ABSTRACT
Verticillium resistance is thought to be mediated by Ve1 protein, which presumably follows a “gene-for-gene” relationship with the V. dahliae Ave1 effector. Because in planta analyses of Ave1 have relied so far on transient expression of the gene in tobacco, this study investigated gene function using stably expressing 35S:Ave1 transgenic tomato. Transgenic Ave1 expression was shown to induce various defense genes including those coding for PR-1 (P6), PR-2 (βbeta-1,3-glucanase) and peroxidases (anionic peroxidase 2, Cevi16 peroxidase). Since a Ve1− tomato cultivar served as germplasm, these results indicate that Ave1 induces these defense genes independently of Ve1.
KEYWORDS: Ave1, avirulence, defense genes, gene expression, gene-for-gene, plant transformation, resistance, tomato, Ve1, Verticillium wilt
One of the most costly plant diseases is vascular wilt, caused by fungi of the genus Verticillium.1-3 By far, the most prevalent are V. dahliae and V. albo-atrum.4,5 These species infect many economically significant crops grown in Canada and throughout the world, including alfalfa, cotton, cucurbits, eggplant, mint, olive, potato, sunflower, strawberry and tomato, as well as many weeds.6-9
Resistance typically results from plant R-proteins recognizing pathogen effector molecules called avirulence factors.10 R-protein activation then leads to a cascade of signaling events, culminating in appropriate defense responses.11-13 In tomato the Ve locus, which consists of two homologous genes (Ve1 and Ve2) that encode putative transmembrane resistance receptor proteins, has been associated with resistance against virulent Verticillium spp.14 However, there is ongoing debate whether only one or both are functional R-proteins.14,15
Kawchuk et al.14 suggested that Ve1 and Ve2 independently confer resistance to virulent Verticillium in potato. On the other hand, Ve1 but not Ve2 was shown to provide Vd1 resistance in MoneyMaker tomato and Arabidopsis.15,16 For the tomato Ve R-proteins, a candidate fungal effector is the Ave1 protein isolated from V. dahliae race 1 strains by high-throughput population genome sequencing.17 Ave1 has been hypothesized to interact with the Ve1 protein, thereby activating downstream defense responses,18 and apparently contributes to fungal virulence.17 Additionally, it can activate a hypersensitive response (HR) by co-expression with Ve1 in tobacco but not in Arabidopsis.18,19 This tobacco HR was exploited recently for mutational analyses of the Ve proteins. The leucine-rich repeat (LRR) domain was revealed to be important for Ve protein function, specifically the eLRR1-eLRR8, eLRR20-eLRR23 and eLRR32-eLRR37 regions.20 The cytoplasmic tail of the Ve1 protein is required to activate immune signaling while its counterpart in Ve2 did not have this function.21 Similar studies utilizing stably expressing tomato transgenics are still lacking and have not been pursued.
To better understand the function of the Ave1 effector in planta, the pFAST-R02-Ave1 construct18 containing the Verticillium Ave1 gene downstream of the 35S CaMV promoter was used to transform tomato plants cv. Craigella GCR26 (Ve1-) using Agrobacterium-mediated transformation modified from an original procedure by McCormick et al.22 The 35S: Ave1 transformants did not look morphologically different from the untransformed controls. Total nucleic acid was extracted from 0.5 g of fresh plant material as previously described23,24 and mRNA levels were determined by RT-PCR analyses as also previously described.25,26 Transgene presence was assessed by PCR amplification of Ave1 (Fig. 1, right panel) with primers IDT510 (5′-GAGCGGATCCTTATATCTGTCTAAATTCG) and IDT511 (5′-GATACAGAATAAAATGCC). RT-PCR amplification of the Ave1 mRNA with the same primers was used to identify a positive line that was used for subsequent analyses of defense-related transcript levels. Gene expression values were normalized against a reference housekeeping gene actin. Statistically significant values were determined based on the Student's t-test (P < 0.05).
Figure 1.
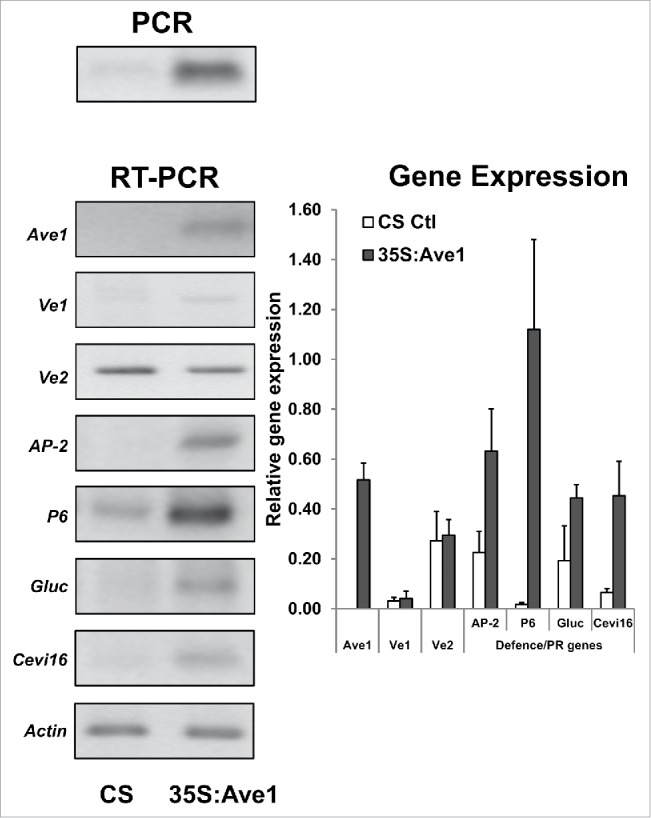
Gene expression in a tomato plant expressing the Verticillium Ave1. The Ave1-pFAST-02 binary vector construct was used to transform Craigella GCR26 tomato plants, as described in the text. Total nucleic acid was prepared from the 35S:Ave1 transgenic line 002 and used as a template for PCR amplification with Ave1-specific primers (upper left panel). Subsequently, the total nucleic acid was used as a template for RT-PCR amplification with primers specific to Ave1, Ve1, Ve2 and key defense genes (lower left panel). PCR and RT-PCR products were fractionated on 2% agarose gels and images were captured using Molecular Analyst software (Bio-Rad). Untransformed plants were used as negative control (CS). Actin was used as the internal control for gene expression. The chart (right panel) summarizes the average transcript levels (±SD) relative to actin for at least three 3 RT-PCR replicates.
As shown in Fig. 1 and in contrast to Ve2 mRNA, Ave1 mRNA is present only in the transformed plant and only traces of mutant Ve1 mRNA were observed. The latter observation is consistent with a premature termination codon,15 which is known to result in nonsense mediated mRNA decay.27 Nevertheless, in the absence of Ve1 protein, the 35S:Ave1 transgenic plant showed upregulation of key defense-related and pathogenesis-related (PR) genes such as anionic peroxidase 2 (AP2), the PR-1 protein P6, βbeta-glucanase and Cevi16 peroxidase (Fig. 1). These genes were chosen because they were representative of genes induced most significantly in Vd1-infected tomato plants.28 PR proteins typically comprise the majority of soluble protein change during the plant defense response.29,30
Overall, our results indicate that the Ave1 effector protein is being perceived by the CS tomato plant resulting in changes in defense gene expression. Since the CS isoline possesses a full-length Ve2 receptor but no full length Ve1 protein,15 these observations emphasize the fact that, at least in tomato, Ave1-induced defense gene expression is independent of Ve1 and raises the possibility that the signal is transduced by another receptor, possibly the functional Ve2 protein.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors would like to thank Dr. Bart Thomma (Wageningen University) for providing the pFAST-R02-Ave1 plasmid.
Funding
This study was supported by NSERC Canada research grants (R.N.N. and J.R.), and Vanier Canada and Ontario Graduate Scholarships (C.D.M.C.).
References
- 1.Tjamos EC, Beckman CH. Vascular wilt diseases of plants. Berlin Heidelberg (Germany: ): Springer-Verlag; 1989; http://dx.doi.org/ 10.1007/978-3-642-73166-2 [DOI] [Google Scholar]
- 2.Luo X, Xie C, Dong J, Yang X, Sui A. Interactions between Verticillium dahliae and its host: vegetative growth, pathogenicity, plant immunity. Appl Microbiol Biotechnol 2014; 98:6921-6932; PMID:24928658; http://dx.doi.org/ 10.1007/s00253-014-5863-8 [DOI] [PubMed] [Google Scholar]
- 3.Klimes A, Dobinson KF, Thomma BP, Klosterman SJ. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu Rev Phytopathol 2015; 53:181-198; PMID:26047557; http://dx.doi.org/ 10.1146/annurev-phyto-080614-120224 [DOI] [PubMed] [Google Scholar]
- 4.Beckman CH. The Nature of Wilt Diseases of Plants. St. Paul(MN: ): The American Phytopathological Society (APS) Press; 1987; http://dx.doi.org/ 10.1086/415877 [DOI] [Google Scholar]
- 5.Barbara DJ, Clewes E. Plant pathogenic Verticillium species: how many of them are there? Mol Plant Pathol 2003; 4:297-305; PMID:20569390; http://dx.doi.org/ 10.1046/j.1364-3703.2003.00172.x [DOI] [PubMed] [Google Scholar]
- 6.Bhat RG, Subbarao KV. Host Range Specificity in Verticillium dahliae. Phytopathology 1999; 89:1218-1225; PMID:18944648; http://dx.doi.org/ 10.1094/PHYTO.1999.89.12.1218 [DOI] [PubMed] [Google Scholar]
- 7.Pegg GF, Brady BL. Verticillium wilts, Ed. 1st Wallingford, Oxon (UK: ): CABI Publishing; 2002; http://dx.doi.org/ 10.1079/9780851995298.0000 [DOI] [Google Scholar]
- 8.Fradin EF, Thomma BPHJ. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 2006; 7:71-86; PMID:20507429; http://dx.doi.org/ 10.1111/j.1364-3703.2006.00323.x [DOI] [PubMed] [Google Scholar]
- 9.Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. Diversity, pathogenicity, and management of verticillium species. Annu Rev Phytopathol 2009; 47:39-62; PMID:19385730; http://dx.doi.org/ 10.1146/annurev-phyto-080508-081748 [DOI] [PubMed] [Google Scholar]
- 10.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 2003; 54:23-61; PMID:14502984; http://dx.doi.org/ 10.1146/annurev.arplant.54.031902.135035 [DOI] [PubMed] [Google Scholar]
- 11.Jones JD, Dangl JL. The plant immune system. Nature 2006; 444:323-329; PMID:17108957; http://dx.doi.org/ 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Liu X, Dai L, Wang G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics 2007; 34:765-776; PMID:17884686; http://dx.doi.org/ 10.1016/S1673-8527(07)60087-3 [DOI] [PubMed] [Google Scholar]
- 13.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 2013; 51:245-266; PMID:23663002; http://dx.doi.org/ 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- 14.Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, et al.. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 2001; 98:6511-6515; PMID:11331751; http://dx.doi.org/ 10.1073/pnas.091114198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fradin EF, Zhang Z, Juarez-Ayala JC, Castroverde CD, Nazar RN, Robb J, Liu CM, Thomma BP. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 2009; 150:320-332; PMID:19321708; http://dx.doi.org/ 10.1104/pp.109.136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 2011; 156:2255-2265; PMID:21617027; http://dx.doi.org/ 10.1104/pp.111.180067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, et al.. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci USA 2012; 109:5110-5115; PMID:22416119; http://dx.doi.org/ 10.1073/pnas.1119623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, van Esse HP, van Damme M, Fradin EF, Liu CM, Thomma BP. Ve1-mediated resistance against Verticillium does not involve a hypersensitive response in Arabidopsis. Mol Plant Pathol 2013; 14:719-727; PMID:23710897; http://dx.doi.org/ 10.1111/mpp.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Thomma BP. Virus-induced gene silencing and Agrobacterium tumefaciens-mediated transient expression in Nicotiana tabacum. Methods Mol Biol 2014; 1127:173-181; PMID:24643561; http://dx.doi.org/ 10.1007/978-1-62703-986-4_14 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Song Y, Liu CM, Thomma BP. Mutational analysis of the Ve1 immune receptor that mediates Verticillium resistance in tomato. PLoS One 2014; 9:e99511; PMID:24911915; http://dx.doi.org/24505431 10.1371/journalpone.0088208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fradin EF, Zhang Z, Rovenich H, Song Y, Liebrand TW, Masini L, van den Berg GC, Joosten MH, Thomma BP. Functional analysis of the tomato immune receptor Ve1 through domain swaps with its non-functional homolog Ve2. PLoS One 2014; 9:e88208; PMID:24505431; http://dx.doi.org/ 10.1371/journal.pone.0088208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 1986; 5:81-84; PMID:24248039; http://dx.doi.org/ 10.1007/BF00269239 [DOI] [PubMed] [Google Scholar]
- 23.Nazar RN, Hu X, Schmidt J, Culham D, Robb J. Potential use of PCR-amplified detection and differentiation of Verticillium wilt pathogens. Physiol Mol Plant Pathol 1991; 39:1-11; http://dx.doi.org/ 10.1016/0885-5765(91)90027-F [DOI] [Google Scholar]
- 24.Robb EJ, Nazar RN. Factors contributing to successful PCR-based diagnostics for potato and other crops In: Marshall G, editor Diagnostics in Crop Production. Surrey (UK: ): British Crop Protection Council; 1996. [Google Scholar]
- 25.Shittu HO, Castroverde CD, Nazar RN, Robb J. Plant-endophyte interplay protects tomato against a virulent Verticillium. Planta 2009; 229:415-426; PMID:18979117; http://dx.doi.org/ 10.1007/s00425-008-0840-z [DOI] [PubMed] [Google Scholar]
- 26.Robb J, Castroverde CDM, Shittu HO, Nazar RN. Patterns of defense gene expression in the tomato-Verticillium interaction. Botany 2009; 87:993-1006; PMID:15315633; http://dx.doi.org/19190664 10.1139/B09-056 [DOI] [Google Scholar]
- 27.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol 2009; 16:107-113; PMID:19190664; http://dx.doi.org/ 10.1038/nsmb.1550 [DOI] [PubMed] [Google Scholar]
- 28.Robb J, Shittu S, Soman KV, Kurosky A, Nazar RN. Arsenal of elevated defense proteins fails to protect tomato against Verticillium dahliae. Planta 2012; 236:623-633; PMID:22481138; http://dx.doi.org/ 10.1007/s00425-012-1637-7 [DOI] [PubMed] [Google Scholar]
- 29.Castroverde CDM, Nazar RN, Robb J. Defense Genes in Tomato. Hauppage (NY: ): Nova Publishers; 2010. [Google Scholar]
- 30.van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 2006; 44:135-162; PMID:16602946; http://dx.doi.org/ 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]


