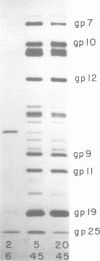
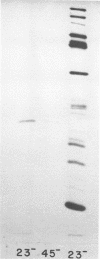
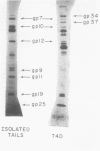
Abstract
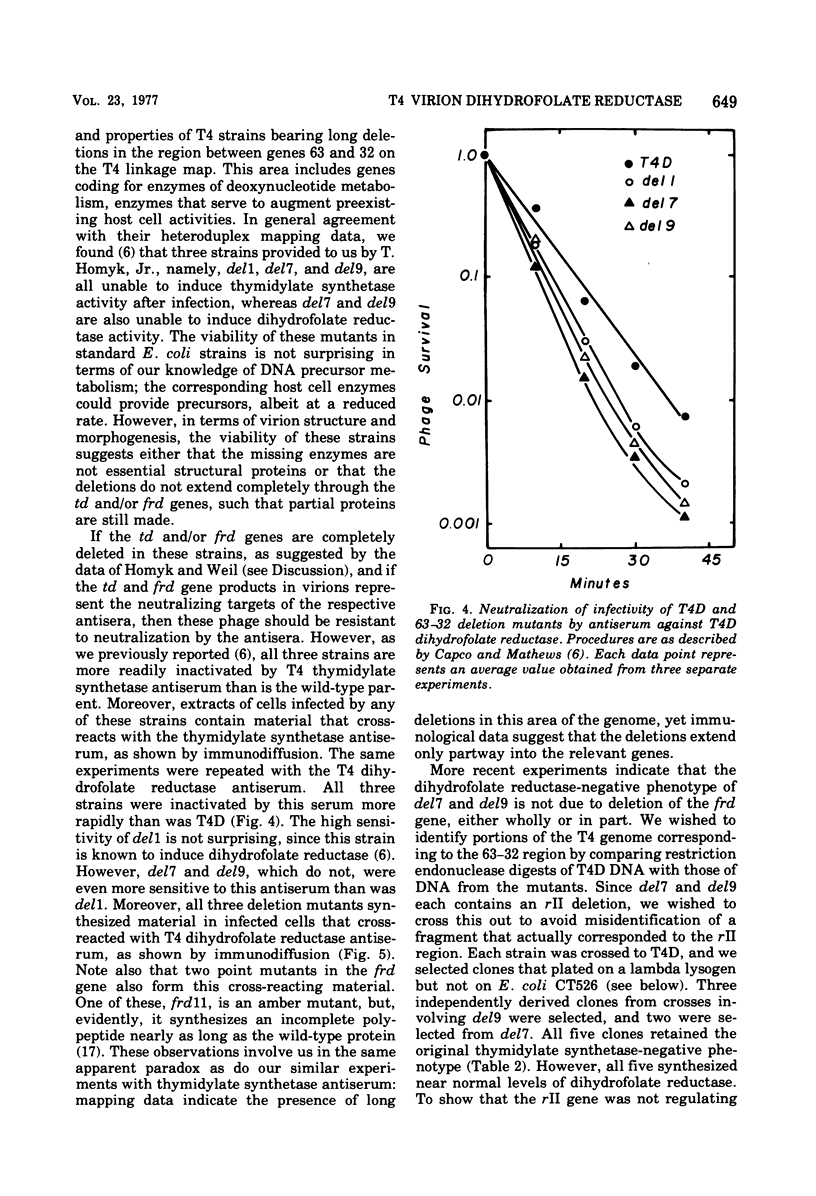
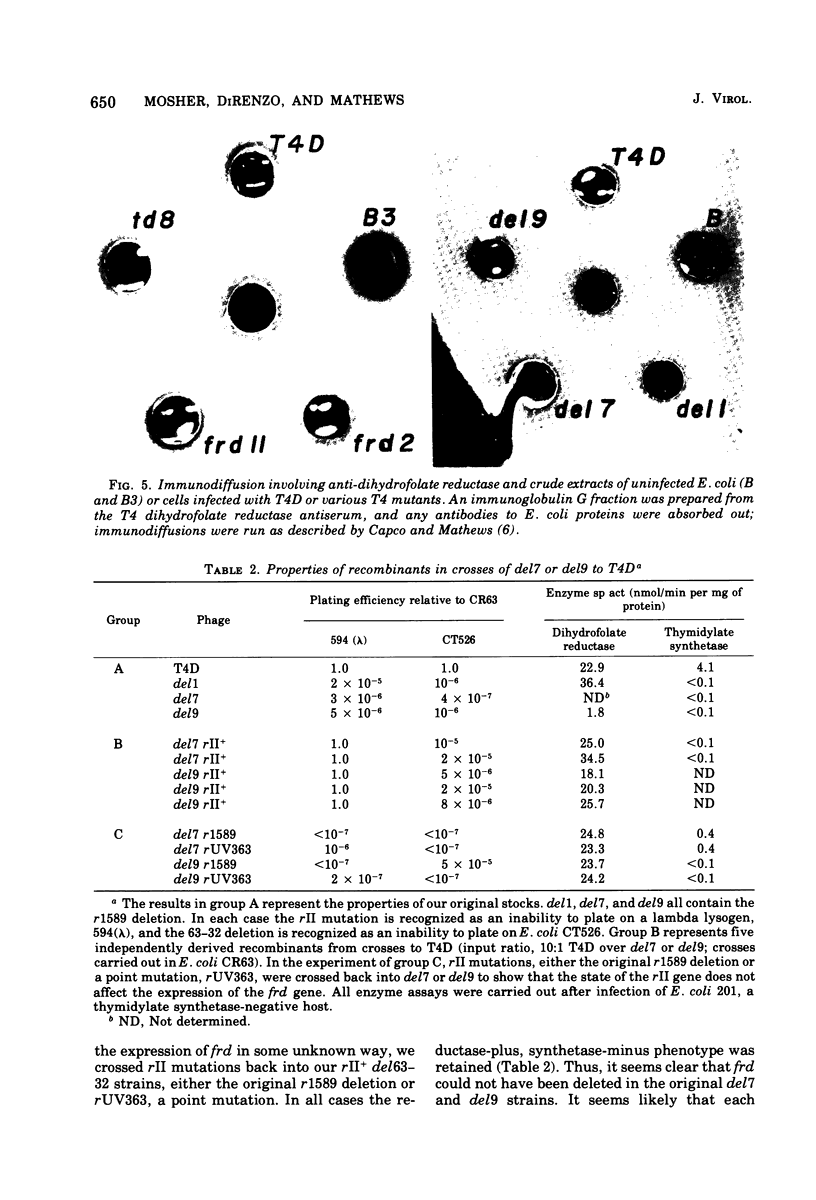
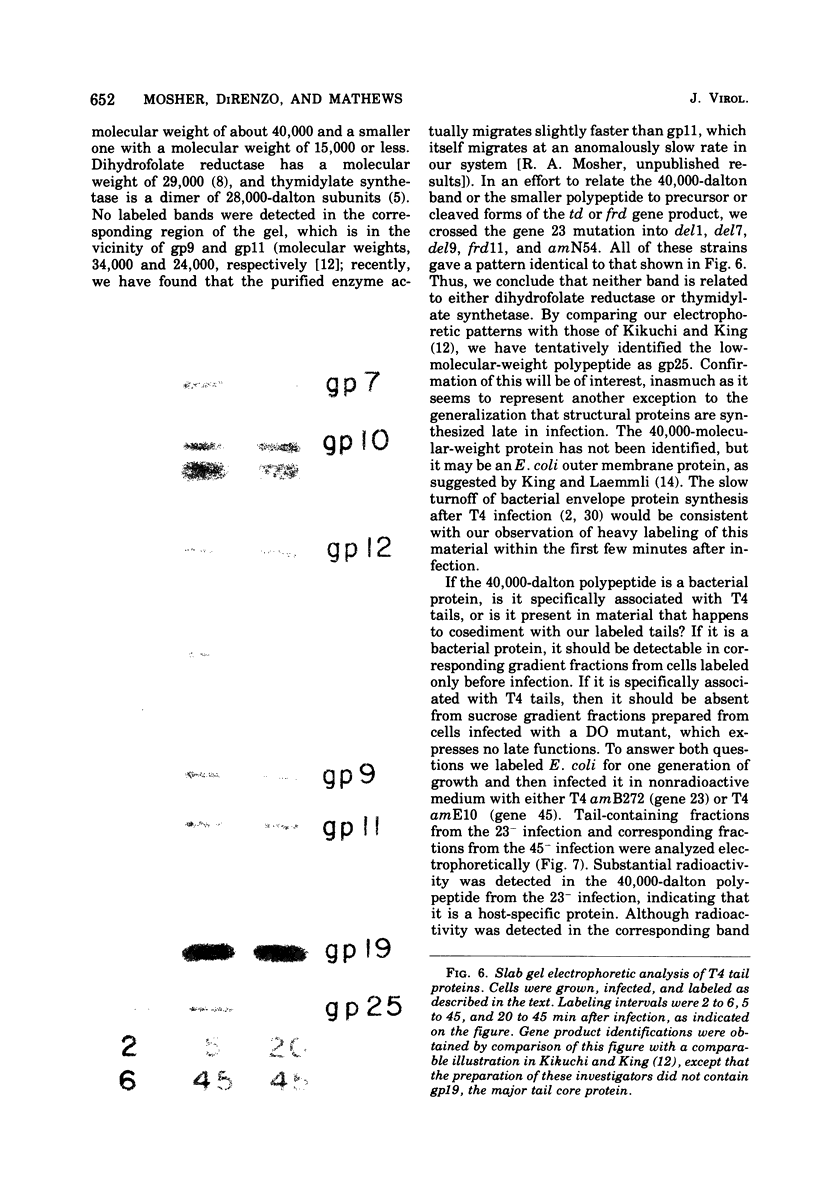
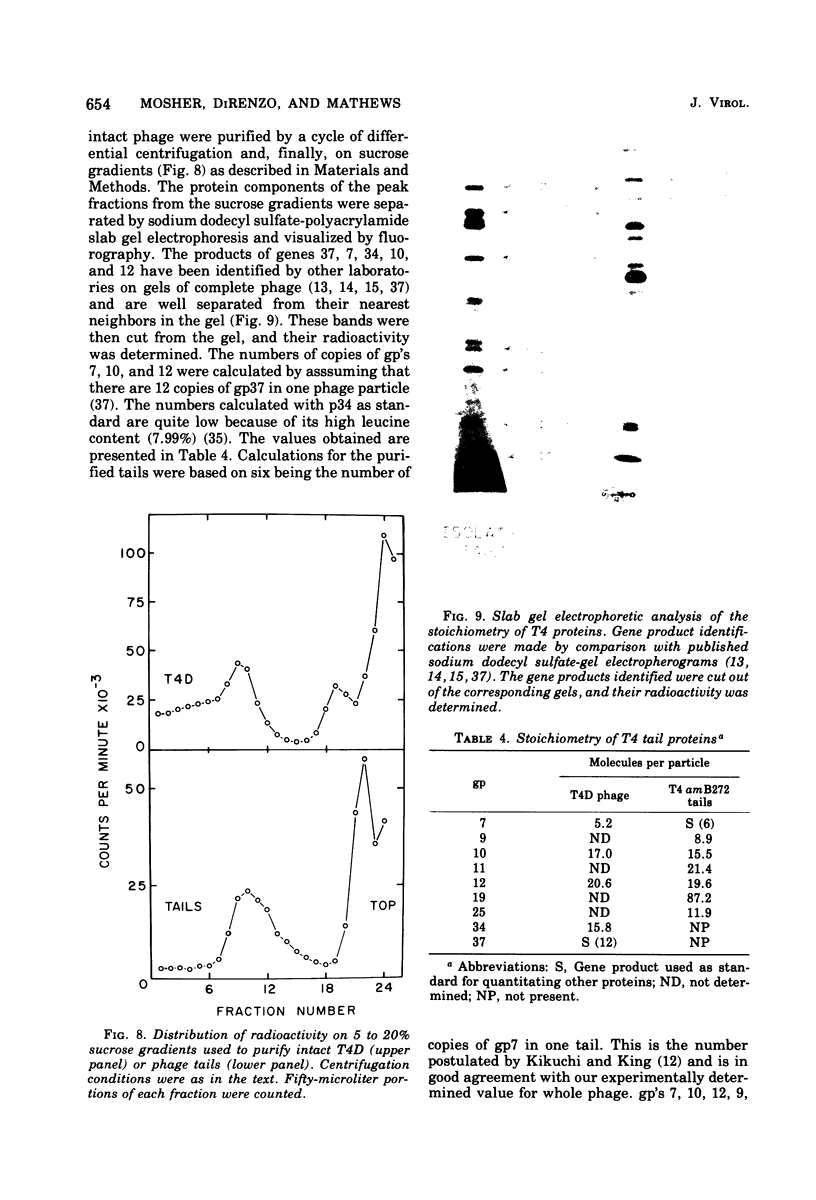
This paper is concerned with the physiological role(s) of T4 phage-coded dihydrofolate reductase, which functions both in DNA precursor metabolism and as a virion protein. (i) We have detected enzyme activity in noninfectious particles produced under restrictive conditions by gene 11 mutants. This supports the conclusion of Kozloff et al. (J. Virol. 16:1401-1408, 1975) that the protein lies in the baseplate, covered by the gene 11 protein. (ii) We have obtained further evidence for virion dihydrofolate reductase as the target for neutralizing activity of T4 dihydrofolate reductase antiserum and as a determinant of the heat lability of the virion. This derives from our observation that the reductases specified by T4B and T4D differ in several properties. (iii) We have investigated several anomalous properties of T4 mutants bearing deletions that reportedly extend into or through the frd gene, which codes for dihydrofolate reductase. Evidence is presented that the deletions in fact do not extend through frd. These strains direct the synthesis of material that cross-reacts with antiserum to homogeneous dihydrofolate reductase. Moreover, they are all quite sensitive to the phage-neutralizing effects of this antiserum. In addition, they are restricted by several of the hospital strains, wild-type strains of Escherichia coli supplied by the California Institute of Technology group. (iv) We have attempted to detect dihydrofolate reductase among early-synthesized proteins present in T4 tails. Two such proteins are seen, one of which is evidently the gene 25 product and one that is a bacterial protein. Quantitation of our electrophoretic technique has allowed determination of the number of molecules of some T4 tail components present per virion. (v) Finally, we have compared the T4 dihydrofolate reductase with the corresponding enzyme specified by two plasmids conferring resistance to trimethoprim (Skold and Widh, J. Biol. Chem. 249:4324-4325, 1974). Although the enzymes are similar in some properties, they differ in several important respects, including immunological activity.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baros A., Witmer H. J. Effect of chloramphenicol and starvation for an essential amino acid on the synthesis and decay of T4 bacteriophage-specific messengers transcribed from early and quasi-late promoters. Arch Biochem Biophys. 1975 Aug;169(2):415–427. doi: 10.1016/0003-9861(75)90183-6. [DOI] [PubMed] [Google Scholar]
- Beckey A. D., Wulff J. L., Earhart C. F. Early synthesis of membrane protein after bacteriophage T4 infection. J Virol. 1974 Oct;14(4):886–894. doi: 10.1128/jvi.14.4.886-894.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Anderson T. F. Effect of host cell wall material on the adsorbability of cofactor-requiring T4. J Virol. 1969 Jul;4(1):94–108. doi: 10.1128/jvi.4.1.94-108.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capco G. R., Krupp J. R., Mathews C. K. Bacteriophage-coded thymidylate synthetase: characteristics of the T4 and T5 enzymes. Arch Biochem Biophys. 1973 Oct;158(2):726–735. doi: 10.1016/0003-9861(73)90567-5. [DOI] [PubMed] [Google Scholar]
- Capco G. R., Mathews C. K. Bacteriophage-coded thymidylate synthetase. Evidence that the T4 enzyme is a capsid protein. Arch Biochem Biophys. 1973 Oct;158(2):736–743. doi: 10.1016/0003-9861(73)90568-7. [DOI] [PubMed] [Google Scholar]
- Dawes J., Goldberg E. B. Functions of baseplate components in bacteriophage T4 infection. I. Dihydrofolate reductase and dihydropteroylhexaglutamate. Virology. 1973 Oct;55(2):380–390. doi: 10.1016/0042-6822(73)90178-5. [DOI] [PubMed] [Google Scholar]
- Erickson J. S., Mathews C. K. T4 bacteriophage-specific dihydrofolate reductase: purification to homogeneity by affinity chromatography. Biochem Biophys Res Commun. 1971 Jun 4;43(5):1164–1170. doi: 10.1016/0006-291x(71)90585-7. [DOI] [PubMed] [Google Scholar]
- FREUNDLICH M., BURNS R. O., UMBARGER H. E. Control of isoleucine, valine, and leucine biosynthesis. I. Multivalent repression. Proc Natl Acad Sci U S A. 1962 Oct 15;48:1804–1808. doi: 10.1073/pnas.48.10.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H. Mutants of bacteriophage T4 unable to induce dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1967 Aug;58(2):584–591. doi: 10.1073/pnas.58.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homyk T., Jr, Weil J. Deletion analysis of two nonessential regions of the T4 genome. Virology. 1974 Oct;61(2):505–523. doi: 10.1016/0042-6822(74)90286-4. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975 Dec 25;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Bacteriophage T4 tail assembly: structural proteins and their genetic identification. J Mol Biol. 1973 Apr 5;75(2):315–337. doi: 10.1016/0022-2836(73)90024-7. [DOI] [PubMed] [Google Scholar]
- King J., Laemmli U. K. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- King J., Mykolajewycz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J Mol Biol. 1973 Apr 5;75(2):339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- Kozloff L. M., Crosby L. K., Lute M. Bacteriophage T4 baseplate components. III. Location and properties of the bacteriophage structural thymidylate synthetase. J Virol. 1975 Dec;16(6):1409–1419. doi: 10.1128/jvi.16.6.1409-1419.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Crosby L. K., Lute M., Hall D. H. Bacteriophage T4 baseplate components. II. Binding and location of bacteriophage-induced dihydrofolate reductase. J Virol. 1975 Dec;16(6):1401–1408. doi: 10.1128/jvi.16.6.1401-1408.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Crosby L. K. Bacteriophage T4 virion baseplate thymidylate synthetase and dihydrofolate reductase. J Virol. 1977 Sep;23(3):637–644. doi: 10.1128/jvi.23.3.637-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Lute M., Crosby L. K., Rao N., Chapman V. A., DeLong S. S. Bacteriophage tail components. I. Pteroyl polyglutamates in T-even bacteriophages. J Virol. 1970 Jun;5(6):726–739. doi: 10.1128/jvi.5.6.726-739.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozloff L. M., Verses C., Lute M., Crosby L. K. Bacteriophage tail components. II. Dihydrofolate reductase in T4D bacteriophage. J Virol. 1970 Jun;5(6):740–753. doi: 10.1128/jvi.5.6.740-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATHEWS C. K., SUTHERLAND K. E. COMPARATIVE BIOCHEMISTRY OF BACTERIAL AND PHAGE-INDUCED DIHYDROFOLATE REDUCTASES. J Biol Chem. 1965 May;240:2142–2147. [PubMed] [Google Scholar]
- Mathews C. K. Biochemistry of deoxyribonucleic acid-defective amber mutant of bacteriophage T4. I. Ribonucleic acid metabolism. J Biol Chem. 1968 Nov 10;243(21):5610–5615. [PubMed] [Google Scholar]
- Mathews C. K. Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. 3. Nucleotide pools. J Biol Chem. 1972 Nov 25;247(22):7430–7438. [PubMed] [Google Scholar]
- Mathews C. K., Crosby L. K., Kozloff L. M. Inactivation of T4D bacteriophage by antiserum against bacteriophage dihydrofolate reductase. J Virol. 1973 Jul;12(1):74–78. doi: 10.1128/jvi.12.1.74-78.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews C. K. Evidence that bacteriophage-induced dihydrofolate reductase in a viral gene product. J Biol Chem. 1967 Sep 25;242(18):4083–4086. [PubMed] [Google Scholar]
- Mathews C. K. Growth of a dihydrofolate reductaseless mutant of bacteriophage T4. J Virol. 1967 Oct;1(5):963–967. doi: 10.1128/jvi.1.5.963-967.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews C. K. Identity of genes coding for soluble and structural dihydrofolate reductases in bacteriophage T4. J Virol. 1971 Apr;7(4):531–533. doi: 10.1128/jvi.7.4.531-533.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C. K. Deoxyribonucleic acid metabolism and virus-induced enzyme synthesis in a thymine-requiring bacterium infected by a thymine-requiring bacteriophage. Biochemistry. 1966 Jun;5(6):2092–2100. doi: 10.1021/bi00870a042. [DOI] [PubMed] [Google Scholar]
- Pollock P. N., Duckworth D. H. Outer-membrane proteins induced by T4 bacteriophage. Biochim Biophys Acta. 1973 Oct 18;322(2):321–328. doi: 10.1016/0005-2795(73)90307-3. [DOI] [PubMed] [Google Scholar]
- Silverstein J. L., Goldberg E. B. T4 DNA injection. II. Protection of entering DNA from host exonuclease V. Virology. 1976 Jul 1;72(1):212–223. doi: 10.1016/0042-6822(76)90324-x. [DOI] [PubMed] [Google Scholar]
- Simon L. D., Swan J. G., Flatgaard J. E. Functional defects in T4 bacteriophages lacking the gene 11 and gene 12 products. Virology. 1970 May;41(1):77–90. doi: 10.1016/0042-6822(70)90056-5. [DOI] [PubMed] [Google Scholar]
- Sköld O., Widh A. A new dihydrofolate reductase with low trimethoprim sensitivity induced by an R factor mediating high resistance to trimethoprim. J Biol Chem. 1974 Jul 10;249(13):4324–4325. [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Takata R., Tsugita A. Morphogenesis of the tail fibre of bacteriophage T4: isolation and characterization of a component (the gene 34 product). J Mol Biol. 1970 Nov 28;54(1):45–57. doi: 10.1016/0022-2836(70)90445-6. [DOI] [PubMed] [Google Scholar]
- Ward S., Dickson R. C. Assembly of bacteriophage T4 tail fibers. 3. Genetic control of the major tail fiber polypeptides. J Mol Biol. 1971 Dec 28;62(3):479–492. doi: 10.1016/0022-2836(71)90149-5. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Function of the bacteriophage T4 transfer RNA's. J Mol Biol. 1973 Mar 15;74(4):753–757. doi: 10.1016/0022-2836(73)90065-x. [DOI] [PubMed] [Google Scholar]