Abstract
The complete mitochondrial genome (mitogenome) of Leucoma salicis (Lepidoptera: Lymantriidae) was sequenced and annotated. It is a circular molecule of 15,334 bp, containing the 37 genes usually present in insect mitogenomes. All protein-coding genes (PCGs) are initiated by ATN codons, other than cox1, which is initiated by CGA. Three of the 13 PCGs had an incomplete termination codon, T or TA, while the others terminated with TAA. The relative synonymous codon usage of the 13 protein-coding genes (PCGs) was consistent with those of published lepidopteran sequences. All tRNA genes had typical clover-leaf secondary structures, except for the tRNASer (AGN), in which the dihydrouridine (DHU) arm could not form a stable stem-loop structure. The A + T-rich region of 325 bp had several distinctive features, including the motif ‘ATAGA’ followed by an 18 bp poly-T stretch, a microsatellite-like (AT)7 element, and an 11-bp poly-A present immediately upstream of tRNAMet. Relationships among 32 insect species were determined using Maximum Likelihood (ML), Neighbor Joining (NJ) and Bayesian Inference (BI) phylogenetic methods. These analyses confirm that L. salicis belongs to the Lymantriidae; and that Lymantriidae is a member of Noctuoidea, and is a sister taxon to Erebidae, Nolidae and Noctuidae, most closely related to Erebidae.
Leucoma salicis is a moth that is mainly distributed in China, Korea and Japan. It is a notorious plant pest and causes considerable economic losses. It typically consumes willow and tea leaves, influencing quality and quantity of tea products1; and damages roadside and garden trees in urban areas. Traditionally, the identification of this species was based on morphological characteristics of adult moths2. However, the moth appears mainly in June to August, the rest of its life go through egg and larva stages (which has no easily identifying morphological features), requiring eggs and larvae to be reared to adult stage for identification, which is time consuming and labor intensive. Molecular methods for identification are under development, including polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP)3. Most previous work on L. salicis has focused on sex pheromone synthesis4, or the nuclear polyhedrosis virus that infects larvae5. Previous studies have not focused on the mitochondrial genome, which can provide systematically-informative information for identification, phylogenetic analysis and evolutionary studies on L. salicis.
Insect mitochondrial DNA (mtDNA) is a double-stranded, circular molecule, ranging in size from 14 to 20 kb. It usually contains a conserved set of 37 genes, including seven NADH dehydrogenase (nad1-nad6 and nad4L), three cytochrome c oxidase (cox1-cox3), two ATPase (atp6 and atp8), one cytochrome b (cob), two ribosomal RNA (rrnL and rrnS), 22 transfer RNA (tRNA) genes, and an adenine (A) + thymine (T)-rich region that contains initiation sites for transcription and replication of the genome6,7. Due to its simple genomic organization, high rate of evolution, and almost unambiguous orthology, mtDNA is typically considered to be an informative molecular marker for species identification and in studies of phylogenetic relationships and population structure8,9.
A better understanding of the lepidopteran mitochondrial genome requires expanded taxon sampling. Lepidoptera contains more than 160,000 described species, classified into 45–48 superfamilies10. Lymantriidae includes about 360 genera and over 2500 species, many of which are agriculturally important. Only eight species have completely-sequenced mitogenomes that are publically available in GenBank, despite the large species diversity in the family. In this study, we sequenced and annotated the complete mitogenome sequence of L. salicis, and compared it with those of other members of Lymantriidae. Our results provide novel methods for species identification of an important pest, as well as phylogenetically-informative sequence data that addresses the position of L. salicis within Noctuoidea.
Results
Geno me organization and composition
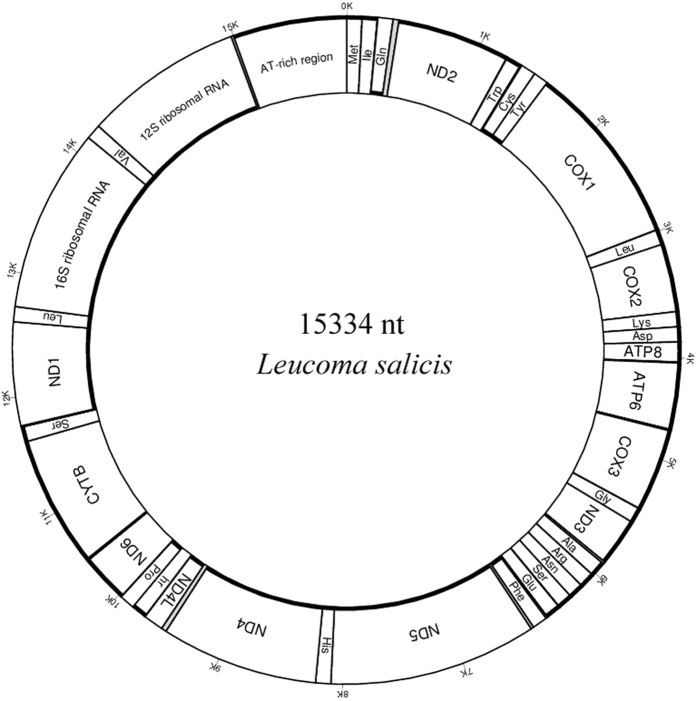
The mitogenome of L. salicis was a circular DNA molecule, 15,334 bp in length (Fig. 1). It contained the typical insect mitogenome set of 22 tRNAs, 13 PCGs (nad1-6, nad4L, cox1-3, cob, atp6 and atp8), two rRNAs (rrnS and rrnL), and the non-coding A + T-rich region (Table 1). Nucleotide composition was highly A + T biased (A: 42.07%, T: 38.57%, G: 7.22%, C: 12.14%; Table 2). Nucleotide BLAST (blastn) of the entire L. salicis mitogenome against GenBank returned sequence identities with closely related species of 79% (Lachana alpherakii), 78% (Euproctis pseudoconspersa), 78% (Gynaephora menyuanensis), and 77% (Lymantria dispar) (Table S1).
Figure 1. Map of the mitogenome of L. salicis.
tRNA genes are labeled according to the IUPAC-IUB three-letter amino acids; cox1, cox2 and cox3 refer to the cytochrome c oxidase subunits; cob refers to cytochrome b; nad1-nad6 refer to NADH dehydrogenase components; rrnL and rrnS refer to ribosomal RNAs.
Table 1. Summary of characteristics of the mitogenome of L. salicis.
| Gene | Direction | Location | Size | Anti codon | Start codon | Stop codon | Intergenic Nucleotides |
|---|---|---|---|---|---|---|---|
| tRNAMet | F | 1–66 | 66 | CAT | — | — | 0 |
| tRNAIle | F | 67–134 | 68 | GAT | — | — | −3 |
| tRNAGln | R | 132–200 | 69 | TTG | — | — | 47 |
| ND2 | F | 248–1233 | 986 | — | ATT | TAA | 3 |
| tRNATrp | F | 1237–1309 | 73 | TCA | — | — | −8 |
| tRNACys | R | 1302–1370 | 69 | GCA | — | — | 0 |
| tRNATyr | R | 1371–1442 | 72 | GTA | — | — | 1 |
| COI | F | 1453–2987 | 1535 | — | CGA | T | 2 |
| tRNALeu(UUR) | F | 2993–3059 | 67 | TAA | — | — | 0 |
| COII | F | 3060–3740 | 681 | — | ATT | T | 0 |
| tRNALys | F | 3741–3811 | 71 | CTT | — | — | 0 |
| tRNAAsp | F | 3812–3878 | 67 | GTC | — | — | 0 |
| ATP8 | F | 3879–4040 | 162 | — | ATA | TAA | −7 |
| ATP6 | F | 4034–4711 | 678 | — | ATG | TAA | 14 |
| COIII | F | 4726–5514 | 789 | — | ATG | TAA | 2 |
| tRNAGly | F | 5517–5582 | 66 | TCC | — | — | 0 |
| ND3 | F | 5583–59336 | 354 | — | ATT | TAA | 10 |
| tRNAAla | F | 5947–6017 | 71 | TGC | — | — | −1 |
| tRNAArg | F | 6017–6084 | 68 | TCG | — | — | 1 |
| tRNAAsn | F | 6086–6152 | 67 | GTT | — | — | 0 |
| tRNASer(AGN) | F | 6153–6219 | 67 | GCT | — | — | 17 |
| tRNAGlu | F | 6237–6301 | 65 | TTC | — | — | 0 |
| tRNAPhe | R | 6302–6368 | 67 | GAA | — | — | 20 |
| ND5 | R | 6389–8103 | 1715 | — | — | — | 8 |
| tRNAHis | R | 8112–8175 | 64 | GTG | — | — | 1 |
| ND4 | R | 8177–9516 | 1341 | — | ATG | TA | 15 |
| ND4L | R | 9532–9825 | 294 | — | — | — | 5 |
| tRNAThr | F | 9831–9897 | 67 | TGT | — | — | 0 |
| tRNAPro | R | 9898–9965 | 68 | TGG | — | — | 2 |
| ND6 | F | 9968–10514 | 547 | — | ATA | TAA | 8 |
| Cytb | F | 10523–11655 | 1133 | — | ATG | TAA | 1 |
| tRNASer(UCN) | F | 11657–11721 | 65 | TGA | — | — | 10 |
| ND1 | R | 11732–12684 | 953 | — | ATT | TAA | 0 |
| tRNALeu(CUN) | R | 12685–12755 | 71 | TAG | — | — | 0 |
| lrRNA | R | 12756–14099 | 1344 | — | — | — | 0 |
| tRNAVal | R | 14100–14169 | 70 | TAC | — | — | 0 |
| srRNA | R | 14170–15009 | 840 | — | — | — | 0 |
| A + T-rich region | — | 15010–15334 | 325 | — | — | — | — |
Table 2. Composition and skew in different lepidopteran mitogenomes.
| Species | Size (bp) | A% | G% | T% | C% | A + T % | AT skewness | GC skewness |
|---|---|---|---|---|---|---|---|---|
| Whole genome | ||||||||
| L. salicis | 15334 | 42.07 | 7.22 | 38.57 | 12.14 | 80.64 | 0.043 | −0.254 |
| C. agnata | 15261 | 39.58 | 7.71 | 41.52 | 11.2 | 81.1 | −0.023 | −0.184 |
| H. cunea | 15481 | 40.58 | 7.55 | 39.81 | 12.06 | 80.39 | 0.009 | −0.229 |
| G. menyuanensis | 15770 | 40.88 | 6.77 | 40.6 | 11.75 | 81.48 | 0.003 | −0.268 |
| L. dispar | 15507 | 40.38 | 7.61 | 39.26 | 12.5 | 79.64 | 0.014 | −0.243 |
| E. pseudoconspersa | 15461 | 40.42 | 7.61 | 39.51 | 12.46 | 79.93 | 0.011 | −0.241 |
| G. argentata | 15337 | 39.64 | 7.56 | 42.05 | 10.75 | 81.69 | −0.029 | −0.174 |
| A. formosae | 15463 | 38.67 | 7.53 | 40.83 | 12.98 | 79.49 | −0.027 | −0.265 |
| P. distinctalis | 15354 | 41.04 | 7.49 | 41.22 | 10.24 | 82.27 | −0.002 | −0.155 |
| L. haraldusalis | 15213 | 40.47 | 7.66 | 41.04 | 10.83 | 81.52 | −0.006 | −0.171 |
| B. thibetaria | 15484 | 42.38 | 7.55 | 37.24 | 12.83 | 79.62 | 0.064 | −0.259 |
| R. menciana | 15301 | 41.42 | 7.82 | 37.45 | 13.31 | 78.86 | 0.050 | −0.259 |
| B. mori | 15666 | 43.09 | 7.31 | 38.26 | 11.34 | 81.35 | 0.059 | −0.216 |
| S. morio | 15299 | 40.64 | 7.58 | 40.53 | 11.26 | 81.17 | 0.0013 | −0.195 |
| S. lechriaspis | 15368 | 39.86 | 7.63 | 41.34 | 11.17 | 81.19 | −0.018 | −0.188 |
| C. benjaminii | 15272 | 40.08 | 7.52 | 40.7 | 11.7 | 80.78 | −0.007 | −0.217 |
| PCG | ||||||||
| L. salicis | 11171 | 42.24 | 7.89 | 37.16 | 12.71 | 79.39 | 0.063 | −0.233 |
| C. agnata | 11238 | 39.12 | 8.37 | 40.79 | 11.72 | 79.91 | −0.020 | −0.166 |
| H. cunea | 11205 | 39.99 | 8.35 | 38.6 | 13.06 | 78.59 | 0.017 | −0.219 |
| G. menyuanensis | 11228 | 40.37 | 7.5 | 39.41 | 12.72 | 79.78 | 0.012 | −0.258 |
| L. dispar | 11236 | 39.52 | 8.44 | 38.18 | 13.62 | 77.71 | 0.017 | −0.234 |
| E. pseudoconspersa | 11187 | 3969 | 8.43 | 38.3 | 13.58 | 77.99 | 0.017 | −0.233 |
| G. argentata | 11203 | 39.05 | 8.29 | 41.27 | 11.38 | 80.33 | −0.027 | −0.157 |
| A. formosae | 11,217 | 38.18 | 8.28 | 39.62 | 13.92 | 77.8 | −0.018 | −0.254 |
| P. distinctalis | 11189 | 40.54 | 8.12 | 40.53 | 10.81 | 81.07 | 0 | −0.142 |
| L. haraldusalis | 11,193 | 39.88 | 8.47 | 40.16 | 11.49 | 80.04 | −0.003 | −0.151 |
| B. thibetaria | 11212 | 41.66 | 8.36 | 35.94 | 14.04 | 77.6 | 0.073 | −0.253 |
| R. menciana | 11225 | 40.97 | 8.58 | 36.12 | 14.33 | 77.1 | 0.063 | −0.251 |
| B. mori | 11177 | 42.93 | 8.16 | 36.64 | 12.28 | 79.57 | 0.079 | −0.201 |
| S. morio | 11179 | 40.28 | 8.27 | 39.56 | 11.89 | 79.84 | 0.009 | −0.179 |
| S. lechriaspis | 11258 | 39.31 | 8.35 | 40.41 | 11.93 | 79.72 | −0.013 | −0.176 |
| C. benjaminii | 11153 | 39.44 | 8.23 | 39.74 | 12.59 | 79.18 | −0.003 | −0.209 |
| tRNA | ||||||||
| L. salicis | 1498 | 43.19 | 6.88 | 40.72 | 9.21 | 83.91 | 0.029 | −0.145 |
| C. agnata | 1477 | 41.23 | 8.19 | 40.22 | 10.36 | 81.45 | 0.012 | −0.117 |
| H. cunea | 1474 | 41.86 | 7.87 | 39.89 | 10.38 | 81.75 | 0.024 | −0.138 |
| G. menyuanensis | 1504 | 41.29 | 7.38 | 41.76 | 9.57 | 83.05 | −0.006 | −0.129 |
| L. dispar | 1466 | 41.41 | 7.98 | 39.5 | 10.91 | 80.9 | 0.024 | −0.155 |
| E. pseudoconspersa | 1466 | 41.41 | 8.19 | 40.18 | 10.23 | 81.58 | 0.015 | −0.111 |
| G. argentata | 1469 | 41.32 | 8.24 | 40.23 | 10.21 | 81.55 | 0.013 | −0.107 |
| A. formosae | 1457 | 40.43 | 7.96 | 40.36 | 11.26 | 80.78 | 0.001 | −0.172 |
| P. distinctalis | 1536 | 42.19 | 8.14 | 39.78 | 9.9 | 81.97 | 0.029 | −0.098 |
| L. haraldusalis | 1451 | 41.08 | 7.86 | 41.42 | 9.65 | 82.49 | −0.004 | −0.102 |
| B. thibetaria | 1478 | 42.08 | 7.85 | 39.24 | 10.83 | 81.33 | 0.035 | −0.160 |
| R. menciana | 1485 | 41.08 | 8.08 | 39.93 | 10.91 | 81.01 | 0.014 | −0.149 |
| B. mori | 1470 | 42.04 | 7.89 | 39.52 | 10.54 | 81.56 | 0.031 | −0.144 |
| S. morio | 1463 | 40.6 | 8.2 | 41.01 | 10.18 | 81.61 | −0.005 | −0.108 |
| S. lechriaspis | 1516 | 41.03 | 7.92 | 41.09 | 9.96 | 82.12 | −0.001 | −0.114 |
| C. benjaminii | 1467 | 40.9 | 8.04 | 40.49 | 10.57 | 81.39 | 0.005 | −0.136 |
| rRNA | ||||||||
| L. salicis | 2184 | 41.39 | 5.04 | 40.8 | 12.77 | 82.19 | 0.007 | −0.434 |
| C. agnata | 2112 | 40.01 | 5.07 | 44.65 | 10.27 | 84.66 | −0.055 | −0.339 |
| H. cunea | 2234 | 42.08 | 4.92 | 42.75 | 10.25 | 84.83 | −0.008 | −0.351 |
| G. menyuanensis | 2311 | 41.89 | 4.28 | 42.84 | 10.99 | 84.73 | −0.011 | −0.439 |
| L. dispar | 2140 | 42.52 | 4.81 | 41.82 | 10.42 | 84.35 | 0.008 | −0.368 |
| E. pseudoconspersa | 2225 | 42.56 | 4.54 | 42.11 | 10.79 | 84.67 | 0.005 | −0.408 |
| G. argentata | 2165 | 40.6 | 4.76 | 45.13 | 9.52 | 85.73 | −0.053 | −0.333 |
| A. formosae | 2163 | 38.93 | 4.72 | 44.85 | 11.51 | 83.77 | −0.071 | −0.418 |
| P. distinctalis | 2174 | 41.31 | 5.34 | 44.02 | 9.34 | 85.33 | −0.032 | −0.272 |
| L. haraldusalis | 2121 | 442.2 | 4.67 | 43.33 | 9.81 | 85.53 | 4.664 | −0.355 |
| B. thibetaria | 2241 | 45.52 | 4.77 | 39.58 | 10.13 | 85.1 | 0.070 | −0.360 |
| R. menciana | 2147 | 43.04 | 4.84 | 40.71 | 11.41 | 83.74 | 0.028 | −0.404 |
| B. mori | 2161 | 43.73 | 4.58 | 41.09 | 10.6 | 84.82 | 0.031 | −0.397 |
| S. morio | 2152 | 41.73 | 4.83 | 43.08 | 10.36 | 84.8 | −0.016 | −0.364 |
| S. lechriaspis | 2160 | 41.71 | 4.95 | 43.84 | 9.49 | 85.56 | −0.025 | −0.314 |
| C. benjaminii | 2132 | 41.7 | 4.88 | 43.76 | 9.66 | 85.46 | −0.024 | −0.329 |
| AT RICH | ||||||||
| L. salicis | 325 | 34.46 | 2.46 | 57.23 | 5.85 | 91.69 | −0.248 | −0.408 |
| C. agnata | 334 | 46.71 | 1.5 | 46.71 | 5.09 | 93.41 | 0.000 | −0.545 |
| H. cunea | 357 | 45.66 | 1.12 | 49.3 | 3.92 | 94.96 | −0.038 | −0.556 |
| G. menyuanensis | 449 | 43.65 | 2.45 | 49.67 | 4.23 | 93.32 | −0.065 | −0.266 |
| L. dispar | 371 | 44.74 | 2.43 | 49.6 | 3.23 | 94.34 | −0.052 | −0.141 |
| E. pseudoconspersa | 388 | 43.56 | 2.32 | 50.26 | 3.87 | 93.81 | −0.071 | −0.250 |
| G. argentata | 340 | 43.24 | 1.47 | 52.06 | 3.24 | 95.29 | −0.093 | −0.376 |
| A. formosae | 482 | 42.95 | 2.9 | 49.79 | 4.36 | 92.74 | −0.074 | −0.201 |
| P. distinctalis | 349 | 46.13 | 1.15 | 49 | 3.72 | 95.13 | −0.030 | −0.528 |
| L. haraldusalis | 310 | 45.81 | 0.97 | 50.32 | 2.9 | 96.13 | −0.047 | −0.499 |
| B. thibetaria | 350 | 44.29 | 2.57 | 48.29 | 4.86 | 92.57 | −0.043 | −0.308 |
| R. menciana | 357 | 43.7 | 3.36 | 47.34 | 5.6 | 91.04 | −0.040 | −0.250 |
| B. mori | 494 | 44.74 | 1.82 | 50.61 | 2.83 | 95.34 | −0.062 | −0.217 |
| S. morio | 316 | 44.3 | 2.53 | 48.42 | 4.75 | 92.72 | −0.044 | −0.305 |
| S. lechriaspis | 441 | 40.36 | 2.49 | 52.38 | 4.76 | 92.74 | −0.130 | −0.313 |
| C. benjaminii | 293 | 46.42 | 3.07 | 45.73 | 4.78 | 92.15 | 0.007 | −0.218 |
Protein-coding genes and codon usage
The PCG region formed 72.9% of the L. salicis mitogenome, and was 11,172 bp long. Nine of 13 PCGs (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6 and cob) were encoded on the H-strand, while the remaining four (nad5, nad4, nad4L and nad1) were encoded on the L-strand. Each PCG was initiated by a canonical ATN codon, except for cox1 (Table 1), which was initiated by a CGA codon. Ten of 13 PCGs used a typical TAA termination codon; but cox1 and cox2 terminated with a single T and nad4 terminated with TA (Table 1).
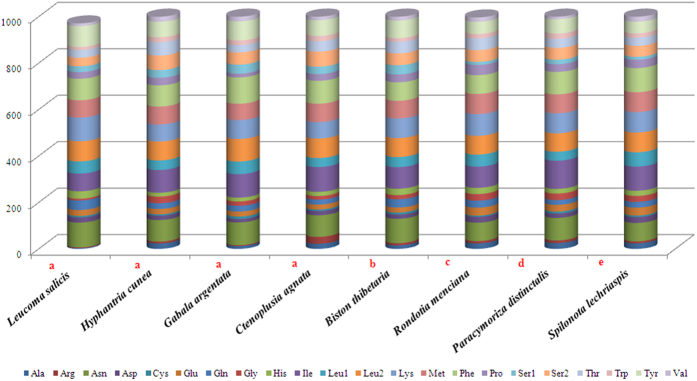
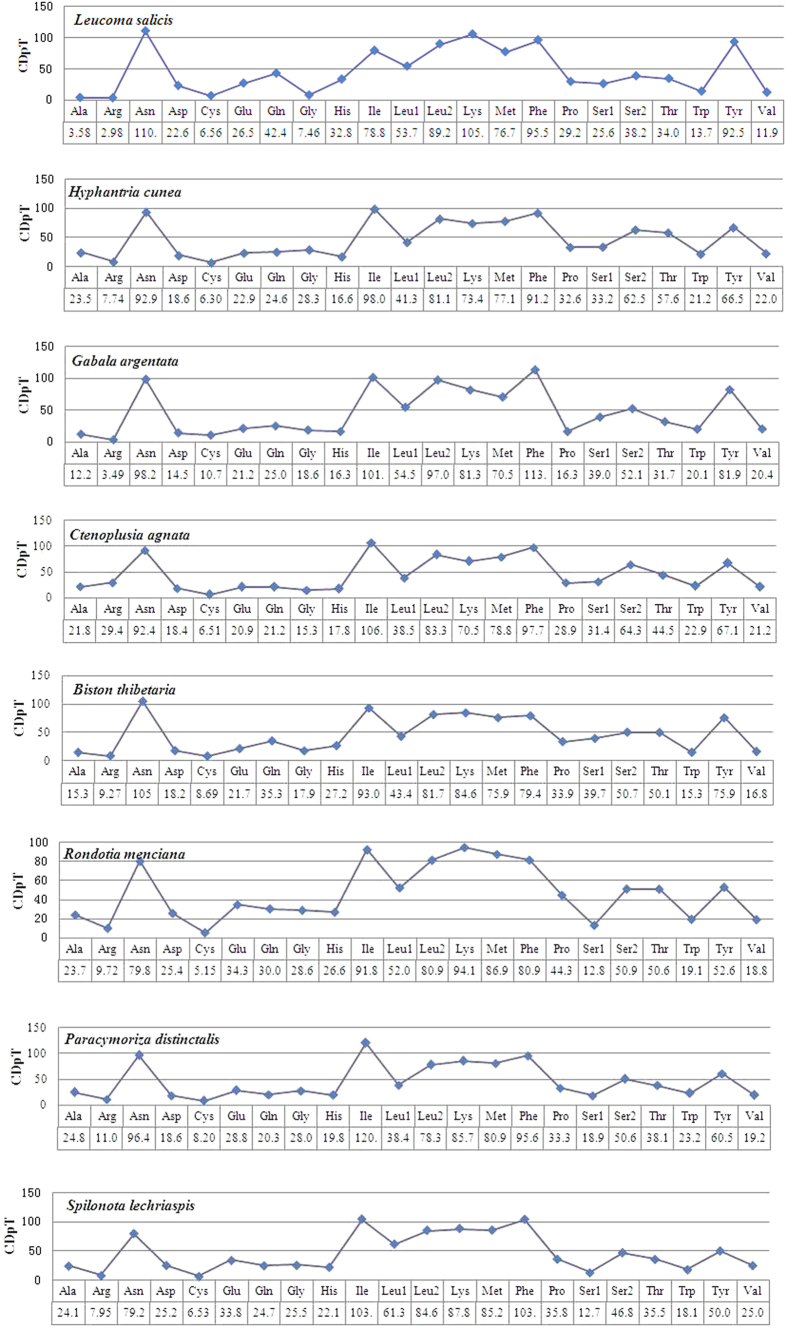
Relative synonymous codon usage (RSCU) analysis of PCGs in L. salicis revealed that the codons encoding Asn, Ile, Leu (UUA, UUG), Lys, Tyr and Phe were the most frequently present, while those encoding Cys and Arg were rare (Fig. 2). In the PCGs of the eight moth species examined, codon distributions and amino acid content were largely consistent among species (Fig. 3). Codons with A or T in the third position were overused in comparison to other synonymous codons: for example, the codons for valine GTC and GTG were rare, while the synonymous codons GTT and GTA were prevalent (Fig. 4). All used codons were present in the PCGs of the L. salicis mitogenome, except for CGC and GGC. This is similar to codon usage in Hyphantria cunea, Spilonota lechriaspis, and Gabala argentata, which respectively lack CGG and CGC, GCG and CGG, and CGG and CGC.
Figure 2. Comparison of codon usage within the mitochondrial genome of members of the Lepidoptera.
Lowercase letters (a,b,c,d and e) above species names represent the superfamily to which the species belongs (a: Noctuoidea, b: Geometroidea, c: Bombycoidea, d: Pyraloidea, e: Tortricoidea).
Figure 3. Codon distribution in members of the Lepidoptera.
CDspT = codons per thousand codons.
Figure 4. Relative Synonymous Codon Usage (RSCU) of the mitochondrial genome of five superfamilies in the Lepidoptera.
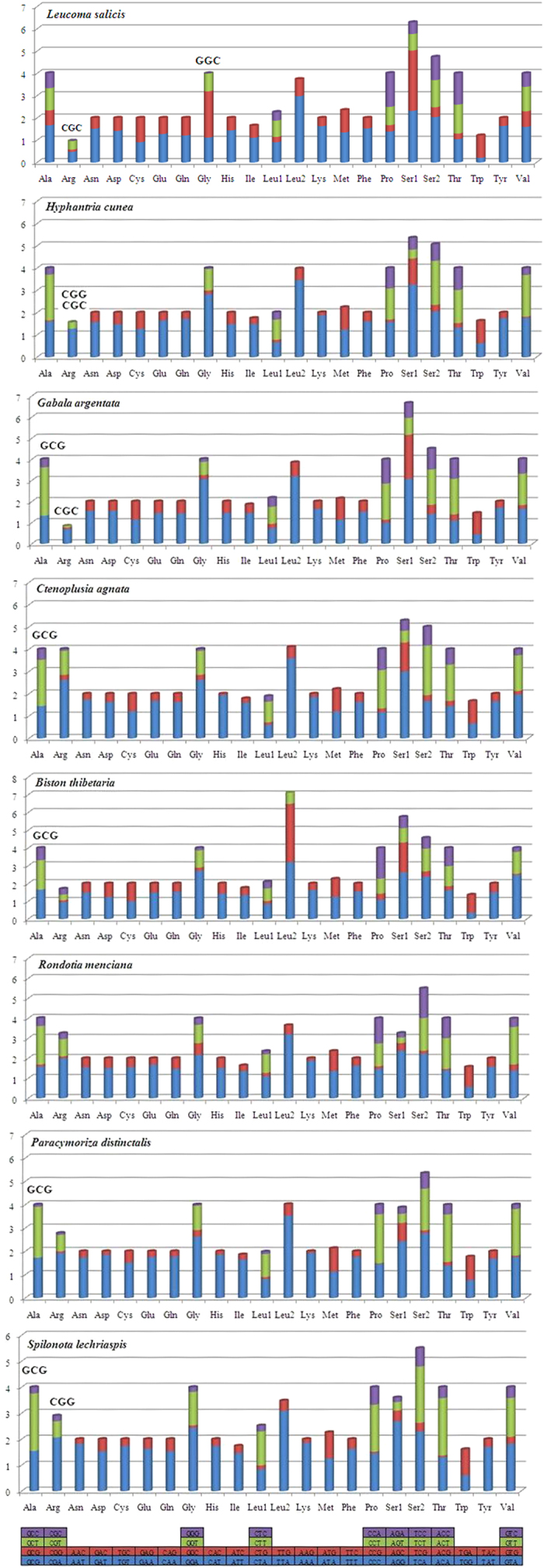
Codon families are plotted on the x-axis. Codons indicated above the bar are not present in the mitogenome.
Ribosomal RNA and transfer RNA genes
The large (rrnL) and small (rrnS) ribosomal RNA subunit genes of L. salicis were located between the tRNALeu1(CUN)/tRNAVal and the tRNAVal/A + Trich regions, respectively (Fig. 1, Table 1). The rrnL gene was 1,344 bp long, while rrnS was 840 bp long. A + T content of the rRNA genes was 83.91%. AT and GC skews were positive (0.029) and negative (−0.144), respectively.
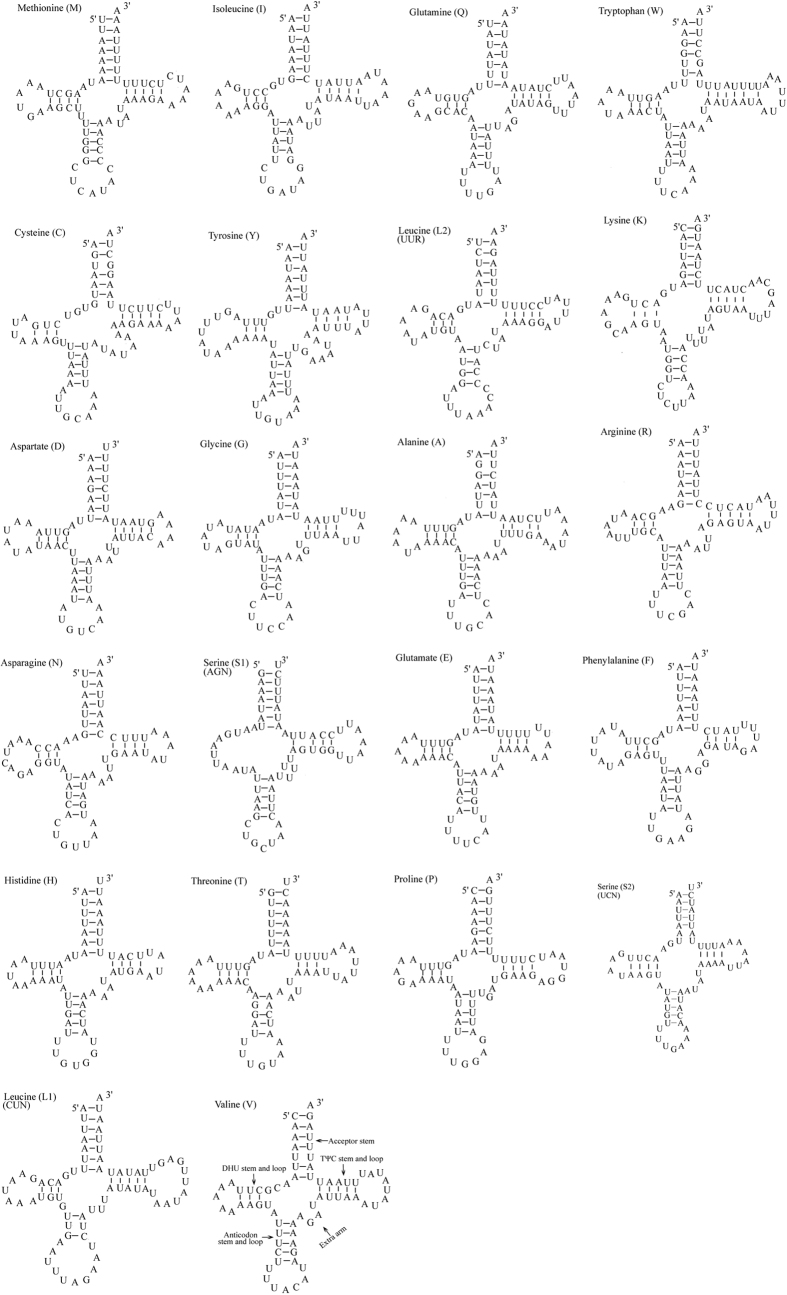
The L. salicis mitogenome included 22 tRNA genes, ranging from 64 bp (tRNAHis) to 73 bp (tRNATrp) long. Of these, 14 genes were encoded on the H-strand and eight on the L-strand (Table 1). The tRNA genes were highly A + T biased (82.19%) with a positive AT-skew (0.007) (Table 2). All the tRNAs possessed a typical clover-leaf secondary structure, except tRNASer(AGN), which lacks the dihydrouridine (DHU) arm and forms a simple loop (Fig. 5). Ten of the tRNA genes were each found to have 11 G-U mismatches in their respective secondary structures, which form a weak bond. Ten U-U mismatches were present in the respective amino acid acceptor stems of tRNAGln, tRNATrp, tRNALeu(UUR), tRNAAla, tRNAThr, tRNALeu(CUN), and tRNAVal (Fig. 5). All tRNA secondary structures of the tRNA genes were calculated using the tRNAscan-SE program.
Figure 5. Predicted secondary structures of the 22 tRNA genes of the L. salicis mitogenome.
Overlapping and intergenic spacer regions
We identified four overlapping gene sequences, varying from 1 bp to 8 bp, making up 19 bp in total. The longest overlapping region was 8 bp between tRNATrp and tRNACys; there was a 7 bp overlap between atp8 and atp6; 3 bp overlap between tRNAIle and tRNAGln, and 1 bp between tRNAAla and tRNAArg (Table 1).
Intergenic spacers were spread over 18 regions, and ranged in length from 1 bp to 47 bp. The longest (47 bp) contained an A + T-rich region and occurred between tRNAGln and nad2. The 10 bp spacer region between tRNASer (UCN) and nad1 included an ‘ATACTAA’ motif (Fig. 6A).
Figure 6.
(A) Alignment of the intergenic spacer region between tRNASer (UCN) and nad1 of several Lepidopteran insects. (B) Features present in the A + T-rich region of L. salicis. The ‘ATATG’ motif is shaded. The poly-A stretch is double underlined, and the poly-T stretch is underlined. The single microsatellite T/A repeat sequence is indicated by dotted underlining.
The A + T-rich region
The 325 bp long A + T-rich region of L. salicis was located between the rrnS and tRNAMet genes (Table 1). A + T content in the A + T-rich region was 91.69%, and both AT (−0.248) and GC (−0.408) skews were negative (Table 2). The A + T-rich region did not contain long repeats, though some short repeating sequences scattered over the entire region were present: an ‘ATAGA’ motif followed by an 18 bp poly-T stretch, a microsatellite-like (AT)7 and a poly-A element upstream of the tRNAMet gene (Fig. 6B).
Phylogenetic relationships
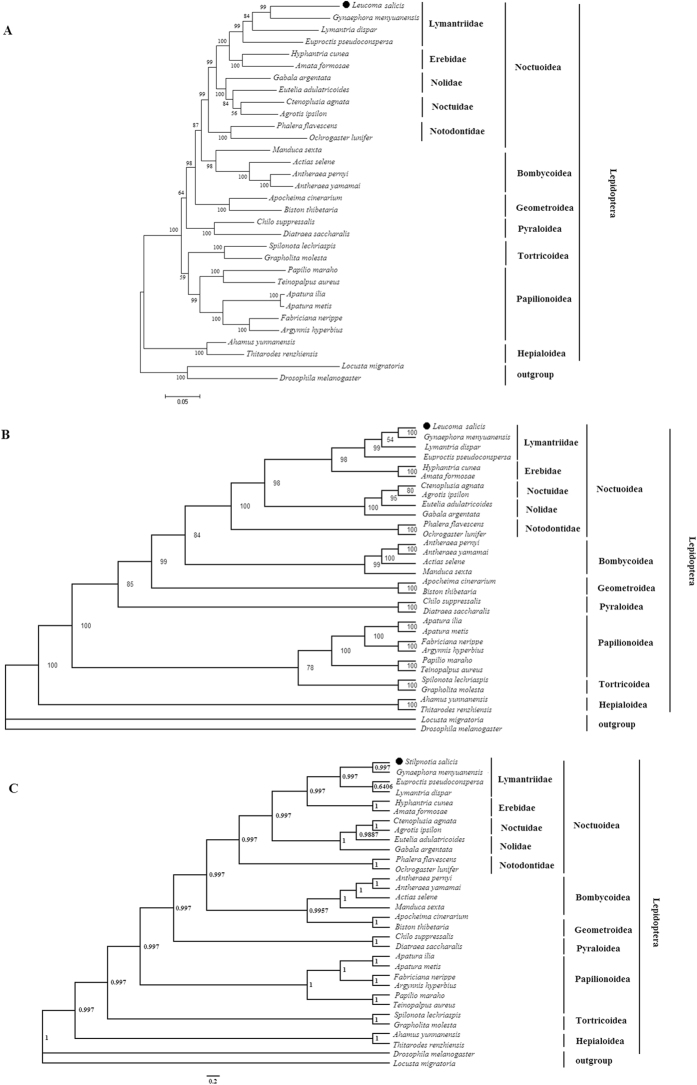
We established phylogenetic relationships among 32 insects (Table 3), based on nucleotide sequences of 13 PCGs, using Maximum Likelihood (ML), Neighbor Joining (NJ) and Bayesian Inference (BI) methods. Species clustered by family (Fig. 7A, B and C). Within Lymantriidae, L. salicis was most closely related to G. menyuanensis. Lymantriidae clustered with Erebidae, while Noctuidae clustered with Nolidae. Noctuoidea was most closely related to Bombycoidea in ML and NJ trees, while in the BI tree Bombycoidea was most closely related to Geometroidea. Papilionoidea and Tortricoidea branched together in ML and NJ methods, but were separated from each other in the BI tree.
Table 3. Details of the lepidopteran mitogenomes used in this study.
| Superfamily | Family | Species | Size (bp) | GenBank No. |
|---|---|---|---|---|
| Bombycoidea | Bombycidae | Bombyx mori | 15666 | KM875545.1 |
| Rondotia menciana | 15301 | KC881286.1 | ||
| Saturniidae | Actias selene | 15,236 | NC_018133 | |
| Antheraea pernyi | 15,566 | AY242996 | ||
| Antheraea yamamai | 15,338 | NC_012739 | ||
| Sphingidae | Sphinx morio | 15299 | KC470083.1 | |
| Manduca sexta | 15,516 | NC_010266 | ||
| Noctuoidea | Lymantriidae | Lymantria dispar | 15507 | GU994783.1 |
| Gynaephora menyuanensis | 15770 | KC185412.1 | ||
| Euproctis pseudoconspersa | 15461 | KJ716847.1 | ||
| Leucoma salicis | 15334 | This study | ||
| Noctuidae | Ctenoplusia agnata | 15261 | KC414791.1 | |
| Agrotis ipsilon | 15,377 | KF163965 | ||
| Nolidae | Eutelia adulatricoides | 15,360 | KJ185131 | |
| Gabala argentata | 15,337 | KJ410747 | ||
| Erebidae | Amata formosae | 15463 | KC513737 | |
| Hyphantria cunea | 15481 | GU592049.1 | ||
| Notodontidae | Phalera flavescens | 15,659 | NC_016067 | |
| Ochrogaster lunifer | 15,593 | NC_011128 | ||
| Geometroidea | Geometridae | Apocheima cinerarium | 15,722 | KF836545 |
| Biston thibetaria | 15,484 | KJ670146.1 | ||
| Pyraloidea | Crambidae | Chilo suppressalis | 15,395 | NC_015612 |
| Diatraea saccharalis | 15,490 | NC_013274 | ||
| Paracymoriza distinctalis | 15354 | KF859965.1 | ||
| Pyralidae | Lista haraldusalis | 15213 | NC_024535 | |
| Tortricoidea | Tortricidae | Spilonota lechriaspis | 15368 | HM204705.1 |
| Grapholita molesta | 15,717 | NC_014806 | ||
| Papilionoidea | Papilionidae | Papilio maraho | 16,094 | NC_014055 |
| Teinopalpus aureus | 15,242 | NC_014398 | ||
| Nymphalidae | Apatura ilia | 15,242 | NC_016062 | |
| Apatura metis | 15,236 | NC_015537 | ||
| Fabriciana nerippe | 15,140 | NC_016419 | ||
| Argynnis hyperbius | 15,156 | NC_015988 | ||
| Hepialoidea | Hepialidae | Thitarodes renzhiensis | 16,173 | NC_018094 |
| Ahamus yunnanensis | 15,816 | NC_018095 | ||
| Hesperioidea | Hesperiidae | Choaspes benjaminii | 15272 | JX101620.1 |
Figure 7.
(A) Tree showing the phylogenetic relationships among 32 species, constructed using Maximum Likelihood with 1000 bootstrap replicates. (B) Neighbor Joining (NJ) tree, with 1000 bootstrap replicates. (C) Tree constructed using Bayesian Inference (BI) MCMC consensus tree, with posterior probabilities shown at nodes. Drosophila melanogaster (NC_025936) and Locusta migratoria (NC_002084) were used as outgroups.
Discussion
At the family level, the length of the L. salicis mitogenome (15,334 bp) is marginally smaller than that of Euproctis pseudoconspersa (15,461 bp), but it falls within the range (15,140–16,173 bp) of other known lepidopteran mitogenomes. Gene order and orientation are the same as in previously-sequenced Lymantriidae. Nucleotide BLAST (blastn) result of the entire mitogenome against closely related species revealed that L. salicis has a high similarity with the Lymantriidae species (77% in L. dispar–79% in L. alpherakii). The conserved regions lie in 22 tRNAs and 13 PCGs, while A + T-rich region varies in these species. These remarkable characteristics have been reported in other lepidopteran species7 and could be used as potential markers for identification at genus and species level in recent molarcular techniques. The highly A + T biased nucleotide composition is within the range of previously sequenced lepidopterans (79.64% in L. dispar–81.48% in G. menyuanensis). The positive AT skew (0.043) observed here, indicating the presence of more As than Ts, is similar to that seen in many lepidopterans, including L. dispar (0.014), Rondotia menciana (0.050), and Biston thibetaria (0.064) (Table 2). It is slightly higher than that of other sequenced mitogenomes in Noctuoidea, including Ctenoplusia agnata (−0.023), G. menyuanensis (0.003) and E. pseudoconspersa (0.011). A similar trend has been observed in other lepidopteran superfamilies such as Bombycoidea, where AT skew varies from 0.001 (Sphinx morio) to 0.059 (Bombyx mori)11. In all sequenced lepidopteran mitogenomes, GC skew ranges from −0.268 in G. menyuanensis to −0.155 in Paracymoriza distinctalis (Table 2). The L. salicis mitogenome is moderately skewed (−0.254), showing the presence of more Cs than Gs.
The AT skew value (0.063) of the protein-coding gene region in the L. salicis mitogenome is higher than that of several previously sequenced mitogenomes. Its negative GC skew (−0.234) is similar to that seen in other animals. Cox1 is thought to initiate with CGA, as found in other lepidopteran insects12,13. Cox1 and cox2 terminate with a single T, while nad4 terminates with TA. Similar results have been documented in several sequenced lepidopteran mitogenomes, including Artogeia melete14, Phthonandria atrilineata15, Ochrogaster lunifer16, H. cunea17 and Amata emma18. The common termination codon TAA is usually created via post-transcriptional polyadenylation19. The relative synonymous codon usage of the 13 protein-coding genes (PCGs) in L. salicis is consistent with those of published lepidopteran sequences. Similarly, codons with A or T in the third codon position being overrepresented relative to other synonymous codons, is consistent with previous observations of lepidopterans9; likewise the absence or underrepresentation of high-GC codons18,20.
The A + T content (83.91%) of rRNA genes is similar to that seen in Lymantriidae (83.05% in G. menyuanensis). The positive AT (0.029) and negative GC (−0.144) skew seen in the L. salicis mitogenome has also been reported in several sequenced lepidopterans (Table 2). For example, H. cunea has a positive AT (0.024) and negative GC (−0.137) skew17; and L. dispar also has positive AT (0.023) and negative GC (−0.155) skew.
The secondary structure of L. salicis tRNASer(AGN) lacks the dihydrouridine (DHU) arm and forms a simple loop. This has also been observed in several other animal mitogenomes21, including those of insects15,22,23. Ten tRNA genes have 11 mismatches in their secondary structures; most of these are located in the acceptor, DHU and anticodon stems. In addition, tRNACys and tRNASer (UCN) contain an A-A mismatch in the anticodon stem. Unmatched base pairs observed in tRNA sequences can be corrected by RNA-editing mechanisms that are well known for arthropod mtDNA24.
Four overlapping sequences occur in the mitogenome of L. Salicis. The 7 bp overlap between atp8 and atp6 has been documented in several other lepidopteran mitogenomes25,26. The 10 bp intergenic spacer region containing an ‘ATACTAA’ motif, between tRNASer (UCN) and nad1, has also been documented in at least nine other species, suggesting that this region is highly conserved among most of the lepidopteran mtDNAs sequenced to date27.
The length of the A + T-rich region of L. salicis (325 bp) is shorter than those of G. menyuanensis (449), L. dispar (371), H. cunea (357) and B. thibetaria (350), and longer than those of Lista haraldusalis (310) and Choaspes benjaminii (293). Extra tRNA-like structures are often found in the A + T-rich region of lepidopteran mitogenomes. For example Antheraea yamamai has tRNASer(UCN)-like and tRNAPhe-like sequences, each with correct anticodon structure and forming a clover-leaf structure, which suggests that they may be functional, though each has several mismatches in both aminoacyl and anticodon stem regions28. Extra tRNA-like structures have not been seen in L. salicis. The presence of multiple tandem-repeat elements is described as being characteristic of insect A + T-rich regions29. Antheraea pernyi has a repeat element of 38 bp tandemly repeated six times25; and Cnaphalocrocis medinalis has a duplicated 25 bp repeat element25,30. Long conspicuous repeats were not observed in the A + T-rich region of L. salicis, though shorter repeating sequences, an ‘ATAGA’ motif and other features were. These characteristic features have each been found in previously sequenced lepidopteran species27,31,32.
In general, the L. salicis mitogenome contains several features in nucleotide composition, structure of tRNAs and PCGs as well as in the A + T rich region. Particularly in advanced technologies like PCR–RFLP methods3 and DNA barcodes33, these similarities and differences between L. salicis and other insects could be used as potential markers in species identification, especially the differences.
Phylogenetic relationships were established using Maximum Likelihood (ML) Neighbor Joining (NJ) and Bayesian Inference (BI) methods. Species clustered in families, and results were broadly consistent with previous work, e.g. Dong et al.26 and Dai et al.34. Results obtained from our analyses also supported the classification proposed by Fibiger and Lafontaine35, including within Lymantriidae a clade comprised of E. pseudoconspersa, L. salicis, L. dispar and G. menyuanensis. The present analysis showed that within Lymantriidae, L. salicis was most closely related to G. menyuanensis, which is consistent with a recent study on E. pseudoconspersa26. Interestingly, L. dispar is more closely related to G. menyuanensis than E. pseudoconspersa in ML and NJ trees (Fig. 7A and B), whereas in the BI consensus tree L. dispar and E. pseudoconspersa branch together with 0.6406 posterior probabilities (Fig. 7C). We conclude from the above results that differences between BI, ML and NJ methods generate different results on the relationship among different Noctuoidea species.
Because most previous classifications of Lymantriidae species have been based on morphological features, the precise position of Lymantriidae within the Noctuoidea is still unclear. Kitching has suggested that the Lymantriidae are the sister group to a paraphyletic Pantheidae, sharing apomorphies such as the presence of secondary setae in first instar larvae36. Zahiri et al. reclassified the Noctuoidea on the basis of molecular analyses, making the group currently named Lymantriinae a subfamily of Erebidae37. Our results suggest that Lymantriidae can be regarded as a sister group to other families (Erebidae, Nolidae and Noctuidae) in the Noctuoidea, being most closely related to Erebidae that is consistent with previous study of Fibiger and Lafontaine (2005) on higher Noctuoidea classification. They placed the Lymantriidae from a position in front of the Nolidae to a position after Arctiidae to reflect the close association of the arctiids and lymantriids, and moved the Nolidae, Arctiidae and Lymantriidae in front of the upgraded family Erebidae so that their close relationship with the “quadrifids’ is better reflected35. It is concluded that further studies are needed on sequencing and characterization of mitogenomes of the family Lymantriidae that will provide insight to classification of Noctuoidea.
At the level of superfamilies, Noctuoidea was closely related to Bombycoidea in our ML and NJ analyses, while in the BI tree, Bombycoidea was closely related to Geometroidea. Papilionoidea and Tortricoidea branched together in ML and NJ trees, but in the BI tree they formed separate branches, more in line with previous studies. Hepialoidea was the sister group to all other superfamilies, as found previously by Salvato et al.16 and Chai et al.38. While several previous studies have been undertaken on mitogenomes of Noctuoidea, relatively little is known about Lymantriidae specifically. Further taxon sampling within Lymantriidae and related families is required to resolve the placement of Lymantriidae in Noctuoidea.
Materials and Methods
Sample collection and mitochondrial DNA extraction
L. salicis larvae were collected from willow trees within the campus of Anhui Agricultural University, Hefei, China. Total genomic DNA was extracted using the Aidlab Genomic DNA Extraction Kit (Aidlab Co., Beijing, China) according to the manufacturer’s instructions. Quality of extracted DNA was assessed by electrophoresis on a 1% agarose gel stained with ethidium bromide.
Primer design, PCR amplification and sequencing
The full mitochondrial genome of L. salicis was PCR amplified in thirteen overlapping fragments, based on primers that were designed from known mitogenomes of Lymantriidae, and synthesized by Invitrogen Co. Ltd. Shanghai, China (Table 4). All PCRs were performed in a 50 μL reaction volume, including 35 μL sterilized distilled water, 5 μL 10 × Taq buffer (Mg2 + ), 4 μL dNTP (25 mM), 1.5 μL DNA, 2 μL of each primer (10 μM) and 0.5 μL (1 unit) Taq (TaKaRa Co., Dalian, China). PCR conditions were as follows: 4 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 40 s at 46–58 °C (Table 4), and 1–3 min (depending on putative length of the fragments) at 72 °C; and then a final extension step of 72 °C for 10 min.
Table 4. Details of the primers used to amplify the mitogenome of L. salicis.
| Primer pair | Primer sequence (5′–3′) | Annealing temperature |
|---|---|---|
| F1 | TAAAAATAAGCTAAATTTAAGCTT | 52 °C |
| R1 | TATTAAAATTGCAAATTTTAAGGA | |
| F2 | AAACTAATAATCTTCAAAATTAT | 46 °C |
| R2 | AAAATAATTTGTTCTATTAAAG | |
| F3 | ATTCTATATTTCTTGAAATATTAT | 46 °C |
| R3 | CATAAATTATAAATCTTAATCATA | |
| F4 | TGAAAATGATAAGTAATTTATTT | 48 °C |
| R4 | AATATTAATGGAATTTAACCACTA | |
| F5 | TAAGCTGCTAACTTAATTTTTAGT | 53 °C |
| R5 | CCTGTTTCAGCTTTAGTTCATTC | |
| F6 | CCTAATTGTCTTAAAGTAGATAA | 48 °C |
| R6 | TGCTTATTCTTCTGTAGCTCATAT | |
| F7 | TAATGTATAATCTTCGTCTATGTAA | 50 °C |
| R7 | ATCAATAATCTCCAAAATTATTAT | |
| F8 | ACTTTAAAAACTTCAAAGAAAAA | 53 °C |
| R8 | TCATAATAAATTCCTCGTCCAATAT | |
| F9 | GTAAATTATGGTTGATTAATTCG | 53 °C |
| R9 | TGATCTTCAAATTCTAATTATGC | |
| F10 | CCGAAACTAACTCTCTCTCACCT | 58 °C |
| R10 | CTTACATGATCTGAGTTCAAACCG | |
| F11 | CGTTCTAATAAAGTTAAATAAGCA | 55 °C |
| R11 | AATATGTACATATTGCCCGTCGCT | |
| F12 | TCTAGAAACACTTTCCAGTACCTC | 52 °C |
| R12 | AATTTTAAATTATTAGGTGAAATT | |
| F13 | TAATAGGGTATCTAATCCTAGTT | 48 °C |
| R13 | ACTTAATTTATCCTATCAGAATAA |
All PCR products were visualized by electrophoresis on a 1.0% TAE agarose gel, and purified using a DNA gel extraction kit (Transgen Co., Beijing, China). The purified PCR fragments were ligated into the T-vector (TaKaRa Co., Dalian, China) and transformed into Escherichia coli DH5α, using the manufacturer’s protocol. Recombinants were cultured overnight at 37 °C on Luria-Bertani (LB) solid medium containing Ampicillin (AMP), isopropylthiogalactoside (IPTG) and 5-bromo-4-chloro-3-indolyl-D-galactopyranoside (X-Gal). White colonies carrying insert DNA were selected, cultured overnight in liquid media, and vector inserts were directly sequenced by Sangon Biotech Co., (Shanghai, China).
Sequence assembly and gene annotation
The complete mtDNA sequence was assembled using the SeqManII program from the Lasergene software package (DNAStar Inc., Madison, USA). Sequence annotation was performed using the NCBI’s web interface for BLAST (http://blast.ncbi.nlm.nih.gov/Blast).
Nucleotide sequences of the PCGs were translated into putative proteins based on insect sequences available in GenBank. Initiation and termination codons were identified using an alignment created in ClustalX version 2.0, with other lepidopteran sequences as references. To describe base composition, we analyzed skew as described by Junqueira39: AT skew = [A − T]/[A + T], GC skew = [G − C]/[G + C]. The relative synonymous codon usage (RSCU) was obtained using MEGA 540.
The tRNA genes were verified using the program tRNAscan-SE with default settings41, in addition to using the alignment to visually identify sequences with the appropriate anticodons capable of folding into the typical clover-leaf secondary structure. In the A + T-rich region, tandem repeats were found with the Tandem Repeats Finder program (http://tandem.bu.edu/trf/trf.html)42.
Phylogenetic analysis
A total of 29 sets of 13 PCG sequences were used to perform phylogenetic analysis, including those of L. salicis. Those from other taxa were downloaded from GenBank, with Drosophila melanogaster (U37541.1)43 and Locusta migratoria (JN858212)44 sequences used as an outgroup. Alignments of the 13 concatenated PCGs were conducted using ClustalX version 2.0. Maximum likelihood (ML) phylogenetic analysis was performed using MEGA 5.0 with Tamura-Nei model40. Neighbor Joining (NJ) distance analysis was performed using PAUP4b1045, and Bayesian Inference (BI) MCMC phylogenetic analysis was performed using MrBayes 3.246. The ML analysis was pseudosampled with 1000 bootstrapped datasets. The NJ analysis was done with 1000 bootstrap replicates. The BI analysis used four chains MCMC, running for 1,000,000 generations, with trees being sampled every 1000 generations. The consensus tree was visualized using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
Additional Information
How to cite this article: Sun, Y.-X. et al. Characterization of the Complete Mitochondrial Genome of Leucoma salicis (Lepidoptera: Lymantriidae) and Comparison with Other Lepidopteran Insects. Sci. Rep. 6, 39153; doi: 10.1038/srep39153 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank the native English speaking scientists of Elixigen Company for editing our manuscript.
Footnotes
Author Contributions Y.X.S., L.W. and C.L.L. designed the research. Y.X.S. and L.W. performed the research. Y.X.S., G.Q.W. and C.Q. analyzed the data. L.S.D., Y.S., M.N.A., and B.J.Z. contributed reagents/materials/analysis tools. Y.X.S. and C.L.L. wrote the paper with other authors.
References
- Zhao Z. L. Fauna Sinica: Insecta, Lepidoptera, Lymantriidae (Chinese Edition). Vol. 30. 281–286 (Science Press, 2003). [Google Scholar]
- Schintlmeister A. The taxonomy of the genus Lymantria Hübner, [1819]: (Lepidoptera: Lymantriidae). Quadrifina. 7, 1–248 (2004). [Google Scholar]
- Arimoto M. & Ren I. Identification of Japanese Lymantria species (Lepidoptera: Lymantriidae) based on PCR-RFLP analysis of mitochondrial DNA. Appl. Entom. Zool. 49, 159–169 (2014). [Google Scholar]
- Muto S. E. & Mori K. Synthesis of all four stereoisomers of leucomalure, components of the female sex pheromone of the satin moth. Leucoma salicis. Cheminform. 7, 1300–1307 (2003). [Google Scholar]
- Strokovskaya L., Ziemnicka J. & Michalik J. Genetic variability of four natural isolates of the Stilpnotia salicis multiple-enveloped nuclear polyhedrosis virus. Acta Biochim. Polonica. 43, 633–638 (1996). [PubMed] [Google Scholar]
- Jiang S. T. et al. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int. J. Biol. Sci. 5, 351–365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel G. S. & Clayton D. A. Mitochondrial transcription initiation. Variation and conservation. J. Biol. Chem. 268, 16083–16086 (1993). [PubMed] [Google Scholar]
- Dowton M., Castro L. R. & Austin A. D. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invertebr. Syst. 16, 345–356 (2002). [Google Scholar]
- Zhang Z. Q. et al. The mitochondrial genome of Dastarcus helophoroides (Coleoptera: Bothrideridae) and related phylogenetic analyses. Gene. 560, 15–24 (2015). [DOI] [PubMed] [Google Scholar]
- Hao J. et al. The Complete Mitochondrial Genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and Its Phylogenetic Implication. Comp. Funct. Genom. 2012, 328049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L. & Zhou Q. M. The complete mitochondrial genome of Bombyx mori strain Huayu (Lepidoptera: Bombycidae). Mitochondr. DNA. 27, 2155–2156 (2016). [DOI] [PubMed] [Google Scholar]
- Dai L. S. et al. The complete mitochondrial genome of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Mitochondr. DNA. 27, 1512–1513 (2016). [DOI] [PubMed] [Google Scholar]
- Liu Q. N. et al. The complete mitochondrial genome of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Mitochondr. DNA. 27, 122–123 (2016). [DOI] [PubMed] [Google Scholar]
- Hong G. et al. The complete nucleotide sequence of the mitochondrial genome of the cabbage butterfly, Artogeia melete (Lepidoptera: Pieridae). Acta Biochim. Biophys. Sinica. 41, 446–455 (2009). [DOI] [PubMed] [Google Scholar]
- Yang L., Wei Z. J., Hong G. Y., Jiang S. T. & Wen L. P. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol. Bio. Rep. 36, 1441–1449 (2009). [DOI] [PubMed] [Google Scholar]
- Salvato P., Simonato M., Battisti A. & Negrisolo E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genom. 9, 331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F. et al. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int. J. Biol. Sci. 6, 172–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. F., Su T. J., Luo A. R., Zhu C. D. & Wu C. S. Characterization of the complete mitochondrion genome of diurnal moth Amata emma (Butler) (Lepidoptera: Erebidae) and its phylogenetic implications. PLoS One. 8, e72410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Montoya J. & Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290, 470–474 (1981). [DOI] [PubMed] [Google Scholar]
- Sun Q. Q. et al. Complete sequence of the mitochondrial genome of the Japanese buff-tip moth, Phalera flavescens (Lepidoptera: Notodontidae). GMR. 11, 4213–4225 (2012). [DOI] [PubMed] [Google Scholar]
- Wolstenholme D. R. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141, 173–216 (1992). [DOI] [PubMed] [Google Scholar]
- Yin J., Hong G. Y., Wang A. M., Cao Y. Z. & Wei Z. J. Mitochondrial genome of the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) and comparison with other Lepidopterans. Mitochondr. DNA. 21, 160–169 (2010). [DOI] [PubMed] [Google Scholar]
- Yin J., Wang A. M., Hong G. Y., Cao Y. Z. & Wei Z. J. Complete mitochondrial genome of Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Mitochondr. DNA. 22, 41–43 (2011). [DOI] [PubMed] [Google Scholar]
- Lavrov D. V., Brown W. M. & Boore J. L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. PNAS. 97, 13738–13742 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Q. et al. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera: Saturniidae). Acta Biochim. Biophys. Sinica. 40, 693–703 (2008). [PubMed] [Google Scholar]
- Dong W. W., Dong S. Y., Jiang G. F. & Huang G. H. Characterization of the complete mitochondrial genome of tea tussock moth, Euproctis pseudoconspersa (Lepidoptera: Lymantriidae) and its phylogenetic implications. Gene. 577, 37–46 (2016). [DOI] [PubMed] [Google Scholar]
- Cameron S. L. & Whiting M. F. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 408, 112–123 (2008). [DOI] [PubMed] [Google Scholar]
- Kim S. R. et al. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol. Bio. Rep. 36, 1871–1880 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang D. X., Szymura J. M. & Hewitt G. M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 40, 382–391 (1995). [DOI] [PubMed] [Google Scholar]
- Arunkumar K. P., Metta M. & Nagaraju J. Molecular phylogeny of silkmoths reveals the origin of domesticated silkmoth, Bombyx mori from Chinese Bombyx mandarina and paternal inheritance of Antheraea proylei mitochondrial DNA. Mol. Phylo. Genet. Evol. 40, 419–427 (2006). [DOI] [PubMed] [Google Scholar]
- Park J. S., Cho Y., Kim M. J., Nam S. H. & Kim I. Description of complete mitochondrial genome of the black-veined white, Aporia crataegi (Lepidoptera: Papilionoidea), and comparison to papilionoid species. J. Asia-Pac. Entomol. 15, 331–341 (2012). [Google Scholar]
- Yukuhiro K., Sezutsu H., Itoh M., Shimizu K. & Banno Y. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol. Biol. Evol. 19, 1385–1389 (2002). [DOI] [PubMed] [Google Scholar]
- Raupach M. J. et al. Molecular species identification of central european ground beetles (coleoptera: carabidae) using nuclear rdna expansion segments and dna barcodes. Front. Zool. 7, 1–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. S. et al. Characterization of the Complete Mitochondrial Genome of Cerura menciana and Comparison with Other Lepidopteran Insects. PLoS One. 10, e0132951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibiger M. & Lafontaine J. D. A review of the higher classification of the Noctuoidea (Lepidoptera) with special reference to the Holarctic fauna. Esperiana. 11, 7–92 (2005). [Google Scholar]
- Kitching I. J. & Rawlins J. E. The Noctuoidea. In: Lepidoptera, Moths and Butterflies Volume 1: Evolution, Systematics and Biogeography. (ed. Kristensen N. P.) 355–401. (Walter de Gruyter, 1999). [Google Scholar]
- Zahiri R. et al. Molecular phylogenetics of Erebidae (Lepidoptera, Noctuoidea). Syst. Entomol. 37, 102–124 (2012). [Google Scholar]
- Chai H. N. & Du Y. Z. The Complete Mitochondrial Genome of the Pink Stem Borer, Sesamia inferens, in Comparison with Four Other Noctuid Moths. Int. J. Mol. Sci. 13, 10236–10256 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira A. C. M. et al. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera : Calliphoridae). Gene. 339, 7–15 (2004). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl. Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucl. Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Goddard J. M., Martin S. C., Fauron C. M. & Wolstenholme D. R. Drosophila mitochondrial DNA: a novel gene order. Nucl. Acids Res. 10, 6619–6637 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. et al. Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol. Ecol. 21, 4344–4358 (2012). [DOI] [PubMed] [Google Scholar]
- Swofford D. L. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. (Sinauer 2003). [Google Scholar]
- Ronquist F. et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 61, 539–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.