Abstract
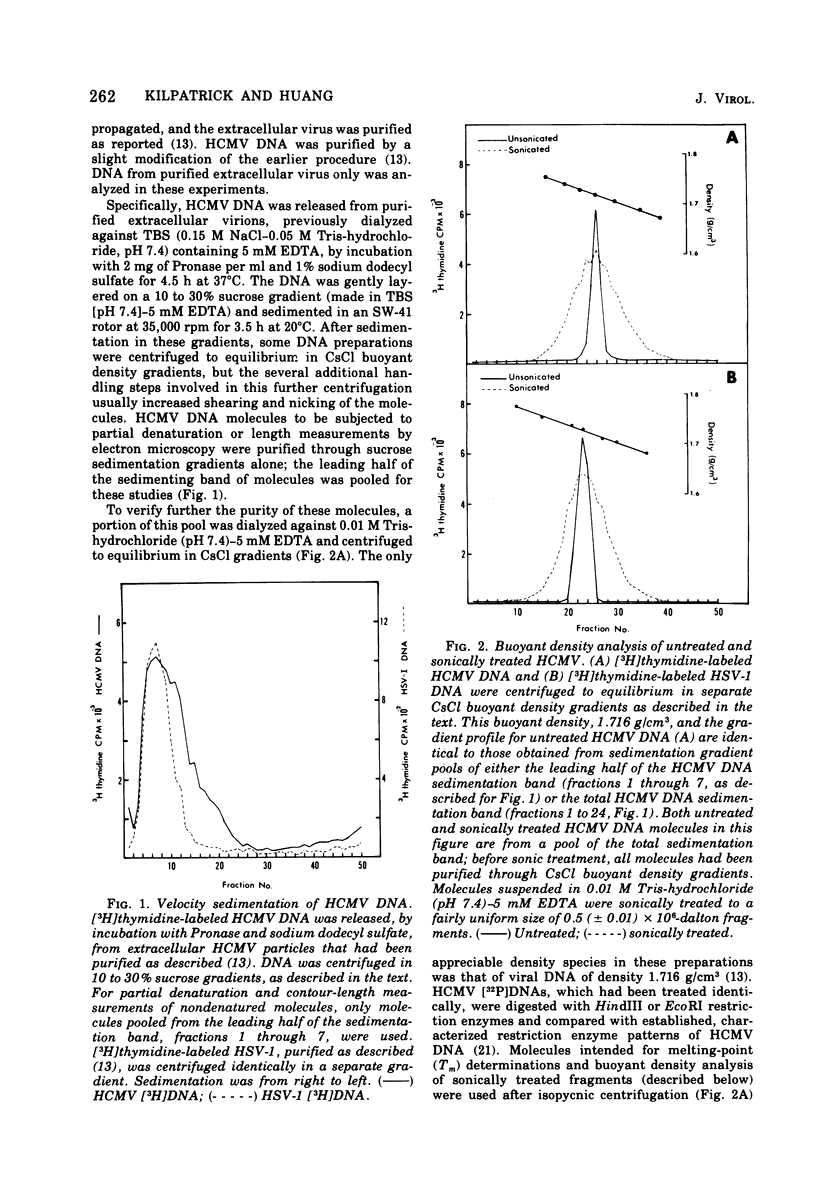
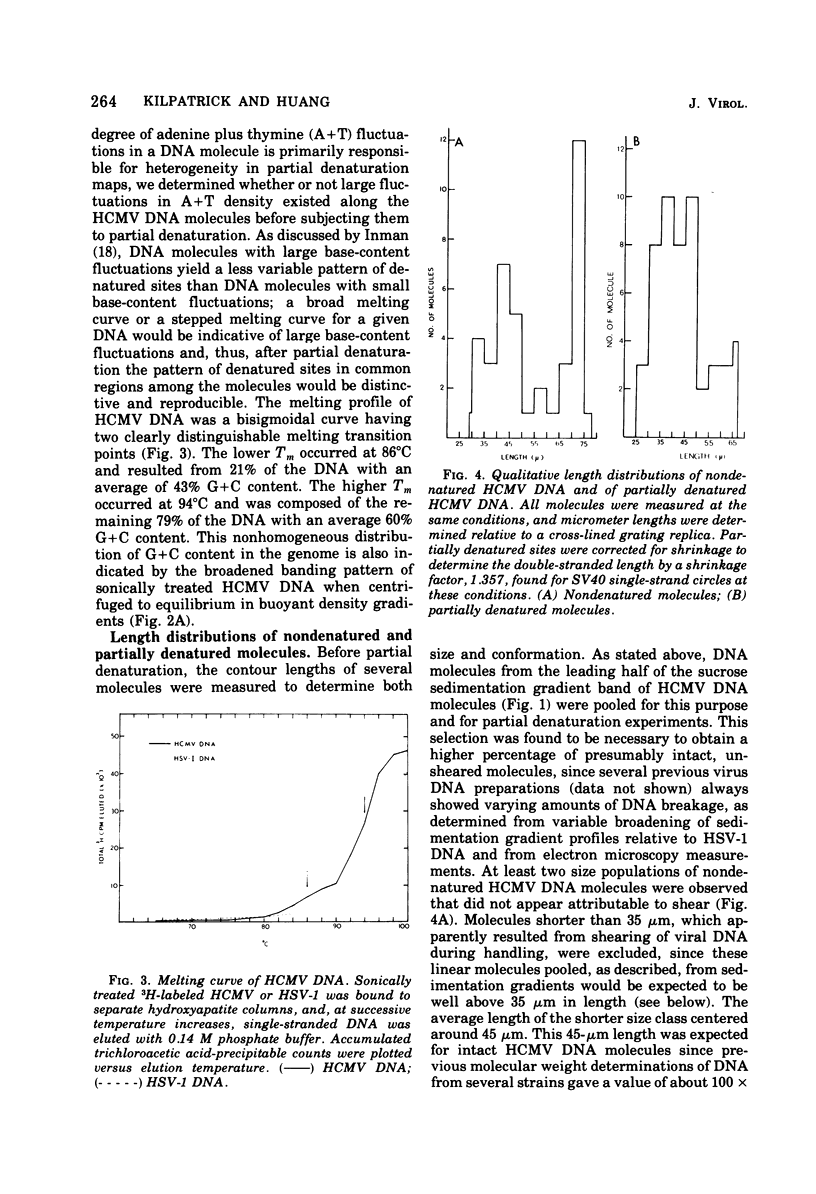
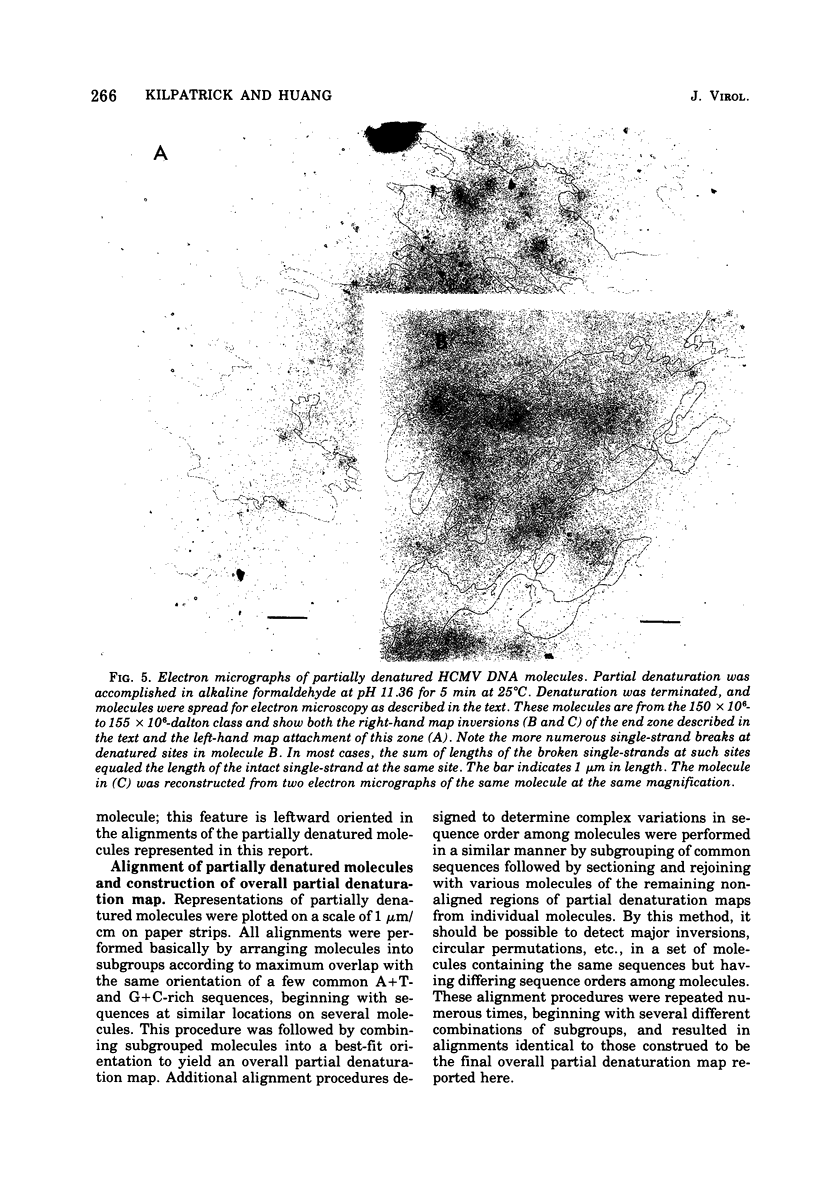
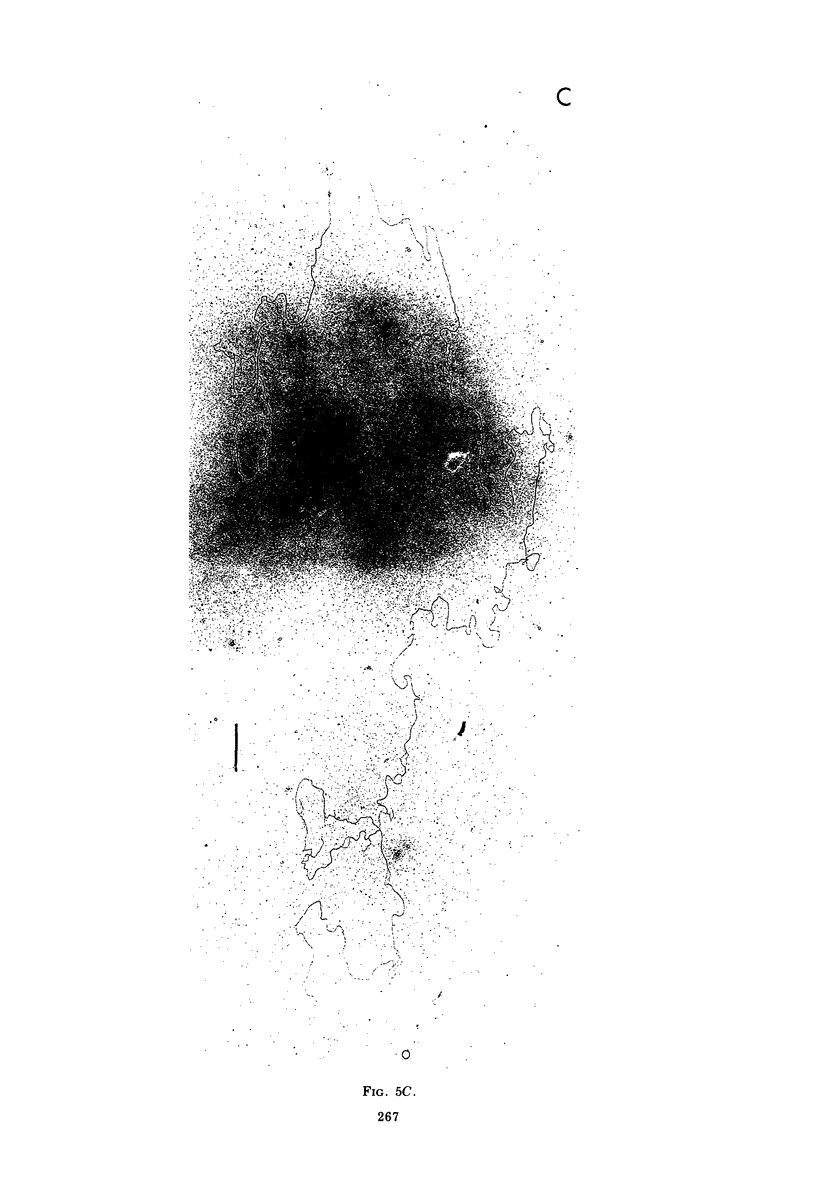
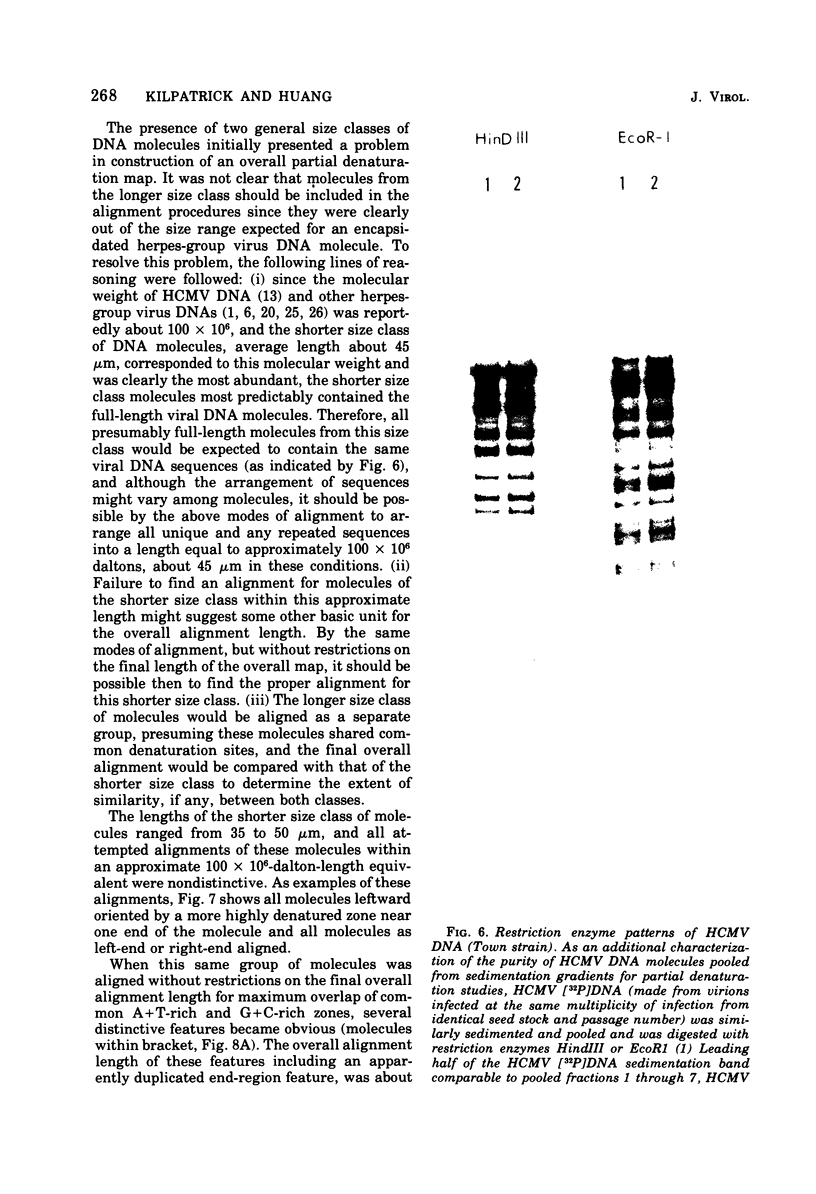
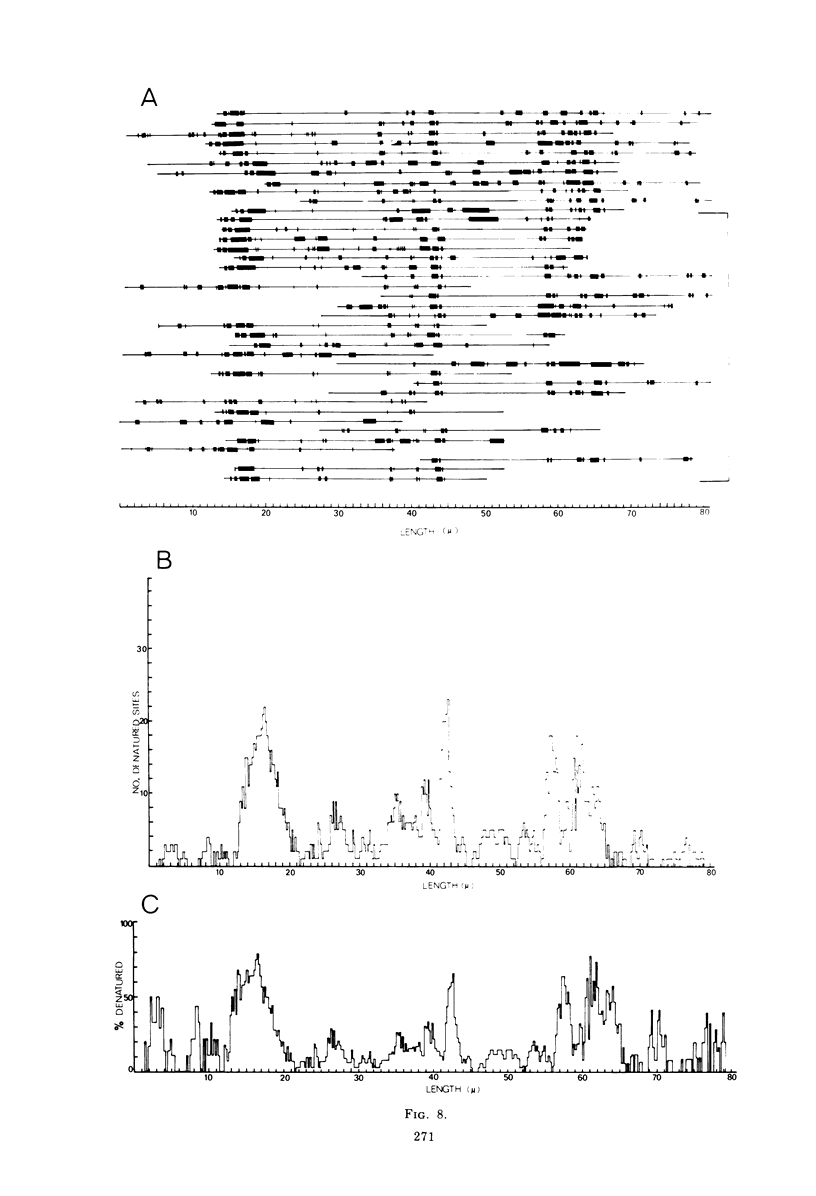
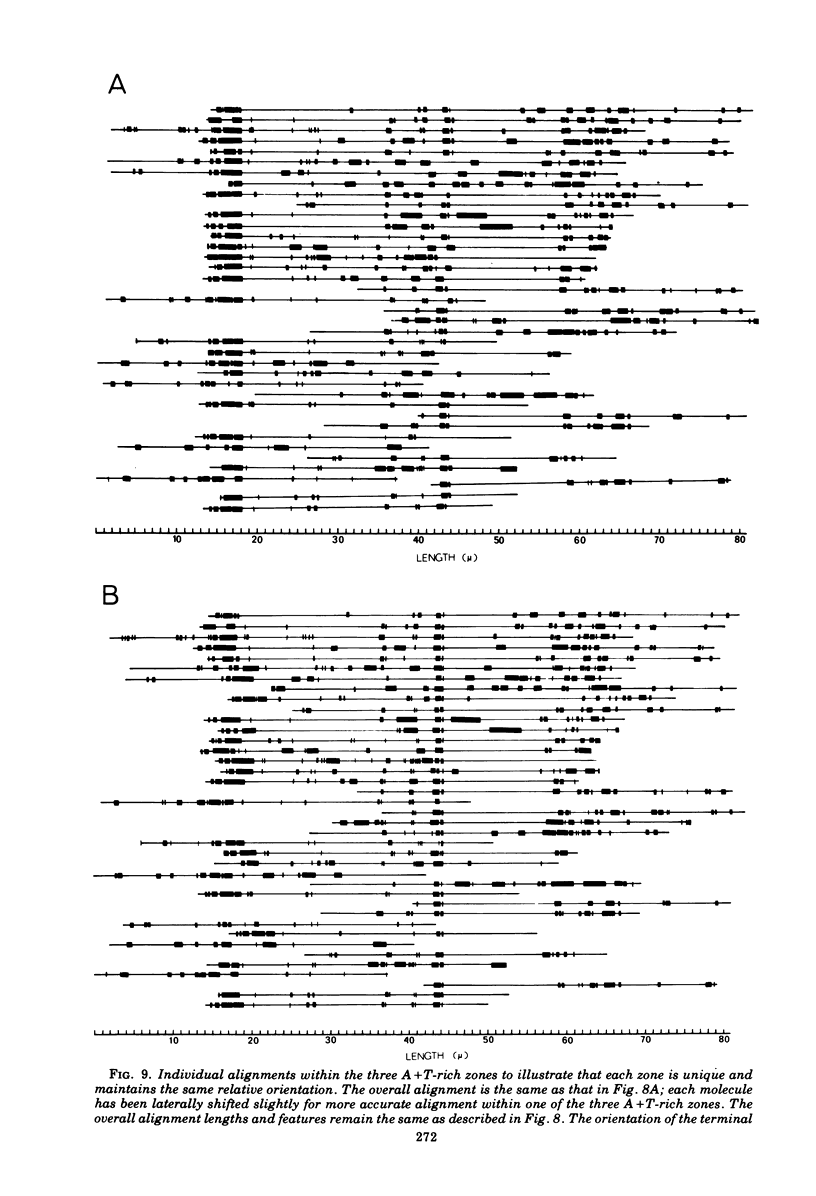
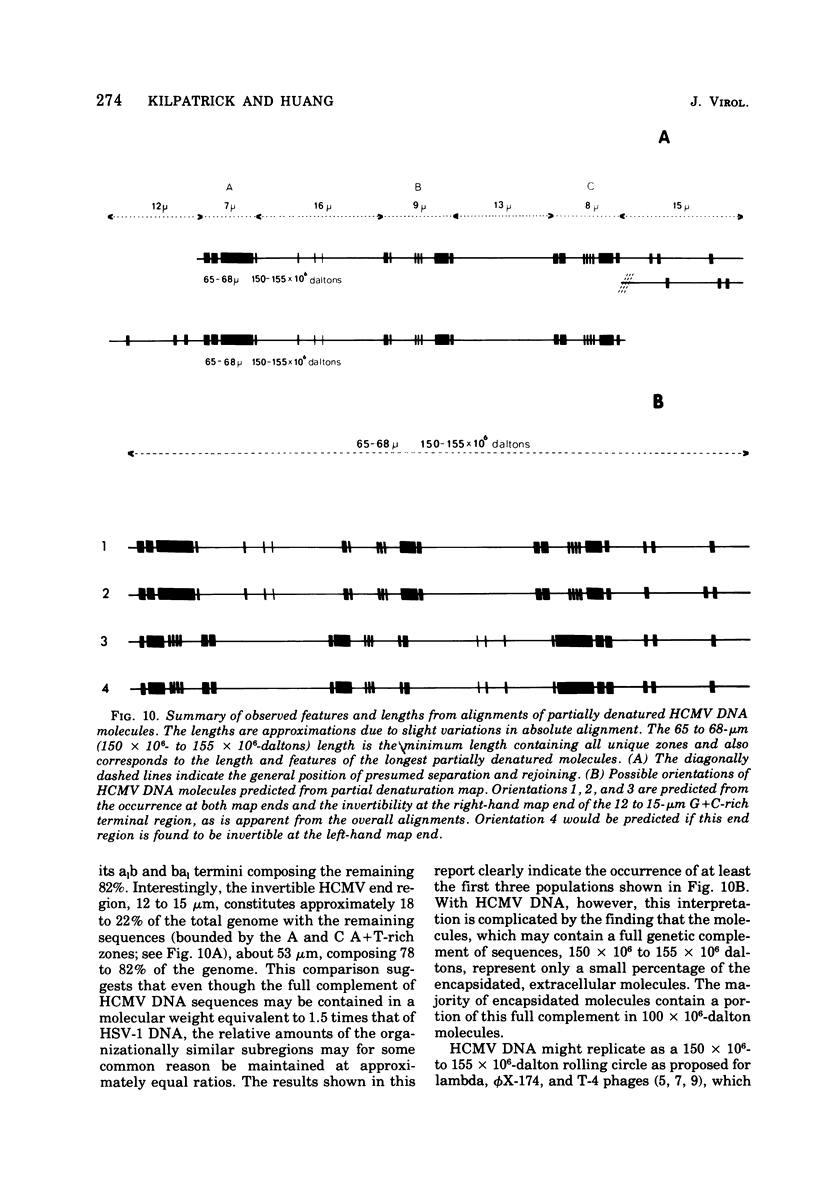
Contour-length measurements of both nondenatured and partially denatured DNA from purified extracellular human cytomegalovirus indicate that more than one size class of viral DNA is encapsidated. In addition to a size class averaging about 100 × 106 daltons, a much less abundant class of larger viral DNA molecules, 150 × 106 to 155 × 106 daltons, was extracted from purified extracellular virus. As predicted by melting-curve analysis, partial denaturation of human cytomegalovirus DNA generates denaturation maps showing distinctive adenine plus thymidine (A+T)-rich and guanine plus cytosine (G+C)-rich localizations. Alignment of partial denaturation maps of both 100 × 106- and 150 × 106- to 155 × 106-dalton molecules from maximum overlap of common A+T- and G+C-rich zones clearly shows six unique zones contained in a length equal to the longest class, 150 × 106 to 155 × 106 daltons. However, various alignments of the smaller class of the molecules within the confines of the approximately 100 × 106-dalton-length equivalent are nondistinctive. Of the six unique A+T- and G+C-rich zones, five are linked in a specific sequence and maintain the same relative orientation; these features indicate the absence of major inversions within these zones. The sixth unique zone may occur at either end of this five-zone series, but it was never found at both ends of the same molecule. Additionally, this terminal zone appears to undergo complete inversions at least at one end of the alignment, and perhaps at both. These data indicate that 150 × 106- to 155 × 106-dalton molecules comprise human cytomegalovirus-specific genetic information.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker Y., Dym H., Sarov I. Herpes simplex virus DNA. Virology. 1968 Oct;36(2):184–192. doi: 10.1016/0042-6822(68)90135-9. [DOI] [PubMed] [Google Scholar]
- Bujard H. Electron microscopy of single-stranded DNA. J Mol Biol. 1970 Apr 14;49(1):125–137. doi: 10.1016/0022-2836(70)90381-5. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dressler D., Wolfson J. The rolling circle for phi X DNA replication. 3. Synthesis of supercoiled duplex rings. Proc Natl Acad Sci U S A. 1970 Sep;67(1):456–463. doi: 10.1073/pnas.67.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Wolf H. Purification and properties of Herpesvirus saimiri DNA. Virology. 1974 Mar;58(1):55–64. doi: 10.1016/0042-6822(74)90140-8. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. DNA replication after T4 infection. Cold Spring Harb Symp Quant Biol. 1968;33:485–493. doi: 10.1101/sqb.1968.033.01.056. [DOI] [PubMed] [Google Scholar]
- Furukawa T., Fioretti A., Plotkin S. Growth characteristics of cytomegalovirus in human fibroblasts with demonstration of protein synthesis early in viral replication. J Virol. 1973 Jun;11(6):991–997. doi: 10.1128/jvi.11.6.991-997.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley P. M., Khoury G., Byrne J. C., Takemoto K. K., Martin M. A. Physical map of the BK virus genome. J Virol. 1975 Oct;16(4):959–973. doi: 10.1128/jvi.16.4.959-973.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Chen S. T., Pagano J. S. Human cytomegalovirus. I. Purification and characterization of viral DNA. J Virol. 1973 Dec;12(6):1473–1481. doi: 10.1128/jvi.12.6.1473-1481.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Kilpatrick B. A., Huang Y. T., Pagano J. S. Detection of human cytomegalovirus and analysis of strain variation. Yale J Biol Med. 1976 Mar;49(1):29–43. [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Newbold J. E., Pagano J. S. Analysis of simian virus 40 DNA with the restriction enzyme of Haemophilus aegyptius, endonuclease Z. J Virol. 1973 Apr;11(4):508–514. doi: 10.1128/jvi.11.4.508-514.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S., Pagano J. S. Human cytomegalovirus. II. Lack of relatedness to DNA of herpes simples I and II, Epstein-Barr virus, and nonhuman strains of cytomegalovirus. J Virol. 1974 Mar;13(3):642–645. doi: 10.1128/jvi.13.3.642-645.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B. Denaturation mapping of DNA. Methods Enzymol. 1974;29:451–458. doi: 10.1016/0076-6879(74)29037-2. [DOI] [PubMed] [Google Scholar]
- Inman R. B. Denaturation maps of the left and right sides of the lambda DNA molecule determined by electron microscopy. J Mol Biol. 1967 Aug 28;28(1):103–116. doi: 10.1016/s0022-2836(67)80081-0. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
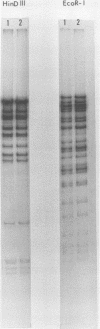
- Kilpatrick B. A., Huang E. S., Pagano J. S. Analysis of cytomegalovirus genomes with restriction endonucleases Hin D III and EcoR-1. J Virol. 1976 Jun;18(3):1095–1105. doi: 10.1128/jvi.18.3.1095-1105.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mulder C., Delius H. Specificity of the break produced by restricting endonuclease R 1 in Simian virus 40 DNA, as revealed by partial denaturation mapping. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3215–3219. doi: 10.1073/pnas.69.11.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoyama M., Pagano J. S. Replication of viral deoxyribonucleic acid and breakdown of cellular deoxyribonucleic acid in Epstein-Barr virus infection. J Virol. 1972 Apr;9(4):714–716. doi: 10.1128/jvi.9.4.714-716.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoyama M., Pagano J. S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972 Aug 9;238(84):169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]