Abstract
HMGA2 is an important chromatin factor that interacts with DNA via three AT-hook domains, thereby regulating chromatin architecture and transcription during embryonic and fetal development. The protein is absent from differentiated somatic cells, but aberrantly re-expressed in most aggressive human neoplasias where it is causally linked to cell transformation and metastasis. DNA-binding also enables HMGA2 to protect cancer cells from DNA-damaging agents. HMGA2 therefore is considered to be a prime drug target for many aggressive malignancies. Here, we have developed a broadly applicable cell-based reporter system which can identify HMGA2 antagonists targeting functionally important protein domains, as validated with the known AT-hook competitor netropsin. In addition, high-throughput screening can uncover functional links between HMGA2 and cellular factors important for cell transformation. This is demonstrated with the discovery that HMGA2 potentiates the clinically important topoisomerase I inhibitor irinotecan/SN-38 in trapping the enzyme in covalent DNA-complexes, thereby attenuating transcription.
INTRODUCTION
The high-mobility group AT-hook 2 (HMGA2) protein is a non-histone chromatin factor that is highly conserved in mammals. It is expressed in embryonic stem cells, during fetal development and in some adult stem cell populations, but it is absent from normal somatic tissues (1–3). Clinical studies showed that HMGA2 is aberrantly re-expressed in most malignant human neoplasias, where the expression level strongly correlates with the degree of malignancy and metastasis (4,5). Furthermore, HMGA2 re-expression is causally linked to cell transformation, epithelial-mesenchymal transition and metastasis, also in the context of cancer stem cells (6–8).
HMGA2 harbors three independent DNA binding domains, so-called AT-hooks, which recognize the minor groove of short, AT-rich duplex sequences with nM affinity. The protein also carries a C-terminal acidic tail which mediates interactions with other proteins and could play a role in regulating DNA-binding (9–11). Through its DNA-binding activity, HMGA2 (similarly to the related HMGA1 protein) modulates chromatin architecture and plays crucial roles globally in the formation of heterochromatic regions, such as telomeres and senescence-associated foci (12–14). DNA-binding is also critical locally for modulation of gene expression in the context of cell differentiation/transformation processes (11,15,16). Furthermore, we have shown that HMGA2 protects cancer cells from DNA damage induced by chemotherapeutic agents via roles in base excision repair (17) and chaperoning of stalled DNA replication forks (18). Both functions require functional AT-hooks for DNA-binding.
The human HMGA2 protein therefore is rapidly emerging as an important drug target for treatment of many aggressive human neoplasias (5,19). Importantly, several recent studies demonstrated a therapeutic benefit of interfering with HMGA2 function(s) through, for example, lentiviral short hairpin RNA (20) or let-7 microRNAs (21). Taken together, the available information warrants the development of cell-based high-throughput compound library screens in order to identify specific HMGA2 antagonists.
In the present study, we present a novel cell-based reporter system, which can easily be adapted for high-throughput screening. We exemplified the broad utility of the system here with the identification of the important anti-cancer drug irinotecan/SN-38 as potential HMGA2 antagonist. This led to the discovery of a functional link between HMGA2 and human topoisomerase I, which can have important implications for the treatment of human malignancies.
MATERIALS AND METHODS
Cell Lines, HMGA2 and Renilla expression vectors
HeLa cells were grown in DMEM with 10% FBS (Life Technologies/GIBCO). HEK 293 cells were cultured in DMEM with 10% FBS (Life Technologies/ GIBCO). HT 1299 cells were grown in RPMI with 10% FBS (Life Technologies/GIBCO).
Expression vectors for wild-type HMGA2, the 23M mutant and HMGA1a/1b were described in (18). Expression vectors for the 123M and the linker 1 deletion mutant were generated by site-directed mutagenesis (Agilent Technologies), using vectors for 23M and wild-type HMGA2 as templates, respectively. The C-terminal truncated HMGA2 was generated by PCR, using an existing prokaryotic expression vector as template. All HMGA2 expression vectors carry a C-terminal Flag-tag. The Renilla luciferase expression vector containing the HSV-TK promoter (pRL-TK) was obtained from Promega. Deletion of the AT-rich stretch from the HSV-TK promoter was achieved via site-directed mutagenesis. Vector sequences were confirmed by sequencing.
Chemicals and reagents
SN-38 was purchased from Abcam; netropsin and irinotecan hydrochloride were purchased from Sigma. Chlorambucil, podofilox and mannitol were from MicroSource Discovery Systems. TopoII inhibitor ICRF-193 [meso-4,4′-(3,2-butanediyl)-bis(2,6-piperazinedione)] was from Sigma.
Western blotting
Primary antibodies were rabbit polyclonal anti-HMGA2 (Cell Signalling, 1:1000), rabbit monoclonal anti-HMGA1 (1:1000; Cell Signaling), and mouse monoclonal anti-FLAG M2 antibodies (1:1000; Sigma Aldrich). Secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit antibodies (1:10 000; Santa Cruz) and polyclonal goat anti-mouse immunoglobulins (1:10 000; Dako).
Renilla luciferase assays and compound testing
For a typical luciferase assay, ∼106 HeLa cells were plated per six-well and co-transfected the following day with 1 μg of both HMGA2 expression and reporter vector using Lipofectamine 2000 (Invitrogen/Life Technologies). About 40 000 cells were transferred the next day per 96-well on a white plate with clear-bottom (COSTAR 3610, Corning) in 150 μl medium. After 24 h recovery, cells were treated for additional 24 h with compounds (dissolved in DMSO), as indicated in the text. Luciferase readings were performed with ‘Renilla Glo’ E2710 kit (Promega) following the manufacturer's instruction on a TECAN Infinite M200 Pro bioluminescence reader.
High-throughput screening
To scale up the cell pool for high-throughput screens, co-transfections were performed in multiple 10 cm dishes using Lipofectamine 2000. After 24 h, cells from identical co-transfections were pooled, and about 10 000 cells were plated per well on a 384-well white plate with clear-bottom (Greiner bio one) using the LABCYTE Echo 550 liquid handler. For compound screening, 10 μM (final assay conc.) of each compound was dispensed from the PHARMAKON 1600 small-compound library (MicroSource Discovery Systems) prior to cell seeding using the Bravo Automated Liquid Handling Platform (Agilent Technologies). Luciferase assays were performed as described above using Infinite M1000 PRO bioluminescence reader (TECAN).
qRT-PCR analysis
Four sets of co-transfections were performed in six-well format, as described in the text. Cells were harvested and total RNA was extracted using TRIzol reagent following a standard protocol (Ambion/Life Technologies). RNA samples were treated with RQ1 DNase (Promega) and RNA quality was subsequently checked via agarose gel electrophoresis. qRT-PCR, using four technical replicates for each sample, was performed using the Power SYBR Green RNA-to-CT 1-Step kit (Invitrogen/Life Technologies) on an Applied Biosystems 7500 Real-Time PCR System. The following primers were used for human Renilla mRNA: fwd, 5'-GTGGGCTCGCTGCAAGCAA-3'; rev, 5'-GCTCTTGCCGGACTTACCCATT-3'. To quantify GAPDH mRNA as control, we used the following primers: fwd, 5′-ACAGCAACAGGGTGGTGGAC-3′; rev, 5′-GACCATTGCTGGGGCTGGTG-3′. As amplification efficiencies for the Renilla and GAPDH primer pairs were similar (1.91 and 1.87, respectively), the ΔΔCT method was used to quantify relative Renilla mRNA expression levels. Statistical significance of the results was established using one-way ANOVA analysis.
Human topoisomerase type I cleavage assay
Human recombinant HMGA2 was purified from BL21 (DE3) Rosetta cells. The purification steps included his-tag affinity chromatography, Tev protease digest, Resource S cation exchange chromatography and size exclusion chromatography. In vitro assays were performed with purified HMGA2 and recombinant human topoisomerase I (PROSPEC) in a buffer containing: 50 mM Tris-Cl, pH 7.5; 100 mM KCl; 1 mM DTT; 10 mM EDTA; 5 μg/ml acetylated BSA (Life Technologies) with or without 24 μM SN-38. For each sample, 300 ng of Renilla reporter plasmid was incubated with various amounts of HMGA2, as indicated in the text, for 5 min at room temperature. DNA relaxation was initiated by the addition of 12 ng topoisomerase I per sample, and incubation was stopped after 30 minutes at 37°C with 0.3% (w/v) SDS. Samples were digested with proteinase K for 20 min at 37°C, and plasmid DNA purified via PCR purification kit (Qiagen). DNA nicking was analyzed through 0.8% agarose gel electrophoresis in 0.5× TBE in the presence of ethidium bromide added to the gel. DNA was visualized under UV and corresponding DNA bands were quantified using ImageJ software.
RESULTS
A robust cell-based reporter assay for transcriptional activation by HMGA2
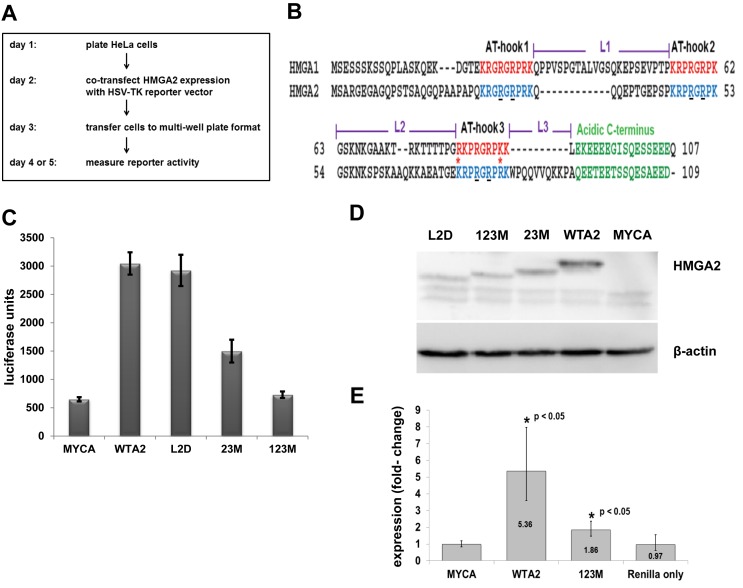
Several reporter systems to study endogenous promoter activation by HMGA2 have previously been reported (22,23). We fortuitously discovered that in HeLa cells, which do not express detectable levels of endogenous HMGA2, C-terminally FLAG-tagged human HMGA2 significantly enhanced Renilla luciferase expression from a reporter gene driven by the herpes simplex virus thymidine kinase (HSV-TK) promoter. We subsequently developed a simple and highly reproducible protocol to measure reporter activity (Figure 1A). In a typical assay, HeLa cells were seeded and co-transfected with HMGA2 expression vector and Renilla reporter vector. Transfected cells were then split into smaller multi-well plates, and reporter assays performed in replicates between 24 and 72 h later.
Figure 1.
Transcriptional activation of HSV-TK promoter depends on functional AT-hooks. (A) Diagram showing the workflow of the reporter system. (B) Sequence alignment of human HMGA1 and HMGA2 proteins, with AT-hooks, linker (L) and C-terminal domains highlighted. Asterisks indicate sequence differences in AT-hook 3 between HMGA1 and HMGA2 (adapted from (9)). The underlined R residues in HMGA2 AT-hooks demarcate glycine substitutions in mutated HMGA2, as described in the text. (C) Representative luciferase reporter assays of co-transfections comparing mock (MYCA), wild-type HMGA2 (WTA2), two AT-hook mutants (23M, 123M) and linker 2 deletion mutant (L2D). We show mean values plus standard deviations of readings from eight replicates per co-transfected plasmids. (D) Western blot analysis of expression of HMGA2 proteins using anti-FLAG antibody and β-actin as loading control. Note that the HMGA2 mutants exhibit a different electrophoretic mobility from the wild-type protein. (E) qRT-PCR analysis of Renilla mRNA after co-transfection with wild type or mutant HMGA2-expression constructs. We used the ΔΔCT method to determine the expression changes from four independent sets of co-transfections, each comparing mock, wild-type HMGA2, mutant HMGA2 and solely transfected Renilla expression vector. Data were normalized to GAPDH mRNA levels. Error bars, mean ± standard deviation. Statistical significance compared to mock transfections as indicated, determined using one-way ANOVA analysis. Nearly identical results were obtained when data were analyzed with Pfaffl's method (not shown).
Using mock expression vector (MYCA) as control, we tested wild-type (WTA2) and three mutant HMGA2 proteins for reporter activation. One mutant, termed 23M, harbored glycine substitutions of two functionally important residues in AT-hooks 2 and 3, leaving the first AT-hook unaltered (Figure 1B). The second mutant, 123M, carried the same substitutions in all three AT-hooks. It is known that these substitutions substantially reduce DNA binding of both HMGA2 and HMGA1 (24,25). The third mutant, L2D, carries a deletion of linker 2, which connects hooks 2 and 3 (Figure 1B).
The results showed that expression of WTA2 compared to MYCA dramatically enhanced luciferase activity (Figure 1C; Supplementary Figure S1A). Western blot analysis using either FLAG or HMGA2 antibodies confirmed expression of recombinant human HMGA2 (Figure 1D and Supplementary Figure S1B, respectively). Furthermore, the results revealed that linker 2 is not critical for full reporter activation by HMGA2 (Figure 1C). Although the level of the overexpressed L2D mutant protein is reduced compared to WTA2 (Figure 1D), it is clear that sufficient recombinant protein is present to achieve reporter activation comparable to WTA2.
The data also revealed that HMGA2-mediated reporter activation critically depends on high affinity DNA-binding of the three AT-hooks. When all three AT-hooks are altered (123M), upregulation of the reporter is either not detectable or significantly reduced to <2-fold (Figure 1C; Supplementary Figure S1A). The presence of the remaining functional AT-hook 1 (23M) also led to a significant drop in reporter expression, but still triggered a 2- to 6-fold increase compared to the mock control (Figure 1C; Supplementary Figure S1A). Western blot analysis confirmed that the expression level of AT-hook mutants were higher than that of the L2D mutant (Figure 1D), which was sufficient for full reporter activation.
The fact that the HMGA2-triggered increase in reporter gene activity almost entirely depended on the presence of all three functional AT-hooks for DNA binding indicated that it might be the result of direct or indirect HSV-TK promoter activation by HMGA2. We tested this hypothesis by qRT-PCR of Renilla mRNA after co-transfection. Compared to controls, expression of wild-type HMGA2 significantly increased the amount of Renilla mRNA in co-transfected cells >5-fold (Figure 1E). In contrast, substitutions in the three AT-hooks increased reporter gene transcription <2-fold. This is in agreement with the modest increase in enzymatic reporter activity observed with 123M (Supplementary Figure S1A) and could be explained by residual low affinity AT-hook DNA-binding of the mutant protein. Taken together, wild-type HMGA2, through DNA-binding, significantly enhanced HSV-TK promoter activity which, in turn, led to dramatic increase in luciferase expression from our reporter system in HeLa cells.
In order to find out whether other cell lines could be used to employ our reporter system, we tested human embryonic kidney (HEK 293T) and human non-small cell lung cancer cells (H1299). The data showed that wild-type HMGA2 was able to enhance expression of the reporter between 3- to 6-fold in both cell lines (Supplementary Figure S2). While this is a significant increase, it falls short of the 10- to 15-fold reporter activation that is achievable with HeLa cells.
Activation of HSV-TK promoter is specific for HMGA2
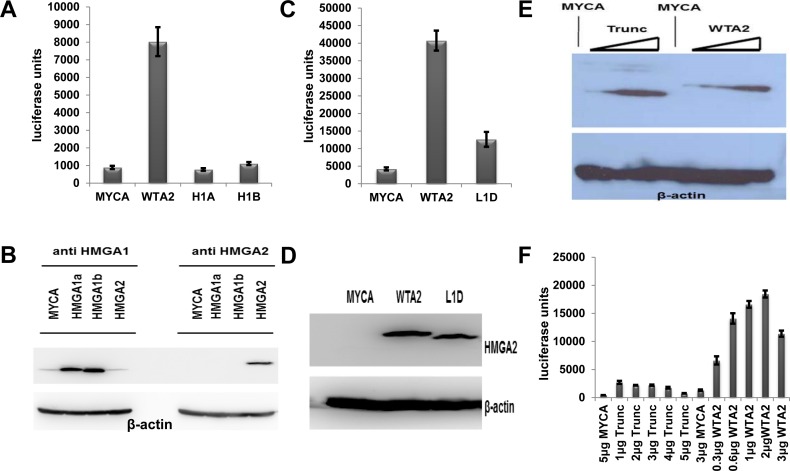
The two closely related human HMGA1 and HMGA2 proteins harbor nearly identical AT-hooks, but they differ in linker domains connecting their AT-hooks (Figure 1B). It was therefore logical to test whether the two main human HMGA1 splice variants, HMGA1a and HMGA1b, activate the HSV-TK promoter.
Following our established co-transfection protocol, the data revealed that neither HMGA1 variant is capable of even modestly activating the HSV-TK promoter (Figure 2A). Western blot analysis confirmed that both HMGA1 variants were highly expressed compared to the low level of endogenous protein detectable in HeLa cells (Figure 2B). Hence, our reporter system is highly specific for HMGA2.
Figure 2.
Characterization of the reporter system. (A) Representative luciferase reporter assays of one of three sets of co-transfections comparing mock, wild-type HMGA2 and HMGA1 splice variants HMGA1a (H1A) and HMGA1b (H1B). We show mean values plus standard deviations of readings from 24 replicates per co-transfected plasmids. (B) Western blot analysis of expression of HMGA1a/1b and HMGA2 using β-actin as loading control. (C) Representative luciferase reporter assays of one of three sets of co-transfections comparing mock, wild-type HMGA2 and a mutant HMGA2 with deleted linker 1 domain (L1D). We show mean values plus standard deviations of readings from 6 replicates per co-transfected plasmids. (D) Western blot analysis of expression of wild-type HMGA2 and L1D mutant HMGA2 using anti-HMGA2 antibodies and β-actin as loading control. (E) Western blot analysis of C-terminal truncated HMGA2 (Trunc) in comparison with wild-type HMGA2 using anti-HMGA2 antibodies and β-actin as loading control. Increasing amounts of expression vector for each protein, as described below in (F), were co-transfected with a fixed amount of reporter vector. (F) Luciferase reporter assays of a set of 12 co-transfections comparing mock, wild-type HMGA2 and truncated HMGA2. We show mean values plus standard deviations of readings from 6 replicates per co-transfected plasmids. The amount (μg) of co-transfected expression vectors is indicated. Note that >4 μg co-transfected DNA induced cytotoxic effects.
HMGA2 linker 1 and the acidic tail are critical for transcriptional activation
In order to test whether HMGA2 protein domains other than the AT-hooks are critical for HSV-TK promoter activation, we deleted the linker 1 domain (L1D) (Figure 1B). The corresponding reporter assays revealed that linker 1 is also critical for full transcriptional activation of the HSV-TK promoter by HMGA2 (Figure 2C). Western blot analysis confirmed similar expression levels for WTA2 and L1D proteins (Figure 2D).
We next asked whether the C-terminal acidic tail of HMGA2, which may play a role in protein-protein interactions and posttranslational modifications (10), is required for transcriptional activation and generated a truncated HMGA2 variant (Trunc) which lacked the entire acidic C-terminus (Figure 1B). We co-transfected various amounts of expression vector for either the truncated or the wild-type protein with a fixed amount of reporter vector. Western blot analysis showed that this resulted in a range of comparable expression levels for both proteins (Figure 2E). The results of corresponding luciferase assays then clearly revealed that the C-terminal tail is absolutely critical for full transcriptional activation. In contrast to the wild-type protein, the observed modest reporter activation triggered by C-terminally truncated HMGA2 did not increase with higher amounts of the protein and reached at best 15% of the level seen with wild-type HMGA2 (Figure 2F). In summary, we conclude that full transcriptional activation of the HSV-TK promoter by HMGA2 depends on (i) the presence of three functional AT-hooks, (ii) a precise arrangement or spacing of AT-hooks 1 and 2 at an unknown DNA target mediated by linker 1 and (iii) the presence of the C-terminal acidic tail.
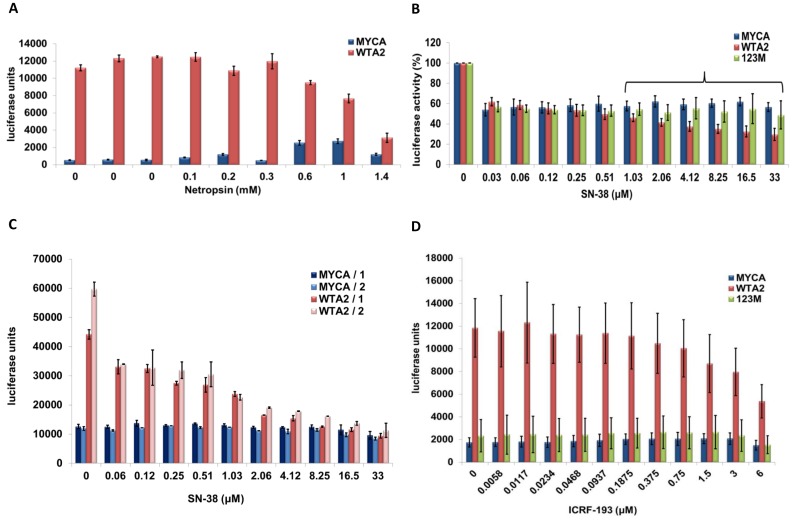
Netropsin inhibits transcriptional activation by HMGA2
We next aimed to validate the utility of our reporter system for high-throughput compound screening and chose the antibiotic netropsin as a test compound. Netropsin, similarly to AT-hooks, binds with high affinity to the minor groove of AT-rich sequences and competes with HMGA2 for DNA-binding (26,27). We followed our established co-transfection protocol and exposed cells to increasing amounts of netropsin for 24 h before luciferase assays were conducted. The results revealed that netropsin was highly effective in antagonizing reporter activation by HMGA2. In contrast, MYCA co-transfected control cells even showed a substantial and reproducible increase in reporter activity at higher netropsin doses (Figure 3A). At 1.4 mM netropsin, HMGA2-triggered activation of the reporter was reduced to <3-fold, compared to the >13-fold activation in the absence of drug. Taken together, netropsin acts as a specific and effective inhibitor for HMGA2 in our reporter assay.
Figure 3.
Netropsin, SN-38 and ICRF-193 are HMGA2 antagonists. (A) Representative luciferase reporter assays of one of three sets of co-transfections with mock or wild-type HMGA2. Cells were exposed to increasing concentrations of netropsin, as indicated, for 24 h before luciferase assays were performed. We show mean values plus standard deviations of readings from four replicates for each netropsin concentration and for three sets of four replicates without netropsin as controls. (B) Luciferase reporter assays were performed with HeLa cells for six independent sets of co-transfections with mock, wild-type HMGA2 or mutant HMGA2 that carried substitutions in all three AT-hooks (123M), as described in the text. In each case, cells were exposed to increasing concentrations of SN-38 for 24 h, and data from two replicates per co-transfected pair of vectors were collected. Mean values from each set of co-transfections were normalized to SN-38-untreated controls. We show the mean values of the combined six normalized data sets with standard deviations. The bracket indicates when statistically significant (P < 0.05) differences between mock and wild-type HMGA2 as well as between mutant HMGA2 and wild-type HMGA2 readings could be determined; Student's t-test. (C) Luciferase reporter assays were performed with H1299 cells for two independent co-transfections with either mock (MYCA) or wild-type HMGA2 (WTA2) in combination with Renilla expression vector. Cells from each co-transfection were plated 24 h later in two rows of 11 wells on a 96-well plate. The next day, SN-38 was titrated in each row, and luciferase activity was measured after additional 24 hours. The bars show mean values with standard deviations from these two luciferase measurements for each co-transfection. (D) Luciferase reporter assays were performed with HeLa cells with mock (MYCA), wild-type HMGA2 (WTA2), or HMGA2 mutant (123M) in combination with Renilla expression vector. Two independent sets of co-transfections were performed. In each set, cells from two six wells (e.g. mock plus Renilla vectors) were combined 24 h after co-transfection and plated in three rows of 12 wells on a 96-plate. Each triplicate of rows was repeated three times, and ICRF-193 was titrated in 24 h later. The bars show mean values with standard deviations from the combined 18 luciferase measurements obtained for each co-transfection after one day of drug exposure.
The IC50 for netropsin determined from three independent co-transfections was at 1.1 mM (Supplementary Figure S3). This is a rather high value and most likely due, in part, to the fact that we co-transfected reporter and HMGA2 expression vectors two days before netropsin challenge. Hence, the drug has to work against an established steady-state level of HMGA2-mediated reporter activation. Furthermore, we transiently overexpress HMGA2 in HeLa cells, which means that high amounts of netropsin are likely required to compete with AT-hook binding to DNA target sites.
High-throughput screening reveals functional interaction between HMGA2 and topoisomerase I
We next adapted our reporter system for high-throughput small-compound screens in a robotic setting with 384-well culture plates. The assays remained highly reproducible with <10% variation between replicates, and full activation of the HSV-TK promoter by HMGA2 tolerated up to 1% of the solvent dimethyl sulfoxide in the culture medium (data not shown).
We screened the PHARMAKON 1600 library which is a collection of 1600 small-compound drugs. Many of them are in medical use and many of their targets have been identified. Based on the collective data obtained from these screens performed at 10 μM drug concentration, employing mock/reporter vector co-transfection as control, we identified the following four candidate HMGA2 antagonists: chlorambucil, podofilox, mannitol and irinotecan hydrochloride. However, except for irinotecan, a specific HMGA2 antagonistic effect could not be validated in follow-up analysis using up-scaled laboratory settings (Supplementary Figure S4; data not shown).
Irinotecan is a human topoisomerase type I inhibitor and prevents re-ligation of DNA strands, which traps the enzyme in a covalent tyrosine–DNA complex (28). After cellular uptake, irinotecan is quickly hydrolyzed into the active compound SN-38 (29). We therefore tested SN-38 and included, in addition to mock co-transfection, mutant HMGA2 which carries glycine substitutions in all three AT-hooks (123M) as control.
A global initial 50% inhibition of reporter activities was observed with SN-38 in every experiment at the lowest SN-38 concentration tested, irrespective of the type of co-transfection (Figure 3B; Supplementary Figure S5). Since this effect was not seen with irinotecan (Supplementary Figure S4D) and appeared to be independent of SN-38 concentration, we ascribe it to impurities in the SN-38 compound sample which affected HeLa cells. Nevertheless, the results clearly showed that in the low μM range, SN-38 significantly and specifically interfered, in a dose-dependent manner, with HMGA2-activated reporter expression. In contrast, reporter control expression remained unaffected up to 33 μM SN-38 (Figure 3B; Supplementary Figure S5). Hence, irinotecan/SN-38 appears to be an antagonist that interferes with HMGA2-mediated transcriptional activation of the HSV-TK promoter.
We next asked whether SN-38-induced inhibition of HMGA2 can be detected in other cell types and employed our reporter system for drug testing in above-mentioned non-small cell lung cancer cell line H1299. The results clearly showed that SN-38 specifically interfered with HMGA2-activated reporter expression. In fact, inhibition appeared to be even more pronounced in H1299 than in HeLa cells, leading to a complete attenuation of reporter activation by HMGA2 at low μM drug concentration (Figure 3C). Hence, SN-38 can function as potent HMGA2 antagonist in different genetic backgrounds.
HMGA2 stabilizes SN-38-induced covalent topoisomerase-DNA complexes in vitro
In order to elucidate how SN-38 functions as HMGA2 antagonist, we first tested whether the compound directly interferes with DNA-binding and performed DNA electrophoretic mobility shift assays using purified HMGA2 and double-stranded, AT-rich DNA fragments. These in vitro assays did not reveal any detectable impact on DNA-binding, even in the high μM SN-38 concentration range (data not shown).
Irinotecan/SN-38 belongs to the camptothecin class of topoisomerase I inhibitors, which block the re-ligation reaction of topoisomerase I (28,29). In a cellular environment, this generates DNA single- and double-strand breaks due to collisions of RNA polymerases and replisomes with the trapped enzyme–DNA complex. Global replication and transcription processes are affected as a consequence, which eventually triggers apoptosis.
More recently discovered modes of irinotecan action also involve accumulation of high levels of torsional strain in transcribed DNA due to impaired DNA strand swiveling within the trapped topoisomerase-DNA complex (30). Torsional strain in the DNA template stalls translocating RNA polymerases even in the absence of collisions between the complex and RNA polymerases due to the build-up of positive supercoiling. Furthermore, topoisomerase I is a positive regulator of RNA polymerase II at promoters, and inhibition by camptothecin interferes with transcription initiation (31,32). We thus hypothesized that the observed attenuation of reporter gene expression in cells expressing HMGA2 may be due to more efficient SN-38-mediated trapping of topoisomerase I in covalent complexes with DNA.
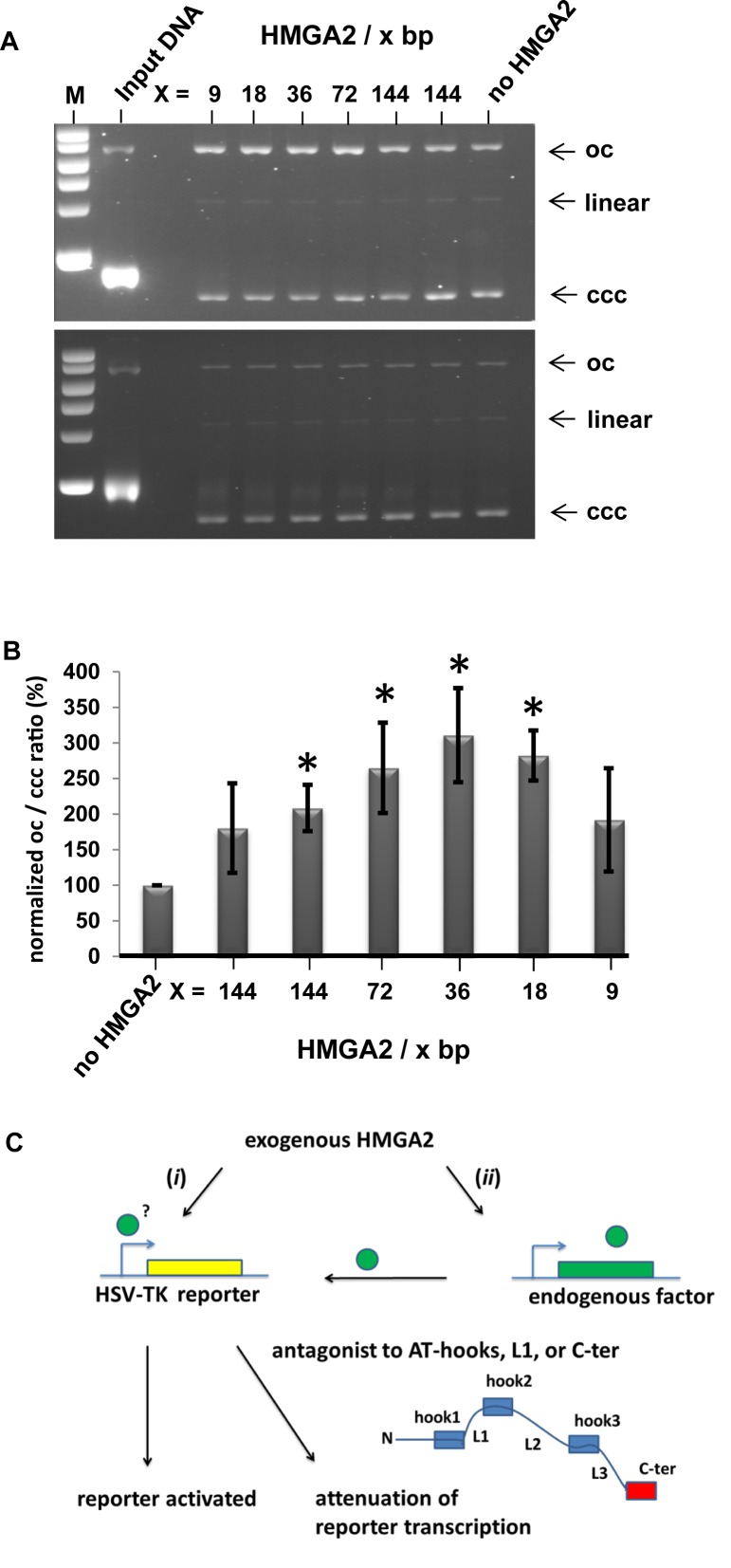
We tested this hypothesis in vitro and incubated negatively supercoiled plasmid DNA with increasing amounts of purified HMGA2 in the presence of SN-38. After addition of human topoisomerase I to initiate supercoil relaxation, reactions were stopped by SDS in order to instantaneously denature the trapped covalent topoisomerase-DNA complexes present at this time point. Purified plasmid DNA was then analyzed through agarose gel electrophoresis in the presence of ethidium bromide. The assay thus measures the occurrence of nicked plasmid DNA (open circle; oc) as a result of topoisomerase trapping in comparison with covalently closed circular DNA (ccc), which result from completed topological relaxation cycles catalyzed by the enzyme without drug interference.
The results clearly revealed that HMGA2 significantly enhanced >3-fold SN-38 efficacy in trapping human topoisomerase I in a cleaved complex with DNA (Figure 4A; top panel; Figure 4B). Without HMGA2, ∼50% of the supercoiled input DNA was converted by the drug into trapped topoisomerase I-DNA complexes, which migrated as oc DNA. The fraction of oc DNA significantly increased when HMGA2 was present. This effect depended on the stoichiometry of HMGA2 to DNA. Importantly, controls showed that HMGA2-triggered complex formation occurred only in the presence of SN-38. Without the drug, HMGA2 does not induce changes in the amount of topoisomerase I-DNA complexes, as indicated by the constant amount of oc DNA fractions in all samples (Figure 4A; bottom panel).
Figure 4.
A combinatorial effect of HMGA2 and SN-38 increased complex formation between topoisomerase I and DNA in vitro. (A) Representative analysis of covalently closed (ccc) and open circular (oc) plasmid DNA after in vitro DNA relaxation reactions with recombinant human topoisomerase I in the presence (top panel) or absence (bottom panel) of SN-38. The two topological DNA forms were resolved via agarose gel electrophoresis in the presence of ethidium bromide. The stoichiometry of HMGA2 per DNA bp in each reaction is indicated. Samples with the lowest amount of HMGA2 were run as duplicates as a result of serial dilution. DNA next to the marker lane (M) is input plasmid DNA, which is more than 95% (-) supercoiled. See text for details. (B) Quantitation of normalized ratios of open circular (oc) to covalently closed (ccc) DNA from four independent experiments. Intensities of corresponding oc and ccc DNA bands were determined by ImageJ, and oc/ccc ratios of reactions without HMGA2 were set as 100% for each experiment. We show mean normalized values plus standard deviations. P values were obtained by comparing the presence of HMGA2 with the control reactions lacking HMGA2. The asterisk indicates statistically significant (P < 0.05) differences. Student's t-test. (C) Summary of the characterization and features of the novel HMGA2 reporter system. See text for details.
Through our novel cell-based reporter system and initial high-throughput compound screening, we provided evidence for a hitherto unknown functional link between HMGA2 and human topoisomerase I, which becomes detectable when topoisomerase I is inhibited by SN-38. Since HMGA2, topoisomerase I and SN-38 bind DNA (9,29), it is very likely that a ternary complex forms on supercoiled DNA in which the trapping of topoisomerase I by the drug is somehow augmented.
We next sought to address the question whether ternary complex formation could involve direct HMGA2-topoisomerase I protein interaction and tested the effect of topoisomerase type II inhibitor ICRF-193 on our reporter system in HeLa cells. Bis-dioxopiperazine ICRF-193, similarly to irinotecan and topoisomerase I, functions as a topoisomerase poison and traps the type II enzyme in a covalent complex with DNA (33,34). We reasoned that if ICRF-193 specifically interferes with reporter activation by HMGA2, it would not only provide further evidence for a functional link between HMGA2 and topoisomerase activity in general, but would also suggest that such a link does not require direct protein-protein interactions and might be mediated by DNA topology. We tested ICRF-193 with wild-type HMGA2, mutant HMGA2 and mock co-transfections as described for SN-38. The result clearly showed that in the low μM range, ICRF-193 significantly and specifically interfered, in a dose-dependent manner, with HMGA2-activated reporter expression (Figure 3D).
DISCUSSION
Our study presented a novel cell-based reporter system that measures transcriptional activation of the HSV-TK promoter by human HMGA2 in co-transfected cells with different genetic background. The system is specific for HMGA2, since HMGA1 completely failed to activate the viral promoter. We consider two, not mutually exclusive modes of HSV-TK promoter activation: (i) direct via binding of HMGA2 to specific sequence elements within the HSV-TK promoter or (ii) indirect by regulating endogenous cellular factor(s) critical for HSV-TK promoter activation (Figure 4C).
We probed into the first possibility and deleted a 10 bp continuous AT-stretch that is present immediately upstream of the first Sp1 transcription factor binding site within the well-studied HSV-TK promoter (35). This sequence stood out as an ideal recognition site for HMGA2. However, its deletion even further enhanced HMGA2-mediated activation of reporter expression (data not shown). To probe into the second possibility, i.e. HMGA2 regulating cellular transcription factor(s) critical for HSV-TK promoter activation, we tested whether ectopic expression of HMGA2 altered endogenous Sp1 protein levels in HeLa cells. Sp1 is an obvious candidate known to play a crucial role in regulating the HSV-TK as well as many cellular promoters (35,36). However, we were unable to detect any changes in Sp1 levels induced by HMGA2 in HeLa cells (data not shown).
Our reporter system can easily be adjusted to high-throughput robotic settings. We demonstrated its utility for compound screening first with netropsin, which is known to compete with AT-hooks for minor groove DNA-binding in vitro (26,37). Netropsin specifically inhibited HMGA2's transcriptional activator function for the HSV-TK promoter in a dose-dependent manner, thus independently confirming the functional importance of AT-hook DNA-binding.
Our cell-based reporter system offers a number of controls in the form of mutant HMGA2 variants and should therefore become a widely applicable, powerful tool for high-throughput compound library screens and validation. Full transcriptional activation of the reporter promoter depends on (i) presence of functional AT-hooks, most importantly hooks 1 and 2, (ii) presence of linker 1, which could either contribute to DNA binding or regulate the three dimensional organization between AT-hooks 1 and 2 on DNA and (iii) presence of the C-terminal tail of the protein. The latter may mediate interactions with cellular proteins important for transcriptional activation. The system therefore could identify HMGA2 antagonists which target these functionally important protein domains (Figure 4C). These antagonists could ultimately interfere with HMGA2's important chromatin and transcriptional control functions in cancer cells and thus become therapeutically beneficial. Perhaps equally important, the system could also identify unknown functional links between HMGA2 and cellular factors.
Our proof-of-concept screen identified such a link. Irinotecan/SN-38 targets DNA topoisomerase I and is currently in clinical use for combination treatment of a number of highly aggressive cancers, in particular metastatic colorectal cancer (38). We discovered that this drug is a potential HMGA2 antagonist and identified a possible mechanism which could explain how it works in combination with HMGA2 and topoisomerase I to interfere with transcription processes. The observed increase in the amount of trapped topoisomerase-DNA complexes triggered by SN-38 in the presence of HMGA2 is also likely to affect DNA transactions other than transcription in human cells. We are currently investigating the mechanistic details and cellular consequences of this combinatorial effect, and especially its impact on malignant cells.
Finally, the discovery that HMGA2, in contrast to the related HMGA1 protein, strongly activated the HSV-TK promoter raises an interesting question: Does HMGA2 play a critical role during productive HSV infection? Although there is, to our knowledge, no direct evidence reported in the literature for such a connection, HMGA2 is known to regulate neural stem cell self-renewal as well as postnatal peripheral neurogenic potential (3,39), Interestingly, HMGA1 appears to co-regulate a latency-active HSV promoter (40). Hence, if HMGA2 is involved in the peripheral nervous system in controlling HSV-TK gene expression during productive viral infection, antagonists identified through our reporter system could also become useful in the context of HSV infection.
Supplementary Material
Acknowledgments
Special thanks go to the Experimental Therapeutics Center (ETC) in Singapore, especially to M.L. Choong and Y.S. Lin, for advice and assistance in performing the high-throughput screen and supplying the PHARMAKON 1600 library. We thank H. Summer for construction of the bacterial expression vector for C-terminal truncated HMGA2 and Z. Ma and C.A. Davey for providing purified HMGA2 and DNA substrates for EMSAs.
Author contribution: P.D. and S.P. designed the study. S.P. performed cell-based, qRT-PCR, Western blot and in vitro assays, constructed expression vectors, and analyzed data. H.Y constructed expression vectors and performed Western blotting. R.I.-N. contributed to qRT-PCR analysis and analyzed data. P.D. wrote the paper.
Footnotes
Present address: Haojie Yu, Harvard University, Department of Stem Cell and Regenerative Biology, 7 Divinity Avenue, Cambridge, MA 02138, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education Academic Research Fund Tier 3 [MOE2012-T3-1-001]. Funding for open access charge: Singapore Ministry of Education Tier 3 grant.
Conflict of interest statement. Sabrina Peter and Peter Droge declare competing financial interests.
REFERENCES
- 1.Rogalla P., Drechsler K., Frey G., Hennig Y., Helmke B., Bonk U., Bullerdiek J. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am. J. Pathol. 1996;149:775–779. [PMC free article] [PubMed] [Google Scholar]
- 2.Gattas G.J., Quade B.J., Nowak R.A., Morton C.C. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;4:316–322. [PubMed] [Google Scholar]
- 3.Nishino J., Kim I., Chada K., Morrison S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleynen I., Van de Ven W.J. The HMGA proteins: a myriad of functions. Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 5.Fedele M., Fusco A. HMGA and cancer. Biochim. Biophys. Acta. 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Droge P., Davey C.A. Do cells let-7 determine stemness? Cell Stem Cell. 2008;2:8–9. doi: 10.1016/j.stem.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Morishita A., Zaidi M.R., Mitoro A., Sankarasharma D., Szabolcs M., Okada Y., D'Armiento J., Chada K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Z., Li X., Wu D., Tang R., Chen R., Xue S., Sun X. Silencing of HMGA2 suppresses cellular proliferation, migration, invasion, and epithelial-mesenchymal transition in bladder cancer. Tumor Biol. 2015;37:7515–7523. doi: 10.1007/s13277-015-4625-2. [DOI] [PubMed] [Google Scholar]
- 9.Pfannkuche K., Summer H., Li O., Hescheler J., Droge P. The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev. 2009;5:224–230. doi: 10.1007/s12015-009-9078-9. [DOI] [PubMed] [Google Scholar]
- 10.Maurizio E., Cravello L., Brady L., Spolaore B., Arnoldo L., Giancotti V., Manfioletti G., Sgarra R. Conformational role for the C-terminal tail of the intrinsically disordered high mobility group A (HMGA) chromatin factors. J. Proteome Res. 2011;10:3283–3291. doi: 10.1021/pr200116w. [DOI] [PubMed] [Google Scholar]
- 11.Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair. 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Déjardin J., Kingston R.E. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra T., Narita M. High-order chromatin structure and the epigenome in SAHFs. Nucleus. 2013;4:23–28. doi: 10.4161/nucl.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah S.N., Kerr C., Cope L., Zambidis E., Liu C., Hillion J., Belton A., Huso D.L., Resar L.M. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One. 2012;7:e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J., Zhang Y., Shi W., Ren C., Liu Y., Pan Y. The critical role of HMGA2 in regulation of EMT in epithelial ovarian carcinomas. Tumour Biol. 2016;37:823–828. doi: 10.1007/s13277-015-3852-x. [DOI] [PubMed] [Google Scholar]
- 16.Winter N., Nimzyk R., Bösche C., Meyer A., Bullerdiek J. Chromatin immunoprecipitation to analyze DNA binding sites of HMGA2. PLoS One. 2011;6:e18837. doi: 10.1371/journal.pone.0018837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summer H., Li O., Bao Q., Zhan L., Peter S., Sathiyanathan P., Henderson D., Klonisch T., Goodman S.D., Droge P. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res. 2009;37:4371–4384. doi: 10.1093/nar/gkp375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H., Lim H.H., Tjokro N.O., Sathiyanathan P., Natarajan S., Chew T.W., Klonisch T., Goodman S.D., Surana U., Dröge P. Chaperoning HMGA2 protein protects stalled replication forks in stem and cancer cells. Cell Rep. 2014;6:684–697. doi: 10.1016/j.celrep.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Rajamani D., Bhasin M.K. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016;8:38–48. doi: 10.1186/s13073-016-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur H., Ali S.Z., Huey L., Hütt-Cabezas M., Taylor I., Mao X.G., Weingart M., Chu Q., Rodriguez F.J., Eberhart C.G., et al. The transcriptional modulator HMGA2 promotes stemness and tumorigenicity in glioblastoma. Cancer Lett. 2016;3835:30255–30263. doi: 10.1016/j.canlet.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busch B., Bley N., Müller S., Glaß M., Misiak D., Lederer M., Vetter M., Strauß H.G., Thomssen C., Hüttelmaier S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016;44:3845–3864. doi: 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattaruzzi G., Altamura S., Tessari M.A., Rustighi A., Giancotti V., Pucillo C., Manfioletti G. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007;35:1751–1760. doi: 10.1093/nar/gkl1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong X., Liu X., Li Y., Cheng M., Wang W., Tian K., Mu L., Zeng T., Liu Y., Jiang X., et al. HMGA2 sustains self-renewal and invasiveness of glioma-initiating cells. Oncotarget. 2016 doi: 10.18632/oncotarget.9744. doi:10.18632/oncotarget.9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel B., Löschberger A., Sauer M., Hock R. Cross-linking of DNA through HMGA1 suggests a DNA scaffold. Nucleic Acids Res. 2011;39:7124–7133. doi: 10.1093/nar/gkr396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill D.A., Pedulla M.L., Reeves R. Directional binding of HMG-I(Y) on four-way junction DNA and the molecular basis for competitive binding with HMG-1 and histone H1. Nucleic Acids Res. 1999;27:2135–2144. doi: 10.1093/nar/27.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao Y., Cui T., Leng F., Wilson W.D. Inhibition of high-mobility-group A2 protein binding to DNA by netropsin: a biosensor-surface plasmon resonance assay. Anal. Biochem. 2008;374:7–15. doi: 10.1016/j.ab.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z., Eguchi-Ishimae M., Yagi C., Iwabuki H., Gao W., Tauchi H., Inukai T., Sugita K., Ishii E., Eguchi M. HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br. J. Haematol. 2015;171:818–829. doi: 10.1111/bjh.13763. [DOI] [PubMed] [Google Scholar]
- 28.Liu L.F., Desai S.D., Li T.K., Mao Y., Sun M., Sim S.P. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 29.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;10:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 30.Koster D.A., Palle K., Bot E.S., Bjornsti M.A., Dekker N.H. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 31.Rialdi A., Campisi L., Zhao N., Lagda A.C., Pietzsch C., Ho J.S., Martinez-Gil L., Fenouil R., Chen X., Edwards M., et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352 doi: 10.1126/science.aad7993. doi:10.1126/science.aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H., et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen L.H., Nitiss K.C., Rose A., Dong J.W., Zhou J.F., Hu T., Osheroff N., Jensen P.B., Sehested M., Nitiss J.L. A novel mechanism of cell killing by anti-topoisomerase II bis-dioxopiperazines. J. Biol. Chem. 2000;275:2137–2146. doi: 10.1074/jbc.275.3.2137. [DOI] [PubMed] [Google Scholar]
- 34.Huang K.C., Gao H., Yamasaki E.F., Grabowski D.R., Liu S., Shen L.L., Chan K.K., Ganapathi R., Snapka R.M. Topoisomerase II poisoning by ICRF-193. J. Biol. Chem. 2001;276:44488–44494. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 35.Jones K.A., Yamamoto K.R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 36.Safe S., Imanirad P., Sreevalsan S., Nair V., Jutooru I. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin. Ther. Targets. 2014;18:759–769. doi: 10.1517/14728222.2014.914173. [DOI] [PubMed] [Google Scholar]
- 37.Alonso N., Guillen R., Chambers J.W., Leng F.A. Rapid and sensitive high-throughput screening method to identify compounds targeting protein-nucleic acids interactions. Nucleic Acids Res. 2015;43:e52. doi: 10.1093/nar/gkv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirstein M.M., Lange A., Prenzler A., Manns M.P., Kubicka S., Vogel A. Targeted therapies in metastatic colorectal cancer: a systematic review and assessment of currently available data. Oncologist. 2014;19:1156–1168. doi: 10.1634/theoncologist.2014-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kishi Y., Fujii Y., Hirabayashi Y., Gotoh Y. HMGA2 regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat. Neurosci. 2012;15:1127–1133. doi: 10.1038/nn.3165. [DOI] [PubMed] [Google Scholar]
- 40.French S.W., Schmidt M.C., Glorioso J.C. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol. Cell Biol. 1996;16:5393–5399. doi: 10.1128/mcb.16.10.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.