Abstract
Transcription initiation is orchestrated by dynamic molecular interactions, with kinetic steps difficult to detect. Utilizing a hybrid method, we aim to unravel essential kinetic steps of transcriptional regulation on the glnAp2 promoter, whose regulatory region includes two enhancers (sites I and II) and three low-affinity sequences (sites III-V), to which the transcriptional activator NtrC binds. By structure reconstruction, we analyze all possible organization architectures of the transcription apparatus (TA). The main regulatory mode involves two NtrC hexamers: one at enhancer II transiently associates with site V such that the other at enhancer I can rapidly approach and catalyze the σ54-RNA polymerase holoenzyme. We build a kinetic model characterizing essential steps of the TA operation; with the known kinetics of the holoenzyme interacting with DNA, this model enables the kinetics beyond technical detection to be determined by fitting the input-output function of the wild-type promoter. The model further quantitatively reproduces transcriptional activities of various mutated promoters. These results reveal different roles played by two enhancers and interpret why the low-affinity elements conditionally enhance or repress transcription. This work presents an integrated dynamic picture of regulated transcription initiation and suggests an evolutionarily conserved characteristic guaranteeing reliable transcriptional response to regulatory signals.
INTRODUCTION
Genetic information is dynamically transcribed with the change of cellular regulatory signals (1–4). Whereas the structural organizations of proteins participating in transcription have been largely determined (5–12), much less is known about the dynamic processes of protein–DNA and protein-protein interactions (12–17). Uncovering such dynamics is not only fundamental to comprehending how transcription is orchestrated, but also essential to interpret the behaviors of gene regulatory networks – due to the resulting complex temporal evolution of transcript numbers (2,4,17–22). Those kinetic steps are hard to detect experimentally, especially when unstable protein complexes and unknown transient interactions are involved (23–26). Recently, the steps of the holoenzyme σ54-RNA polymerase (σ54RNAP) associating with promoter DNA have been dissected (27,28). Nevertheless, how transcriptional activators interact with the cis-regulatory elements and σ54RNAP to control transcription initiation remains unclear. Here, we address this issue in terms of activity from the glnAp2 promoter of Escherichia coli, which is the most extensively studied σ54-depedent promoter.
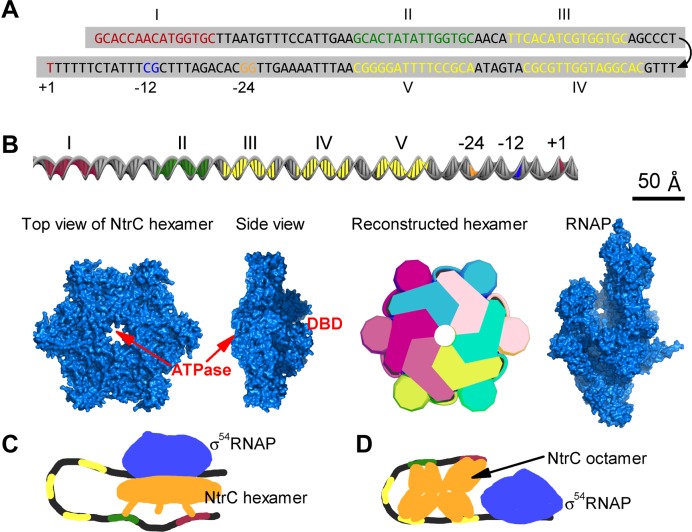
glnAp2 transcription is activated by NtrC in response to nitrogen limitation (29–33). NtrC molecules are dimeric in their inactive state. Upon activation, NtrC dimers are phosphorylated and bind to two enhancers centered at −140 (site I) and −108 (site II) relative to the transcription start site (21,34–37) (Figure 1A and B). The bound dimers have much lower mobility and nucleate free dimers to form NtrC hexamers (38). σ54RNAP binds to the −24–−12 region at one face of the double helix (11). NtrC hexamers catalyze σ54RNAP via DNA looping; the catalysis reaction takes place at approximately −12 region and the edge of the central pore of NtrC hexamer (10,11,29,38). Such a regulatory mode—activators at remote enhancers direct transcription initiation though DNA looping—is similar to that in eukaryotes. Additionally, there are three low-affinity binding sites for NtrC, which are separately centered at −89, −66 and −45 (sites III-V) (21,33,39). These sites are rarely occupied at low and intermediate concentrations of NtrC dimers. Low-affinity sequences also widely exist in eukaryotes, with the function largely unclear. Uncovering the transcriptional regulation on glnAp2 is thus promising to provide general insights.
Figure 1.
Components of the transcription apparatus on the glnAp2 promoter. (A) Sequence of the promoter DNA. Enhancers I and II, low-affinity elements (sites III-V), the −24–−12 region and transcription start site are marked. (B) Size comparison of the DNA, NtrC hexamer and RNAP. A scale bar is shown in the top right. ATPase is the active center of NtrC hexamer; around the pore at the opposite side are the three DNA binding domains (DBDs). Six monomers are differently colored in a reconstructed hexamer. (C) A traditional model suggested that an NtrC hexamer simultaneously bound to the two enhancers drove transcription initiation. (D) Another model suggested that the five binding sites collectively nucleated NtrC dimers to form an octamer that catalyzes the holoenzyme.
Although the transcription apparatus (TA) on the glnAp2 promoter only involves NtrC, σ54RNAP and promoter DNA, it exhibits complicated transcriptional activities (21,22,40). If the low-affinity sites are all substituted with the sequences that do not bind any protein, glnAp2 is transcribed at ∼45% of the wild-type level; but the transcriptional level is further lowered to ∼22% if these sites are substituted with the enhancer sequences. Moreover, the low-affinity sites act to repress transcription at high concentrations of NtrC dimers. These characteristics cannot be fully accounted for by traditional views (Figure 1C and D). The notion that transcription initiation is activated by an NtrC hexamer spanning the two enhancers fails to explain why the low-affinity sites can promote transcriptional output (35,41). Structurally, the binding of NtrC to the low-affinity sites enhances the bending rigidity of DNA, rather than significantly bend DNA as the integration host factor (7), and thus prevents the hexamers at enhancers from contacting the holoenzyme (29,36,42). An alternative postulation was that NtrC dimers could constitute a huge octamer at the five sites to activate transcription (22) (Figure 1D); but this is inconsistent with the structural basis of activation—the holoenzyme is catalyzed at the edge of the central pore of NtrC hexamer (10,11,29,38). All these suggest that it is necessary to revisit the role for the low-affinity sites in transcriptional regulation and to unravel the kinetics of NtrC interacting with the five cis-regulatory elements.
By three-dimensional structure reconstruction, we first explore all possible architectures and conformational transitions of the TA. Although NtrC hexamers at either enhancer are capable of catalyzing the holoenzyme, the main regulatory mechanism involves the II-V bridging mediated by NtrC oligomers at enhancer II. This unstable DNA bridging is also the structural basis for the low-affinity sites to promote transcriptional output. We then construct a model characterizing how the TA dynamically operates; the model is validated by its ability to quantitatively recapitulate transcriptional activities from various mutated promoters. The kinetic features of key molecular interactions are also unraveled. The proposed dynamic mechanisms for transcriptional regulation exhibit strong robustness to various perturbations. Since glnAp2 transcription is regulated in a mode similar to that in eukaryotes, the unexpected findings such as the key roles played by low-affinity sequences may be of wide implications.
MATERIALS AND METHODS
Molecular structure
To reveal how NtrC interacts with the five binding sites and controls transcription initiation, the molecular surface of NtrC hexamer and standard B-DNA double helix were reconstructed using the software 3ds Max (Figure 1B). The dimensions of the standard B-DNA double helix in solution are 34 Å per helical turn, 24 Å in diameter, 22 Å across the major groove and 12 Å across the minor groove. The NtrC hexamer was reconstructed based on its X-ray structure (38). Its DNA binding domains (DBDs) were not reconstructed to the surface for accuracy since they may detach from the main ring. The slightly raised part surrounding the central pore was also not reconstructed for simplicity. The X-ray structure of RNAP (PDB id: 1IW7) was used in structural analysis. Referring to the conformations of NtrC's DBD interacting with DNA (29,42) and of the holoenzyme catalyzed by NtrC at the core promoter (11,38), we examined all possible structural organizations together with their conformational changes. The surface structure of NtrC hexamer changes very slightly when catalyzing the holoenzyme, and there are no other conformational changes (38). The spatial arrangement of NtrC hexamers on the DNA can thus be evaluated with sufficient accuracy. Possible conformations of DNA bridging are presented in Supplemental S1.
Numeric calculations
Entropy theory and the Jacobson & Stockmayer function were employed to evaluate the timescale of DNA looping (22,43–45). The standard Gillespie method was used to fit the experimental data and perform numerical simulations (46,47). Details of all analytical and numerical calculations are available in Supplemental S2 and S3. All statistical analyses were based on sufficient sample data.
RESULTS
Configurations of the TA underlying transcription initiation
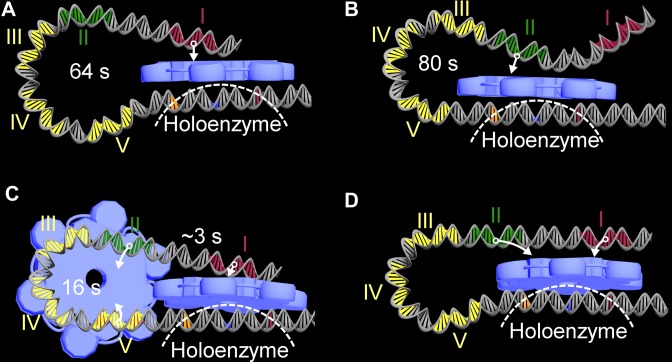
Based on structure reconstruction, we first analyze whether an NtrC hexamer nucleated at any binding site can approach and catalyze the holoenzyme. Obviously, a hexamer at enhancer I or II can contact the holoenzyme via DNA looping (Figure 2A and B). Nevertheless, it becomes difficult for those enhancer-bound hexamers to approach the holoenzyme at high NtrC concentrations, since the bending rigidity of intervening DNA is strengthened due to the occupancy of low-affinity sites (29,36,42). In contrast, hexamers at any low-affinity site hardly contact the holoenzyme. This is interpreted as follows. Given a hexamer formed at site III, sites IV and V are also occupied at least by NtrC dimers which block DNA bending. A hexamer at site IV is similarly hindered from contacting the holoenzyme because of site V. The active center of a hexamer at site V cannot reach the -12 region because of short intervening DNA. Together, at high concentrations hexamers at low-affinity sites not only fail to stimulate transcription initiation but also hinder hexamers at enhancers from contacting the holoenzyme. If the low-affinity sites are substituted with enhancer sequences, such a repressive effect occurs even at low concentrations, leading to a reduction in transcriptional levels (22).
Figure 2.
Configurations of the transcription appratus (TA) for directing transcription initiation. (A) A hexamer at enhancer I gets close to the holoenzyme via DNA looping. (B) A hexamer at enhancer II approaches the holoenzyme. (C) The II-V bridging facilitates enhancer I-mediated transcription initiation. (D) A hexamer simultaneously binding to enhancers I and II approaches the holoenzyme; this configuration hardly occurs. The DBDs and flexible peptide chain of NtrC are denoted by arrows. The holoenzyme is simply outlined by a dashed line. The looping of glnAp2 promoter is due to intrinsic bending (58). It separately takes ∼64 s and 80 s on average for a hexamer at enhancer I or II to encounter the holoenzyme. It takes ∼16 s to form the II-V bridging, whose average duration is ∼55 s, and during which it takes ∼3 s for a hexamer at enhancer I to approach the holoenzyme.
We then probe whether any two sites can be bridged by an NtrC oligomer (a hexamer or a tetramer that exists during the formation of a hexamer) and whether a hexamer spanning two sites can catalyze transcription initiation. This is based on the consideration that two sites may be simultaneously bound by two DBDs of an NtrC oligomer if topologically and spatially favorable. The possible bridging manners fall into three categories, i.e. enhancer–low-affinity site (including I-III, I-IV, I-V, II-III, II-IV and II-V), low-affinity site–low-affinity site (including III-IV, III-V and IV-V) and enhancer–enhancer (I-II) bridging.
All the bridging conformations in the first two categories are rather unstable since they involve the low-affinity sites and unstable NtrC oligomers (38). We analyze each possible conformation in terms of its 3D structure, stability, dependence on NtrC concentration and potential influence on transcriptional output (see Supplemental S1 and Supplementary Table S1). It turns out that these conformations rarely occur and do not significantly affect transcriptional dynamics except the II-V bridging. The II-V bridging exactly constrains enhancer I in the vicinity of the -12 region, facilitating the hexamer at enhancer I to rapidly find and catalyze the holoenzyme (Figure 2C). The II-V bridging is the only rational architecture underlying the contribution of low-affinity sites to elevated transcriptional output. Of note, when the low-affinity sites are unoccupied, the II-V bridging forms when an enhancer II-bound NtrC oligomer encounters site V; the II-V bridging rarely forms at high concentrations because of the occupancy of sites III and IV.
The two enhancers may be transiently bridged by an NtrC tetramer rather than a hexamer. A previous structural study showed that for a hexamer to span the two enhancers, rather high energy is required to severely bend or even twist the DNA (38) (Figure 2D; Supplementary Table S1). Another study revealed that the putative cooperative binding of NtrC to the two enhancers is independent of the conformational change of DNA and lies outside the DBD (34). Thus, the two enhancers cannot be simultaneously bound by an NtrC hexamer. If the cooperativity exists, an eligible speculation could be that the oligomerization domain of a bound dimer helps recruit another free dimer that then dissociates and binds to the other enhancer (40,48). That is, the two enhancers may be bridged by an NtrC tetramer very transiently.
In summary, there exist three configurations of the TA allowing for transcription initiation, i.e. an NtrC hexamer at enhancer I or II catalyzes the holoenzyme via DNA looping, and the II-V bridging facilitates the enhancer I-mediated transcription initiation. At low and intermediate NtrC concentrations, NtrC oligomers (mainly hexamers as seen later) at enhancer II are topologically and spatially favored to engage in the II-V bridging. At high concentrations, the occupancy of low-affinity sites represses transcription by hindering both the formation of II-V bridging and interplay of hexamers and the holoenzyme.
Modeling how the TA dynamically operates
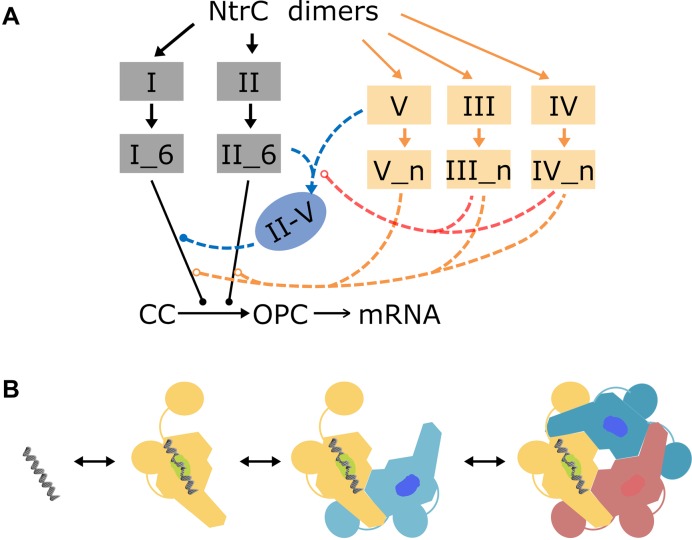
The above analyses also suggest a minimal kinetic model for how the TA operates, which comprises the essential pathways of conformational changes (Figure 3A). In brief, an NtrC dimer bound to DNA acts as a nucleus condensing free dimers to form a tetramer and then a hexamer. NtrC hexamers at either enhancer stimulate transcription initiation when they contact the holoenzyme. NtrC oligomers at enhancer II also tend to approach site V, bridging sites II and V. The II-V bridging shortens the time required for a hexamer at enhancer I to search the holoenzyme. The occupancy of low-affinity sites hinders DNA looping. To reveal the kinetics of essential conformational changes, we make the following simplifications. Each binding site may be vacant, bound by an NtrC dimer, tetramer or hexamer, and the conversion between these states is taken into account (Figure 3B). We need not consider whether there exist other NtrC oligomeric structures since NtrC is recruited in units of a dimer and only hexamers catalyze the holoenzyme (11,38). We also ignore the rather small affinity difference between the two enhancers and that among the three low-affinity sites (39,40). Additionally, at high NtrC concentrations a very small number of free hexamers may form and stimulate transcription without binding to DNA (40,49); this minor effect is also neglected.
Figure 3.
Essentials of the model for NtrC-regulated glnAp2 transcription initiation. (A) NtrC dimers bind to enhancers I and II and low-affinity sites III-V. NtrC hexamers formed at enhancers (denoted by I_6 and II_6) stimulate the transition from the posterior closed complex ‘CC’ to the open complex ‘OPC’. Mainly via the pathway that an NtrC tetramer/hexamer at enhancer II binds to site V, the II-V bridging forms facilitating the enhancer I-mediated transcription initiation (denoted by blue lines). The occupation of low-affinity sites by an NtrC dimer, tetramer or hexamer (denoted by n, n = 2, 4 or 6) at high concentrations increases the DNA rigidity, thus hindering the formation of II-V bridging and association of enhancer-bound hexamers with the closed complex (denoted by red and orange dashed lines). Solid and open circles separately denote the promotion and inhibition of molecular interactions. (B) For each binding site, its state converts stochastically among being vacant, being bound by an NtrC dimer, tetramer or hexamer (dimers in a tetramer/hexamer are differently colored).
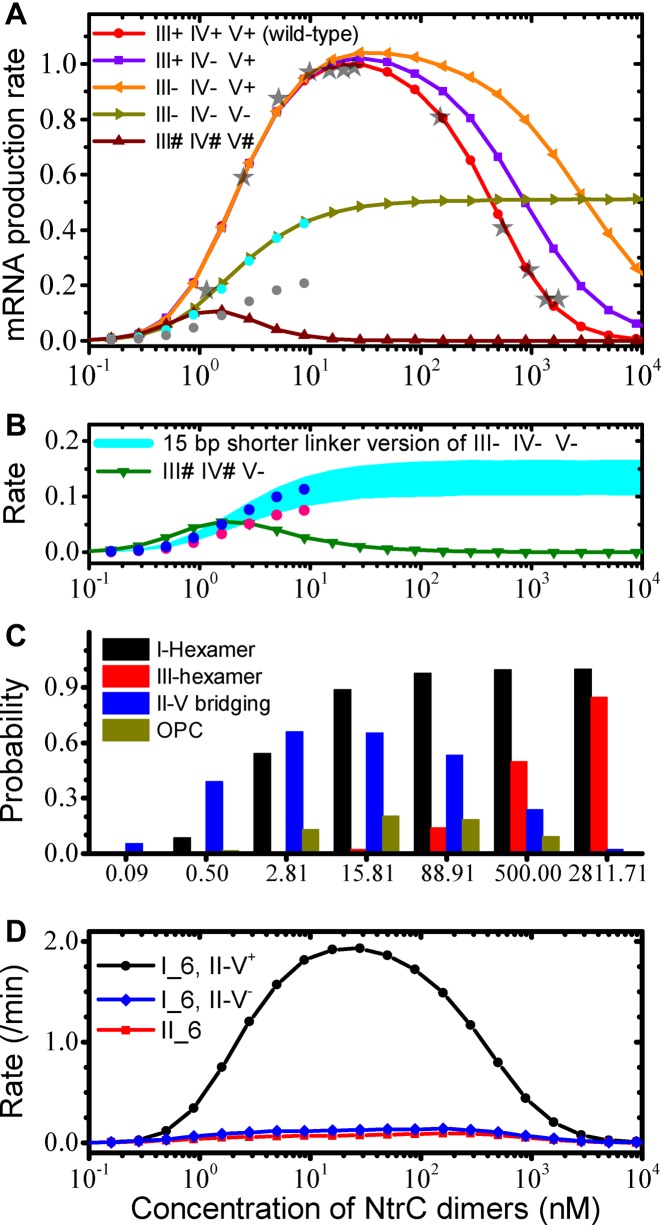
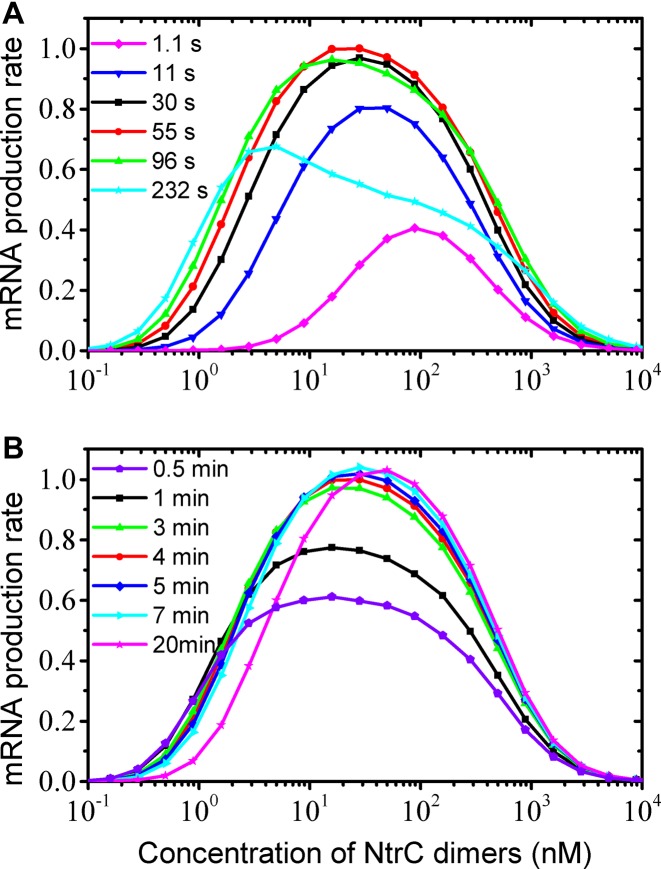
Owing to the simplicity of the TA and rather limited reaction types (detailed reaction steps are shown in Supplementary Figure S1 and Supplementary Table S2), the reaction rate constants can be determined as follows. Presumably, the kinetic data on the holoenzyme interacting with the −24–−12 region measured at glnALG are applicable to glnAp2 (27), since the two promoters share the same core promoter together with the neighboring sequences. Then, the few remaining rate constants can be estimated by fitting the input-output function of the wild-type glnAp2 promoter (21,22,40) (Supplemental S2 and S3). The exact fitting is shown in Figure 4A (for the experimental data see Figure 3 in (21) and Figure 6 in (40)). As the concentration of NtrC dimers, C, rises, the average rate of mRNA production, R, gradually rises and drops. R changes slightly in the range of 20–50 nM, with a maximum around 30 nM. Half-maximal production rates are separately at 2 nM and 400 nM. In the following, we further test the validity and robustness of this model.
Figure 4.
Reproduced transcriptional activities and kinetics of molecular interactions. (A) The average rate of mRNA production versus the concentration of NtrC dimers on the wild-type or mutated glnAp2 promoter. The data are normalized by setting the maximal rate in the wild-type case to 1.0. The 45% and 22% of the rate at low concentrations in the wild-type case are denoted by cyan and gray dots, respectively. The experimental data from Figure 3 in (21) and Figure 6 in (40) are also rescaled and shown by gray stars. (B) The average rate of mRNA production on two mutated glnAp2 promoters. The 12% and 8% of the rate at low concentrations in the wild-type case are denoted by blue and pink dots, respectively. (C) Probabilities of the promoter in specific states at various NtrC concentrations. The states shown correspond to the promoter with enhancer I occupied by NtrC hexamers, with site III occupied by hexamers, with sites II and V bridged, and in the OPC state. (D) The average number of mRNAs initiated per minute from the wild-type promoter at various NtrC concentrations. ‘I_6, II-V+’ and ‘I_6, II-V−’ denote the rate of mRNA production stimulated by enhancer I-bound hexamers in the presence or absence of II-V bridging, respectively, while ‘II_6’ denotes that by enhancer II-bound hexamers.
Validity and robustness of the model
With the reaction rate constants obtained above, the model quantitatively reproduces transcriptional activities from various types of mutated promoters (Figure 4A and B). In Case 1, the three low-affinity sites are all substituted with sequences without similarity to the enhancers (III- IV- V-), i.e. these sites do not associate with NtrC anymore. We thus set the affinity of the low-affinity sites for NtrC to 0. Consequently, at low and intermediate concentrations R is ∼45% of that in the wild-type case, quantitatively in agreement with the experimental data (cf. Figure 5 in (22)). The model further predicts that R nearly remains unchanged for C ≥ 100 nM.
Figure 5.
Proper instability of both the II-V bridging and NtrC hexamer is crucial for effective transcriptional regulation. The data are collected from the wild-type promoter. (A) Dependence of the R-C curve on the half-life of II-V bridging. The optimal duration of II-V bridging is 55 s. (B) Dependence of the R-C curve on the half-life of NtrC hexamer. The most effective transcriptional regulation—both in enhancing and repressing transcription—requires a half-life of 3–5 min.
In Case 2, the three low-affinity sites are all substituted with the enhancer sequences (III# IV# V#). We set the affinity of the low-affinity sites for NtrC to that of the enhancers. Notably, R first rises and then drops to zero quickly with increasing C. At low concentrations, R is around ∼22% of that in the wild-type case, also quantitatively consistent with the data (cf. Figure 5 in (22)).
In Case 3, either one or both of sites III and IV are mutated to sequences unable to bind any protein (III- IV+ V+ or III+ IV- V+ or III- IV- V+). The binding affinity of the low-affinity sites for NtrC is altered accordingly. Compared with the wild-type case, here R is the same for C ≤ 20 nM but becomes larger for C > 20 nM; R is higher in the III- IV- V+ case than in the III+ IV- V+ case. These features agree well with the experimental observations (21). Concretely, Atkinson et al. obtained two groups of data at high concentrations (cf. Table 2 in (21)). One showed that R is ∼125% in the III- IV+ V+ or III+ IV- V+ case, while R is ∼154% in the III- IV- V+ case. The other showed that the two values are separately ∼224% and ∼406%. The authors speculated that the first group was obtained at lower concentration than the second. Our data are quantitatively consistent with those results and reveal that the corresponding concentrations are ∼300 nM and ∼1200 nM. This consistence also indicates that site V has a critical role in governing transcriptional dynamics.
In Case 4, the promoter is mutated as in Case 1 (i.e. III- IV- V-), but the region comprising the three low-affinity sites is shortened by 15 bp. The DNA double helix makes one complete turn along its axis every ∼11.1–11.2 bp in vivo (50,51). The orientations of the two enhancers, toward which NtrC's DBDs insert into the major grooves, thus form an angle of ∼47° (29,36,42). The reduction of 15 bp means that the orientations of both the enhancers are rotated to the opposite side by an angle of ∼124°. Of note, according to B-DNA with 10.4 bp/turn in solution, the two angles above are separately ∼28° and ∼159°. Thus, the central pores of hexamers at either enhancer no longer smoothly and precisely get contact with the catalysis site of the holoenzyme. Previous studies inferred that such an orientation change of ∼124° results in a decrease of ∼80–88% in transcriptional levels; a change of ∼159° leads to a similar decrease (cf. Figure 4C in (22) and Figure 2A in (52)). In other words, the time required for hexamers at enhancers to find the catalysis site is increased by ∼4- to 7.3-fold on average. Given the shortened DNA, the time is additionally decreased by one-fifth on average (based on Equation 11 in Supplementary Material S2). Thus, the searching time for hexamers at enhancer I and II to find the catalysis site is separately ∼256–427 s and 320–533 s. Consequently, R is ∼9–14% of that in the wild-type case (Figure 4B), consistent with 12% by experiment (cf. Figure 5 in (22)).
In Case 5, sites III and IV are substituted with the enhancer sequences, and site V is mutated unable to bind any protein (III# IV# V-). It might be expected that R would be greater than that in Case 2 (III# IV# V#), since the enhanced bending rigidity of DNA due to NtrC binding is less prominent here. Unexpectedly, R is only ∼8% of that in the wild-type case (Figure 4B), which is quantitatively consistent with the experimental observation on a similarly mutated promoter (4OP, cf. Figure 5 in (22)). This result can be explained in terms of contributions to transcription initiation by various TA configurations. Without incorporating transcription initiations in the presence of II-V bridging, the rate of mRNA production in Case 5 would be indeed higher than that in Case 2 (Supplementary Figure S2A). The higher transcriptional output in Case 2 is due to the II-V bridging, which seldom forms but is rather stable once formed (Supplementary Figure S2B–D). These results should exclusively confirm the existence of II-V bridging.
We further examine whether the operation mechanism of the TA is robust to various perturbations (such as fluctuations in temperature and concentrations of cellular molecules) that affect the rates of biochemical reactions. To this end, independent Gaussian white noise is added to each rate constant, with the standard deviation being 20% of its default value (see Supplementary Material S4). Compared with the case without noise, here the average transcription rate becomes smaller, but the relative dependence of R on C in both the wild-type and mutation cases is almost unchanged (Supplementary Figure S3). Further in-depth analyses not only support such strong robustness, but also verify the kinetic features reported above (Supplementary Figures S4–S8). We also predict the transcriptional activities in two cases where either of two enhancers is mutated to a sequence that does not bind any protein – this can be used for further testing our model (Supplementary Figure S9).
Dynamic characteristics of molecular interactions
The above results suggest that the current model captures the microscopic mechanisms for the TA operation. Here, we summarize the dynamic nature of key molecular interactions (Table 1). NtrC dynamically binds to and dissociates from the enhancers and low-affinity sites. The average time of its DBD in association with an enhancer and a low-affinity site is ∼12 min and 72 s, respectively. The probabilities of NtrC hexamers formed at these sites gradually rise to saturation with increasing NtrC concentration (Figure 4C). Half the maximal occupation of an enhancer and a low-affinity site by hexamers appears at ∼2 nM and ∼400 nM, respectively, and the occupation probabilities nearly remain unchanged separately for C > 100 nM and C > 10 μM.
Table 1. Kinetic nature of molecular interactions.
| Average lifetime of NtrC hexamer | ∼4 min |
| Average time of NtrC's DBD in association with an enhancer | ∼12 min |
| Average time of NtrC's DBD in association with a low-affinity site | ∼72 s |
| Searching time for hexamers at enhancer I to encounter a holoenzyme | 3 s, 64 s, 128 s, 640 s and +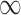 – separately corresponding to the cases where the II-V bridging is present, none of, one of, two of and all of sites III-V is/are bound by NtrC – separately corresponding to the cases where the II-V bridging is present, none of, one of, two of and all of sites III-V is/are bound by NtrC |
| Searching time for hexamers at enhancer II to encounter a holoenzyme | 80 s, 160 s, 640 s, +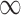 – separately corresponding to the cases where none, one, two and all of sites III-V is/are bound by NtrC – separately corresponding to the cases where none, one, two and all of sites III-V is/are bound by NtrC |
| Searching time for hexamers at enhancer II to encounter site V | ∼16 s, 160 s, +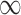 – separately corresponding to the cases where neither, either and both of sites III and IV is/are bound by NtrC – separately corresponding to the cases where neither, either and both of sites III and IV is/are bound by NtrC |
| Average lifetime of II-V bridging | ∼55 s |
| Half the maximal occupation of an enhancer | ∼2 nM of NtrC dimers |
| Half the maximal occupation of a low-affinity site | ∼400 nM of NtrC dimers |
When none of the low-affinity sites is occupied, it takes ∼64 s on average for a hexamer at enhancer I to approach the holoenzyme within the posterior closed complex and ∼80 s for a hexamer at enhancer II (Table 1). On the other hand, it takes only ∼16 s for a hexamer at enhancer II to encounter site V; notably, this is also the predominant pathway leading to the II-V bridging, compared with that mediated by a tetramer (Supplementary Table S3). Thus, most hexamers at enhancer II are engaged in bridging sites II and V, rather than stimulate transcription initiation. Once formed, the II-V bridging lasts about 55 s, during which it takes only several seconds for a hexamer at enhancer I to contact the holoenzyme. Most mRNA production is induced by hexamers at enhancer I in the presence of II-V bridging (Figure 4D).
Large deviations from the optimal lifetime of II-V bridging, either too short or too long, deteriorate effective transcriptional regulation (Figure 5A). The existence of an optimal lifetime suggests the importance of low affinity of site V for NtrC and instability of NtrC hexamer. Otherwise, if the II-V bridging existed stably, it would be required that site V have a high affinity for NtrC and NtrC hexamer be stable. Accordingly, at low and intermediate concentrations site V would be bound by NtrC hexamers, inhibiting both the formation of II-V bridging and transcription initiation. The optimal lifetime of a hexamer is 4 min; beyond the range of ∼3–5 min, effective transcriptional modulation is also disrupted (Figure 5B).
Although the II-V bridging effectively enhances transcription, it is not straightforward to judge whether it plays a role by counting transcript numbers in individual cells, because mRNAs are produced in bursts (Supplementary Figure S10A). Nevertheless, the dynamics of transcription initiations via the II-V bridging present a unique signature. When the II-V bridging is in place, transcription initiation is faster, implying that more closely spaced polymerases get into elongation successively and hence a sharper burst appears. Such characteristics may be justified by using RNA labeling technologies such as the MS2 system and single-molecule fluorescence in situ hybridization (FISH) (53–56). Of note, measurements with high resolution are required because of the instability of II-V bridging. On the other hand, the distribution of transcript numbers from the wild-type promoter over a cell population is shown in Supplementary Figure S10B. For 10 nM ≤ C ≤ 100 nM, the distribution does not change markedly and nearly obeys a normal distribution. At low or high concentrations, most cells produce fewer mRNAs. These characteristics underline the importance of transcriptional dynamics itself in determining transcriptional output.
DISCUSSIONS
The kinetics of the holoenzyme σ54RNAP interacting with promoter DNA was revealed using a single-molecule fluorescence technology called CoSMoS (27). The current work proposes a complementary approach, which fills the gap across studies on molecular structures, reaction kinetics and transcriptional activities in comprehending transcriptional dynamics. We present an integrative picture of how transcription initiation is regulated by transcriptional activators and cis-regulatory elements on the glnAp2 promoter. While NtrC hexamers at either enhancer can stimulate transcription initiation, the main regulatory mode involves their cooperation. At low and intermediate NtrC concentrations, it is structurally and topologically favorable for a hexamer at enhancer II to bridge sites II and V via one of its free DBDs. This transient bridging greatly facilitates the interplay between NtrC hexamers at enhancer I and the holoenzyme, underlying the contribution of low-affinity sites to elevated transcriptional output. At high concentrations, the three low-affinity sites are occupied, hindering DNA looping and leading to a drop in transcriptional levels. The unexpected implications of this work are as follows:
In dynamically regulating transcriptional output, the low-affinity cis-regulatory elements can exert a marked influence. Although the topology, torsion and rigidity of DNA are altered transiently due to unstable association of proteins with low-affinity sites, the inherent nonlinear features of molecular interactions substantially affect transcriptional output. Low-affinity sequences also exist widely in eukaryotic genomes, but little attention was paid to their functions. It is expected that more functions of low-affinity sites would be found. This work also suggests that the instability of NtrC hexamer and II-V bridging is crucial for effective transcriptional regulation.
The roles played by the two enhancers are quite different on the wild-type promoter. Whereas enhancer I-bound hexamers catalyze the holoenzyme, enhancer II-bound hexamers are mainly engaged in bridging the DNA. Notably, there also exist several or more enhancer elements in the regulatory region of higher eukaryotic genes. Likely, the presence of multiple enhancers is not simply for cooperatively recruiting activators; each may also perform individual functions, constrained by topological and structural factors such as the orientation of major DNA grooves.
A clue may be inferred as to evolution of the dynamic mechanism of regulated transcription initiation. Previously, a generic model for how the eukaryotic TA operates dynamically showed that the temporal tethering of a distant enhancer to the surrounding area of a core promoter enables the most efficient conversion of regulatory signals into the rate of mRNA production (4). A more recent study reported that such tethering is already formed before the arrival of specific cellular signaling (57). The current study revealed the similar feature in transcriptional regulation in prokaryotes. Thus, such a characteristic may be a conserved evolutionary choice.
Supplementary Material
Acknowledgments
The authors thank Dr Jian Zhang and Dr Jun Wang for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2013CB834104]; National Natural Science Foundation of China [11175084, 31361163003, 81421091]. Funding for open access charge: Ministry of Science and Technology of China [2013CB834104].
Conflict of interest statement. None declared.
REFERENCES
- 1.Weake V.M., Workman J.L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 2.Gregor T., Tank D.W., Wieschaus E.F., Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locke J.C.W., Young J.W., Fontes M., Jimenez M.J.H., Elowitz M.B. Stochastic pulse regulation in bacterial stress response. Science. 2011;334:366–369. doi: 10.1126/science.1208144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Liu F., Wang W. Dynamic mechanism for the transcription apparatus orchestrating reliable responses to activators. Sci. Rep. 2012;2:422. doi: 10.1038/srep00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornberg R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wigneshweraraj S., Bose D., Burrows P.C., Joly N., Schumacher J., Rappas M., Pape T., Zhang X., Stockley P., Severinov K., et al. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 8.Sharma A., Leach R.N., Gell C., Zhang N., Burrows P.C., Shepherd D.A., Wigneshweraraj S., Smith D.A., Zhang X., Buck M., et al. Domain movements of the enhancer-dependent sigma factor drive DNA delivery into the RNA polymerase active site: insights from single molecule studies. Nucleic Acids Res. 2014;42:5177–5190. doi: 10.1093/nar/gku146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plaschka C., Lariviere L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F., et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty A., Wang D., Ebright Y.W., Korlann Y., Kortkhonjia E., Kim T., Chowdhury S., Wigneshweraraj S., Irschik H., Jansen R., et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bose D., Pape T., Burrows P.C., Rappas M., Wigneshweraraj S.R., Buck M., Zhang X. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine M., Cattoglio C., Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dangkulwanich M., Ishibashi T., Bintu L., Bustamante C. Molecular mechanisms of transcription through single-molecule experiments. Chem. Rev. 2014;114:3203–3223. doi: 10.1021/cr400730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darzacq X., Yao J., Larson D.R., Causse S.Z., Bosanac L., de Turris V., Ruda V.M., Lionnet T., Zenklusen D., Guglielmi B., et al. Imaging transcription in living cells. Annu. Rev. Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hager G.L., McNally J.G., Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj A., van Oudenaarden A. Nature, nature, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munsky B., Neuert G., van Oudenaarden A. Using gene expression noise to understand gene gegulation. Science. 2012;336:183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G.W., Xie X.S. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Liu F., Li J., Wang W. Reconciling the concurrent fast and slow cycling of proteins on gene promoters. J. R. Soc. Interface. 2014;11:20140253. doi: 10.1098/rsif.2014.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atkinson M.R., Pattaramanon N., Ninfa A.J. Governor of the glnAp2 promoter of Escherichia coli. Mol. Microbiol. 2002;46:1247–1257. doi: 10.1046/j.1365-2958.2002.03211.x. [DOI] [PubMed] [Google Scholar]
- 22.Lilja A.E., Jenssen J.R., Kahn J.D. Geometric and dynamic requirements for DNA looping, wrapping and unwrapping in the activation of E. coli glnAp2 transcription by NtrC. J. Mol. Biol. 2004;342:467–478. doi: 10.1016/j.jmb.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 23.Sepúlveda L.A., Xu H., Zhang J., Wang M., Golding I. Measurement of gene regulation in individual cells reveals rapid switching between promoter states. Science. 2016;351:1218–1222. doi: 10.1126/science.aad0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusk N. Transcription factors without footprints. Nat. Methods. 2014;11:988–989. doi: 10.1038/nmeth.3128. [DOI] [PubMed] [Google Scholar]
- 25.Sung M.-H., Guertin M.J., Baek S., Hager G.L. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol. Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gourse R.L., Landick R. CoSMoS unravels mysteries of transcription initiation. Cell. 2012;148:635–637. doi: 10.1016/j.cell.2012.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman L.J., Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Revyakin A., Grimm J.B., Lavis L.D., Tjian R. Single-molecule tracking of the transcription cycle by sub-second RNA detection. Elife. 2014;3:e01775. doi: 10.7554/eLife.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidangos N., Maris A.E., Young A., Hong E., Pelton J.G., Batchelor J.D., Wemmer D.E. Structure, function and tethering of DNA-binding domains in σ54 transcriptional activators. Biopolymers. 2013;99:1082–1096. doi: 10.1002/bip.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 31.Rombel I., North A., Hwang I., Wyman C., Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harb Symp. Quant. Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 32.Merrick M.J., Edwards R.A. Nitrogen control in bacteria. Microbiol. Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninfa A.J., Reitzer L.J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 34.Porter S.C., North A.K., Wedel A.B., Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 35.Wyman C., Rombel I., North A.K., Bustamante C., Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 36.Pelton J.G., Kustu S., Wemmer D.E. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 1999;292:1095–1110. doi: 10.1006/jmbi.1999.3140. [DOI] [PubMed] [Google Scholar]
- 37.Bush M., Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Carlo S., Chen B., Hoover T.R., Kondrashkina E., Nogales E., Nixon B.T. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirschman J., Wong P.K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss V., Claverie-Martin F., Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bondarenko V., Liu Y., Ninfa A., Studitsky V.M. Action of prokaryotic enhancer over a distance does not require continued presence of promoter-bound σ54 subunit. Nucleic Acids Res. 2002;30:636–642. doi: 10.1093/nar/30.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidangos N.K., Heideker J., Lyubimov A., Lamers M., Huo Y., Pelton J.G., Ton J., Gralla J., Berger J., Wemmer D.E. DNA recognition by a σ54 transcriptional activator from Aquifex aeolicus. J. Mol. Biol. 2014;426:3553–3568. doi: 10.1016/j.jmb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiggins P.A., van der Heijden T., Moreno-Herrero F., Spakowitz A., Phillips R., Widom J., Dekker C., Nelson P.C. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006;1:137–141. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J., Lin M., Chen R., Wang W., Liang J. Discrete state model and accurate estimation of loop entropy of RNA secondary structures. J. Chem. Phys. 2008;128:125107. doi: 10.1063/1.2895050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobson H., Stockmayer W.H. Intramolecular reaction in polycondensations. I. The theory of linear systems. J. Chem. Phys. 1950;18:1600–1606. [Google Scholar]
- 46.Gillespie D.T. A general method for numerically simulating stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 1976;22:403–434. [Google Scholar]
- 47.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 48.Sevenich F.W., Langowski J., Rippe K., Weiss V. DNA binding and oligomerization of NtrC studied by fluorescence anisotropy and fluorescence correlation spectroscopy. Nucleic Acids Res. 1998;26:1373–1381. doi: 10.1093/nar/26.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck L.L., Smith T.G., Hoover T.R. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 2007;15:530–537. doi: 10.1016/j.tim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Haykinson M.J., Johnson R.C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D.H., Schleif R.F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc. Natl. Acad. Sci. U.S.A. 1989;86:476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huo Y.X., Tian Z.X., Rappas M., Wen J., Chen Y.C., You C.H., Zhang X., Buck M., Wang Y.P., Kolb A. Protein-induced DNA bending clarifies the architectural organization of the σ54-dependentglnAp2promoter. Mol. Microbiol. 2006;59:168–180. doi: 10.1111/j.1365-2958.2005.04943.x. [DOI] [PubMed] [Google Scholar]
- 53.Golding I., Paulsson J., Zawilski S.M., Cox E.C. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 54.Zenklusen D., Larson D.R., Singer R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corrigan A.M., Tunnacliffe E., Cannon D., Chubb J.R. A continuum model of transcriptional bursting. Elife. 2016;5:e13051. doi: 10.7554/eLife.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tantale K., Mueller F., Kozulic-Pirher A., Lesne A., Victor J.M., Robert M.C., Capozi S., Chouaib R., Bäcker V., Mateos-Langerak J., et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carmona M., Claverie-Martin F., Magasanik B. DNA bending and the initiation of transcription at σ54-dependent bacterial promoters. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9568–9572. doi: 10.1073/pnas.94.18.9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.