Abstract
Genotype imputation is a key component of genetic association studies, where it increases power, facilitates meta-analysis, and aids interpretation of signals. Genotype imputation is computationally demanding and, with current tools, typically requires access to a high-performance computing cluster and to a reference panel of sequenced genomes. Here we describe improvements to imputation machinery that reduce computational requirements by more than an order of magnitude with no loss of accuracy in comparison to standard imputation tools. We also describe a new web-based service for imputation that facilitates access to new reference panels and greatly improves user experience and productivity.
After study samples are genotyped on an array, typically assaying 300,000–1,000,000 single-nucleotide variants (SNVs), imputation finds haplotype segments that are shared by study individuals and a reference panel of sequenced genomes, such as those from the 1000 Genomes Project1 or recent population sequencing studies2–4. Imputation accurately assigns genotypes at untyped markers, improving genome coverage, facilitating comparison and combination of studies that use different marker panels, increasing power to detect genetic association, and guiding fine-mapping5,6.
Imputation accuracy increases with the number of haplotypes in the reference panel of sequenced genomes7–9, particularly for rare (minor allele frequency (MAF) < 0.5%) and low-frequency (0.5% < MAF < 5%) variants. These rare and low-frequency variants include most loss-of-function alleles10 and other high-impact variants that are key for genotype-based callback and focused studies of natural knockout alleles11–13.
Large reference panels, such as the one developed by the Haplotype Reference Consortium14 (HRC), extend accurate imputation to variants with frequencies of 0.1–0.5% or less and already include thousands of putative loss-of-function alleles. The HRC panel combines sequence data across >32,000 individuals from >20 medical sequencing studies and is cumbersome to access directly as a result of participant privacy protections in individual studies and the large volumes of data involved. Imputing 1,000 genome-wide association study (GWAS) samples using the HRC reference set requires ~2 years on a single CPU, or 1 week on a 100-core cluster, using minimac2 (ref. 8).
Here we present new algorithms for genotype imputation that increase computational efficiency with no loss of accuracy by leveraging local similarities between sequenced haplotypes. We also present a new web-based imputation service that greatly simplifies analysis, eliminates the need for cumbersome data access agreements, and so allows users to devote their time to other essential tasks.
The methods described here provide an extremely efficient strategy for genotype imputation. Together, they ensure accurate imputation, while reducing computational requirements and user time. Our implementation supports reference panels composed of hundreds of thousands of haplotypes and is freely available, enabling others to build on our work.
To illustrate the potential benefits of these improved methods for imputation, we first used computer simulation (Online Methods and Supplementary Table 1). We found substantial gains in imputation accuracy between imputed genotypes and the true simulated genotypes as panel size increased. For variants with MAF of 0.01–0.1%, average imputation r2 values increased from 0.41 to 0.79 when reference panels grew from n = 1,000 to n = 20,000 individuals, a near doubling in effective sample size for association (which scales as r2)15.
Imputation into current large GWAS data sets, which total millions of samples, can quickly and cost-efficiently identify carriers of many interesting rare and low-frequency variants. For example, with a reference panel of 20,000 individuals, our simulations estimate the probability that individuals identified as rare allele carriers through imputation actually carry the allele is ~84% (MAF of 0.01–0.1%; Supplementary Table 1). If desired, genotypes for imputation-identified carriers can be validated through Sanger sequencing or other targeted assays before callback phenotyping or other follow-up analyses.
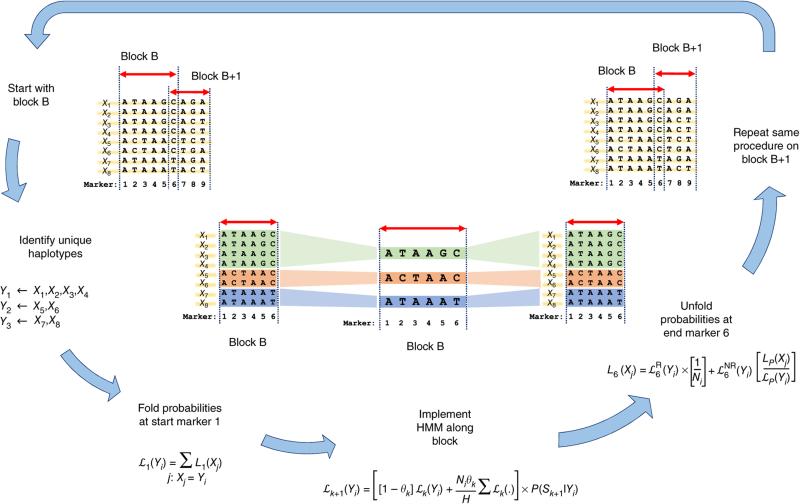
To enable many researchers to use larger reference panels and so benefit from this potential for improved power and targeted analyses for carriers of rare variants, we devised a new, faster imputation algorithm. This algorithm is based on a ‘state space reduction’ of the hidden Markov models (HMMs) describing haplotype sharing; it exploits similarities among haplotypes in small genomic segments to reduce the effective number of states over which the HMM iterates (Fig. 1 and Online Methods). Similar ideas are a fundamental part of haplotype sharing analyses and have been used in Beagle16, SHAPEIT17,18, and other genomics tools19–21 to improve computational performance. Whereas methods such as SHAPEIT are designed to work best in settings where there are few missing genotypes, our own algorithm works well with data where most genotypes are missing and need to be imputed. Our model divides the genome into consecutive blocks and iterates only over the unique haplotypes in each genomic block. It then uses a reversible mapping function that can reconstruct exactly the state space used by minimac9 and IMPUTE2 (ref. 22). Two important features of the algorithm are that it yields exactly the same results as more cumbersome analyses in the original state space and that it remains computationally efficient in the presence of missing data (which is essential for imputation). We implemented this method in the C++ package minimac3.
Figure 1.
Overview of state space reduction. We consider a chromosome region with M = 9 markers and H = 8 haplotypes: X1, X2, ..., X8. We break the region into consecutive genomic segments (blocks) and start by analyzing block B from marker 1 to marker 6. In block B, we identify U = 3 unique haplotypes: Y1, Y2, and Y3 (colored in green, red, and blue, respectively). Given we know the left probabilities of the original state space at marker 1 (that is, L1(X1), ..., L1(X8)), we fold them to get the left probabilities of the reduced state space at marker 1: , , and . We implement HMM on the reduced state space (Y1, Y2, and Y3) from marker 1 to marker 6 to get , , and . We next unfold the left probabilities of the reduced state space at marker 6 to obtain the left probabilities of the original state space: L6(X1), ..., L6(X8). We repeat this procedure on the next block, starting with L6(X1), ..., L6(X8), to finally obtain L9(X1), ..., L9(X8).
We compared run time and memory requirements for minimac3 against those for minimac2 (ref. 8), Beagle 4.1 (ref. 16), and IMPUTE2 (ref. 9) by carrying out genotype imputation into 100 individuals of European ancestry using reference panels of 1,091 to 32,390 sequenced individuals (Table 1 and Online Methods). For minimac2 and IMPUTE2, memory and run time increased linearly with panel size. The newest Beagle 4.1 was substantially faster, and its run time increased slightly less than linearly with panel size. minimac3 consistently outperformed all alternatives: increasing panel size ~30-fold from 1,091 to 32,390 samples increased memory requirements sixfold and run time eightfold. For this largest panel, minimac3 was twice as fast as Beagle 4.1, 29 times faster than minimac2, and 30 times faster than IMPUTE2 and reduced memory usage by 72%, 94%, and 97%, respectively (Table 1).
Table 1.
Comparison of minimac3, minimac2, IMPUTE2, and Beagle 4.1
| Reference panel | Number of samples | minimac3 | minimac2 | IMPUTE2 | Beagle 4.1 |
|---|---|---|---|---|---|
|
Time (in CPU-hours)
|
|||||
| 1000G Phase 1 | 1,092 | 4 | 27 | 34 | 5 |
| AMD | 2,074 | 9 | 59 | 73.5 | 9 |
| 1000G Phase 3 | 2,504 | 6 | 61 | 78 | 9 |
| SardiNIA | 3,489 | 7 | 85 | 108 | 11 |
| COMBINED | 9,341 | 17 | 236 | 288 | 31 |
| Mega | 11,845 | 21 | 304 | 364 | 40 |
| HRC v1.1 | 32,390 | 31 | 925 | 951 | 128 |
|
Memory (in CPU-GB)
|
|||||
| 1000G Phase 1 | 1,092 | 0.09 | 0.34 | 0.91 | 0.51 |
| AMD | 2,074 | 0.14 | 0.62 | 1.58 | 0.39 |
| 1000G Phase 3 | 2,504 | 0.13 | 0.75 | 1.88 | 0.56 |
| SardiNIA | 3,489 | 0.13 | 1.03 | 2.55 | 0.46 |
| COMBINED | 9,341 | 0.28 | 2.73 | 6.57 | 0.41 |
| Mega | 11,845 | 0.33 | 3.51 | 8.28 | 0.43 |
| HRC v1.1 | 32,390 | 0.55 | 9.31 | 22.08 | 1.98 |
|
Imputation accuracy (mean r2), MAF = 0.0001–0.5%
|
|||||
| 1000G Phase 1 | 1,092 | 0.45 | 0.45 | 0.43 | 0.42 |
| AMD | 2,074 | 0.54 | 0.54 | 0.51 | 0.52 |
| 1000G Phase 3 | 2,504 | 0.52 | 0.52 | 0.49 | 0.52 |
| SardiNIA | 3,489 | 0.55 | 0.55 | 0.53 | 0.54 |
| COMBINED | 9,341 | 0.76 | 0.76 | 0.74 | 0.76 |
| Mega | 11,845 | 0.76 | 0.76 | 0.74 | 0.76 |
| HRC v1.1 | 32,390 | 0.77 | 0.77 | 0.75 | 0.77 |
|
Imputation accuracy (mean r2), MAF = 0.5–5%
|
|||||
| 1000G Phase 1 | 1,092 | 0.77 | 0.77 | 0.76 | 0.73 |
| AMD | 2,074 | 0.82 | 0.82 | 0.80 | 0.80 |
| 1000G Phase 3 | 2,504 | 0.79 | 0.79 | 0.78 | 0.79 |
| SardiNIA | 3,489 | 0.79 | 0.79 | 0.78 | 0.80 |
| COMBINED | 9,341 | 0.89 | 0.89 | 0.88 | 0.89 |
| Mega | 11,845 | 0.89 | 0.89 | 0.88 | 0.89 |
| HRC v1.1 | 32,390 | 0.90 | 0.90 | 0.89 | 0.90 |
|
Imputation accuracy (mean r2), MAF = 5–50%
|
|||||
| 1000G Phase 1 | 1,092 | 0.96 | 0.96 | 0.95 | 0.95 |
| AMD | 2,074 | 0.96 | 0.96 | 0.96 | 0.96 |
| 1000G Phase 3 | 2,504 | 0.96 | 0.96 | 0.96 | 0.96 |
| SardiNIA | 3,489 | 0.96 | 0.96 | 0.96 | 0.96 |
| COMBINED | 9,341 | 0.97 | 0.97 | 0.97 | 0.97 |
| Mega | 11,845 | 0.97 | 0.97 | 0.97 | 0.97 |
| HRC v1.1 | 32,390 | 0.98 | 0.98 | 0.98 | 0.98 |
This table compares the imputation accuracy, run time, and memory required to impute 100 whole genomes using different reference panels and imputation tools (Online Methods). Run time is interpolated from analysis on chromosome 20. All four tools were run on 5-Mb chunks with 1-Mb overlap (13 chunks in serial or chromosome 20, yielding a total of 227,925 variants). minimac3, minimac2, and IMPUTE2 were run with precalculated recombination and error estimates. minimac3 and Beagle 4.1 were run with their input file formats (m3vcf and bref, respectively), while minimac2 and IMPUTE2 were run on VCF files. The number of variants in the three MAF bins is 32,945 (0.0001–0.5%), 70,016 (0.5–5%), and 104,751 (5–50%). The best results for each reference panel (lowest run time, lowest memory, or highest mean r2) are highlighted in bold.
In this comparison, all programs were run on a single thread although, because imputation is trivially parallelizable, all can benefit from additional CPUs (for example, by imputing multiple chromosomes, chromosome segments, or samples in a single or multiple parallel invocations of the program). A comparison of multiple-threaded run times for Beagle 4.1 and minimac3 is presented in Supplementary Table 2.
We compared imputation quality across the four methods by calculating the squared correlation coefficient (r2) between imputed allele dosages and masked genotypes (Table 1 and Online Methods). minimac, minimac2, and minimac3 are based on the same mathematical model and hence gave identical results. minimac3 slightly outperformed Beagle 4.1 and IMPUTE2, particularly for rare variants (MAF = 0.0004–0.5%) where, with 3,489 reference samples, IMPUTE2 attained r2 = 53.3%, Beagle 4.1 attained r2 = 54.3%, and minimac3 attained r2 = 55.5%. All methods demonstrated improved imputation quality with increasing panel size. For example, for minimac3, the imputation quality of rare variants increased from r2 = 45.3% to r2 = 77.2% when panel size increased from 1,092 to 32,390.
The complexity of the minimac3 algorithm depends on the number of unique haplotypes in each genomic segment and the total number of such segments in the reference panel (Fig. 1). As a result, minimac3 scales better than linearly over our range of reference panel sizes. For example, increasing the simulated reference panel from 1,000 to 20,000 individuals (20-fold) increased memory and CPU requirements sevenfold (Online Methods). In real data, increasing the panel size from 1,092 (1000 Genomes Project Phase 1, ~27 million variants) to ~33,000 (HRC, ~40 million variants) (>40-fold increase in number of genotypes) increased run time only tenfold (5.3 h versus 51.3 h). When we ran the same analysis on the same set of markers (~22 million variants common to both panels), the run time increased 8.5-fold (4.15 h versus 31.1 h).
Our state space reduction also provides an efficient way to represent haplotype data, substantially reducing file size relative to the now standard VCF format. The relative efficiency of the representation reflects population genetics: for example, data from European-ancestry samples can be compressed more efficiently than data from African-ancestry samples (Supplementary Table 3). We adapted the VCF format to allow for these efficiencies, resulting in the m3vcf format that stores only one copy of each unique haplotype in a genomic segment (Online Methods). For the HRC data set with >60,000 haplotypes, uncompressed m3vcf files are ~97% smaller than uncompressed VCF files; gzip-compressed m3vcf files are ~86% smaller than gzip-compressed VCF files. Our minimac3 distribution includes simple utilities to manipulate m3vcf files.
Continued computational improvements in imputation tools are necessary for imputation to remain practical as millions of samples are genotyped and reference panels increase in size to tens of thousands of sequenced genomes. An additional burden of imputation and other genomics tools is the requirement for users to master and manage large high-performance computing clusters and associated tools.
To reduce these burdens, we incorporated minimac3 in a cloud-based imputation server, which combines minimac3, the MapReduce paradigm23, and a user-friendly interface. Behind the scenes, the server divides reference and target data sets into small chromosome segments that are processed in parallel. Results are collected and pasted together so that the process is seamless to end-users. The server uses Apache Hadoop MapReduce for low-level tasks, such as parallelization and distribution of jobs, and the Cloudgene24 workflow system to drive the user interface (Supplementary Fig. 1). It automatically performs quality checks (verifying strand orientation, allele labeling, file integrity, MAF distribution, and per-sample and per-variant missingness, among others; Supplementary Fig. 2). If no major problems are encountered, samples are phased (if this has not been done already) and then imputed using one of the currently available reference panels: HRC14, 1000 Genomes Project Phase 1 (refs. 25,26) or Phase 3 (ref. 1), HapMap Phase 2 (ref. 27), or African-ancestry genomes from CAAPA. Data can be uploaded directly or by specifying a remote secure file transfer protocol (sftp) location. Data transfers are encrypted, and input data are deleted after processing. We require each user to agree not to try to identify the underlying panel. As imputation proceeds, users are provided feedback on progress, summary reports, e-mail notification, and a download link for the imputed data. Results are encrypted with a one-time password available only to the user and deleted after 7 d (Online Methods). More than 4.5 million genomes have been processed by >1,200 users since the service was announced at the American Society of Human Genetics annual meeting in October 2014.
Here we have described improved computational methods, resources, and interfaces to enable researchers to rapidly impute large numbers of samples, without first becoming experts in the intricacies of imputation software and cluster job management, and to conveniently access large reference panels of sequenced individuals, such as those from the HRC. To make imputation highly scalable, we reengineered the core algorithms in our minimac imputation engine. Our state space reduction provides a computationally efficient solution for genotype imputation with no loss in imputation accuracy and enables the use of large reference panels. Our cloud-based interface simplifies analysis steps and can be adapted to other analysis needs, such as linkage-disequilibrium-aware genotype calling from low-coverage sequence data. This trend, where cutting-edge software, large data, computational power, and a friendly interface are packaged together, will become increasingly important as genomic data sets increase in size and complexity.
ONLINE METHODS
Simulations
We simulated haplotypes for a three-population coalescent model using the program ms8. We chose a demographic model consistent with patterns of diversity observed in European-ancestry samples28.
Imputation method with state space reduction
Here we describe the state space reduction that uses the similarity between haplotypes in small genomic segments to reduce computational complexity. We recommend first reading a description of the original minimac algorithm9. Consider a reference panel with H haplotypes and a genomic segment bounded by markers P and Q. Let U ≤ H be the number of distinct haplotypes in the block.
Label the original haplotypes as X1, X2, ..., XH and the distinct unique haplotypes as Y1, Y2, ..., YU. For example, in Figure 1, block B bounded by markers P = 1 and Q = 6 has U = 3 distinct haplotypes. Let and denote the left probabilities for the original states and reduced states at marker k (ref. 29). Assuming we know LP(X1), ..., LP(XH), equation (1) allows us to obtain for each distinct haplotype.
| (1) |
In this reduced state space, we modify the Baum–Welch forward equations30 to obtain recursively for k = P + 1, P + 2, ...., Q.
| (2) |
In equation (2), θk denotes the template switch probability between markers k and k+1 (analogous to a recombination fraction), Sk+1 is the genotype in the study sample, P(Sk+1|Yi) is the genotype emission probability, and Ni is the number of haplotypes matching Yi in the original state space (for example, in Fig. 1, N1 = 4, N2 = 2, and N3 = 2). Once we obtain values for all the reduced states, we use them to calculate at the final block boundary, enabling us to transition between blocks. To accomplish this, we split probability into two parts, and , where denotes the left probability at marker Q when no template switches occur between markers P and Q and denotes the probability when at least one switch occurs. This leads to equation (3) (where i is such that Yi = Xj)
| (3) |
Similar equations can be derived for the right probabilities Rk(.) and . Once we have the left and right probabilities for all the reduced states, the posterior probabilities for a template including any allele of interest at marker k can be calculated within the reduced state space as
| (4) |
Computational complexity and optimal allocation
Methods that perform phasing and imputation simultaneously (for example, MaCH29 and IMPUTE31) have a computational cost proportional to the number of study samples (N), the number of genotyped markers in the reference panel (M), and the square of the number of reference haplotypes (H2), or in total O(NMH2). In the context of prephasing, as in minimac and IMPUTE2 (ref. 9), this computational cost is reduced to O(NMH).
For imputation using mimimac3, we break up a chromosome into K consecutive segments. If Ui denotes the number of unique haplotypes and Mi denotes the number of markers in segment i, then complexity is
The second term accounts for the complexity of transitions between blocks, which occur in the original state space. Thus, although very short segments could reduce the number of unique haplotypes per segment (Ui) and complexity measured by the first term, such segments would also increase the total number of segments (K) and complexity measured by the second term. An optimal allocation of genomic regions must balance these two goals.
We implement a recursive dynamic programming algorithm to find the optimal allocation of the genomic segments, as a brute force approach is not feasible (~2M–1 alternatives). We assume that the optimal complexity of imputation until marker i < M is denoted by C(i) and calculate C(M) recursively as
| (5) |
In equation (5), C(i) is the optimal cost for imputation from marker 1 to marker i and U(i,M) is the number of unique haplotypes between marker i and marker M (inclusive). This expression requires at most M2 comparisons; this number can be further reduced because we do not need to consider large segments, as the unique number of haplotypes in large segments will be close to the total number of haplotypes.
Parameter estimation under reduced state space
We implemented both the expectation–maximization algorithm and Monte Carlo Markov chain (MCMC) sampling to estimate θ for adjacent marker pairs and ε for each marker. θ is the template switching rate, which reflects a combination of population recombination rates and relatedness between the samples. ε is the error parameter, which reflects a combination of genotyping error, gene conversion events, and recurrent mutation (for details, see refs. 9,29).
Comparison of minimac2, minimac3, IMPUTE2, and Beagle 4.1
We evaluated the performance of minimac3 (v1.0.14) in comparison to the three most commonly used imputation tools: minimac2 (v2014.9.15), IMPUTE2 (v2.3.1), and Beagle 4.1 (v22Feb16) (Table 1). We combined chromosome 20 data across multiple whole-genome sequencing studies to generate large reference panels. We compared results for the following seven reference panels: (i) 1000G Phase 1: 1,092 individuals from 1000 Genomes Project Phase 1 (refs. 25,26), (ii) AMD: 2,074 individuals sequenced for study of age-related macular degeneration32, (iii) 1000G Phase 3: 2,504 individuals from 1000 Genomes Project Phase 3 (ref. 1), (iv) SardiNIA: 3,489 individuals from the SardiNIA project4, (v) COMBINED: 9,341 individuals combined together from AMD, SARDINIA, the BRIDGE study of bipolar disorder (L.J.S., unpublished data) (2,464 samples), and the Minnesota Twins study33 (1,314 samples), (vi) Mega: 11,845 individuals obtained by merging COMBINED and G1KP3, and (vii) HRC v1.1: 32,390 individuals from HRC14. To mimic a GWAS, we selected 25 unrelated individuals each from AMD, SardiNIA, BRIDGE Study, and Minnesota Twins and masked all variants except those typed on the Illumina Duo 1M chip (resulting in ~20,000 genotyped variants for chromosome 20). To evaluate imputation accuracy, we estimated the squared Pearson correlation coefficient (r2) between the imputed genotype probabilities and genotype calls from sequence data. We evaluated imputation accuracy at the 227,925 variants that were present in all the respective data sets and had MAF of at least 0.00005 in all contributing studies. For each of the combinations of the four imputation methods and seven reference panels, we recorded the average imputation accuracy, total computational time, and physical memory required to impute 100 GWAS individuals.
Imputation server architecture
The Michigan Imputation Server implements the whole-genotype imputation workflow using the MapReduce programming model for efficient parallelization of computationally intensive tasks. We use the open source framework Hadoop to implement all workflow steps. Maintenance of the server, including node configuration (for example, amount of parallel tasks, memory for each chunk, and monitoring of all nodes), is achieved using the Cloudera Manager. During cluster initialization, reference panels, genetic maps, and software packages are distributed across all cluster nodes using the Hadoop file system HDFS. The imputation workflow itself consists of two steps: first, we divide the data into non-overlapping chunks (here, chromosome segments of 20 Mb). Second, we run an analysis (here, quality control or phasing and imputation) in parallel across chunks. To avoid edge effects, 5 Mb for phasing and 500 kb for imputation are added to each chunk. Finally, all results are combined to generate an aggregate final output.
Genotype imputation can be implemented with MapReduce, as the computationally expensive whole-genome calculations can be split into independent chromosome segments. Our imputation server accepts phased and unphased GWAS genotypes in VCF file format. File format checks and initial statistics (numbers of individuals and SNVs, detected chromosomes, unphased/phased data set, and number of chunks) are generated during the preprocessing step. Then, the submitted genotypes are compared to the reference panel to ensure that alleles, allele frequencies, strand orientation, and variant coding are correct. In this first MapReduce analysis, the map function calculates the VCF statistics for each file chunk, and the reducer summarizes the results and forwards only chunks that pass quality control to the subsequent imputation step (Supplementary Fig. 2). The MapReduce imputation step constitutes a map-only job. This means that no reducer is applied and each mapper imputes genotypes using minimac3 on the previously generated chunk. If the user has uploaded unphased genotypes, the data are prephased with one of the available phasing engines: Eagle 2, HAPI-UR34, or SHAPEIT17. A post-processing step generates a zipped and indexed VCF file (using bgzip and tabix35) for each imputed chromosome. To minimize the input/output load, the reference panel is distributed across available nodes in the cluster using the distributed cache feature of Hadoop. To ensure data security, imputation results are encrypted on the fly using a one-time password. All result files and reports can be viewed or downloaded via the web interface.
The imputation server workflow has been integrated into Cloudgene24 to provide a graphical user interface. Cloudgene is a high-level workflow system for Apache Hadoop designed as a web application using Bootstrap, CanJs, and JQuery. On the server side, all necessary resources are implemented in Java using the RESTful web framework Restlet. The Cloudgene API provides methods for the execution and monitoring of MapReduce jobs and can be seen as an additional layer between Hadoop and the client. The imputation server is integrated into Cloudgene using the provided workflow definition language and its plugin interface. On the basis of the workflow information, Cloudgene automatically renders a web form for all required parameters to submit individual jobs to the Cloudgene server. The server communicates and interacts with the Hadoop cluster and receives feedback from currently executing jobs. Client and server communicate by asynchronous HTTP requests (AJAX) with JSON as an interchange format. All transmissions between server and client are encrypted using SSL (Secure Socket Layer).
Parameter estimation study
Parameter estimates for the reference panel can be precalculated and saved to speed up the imputation process. To examine the importance of GWAS panel individuals during the parameter estimation step, we used 938 unrelated individuals from 53 worldwide populations from the Human Genome Diversity Panel36. We compared the imputation accuracy across three parameter estimation methods: constant parameters (θ = 0.001 and ε = 0.01), reference panel only for updating the parameters using a leave-one-out method, and reference and GWAS panels for updating. The results of imputation accuracy evaluated on ~6,000 masked variants from chromosomes 20–22 on the ExomeArray are shown in Supplementary Figure 3. We see that updating the parameters results in increased imputation accuracy in comparison to constant estimates (especially for European samples, where imputation r2 increases from 0.35 to 0.45 in the lowest MAF bin). However, including the target panel (along with the reference panel) typically produced only a very small improvement in imputation accuracy.
Optimized file structure for large reference panels
The idea of state space reduction can be applied not only to improve HMM implementation efficiency but also to store large reference panels using less disk space. We introduce the m3vcf (minimac3 VCF) format, which is compatible with the Variant-Call Format (VCF) format. m3vcf files save each genomic segment in series where each segment has the list of bi- and multiallelic variants in order along with the unique haplotypes at these variants and a single line at the beginning of the block that describes which individual maps to which unique haplotype. This format reduces disk space requirements because it saves only the unique haplotypes at each block rather than all the haplotypes. The way in which the unique haplotypes are ordered (along columns) creates long runs of 0's and 1's (as they are ordered lexicographically from the first variant to the last variant) and is thus even more helpful in disk space reduction when using standard file compression methods such as gzip.
We calculated the order of disk space saved using m3vcf files in comparison to the usual VCF files (in both unzipped and zipped formats) and found that, for 1000 Genomes Project Phase 1 with ~1,000 reference samples, we save 60% of disk space using zipped m3vcf files in comparison to zipped VCF files and 93% when compared across unzipped formats. The saving is even greater for larger panels. For example, for the HRC reference panel with ~33,000 samples, we save ~84% and 98% of disk space using zipped and unzipped m3vcf files, respectively (Supplementary Table 4).
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge D. Hinds for assistance with minimac3 code optimizations and A.L. Williams for providing HAPI-UR. We acknowledge support from National Institutes of Health grants HG007022 and HL117626 (G.R.A.), HG000376 (M.B.), and R01DA037904 (S.I.V.), Austrian Science Fund (FWF) grant J-3401 (C.F.), and the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement 602133 (L.F. and S.S.). This work was also supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (D. Schlessinger).
Footnotes
AUTHOR CONTRIBUTIONS
S.D., L.F., S.S., G.R.A., and C.F. designed the methods and experiments. C.S., A.E.L., A.K., S.I.V., E.Y.C., S.L., M.M., D. Schlessinger, P.-R.L., D. Stambolian, W.G.I., A.S., L.J.S., F.C., F.K., and M.B. provided data or tools. S.D., G.R.A., and C.F. wrote the first draft. All authors contributed critical reviews of the manuscript during its preparation.
URLs. minimac3, instructions, and source code are available from http://genome.sph.umich.edu/wiki/minimac3. Imputation on our server is available from https://imputationserver.sph.umich.edu/. Cloudera manager, http://www.cloudera.com/; CAAPA project, https://www.nhlbiwgs.org/group/bags-asthma; Eagle 2, https://data.broadinstitute.org/alkesgroup/Eagle/; Beagle, https://faculty.washington.edu/browning/beagle/beagle.html; HAPI-UR, https://code.google.com/archive/p/hapi-ur/; SHAPEIT2, https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html; IMPUTE2, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 3.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 4.Sidore C, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 7.Pistis G, et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. 2015;23:975–983. doi: 10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen JC, Boerwinkle E, Mosley TH, Jr., Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 12.Stitziel NO, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N. Engl. J. Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016 doi: 10.1038/ng.3643. http://dx.doi.org/10.1038/ng.3643. [DOI] [PMC free article] [PubMed]
- 15.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 18.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 19.Paul JS, Song YS. Blockwise HMM computation for large-scale population genomic inference. Bioinformatics. 2012;28:2008–2015. doi: 10.1093/bioinformatics/bts314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 21.Markianos K, Daly MJ, Kruglyak L. Efficient multipoint linkage analysis through reduction of inheritance space. Am. J. Hum. Genet. 2001;68:963–977. doi: 10.1086/319507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean J, Ghemawat S. Mapreduce: simplified data processing on large clusters. Commun. ACM. 2008;51:107–113. [Google Scholar]
- 24.Schönherr S, et al. Cloudgene: a graphical execution platform for MapReduce programs on private and public clouds. BMC Bioinformatics. 2012;13:200. doi: 10.1186/1471-2105-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 28.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baum LE, Petrie T, Soules G, Weiss N. A maximization technique occurring in statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat. 1970;41:164–171. [Google Scholar]
- 31.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vrieze SI, et al. In search of rare variants: preliminary results from whole genome sequencing of 1,325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014;51:1309–1320. doi: 10.1111/psyp.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams AL, Patterson N, Glessner J, Hakonarson H, Reich D. Phasing of many thousands of genotyped samples. Am. J. Hum. Genet. 2012;91:238–251. doi: 10.1016/j.ajhg.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.