Abstract
Specific structures in mRNA modulate translation rate and thus can affect protein folding. Using the protein structures from two eukaryotes and three prokaryotes, we explore the connections between the protein compactness, inferred from solvent accessibility, and mRNA structure, inferred from mRNA folding energy (ΔG). In both prokaryotes and eukaryotes, the ΔG value of the most stable 30 nucleotide segment of the mRNA (ΔGmin) strongly, positively correlates with protein solvent accessibility. Thus, mRNAs containing exceptionally stable secondary structure elements typically encode compact proteins. The correlations between ΔG and protein compactness are much more pronounced in predicted ordered parts of proteins compared to the predicted disordered parts, indicative of an important role of mRNA secondary structure elements in the control of protein folding. Additionally, ΔG correlates with the mRNA length and the evolutionary rate of synonymous positions. The correlations are partially independent and were used to construct multiple regression models which explain about half of the variance of protein solvent accessibility. These findings suggest a model in which the mRNA structure, particularly exceptionally stable RNA structural elements, act as gauges of protein co-translational folding by reducing ribosome speed when the nascent peptide needs time to form and optimize the core structure.
INTRODUCTION
The primary function of a mRNA is to encode the sequence of a specific protein. However, an mRNA is not an abstract sequence of codons but rather a molecule with its own complex structure various features of which can be subject to selection. In particular, mRNA molecules form secondary structure elements (stems and loops) of broadly varying stability that can affect both the stability of the mRNA and the speed and fidelity of translation. Although much attention had focused on the initiation step as a major determinant of translation rate, multiple studies over nearly two decades have made it clear that elongation also plays an important role in the regulation of translation and co-translational protein folding (1–3). In particular, it has been shown that in some proteins, α-helices and β-strands are flanked by strong signals in the mRNA sequence (4,5). Different protein structures have been reported to correlate with distinct patterns of synonymous codon usage in the respective mRNAs (6,7). More generally, it has been proposed that different protein secondary structures are encoded by mRNA sequences with distinct properties. For example, α-helices in Escherichia coli and human proteins appear to be preferentially encoded by ‘fast’ mRNA regions, i.e. those enriched in optimal codons, whereas ‘slow’ regions often code for β-strands and loops (8–10).
Recent work, in particular ribosome profiling experiments, has clearly demonstrated that translation speed is far from being uniform either among different mRNAs or along a single mRNA molecule (11,12). Structural features of an mRNA, in particular segments with a stable secondary structure, as well as specific protein sequences within a nascent polypeptide (arrest sequences), cause translating ribosomes to pause or stall (13,14). For example, positively charged amino acid residues and proline-rich sequences have been reported to substantially affect the rate of translation and hence protein production, apparently by partially blocking the ribosomal exit channel (14–18). It has been shown that in the yeast Saccharomyces cerevisiae, positively charged amino acid residues in the growing translated peptide interact with the negatively charged inner surface of the ribosomal exit tunnel and slow down translation (15). Ribosome stalling caused by the ‘arrest peptides’ that block the ribosomal exit channel affects the structure of the mRNA, resulting in specific biological effects including regulation of protein production, maturation or localization (19). Notably, it has been shown that to overcome stalling at polyproline sequences, the ribosome requires a specific translation elongation factor (17). The availability of tRNAs also can affect the rate of elongation of nascent polypeptide chains (20). However, the actual contributions of each of these potential mechanisms to the control of translation speed remain uncertain and controversial (21–23).
Is the efficiency of translation and/or protein folding encoded in the mRNA sequence, and if so, how? The 5′-terminal 30–50 nucleotides of the protein-coding regions in prokaryotes and eukaryotes are generally devoid of stable secondary structure that conceivably could impede mRNA interaction with the ribosomes (24–27). However, 5′-terminal parts of coding regions also contain structural elements that function as various translation regulatory signals. For example, the secondary structure in the region between codons 14 and 34 downstream of the start codon has been variously proposed to facilitate the recognition of the start codon and/or to prevent ribosomal jamming (13,28–30). Furthermore, it has been shown that the 5′-terminal regions of mRNAs form a ‘translational ramp’ which is translated substantially slower than the rest of the coding region (31,32).
The relationships between mRNA folding and translation appear to be complex and involve opposite effects. Numerous, independent experiments indicate that stable secondary structure elements in mRNA decrease the rate of translation, especially in vitro (33–36). However, in an apparent contradiction to these findings, recent progress in the experimental determination of RNA secondary structures has led to the demonstration of significant positive correlations between mRNA folding (the prevalence of stable structures) and protein abundance (37–39). These orthogonal findings imply that multiple, still poorly understood mechanisms that involve interactions between ribosomes and different structural elements in translated mRNAs differentially modulate the rate and efficiency of translation (24,27,40).
A pronounced periodic pattern of mRNA secondary structure, stability and nucleotide base-pairing has been identified in coding regions of diverse eukaryotes (24,41,42). Although all codon positions are important for the formation of the secondary structure of mRNA, synonymous positions that are free from selection on protein sequence make the greater contribution to the evolution of mRNA secondary structures and thus to the regulation of translation. Periodicity is most pronounced in highly expressed genes (24,43). Thus, synonymous positions appear to be subject to RNA-level selection maintaining a stable mRNA secondary structure that is likely to be important for transcript stability and translation and appears to be more common than previously assumed (24,27). Recent experimental evidence and analysis of mRNA structure and evolutionary conservation suggest a trade-off between selective pressures acting at the RNA and protein levels (27,44).
The abundance of a protein is related to the abundance of its mRNA, translation rate and degradation rate. Assuming a constant mRNA level, the translation rate is expected to positively correlate with protein abundance, whereas the degradation rate is expected to show a negative correlation. Synonymous substitutions can affect translation by facilitating the formation of stable stem-loop structures that substantially slow down translation initiation and/or ribosome translocation, or conversely, by loosening mRNA secondary structures and eliminating obstacles to speedy translation (27,45). Indeed, mRNA structure-dependent changes in translation rates can dramatically affect protein abundance and cause major phenotypic effects including human disease (46).
Additionally, several recent studies have demonstrated that variations in translation speed mediated by mRNA secondary structure can lead to changes in post-translational modifications of the nascent polypeptide, a level of protein regulation previously believed not to be connected with the RNA level regulation (27). In particular, translation-dependent regulation of post-translational protein arginylation mediated by synonymous codon usage has been demonstrated for the purine nucleotide biosynthesis enzyme PRPS2 (47) and for actins (48).
A rapidly growing body of experimental data indicates that folding of many if not most proteins is predominantly co-translational, i.e. individual protein domains fold before the synthesis of the respective polypeptide chain is complete (49–53). Recently, the process of co-translational folding of several proteins has been dissected experimentally (54–57). For example, cystic fibrosis transmembrane conductance regulator has been shown to fold in discrete steps, namely sequential compaction of the N-terminal, α-helical and α/β-core domains. The sequence of these events is critical for the overall folding completion as premature α-helical domain folding hampered the subsequent formation of the core domain. The synthesis of this particular protein is facilitated by intrinsic folding propensity modulation in three distinct ways: delaying α-subdomain compaction, facilitating β-strand intercalation and optimizing translation kinetics via codon usage (58).
Evidence that folding of at least some proteins is modulated by translational pauses caused by mRNA secondary structure has been reported, for example, for the coat protein of the RNA bacteriophage MS2 (59,60). Synonymous single-nucleotide polymorphisms within the same gene that affect the secondary structure of mRNA can create variations in translation speed, leading to dramatic differences in protein folding between individuals. A striking example of this effect involves synonymous polymorphisms in the multidrug resistance 1 (MDR1 or ABCB1) gene which affect protein folding and as a result substantially alter the conformation and function of the multidrug transporter (61,62).
Given the wealth of observations on the functional importance of the mRNA secondary structure and its multiple contributions to the control of translation and protein folding, we sought to investigate potential connections between the structures of an mRNA and the encoded protein. By directly comparing the predicted mRNA structure with the experimentally determined structures of proteins from several eukaryotes and prokaryotes, we show that mRNA regions containing stable secondary structure elements typically encode compact protein domains and large proteins. These findings suggest a model in which the mRNA structure acts as a gauge of co-translational protein folding by reducing ribosome speed when the nascent peptide needs extra time to form and optimize the core structure.
MATERIALS AND METHODS
The transcriptome data set
For the present analysis, we selected 12 model species, six eukaryotes and six prokaryotes, including a mammal: Homo sapiens (HSA), an insect: Drosophila melanogaster (DME), a worm: Caenorhabditis elegans (CEL), three fungi: Saccharomyces cerevisiae (SCE), Aspergillus orizae (ASP) and Neurospora crassa (NEU), three bacteria: Bacillus subtilis (BSU), E. coli (ECO), Deinococcus radiodurans (DEI) and three archaea: Methanosarcina marzei (MET), Haloferax volcanii (HAL), Thermococcus gammatolerans (TGA). The mRNA sequences and the sequences of the encoded proteins from these organisms were extracted from the RefSeq database (63) (see Supplementary Dataset 1).
The protein structure data set
The protein structure data set was constructed as previously described (64). Briefly, the protein sequences encoded by mRNAs in the transcriptome data set were used as queries to search the Protein Data Bank (65) for the corresponding protein structures using BLASTP (66). For further analysis, the protein structures from the four best covered species, H. sapiens, S. cerevisiae, E. coli and B. subtilis, were selected. In addition, to include a hyperthermophilic archaeon, protein structures corresponding to sequences with >70% sequence identity with T. gammatolerans protein sequences were included. Protein structures were isolated by single chain, curated (64) and side chains were rebuilt as needed with SCWRL (67) (see Supplementary Dataset 1).
Gene orthology and estimation of evolutionary rates
Orthologous sequences were extract from the SensuStricto database (68) for S. cerevisiae (Saccharomyces paradoxus, Saccharomyces mikitae, Saccharomyces kudriavzevii and Saccharomyces bayanus var. uravum), the ATGC database (69) for Thermococcus gammatolerans (T. AM4_uid54) and the OMA database (70) for the other species. The orthologous gene pairs were extracted from OMA for the following pairs of species: Caenorhabditis elegans and Caenorhabditis briggsae, Drosophila melanogaster and Drosophila pseudoobscura, H. sapiens and Mus musculus, Bacillus subtulis and Bacillus subtilis subsp. Spizizenii, E. coli and Salmonella typhi, Aspergillus oryzae and Aspergillus flavus, Neurospora crassa and Neurospora tetrasperma, Methanosarcina mazei and Methanosarcina acetivorans, Haloferax volcanii and Haloferax mediterranei, Deinococcus radiodurans and Deinococcus deserti, Thermococcus gammatolerans and Thermococcus AM4 iu54 735.
The nucleotide sequence alignments of orthologous coding sequences were obtained by backtracking the amino acid sequence alignments which were constructed using MUSCLE (71). Evolutionary rates for synonymous and non-synonymous positions (dS and dN, respectively) were estimated with the PAML software (72) using the Maximum Likelihood method for pairs of species or the phylogenetic tree of S. cerevisiae from SensuStricto. The dN and dS values >3 and <0.001, which result in unreliable estimation of evolutionary rates, were excluded from the calculations.
Protein abundance
Protein abundance data were extracted from the PAX database (73) using integrated data sets http://pax-db.org/dao/4932-S.cerevisiae_whole_organism-integrated_dataset.txt for S. cerevisiae, http://pax-db.org/dao/6239-C.elegans_whole_organism-integrated_dataset.txt for C. elegans, http://pax-db.org/dao/7227-D.melanogaster_whole_organism-integrated_dataset.txt for D. melanogaster, http://pax-db.org/dao/9606-H.sapiens_whole_organism-integrated_dataset.txtfor H. sapiens, http://pax-db.org/dao/511145-E.coli_whole_organism-integrated_dataset.txt for E. coli and http://pax-db.org/dao/224308-Spectral_counting_B.subtili_Chi_MCP_2011.txt for B. subtilis. We used the relative abundance, by taking the log value of protein abundance and scaling it between 0 and 1 for each species.
Translation efficiency data (RNAseq and Riboseq)
Translation efficiency (TE) data were extracted from recent studies that include both ribosome profiling and RNA-seq experiments for the same cell condition. The data set for E. coli includes four conditions: lysogenic broth (LB), heated medium (heat), minimal medium (MM) and osmotic medium (OSM). The data set for S. cerevisiae which also includes four conditions: Yeast extract peptone dextrose (YPD), no cyclohexamide (NOCHX), diamide and rapamycin, and a dataset for H. sapiens which contains three sets obtains during G2, G1 and mitose cell phase. Translation efficiency (TE) was estimated in the same way for all datasets, as the ratio between the ribosome abundance for a given mRNA and the abundance of the mRNA itself. The ribosome and mRNA abundances were estimated in reads per kilobase per million mapped reads (RPKM) from the CDS positions (12).
The TE data sets, based on the profiles of ribosome density with single nucleotide resolution and RNA-seq data, were obtained for E. coli (74), yeast (75) and human (76). Additionally, protein abundance data were extracted from the PAX database for the same species (http://pax-db.org/#!home).
Estimation of mRNA folding energy
mRNA folding energy was estimated using a customized version of the Afold software (77); only coding regions of the mRNAs were folded. Afold estimates the free energy of folding-unfolding (ΔG) for 30 nucleotide segments of an mRNA by calculating of the difference between optimal free energies of mRNA foldings with paired and completely unpaired states of the given segment. The segment length corresponds to the size of the ribosomal footprint (11), so that ΔG is the energetic cost of making a completely unpaired segment accessible to the ribosome. Afold scans the entire coding sequence of an mRNA and processes all overlapping 30 nucleotide windows. The mean mRNA folding energy (ΔG mean) is the average folding energy of all windows along the complete mRNA sequence. The minimum (ΔGmin) and maximum (ΔGmax) values of ΔG among all the 30-nt windows were also used as measures of the local mRNA stability (Supplementary Figure S1). Taking into account numerous genome-wide comparisons between theoretically predicted and experimentally verified (in vitro and in vivo) mRNA folding with stable level of pairing and periodic patterns of pairing in coding regions (24,41–43,78–81), we estimated mRNA stability using previously described tools that have been successfully applied to the analysis of multiple mammalian and prokaryotic genomes (24,27,77).
Estimation of protein solvent accessibility
Protein solvent accessibility was estimated using the Naccess software (82). From a protein structure, Naccess extracts the relative solvent accessibility (RSA) of each amino acid side chain. Briefly, RSA corresponds to the surface percentage of the side chain that a residue exposes to the solvent within the given protein structure normalized by the surface percentage that is exposed when the same residue X is in an Ala-X-Ala tripeptide (see (78) for more details). A protein solvent accessibility (ACC) is computed as the mean RSA over all residues. To estimate the RSA of disordered parts of the protein, we extracted the RSA value computed from the whole protein for each residue belonging to a disordered segment and calculated the mean value. Thus, the RSA of the disordered parts of the protein is the contribution of the solvent accessibility value of the disordered parts to the RSA of the complete protein.
Partitioning of ordered and disordered regions of proteins
Ordered and disordered parts of proteins were predicted using the SEGHCA software (83). This software has the ability to detect absolute disorder, i.e. those portions of the protein that remain unstructured under any conditions. From the protein sequence, SEGHCA delineates segments with high density of hydrophobic clusters that typically correspond to regular secondary structures. These segments possess the ability to fold via hydrophobic interactions and are thus taken to correspond to the ordered part of the protein. The disordered segments are then predicted by subtraction. These segments are considered ‘absolute disorder’, i.e. as being unable to get structured via hydrophobic interactions under any conditions. The ordered and disordered segments were concatenated separately to analyze their respective features. In addition, as a control, disordered segments were predicted using IUPRED, a widely used protein disordered predictor (84).
Multiple linear regression and prediction of protein solvent accessibility
All statistical analyses were performed using R cran and specifically the dplyr, tydir, ggplot2 and leaps libraries. Correlation and partial correlation analyses were performed using Spearman rank correlation. Statistical significance was reported using probability values (P-values); P-value below 0.05 were considered significant and displayed in bold in Sup. tables, in figures ** and * denote, respectively, P-values below 0.005 and 0.05. To identify stable combinations of parameters to explain the ACC variable and to evaluate relationships between structural features of mRNAs and proteins, and evolutionary rates, different multiple regression models were generated and cross-validated using all combinations of for variables, namely ΔG (local mRNA stability), protein size (length of the coding region), GC content, dS (synonymous substitution rate) and dN (non-synonymous substitution rate). The three model selection approach (stepwise, forward, and backward) was employed to select the variables that can best explain the variance of ACC.
RESULTS
mRNA stability and compactness of the encoded protein
We first investigated the dependency between the predicted mRNA structure and the compactness of the encoded protein. The mRNA folding energy was estimated as the mean of the local free energy estimates (ΔGmean) computed for each 30 nucleotide (nt) window along the complete mRNA molecule (see Materials and Methods). The 30-nt window size corresponds to the ribosomal footprint, i.e. the length of the linear RNA segment covered by the ribosome during translation (11). We additionally used the minimum and maximum values of ΔG among all 30-nt windows (ΔGmin and ΔGmax, respectively) to assess potential links between protein compactness and the folding energy of the most stable and the least stable portions of the messenger. Protein compactness, which is defined as the inverse of protein solvent accessibility (ACC), quantifies the fraction of exposed (or buried) residues in the structure of a given protein, was estimated from the protein structure (see Materials and Methods). In proteins with a high ACC value, i.e. small core proteins, most of the amino acid residues are exposed to the solvent (64,85,86). Conversely, in large core, compact proteins with low ACC, most of the residues are buried.
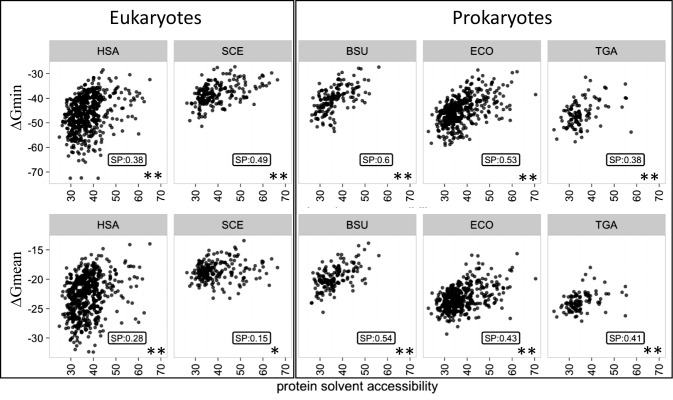
When the estimated mRNA folding energy was compared to protein accessibility, a highly significant positive correlation between ΔGmin and ACC was observed for all organisms (Spearman correlation coefficient [SP] between 0.38 and 0.6 [hereinafter, when describing correlations, we use the Spearman correlation coefficient unless otherwise indicated]; P-value < 0.005) (Figure 1 and Supplementary Table S1), and a somewhat lower but also significant (with the exception of yeast) positive correlation was found for ΔGmean (between 0.15 and 0.54; P-value < 0.05), whereas only weak negative correlation was detected for ΔGmax (Supplementary Table S1). Thus, stable mRNAs, especially those that contain at least one highly stable region (low ΔGmin), typically encode compact, large core proteins. Notably, the correlation between ΔGmean and ACC was much stronger in prokaryotes (between 0.41 and 0.54) compared to eukaryotes (between 0.15 to 0.28), whereas the difference between the correlation coefficients for ΔGmin was much less pronounced (between 0.38 and 0.49 in Prokaryotes and between 0.38 and 0.60 in Eukaryotes) (Supplementary Table S1).
Figure 1.
Correlation between mRNA stability and the compactness of the encoded protein. mRNA stability (folding energy) was estimated from the most stable segment (ΔGmin) or the average of all segments (ΔGmean) and is expressed in kcal/mol. Protein compactness was estimated from the protein solvent accessibility expressed as percent of a residue surface exposed to the solvent. HSA, SCE, BSU, ECO, TGA denote, respectively, H. sapiens, S. cerevisiae, B. subtilis, E. coli and T. gammatolerans. The structural data set was analyzed. The Spearman's rank correlation coefficient (SP) and the associated P-value (PV) are indicated for each plot.
To control for the robustness of the observed correlations to potential biases in the analyzed structural data set, we removed the protein superfamily redundancy using the SCOP annotation to generate a data set containing a single representative of each superfamily (selected randomly whenever several proteins belonged to the same superfamily) from each organism (87). The comparison of ΔGmin to ACC for this ‘pruned’ data set yielded results closely similar to those obtained with the complete data set (Supplementary Table S1) (between 0.39 and 0.60; P-value < 0.005). We then removed the structural redundancy at the protein fold level, and despite the loss of many structures, observed a significant positive correlation between ΔGmin and ACC (Supplementary Table S1) (between 0.36 and 0.63; P-value < 0.05). Thus, the observed link between the RNA folding anergy and protein solvent accessibility (compactness) is a robust feature that is not appreciably affected by biases that might exist in our protein structure data set.
Taking into account the expected strong negative correlation between ΔGmean and mRNA GC content (between −0.69 to −0.92; P-value < 0.005), which indeed was observed in all species (Supplementary Table S2), we controlled the dependency between ΔGs (ΔGmean and ΔGmin) and ACC for the GC-content. After controlling for GC, the correlation between ΔGmean and ACC in eukaryotes becomes as high as it is in prokaryotes (between 0.34 and 0.41; P-value < 0.005) (Supplementary Table S1). This effect was also detectable, albeit to a much lesser extent, in the case of ΔGmin (Supplementary Table S1). Thus, in eukaryotes, GC is a strong suppressor (modulator) of the relationship between ΔGmean and ACC, in part, probably, due to the much higher characteristic variance of the GC content along the eukaryotic mRNA sequences (Supplementary Figure S2). Additionally, we controlled the dependency between ΔGs (ΔGmean and ΔGmin) and ACC for amino acid content and codon usage. After controlling for amino acid content, the correlation remains significant (except in S. cerevisiae) (Supplementary Table S1). We then grouped the amino acids according to their physicochemical characteristics (hydrophobic: ILVM, aromatic: WFYH, polar: QNSTC, positively charged: RK, negatively charged: DE; small: PGA) and controlled the correlation between ΔG and ACC for the frequencies of these groups. Again, a positive and significant correlation was observed (except in S. cerevisiae) (Supplementary Table S1). Similarly, after controlling for codon frequencies, we still found a significant positive correlation (except in S. cerevisiae and in T. gammatolerans using ΔGmin). Taking into account all these controls, the present analysis reveals a universal relationship between mRNA and protein structures that is most pronounced for a local characteristic of the mRNA structure, i.e. folding energy of the most stable element (ΔGmin), and is largely independent of the features of mRNA and protein sequences.
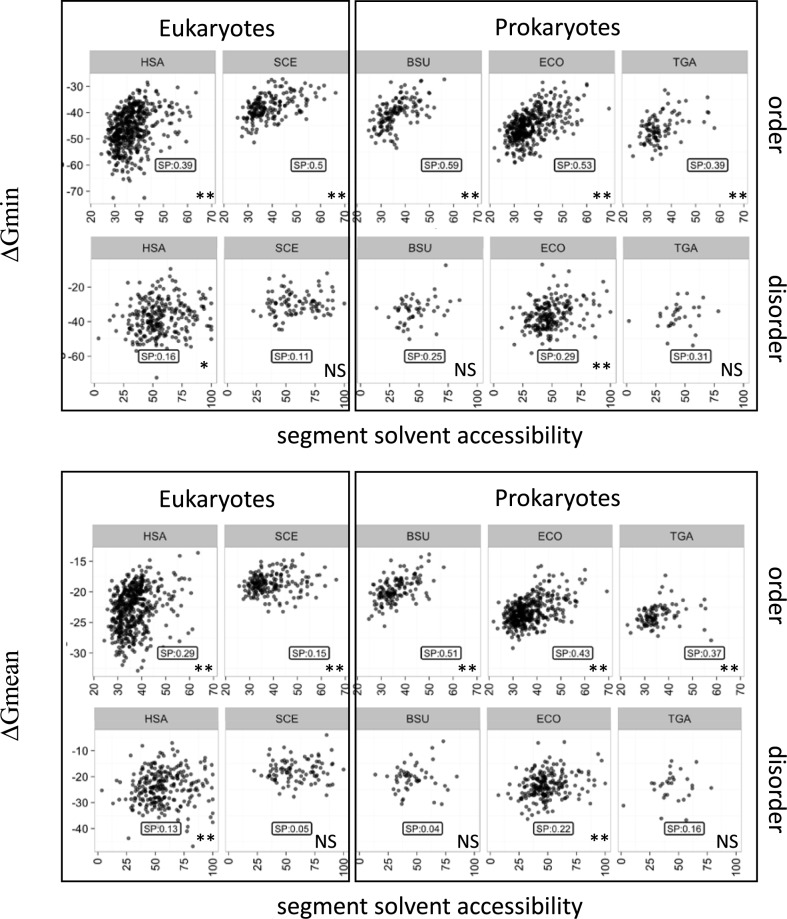
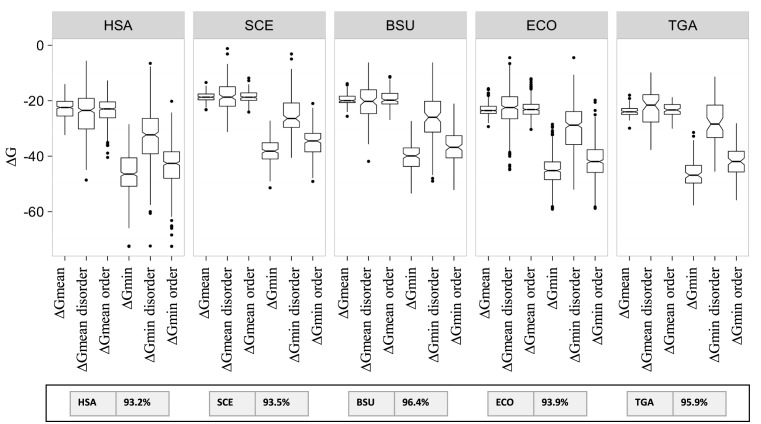
We then partitioned the protein sequences into predicted ordered and predicted absolute disordered regions using the SEGHCA software (see Methods). As expected, on average, the predicted disordered regions are significantly more accessible to the solvent than the ordered regions (Supplementary Figure S3). The correlations between ΔGmean or ΔGmin and ACC were then measured separately for the concatenated ordered and disordered segments. In all 5 analyzed organisms, the correlation for the ordered regions was roughly the same as for the complete sequences (between 0.15 and 0.51 using ΔGmean; P-value < 0.05; between 0.39 and 0.59 using ΔGmin; P-value < 0.005) whereas the disordered segments showed virtually no correlation (Figure 2 and Supplementary Table S3). In order to ascertain the robustness of these observations, the analysis was reproduced using the IUPRED software, a widely used disordered predictor, with similar results (Supplementary Table S3). These observations are consistent with the possibility that the link between the mRNA folding energy and protein compactness has to do with the folding of protein domains. A comparison of the ΔGmean and ΔGmin values in the predicted ordered and disordered regions of proteins showed that in the ordered regions, ΔGmin was much lower than in the disordered regions whereas the difference between the ΔGmean values was considerably less pronounced (Figure 3). Furthermore, as one could expect, about 95% of the 30 nt segments associated with the ΔGmin values for the complete mRNA sequences were located in regions coding for predicted ordered parts of proteins (see the Inset in Figure 3).
Figure 2.
Correlation between mRNA stability and solvent accessibility of the predicted ordered and disordered parts of proteins. mRNA stability and solvent accessibility were estimated as indicated for Figure 1. The designations are as in Figure 1.
Figure 3.
Distributions of the ΔG values in complete coding regions and in sequences encoding predicted ordered and disordered parts of the protein. The inset shows the percentage of the segments associated with ΔGmin that fall into the regions encoding predicted ordered portions of proteins in each organism. HSA, SCE, BSU, ECO, TGA denote, respectively, H. sapiens, S. cerevisiae, B. subtilis, E. coli and T. gammatolerans. The structural data set was analyzed.
The mRNA folding energy was also estimated for complete sequences as well as for sequences encoding predicted ordered and disordered segments using a smaller, 3-nt window, and the obtained ΔG values were compared to ACC. The results closely recapitulated the trends observed with the 30 nucleotide window (Supplementary Table S4), in agreement with the conclusion that the connection between protein compactness and mRNA folding is a local phenomenon.
The relationship between mRNA and protein structures is partially determined by GC content and protein size
The folding energy of a mRNA obviously depends on the GC content whereas protein compactness can depend on the length of the polypeptide chain (protein or mRNA size), potentially, in a complex manner. Indeed, as indicated above, we found that, for eukaryotes, the correlation between ΔGmean and ACC strongly depends on the GC content of the mRNA. Therefore, we systematically examined the effect of the GC content and the length of the coding sequence on the observed dependencies between mRNA and protein structures.
The GC content (GC), mRNA length (size) and mRNA folding energy (ΔGmean and ΔGmin) were compared for several prokaryotes and eukaryotes (in this case, the analysis did not have to be limited to proteins with available structures, so the number of organisms could be increased). A strong negative correlation between ΔGmean and the GC content of mRNAs was observed in both prokaryotes and eukaryotes (between −0.59 and −0.92; P-value < 0.005), and a slightly weaker correlation was observed for ΔGmin (between −0.23 and −0.71; P-value < 0.005) (Supplementary Table S2). Thus, as expected, GC-rich mRNAs are typically more stable and form compact structures.
The mRNA folding energy and protein size were strongly, negatively correlated in all organisms, especially when ΔGmin was used as the local measure of folding energy (between −0.43 and −0.66; P-value < 0.005) (Supplementary Table S2). Similar to the relationship between ΔG and ACC described above, the difference between prokaryotes and eukaryotes was notably more pronounced for ΔGmean than for ΔGmin (Supplementary Table S2). The relationship between ΔG and the coding sequence length was substantially stronger for the ordered segments of proteins than for the disordered segments (Supplementary Table S5). Altogether, these observations show that mRNAs encoding large proteins are typically GC-rich and accordingly highly structured, and contain local elements of exceptional stability.
Selection pressure at the mRNA and protein levels
The rate of protein sequence evolution generally depends on the level of functional constraint (88) but the contributions of different types of constraints are not fully understood although it is well established that buried amino acid positions evolve significantly slower than exposed positions (64,85,86). A significant negative correlation was observed between ACC and synonymous and non-synonymous evolutionary rates (ERs) (dS and dN) in all species (except dS in human) (between −0.17 and −0.30; P-value < 0.005 for dN and between −0.24 and −0.33; P-value < 0.005 for dS) (Supplementary Table S6). Thus, large core, compact proteins on average evolve faster than small core proteins, conceivably, because the former are more robust to deleterious effects of mutations, in agreement with previous findings (64,85,86). We also repeated the analysis of ACC versus dS and dN separately for the predicted ordered and disordered segments of proteins and observed a more pronounced relationship between ACC and ERs in the ordered compared to disordered parts of the protein (Supplementary Table S6) in agreement with previous findings (44).
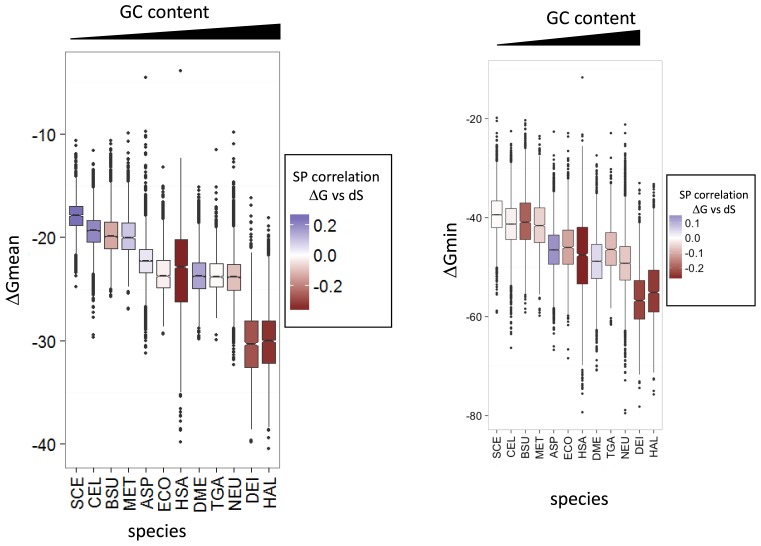
We further compared the estimated mRNA folding energy to the synonymous and non-synonymous ERs (dS and dN, respectively) and the protein-level selection pressure measured as dN/dS. As observed previously, in all species, dN and dS are significantly correlated (Supplementary Table S7) indicating that selection pressure applies similarly at the protein and mRNA levels albeit at substantially different strengths. Comparative analysis of ERs and the mRNA folding energy showed a significant positive correlation in organisms with high GC content, for example, H. volcanii and D. radiodurans. In contrast, S. cerevisiae and C. elegans, the most AT–rich of the analyzed genomes, showed a significant negative correlation with ΔGmean, whereas the genomes with an intermediate GC content showed little correlation (Figure 4 and Supplementary Table S7). Thus, in organisms with GC-rich genomes, stable mRNA regions generally evolve fast, whereas in organisms with AT-rich genomes, they evolve slowly compared to less stable regions. A weak to moderate positive correlation was detected between dN/dS (measure of protein-level selection) and ΔG for all organisms except for those with the highest GC content (Supplementary Table S7). Thus, highly conserved proteins show a tendency to be encoded by stable mRNAs except for those organisms in which RNA is generally highly structured due to the high GC content. Taken together, these findings suggest that selection acts to optimize rather than minimize or maximize mRNA stability.
Figure 4.
Distributions of mRNA folding energy in the analyzed organisms. Species are ordered following their transcriptome GC content and the Spearman's rank correlation coefficient between ΔGmean and synonymous mutation (dS) is mapped on the boxes as color gradient. SCE, CEL, BSU, MET, ASP, ECO, HSA, DME, TGA, NEU, DEI, HAL denote, respectively, S. cerevisiae, C. elegans, B. subtilis, M. mazei, A. orizae, E. coli, H. sapiens, D. melanogaster, T. gammatolerans, D. radiodurans, H. volvanii. The transcriptome data set was analyzed.
Translation efficiency, protein abundance, protein compactness and mRNA structure
The observations presented above, on the correlation between mRNA folding energy (ΔG) and ACC, suggest that mRNA stability could significantly contribute to protein folding and compactness. One possible model is that the ribosome requires extra time to unwind stable secondary structure elements in the mRNA, providing an opportunity for the protein domains to fold properly, which is particularly important for compact (large core) and large proteins. This model appears to be compatible with the available data on ribosome stalling triggered by specific mRNA structures such as pseudoknots or stable hairpins (89,90). To assess this model more directly, we compared the stability of the analyzed mRNAs to the experimentally determined translation efficiency and protein abundance.
Translation efficiency (TE) is quantified by the ratio of the abundance of ribosomes associated with the given mRNA to the abundance of the mRNA itself (see Materia;s and Methods) obtained, respectively, from ribosome profiling and RNA-seq experiments (12). We found that the experimentally measured protein abundance positively correlates with translation efficiency under 4 different conditions in E. coli and yeast and in all three phases of the cell cycle in humans (between 0.15 and 0.60; P-value < 0.005) (see Materials and Methods) (Supplementary Table S8). Thus, as could be expected, abundant proteins are translated fast under various conditions in both prokaryotes and eukaryotes. We also detected a positive correlation between protein abundance and solvent accessibility (except for T. gammatolerans) (Supplementary Table S9), i.e. on average, abundant proteins are less compact than proteins produced in lower quantities. Taking into account that abundant proteins are less compact and are translated faster than proteins of lower abundance, a positive correlation between protein abundance and ΔG could be expected. However, in agreement with previous reports (37) and similar to the case of evolutionary rates and protein-level selection discussed above, no universal relationship between protein abundance and ΔG was observed (Supplementary Table S10). A negative correlation was detected in AT-rich species, such as yeast and C. elegans (−0.29; P-value < 0.005), but a positive correlation was found in more GC-rich organisms, such as E. coli (0.19; P-value < 0.005); these correlations were independent of other tested variables, in particular mRNA length and protein size (Supplementary Table S10). Along with the results in the previous section, these observations point to evolutionary optimization of ΔG.
For E. coli, in general, ΔG is a better predictor of TE than ACC, and correlation between TE and ΔG is significant for all four conditions. Taking into account that correlation between TE and ΔG is evident only in the G2 phase of the cell cycle in human whereas in yeast there was no correlation for any conditions in yeast, we further examined the connection between the GC content and TE in different organisms. In all cases with a significant positive correlation between TE and ΔG, there was a negative correlation between GC content and TE, whereas the other organisms showed a positive correlation (Supplementary Table S8). After controlling for GC content, a significant positive correlation between ΔG and TE was observed for all organisms and all conditions (Supplementary Table S11). This result is in agreement with a model under which stable mRNAs are translated slowly (with a low TE), allowing more time for the encoded compact and/or large proteins to fold co-translationally.
Prediction of protein features from the mRNA characteristics using multiple regression models
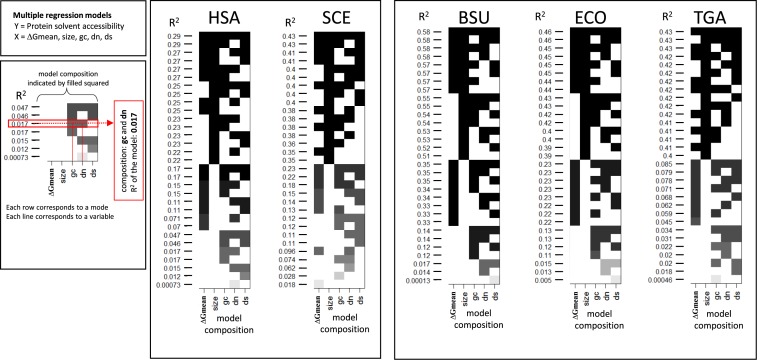
An attempt to extend the partial correlation analysis to all variables employed here, namely GC content, protein size, dN, dS, protein abundance, and fractions of each of 6 groups of amino acids (see above) failed to produce compelling results as the magnitude of the correlation between the mRNA folding energy (ΔGmean or ΔGmin) and protein compactness (solvent accessibility) substantially dropped, most likely due to over-fitting (see Supplementary Table S12). Such an outcome is expected with a large number of variables and a limited number of data points (91,92).
Given these complex relationships between different variables related to mRNA and protein structures, we turned to the analysis multiple linear regression (MLR) models in order to assess the ability of different combinations of these variables to account for the variance of ACC (see Materials and Methods). The models included combinations of five variables: ΔG (either ΔGmean or ΔGmin), coding region length (size), GC content, and ERs (dS and dN) (see Materials and Methods for further details). On average, ΔGmin was a slightly better predictor of ACC than ΔGmean. Among the individual variables, the strongest correlation with protein compactness was observed for the length of the coding region, especially for prokaryotes, followed by ΔG (Figure 5). In most cases, this pair of variables possessed the greatest explanatory power compared to other pairs in both prokaryotes and eukaryotes (Figure 5). Evolutionary rate in synonymous positions (dS) also significantly increased the prediction power in most organisms, particularly in eukaryotes. In prokaryotes, dS alone does not significantly correlate with ACC but increases the prediction power of MLR models when added to the strongest pair of variables, namely size and ΔG (Figure 5).
Figure 5.
Multiple linear regression analysis. The vertical axis shows the percentage of the protein solvent accessibility variance (R2 values) that explained by models that include different combinations of variables as indicated by shaded squares in the matrix. Each model combine different parameters indicated by filled squares on the same row and associated the R2 value on the left of the row. Overfitting was tested for models using 5 variables (see Figure 6 and corresponding section). HSA, SCE, BSU, ECO, TGA denote, respectively, H. sapiens, S. cerevisiae, B. subtilis, E. coli and T. gammatolerans. The structural data sets were analyzed.
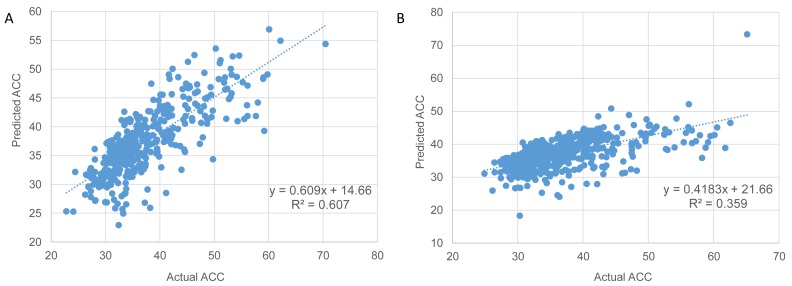
We further explored the predictive power of the MLR models for the two organisms, E. coli and H. sapiens (Figure 6A and B), where sufficient expression data were available. Using the cross-validation approach, random training subsets were selected to include approximately one third of the gene loci available for each organism. The remaining two thirds of genes from each organism were used as validation sets. Cross validation for the models with the largest number of variables (Figures 5 and 6) and comparison of the adjusted correlation coefficients showed similar R2 values for the training and validation data sets indicating that there was no appreciable overfitting in our models with three parameters (size, ΔG and dS). Root mean squared error (RMSE) values support the conclusion that the optimal model should include these three variables (Supplementary Figure S4) whereas the remaining two variables (dN and GC-content) should be discarded.
Figure 6.
Prediction of protein solvent accessibility for proteins with known structures in E. coli (A) and H. sapiens (B). The plot of multiple linear regression (MLR) predictions versus the actual ACC estimations based on the known structural data of the proteins. Spearman correlation (R2 values) is 0.602 in E. coli (A) and 0.4 in human (B). The actual ACC values were estimated from known protein structures by extraction of the relative solvent accessibility (RSA) of each side chain residues. A protein solvent accessibility is computed as the mean RSA over all residues (see Materials and Methods). Values of predicted average solvent accessibility were estimated using a MLR model including the following mRNA features: (i) size (log) − the length of the coding region (P-values < 0.005), (ii) ΔGmin, free energy of mRNA folding (P-values < 0.005) and (iii) dS (log), synonymous evolutionary rate, which was estimated for E. coli versus S. typhi (A) and human vs mouse (B) orthologous gene pairs (P-values < 0.05). All three features significantly contribute to the model (P values show above); their coefficients are significantly different from 0.
Three variables, namely the size (length) of the coding region (P value < 0.005), ΔG (ΔGmin or ΔGmean) of mRNA folding (P value < 0.005) and dS (P value < 0.05), showed independent predictive power for ACC in all organisms. The plot of predicted (evaluated by 3-fold cross validation) versus the actual ACC values for the E. coli validation set using the three variables combined is shown in Figure 6A. In terms of the predictive power, the MLR models, which yield R values in the range between 0.59 and 0.75 for different organisms, perform better for prokaryotes (Figure 6A and B). The predictive power of our model for E. coli is comparable to that of the previous models based on protein features, including predicted protein secondary structure, native disorder and physicochemical propensities (93,94). Thus, protein compactness can be predicted with considerable accuracy using mRNA features only, specifically, length, ΔG and dS. Adding TE and protein abundance to the models did not enhance the prediction power.
We additionally generated multiple linear regression models using independent randomly chosen data sets to explore the connections between all analyzed features of mRNAs and proteins (ΔG, ACC, GC content, mRNA size, dN, dS, protein abundance) and their contributions to the variance of TE (Supplementary Figure S5). The MLR models demonstrate that, in E. coli, the combination of three features (ΔG, abundance and dS) yields stable predictions for all four conditions albeit with limited predictive power. Protein abundance, in general, performs better as a predictor of TE and could be used to replace ACC in models. In E. coli, in general, ΔG is a better predictor of TE than ACC but the contributions of these two variables are independent. In human, the model with three independent variables (ΔG, size and abundance) was the best predictor of TE for G2. Rates of evolution did not add much to the prediction in the MLR models in eukaryotes.
Taken together, the results of the MLR analysis indicate that protein solvent accessibility (a measure of compactness) can be predicted with considerable accuracy using only mRNA features, namely length of the coding region, ΔG and dS.
DISCUSSION
The findings presented here reveal a universal connection between protein compactness (measured as ACC) and local free energy of mRNA folding (ΔGmean or ΔGmin) which is a measure of RNA stability. In all organisms for which we could collect sufficient structural information, compact proteins with large cores are encoded by RNA molecules that are, on average, more stable at the local level than those encoding small-core, less compact proteins. In principle, this relationship could reflect different types of connections between mRNA and the protein it encodes, e.g. the high folding potential of the mRNA could reflect specific requirements of mRNA stability or translation fidelity for compact proteins. However, we obtained a specific clue as to the likely driving force behind the link. The correlation between ΔGmin or ΔGmean and protein compactness was found to be pronounced in the mRNA segments that encode predicted ordered (structured) portions of proteins but was much weaker or non-existent in the regions predicted to encode disordered (unstructured) parts (Figure 2). This observation implies that ΔG of the coding RNA sequence matters for the portions of a protein that have to fold co-translationally but not for the disordered portions.
Throughout this work, we compare global measures of protein compactness to local measures of RNA stability, namely ΔGmin or ΔGmean values, in a 30-nt window covered by the ribosome during translation. Moreover, we found that ΔGmin is a stronger correlate of protein compactness than ΔGmean, suggesting that many mRNAs contain exceptionally stable elements which could function as distinct regulatory devices by slowing down translation and hence facilitating co-translational protein folding. In agreement with the proposed role of exceptionally stable secondary structure elements in mRNA as regulators of protein folding, we found that the segments corresponding to ΔGmin are almost always located within the portion of the mRNA that encodes the structured part of the protein.
In addition to the connection between ΔG and protein compactness, we also detected a negative correlation between ΔG and protein length, and again, this correlation was found to be much more pronounced for the predicted structured segments than for the non-structured ones. This finding is compatible with the substantial contribution of the mRNA structure to protein folding because longer protein sequences on average take more time to fold than shorter ones.
The above observations imply that in the portions of mRNAs that encode structured parts of proteins, RNA stability should be subject to purifying selection. The magnitude of this selection pressure can be predicted to positively correlate with the length of the structured portion of the protein and the relative core size. Such selection would primarily affect synonymous positions in which the RNA-level selection is largely decoupled from the protein-level selection. In agreement with these predictions, we indeed detected the expected dependencies between dS, mRNA stability and protein compactness. Multiple regression analysis showed that the contributions of ΔG, protein length and dS to the prediction of protein compactness (surface accessibility) are partially independent. Thus, there seems to be a significant length-independent component in protein compactness, and furthermore, stability might not be the only feature of mRNA structure that is subject to selection related to protein folding. The MLR models analyzed here show that these three features jointly account for about half of the variance in protein surface accessibility (at least in prokaryotes) which is indicative of a robust link between mRNA and protein structures.
The hypothesis that highly structured elements in mRNA function as gauges of protein folding is compatible with a considerable body of experimental evidence indicating that synonymous changes or polymorphic variants affecting the stability of highly structured mRNA elements can dramatically change translation speed, inhibit translation and influence co-translational protein folding (27,61,95,96). These highly stable elements even appear to play different roles in post-translational modifications and protein functions, for example, in actin (48) and prosphorybosyl pyrophosphate synthase (47). Correlations between protein and mRNA structures have been recently demonstrated for several specific cases of mRNAs with experimentally characterized structures (78,97,98). Furthermore, extensive observations have been reported connecting the evolutionarily conserved patterns of optimal and non-optimal codons with elements of protein secondary structure and, by inference co-translational folding (54,99,100). Together with the findings on the apparent role of mRNA secondary structure elements reported here, these results indicate that there multiple ways in which mRNA sequence and structure can be subject to selection driven by the requirements of protein folding optimization. The mRNA appears to contain not only a second layer of information as has been recently proposed (101) but perhaps, multiple layers of subtle, entangled signals.
Throughout this analysis, we observed substantial differences between prokaryotes and eukaryotes. All observed correlations as well as the explanatory power of the MLR models are weaker in eukaryotes compared to prokaryotes, which is likely to be due to the generally greater power of selection in the large prokaryotic populations compared to the weaker selection in the smaller populations of eukaryotes (102,103). More specifically, the correlation between ΔGmean and ACC was much weaker in eukaryotes than in prokaryotes whereas in the case of ΔGmin, the difference was substantially smaller. When the comparison was controlled for the GC content, the correlation for ΔGmean in eukaryotes substantially increased. The causes of the suppressing effect of the GC content on the ΔGmean versus compactness correlation in eukaryotes are not entirely clear. We suspect that one of the contributing factors is the high variability of the GC content along the mRNA and of the transcript size in eukaryotes compared to prokaryotes (Supplementary Figure S2). Another likely relevant factor is the greater content of disordered segments in eukaryotic proteins. Disordered segments tend to be enriched for polar amino acids, glycine and proline, which are encoded by GC-rich codons (Supplementary Figure S6), which contribute to the formation of stable local mRNA structures and thus suppress the link between ΔGmean and ACC. Furthermore, in eukaryotes, translation and protein folding do not occur co-transcriptionally as they do in prokaryotes (104,105), and therefore there is a greater potential for functionally relevant long range interactions in eukaryotic mRNAs. Nevertheless, the finding that ΔGmin is the strongest correlate of ACC in all cases implies that specific, highly stable secondary structure elements in mRNAs function as distinct devices to regulate protein folding in prokaryotes and eukaryotes alike.
CONCLUSIONS
The findings presented here reveal strong, universal connections between the structures of an mRNA and the encoded protein. The observation that these correlations are pronounced for predicted ordered parts of proteins but are much weaker or non-existent in the predicted disordered parts suggests that mRNA stability affects co-translational protein folding. Furthermore, we found that the folding energy of the most stable segment of an mRNA correlates with protein compactness stronger than the mean folding energy. These observations lead to the experimentally testable model in which elements with highly stable secondary structure that are typically located in the portions of mRNAs that encode structured protein parts, function as control devices for co-translational protein folding. This hypothesis is compatible with the demonstration of the RNA-level selection which correlates with RNA folding energy and protein compactness. The connection between mRNA and protein structures is more pronounced in prokaryotes compared to eukaryotes, conceivably due to the greater power of selection in the former.
Supplementary Material
Acknowledgments
We thank Koonin group members for useful discussions.
Author contributions: G.F. and S.A.S. performed research; G.F., A.Y.O., S.A.S. and E.V.K. analyzed the results; G.F., S.A.S. and E.V.K. wrote the manuscript which was read, edited and approved by all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural funds of the US Department of Health and Human Services (to the National Library of Medicine). The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gingold H., Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter J.D., Coller J. Pausing on Polyribosomes: Make Way for Elongation in Translational Control. Cell. 2015;163:292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunak S., Engelbrecht J. Protein structure and the sequential structure of mRNA: alpha-helix and beta-sheet signals at the nucleotide level. Proteins. 1996;25:237–252. doi: 10.1002/(SICI)1097-0134(199606)25:2<237::AID-PROT9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 5.Oresic M., Shalloway D. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol. 1998;281:31–48. doi: 10.1006/jmbi.1998.1921. [DOI] [PubMed] [Google Scholar]
- 6.Adzhubei I.A., Adzhubei A.A., Neidle S. An Integrated Sequence-Structure Database incorporating matching mRNA sequence, amino acid sequence and protein three-dimensional structure data. Nucleic Acids Res. 1998;26:327–331. doi: 10.1093/nar/26.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie T., Ding D. The relationship between synonymous codon usage and protein structure. FEBS Lett. 1998;434:93–96. doi: 10.1016/s0014-5793(98)00955-7. [DOI] [PubMed] [Google Scholar]
- 8.Thanaraj T.A., Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996;5:1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thanaraj T.A., Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia M., Luo L., Liu C. Statistical correlation between protein secondary structure and messenger RNA stem-loop structure. Biopolymers. 2004;73:16–26. doi: 10.1002/bip.10496. [DOI] [PubMed] [Google Scholar]
- 11.Ingolia N.T. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuller T., Veksler-Lublinsky I., Gazit N., Kupiec M., Ruppin E., Ziv-Ukelson M. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol. 2011;12:R110. doi: 10.1186/gb-2011-12-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dana A., Tuller T. Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput. Biol. 2012;8:e1002755. doi: 10.1371/journal.pcbi.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J., Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ude S., Lassak J., Starosta A.L., Kraxenberger T., Wilson D.N., Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 17.Lassak J., Wilson D.N., Jung K. Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol. Microbiol. 2015;99:219–235. doi: 10.1111/mmi.13233. [DOI] [PubMed] [Google Scholar]
- 18.Charneski C.A., Hurst L.D. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito K., Chiba S. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 20.Dana A., Tuller T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 2014;42:9171–9181. doi: 10.1093/nar/gku646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artieri C.G., Fraser H.B. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koutmou K.S., Schuller A.P., Brunelle J.L., Radhakrishnan A., Djuranovic S., Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. Elife. 2015;4 doi: 10.7554/eLife.05534. doi:10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Requiao R.D., de Souza H.J., Rossetto S., Domitrovic T., Palhano F.L. Increased ribosome density associated to positively charged residues is evident in ribosome profiling experiments performed in the absence of translation inhibitors. RNA Biol. 2016;13:561–568. doi: 10.1080/15476286.2016.1172755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabalina S.A., Ogurtsov A.Y., Spiridonov N.A. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;34:2428–2437. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu W., Zhou T., Wilke C.O. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shabalina S.A., Spiridonov N.A., Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 30.Tuller T., Zur H. Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res. 2015;43:13–28. doi: 10.1093/nar/gku1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 32.Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen J.D., Lancaster L., Hodges C., Zeri A.C., Yoshimura S.H., Noller H.F., Bustamante C., Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Q., Ryvkin P., Li F., Dragomir I., Valladares O., Yang J., Cao K., Wang L.S., Gregory B.D. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 36.Locker N., Chamond N., Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zur H., Tuller T. Strong association between mRNA folding strength and protein abundance in S. cerevisiae. EMBO Rep. 2012;13:272–277. doi: 10.1038/embor.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park C., Chen X., Yang J.R., Zhang J. Differential requirements for mRNA folding partially explain why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E678–E686. doi: 10.1073/pnas.1218066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorochowski T.E., Ignatova Z., Bovenberg R.A., Roubos J.A. Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res. 2015;43:3022–3032. doi: 10.1093/nar/gkv199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao Y., Liu H., Liu Y., Tao S. Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:4813–4822. doi: 10.1093/nar/gku159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E., et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Incarnato D., Neri F., Anselmi F., Oliviero S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 44.Shabalina S.A., Ogurtsov A.Y., Spiridonov N.A., Koonin E.V. Evolution at protein ends: major contribution of alternative transcription initiation and termination to the transcriptome and proteome diversity in mammals. Nucleic Acids Res. 2014;42:7132–7144. doi: 10.1093/nar/gku342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotkin J.B., Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaney J.L., Clark P.L. Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 2015;44:143–166. doi: 10.1146/annurev-biophys-060414-034333. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F., Patel D.M., Colavita K., Rodionova I., Buckley B., Scott D.A., Kumar A., Shabalina S.A., Saha S., Chernov M., et al. Arginylation regulates purine nucleotide biosynthesis by enhancing the activity of phosphoribosyl pyrophosphate synthase. Nat. Commun. 2015;6:7517. doi: 10.1038/ncomms8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Saha S., Shabalina S.A., Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardesty B., Tsalkova T., Kramer G. Co-translational folding. Curr. Opin. Struct. Biol. 1999;9:111–114. doi: 10.1016/s0959-440x(99)80014-1. [DOI] [PubMed] [Google Scholar]
- 50.Hardesty B., Kramer G. Folding of a nascent peptide on the ribosome. Prog. Nucleic Acids Res. Mol. Biol. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhang G., Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Holtkamp W., Kokic G., Jager M., Mittelstaet J., Komar A.A., Rodnina M.V. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015;350:1104–1107. doi: 10.1126/science.aad0344. [DOI] [PubMed] [Google Scholar]
- 53.Rodnina M.V., Wintermeyer W. Protein elongation, co-translational folding and targeting. J. Mol. Biol. 2016;428:2165–2185. doi: 10.1016/j.jmb.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien E.P., Ciryam P., Vendruscolo M., Dobson C.M. Understanding the influence of codon translation rates on cotranslational protein folding. Acc. Chem. Res. 2014;47:1536–1544. doi: 10.1021/ar5000117. [DOI] [PubMed] [Google Scholar]
- 55.Bustamante C.J., Kaiser C.M., Maillard R.A., Goldman D.H., Wilson C.A. Mechanisms of cellular proteostasis: insights from single-molecule approaches. Annu. Rev. Biophys. 2014;43:119–140. doi: 10.1146/annurev-biophys-051013-022811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldman D.H., Kaiser C.M., Milin A., Righini M., Tinoco I., Jr, Bustamante C. Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puglisi J.D. Protein synthesis. The delicate dance of translation and folding. Science. 2015;348:399–400. doi: 10.1126/science.aab2157. [DOI] [PubMed] [Google Scholar]
- 58.Kim S.J., Yoon J.S., Shishido H., Yang Z., Rooney L.A., Barral J.M., Skach W.R. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 59.Guisez Y., Robbens J., Remaut E., Fiers W. Folding of the MS2 coat protein in Escherichia coli is modulated by translational pauses resulting from mRNA secondary structure and codon usage: a hypothesis. J. Theor. Biol. 1993;162:243–252. doi: 10.1006/jtbi.1993.1085. [DOI] [PubMed] [Google Scholar]
- 60.Komar A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 62.Fung K.L., Pan J., Ohnuma S., Lund P.E., Pixley J.N., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res. 2014;74:598–608. doi: 10.1158/0008-5472.CAN-13-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faure G., Koonin E.V. Universal distribution of mutational effects on protein stability, uncoupling of protein robustness from sequence evolution and distinct evolutionary modes of prokaryotic and eukaryotic proteins. Phys. Biol. 2015;12:035001. doi: 10.1088/1478-3975/12/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q., Canutescu A.A., Dunbrack R.L., Jr SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scannell D.R., Zill O.A., Rokas A., Payen C., Dunham M.J., Eisen M.B., Rine J., Johnston M., Hittinger C.T. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 (Bethesda) 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Novichkov P.S., Ratnere I., Wolf Y.I., Koonin E.V., Dubchak I. ATGC: a database of orthologous genes from closely related prokaryotic genomes and a research platform for microevolution of prokaryotes. Nucleic Acids Res. 2009;37:D448–D454. doi: 10.1093/nar/gkn684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altenhoff A.M., Skunca N., Glover N., Train C.M., Sueki A., Pilizota I., Gori K., Tomiczek B., Muller S., Redestig H., et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 73.Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S.P., Hengartner M.O., von Mering C. PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell Proteomics. 2012;11:492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bartholomaus A., Fedyunin I., Feist P., Sin C., Zhang G., Valleriani A., Ignatova Z. Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. A Math. Phys. Eng. Sci. 2016;374 doi: 10.1098/rsta.2015.0069. doi:10.1098/rsta.2015.0069. [DOI] [PubMed] [Google Scholar]
- 75.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanenbaum M.E., Stern-Ginossar N., Weissman J.S., Vale R.D. Regulation of mRNA translation during mitosis. Elife. 2015;4 doi: 10.7554/eLife.07957. doi:10.7554/eLife.07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogurtsov A.Y., Shabalina S.A., Kondrashov A.S., Roytberg M.A. Analysis of internal loops within the RNA secondary structure in almost quadratic time. Bioinformatics. 2006;22:1317–1324. doi: 10.1093/bioinformatics/btl083. [DOI] [PubMed] [Google Scholar]
- 78.Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W., Jr, Swanstrom R., Burch C.L., Weeks K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwok C.K., Ding Y., Tang Y., Assmann S.M., Bevilacqua P.C. Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 2013;4:2971. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 80.Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doerr A. The in vivo RNA structurome. Nat Methods. 2014;11:11. doi: 10.1038/nmeth.2799. [DOI] [PubMed] [Google Scholar]
- 82.Hubbard S.J., Thronton J.M. Department of Biochemistry and Molecular Biology. London: University College; 1993. NACCESS.https://sbgrid.org/software/titles/naccess [Google Scholar]
- 83.Faure G., Callebaut I. Comprehensive repertoire of foldable regions within whole genomes. PLoS Comput. Biol. 2013;9:e1003280. doi: 10.1371/journal.pcbi.1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dosztanyi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 85.Franzosa E.A., Xia Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol. Biol. Evol. 2009;26:2387–2395. doi: 10.1093/molbev/msp146. [DOI] [PubMed] [Google Scholar]
- 86.Franzosa E.A., Xia Y. Independent effects of protein core size and expression on residue-level structure-evolution relationships. PLoS ONE. 2012;7:e46602. doi: 10.1371/journal.pone.0046602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fox N.K., Brenner S.E., Chandonia J.M. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koonin E.V., Wolf Y.I. Constraints and plasticity in genome and molecular-phenome evolution. Nat. Rev. Genet. 2010;11:487–498. doi: 10.1038/nrg2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Somogyi P., Jenner A.J., Brierley I., Inglis S.C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lopinski J.D., Dinman J.D., Bruenn J.A. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Altman D.G. Practical Statistics for Medical Research (Chapman & Hall/CRC Texts in Statistical Science) 1st edn. NY: Chapman & Hall/CRC; 1991. [Google Scholar]
- 92.Draper N.R., Smith H. Applied Regression Analysis. 3rd edn. NY: Wiley Belsley; 1998. [Google Scholar]
- 93.Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J., Chen W., Sun P., Zhao X., Ma Z. Prediction of protein solvent accessibility using PSO-SVR with multiple sequence-derived features and weighted sliding window scheme. BioData Min. 2015;8:3. doi: 10.1186/s13040-014-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nackley A.G., Shabalina S.A., Tchivileva I.E., Satterfield K., Korchynskyi O., Makarov S.S., Maixner W., Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 96.Komar A.A. Silent SNPs: impact on gene function and phenotype. Pharmacogenomics. 2007;8:1075–1080. doi: 10.2217/14622416.8.8.1075. [DOI] [PubMed] [Google Scholar]
- 97.Sanjuan R., Borderia A.V. Interplay between RNA structure and protein evolution in HIV-1. Mol. Biol. Evol. 2011;28:1333–1338. doi: 10.1093/molbev/msq329. [DOI] [PubMed] [Google Scholar]
- 98.Tang Y., Assmann S.M., Bevilacqua P.C. Protein structure Is related to RNA structural reactivity in vivo. J. Mol. Biol. 2016;428:758–766. doi: 10.1016/j.jmb.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 99.Spencer P.S., Siller E., Anderson J.F., Barral J.M. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pechmann S., Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramos S.B., Laederach A. Molecular biology: A second layer of information in RNA. Nature. 2014;505:621–622. doi: 10.1038/505621a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. U.S.A. 2007;104(Suppl. 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lynch M., Conery J.S. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 104.Grundy F.J., Henkin T.M. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit. Rev. Biochem. Mol. Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 105.French S.L., Santangelo T.J., Beyer A.L., Reeve J.N. Transcription and translation are coupled in Archaea. Mol. Biol. Evol. 2007;24:893–895. doi: 10.1093/molbev/msm007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.