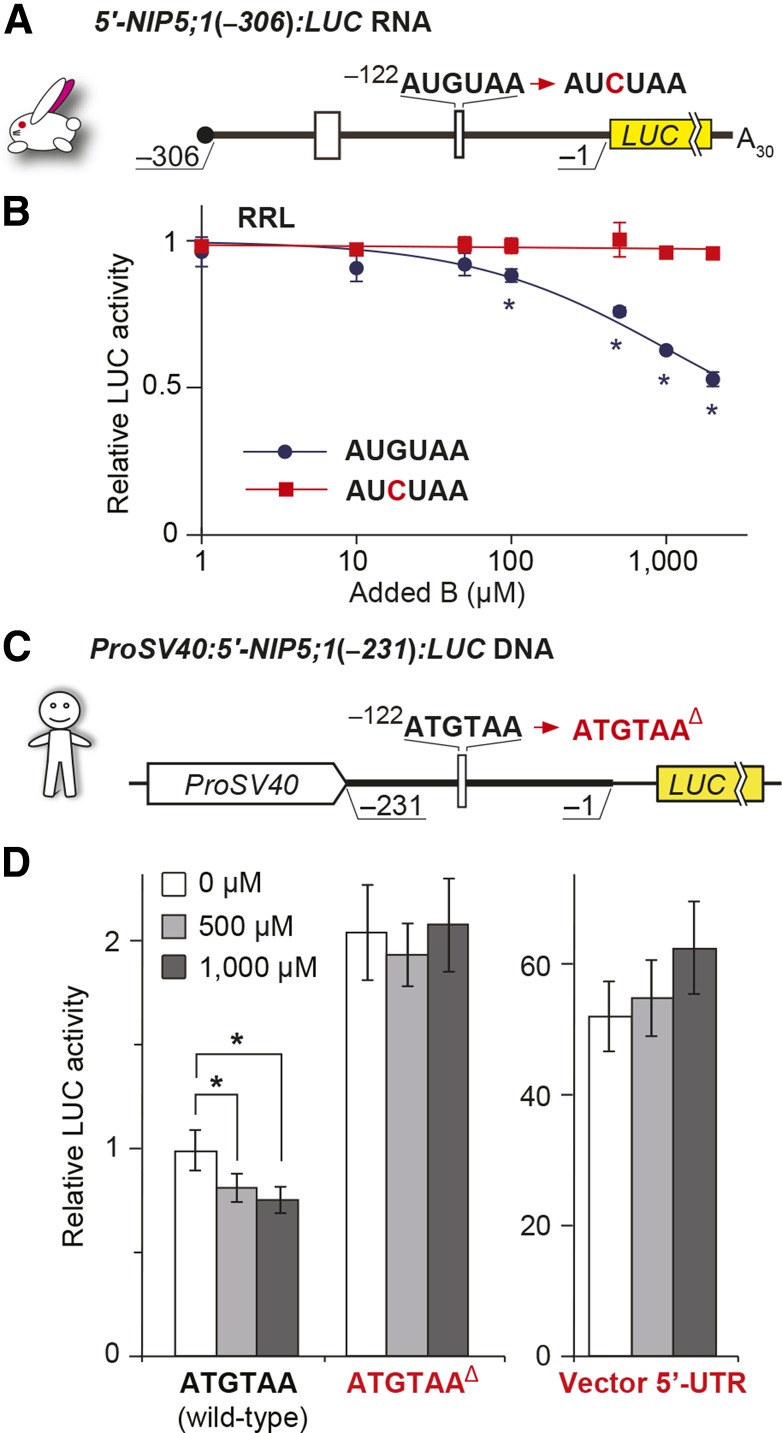
To examine whether this regulatory mechanism also functions in animals, we conducted in vitro translation and transfection experiments. For the in vitro translation study, we used a rabbit reticulocyte lysate (RRL) in vitro translation system (Figures 10A and 10B). The results demonstrate that B-dependent suppression of reporter activity was observed in RRL. To determine if the regulatory mechanism is also active in animal cells, HeLa cells were transfected with a plasmid containing NIP5;1 5′-UTR (−231 to −1 nucleotides) fused to the LUC reporter gene driven by the SV40 early promoter [ProSV40:5′-NIP5;1(−231):LUC] (Figure 10C). When the cells were transfected with the construct carrying −122AUG-stop, reporter activity was reduced to ∼80% under high B conditions compared with the activity without B addition, although the extent of the reduction was less than that observed in plants, presumably due to the low B concentration in animal cells (Ayres and Hellier, 1998). The deletion of the ATGTAA sequence abolished the response to high B conditions (Figure 10D). These results suggest that B-dependent regulation through AUG-stop is also functional in animals and that regulation through AUG-stop is a ubiquitous system.
Figure 10.
B-Dependent Downregulation through the AUG-Stop of NIP5;1 in Animal Systems.
(A) Schematic representation of 5′-NIP5;1(−306):LUC RNA in rabbit. Open boxes represent uORFs.
(B) 5′-NIP5;1(−306):LUC RNA was translated in RRL, and means ± sd of relative LUC activities (n = 3) are shown. Asterisks indicate significant reductions in RNA carrying −122AUGUAA (P < 0.05).
(C) Schematic representation of ProSV40:5′-NIP5;1(−231):LUC RNA. The thick line represents the sequence derived from the NIP5;1 5′-UTR and the thin line represents the 35-nucleotide linker sequence.
(D) Effect of B on reporter activities in transfected HeLa cells. Transfected HeLa cells were incubated under various B conditions, and means ± sd of relative LUC activities (n = 6) are shown. Asterisks indicate significant reductions in +B compared with no supplementation of B (P < 0.05). The “vector 5′-UTR” negative control carries only the vector’s 5′-UTR sequence.
Negative Selection Pressure against AUG-Stop in Plant 5′-UTRs
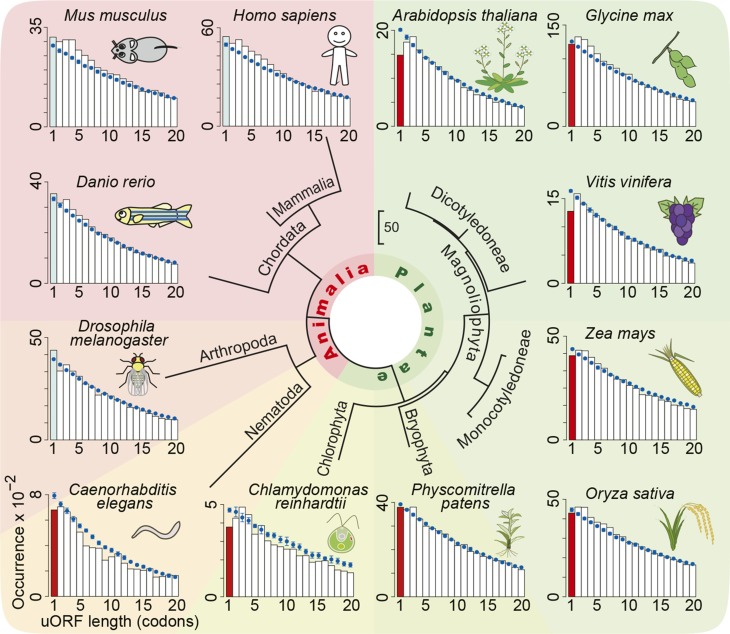
Because AUG-stop consists of only six nucleotides, it frequently appears in the genome. If AUG-stop functions in the regulation of gene expression, the presence of AUG-stop at a high frequency in the 5′-UTR may not be beneficial to the organism. The frequency of AUG-stops is lower than that expected from random occurrence in Arabidopsis (von Arnim et al., 2014).
To investigate the frequency of AUG-stop within the 5′-UTRs of various species, we conducted in silico analysis using whole genome sequences and annotations of 12 organisms available in public databases. We calculated the length of every uORF in individual organisms and compared the distribution with that obtained after randomization of the 5′-UTR sequences (Figure 11). In all of the plant species examined, the frequency of AUG-stops (i.e., uORF with one amino acid in length in Figure 11) was lower than that expected from random occurrence. This was not the case in animals, with the exception of soil-borne nematodes. It is intriguing that Chlamydomonas reinhardtii and zebra fish (Danio rerio) share similar habitats in that they both live in paddy fields, whereas Chlamydomonas shows bias against AUG-stop but zebra fish does not. B concentrations in cells are generally higher in plants than in animals (Ayres and Hellier, 1998), and the former depends largely on the soil B concentrations. Undesired B responses may exert negative selection pressure on the occurrence of AUG-stops in organisms that are exposed to soil B conditions leading to reasonable B accumulation in cells. It is intriguing that nematode is the only animal examined in this study that has a reduced frequency of AUG-stops. B concentrations in soil solution can fluctuate due to water conditions via a dilution effect.
Figure 11.
Distribution of uORF Lengths in Various Eukaryotic Species.
Distributions of uORF lengths in the annotated transcripts are shown in bar plots. The calculated distributions of uORF lengths after randomization of 5′-UTR sequences are shown in blue (mean ± sd). Frequency of AUG-stops lower than expected is shown in red.
DISCUSSION
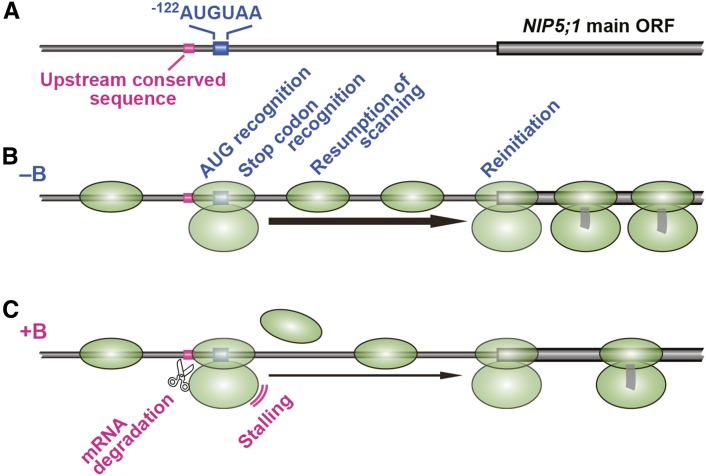
In this study, we identified AUGUAA as the sequence necessary for B-dependent regulation of NIP5;1 transcript accumulation. We then demonstrated that ribosome stalling at AUG-stops in the 5′-UTR of NIP5;1 was enhanced under high B conditions and that the stalling was coupled with mRNA degradation. An upstream conserved sequence of AUG-stop required for enhancing mRNA degradation was identified. AUG-stop is a minimum ORF, consisting of only initiation and termination codons; this process is illustrated in Figure 12.
Figure 12.
Model of NIP5;1 Expression Regulation through B-Dependent Ribosome Stalling at AUG-Stop.
(A) Schematic representation of −122AUGUAA to the NIP5;1 main ORF region. The upstream conserved sequence, −122AUGUAA, and the main ORF of NIP5;1 are shown.
(B) Under the low B conditions (−B), the 40S subunit (the 43S preinitiation complex) scans mRNA and finds the AUG of AUGUAA where it assembles into a 80S ribosome. The ribosome dissociates, but some of the 40S subunit remains on the mRNA and resumes scanning for the downstream AUG. The 80S ribosome is reassembled and reinitiates translation at the AUG codon of the main ORF.
(C) Under the high B conditions (+B), the ribosome mainly stalls at the AUGUAA. The stalling inhibits resumption of scanning by the 40S subunit, thereby reducing the possibility of reinitiation of translation at the main ORF. An mRNA degradation event is induced near the 5′-edge of the stalled ribosome, and translation of the main ORF is no longer possible. The upstream conserved sequence is responsible for enhancing the mRNA degradation.
Sensing the Cytoplasmic B Concentration Required to Regulate NIP5;1 Expression
NIP5;1 encodes a transporter that is necessary under low B conditions but is undesirable under high B conditions (Figure 9; Tanaka et al., 2011). The B-dependent regulation of NIP5;1 expression is thus important for maintaining B homeostasis and growth under a wide range of soil B concentrations. In vitro translation systems allow translation reactions that occur in the cytoplasm to be investigated. In this study, B-dependent regulation was observed in both the WGE and RRL in vitro translation systems, suggesting that the expression of NIP5;1 is regulated in response to cytoplasmic B concentrations, not to extracellular B concentrations after signal transduction.
A number of plant mineral nutrient transporters are regulated under different nutritional conditions, which are considered to represent adaptive responses to nutritional conditions. However, except for NRT1.1, a nitrate transporter that responds to extracellular nitrate concentrations (Ho et al., 2009), it is not clear whether gene regulation occurs in response to external or internal nutrient concentrations. The results of this study indicate that, in the case of NIP5;1, the cytoplasmic B concentration is sensed during translation. Sensing internal (cytosolic) nutrient conditions is probably more beneficial than detecting external concentrations from the viewpoint that most of biological reactions occur within the cell. However, the detection of external nutrient concentrations may have a different benefit, enabling transporters to respond to environmental changes in advance. B is relatively membrane permeable (Takano et al., 2006), which allows intracellular B concentrations to be rapidly influenced by external concentrations. Given such circumstances, sensing the internal concentrations of B may be more beneficial for plants than sensing extracellular B concentrations.
Effect of B on AUG-Stop-Mediated Ribosome Stalling and Reinitiation of Translation
Our study demonstrated two effects of B on NIP5;1 expression. One is B-dependent ribosome stalling (Figure 3E), followed by mRNA degradation (Figure 3D). The other is the possible inhibition of reinitiation (Figure 4B), but not leaky scanning (Figure 1B). In the experiments described in Figure 4B, the sequence around the AUG-stop was common among constructs; thus, it is very unlikely that the extent of ribosome stalling at AUG-stop under high B conditions is different among constructs, which makes it likely that a high B level negatively regulates the process of reinitiation. In yeast, the efficiency of reinitiation is higher for the uORFs with fewer codons (Rajkowitsch et al., 2004). It is reasonable to assume that the efficiency of reinitiation is high for AUG-stop, a uORF with the fewest codons, and that such a process may be the target of regulation under high B conditions. It is not clear how B affects reinitiation, but it is possible that ribosomes stalled on AUG-stop under high B conditions are in a state that does not allow efficient resumption of scanning for the downstream AUG (Figure 12).
Our findings allow us to consider possible mechanisms of B action. The disruption of the Kozak sequence at AUG-stop resulted in higher reporter activity under both high and low B conditions (Figure 1B). The disruption of the Kozak sequence at AUG-stop adversely affected the recognition of AUG in AUG-stop, but not that of the main ORF. The observation that the disruption of the Kozak sequence at AUG-stop affects the translation of the main ORF suggests that the AUG codon of AUG-stop is recognized as a start codon. This recognition could be affected by B, although other possibilities, including the possible effects of B on the efficiency of termination, cannot be excluded.
These data do not allow us to discuss the molecular target(s) of B action. However, the finding that the B-response is recapitulated in the in vitro translation systems suggests that boron or boric acid acts on the translation machinery. In this regard, it is worth noting that boric acid forms a complex with the cis-diol group (Power and Woods, 1997; Amaral et al., 2008), two hydroxyl groups positioned in cis-conformation, which is found in the 3′-end ribose moiety of RNA, including the cis-diol groups in rRNAs and deacylated tRNA produced after the peptydiltransfer reaction, as well as upon termination of translation at the stop codon.
Degradation of mRNA Is Associated with B-Dependent Ribosome Stalling at AUG-Stop
Ribosome stalling through AUG-stops was accompanied by mRNA degradation. One of the possible pathways of degradation is NMD (Amrani et al., 2006); however, NMD mutant analysis indicated that NMD does not function in the B-dependent regulation of NIP5;1 (Supplemental Figure 4). Considering that the induction of NMD by uORFs in plants is efficient if the uORF encodes a peptide longer than 34 amino acids (Nyikó et al., 2009), it is reasonable to assume that a mechanism other than NMD is responsible for AUG-stop-mediated mRNA degradation.
In this study, we identified an upstream conserved sequence that is responsible for promoting mRNA degradation through AUG-stop (Figure 8; Supplemental Figure 11). The sequence covers the position of the 5′-edge of the ribosome stalled at AUG-stop where the 5′-ends of the mRNA degradation intermediates are located. This sequence does not affect the efficiency of B-dependent ribosome stalling, suggesting that the upstream conserved sequence affects mRNA degradation at a step(s) after ribosome stalling. The presence of a sequence that affects mRNA degradation suggests the involvement of a ribonuclease that recognizes the upstream conserved sequence directly or indirectly.
AUG-Stop-Mediated B-Dependent Regulation Is Common in Eukaryotes
Although only a subset of genes containing AUG-stops in their 5′-UTRs were regulated by ribosome stalling through AUG-stops, we discovered at least three genes, NIP5;1, SKU5, and ABS2/NGAL1, that are regulated by this mechanism in Arabidopsis. Additionally, the introduction of AUG-stop into non-B-responsive genes conferred B-dependent suppression of gene expression (Figure 5; Supplemental Figure 11). These findings suggest that AUG-stop-mediated B-dependent regulation is a common mechanism in Arabidopsis. Moreover, the fact that PARE lists mRNA decay intermediates at 12 to 13 nucleotides upstream of AUGUAA in rice and maize orthologs of Arabidopsis NIP5;1 (Supplemental Figure 3) suggests that the same mechanism functions in rice and maize. Therefore, the AUG-stop-mediated B-dependent regulatory mechanism operates in the plant kingdom.
B-dependent gene regulation through AUG-stop in NIP5;1 5′-UTR was also observed in the RRL in vitro translation system and in HeLa cells (Figure 10), although the B-response was weaker than in the Arabidopsis systems. A trans-acting factor(s) may be involved in this regulation, and a plant-specific factor(s) may be more effective than the animal one. Our results suggest that AUG-stop-mediated gene regulation in response to B is not limited to plants but is a common system in eukaryotes.
METHODS
Plasmids Used for Transient Assay
pMT101 (Supplemental Table 3) carries Pro35S, −306 to −1 nucleotides (relative to the translation start site of the main ORF of NIP5;1) of the wild-type ProNIP5;1 5′-UTR [5′-NIP5;1(−306)] and the LUC reporter gene. To construct pMT101, the 5′-NIP5;1(−306) region was amplified by PCR using pMW1 (Takano et al., 2006), which carries the wild-type NIP5;1 5′-UTR, as a template with primers MT1f and MT12r (Supplemental Table 4). The amplified DNA fragment was digested with BamHI and AvrII and inserted into pBI221-LUC+ (Matsuo et al., 2001) between the BamHI and AvrII sites.
pMT104, pMT105, pMT107, pMT108, pMT109, pMT110, pMT148, pMT151, pMT152, pMT153, and pMT154 (Supplemental Table 3), each of which carries a different mutation in and around the −122AUG-stop in NIP5;1 5′-UTR, were generated by inserting the −306- to −1-nucleotide fragment with different mutations into pBI221-LUC+. For pMT104, pMT105, pMT107, pMT108 pMT109, pMT151, pMT152, pMT153, and pMT154, DNA fragments with mutations were generated by overlap extension PCR (Ho et al., 1989) with the internal primers listed in Supplemental Table 4 and the flanking primers MT1f and MT12r using pMW1 as a template. For pMT110 and pMT148, the same procedure was used except that pMT109 and pMT101 were used as a template, respectively. The resulting DNA fragments with mutations were inserted into pBI221-LUC+ in the same way as for the construction of pMT101.
pMT141, pMT142, and pMT143 (Supplemental Table 3), which have deletions of −41, −65, and −95 nucleotides, respectively, upstream from the translation start site of NIP5;1, were generated by inserting DNA fragments with deletions. The DNA fragments were generated by PCR using pMW1 as a template with primers listed in Supplemental Table 4. The PCR-amplified fragments were inserted into pBI221-LUC+ in the same way as for the construction of pMT101. The plasmids described above contain 18-nucleotide (5′-GGGGACTCTAGAGGATCC-3′) and 15-nucleotide (5′-CCTAGGAAGCTTTCC-3′) linkers derived from pBI221-LUC+ at the 5′ and 3′, respectively, of NIP5;1 5′-UTR. pBI221-LUC+ was used as the Vector 5′-UTR negative control (Supplemental Table 3). These plasmids were used for transfection experiments in Arabidopsis thaliana suspension cells.
The plasmid pKM75 (Supplemental Table 3) carries a RLUC reporter gene under the control of Pro35S and was used as an internal control for transfection experiments in Arabidopsis suspension cells. To construct pKM75, the PCR fragment amplified from pIE0 (Ebina et al., 2015) with primers 1832f and 1801r (Supplemental Table 4) was digested with XbaI and BstBI and inserted into pIE0 between the XbaI and BstBI sites to replace the 5′-UTR. The resulting RLUC gene in pKM75 carries additional Met-Val codons at its N terminus.
For transient assay in HeLa cells, we used a −231- to −1-nucleotide fragment of the NIP5;1 5′-UTR that contains only one uORF, −122AUGUAA, to simplify the 5′-UTR structure to avoid the possible masking effects of other uORFs in animal systems. pMT139 and pMT140 (Supplemental Table 3) carry ProSV40, −231 to −1 nucleotides of NIP5;1 5′-UTR [5′-NIP5;1(−231)] with and without −122AUGUAA, respectively, and the LUC reporter gene. The pRL-SV40 (Promega) carries ProSV40 and the RLUC reporter gene and was used as an internal control. For pMT139 and pMT140, the 5′-NIP5;1(−231) region was amplified by PCR using pMT101 and pMT117 (see below), respectively, as templates with primers listed in Supplemental Table 4. pMT101 and pMT117 carry wild-type NIP5;1 5′-UTR and NIP5;1 5′-UTR without AUG-stop, respectively. The amplified fragments were digested with HindIII and inserted into the corresponding HindIII site of pGL3 promoter vector (Promega). The plasmids thus constructed contain a 35-nucleotide linker (5′-AAGCTTGGCATTCCGGTACTGTTGGTAAAGCCACC-3′) between the NIP5;1 5′-UTR and the LUC reporter gene. The 3′ end of the linker carries an optimized Kozak sequence for the reporter ORF.
Plasmids Used for in Vitro Translation
pMT131 (Supplemental Table 3) carries −306 to −1 nucleotides of the wild-type NIP5;1 5′-UTR [5′-NIP5;1(−306)] and the LUC reporter gene. For the construction of pMT131, the 5′-NIP5;1(−306) region was amplified by PCR using pMW1 as a template with primers MT51f and MT34r (Supplemental Table 4). The amplified DNA fragment was digested with XbaI and NcoI and inserted into the pSP64 poly(A) vector (Promega)-based plasmid pMI21 (Chiba et al., 2003) between the XbaI and NcoI sites.
pMT132, pMT144, and pMT156 (Supplemental Table 3), which carry different mutations in and around the −122AUG-stop in NIP5;1 5′-UTR, were generated by inserting −306- to −1-nucleotide fragments with different mutations into pMI21. To construct pMT132, a DNA fragment with the corresponding mutation was amplified by PCR using pMT105 as a template with primers MT51f and MT34r (Supplemental Table 4). pMT105 carries a mutation in −122AUG-stop in the NIP5;1 5′-UTR (Supplemental Table 3). For pMT144, a DNA fragment with mutations was generated by overlap extension PCR with the internal primers listed in Supplemental Table 4 and the flanking primers MT51f and MT34r using pMT131 as a template. To construct pMT156, DNA fragments with mutations were generated by overlap extension PCR with the internal primers listed in Supplemental Table 4 and the flanking primers MT51f and MT34r using pMW1 as a template. The amplified DNA fragments were inserted into pMI21 in the same way as for the construction of pMT131.
pMT114 (Supplemental Table 3) carries a −558- to −1-nucleotide fragment of the wild-type NIP5;1 5′-UTR [5′-NIP5;1(−558)] and the LUC reporter gene. To construct pMT114, the 5′-NIP5;1(−558) region was amplified by PCR using pMW1 as a template with primers MT25f and MT34r (Supplemental Table 4). The amplified DNA fragment was inserted into pMI21 in the same way as for the construction of pMT131.
pMT115, pMT117, and pMT116 (Supplemental Table 3) carry deletions of either of the −312AUG-stop and −122AUG-stop or both in the NIP5;1 5′-UTR(−558), respectively. For pMT115 and pMT117, DNA fragments with corresponding mutations were generated by overlap extension PCR with the internal primers listed in Supplemental Table 4 and the flanking primers MT25f and MT34r using pMW1 as a template. For pMT116, the same procedure was followed except that pMT115 was used as a template. The amplified DNA fragments were inserted into pMI21 in the same way as for the construction of pMT131.
pMT125, pMT127, pMT129, pMT161, pMT163, and pMT157 (Supplemental Table 3) carry 5′-UTRs of MOT2, SKU5, ABS2/NGAL1, CPuORF47, At4g19370, and Dof1.8, respectively, and the LUC reporter gene. To construct pMT125, pMT127, pMT129, pMT161, pMT163, and pMT157, the 5′-UTRs of corresponding genes were amplified by PCR using Arabidopsis genomic DNA as a template with primers listed in Supplemental Table 4. The amplified DNA fragments were inserted into pMI21 in the same way as for the construction of pMT131.
pMT126, pMT128, pMT130, pMT162, and pMT158 (Supplemental Table 3) carry a mutation of AUG-stop in the 5′-UTR of MOT2, SKU5, ABS2/NGAL1, CPuORF47, and Dof1.8, respectively. To construct pMT126, pMT128, pMT130, pMT162, and pMT158, DNA fragments with corresponding mutations were generated by overlap extension PCR with the internal primers listed in Supplemental Table 4 and the flanking primers MT32f and MT41r, MT43f and MT47r, MT45f and MT49r, MT52f and MT59, and MT87f and MT95, respectively, using Arabidopsis genomic DNA as a template. The amplified DNA fragments were inserted into pMI21 in the same way as for the construction of pMT131.
pMI27 (Chiba et al., 2003; Supplemental Table 3) carries a RLUC reporter gene in pSP64 poly(A) vector. pMI27 was used as an internal control for the in vitro translation experiments.
Construction of Transgenic Plants
pMT100 (Supplemental Table 3) carries Pro35S, −306 to −1 nucleotides of the wild-type NIP5;1 5′-UTR [5′-NIP5;1(−306)] and the GUS reporter gene, and was described as P35SUTR+7-GUS by Tanaka et al. (2011). pMT113 and pMT155 (Supplemental Table 3), which carries a mutation in the −122AUG-stop in NIP5;1 5′-UTR, were generated by inserting −306 to −1 nucleotides with a mutation in AUGUAA into pMDC140 (Curtis and Grossniklaus, 2003). To construct pMT113 and pMT155, the 5′-NIP5;1(−306) region was amplified by PCR using pMT104 and pMT152, respectively, as a template with primers MT23f and MT24r (Supplemental Table 4). pMT104 and pMT152 carry a mutation in −122AUG-stop in the NIP5;1 5′-UTR (Supplemental Table 3). The amplified fragment was subcloned into the pENTR/D-TOPO vector via the TOPO cloning reaction (Invitrogen). The 5′-NIP5;1(−306) region was then subcloned into pMDC140 using the Gateway system (Curtis and Grossniklaus, 2003). The Gateway vector contains a 37-nucleotide linker (5′-TCTAGAACTAGTTAATTAAGAATTATC-3′) and a 59-nucleotide linker (5′-TTGATAGCTTGGCGCGCCTCGACTCTAGAGGATCGATCCCCGGGTACGGTCAGTCCCTT-3′) at the 5′ and 3′, respectively, of NIP5;1 5′-UTR.
Transformation of Arabidopsis plants was performed using the floral-dip method (Clough and Bent, 1998). Homozygous T3 generation plants were established and used for analyses. Transgenic lines #4, #8, and #10 refer to independently transformed lines carrying Pro35S:5′-NIP5;1(−306):GUS with a mutation in the AUG codon of −122AUG-stop. Transgenic lines #2, #18, and #19 refer to independently transformed lines carrying Pro35S:5′-NIP5;1(−306):GUS with a mutation in the stop codon of −122AUG-stop. The transgenic line WT#8, which carries Pro35S:5′-NIP5;1(−306):GUS with wild-type −122AUG-stop, has been described previously (Tanaka et al., 2011).
pMT145 (Supplemental Table 3) carries ProNIP5;1 (−1669 to −559), the NIP5;1 5′-UTR (−558 to −1) with deletion of −312AUG-stop region, and the NIP5;1 (+1 to +3578). To construct pMT145, a DNA fragment with the corresponding mutations was generated by overlap extension PCR with the internal primers MT26f and MT35r and the flanking primers MT78f and MT79r using pYK02 (Kato et al., 2009) as a template. The amplified DNA fragment was digested with SalI and inserted into a pTbar vector-based pPZP212 (Hajdukiewicz et al., 1994) at the SalI site.
pMT146 and pMT147 (Supplemental Table 3), which carry different mutations in and around the −122AUG-stop in NIP5;1 5′-UTR, were generated by inserting −1669 to +3578 nucleotides in the NIP5;1 region with different mutations. To construct pMT146 and pMT147, DNA fragments with corresponding mutations were generated by overlap extension PCR with the internal primers MT27f and MT36r, and MT73f and MT74r, respectively and with the flanking primers MT78f and MT79r using pMT145 as a template. The amplified DNA fragments were inserted into pPZP212 in the same way as for the construction of pMT145.
After transformation of Arabidopsis, homozygous T3 generation plants were established and used for analyses. Transgenic lines WT#15 and WT#18 refer to independently transformed lines carrying the wild-type −122AUG-stop in NIP5;1 5′-UTR. Transgenic lines #6 and #11 refer to independently transformed lines carrying deletion of −122AUG-stop in NIP5;1 5′-UTR, and transgenic lines #12 and #16 refer to independently transformed lines carrying the wild-type −122AUG-stop in NIP5;1 5′-UTR carrying a mutation in the upstream conserved sequence (14 to 17 nucleotides upstream of −122AUG-stop).
B Conditions
Boric acid was used for B treatments, with different boric acid concentrations used depending on the organism and experimental system. Responses of different organisms to the range of B concentrations vary widely, and we selected B concentrations depending on the response of organisms to different B concentrations. In experiments using intact plants, 0.3 µM B and 100 µM B conditions were used for the plant treatments as low B (−B) and high B (+B) conditions, respectively. This is based on the reports by Takano et al. (2006). In transfection assay using Arabidopsis MM2d suspension cells, we first examined reporter activities under 1, 100, and 500 µM B conditions. The reporter activities were reduced to ∼70% and 50% under 100 and 500 µM B conditions, respectively, compared with 1 µM B condition (Supplemental Figure 13). Since the B-dependent downregulation was weaker under 100 µM B conditions than under 500 µM B conditions, 1 µM B and 500 µM B were selected as low B (−B) and high B (+B) conditions, respectively. For the experiments using HeLa cells, for the low B condition, B was not applied, as B is not an essential element for animal cells. For the high B conditions, 500 and 1000 µM B were applied to HeLa cells during the incubation period after transfection. Since the B response in RRL (Figure 10A) is relatively weak compared with that in WGE (Figure 3B) in the in vitro translation assay, we included 1000 µM B conditions to ascertain B-excess conditions. For in vitro assays including primer extension and toeprint analyses, 0 µM B and 300 µM B were used as low B (−B) and high B (+B) conditions, respectively. This is based on the results of reporter activities in the in vitro translation assay (Figure 3B), in which the reporter activities were reduced to 79% and 50% under 100 µM B and 500 µM B conditions, respectively, compared 0 µM B. To avoid excess B concentrations in WGE for primer extension and toeprint analyses, we selected 300 µM B, which is the average of 100 and 500 µM.
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia (Col-0) was used as a wild-type plant in this study, except for Figure 6F, in which ecotype Wassilewskija was used as a wild-type plant for corresponding T-DNA insertion lines. Transgenic plants carrying Pro35S:5′-NIP5;1(−306):GUS and ProNIP5;1:5′-NIP5;1(−558):NIP5;1 with or without mutations in the NIP5;1 5′-UTR were generated as described above. For primer extension analysis, Arabidopsis plants were grown on hydroponic culture medium (Takano et al., 2005) containing 1% (w/v) sucrose and 0.3 μM B solidified with 0.15% (w/v) gellan gum (Wako Pure Chemicals) for 28 d and then transferred to hydroponic culture medium containing the same concentration of B for 1 d at 22°C under short-day conditions (10 h light/14 h dark with fluorescent lamps, 100 to 160 μmol photons m−2 s−1). The plants were then transferred to 0.3 µM or 100 µM B for 10 min before RNA extraction. For the GUS assay, qRT-PCR analyses and root length measurement, plants were grown on hydroponic culture medium containing 1% (w/v) sucrose and 0.3, 100, or 3000 μM B solidified with 1% (w/v) gellan gum at 22°C under long-day conditions (16-h-light/8-h-dark cycle) for 10 to 14 d. For microarray analysis, plants were grown on hydroponic culture medium containing 1% (w/v) sucrose and 100 μM B solidified with 1% (w/v) gellan gum at 22°C under long-day conditions for 12 d and then transferred to 0.3 or 100 µM B conditions for 2 d. For mRNA half-life measurement, plants were grown on hydroponic culture medium containing 1% (w/v) sucrose and 0.3 or 100 μM B solidified with 1% (w/v) gellan gum at 22°C under long-day conditions for 10 d, followed by transfer to hydroponic culture medium containing 0.3 or 100 µM B for 10 min. At least three independently grown plant samples were examined in all experiments.
Transient Expression Analysis of Arabidopsis Suspension Cell Cultures and Reporter Assay
Arabidopsis MM2d suspension cells (Menges and Murray, 2002) were cultured in LS medium (Nagata et al., 1992) at 26°C. To prepare protoplasts, MM2d cells were suspended in LS medium containing 1% (w/v) cellulase Onozuka RS (Yakult Pharmaceutical Industry), 0.5% (w/v) pectolyase Y23 (Seishin Pharmaceutical), and 0.4 M mannitol and incubated at 26°C with gentle shaking until the suspension became turbid with protoplasts (∼3 h). The protoplasts were washed five times with wash buffer (0.4 M mannitol, 5 mM CaCl2, and 12.5 mM NaOAc, pH 5.8) and suspended in electroporation buffer (5 mM MES, 70 mM KCl, and 0.3 M mannitol, pH 5.8). Electroporation was performed as described previously (Ishikawa et al., 1993) with some modifications. Ten micrograms each of tester plasmid carrying the LUC reporter and internal control plasmid pKM75, carrying the RLUC reporter, were gently mixed with 1.5 × 106 protoplasts in 500 μL of electroporation buffer in an electroporation cuvette with a 4-mm electrode distance. Electroporation was performed using an Electro Cell Manipulator 600 (BTX) with voltage, capacitance, and resistance settings at 190 V, 100 µF, and 480 Ω, respectively. The protoplasts were kept on ice for 30 min and then incubated at 25°C for 5 min, centrifuged (60g, 2 min at 25°C) and resuspended in 1 mL of LS medium containing 0.4 M mannitol with 500 or 1 µM B. After 2 d of incubation at 23°C in the dark, the protoplasts were disrupted as described previously (Chiba et al., 1999), and LUC and RLUC activities were assayed using a PicaGene Dual-Luciferase Assay kit (PG-DUAL SP; Toyo Ink). LUC activity was normalized with RLUC activity to obtain relative LUC activity.
Transient Expression of HeLa Cell Cultures and Reporter Assay
HeLa cells were cultured in complete Dulbecco’s modified eagle’s medium (Nissui) supplemented with 10% fetal bovine serum. Plasmids were transfected into HeLa cells with Lipifectamine 2000 (Invitrogen). HeLa cells were transfected with the plasmid constructs carrying LUC reporter, along with internal control plasmid carrying RLUC reporter, in the presence of 500 or 1000 µM B (high B conditions) or without B (low B condition). Twenty-four hours after transfection, the cells were lysed and LUC activities were measured. LUC and RLUC assays were performed with the Dual-Luciferase Reporter Assay System (Promega). LUC activities were normalized with the RLUC activities of the cotransfected internal controls to obtain relative LUC activities.
In Vitro Transcription
DNA templates in pSP64 poly(A) vector were linearized with EcoRI and purified using a QIAquick Nucleotide Removal kit (Qiagen). In vitro transcription was performed using an AmpliCap SP6 High Yield Message Maker kit (Epicentre Technologies) in the presence of cap analog m7G[5′]ppp[5′]GTP (Epicentre Technologies) as described previously (Chiba et al., 2003). Following in vitro transcription, RNA was purified using an RNeasy mini kit (Qiagen) and was poly(A)-selected using a GenElute mRNA miniprep kit (Sigma-Aldrich) prior to the in vitro translation reactions.
In Vitro Translation and Reporter Assays
RNA was denatured for 5 min at 67°C and rapidly chilled on ice water. Each reaction mixture (10 μL) contained 5 μL WGE (Promega), 0.8 μL of 1 mM amino acid mixture lacking methionine (Promega), 80 µM l-methionine, 1 unit µL−1 RNasin (Promega), 2 fmol µL−1 tester RNA carrying LUC reporter, 0.2 fmol µL−1 internal control RNA carrying RLUC reporter, and various concentrations of boric acid or 500 µM sodium chloride, potassium dihydrogen phosphate, and potassium nitrate. All reactions were performed at 25°C for 120 min. In vitro translation in RRL was performed with a protocol similar to WGE with a few modifications. Each reaction mixture (10 µL) contained 6.6 μL RRL (Promega), 0.2 μL of 1 mM amino acid mixture lacking methionine, 80 µM l-methionine, 1 unit µL−1 RNasin, 70 mM KOAc, 2 fmol µL−1 LUC RNA, 0.2 fmol µL−1 RLUC RNA, and various concentrations of B. All reactions were performed at 30°C for 120 min. LUC and RLUC activities were assayed using a Dual Luciferase reporter assay kit, and the LUC activity was normalized with RLUC activity to obtain relative LUC activity. The relative LUC activity was then normalized with the data obtained without addition of B. For the sake of interpolation, three-parameter log-logistic function,
with optimum parameters, a, b, and c, was applied and shown in each graph. The concentration of B added to the reaction mixture is represented by x.
Primer Extension Analysis
The primer extension analysis of mRNA shown in Figure 2B was performed using poly(A) RNA extracted from Arabidopsis plants grown under 100 or 0.3 µM B, or from plants grown under 0.3 µM B and transferred to 100 µM B for 10 min. Primer-1 (Supplemental Table 4) was used. Poly(A) RNA (500 ng) was annealed with 32P-labeled primer in 40 μL of annealing buffer (Chiba et al., 2003) at 58°C for 2 h, and the reverse transcription reaction was performed using Thermoscript RNase H− reverse transcriptase (Invitrogen) at 58°C for 60 min.
Primer extension analysis of RNA following in vitro translation was performed as described previously (Chiba et al., 2003), using RNA extracted from an in vitro translation reaction without RLUC RNA. To detect primer extension signals corresponding to −312AUG-stop in Supplemental Figure 6, the 32P-labeled Primer-2 (Supplemental Table 4) was used. For other primer extension analyses, 32P-labeled ZW4 primer (Wang and Sachs, 1997) (Supplemental Table 4), which anneals with the LUC coding region, was used. Following phenol-chloroform extraction, the samples were separated on a 6% polyacrylamide/7 M urea gel. DNA sequence ladders were prepared using the DNA Cycle Sequencing System (Promega). The intensities of primer extension signals are presented after normalizing with those of the full-length mRNA, and the relative primer extension signals were then normalized with those under −B conditions. The sequence ladder was generated using the same primer and the RNA construct carrying AUGUAA sequence as a template.
Toeprint Analysis
In vitro translation in WGE was performed using 200 fmol μL−1 LUC RNA, but without RLUC RNA, in a 20 μL reaction mixture at 25°C in the presence or absence of 300 µM B for 30 min. Following in vitro translation, toeprint analysis was performed as described previously (Onouchi et al., 2005) using SuperScript III RNase H− reverse transcriptase (Invitrogen). To detect toeprint signal corresponding to −312AUG-stop, 32P-labeled Primer-2 was used (Supplemental Figure 6). For other toeprint analyses, 32P-labeled ZW4 primer was used. The prereaction mixture (16.4 µL) contained 4 μL 5× first-strand buffer (Invitrogen), 2 μL 2.5 mM deoxynucleotide triphosphate, 1 μL 0.1 M DTT, and 20 units of RNasin, with or without 1 μL of 10 mg mL−1 CHX, and was placed on ice. One microliter of in vitro translation reaction mixture was added to the prereaction mixture and placed on ice for 2 min. Two microliters of 5′-32P-labeled primer (0.2 µM) was added to the reaction mixture and incubated at 37°C for 3 min. After adding 10 units reverse transcriptase, the reaction mixture (20 µL) was incubated at 37°C for 30 min. Following phenol-chloroform extraction, the products were separated on a 6% polyacrylamide/7 M urea sequencing gel. The toeprint signals are presented as for the primer extension experiment. Representative results of triplicate experiments are presented. The sequence ladder was generated as in primer extension analyses.
Measurement of B Concentration in WGE
The B concentration in WGE was determined using inductively coupled plasma-mass spectrometry (ICP-MS) (SPQ-9000; Seiko Instruments) as previously described (Takano et al., 2006). WGE (100 μL) was digested with nitric acid and the digest was dissolved in 1 mL of 0.08 M nitric solution, but the signal intensities from ICP-MS analysis corresponding to both 10B and 11B were not distinguishable from the background. In our ICP-MS analysis, the detection limit for boric acid in solution is in the range of 0.05 µM, suggesting that the concentration of boric acid in WGE we used in our study was below 0.005 µM. This is much lower than the boric acid concentrations we used in our study.
Fluorometric Assays of GUS Activity
For GUS assays, 5 to 10 transgenic Arabidopsis seedlings were combined, and GUS activity was assayed as described (Jefferson et al., 1987).
Quantification of Transcript Accumulation by Quantitative RT-PCR
Arabidopsis plants were grown on solid medium containing 0.3 or 100 µM B for 10 d. Total RNA from root samples was extracted using an RNeasy Plant Mini kit (Qiagen), and 500 ng was reverse-transcribed in a 20-μL reaction mixture using an ExScript RT reagent kit (Takara Bio) with oligo-dT16 primers. The cDNA was amplified by qRT-PCR using a Thermal Cycler Dice Real Time System TP800 (Takara Bio) with a SYBR Premix Ex Taq kit (Takara Bio). The primers used for qRT-PCR analysis are listed in Supplemental Table 4. We used three reference genes (eEF1α, Actin10, and Ubq10) in the initial phase of the experiments (Supplemental Table 5). Since similar data were obtained with the three genes, we used eEF1α as a reference gene for subsequent analyses. Quantification was performed on three independently grown plant materials (n = 3).
Half-Life Measurement of mRNA
mRNA half-lives were determined as described previously (Tanaka et al., 2011). Briefly, Arabidopsis plants grown on solid medium for 10 d were preincubated with hydroponic culture medium containing 0.3 or 100 µM B for 10 min. Following preincubation, 3′-deoxyadenosine (cordycepin) (Funakoshi) was added to the hydroponic culture medium at a final concentration of 0.6 mM (t = 0 min), and the sample was immediately vacuum infiltrated for 45 s. Root samples were harvested at 0, 10, 30, and 60 min and frozen in liquid nitrogen. Total RNA was isolated and analyzed by qRT-PCR. The amount of RNA remaining at each time point was determined relative to the amount at the zero time point. The mRNA half-lives were determined by fitting the remaining amounts of the mRNA to a linear line after log conversion by the least squares method.
Microarray Analysis
Total RNA was prepared from Col-0 roots of 12-d-old seedlings that were grown under 100 µM B conditions for 10 d and then transferred to 0.3 or 100 µM B conditions for 2 d. Microarray analysis was performed with a GeneChip Arabidopsis ATH1 genome array (Affymetrix). From the raw data, 15,074 genes with quality flag value of “present” for both 0.3 and 100 µM B samples were selected. A list of genes whose 5′-UTR sequences contain AUG-stops was generated from the Arabidopsis Information Resource version 10 (TAIR10) database (Lamesch et al., 2012) using a custom R script (R version 3.2.2).
Degradome Data Processing
Degradome sequencing data sets from Arabidopsis, rice (Oryza sativa), and maize (Zea mays) were downloaded from the PARE database (German et al., 2008). The data sets used were as follows: inflorescence tissue of Arabidopsis (wild-type Col-0 and xrn4 mutant) grown on soil (GSE11094; GSM280227); rice (Oryza sativa cv Nipponbare) seedlings gown in hydroponic culture for 3 weeks (GSE17398; GSM434596); and ears of maize (Zea mays, inbred line B73) at stage IV grown in a controlled environment (GSE47837; GSM1160282). In the data sets, abundances of the decay intermediates were expressed in transcripts per 10 million via normalization with the number of total genome-matched reads (except for those mapped to t/r/sn/snoRNA). The relevant region in the 5′-UTR of each gene of interest was plotted using custom R scripts.
Sequence Logo
For B-responsive genes, NIP5;1, ABS2/NGAL1 and SKU5 of Arabidopsis, and OsNIP3;1 and ZmNIP3;1, rice and maize orthologs of Arabidopsis NIP5;1, respectively, were used. For non-B-responsive genes, 100 genes whose FC+B/−B values were close to 1.0 were used. Sequence logos were generated with the Sequence_logo_v03.R package using the pooled sequences.
Ribosome Footprint Data Analysis
The ribosome stalling frequency for every 6-nucleotide sequence (combination of two codons) in the 5′-UTR was estimated from a public ribosome footprint data set of Arabidopsis whole seedlings (Juntawong et al., 2014; accession SRR966474, RF#2). The FASTQ file was processed using a custom R script to remove the adapter sequence and was mapped to Arabidopsis genome sequence (TAIR10) by tophat2 algorithm with the default parameter settings. The resultant SAM file was subjected to further analysis with custom R scripts.
As a criterion for ribosome stalling at each combination of two codons, the number of reads that start from 15 nucleotides upstream of codon 1, which corresponds to the 5′-end of the ribosome stalled with the P-site on codon 1, was counted and normalized by average read count of the mRNA to cancel the read depth variation caused by different expression levels (designated as ribosome arrest value [RAV]). RAV was calculated for each instance of any combinations of two codons in 5′-UTRs, and the number of instances that satisfy RAV > 2 was tabulated.
In Silico Analysis of uORF Length Distribution in Various Organisms
The distribution of uORF lengths in various organisms was calculated using a custom R script. The algorithm searches 5′-UTR sequences of each transcript to find all ATGs and the nearest downstream in-frame stop codon (TAA, TAG, and TGA), and output is the length of each uORF. The distribution without bias was estimated by repeating the same calculation 10 times with randomization of 5′-UTR sequences and rescaling the total uORF number to that of the actual ones. As input data, masked genomic DNA sequence (sequence type “dna_rm”) and gene annotation files were downloaded from the public Ensembl database. For animals, FASTA and GTF files were downloaded from ftp://ftp.ensembl.org (release-77), and for plants, FASTA and GFF3 files were downloaded from ftp://ftp.ensemblgenomes.org (release-23).
Phylogenetic Analysis
The phylogenetic tree in Figure 11 was generated using MEGA6 software (Tamura et al., 2013) by applying the neighbor-joining method (Saitou and Nei, 1987) to 18S rRNA sequences for each organism, with the phylogenetic analysis parameters “Test of Phylogeny”, “None”, “Substitution Model-Model/Method”, “No. of difference”, “Substitution Model-Substitutions to Include”, “d: Transitions + Transversions”, “Rates and Patterns-Rates among Sites”, “Uniform rates”, “Data Subset to Use-Gaps/Missing Data Treatment”, and “Complete deletion.” The resulting alignment is provided in Supplemental File 1. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the number of differences method (Nei and Kumar, 2000) and are in the units of the number of base differences per sequence. Those positions that contain gaps and missing data in any of the organisms tested were eliminated. A total of 1667 nucleotide were analyzed.
Statistical Methods
To evaluate B-dependent reductions in reporter activity and mRNA accumulation levels, statistical analyses were performed using two-tailed two-sample Student’s t tests. To evaluate root length differences, statistical analyses were performed using Tukey’s test. To evaluate changes in signal intensities in primer extension and toeprint experiments, the signal intensities were normalized with the respective full-length signals. The values obtained under +B conditions were divided with those under −B conditions and were used for one-tailed one-sample Student’s t tests after log conversion. In all figures, the error bars represent standard deviations.
Accession Numbers
Microarray data from this article have been deposited in NCBI’s Gene Expression Omnibus data library under accession number GSE52208. PARE data from this article can be found in the Gene Expression Omnibus data library under the following accession numbers: Arabidopsis, GSE11094; rice, GSE17398; and maize, GSE47837. Ribosome profiling data from this article can be found in DRASearch under accession number SRR966474. Arabidopsis sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL/DDBJ databases under the following accession numbers: NIP5;1, At4g10380; CPuORF47, At5g03190; ABS2/NGAL1, At2g46080 and At4g19370; SKU5, At4g12420; MOT2, At1g80310; Dof1.8, At1g64620; OsNIP3;1, Os10g36924; and ZmNIP3;1, GRMZM2G176209.
Supplemental Data
Supplemental Figure 1. General Effects of uORF on Translation of the Main ORF.
Supplemental Figure 2. Positions of uORFs, RNA Sequence of NIP5;1 5′-UTR, and a Portion of the NIP5;1 Coding Region.
Supplemental Figure 3. Positions of 5′-Ends of mRNA Decay Intermediates in the 5′-UTRs of NIP5;1 and Its Rice and Maize Orthologs.
Supplemental Figure 4. B Dependence of NIP5;1 mRNA Accumulation and Half-Lives in NMD Mutants.
Supplemental Figure 5. The Extra Bands in the Primer Expression Assay Do Not Respond to B Conditions.
Supplemental Figure 6. Both −312AUG-Stop and −122AUG-Stop Are Responsive to B in a Wheat Germ Extract in Vitro Translation System.
Supplemental Figure 7. B-Dependent Toeprint Signals Are Strengthened by CHX Treatment.
Supplemental Figure 8. B-Dependent Downregulation Is Conferred by Introduction of AUG-Stop into Dof1.8 5′-UTR.
Supplemental Figure 9. Tendency of Two Codon Combinations to Trigger Ribosome Arrest in 5′-UTRs.
Supplemental Figure 10. Role of AUG-stop in B-Dependent Downregulation of ABS2/NGAL1.
Supplemental Figure 11. B-Dependent mRNA Degradation Is Enhanced by Introduction of the Upstream Conserved Sequence into MOT2 5′-UTR.
Supplemental Figure 12. Nine-Nucleotide Region Downstream of AUGUAA Is not Involved in B-Dependent Downregulation in the 5′-UTR of NIP5;1 in the in Vitro Translation Assay.
Supplemental Figure 13. B-Dependent Downregulation of NIP5;1 under Different B Conditions.
Supplemental Table 1. List of B-Responsive Genes Carrying AUGUAA in Their 5′-UTRs.
Supplemental Table 2. Sequences around AUG-Stops of B-Responsive Genes.
Supplemental Table 3. Plasmids Used in This Study and the Primers Used to Construct the Plasmids.
Supplemental Table 4. Primers Used in This Study.
Supplemental Table 5. Relative NIP5;1 mRNA Levels Obtained Using Different Reference Genes.
Supplemental File 1. Alignment Used to Produce the Phylogenetic Tree in Figure 11.
Supplementary Material
Acknowledgments
We thank R. Beckmann, D.N. Wilson, T. Kamiya, and J. Takano for critical reading of the manuscript and valuable discussions. Arabidopsis uORF information and Arabidopsis upf1 and upf3 mutant seeds were provided by K. Mineta and Y. Watanabe, respectively. T-DNA insertion lines were provided by the ABRC. We thank Y. Kawara, S. Oyama, H. Nagano, and Y. Ohashi for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research (Nos. 22119006 and 15H01525 to S.N. and No. 25221202 to T.F.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We used the Radioisotope Laboratory at the Graduate School of Agriculture, Hokkaido University.
AUTHOR CONTRIBUTIONS
M.T., Y. Yamashita, Y.C., T.A., H.O., S.N., and T.F. designed the experiments. M.T., H.O., K. Murota, K. Miwa, Y. Yamazumi, Y. Yamashita, and M.Y.H. performed the experiments. N.S. conducted in silico analysis. M.T., K. Miwa, N.S., M.Y.H., Y.C., H.O., S.N., and T.F. analyzed data. M.T., N.S., K. Miwa, Y. Yamashita, H.O., S.N., and T.F. wrote the manuscript.
Glossary
- ORF
open reading frame
- uORF
upstream ORF
- UTR
untranslated region
- NMD
nonsense-mediated mRNA decay
- WGE
wheat germ extract
- CHX
cycloheximide
- ICP-MS
inductively coupled plasma-mass spectrometry
- RAV
ribosome arrest value
References
- Amaral A.F., Marques M.M., da Silva J.A.L., da Silva J.J.R.F. (2008). Interactions of D-ribose with polyatomic anions, and alkaline and alkaline-earth cations: possible clues to environmental synthesis conditions in the pre-RNA world. New J. Chem. 32: 2043–2049. [Google Scholar]
- Amrani N., Sachs M.S., Jacobson A. (2006). Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7: 415–425. [DOI] [PubMed] [Google Scholar]
- Arciga-Reyes L., Wootton L., Kieffer M., Davies B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47: 480–489. [DOI] [PubMed] [Google Scholar]
- Ayres D.C., Hellier D.G. (1998). Boron. In Dictionary of Environmentally Important Chemicals. (London: Blackie Academic and Professional; ), pp. 64–65. [Google Scholar]
- Blevins D.G., Lukaszewski K.M. (1998). Boron in plant structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 481–500. [DOI] [PubMed] [Google Scholar]
- Bonilla I., Garcia-González M., Mateo P. (1990). Boron requirement in cyanobacteria : its possible role in the early evolution of photosynthetic organisms. Plant Physiol. 94: 1554–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.H., Bellaloui N., Wimmer M.A., Bassil E.S., Ruiz J., Hu H., Pfeffer H., Dannel F., Römheld V. (2002). Boron in plant biology. Plant Biol. (Stuttg.) 4: 205–223. [Google Scholar]
- Chen X., Schauder S., Potier N., Van Dorsselaer A., Pelczer I., Bassler B.L., Hughson F.M. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415: 545–549. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Ishikawa M., Kijima F., Tyson R.H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R.M., Naito S. (1999). Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science 286: 1371–1374. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S. (2003). S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child S.J., Miller M.K., Geballe A.P. (1999). Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 274: 24335–24341. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembitsky V.M., Smoum R., Al-Quntar A.A., Ali H.A., Pergament I., Srebnik M. (2002). Natural occurrence of boron-containing compounds in plants, algae and microorganisms. Plant Sci. 163: 931–942. [Google Scholar]
- Ebina I., et al. (2015). Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 43: 1562–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort D.J., Propst T.L., Stover E.L., Strong P.L., Murray F.J. (1998). Adverse reproductive and developmental effects in Xenopus from insufficient boron. Biol. Trace Elem. Res. 66: 237–259. [DOI] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M.S. (2005). Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20: 449–460. [DOI] [PubMed] [Google Scholar]
- Gasber A., Klaumann S., Trentmann O., Trampczynska A., Clemens S., Schneider S., Sauer N., Feifer I., Bittner F., Mendel R.R., Neuhaus H.E. (2011). Identification of an Arabidopsis solute carrier critical for intracellular transport and inter-organ allocation of molybdate. Plant Biol (Stuttg) 13: 710–718. [DOI] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Brown C.M., Chisnall M.A., Waterhouse P.M., Macknight R.C. (2016). The emerging world of small ORFs. Trends Plant Sci. 21: 317–328. [DOI] [PubMed] [Google Scholar]
- Hieno A., et al. (2014). ppdb: plant promoter database version 3.0. Nucleic Acids Res. 42: D1188–D1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Hood H.M., Neafsey D.E., Galagan J., Sachs M.S. (2009). Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu. Rev. Microbiol. 63: 385–409. [DOI] [PubMed] [Google Scholar]
- Hori K., Watanabe Y. (2005). UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 43: 530–540. [DOI] [PubMed] [Google Scholar]
- Ishikawa M., Naito S., Ohno T. (1993). Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J. Virol. 67: 5328–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C.P., Zhou H., Huang X., Chiang V.L. (1997). Context sequences of translation initiation codon in plants. Plant Mol. Biol. 35: 993–1001. [DOI] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabu M., Akosman M.S. (2013). Biological effects of boron. Rev. Environ. Contam. Toxicol. 225: 57–75. [DOI] [PubMed] [Google Scholar]
- Kato Y., Miwa K., Takano J., Wada M., Fujiwara T. (2009). Highly boron deficiency-tolerant plants generated by enhanced expression of NIP5;1, a boric acid channel. Plant Cell Physiol. 50: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1987). Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 7: 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2001). Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 29: 5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummheuer J., Johnson A.T., Hauber I., Kammler S., Anderson J.L., Hauber J., Purcell D.F., Schaal H. (2007). A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 363: 261–271. [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanoue L., Trollinger D.R., Strong P.L., Keen C.L. (2000). Functional impairments in preimplantation mouse embryos following boron deficiency. FASEB J. 14A: 539. [Google Scholar]
- Li Y.F., Zheng Y., Addo-Quaye C., Zhang L., Saini A., Jagadeeswaran G., Axtell M.J., Zhang W., Sunkar R. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62: 742–759. [DOI] [PubMed] [Google Scholar]
- Liu H., et al. (2014). Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N., Minami M., Maeda T., Hiratsuka K. (2001). Dual luciferase assay for monitoring transient gene expression in higher plants. Plant Biotechnol. 18: 71–75. [Google Scholar]
- Menges M., Murray J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212. [DOI] [PubMed] [Google Scholar]
- Morris D.R., Geballe A.P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Nemoto Y., Haswzawa S. (1992). Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int. Rev. Cytol. 132: 1–30. [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Nyikó T., Sonkoly B., Mérai Z., Benkovics A.H., Silhavy D. (2009). Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 71: 367–378. [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Eberhard S., Albersheim P., Darvill A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849. [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. (2004). Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55: 109–139. [DOI] [PubMed] [Google Scholar]
- Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S. (2005). Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 19: 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power P.P., Woods W.G. (1997). The chemistry of boron and its speciation in plants. Plant Soil 193: 1–13. [Google Scholar]
- Rajkowitsch L., Vilela C., Berthelot K., Ramirez C.V., McCarthy J.E.G. (2004). Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335: 71–85. [DOI] [PubMed] [Google Scholar]
- Rowe R.I., Eckhert C.D. (1999). Boron is required for zebrafish embryogenesis. J. Exp. Biol. 202: 1649–1654. [DOI] [PubMed] [Google Scholar]
- Sachs M.S., Wang Z., Gaba A., Fang P., Belk J., Ganesan R., Amrani N., Jacobson A. (2002). Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26: 105–114. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.A., Dugger W.M. (1981). Cellular changes during boron deficient culture of the diatom Cylindrotheca fusiformis. Physiol. Plant. 51: 111–117. [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., von Wirén N., Fujiwara T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18: 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Takano J., Chiba Y., Lombardo F., Ogasawara Y., Onouchi H., Naito S., Fujiwara T. (2011). Boron-dependent degradation of NIP5;1 mRNA for acclimation to excess boron conditions in Arabidopsis. Plant Cell 23: 3547–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama-Kadokura N., Murakami K., Takemoto M., Koyanagi N., Murota K., Naito S., Onouchi H. (2014). Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol. 55: 1556–1567. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Jia Q., Vaughn J.N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214: 1–12. [DOI] [PubMed] [Google Scholar]
- Wang Z., Sachs M.S. (1997). Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 17: 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warington K. (1923). The effect of boric acid and borax on the broad bean and certain other plant. Ann. Bot. (Lond.) 37: 629–672. [Google Scholar]
- Woods W.G. (1996). Review of possible boron speciation relating to its essentiality. J. Trace Elem. Exp. Med. 9: 153–163. [Google Scholar]
- Yamashita Y., Kadokura Y., Sotta N., Fujiwara T., Takigawa I., Satake A., Onouchi H., Naito S. (2014). Ribosomes in a stacked array: elucidation of the step in translation elongation at which they are stalled during S-adenosyl-L-methionine-induced translation arrest of CGS1 mRNA. J. Biol. Chem. 289: 12693–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7: 555–560. [DOI] [PubMed] [Google Scholar]
- Yoine M., Nishii T., Nakamura K. (2006a). Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 47: 572–580. [DOI] [PubMed] [Google Scholar]
- Yoine M., Ohto M.A., Onai K., Mita S., Nakamura K. (2006b). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 47: 49–62. [DOI] [PubMed] [Google Scholar]
- Zhou F., Roy B., von Arnim A.G. (2010). Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.