Abstract
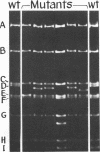
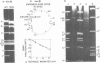
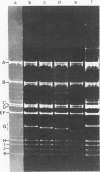
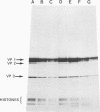
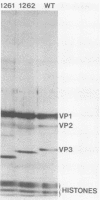
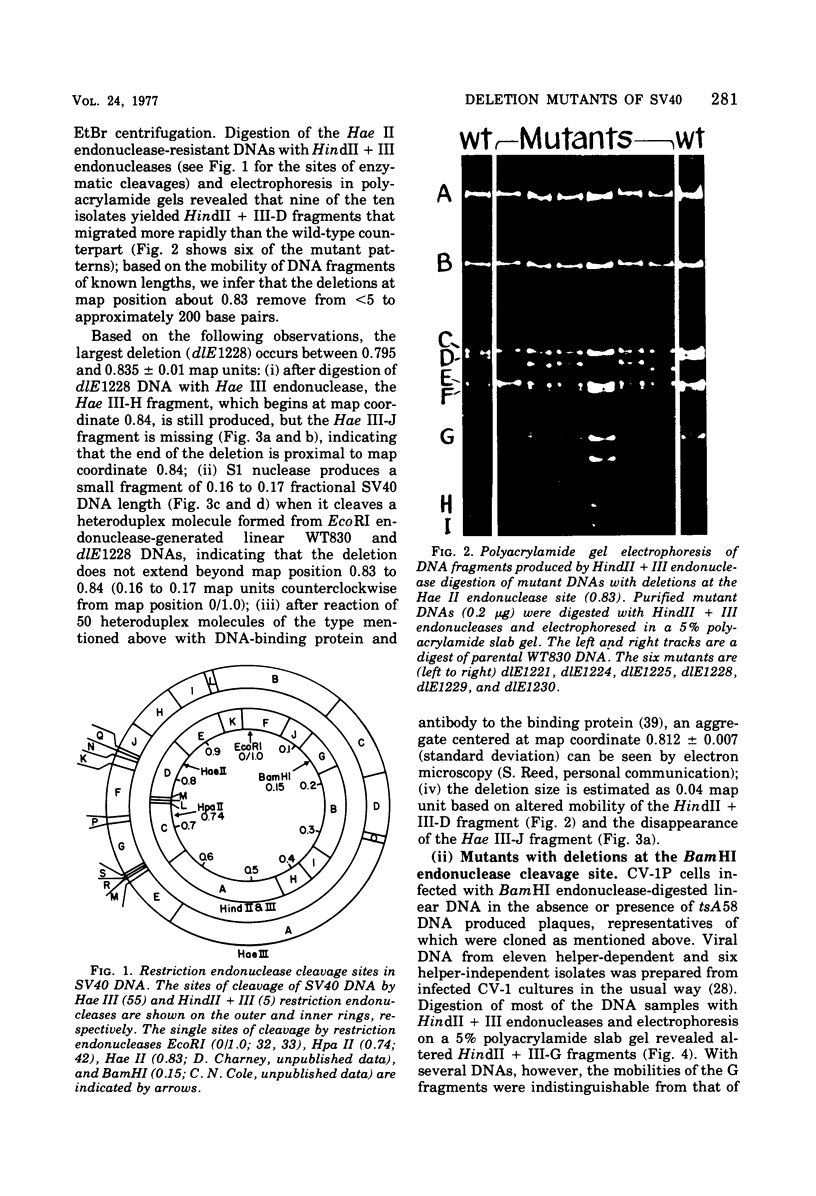
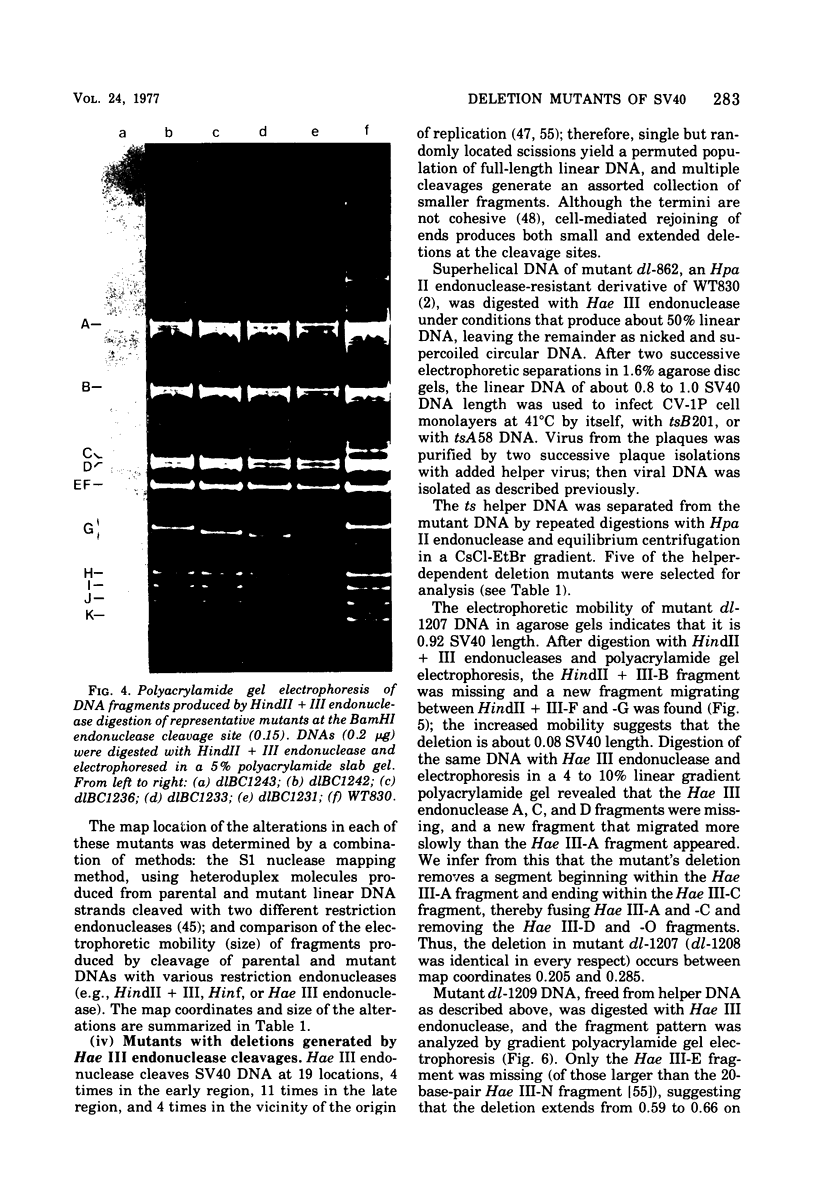
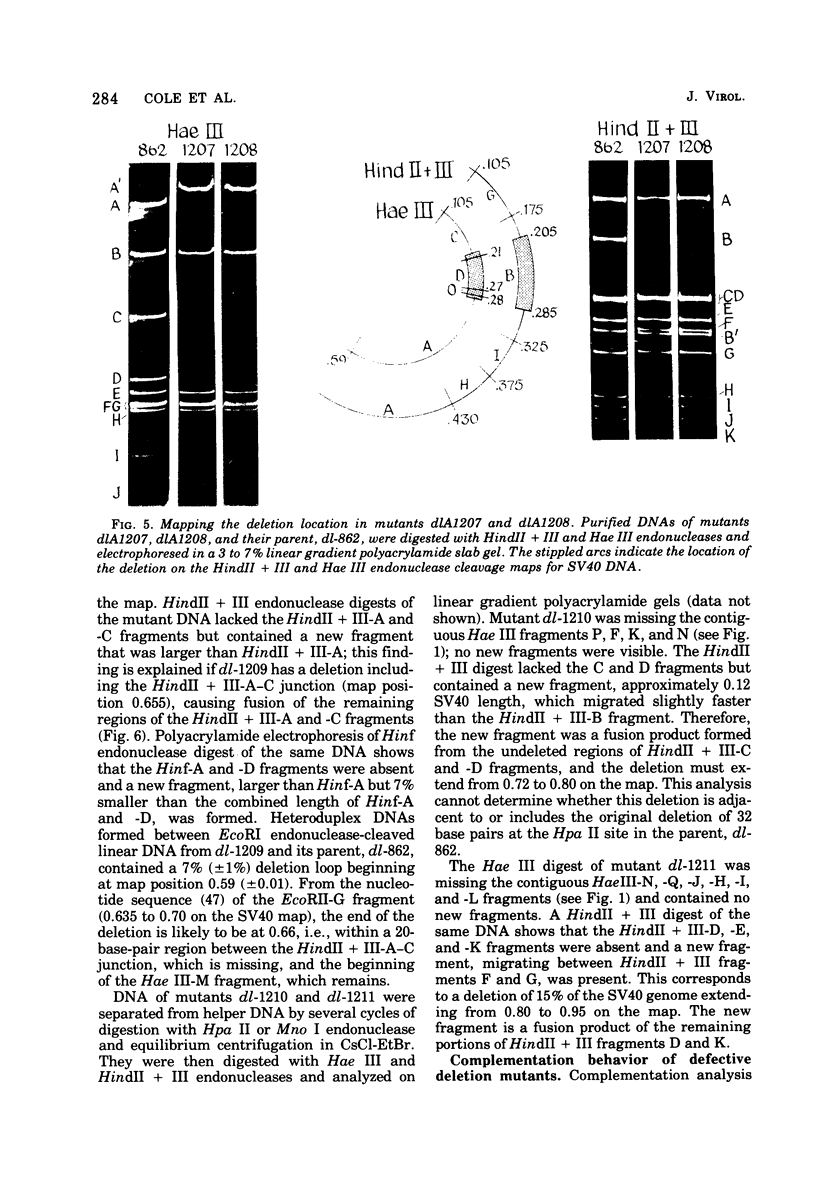
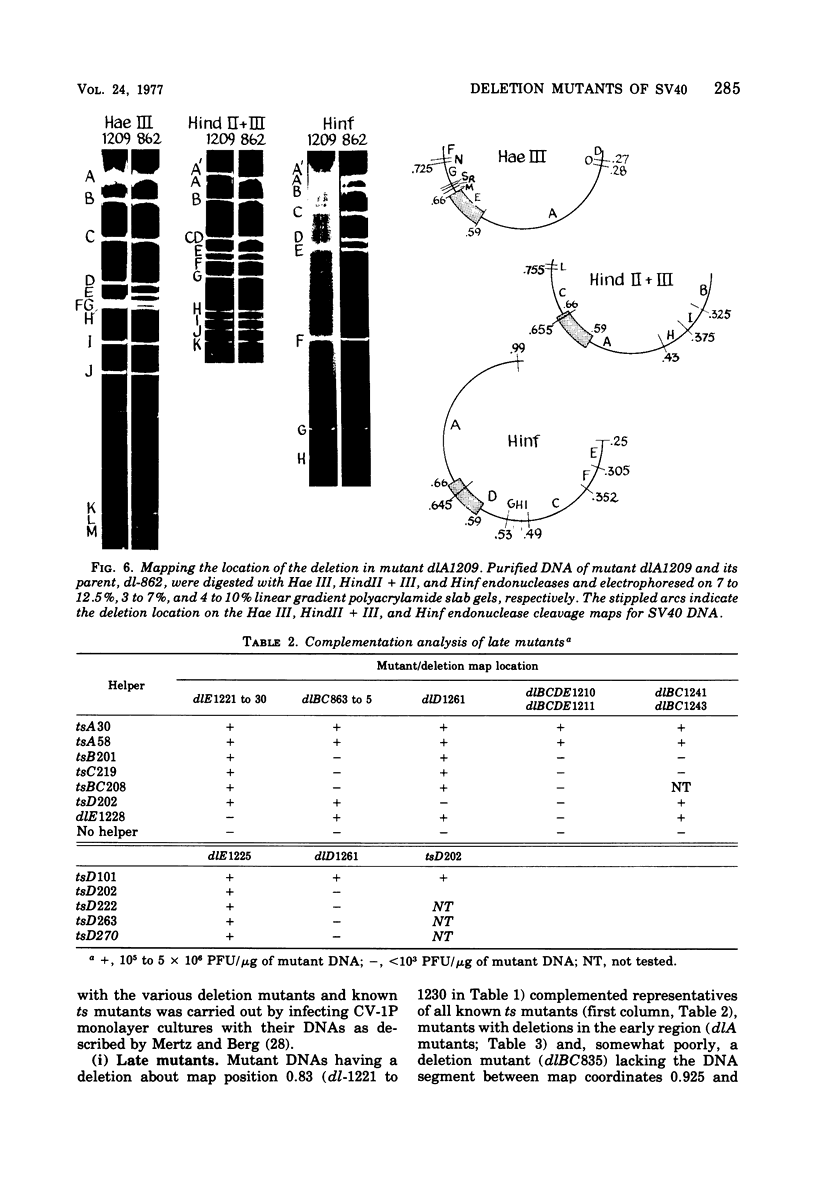
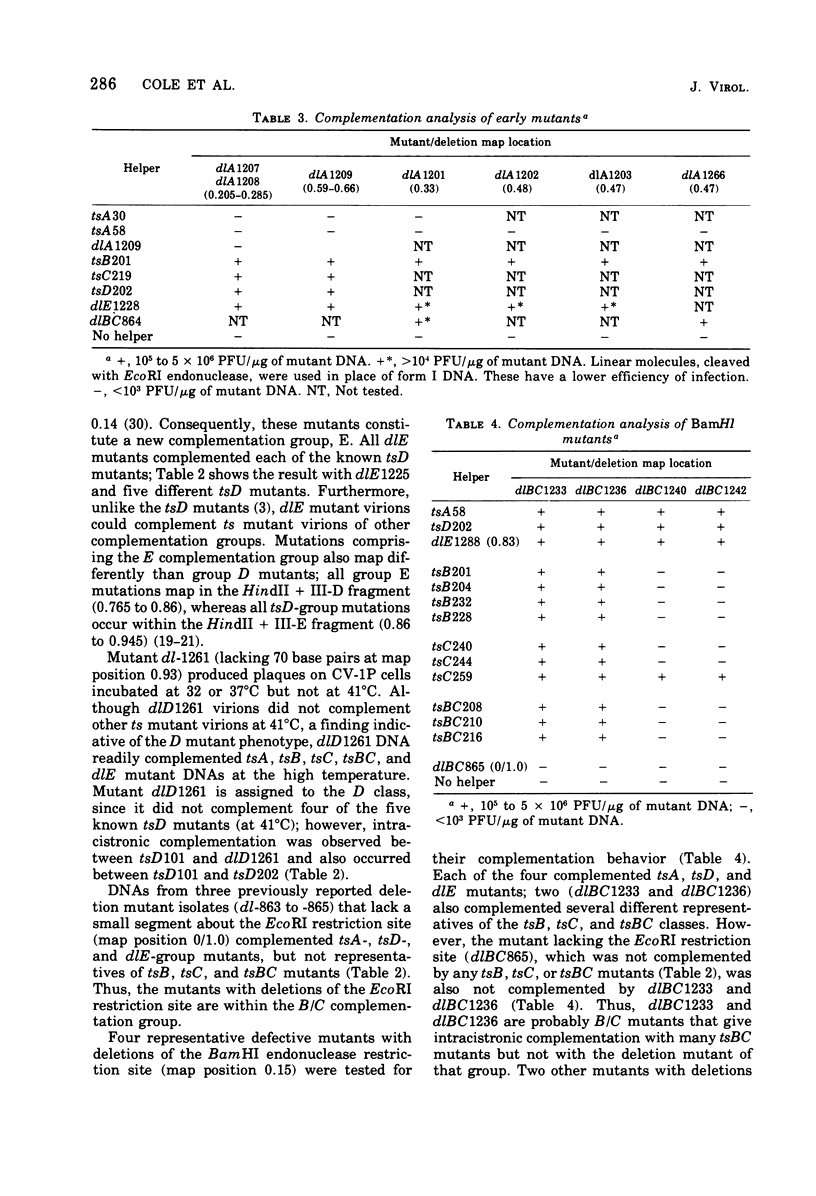
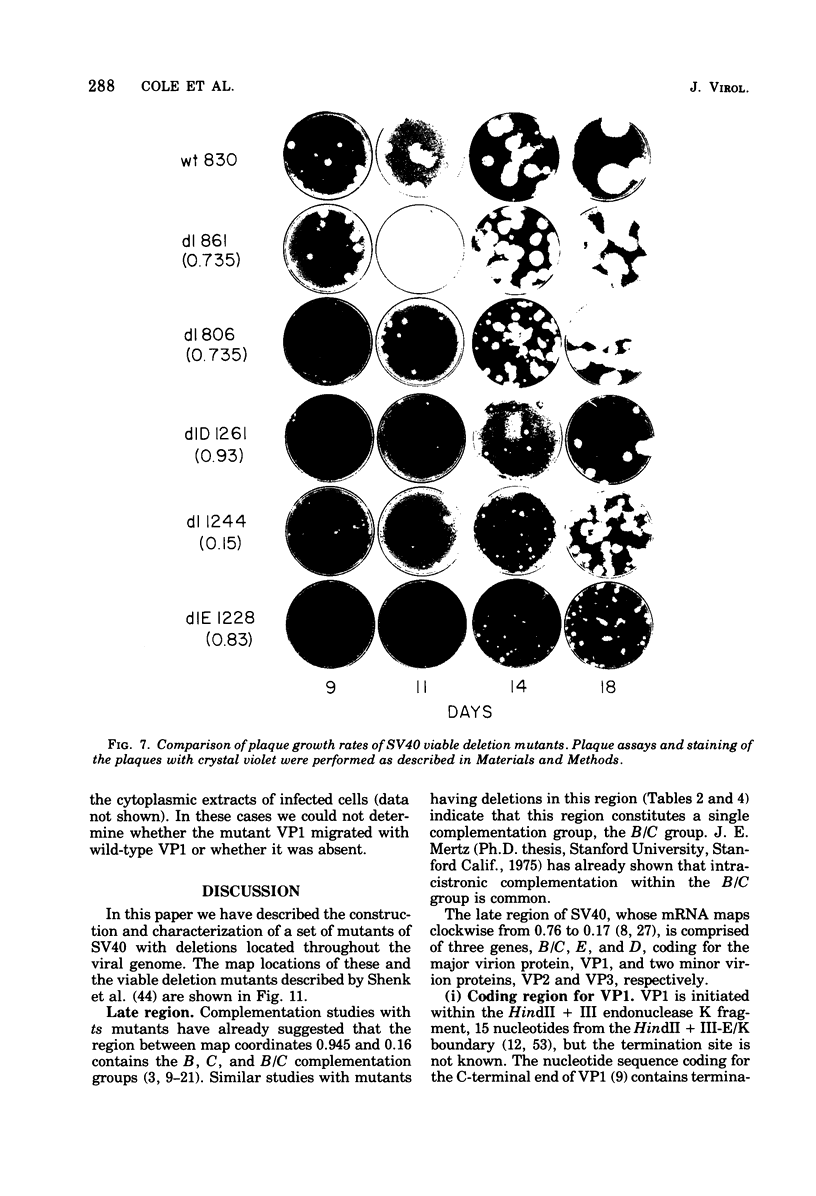
Mutants of simian virus 40 (SV40), with deletions ranging in size from fewer than 3 to 750 base pairs located throughout the SV40 genome, were obtained by infecting CV-1P cells with linear SV40 DNA and DNA of an appropriate helper virus. The linear DNA was obtained by complete cleavage of closed circular DNA with Hae II or Bam HI endonuclease or partial cleavage with either Hae III endonuclease or nuclease S1, followed, in some cases, by mild digestion with phage λ 5′ -exonuclease. The following mutants with deletions in the late region of the SV40 genome were obtained and characterized. Ten, containing deletions at the Hae II endonuclease site (map location 0.83), define a new genetic complementation group, E, grow extremely slowly without helper virus, and cause alterations only in VP2. Two mutants with deletions in the region 0.92 to 0.945 affect both VP2 and VP3, demonstrating that VP3 shares sequences with the C-terminal portion of VP2. The mutant with a deletion at 0.93 is the first deletion mutant in the D complementation group and is also temperature sensitive; the mutant with a deletion at 0.94 is viable and grows normally. Three mutants with deletions at the EcoRI endonuclease site (0/1.0) and eleven with deletions at the BamHI endonuclease site (0.15) fall into the B/C complementation group. Six additional mutants with deletions at the BamHI endonuclease site are viable, growing more slowly than wild type. VP1 is the only polypeptide affected by mutants in the B/C group. A mutant with a deletion of the region 0.72 to 0.80 has a polar effect, failing to express the E, D, and B/C genes. Mutants with deletions in the early region (0.67 counterclockwise to 0.17) at 0.66 to 0.59, 0.48, 0.47, 0.33, and 0.285 to 0.205 are all members of the A complementation group. Thus, the A gene is the only viral gene in the early region whose expression is necessary for productive infection of permissive cells. Since mutants with deletions in the region 0.59 to 0.54 are viable, two separate regions are essential for expression of the gene A function: 0.66 to 0.59 and 0.54 to 0.21. Mutants with deletions at 0.21 and 0.18 are viable. Approximate map locations of SV40 genes and possible models for their regulation are discussed.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binstock J. F., Pramanik A., Schulz H. Isolation of a multi-enzyme complex of fatty acid oxidation from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):492–495. doi: 10.1073/pnas.74.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Shenk T. E., Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Martin R. G. Complementation analysis of simian virus 40 mutants. J Virol. 1974 May;13(5):1101–1109. doi: 10.1128/jvi.13.5.1101-1109.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Martin R. G. Products of complementation between temperature-sensitive mutants of simian virus 40. J Virol. 1975 Jan;15(1):127–136. doi: 10.1128/jvi.15.1.127-136.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Dhar R., Subramanian K. N., Pan J., Weissman S. M. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. IV. Localization of the SV40 DNA complementary to the 5' ends of viral mRNA. J Biol Chem. 1977 Jan 10;252(1):368–376. [PubMed] [Google Scholar]
- Dhar R., Subramanian K. N., Pan J., Weissman S. M. Structure of a large segment of the genome of simian virus 40 that does not encode known proteins. Proc Natl Acad Sci U S A. 1977 Mar;74(3):827–831. doi: 10.1073/pnas.74.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., Zain S., Weissman S. M., Pan J., Subramanian K. Nucleotide sequences of RNA transcribed in infected cells and by Escherichia coli RNA polymerase from a segment of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1974 Feb;71(2):371–375. doi: 10.1073/pnas.71.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M. K., Huang E. S., Pagano J. S. Structural polypeptides of simian virus 40. J Virol. 1971 May;7(5):635–641. doi: 10.1128/jvi.7.5.635-641.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M., Marty L., Suarez F. Capsid proteins of Simian virus 40. Biochem Biophys Res Commun. 1970 Jul 13;40(1):97–102. doi: 10.1016/0006-291x(70)91051-x. [DOI] [PubMed] [Google Scholar]
- HOLLAND J. J., McLAREN L. C. Improved method for staining cell monolayers for virus plaque counts. J Bacteriol. 1959 Oct;78:596–597. doi: 10.1128/jb.78.4.596-597.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Howley P., Nathans D., Martin M. Posttranscriptional selection of simian virus 40-specific RNA. J Virol. 1975 Feb;15(2):433–437. doi: 10.1128/jvi.15.2.433-437.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. A map of temperature-sensitive mutants of simian virus 40. Virology. 1975 Jul;66(1):70–81. doi: 10.1016/0042-6822(75)90179-8. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Deletion mutants of simian virus 40 generated by enzymatic excision of DNA segments from the viral genome. J Mol Biol. 1974 Oct 15;89(1):179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Mapping temperature-sensitive mutants of simian virus 40: rescue of mutants by fragments of viral DNA. Virology. 1974 Aug;60(2):466–475. doi: 10.1016/0042-6822(74)90340-7. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Mapping the genes of simian virus 40. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):53–60. doi: 10.1101/sqb.1974.039.01.010. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. The B/C gene of simian virus 40. Virology. 1976 Dec;75(2):335–345. doi: 10.1016/0042-6822(76)90032-5. [DOI] [PubMed] [Google Scholar]
- Lavi S., Winocour E. Acquisition of sequences homologous to host deoxyribonucleic acid by closed circular simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):309–316. doi: 10.1128/jvi.9.2.309-316.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. N., Brockman W. W., Nathans D. Evolutionary variants of simian virus 40: cloned substituted variants containing multiple initiation sites for DNA replication. Virology. 1975 Jul;66(1):53–69. doi: 10.1016/0042-6822(75)90178-6. [DOI] [PubMed] [Google Scholar]
- Little J. W., Lehman I. R., Kaiser A. D. An exonuclease induced by bacteriophage lambda. I. Preparation of the crystalline enzyme. J Biol Chem. 1967 Feb 25;242(4):672–678. [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Viable deletion mutants of simian virus 40: selective isolation by means of a restriction endonuclease from Hemophilus parainfluenzae. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4879–4883. doi: 10.1073/pnas.71.12.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E., Carbon J., Herzberg M., Davis R. W., Berg P. Isolation and characterization of individual clones of simian virus 40 mutants containing deletions duplications and insertions in their DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):69–84. doi: 10.1101/sqb.1974.039.01.012. [DOI] [PubMed] [Google Scholar]
- Middleton J. H., Edgell M. H., Hutchison C. A., 3rd Specific fragments of phi X174 deoxyribonucleic acid produced by a restriction enzyme from Haemophilus aegyptius, endonuclease Z. J Virol. 1972 Jul;10(1):42–50. doi: 10.1128/jvi.10.1.42-50.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3365–3369. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder C., Delius H. Specificity of the break produced by restricting endonuclease R 1 in Simian virus 40 DNA, as revealed by partial denaturation mapping. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3215–3219. doi: 10.1073/pnas.69.11.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer H. L. Synthesis and assembly of simian virus 40. I. Differential synthesis of intact virions and empty shells. J Virol. 1972 Jan;9(1):41–51. doi: 10.1128/jvi.9.1.41-51.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C. L., Aviv H., Paterson B. M., Roberts B. E., Rozenblatt S., Revel M., Winocour E. Cell-free translation of messenger RNA of simian virus 40: synthesis of the major capsid protein. Proc Natl Acad Sci U S A. 1974 Feb;71(2):302–306. doi: 10.1073/pnas.71.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C., Gilboa E., Revel M., Winocour E. Cell-free translation of simian virus 40 early messenger RNA coding for viral T-antigen. Proc Natl Acad Sci U S A. 1977 Feb;74(2):457–461. doi: 10.1073/pnas.74.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Alwine J. C. An analysis by electron microscopy of early simian virus 40 RNA from a tsA mutant. Cell. 1977 Jul;11(3):523–531. doi: 10.1016/0092-8674(77)90070-8. [DOI] [PubMed] [Google Scholar]
- Robb J. A., Martin R. G. Genetic analysis of simian virus 40. 3. Characterization of a temperature-sensitive mutant blocked at an early stage of productive infection in monkey cells. J Virol. 1972 Jun;9(6):956–968. doi: 10.1128/jvi.9.6.956-968.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Rozenblatt S., Mulligan R. C., Gorecki M., Roberts B. E., Rich A. Direct biochemical mapping of eukaryotic viral DNA by means of a linked transcription-translation cell-free system. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2747–2751. doi: 10.1073/pnas.73.8.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K., Collins J. K., Tegtmeyer P., Ozer H. L., Lai C. J., Nathans D. Identification of simian virus 40 protein A. J Virol. 1977 Feb;21(2):636–646. doi: 10.1128/jvi.21.2.636-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Subramanian K. N., Dhar R., Weissman S. M., Ghosh P. K. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. II. Sequences of T1 and pancreatic ribonuclease digestion products of RNA transcripts prepared from subfragments of EcoRII-G. J Biol Chem. 1977 Jan 10;252(1):340–354. [PubMed] [Google Scholar]
- Subramanian K. N., Dhar R., Weissman S. M. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. III. Construction of the total sequence of EcoRII-G fragment of SV40 DNA. J Biol Chem. 1977 Jan 10;252(1):355–367. [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K. K., Kirschstein R. L., Habel K. Mutants of simian virus 40 differing in plaque size, oncogenicity, and heat sensitivity. J Bacteriol. 1966 Oct;92(4):990–994. doi: 10.1128/jb.92.4.990-994.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Ozer H. L. Temperature-sensitive mutants of simian virus 40: infection of permissive cells. J Virol. 1971 Oct;8(4):516–524. doi: 10.1128/jvi.8.4.516-524.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Ozer H. L. Temperature-sensitive mutants of simian virus 40: infection of permissive cells. J Virol. 1971 Oct;8(4):516–524. doi: 10.1128/jvi.8.4.516-524.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Voorde A., Contreras R., Rogiers R., Fiers W. The initiation region of the SV40 VP1 gene. Cell. 1976 Sep;9(1):117–120. doi: 10.1016/0092-8674(76)90057-x. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Van de Voorde A., Fiers W. Cleavage map of the simian-virus-40 genome by the restriction endonuclease III of Haemopholus aegyptius. Eur J Biochem. 1976 Jan 2;61(1):101–117. doi: 10.1111/j.1432-1033.1976.tb10002.x. [DOI] [PubMed] [Google Scholar]
- Yoshiike K. Studies on DNA from low-density particles of SV40. I. Heterogeneous defective virions produced by successive undiluted passages. Virology. 1968 Mar;34(3):391–401. doi: 10.1016/0042-6822(68)90059-7. [DOI] [PubMed] [Google Scholar]