Abstract
Background and aims Few studies have evaluated the genetic structure and evolutionary history of wild varieties of important crop species. The wild papaya (Carica papaya) is a key element of early successional tropical and sub-tropical forests in Mexico, and constitutes the genetic reservoir for evolutionary potential of the species. In this study we aimed to determine how diverse and structured is the genetic variability of wild populations of C. papaya in Northern Mesoamerica. Moreover, we assessed if genetic structure and evolutionary history coincide with hypothetized (1) pre-Pleistocene events (Isthmus of Tehuantepec sinking), (2) Pleistocene refugia or (3) recent patterns.
Methods We used six nuclear and two chloroplast (cp) DNA markers to assess the genetic diversity and phylogeographical structure of 19 wild populations of C. papaya in its natural distribution in Northern Mesoamerica.
Key Results We found high genetic diversity (Ho = 0·681 for nuclear markers, and h = 0·701 for cpDNA markers) and gene flow between populations of C. papaya (migration r up to 420 km). A lack of phylogeographical structure was found with the cpDNA markers (NST < GST), whereas a recent population structure was inferred with the nuclear markers. Evidence indicates that pre-Pleistocene events or refugia did not play an important role in the genetic structuring of wild papaya.
Conclusions Because of its life history characteristics and lack of an ancient phylogeographical structure found with the cpDNA markers, we suggest that C. papaya was dispersed throughout the lowland rain forests of Mexico (along the coastal plains and foothills of Sierras). This scenario supports the hypothesis that tropical forests in Northern Mesoamerica did not experience important climate fluctuations during the Pleistocene, and that the life history of C. papaya could have promoted long-distance dispersal and rapid colonization of lowland rainforests. Moreover, the results obtained with the nuclear markers suggest recent human disturbances. The fragmentation of tropical habitats in Northern Mesoamerica appears to be the main driver of genetic structuring, and the major threat to the dispersion and survival of the species in the wild.
Keywords: Carica papaya, Northern Mesoamerica, nuclear microsatellites, chloroplast DNA, genetic diversity, genetic structure, phylogeography, gene flow, barriers, habitat fragmentation, wild papaya
INTRODUCTION
Mexico is one of the highest ranked countries in terms of biodiversity, comprising approx. 12 % of the world’s biota (Toledo and Ordóñez, 1993). Its biological diversity is due partially to its geographical position between the Nearctic and the Neotropics, and to its highly complex topography that has given rise to a diverse combination of environmental conditions that host numerous ecosystems (Coates and Obando, 1996). South-eastern Mexico covers a large part of the Mesoamerican territory (hereafter Northern Mesoamerica), one of Earth’s most important biodiversity ‘hotspots’ (Myers et al., 2000).
Genetic structure and genetic divergence within and among populations of tropical trees in Mesoamerica result from a number of contemporary and historical factors acting at different temporal and spatial scales. For tropical forest species a variety of scenarios can be expected to explain the structuring of their genetic diversity. Tropical forests date back to Cretaceous times (Davis et al., 2005) and some authors believe that these have remained relatively stable since the climatic fluctuations of the Pliocene–Pleistocene (Colinvaux et al., 1996; Fine and Ree, 2006), preventing genetic structuring of tropical species. However, longer periods of climatic stability in the tropics may reflect range disjunctions around geological barriers that are much older than the Pleistocene vegetation changes that shaped genetic patterns at higher latitudes (Dick, 2010). Other authors suggest that the areas of concentrated species diversity and species endemism in mountainous regions of tropical America represent zones that acted as tropical refugia for rain forest taxa during the cool, dry Pleistocene period; these Pleistocene refugia may have received high rainfall and remained continually warm while savannas and tropical dry forests expanded (Haffer, 1969; Graham, 1973; Toledo, 1982; Prance, 1982; Pennington et al., 2000), leaving a high genetic structure signature in populations from different refugia. In addition, because most tropical tree species occur at low population densities, they may be more susceptible to genetic drift, showing higher inbreeding than the relatively common tree species studied in temperate and boreal forests (Fedorov, 1966). Moreover, lowland tropical plants are thought to be physiologically sensitive to mild stress of drought and cold, especially to low temperatures associated with increasing elevations (Janzen, 1967). This could promote a latitudinal gradient with a stronger phylogeographical structure in tropical lowland species than in temperate plants (Dick and Heuertz, 2008), and several studies have documented phylogeographical structure of Neotropical trees sampled around geographical barriers (Aide and Rivera, 1998; Cavers et al., 2003; Dick et al., 2003; Novick et al., 2003), and higher FST values in tropical trees than in trees from temperate and/or boreal forests (Dick and Heuertz, 2008). Finally, current environmental features may drive patterns of gene flow and genetic structure of tropical plants (Poelchau and Hamrick, 2012); in particular, habitat fragmentation in recent times may also have played a role in structuring genetic diversity (Twyford et al., 2013).
In Northern Mesoamerica two historical events are known to have reduced dispersal and generated a spatial structuring of genetic diversity in different species: the Isthmus of Tehuantepec (pre-Pleistocene event) and the Pleistocene glacial refugia (Bryson et al., 2011; Gutiérrez-Rodríguez et al., 2011; Rodríguez-Gómez et al., 2013). The Isthmus of Tehuantepec forms a narrow piece of lowland that separates the south-central Mexican highlands (Sierra de Juárez and Sierra Madre del Sur) from the highlands of Chiapas (Ornelas et al., 2010). It has been suggested that the isthmus was a seaway for much of the Pliocene (Morrone, 2006), and therefore a major barrier to gene flow during this period (Twyford et al., 2013). Moreover, if the Pleistocene refugia theory is correct, some regions in south-eastern Mexico might have acted as Pleistocene refuges in the past, as suggested by Toledo (1982). Thus, for tropical species in Northern Mesoamerica, we could expect the following scenarios: high levels of population differentiation among populations of tropical species inhabiting different regions proposed as Pleistocene refugia; a genetic subdivision west and east of the Isthmus of Tehuantepec; and high genetic structure promoted by recent anthropogenic disturbances that have modified the Northern Mesoamerican landscape. The current rate of deforestation and habitat fragmentation in Northern Mesoamerica is high enough to interrupt potential gene flow and endanger the existence of tree species (Novick et al., 2003).
There is a relative dearth of detailed phylogeographical information on Neotropical lowland plant species (but see: Cavers et al., 2003; Novick et al., 2003; Poelchau and Hamrick, 2012; Twyford et al., 2013), even though lowland tropical vegetation covers most of the Mesoamerican landscape. Although the vast majority of the world’s tree diversity lies in the tropics, most of the evolutionary history and genetic knowledge comes from temperate and boreal forests, with a marked emphasis on pines, poplars and oaks (Dick, 2010). In Northern Mesoamerica, phylogeographical studies have focused mainly on montane temperate species or tropical species inhabiting cloud forest (Jaramillo Correa et al., 2008; Cavender-Bares et al., 2011; Gutiérrez-Rodríguez et al., 2011; Ornelas et al., 2013; Ruíz-Sánchez and Ornelas, 2014). Moreover, Mesoamerica has been recognized as one of the centres of origin of many domesticated plant species (Vavilov, 1926), so natural populations of important crops species and their wild relative species inhabit this region. In spite of their importance, few studies have addressed the phylogeographical history and genetic structure and diversity of wild populations of important tropical crop species (Fukunaga et al., 2005; Londo et al., 2006; Serrano-Serrano et al., 2010).
Wild papaya (Carica papaya) is a tropical nomadic pioneer tree that occurs naturally in lowland tropical and sub-tropical forests of Mesoamerica. The cultivated varieties of C. papaya represent the third most cultivated tropical crop worldwide (FAO, 2012). Natural populations of C. papaya occur in Northern Mesoamerica; however, the wide geographical distribution of wild populations and the lack of collections in many areas of the region have prevented a precise assessment of their diversity and genetic structure for conservation purposes. Few studies have evaluated the genetic structure and evolutionary history of wild varieties of important crops, with relevance to conservation and management. Moreover, Mesoamerica (particularly Mexico) represents one of the regions where Vavilov (1926) suggested that many cultivated species originated, diversified and were domesticated, such as maize, tomato, beans, cotton, cacao and vanilla. Thus, the aims of the present study were (1) to assess the actual state of natural undomesticated populations of C. papaya regarding its distribution and genetic diversity in Northern Mesoamerica, using nuclear and chloroplast markers; (2) to evaluate the genetic structure and evolutionary history in its Mesoamerican distribution and whether it is consistent with three hypothetical scenarios: pre-Pleistocene events (sea-level oscillations in the Isthmus of Tehuantepec), patterns of ancient Pleistocene refugia or more recent events; (3) to define the possible barriers to gene flow in recent and ancestral times; and (4) to estimate recent migration rates. Finally, in light of our results we discuss some conservation remarks for this important species.
MATERIALS AND METHODS
Study system
Carica papaya L. is a tropical, nomadic, short-lived tree (Fig. 1) that belongs to the family Caricaceae and is the only member of the genus after removal from the Vasconcella group, until recently considered as a section within the genus Carica (Badillo, 2000). Carica papaya is part of a small clade confined to Mexico and Guatemala that also includes three perennial herbs (Jarilla chocola, J. heterophylla and J. nana) and a treelet with spongy thin stems (Horovitzia cnidoscoloides) (Carvalho and Renner, 2012).
Fig. 1.
(A) Natural population of Carica papaya at El Cielo, Tamaulipas, Mexico. (B) Female individual of C. papaya with fruits. (C) Male inflorescence of a wild C. papaya plant. (D) Female flower of a wild C. papaya plant. (E) Mature fruits of a wild C. papaya plant.
There are many cultivated varieties of papaya that differ in traits such as fruit size, colour and flavour, and tree size. Nowadays, only two varieties are cultivated in Mexico: ‘Maradol’, introduced from Cuba, and ‘Mexicana/Amarilla’, domesticated in Mexico (SAGARPA, 2009). The two main differences between wild and domesticated papayas relate to flower morphology and fruit size. Wild (undomesticated) populations of papaya are characterized by a strictly dioecious breeding system (Chávez-Pesqueira et al., 2014) (rather than being trioecious like most varieties of cultivated papaya, where hermaphroditic individuals are preferred); and female trees produce small (no more than 15 cm in diameter), seedy fruits with a thin mesocarp and almost without pulp (Carvalho and Renner, 2012) (Fig. 1). Sexual expression in C. papaya is genetically determined by a pair of homomorphic sex chromosomes (Yu et al., 2007). Carica papaya is pollinated by Lepidoptera, mainly by sphingid hawk moths (Vega-Frutis & Guevara, 2009). Wild C. papaya blooms and fruits year round; the fruits are consumed and probably dispersed mainly by birds and small mammals that climb the stem to consume the fruits (our pers. observ.).
The geographical distribution of the C. papaya clade and the occurrence of wild papayas in Mesoamerica are consistent with a domestication of papaya in that region (Carvalho and Renner, 2012). Carica papaya diverged from its sister clade approx. 25 Mya. The biogeographical history of Caricaceae involves long-distance dispersal from Africa to Central America 35 Mya, and expansion across the Panamanian land bridge between 27 and 19 Mya (Carvalho and Renner, 2012).
Wild papayas inhabit many parts of south-eastern Mexico, usually associated with lowland humid and sub-humid tropical forests (Paz and Vázquez-Yanes, 1998). In many cases, wild varieties are also found in home gardens of ethnic groups from south-eastern Mexico. In some regions such as Yucatan (Terán and Rasmussen, 1995), San Luis Potosí and Veracruz in Mexico, people use wild papaya fruits to make sweet glaze (our pers. observ.). In tropical rain forests wild papaya behaves like a typical fast-growing, short-lived, pioneer tree; they establish rapidly and grow only in recent and relatively large canopy light-gaps in mature forest and in early secondary forest (Chávez-Pesqueira et al., 2014). Wild papaya is relatively abundant in recent large, man-made, clearings that are not ploughed (called acahuales). Large natural forest gaps 1–5 years old can sometimes contain several wild papaya trees (Paz and Vázquez-Yanes, 1998). In Mexico wild papayas occur within pollen/seed dispersal range from cultivars (our pers. observ.)
Sampling
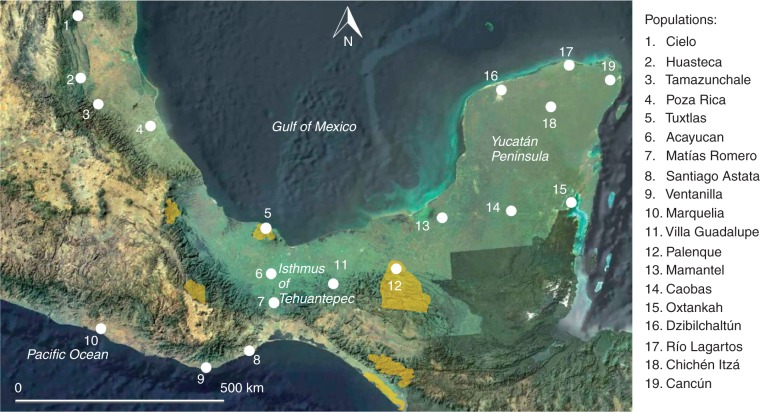
A total of 355 individuals from 19 populations were sampled, covering the natural distribution of C. papaya in Mexico (Fig. 2). Between 11 and 35 individuals were sampled in each population (Table 1). To find these populations, extensive searches were done throughout tropical and subtropical regions of south-eastern Mexico. Populations were chosen and collected only if they were strictly wild (dioecious individuals, small fruits), distant of human populations and had a significant number of individuals to sample.
Fig. 2.
Distribution of the 19 sample locations of wild Carica papaya in Northern Mesoamerica. Yellow areas represent the proposed Pleistocene refugia for Mexico (Toledo, 1982).
Table 1.
Population number, sample sites, geographical location and sample size (n) of wild populations of Carica papaya in its natural distribution in Northern Mesoamerica
| Population no. | Sample site | Geographical location | n |
|---|---|---|---|
| 1 | Cielo, Tamaulipas | 23°01′01·60″N, 99°07′31·50″W | 35 |
| 2 | Huasteca, San Luis Potosí | 21°50′35·20″N, 99°09′06·50″W | 20 |
| 3 | Tamazunchale, San Luis Potosí | 21°14′36·80′′N, 98°44′36·90′′W | 30 |
| 4 | Poza Rica, Veracruz | 20°28′49·61′′N, 97°39′22·21′′W | 16 |
| 5 | Tuxtlas, Veracruz | 18°34′56·94′′N, 95°04′44·00′′W | 30 |
| 6 | Acayucan, Veracruz | 17°30′51·25″N, 95°05′12·75″W | 25 |
| 7 | Matías Romero, Oaxaca | 16°54′24·42′′N, 95°00′35·64′′W | 30 |
| 8 | Santiago Astata, Oaxaca | 15°59′01·10′′N, 95°39′02·17′′W | 11 |
| 9 | Ventanilla, Oaxaca | 15°40′43·15′′N, 96°34′10·81′′W | 30 |
| 10 | Marquelia, Guerrero | 16°34′51·26′′N, 98°47′17·21′′W | 23 |
| 11 | Villa Guadalupe, Tabasco | 17°22′10·78′′N, 93°37′13·35′′W | 30 |
| 12 | Palenque, Chiapas | 17°29′39·98′′N, 92°01′09·13′′W | 26 |
| 13 | Mamantel, Campeche | 18°32′49·58′′N, 91°05′09·00′′W | 30 |
| 14 | Caobas, Campeche | 18°30′47·65″N, 89°28′15·21″W | 32 |
| 15 | Oxtankah, Quintana Roo | 18°34′58·40′′N, 88°14′40·06′′W | 25 |
| 16 | Dzibilchaltún, Yucatán | 21°05′28·34″N, 89°35′51·00″W | 28 |
| 17 | Río Lagartos, Yucatán | 21°31′02·66′′N, 88°08′25·62′′W | 27 |
| 18 | Chichén Itzá, Yucatán | 20°40′56·17″N, 88°34′06·53″W | 26 |
| 19 | Cancún, Quintana Roo | 20°54′33·61″N, 87°25′51·62″W | 18 |
DNA extraction, amplification and sequencing
DNA was extracted using the CTAB method (Doyle and Doyle, 1987) with some modifications. Nuclear and chloroplast markers exhibit different modes of inheritance that help to elucidate the evolutionary history of species. DNA microsatellites show high variability and allow genome-wide information that reflects both contemporary pollen and seed dispersal. In contrast, chloroplast DNA (cpDNA) is maternally inherited in many plants (including C. papaya) and is dispersed only by seeds, thus providing information about ancient seed dispersal and reflecting a different evolutionary history from nuclear DNA because of its small effective population size. Because of this, we used both nuclear (DNA microsatellites) and chloroplast (cpDNA) markers to assess the genetic structure and phylogeographical history of natural populations of C. papaya.
Six microsatellite primers from Ocampo et al. (2006), previously used for wild C. papaya (Chávez-Pesqueira et al., 2014), were amplified using a Multiplex approach. Each multiplex (10 μL) contained 20 ng of DNA template (2 μl), 0·2 μm fluorescently labelled forward primers, 0·2 μm reverse primers, RNase/DNase-free water and Reaction Mix (1×). Amplifications reactions were carried on a Veriti 96-well thermal cycler. The PCRs were performed through touchdown reactions, starting with an initial heat activation at 95 °C for 10 min followed by 31 cycles with a denaturation at 94 °C for 1 min, an annealing for 1 min and 1 min of extension at 72 °C. Annealing cycling temperature began at 57 °C dropping one degree every cycle until reaching 51 °C; this temperature was held for six cycles, followed by two stages of 12 cycles, at 55 and 54 °C, respectively. PCR products were run on ABI Prism 310 and ABI 3730xl automated capillary sequencers, and allele sizes were scored manually using LIZ-500 size standard in GeneMarker v2.4.0 (SoftGenetics, LLC, State College, PA, USA).
Additionally, nine chloroplast markers (ndhJ-trnF, trnQ-5′rps16, ndhF-rp132, psbJ-petA, trnH-psbA, trnL-F, rbcL800f-600r, matKF1-R1 and rpl20-rps12), as well as the internal transcribed spacer (ITS1–2), were tested for amplification and sequence variation in C. papaya. trnH-psbA and rpl20-rps12 were chosen because of their high amplification quality and variability among populations of C. papaya. The remaining tested markers were poorly amplified or were invariant across populations. PCRs were performed in a volume of 15 μL, containing 1 μL DNA, 2 μL buffer, 1·25 μL MgCl2 (1·25 mm), 2 μL dNTP and 1 μL of each primer. The amplification condition for trnH-psbA was 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, 56 °C for 1 min and 72 °C for 2 min, and a final extension at 72 °C for 8 min. For rpl20-rps12 the same conditions were used, but the annealing temperature was set to 53 °C. PCR products were visualized via 1 % agarose gel electrophoresis. All forward and reverse strands were sequenced and then edited in Sequencher 5.0 (Gene Codes Corp., Ann Arbor, MI, USA); nucleotide substitutions, indels (i.e. insertions or deletions) and inversions were visually checked. Sequences were aligned online with MAFFT version 7 (Katoh and Standley, 2012). The sequences reported in this study are available from GenBank (accession numbers: KX451371–KX451661 for psbA-trnH and KX451662–KX451837 for rpl20-rps12).
Data analyses for nuclear microsatellites
Parameters of genetic diversity for each population [percentage of polymorphic loci (%P), the number of alleles (A), observed heterozygosity (Ho), expected heterozygosity (He) and inbreeding coefficient (f)] were calculated using GenAlex 6.5 (Peakall and Smouse, 2012). We used Microchecker 2.2.3 (van Oosterhout et al., 2004) to detect null alleles.
The genetic structure of populations was inferred using Geneland (version 4.0; Guillot et al., 2008) in R (R Development Core Team, 2011). We preferred Geneland over Structure because Geneland includes a null allele model and, in particular, because it takes into account the spatial distribution of populations. Geneland determines the number of population subdivisions for multilocus genotypic data using a Bayesian procedure, considering spatial proximity when assigning individuals to clusters. Ten independent runs were conducted for each analysis using 1 000 000 Markov chain Montel Carlo (MCMC) iterations with a thinning value of 100. Uncorrelated and null allele model options were activated. The number of genetic clusters was set to unknown, but the maximum possible number of clusters was limited to 19 to offer a large enough search space for the MCMC algorithm. Geographical coordinates were decimal degrees of the sampling locations with an uncertainty of 400 km (based on the long migration rates found with BAYESASS among populations; see Results). MCMC post-processing was done with a burn-in of 1000 iterations, and the average posterior probability was used to select the best-suited run.
We further evaluated the genetic differentiation of populations by assuming a step-wise mutation model with Slatkin’s RST (Slatkin, 1995). Partitioning of genetic variability within and among population groups was tested by analysis of molecular variance (AMOVA; Excoffier et al., 1992) among the genetic clusters derived from Geneland, and between the populations east and west from the Isthmus of Tehuantepec, using GenAlex 6.5 (Peakall and Smouse, 2012). In addition, the isolation-by-distance model (Wright, 1943) was tested using a Mantel test (Mantel, 1967), to determine whether matrices of pairwise population genetic distances [RST/(1 − RST)] were correlated to the logarithm of the Euclidean geographical distances. We calculated Mantel’s correlation coefficients (r) using the ade4 package (Dray and Dufour, 2007) in R (R Development Core Team, 2011). The statistical significance of the estimators was determined with 9999 permutations.
The geographical location of genetic discontinuities among populations was assessed with the Monmonier’s maximum difference algorithm implemented in BARRIER version 2.2 (Manni et al., 2004). This program first creates a map of the sampling locations from geographical coordinates. From a matrix of pairwise genetic distances (FST) between populations, barriers are then represented on the map by identifying the edges of polygons where the maximum distances occur. Population pairwise FST comparisons were calculated using ARLEQUIN version 3.5.1.2 (Excoffier et al., 2005). To obtain statistical confidence values for the barriers, 100 replicates of both distance matrices were calculated by resampling individuals within populations.
The program BAYESASS edition 3 (Wilson and Rannala, 2003) was used to estimate short-term migration rates. BAYESASS uses a genetic assignment to estimate short-term dispersal rates, providing an estimate of migration rates over the past two generations. We performed five runs (each with different starting seed value) of 10 million generations, with a burn-in of 1 million generations, and sampled the chain every 2000 generations.
Data analyses of cpDNA
Haplotype and nucleotide diversities, as well as neutrality test statistics of Tajima’s D (Tajima, 1989) and Fu’s F (Fu, 1997) were calculated for each population using ARLEQUIN version 3.5.1.2 (Excoffier et al., 2005). Tajima’s D takes into account the genetic diversity and the number of variable sites in a sequence, to test for demographic range expansion. Significant D values can be due to bottlenecks, selective effects, population expansion or heterogeneity of mutation rates (Tajima, 1996). Fu’s F (Fu, 1997) uses information of the distribution of haplotypes to test for demographic expansion and it is more sensitive to population growth than Tajima’s D. Large negative F values generally are indicative of sudden and significant population growth.
We were not able to concatenate the two studied regions of cpDNA because of inconsistency in the amplification of many individuals. The genealogical relationship between haplotypes was estimated independently for trnH-psbA and rpl20-rps12 sequences using statistical parsimony in TCS 1.2.1 (Clement et al., 2000), using an algorithm for cladograms estimated by maximum parsimony, with indels coded as a fifth state.
Spatial structuring of variation in psbA-trnH was examined using spatial analysis of molecular variance (SAMOVA; Dupanloup et al., 2002), to infer population structure without any prior knowledge. SAMOVA attempts to identify groups of locations that are geographically homogeneous and genetically differentiated from each other, maximizing the proportion of total genetic variance due to differences between groups of populations (FCT). We considered values of K (group number) between 2 and 19 using 100 initial conditions for each run. AMOVAs (Excoffier et al., 1992) for psbA-trnH were performed to determine how genetic variability is distributed within and among localities, and between two pre-defined genetic groups: east and west of the Isthmus of the Tehuantepec. A total of 1000 permutations were performed for each AMOVA. A pattern of isolation by distance (Wright, 1943) was evaluated for the chloroplast marker psba-trnH, which can indicate restricted seed dispersal, with a Mantel test in R (package ade4; Dray and Dufour, 2007). We compared FST pairwise genetic distance among populations and the logarithm of the Euclidean geographical distances. The statistical significance of the estimators was determined with 9999 permutations.
Because nuclear microsatellites reflect a recent history of populations, we also used BARRIER version 2.2 (Manni et al., 2004) for the psba-trnH region to account for more ancient barriers to gene flow.
Genetic differentiation among populations was estimated by computing a distance matrix based on the number of mutational steps between haplotypes (NST) and by using haplotype frequencies (GST). The occurrence of phylogeographical structure was inferred by testing for significant differences between GST and NST using PERMUT 2.0 (Pons and Petit, 1996) with 1000 permutations. In contrast to GST, NST considers sequence differences between the haplotypes. Thus, NST > GST indicates that closely related haplotypes are observed more often in a given geographical area than would be expected by chance (Pons and Petit 1996).
RESULTS
Nuclear microsatellite diversity and population structure
All loci were polymorphic and moderate to high values of neutral genetic diversity were estimated in all natural populations of C. papaya (Ho values from 0·409 to 0·783 and He values from 0·634 to 0·806) (Table 2). Populations 3, 4, 6, 7, 11, 13, 14 and 15 (Tamazunchale, Poza Rica, Acayucan, Matías Romero, Villa Guadalupe, Mamantel, Caobas and Oxtankah) showed He values higher than 0·7. In contrast, populations 5, 8 and 19 (Tuxtlas, Santiago Astata and Cancún) showed the lowest values. Furthermore, f values were, in general, close to 0 but with positive and negative values, implying populations with excess (–) and deficiency (+) of heterozygotes (Table 2) at microsatellite loci. In particular, the Oxtankah population (population 15) showed the lowest value (f = −0·100) and Tuxtlas population (population 5) the highest (0·100). We detected null alleles in all loci but their estimated frequency was relatively low (<10 %) (Microchecker 2.2.3; van Oosterhout et al., 2004) .
Table 2.
Genetic variability (±s.d.) at DNA microsatellite loci in 19 populations of Carica papaya along its natural distribution in Mexico. n = simple size; %P = polymorphic loci percentage; A = allele number; Ho = observed heterozygosity; He = expected heterozygosity; f = inbreeding coefficient
| No. | Population | n | %P | A | Ho | He | f |
|---|---|---|---|---|---|---|---|
| 1 | Cielo | 21 | 100 | 6·167 (0·946) | 0·667 (0·082) | 0·655 (0·076) | −0·017 (0·039) |
| 2 | Huasteca | 16 | 100 | 6·167 (0·703) | 0·698 (0·030) | 0·710 (0·450) | 0·006 (0·044) |
| 3 | Tamazunchale | 15 | 100 | 5·500 (0·617) | 0·744 (0·036) | 0·720 (0·047) | −0·041 (0·028) |
| 4 | Poza Rica | 17 | 100 | 7·667 (0·760) | 0·735 (0·062) | 0·806 (0·009) | 0·089 (0·073) |
| 5 | Tuxtlas | 20 | 100 | 6·167 (0·477) | 0·600 (0·058) | 0·671 (0·018) | 0·100 (0·090) |
| 6 | Acayucan | 20 | 100 | 7·167 (0·601) | 0·742 (0·024) | 0·726 (0·026) | −0·025 (0·037) |
| 7 | Matías Romero | 19 | 100 | 7·667 (0·989) | 0·711 (0·033) | 0·740 (0·030) | 0·034 (0·050) |
| 8 | Santiago Astata | 11 | 100 | 4·167 (0·307) | 0·409 (0·080) | 0·654 (0·029) | 0·387 (0·110) |
| 9 | Ventanilla | 18 | 100 | 6·167 (0·401) | 0·630 (0·031) | 0·688 (0·028) | 0·083 (0·030) |
| 10 | Marquelia | 19 | 100 | 4·500 (0·428) | 0·693 (0·066) | 0·656 (0·036) | −0·064 (0·093) |
| 11 | Villa Guadalupe | 18 | 100 | 7·167 (0·792) | 0·750 (0·087) | 0·762 (0·036) | 0·035 (0·083) |
| 12 | Palenque | 18 | 100 | 6·667 (0·333) | 0·667 (0·059) | 0·705 (0·046) | 0·059 (0·042) |
| 13 | Mamantel | 20 | 100 | 8·000 (1·095) | 0·783 (0·069) | 0·788 (0·021) | 0·013 (0·070) |
| 14 | Caobas | 20 | 100 | 7·833 (0·792) | 0·758 (0·033) | 0·748 (0·030) | −0·019 (0·050) |
| 15 | Oxtankah | 20 | 100 | 6·667 (0·715) | 0·733 (0·063) | 0·669 (0·046) | −0·100 (0·070) |
| 16 | Dzibilchaltún | 20 | 100 | 5·500 (0·563) | 0·692 (0·054) | 0·655 (0·018) | −0·054 (0·074) |
| 17 | Río Lagartos | 20 | 100 | 5·667 (0·615) | 0·658 (0·082) | 0·652 (0·052) | −0·004 (0·100) |
| 18 | Chichén Itzá | 23 | 100 | 6·617 (0·477) | 0·692 (0·046) | 0·690 (0·039) | −0·005 (0·041) |
| 19 | Cancún | 20 | 100 | 5·167 (0·601) | 0·583 (0·076) | 0·634 (0·077) | 0·075 (0·055) |
| Mean | 355 | 100 | 6·325 (0·175) | 0·681 (0·015) | 0·701 (0·010) | 0·029 (0·017) |
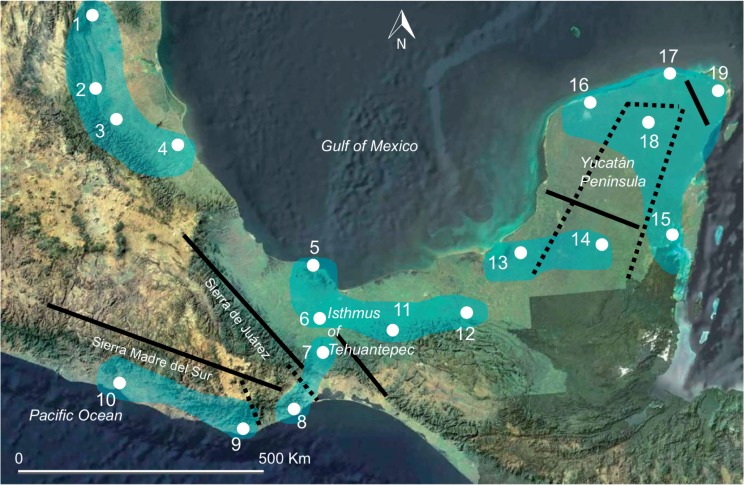
Six clusters were found by posterior cluster membership (GENELAND; K = 6): (1) Cielo, Huasteca, Tamazunchale and Poza Rica (populations 1, 2, 3 and 4); (2) Ventanilla and Marquelia (populations 9 and 10); (3) Tuxtlas, Acayucan, Villa Guadalupe and Palenque (populations 5, 6, 11 and 12); (4) Matías Romero and Santiago Astata (populations 7 and 8); (5) Mamantel and Caobas (populations 13 and 14); and (6) Oxtankah, Dzibichaltún, Río Lagartos, Chichén Itzá and Cancún (populations 15, 16, 17, 18 and 19) (Fig. 3). The inferred clusters contained closely distributed populations, with the exception of Oxtankah (population 15), which was grouped with the northern populations of the Yucatán Peninsula.
Fig. 3.
The six genetic clusters (in blue) inferred from GENELAND using DNA microsatellite loci of natural populations of Carica papaya in Northern Mesoamerica. Solid black lines represent the location of the most probable barriers obtained with BARRIER for DNA microsatellite loci, and dotted black lines for the psbA-trnH chloroplast region.
Genetic differentiation among all populations was moderate (FST = 0·148 and RST = 0·149) whereas differentiation among Geneland clusters was lower (FST = 0·112 and RST = 0·082). Hierarchical partitioning of molecular variance (AMOVA) for the Geneland groups revealed that the highest proportion of variance was located within populations (84·45 %; P < 0·0001) and lower proportions among populations within groups (10·64 %; P < 0·0001) or among groups (4·91 %; P = 0·0293). For the east/west partition, we found the same pattern. Pairwise genetic differentiation was significantly correlated to geographical distance, according to the Mantel test for all sites (r = 0·171, P = 0·0189).
BARRIER suggested that the largest genetic breaks were in many cases concordant with mountainous areas (Fig. 3), with the Sierra Madre del Sur and the Sierra de Juárez Oaxaca being the most prominent barriers for C. papaya dispersion. Interestingly, some barriers were identified in the Yucatán Peninsula, between the Cancún population and its surroundings and north from the Mamantel and Caobas populations, although no visible physical barriers are present in this area.
Recent migration rates among populations, as estimated with BAYESASS, showed moderate recent migration rates among populations that were separated by as much as approx. 420 km [between Poza Rica (population 4) and Acayucan (population 6)] (Table 3). Three populations [Río Lagartos (population 17), Acayucan (population 6) and Tuxtlas (population 5)] were the principal source with higher migration rates than other populations. Migration rates between Río Lagartos (population 17) and Chichén Itzá (population 18) (migration rate = 0·1955), Río Largartos and Oxtankah (population 15) (migration rate = 0·1834), and Río Lagartos and Caobas (population 14) (migration rate = 0·1725) were the highest. Some populations such as Cancún, Cielo, Marquelia, Río Lagartos and Tuxtlas did not receive significant migration from any other population. In addition, 12 populations did not represent important sources of gene flow (Caobas, Chichén Itzá, Dzibilchaltún, Huasteca, Mamantel, Marquelia, Matías Romero, Oxtankah, Poza Rica, Santiago Astata, Tamazunchale and Villa Guadalupe).
Table 3.
Recent migration rates (±s.d.) and distance (km) between recipient and source populations of Carica papaya in its natural distribution in Northern Mesoamerica; migration rates higher than 0·05 are reported
| Recipient population | Source population | Migration rate (s.d.) | Distance (km) |
|---|---|---|---|
| Acayucan | Tuxtlas | 0·1439 (0·0311) | 112·48 |
| Caobas | Río Lagartos | 0·1725 (0·0286) | 359·10 |
| Chichén Itzá | Río Lagartos | 0·1955 (0·0247) | 98·64 |
| Dzibilchaltún | Cancún | 0·1657 (0·0282) | 228·13 |
| Huasteca | Cielo | 0·1640 (0·0283) | 132·34 |
| Mamantel | Palenque | 0·0574 (0·0644) | 158·15 |
| Matías Romero | Acayucan | 0·1079 (0·0538) | 74·11 |
| Oxtankah | Río Lagartos | 0·1834 (0·0272) | 323·60 |
| Palenque | Tuxtlas | 0·1340 (0·0391) | 341·62 |
| Poza Rica | Acayucan | 0·0568 (0·0329) | 423·50 |
| PozaRica | Cielo | 0·0674 (0·0301) | 322·11 |
| Santiago Astata | Ventanilla | 0·0556 (0·0254) | 107·16 |
| Tamazunchale | Cielo | 0·1601 (0·0307) | 205·57 |
| Ventanilla | Acayucan | 0·1299 (0·0329) | 251·69 |
| Villa Guadalupe | Tuxtlas | 0·1696 (0·0287) | 204·20 |
Chloroplast genetic diversity and structure
We obtained 291 cpDNA sequences for the psbA-trnH region and 176 for the rpl20-rps12 region. No individuals of the Huasteca population amplified for rpl20-rps12. The psbA-trnH region was 423 bp long with 34 variable sites, 20 substitutions and 14 indels, while the rpl20-rps12 region was 721 bp long with 15 variable sites. Indels more than 2 bp long were treated as a fifth character state. For psbA-trnH, an inversion of 5 bp was also treated as a fifth character state. Haplotype diversity (h) for psbA-trnH was high for most localities, ranging from 0·307 to 0·934, reflecting the presence of different haplotypes within each site (Table 4). Nucleotide diversity (π) was moderate (0·0009–0·0211) for most populations, indicating some variation between sequences within the same population (Table 4). Within-population average haplotype diversity (hs) was 0·701 (0·089) and nucleotide diversity (πs) was 0·014 (0·005). For rpl20-rps12, h and π were 0 for ten out of the 18 populations, reflecting very low within-site diversity and the lack of variation between sequences from the same population. Only the Tamazunchale population showed a moderate haplotype and nucleotide diversity (h = 0·6762, π = 0·0022). Stochasticity in polymorphisms, i.e. fewer mutations producing homoplasy, could explain the differences in variation between the two cpDNA markers. Discordant patterns may arise from hybridization/introgression between different species (or even between highly differentiated populations in time and space), by homoplasy or even heteroplasmy (reviewed by Wheeler et al., 2014). Yet for papaya, little is known about these processes. Moreover, the psbA-trnH region has been recognized to show high levels of variation compared to other cpDNA loci and therefore has been widely used for barcoding (Kress et al., 2005).
Table 4.
Population size (n), number of haplotypes, number of polymorphic sites, haplotype diversity (h) ± s.d., nucleotide diversity (π) ± s.d., Tajima’s D and Fu’s F for the plastid trnH-psbA marker in 19 wild Carica papaya populations in Northern Mesoamerica; Tajima’s D and Fu’s F (P-values are not shown because in all cases P > 0·1)
| Population | n | Haplotype no. | Polymorphic sites | h | π | D | F |
|---|---|---|---|---|---|---|---|
| 1. Cielo | 18 | 3 | 13 | 0·3072±0·1316 | 0·0062±0·0038 | −1·1157 | 4·0306 |
| 2. Huasteca | 14 | 9 | 13 | 0·9341±0·0448 | 0·0152±0·0086 | 2·2854 | −0·6617 |
| 3. Tamazunchale | 15 | 4 | 14 | 0·7333±0·0669 | 0·0159±0·0089 | 2·1334 | 5·9607 |
| 4. Poza Rica | 13 | 4 | 13 | 0·7692±0·0724 | 0·0164±0·0092 | 2·3307 | 5·3750 |
| 5. Tuxtlas | 11 | 6 | 7 | 0·8364±0·0887 | 0·0052±0·0035 | −1·0291 | −1·2888 |
| 6. Acayucan | 17 | 8 | 21 | 0·8382±0·0675 | 0·0211±0·0115 | 0·6628 | 2·1509 |
| 7. Matías Romero | 12 | 4 | 16 | 0·7121±0·1053 | 0·0128±0·0075 | −0·2247 | 3·9722 |
| 8. Santiago Astata | 10 | 3 | 2 | 0·5111±0·1643 | 0·0013±0·0013 | −1·1117 | −0·5938 |
| 9. Ventanilla | 15 | 9 | 18 | 0·9048±0·0544 | 0·0192±0·0106 | 1·2114 | 0·3064 |
| 10. Marquelia | 20 | 5 | 17 | 0·7526±0·0615 | 0·0091±0·0053 | −0·2945 | 2·8275 |
| 11. Villa Guadalupe | 14 | 6 | 7 | 0·7912±0·0894 | 0·0061±0·0039 | 0·9683 | −0·2864 |
| 12. Palenque | 19 | 8 | 18 | 0·8304±0·0657 | 0·0113±0·0064 | −0·5600 | 0·4672 |
| 13. Mamantel | 19 | 4 | 5 | 0·5556±0·1030 | 0·0047±0·0031 | 0·3456 | 1·7388 |
| 14. Caobas | 15 | 5 | 6 | 0·8095±0·0589 | 0·0050±0·0033 | −0·0290 | 0·3408 |
| 15. Oxtankah | 18 | 5 | 6 | 0·7190±0·0910 | 0·0068±0·0042 | 1·5282 | 1·5639 |
| 16.Dzibilchaltún | 16 | 3 | 2 | 0·4250±0·1326 | 0·0012±0·0012 | −0·3301 | −0·2898 |
| 17. Río Lagartos | 16 | 6 | 6 | 0·8417±0·0534 | 0·0071±0·0043 | 2·0978 | 0·4179 |
| 18. Chichén Itzá | 10 | 4 | 5 | 0·6444±0·1518 | 0·0031±0·0023 | −0·8222 | −0·3120 |
| 19. Cancún | 19 | 2 | 1 | 0·4094±0·1002 | 0·0009±0·0010 | 0·7937 | 1·0079 |
| Mean | 5·16 | 10 | 0·7013±0·0896 | 0·0149±0·0049 | 0·4654±1·206 | 1·4067±2·1249 | |
| Total | 291 | 50 | 30 |
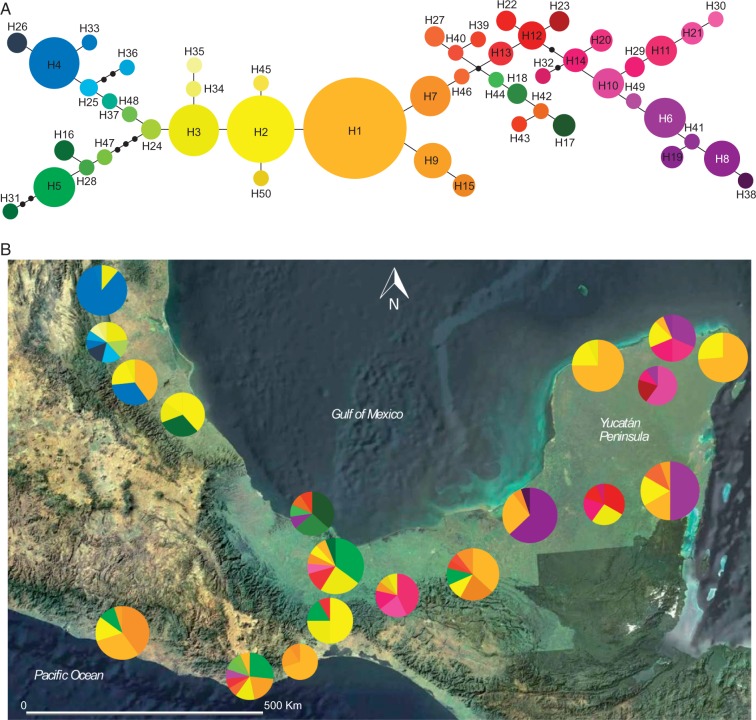
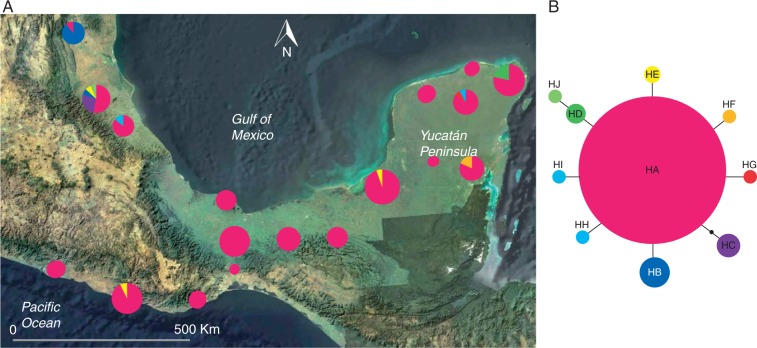
The statistical parsimony network of natural populations of C. papaya in Northern Mesoamerica recovered 50 haplotypes for the psbA-trnH region and ten for the rpl20-rps12 region (Figs 3 and 4). For psbA-trnH, ten out of 50 haplotypes were shared between populations, and 40 were private. The most widespread haplotypes were H1, H2 and H3, which were found in 11, ten and ten populations, respectively, and were shared between distant populations. Huasteca (population 2) and Tuxtlas (population 5) were the populations with more private haplotypes, seven (H24, H25, H26, H34, H35, H36, H37) and six (H17, H18, H42, H43, H44, H45), respectively (Fig. 4). For rpl20-rps12, less variation was found, and most individuals (86 %) bore haplotype HA, while six out of nine haplotypes were private. We found that Tamazunchale (population 3) was the population with the highest number of haplotypes (five), and was also the one with more private haplotypes, together with Chichén Itzá (population 18) (HC and HJ for Tamazunchale and HG and HH for Chichen Itzá) (Fig. 5). Due to low variation at rpl20-rps12, this marker was not analysed further.
Fig. 4.
(A) Statistical parsimony haplotype network for the psbA-trnH chloroplast region of Carica papaya. The size of circles is proportional to the frequency of each haplotype, and small black circles represent non-sampled haplotypes. (B) Geographical distribution of 19 natural populations of C. papaya cpDNA haplotypes in south-eastern Mexico. Pie charts represent the haplotypes found in each sampling locality. The area of the pie chart represents the size of the population and the size of sections is proportional to the haplotypic frequency.
Fig. 5.
(A) Statistical parsimony haplotype network for the rpl20-rps12 chloroplast region of Carica papaya. Haplotype designations in the network correspond to those in Table 4. The size of circles is proportional to the frequency of each haplotype, and small black circles represent non-sampled haplotypes. (B) Geographical distribution of 19 natural populations of C. papaya cpDNA haplotypes in south-eastern Mexico. Pie charts represent the haplotypes found in each sampling locality. The area of the pie chart represents the size of the population and the size of sections is proportional to the haplotypic frequency.
The analysis of spatial genetic structure for psbA-trnH using SAMOVA showed that the highest FCT value was for K = 2; however, one group was formed by only one population (population 1). The K with the highest FCT increment was 9, but again, some groups included only one sampling site. When the AMOVA was performed between the populations east and west of the Isthmus of Tehuantepec, we found that most variation was contained within stands (51·2 %; P < 0·0001), followed by among populations within groups (34·9 %; P < 0·0001) and finally among groups (13·9 %; P = 0·0117). A pattern of isolation by distance was also detected (Mantel test; r = 0·1663, P = 0·0255).
We assessed the potential of genetic breaks that could suggest more ancient barriers among populations, in comparison with the nuclear microsatellite data, using BARRIER for psbA-trnH (Fig. 3). As for the nuclear markers, the most likely barriers were found in the Yucatán Peninsula separating Chichén Itzá (population 18) and Caobas (population 14) from all populations nearby. BARRIER also detected two barriers in southern Oaxaca, isolating the Santiago Astata population (population 8).
The results from PERMUT showed that the level of NST (0·224, s.e. 0·0410) was not significantly higher (P > 0·05) than GST (0·273, s.e. 0·0571), indicating a lack of phylogeographical structure for C. papaya.
Finally, demographic analyses for psbA-trnH showed no significant expansion for any of the populations of C. papaya (Table 4).
DISCUSSION
Evolutionary research of lowland tropical tree species is at an early stage of development (Dick and Heuertz, 2008). For C. papaya, we found contrasting results between neutral (nuclear microsatellites) and chloroplast (cpDNA) markers. Microsatellite data are known to resolve patterns of recent gene flow and pollen dispersal, due to their high mutational rate, whereas chloroplast markers tend to reflect ancient patterns because lack of recombination, low effective population size and their conservative mutation rate (Provan et al., 2001). Our results suggest a lack of phylogeographical structure for C. papaya inferred by cpDNA, but a recent structuring derived from the microsatellite data.
Isthmus of Tehuantepec and Pleistocene refugia hypothesis
We evaluated whether two hypothetical biogeographical events documented for other tropical species have influenced the genetic structure of C. papaya in Northern Mesoamerica: the Isthmus of Tehuantepec and putative Pleistocene glacial refugia. As a result of the several sea-level oscillations and continental uplifts that took place in the Isthmus of Tehuantepec during the Pliocene (Lambeck and Chappell, 2001), isolation and reductions to gene flow among populations of lowland tropical species located at each side of the isthmus could be expected. However, we found little evidence of a genetic break corresponding to the isthmus as indicated by the low FCT value in the AMOVA for both markers. Furthermore, the genetic relationships depicted by the haplotype network obscure the role of the Isthmus of Tehuantepec as a biogeographical barrier as it supports that populations between west and east share haplotypes. Finally, with both nuclear and chloroplast genetic data, no genetic break was detected in the Isthmus of Tehuantepec that could suggest an ancient or recent barrier in the zone. Probably, because of the pioneer/nomadic behaviour of wild C. papaya, and its short life cycle, populations could have established and expanded rapidly after the Isthmus of Tehuantepec arose, erasing any sign of past genetic break. Another study has shown similar results; Twyford et al. (2013) found low FCT values between populations east and west from the isthmus for Begonia heracleifolia, a neotropical lowland plant. In contrast, Gutiérrez-Rodríguez et al. (2011) found a phylogeographical break at the Isthmus of Tehuantepec, with private haplotypes at either side of the isthmus for Palicourea padifolia, a cloud forest herb. For a hummingbird species, Amazilia cyanocephala, Rodríguez-Gómez et al. (2013) found two genetic groups separated by the Isthmus of Tehuantepec in the late Pleistocene, with the split occurring in the presence of gene flow. It is interesting to note that neither lowland species (C. papaya and B. heracleifolia) showed genetic signals by the Isthmus of Tehunatepec, and in contrast the shrub P. padifolia and the hummingbird, both from cloud forests, showed a genetic break. This suggests that this event, in contrast to species that inhabit cloud forest at higher altitudes, did not represent a strong barrier for gene flow of lowland tropical species and that these species were able to recolonize once the isthmus emerged.
On the other hand, we could not test properly the Pleistocene refugia hypothesis given that we did not find natural populations of C. papaya in most of the proposed refugia in Mexico (La Lacandona, Soconusco, Los Tuxtlas, Sierra de Juárez and Córdoba) (Fig. 2); we found wild papaya populations only at the Los Tuxtlas region (population 5) and at the La Lacandona rain forest [population 12 (Palenque) lies in the region of La Lacandona rain forest]. However, we did find signs of genetic structuring in the Tuxtlas population, which bore many private haplotypes for the chloroplast marker, suggesting a long-term accumulation of variation and therefore a possible refugia role of this region. Despite this, few studies have found evidence supporting the refugial hypothesis, particularly for lowland tree species. For temperate and tropical cloud forests, evidence has suggested that the great endemism of Mesoamerican highlands is the result of persistence throughout glacial cycles of the relict montane taxa that survived through the interglacials rather than the persistence of relict lowland tropical species that migrated to the highlands (Colinvaux et al., 2000). However, Gutiérrez-Rodríguez et al. (2011) did not find evidence of genetic structure by Pleisotcene refugia in the cloud forest species P. padifolia. For tropical lowland species like C. papaya, responses to historical climatic fluctuations may depend on species-specific adaptations, therefore making the identification of refugia for complex tropical species assemblages difficult (Poelchau and Hamrick, 2012). Generalizations about refugia may apply only for species with similar ecological preferences, rather than at broad taxonomic scales (Twyford et al., 2013).
Genetic diversity and structure
Overall, we found high values of genetic diversity. With the microsatellite data, we found values of observed heterozygosity (Ho) above 0·7 for many populations across the entire distribution of wild papaya in Mexico. Only three populations showed values of Ho below 0·6: Santiago Astata (population 8), Tuxtlas (population 5) and Cancún (population 19). Santiago Astata has the smaller population size and the highest inbreeding coefficient (f = 0·387). Wild C. papaya combines small population sizes (M.C.-P., pers. observ.) and strict dioecy, which can compromise their amount and maintenance of genetic diversity in the long term. The Los Tuxtlas population lies in a very fragmented rain forest area that could affect genetic contact among sub-populations, thus decreasing genetic diversity, as previously shown (Chávez-Pesqueira et al., 2014). A similar scenario could be the case for the Cancún population, which lies near to a human-populated and tourist area.
Regarding the chloroplast genetic data, we found high variability for psbA-trnH. The high level of haplotype diversity (h = 0·7013) was surprising, as single haplotypes may be expected to be fixed by genetic drift in the typical small populations of C. papaya. However, the papaya populations may have acted as a meta-population, maintaining haplotype diversity via extinction–colonization dynamics (Ray, 2001) if dispersal was effective in the past. The most frequent haplotypes (H1, H2 and H3) were widely represented across the C. papaya range although 40 haplotypes (80 %) were private. In particular, Huasteca and Tuxtlas were the populations with more private haplotypes, suggesting particular cases of ancestral polymorphisms.
Genetic structure for the nuclear microsatellites was associated with six groups. All groups were geographically close, showing moderate values of genetic differentiation among them. For the chloroplast marker, we found an almost lack of phylogeographical structure, suggesting ancient seed dispersal across the distribution of C. papaya in Northern Mesoamerica. This could be explained by a more effective dispersal of fruits. Although wild papayas are mainly pollinated by nocturnal sphingids, which are known to be efficient, and long-distance pollinators (Dafni, 1992), moving pollen as far as 20 km (Amorim et al., 2014), fruits may have longer dispersal. Fruits are important resources for frugivores, mainly small mammals and birds (Chávez-Pesqueira et al., 2014), which can cross fragmented habitats efficiently (Diffendorfer et al., 1995; Levey et al., 2005). We could expect that, in the past, tropical and sub-tropical forests in Northern Mesoamerica were continuous, allowing efficient dispersal between populations of wild papaya, whereas now, fragmentation of the natural habitat of wild papaya may decrease the efficiency of dispersers with low mobility. Moreover, although we found many private haplotypes for natural populations of C. papaya, which could suggest a high phylogeographical structure, the most common haplotypes were widely represented among populations and covered most of the distribution of this species in Northern Mesoamerica. This occurrence of widespread plastid haplotypes in the range of C. papaya is consistent with the effective seed dispersal in the past that we propose.
Weak population genetic structure spanning tropical America has been also reported for pioneer, drought-tolerant trees such as Ceiba pentandra (Dick et al., 2007), Cordia alliodora (Rymer et al., 2013), Jacaranda copaia (Scotti-Saintagne et al., 2013), Trema micrantha (Dick et al., 2013) and Begonia heracleifolia (Twyford et al., 2013), suggesting an apparent association between drought tolerance and low levels of phylogeographical structure (Honorio Coronado et al., 2014). This suggests that populations survived in situ through historical climatic fluctuations, having enough time to differentiate by drift and selection for local adaptation, and limited dispersal of the accumulated new mutations across the range of the species. Carica papaya is a light-demanding tree that tolerates sub-tropical forest conditions, and can tolerate drought to some extent. This could explain, in part, the lack of phylogeographical structure we found for wild papaya.
Barriers to gene flow and migration rates
Important mountain chains were recognized as the most likely barriers to gene flow for the nuclear microsatellites. In fact, the Sierra Madre del Sur, Sierra Madre Oriental and Sierra de Oaxaca greatly exceed the current elevational limits of lowland rain forest trees (1000 m a.s.l. for C. papaya). For Cancún the inferred barrier may be due to the species low genetic diversity and anthropogenic fragmentation rather to a physiographical feature preventing gene flow. For the chloroplast marker, no geographical features such as mountain chains were recognized as barriers. The barriers inferred were mainly in the Yucatán Peninsula, suggesting isolation of the Chichén Itzá and Caobas populations; although there are no actual climatic or geographical events associated with this isolation, palaeobotanical studies have documented dramatic changes in the vegetation cover of the peninsula along its biogeographical history. For instance, extensive savannas existed in what is now covered with tropical rain forest (Vázquez-Domínguez and Arita, 2010). Moreover, barriers could arise not only if gene flow is interrupted but also if populations are undergoing an adaptive isolation process in which migrants are selected against.
In contrast to the barrier analysis, migration rates obtained from microsatellite data showed that, in recent times, the Yucatán Peninsula may not represent a barrier and that the Río Lagartos population, the most northern population in the Yucatán Peninsula, is an important source of variation for the region. In general, high migration rates were found between populations separated by as far as 420 km, suggesting that wild C. papaya has long dispersal ability. However, we did not find significant migration rates between populations separated by mountainous chains, thus validating them as important gene flow barriers for this species. Furthermore, non-sampled populations of wild papaya, or even cultivars, could act as gene flow corridors, contributing to the high migration rates we found. Yet, we do not know to what extent the movement of seeds or disturbance by ancient human cultures in the region have played a role in the genetic structure of C. papaya.
Conservation remarks
Wild C. papaya is a key element of early-successional tropical and sub-tropical forests in Mexico, and represents the genetic reservoir for the evolutionary potential of the species (Chávez-Pesqueira et al., 2014). Although we found overall high levels of genetic variability, we also found that natural populations of this species are becoming structured, probably because of human disturbance to its natural habitat. This result is extremely important for generating adequate conservation strategies for this important crop species. Moreover, the centre of origin for papaya has not been appropriately defined. It has been generally suggested that papaya originated somewhere in Mesoamerica (Vavilov, 1926; Storey, 1976). We found high genetic diversity in the Isthums of Tehuantepec zone for the plasmid marker, as well as highest number of haplotypes, suggesting a hotspot of diversity in that region. We propose, based on these data, a tentative origin of the species in that region. In addition, papaya is most closely related to four species from southern Mexico and Guatemala (Carvalho and Renner, 2012), suggesting that C. papaya could have originated in southern Mexico. Moreover, this plant has been proposed as being domesticated in this area by the Mayans (Carvalho and Renner, 2012). Finally, when comparing our estimates of genetic diversity with the few other studies of natural populations of papaya, both in Costa Rica (Coppens d’Eeckenbrugge et al., 2007; Brown et al., 2012), we found higher values in Mexico. However, these Costa Rican natural populations are considered possible feral plants, i.e. descended from domesticated plants, which may explain their lower diversity derived by domestication genetic bottlenecks (Doebley et al., 2006).
Although mating between wild and cultivated papayas has not been assessed in Mexico, possible ‘hybrids’ have been seen in tropical zones in the country (our pers. observ.) and evidence exists from Costa Rica for gene flow between feral and cultivated plants (Brown et al., 2012). Crosses between wild and domesticated papaya threaten the maintenance of the natural genetic pool of the species. Indeed, this genetic diversity forms the capital for current and future improvements to crop plants (Chávez-Pesqueira et al., 2014). Thus, our results point to potential problems that in situ conservation of genetic pools may face, and also provide an important warning regarding the cultivation of transgenic papayas in Mexico. While no transgenic papayas have been established in Mexico thus far, there are attempts to do so (Silva-Rosales et al., 2010). The high migration rates, in combination with the reproductive characteristics of papaya, warn against the ecological and evolutionary effects of domesticated genes and transgenes contaminating wild populations.
CONCLUSIONS
Our study has revealed contrasting results depending on the genetic marker used. Given the life history characteristics of wild C. papaya, and the lack of an ancient phylogeographical structure, we suggest that tropical forests in Northern Mesoamerica did not suffer important climate fluctuations, as other authors have suggested in relation to the refugial hypothesis (Colinvaux et al., 1996, 2000; Fine and Ree, 2006). Our results also suggest that the life history of C. papaya promoted its long dispersal and rapid colonization of lowland rain forests, thus maintaining genetic diversity throughout its range. However, recent human disturbance, mainly the fragmentation of tropical habitats in Northern Mesoamerica, appear to represent a threat to its dispersion and therefore to its genetic diversity and structure.
Further research in lowland tropical species of Mesoamerica is necessary to understand the present distribution of genetic variability of species inhabiting this ecosystem, and their phylogeographical history. Moreover, assessing the actual situation of wild varieties and relatives of crop species is fundamental to ensure the maintenance of their genetic reservoirs and evolutionary potential.
ACKNOWLEDGEMENTS
We thank R. Tapia-López for assistance in obtaining genetic data, and the members of the Laboratorio de Genética Ecológica y Evolución for logistical support and field assistance. We thank Pilar Suárez-Montes, Diego Carmona, Marisol De la Mora and Jorge Juárez for their help during the field trips. We are very grateful to Juan Pablo Jaramillo-Correa for his valuable and insightful help through the whole process of this study. Finally, we thank Daniel Piñero, Mauricio Quesada, Antonio González, Alejandro Casas and Alejandra Vázquez-Lobo for suggested improvements to the manuscript. This work was supported by a grant of the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica [PAPIIT IN 215111-3 awarded to J.N.F.] and a grant of Comisión nacional para el conocimiento y uso de la biodiversidad [CONABIO DGAP003/WQ003/15 awarded to M.C.P]. This paper constitutes a partial fulfillment of the Graduate Program in Biological Sciences of the National Autonomous University of Mexico (UNAM) for M.C.P., who acknowledges a scholarship and financial support provided by the National Council of Science and Technology (CONACYT).
LITERATURE CITED
- Aide TM, Rivera E. 1998. Geographic patterns of genetic diversity in Poulsenia armata (Moraceae): implications for the theory of Pleistocene refugia and the importance of riparian forest. Journal of Biogeography 25: 695–705. [Google Scholar]
- Amorim FW, Wyatt GE, Sazima M. 2014Low abundance of long-tongued pollinators leads to pollen limitation in four specialized hawkmoth-pollinated plants in the Atlantic rain forest, Brazil. Naturwissenschaften 101: 893–905. [DOI] [PubMed] [Google Scholar]
- Badillo VM. 2000. Carica L. vs Vasconcella St. Hil. (Caricaceae): con la rehabilitación de este último. Ernstia 10:74–79. [Google Scholar]
- Brown JE, Bauman JM, Lawrie JF, Rocha OJ, Moore RC. 2012. The structure of morphological and genetic diversity in natural populations of Carica papaya (Caricaceae) in Costa Rica. Biotropica 44: 179–188. [Google Scholar]
- Bryson RW, García‐Vázquez UO, Riddle BR. 2011. Phylogeography of Middle American gophersnakes: mixed responses to biogeographical barriers across the Mexican Transition Zone. Journal of Biogeography 38: 1570–1584. [Google Scholar]
- Carvalho FA, Renner SS. 2012. A dated phylogeny of the papaya family (Caricaceae) reveals the crop’s closest relatives and the family’s biogeographic history. Molecular Phylogenetics and Evolution 65: 46–53. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Gonzalez-Rodriguez A, Pahlich A, Koehler K, Deacon N. 2011. Phylogeography and climatic niche evolution in live oaks (Quercus series Virentes) from the tropics to the temperate zone. Journal of Biogeography 38: 962–981. [Google Scholar]
- Cavers S, Navarro C, Lowe A. 2003. Chloroplast DNA phylogeography reveals colonization history of a Neotropical tree, Cedrela odorata L., in Mesoamerica. Molecular Ecology 12: 1451–1460. [DOI] [PubMed] [Google Scholar]
- Chávez-Pesqueira M, Suárez-Montes P, Castillo G, Núñez-Farfán J. 2014. Habitat fragmentation threatens wild populations of Carica papaya (Caricaceae) in a lowland rainforest. American Journal of Botany 101: 1092–1101. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1660. [DOI] [PubMed] [Google Scholar]
- Coates AG, Obando JA. 1996. The geologic evolution of the Central American Isthmus In: Jackson JBC, Budd AF, Coates A, eds. Evolution and environment in tropical America. Chicago: The University of Chicago Press, 21–52. [Google Scholar]
- Colinvaux PA, De Oliveira PE, Moreno JE, Miller MC, Bush MB. 1996. A long pollen record form lowland Amazonia: forest and cooling in glacial times. Science 274: 85–88. [Google Scholar]
- Colinvaux PA, De Oliveira PE, Bush MB. 2000. Amazonian and neotropical plant communities on glacial time-scales: the failure of the aridity and refue hypothesis. Quaternary Science Reviews 19: 141–169. [Google Scholar]
- Coppens d’Eeckenbrugge G, Restrepo MT, Jiménez D, Mora E. 2007. Morphological and isozyme characterization of common papaya in Costa Rica. Acta Horticulturae 740: 109–120. doi:10.17660/ActaHortic.2007.740.11. [Google Scholar]
- Dafni A. 1992. Pollination ecology. Oxford: Oxford University Press. [Google Scholar]
- Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. 2005. Explosive radiation of Malpighiales supports a mid-Cretaceous origin of modern tropical rain forests. The American Naturalist 165: 36–65. [DOI] [PubMed] [Google Scholar]
- Dick CW. 2010. Phylogeography and populations structure of tropical trees. Tropical Plant Biology 3: 1–3. [Google Scholar]
- Dick CW, Heuertz M. 2008. The complex biogeographic history of a widespread tropical tree species. Evolution 62: 2760–2774. [DOI] [PubMed] [Google Scholar]
- Dick CW, Abdul-Salim K, Bermingham E. 2003. Molecular systematic analysis reveals cryptic tertiary diversification of a widespread tropical rainforest tree. The American Naturalist 162: 691–703. [DOI] [PubMed] [Google Scholar]
- Dick CW, Bermingham E, Lemes MR, Gribel R. 2007. Extreme long-distance dispersal of the lowland tropical rainforest tree Ceiba pentandra L. (Malvaceae) in Africa and the Neotropics. Molecular Ecology 16: 3039–3049. [DOI] [PubMed] [Google Scholar]
- Dick CW, Lewis SL, Maslin M, Bermingham E. 2013. Neogene origins and implied warmth tolerance of Amazon tree species. Ecology and Evolution 3: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffendorfer JE, Gaines MS, Holt RD. 1995. Habitat fragmentation and movements of three small mammals (Sigmodon, Microtus, and Peromyscus). Ecology 76: 827–839. [Google Scholar]
- Doebley J, Gaut B, Smith B. 2006. The molecular genetics of domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doyle J, Doyle J. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22: 1–20. [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. 2002. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology 11: 2571–2581. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- FAO. 2012. Crop production. http://faostat.fao.org/site/567/default.aspx#ancor.
- Fedorov AA. 1966. The structure of the tropical rain forest and speciation in the humid tropics. Journal of Ecology 54: 1–11. [Google Scholar]
- Fine PVA, Ree R. 2006. Evidence for a time-integrated species-area effect on the latitudinal gradient in tree diversity. The American Naturalist 168: 796–804. [DOI] [PubMed] [Google Scholar]
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Hill J, Vigouroux Y, et al. 2005. Genetic diversity and population structure of Teosinte. Genetics 169: 2241–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A. 1973. History of the arborescent temperate element in the northern Latin American biota In: Graham A, ed. Vegetation and vegetational history of northern Latin America. New York: Elsevier Scientific Publishing Co, 301–314. [Google Scholar]
- Guillot G, Santos F, Estoup A. 2008. Analysing georeferences population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24: 1406–1407. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Rodríguez C, Ornelas JF, Rodríguez-Gómez F. 2011. Chloroplast DNA phylogeography of a distylous shrub (Palicourea padifolia, Rubiaceae) reveals past fragmentation and demographic expansion in Mexican cloud forests. Molecular Phylogenetics and Evolution 61:603–615. [DOI] [PubMed] [Google Scholar]
- Haffer J. 1969. Speciation in Amazonian forest birds. Science 165: 131–136. [DOI] [PubMed] [Google Scholar]
- Honorio Coronado EN, Dexter KG, Poelchau MF, Hollingsworth PM, Phillips OL, Pennington RT. 2014. Ficus insipida subsp. insipida (Moraceae) reveals the role of ecology in the phylogeography of widespread Neotropical rain forest tree species. Journal of Biogeography 41: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DH. 1967. Synchronization and sexual reproduction of trees with the dry season in Central America. Evolution 20: 249–275. [DOI] [PubMed] [Google Scholar]
- Jaramillo Correa JP, Aguirre-Planter E, Khasa DP, et al. 2008. Ancestry and divergence of subtropical montane forest isolates: molecular biogeography of the genus Abies (Pinaceae) in southern México and Guatemala. Molecular Ecology 17: 2476–2490. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2012. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. 2005. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeck K, Chappell J. 2001. Sea level change through the last glacial cycle. Science 292: 679–686. [DOI] [PubMed] [Google Scholar]
- Levey DJ, Bolker BM, Tewksbury JJ, Sargent S, Haddad NM. 2005. Effects of landscape corridors on seed dispersal by birds. Science 309: 146–148. [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang Y, Hung K, Chiang T, Schaal BA. 2006. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proceedings of the National Academy of Sciences of the United States of America 103: 9578–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni F, Guérard E, Heyer E. 2004. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonieŕs algorithm. Human Biology 76: 173–190. [DOI] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Research 27: 209–220. [PubMed] [Google Scholar]
- Morrone JJ. 2006. Biogeographic areas and transitions zones of Latin America and the Caribbean islands based on panbiogeographic and cladistic analyses of the entomofauna. Annual Review of Entomology 51: 467–494. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- Novick RR, Dick CW, Lemes MR, Navarro C, Caccone A, Bermingham E. 2003. Genetic structure of Mesoamerican populations of big-leaf mahogany (Swietenia macrophylla) inferred from microsatellite analysis. Molecular Ecology 12: 2885–2893. [DOI] [PubMed] [Google Scholar]
- Ocampo J, Dambier D, Ollitrault P, et al. 2006. Microsatellite markers in Carica papaya L.: isolation, characterization and transferability to Vasconcellea species. Molecular Ecology Notes 6: 212–217. [Google Scholar]
- Ornelas JF, Ruiz-Sánchez E, Sosa E. 2010. Phylogeography of Podocarpus matudae (Podocarpaceae): pre-Quaternary relicts in Northern Mesoamerican cloud forests. Journal of Biogeography 37: 2384–2396. [Google Scholar]
- Ornelas JF, Sosa V, Ornelas JF, et al. 2013. Comparative phylogeographic analyses illustrate the complex evolutionary history of threatened cloud forests of Northern Mesoamerica. PLoS One 8:e56283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz H, Vázquez-Yanes C. 1998. Comparative seed ecophysiology of wild and cultivated Carica papaya trees from a tropical rain forest region in Mexico. Tree Physiology 18: 277–280. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington RT, Prado DE, Pendry CA. 2000. Neotropical seasonally dry forests and Quaternary vegetation changes. Journal of Biogeography 27: 261–273. [Google Scholar]
- Poelchau MF, Hamrick JL. 2012. Differential effects of landscape-level environmental features on genetic structure in three codistributed tree species in Central America. Molecular Ecology 21: 4970–4982. [DOI] [PubMed] [Google Scholar]
- Pons O, Petit RJ. 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prance GT. 1982. Biological diversification in the tropics. New York: Columbia University Press. [Google Scholar]
- Provan J, Powell W, Hollingsworth PM. 2001. Chloroplast microsatellites: new tools for studies in plants ecology and evolution. Trends in Ecology and Evolution 16: 142–147. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2011. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; http://www.R-project.org. [Google Scholar]
- Ray C. 2001. Maintaining genetic diversity despite local extinctions: effects of population scale. Biological Conservation 100: 3–14. [Google Scholar]
- Rodríguez-Gómez F, Gutiérrez-Rodríguez C, Ornelas JF. 2013. Genetic, phenotypic and ecological divergence with gene flow at the Isthmus of Tehuantepec: the case of the azure-crowned hummingbird (Amazalia cyanocephala). Journal of Biogeography 40: 1360–1373. [Google Scholar]
- Ruíz-Sanchez E, Ornelas F. 2014. Phylogeography of Liquidambar styraciflua (Altingiaceae) in Mesoamerica: survivors of a Neogene widespread temperate forest (or cloud forest) in North America? Ecology and Evolution 4: 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymer PD, Dick CW, Vendramin GG, Buonamici A, Boshier D. 2013. Recent phylogeographic structure in a widespread ‘weedy’ Neotropical tree species, Cordia alliodora (Boraginaceae). Journal of Biogeography 40: 693–706. [Google Scholar]
- Serrano-Serrano ML, Hernández-Torres J, Castillo-Villamizar G, Debouck DG, Chacón Sánchez MI. 2010. Gene pools in wild Lima bean (Phaseolus lunatus L.) from the Americas: evidence for an Andean origin and past migrations. Molecular Phylogenetics and Evolution 54: 76–87. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey WB. 1976. Papaya In: Simmonds NW, ed. Evolution of crop plants. London: Longman, 21–24. [Google Scholar]
- Tajima F. 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1996. The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terán S, Rasmussen CH. 1995. Genetic diversity and agricultural strategy in 16th century and present-day Yucatecan milpa agriculture. Biodiversity and Conservation 4: 363–381. [Google Scholar]
- Toledo VM. 1982. Pleistocene changes of vegetation in tropical Mexico In: Prance GT, ed. Biological diversification in the Tropics: Proceedings of the Fifth International Symposium of the Association for Tropical Biology, Caracas. New York: Columbia University Press, 93–111. [Google Scholar]
- Toledo VM, Ordóñez MJ. 1993. The biodiversity scenario of Mexico: a review of terrestrial habitats In: Ramamoorthy TP, Bye TP, Lot A, Fa JE, eds. Biological diversity of Mexico: origins and distribution. New York: Oxford University Press, 757–777. [Google Scholar]
- Twyford AD, Kidner CA, Harrison N, Ennos R. 2013. Population history and seed dispersal in widespread Central American Begonia species (Begoniaceae) inferred from plastome-derived microsatellite markers. Botanical Journal of the Linnean Society 17: 260–276. [Google Scholar]
- SAGARPA. 2009. Estudio de oportunidades de Mercado e inteligenica commercial internacional de la papaya Mexicana e identificacion de necesidades de infraestructura logistica. http://www.sagarpa.gob.mx/agronegocios/Documents/Estudios_promercado/PAPAYA2009.pdf. (last accessed 12 January 2016).
- Scotti-Saintagne C, Dick CW, Caron H, et al. 2013. Phylogeography of a species complex of lowland Neotropical rain forest trees (Carapa, Meliaceae). Journal of Biogeography 40: 676–692. [Google Scholar]
- Silva-Rosales L, González de León D, Guzmán-González S, Chauvet M. 2010. Why there is no transgenic papaya in Mexico. Transgenic Plant Journal 4: 45–51. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. [Google Scholar]
- Vavilov NI. 1926. The centres of origin of cultivated plants. Bulletin of Applied Botany of Genetics and Plant Breeding 16: 1–248. [Google Scholar]
- Vázquez-Domínguez E, Arita HT. 2010. The Yucatan península: biogeographical story 65 million years in the making. Ecography 33: 212–219. [Google Scholar]
- Vega-Frutis R, Guevara R. 2009. Different arbuscular mycorrhizal interactions in male and female plants of wild Carica papaya L. Plant Soil 322: 165–176. [Google Scholar]
- Wheeler GL, Dorman HE, Buchanan A, Challangundla L, Wallace LE. 2014. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Applications in Plant Sciences 2: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Hou S, Hobza R, Feltus FA, et al. 2007. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Molecular Genetics and Genomics 278: 177–185. [DOI] [PubMed] [Google Scholar]