Abstract
To understand the epigenetic regulation of transcriptional response of macrophages during early-stage M. tuberculosis (Mtb) infection, we performed ChIPseq analysis of H3K4 monomethylation (H3K4me1), a marker of poised or active enhancers. De novo H3K4me1 peaks in infected cells were associated with genes implicated in host defenses and apoptosis. Our analysis revealed that 40% of de novo regions contained human/primate-specific Alu transposable elements, enriched in the AluJ and S subtypes. These contained several transcription factor binding sites, including those for members of the MEF2 and ATF families, and LXR and RAR nuclear receptors, all of which have been implicated in macrophage differentiation, survival, and responses to stress and infection. Combining bioinformatics, molecular genetics, and biochemical approaches, we linked genes adjacent to H3K4me1-associated Alu repeats to macrophage metabolic responses against Mtb infection. In particular, we show that LXRα signaling, which reduced Mtb viability 18-fold by altering cholesterol metabolism and enhancing macrophage apoptosis, can be initiated at response elements present in Alu repeats. These studies decipher the mechanism of early macrophage transcriptional responses to Mtb, highlighting the role of Alu element transposition in shaping human transcription programs during innate immunity.
INTRODUCTION
Transcription factors (TFs), and the gene transcription programs they control, integrate the molecular events and signaling pathways controlling the state of a cell under a variety of physiological and pathophysiological conditions. In this regard, expression profiling studies revealed that infection of macrophages with the intracellular pathogen Mycobacterium tuberculosis (Mtb) induced widespread changes in host gene transcription within 24 h (1). These early host responses are particularly important as epidemiologic evidence supported by genetic studies suggests that Mtb can be eliminated before establishment of long-term infection (2). Here, for the first time in Mtb-infected cells, we have integrated the results of transcriptomic analyses (1) with ChIPseq profiles of histone H3 lysine 4 monomethylation (H3K4me1), a marker of poised or active enhancers (3,4). Remarkably, binding sites for several TFs highly expressed during Mtb infection were enriched in de novo H3K4me1 regions only if the data were not filtered to remove repetitive elements. These transcription factor binding sites (TFBS) include motifs recognized by members of the activating transcription factor (ATF) and myocyte enhancer factor 2 (MEF2) families, and nuclear receptors liver X receptors (LXRs) and retinoic acid receptors (RARs), all of which have been implicated in macrophage survival and cellular responses to stress or infection (5–8). These motifs were imbedded specifically in the AluJ and AluS subtypes of the Alu repeat family of transposable elements (TE) (9).
Alu repeats are ancestral short interspersed elements (SINEs), whose original insertion in genomic sequences appears to have occurred shortly after the dawn of the primate lineage. They are ∼300bp long and are derived from the 7SL RNA gene, which encodes a component of the protein signal recognition complex. Alu repeats account for 11% of human/primate genomes (4), and AluJ and AluS sequences are the most frequent subtypes of the >1 million Alu repeats distributed throughout the genome. The J and S subtypes were amplified early in primate evolution and thus represent classes of old Alu repeats (10). Compared to younger elements, they are rich in CpG dinucleotides, which mutate rapidly and contribute a substantial portion of the single nucleotide polymorphisms in the human genome (4). There is ample evidence that Alu transposition, recombination, and expansion have contributed to genome evolution and changes to gene regulation (11,12). Alu repeats contain motifs recognized by several transcription factors (TFs) including SP1, p53, c-MYC, ANRIL, NF-κB (13), and previous work has suggested that they can function as enhancers (14).
Here, because of its importance in control of Mtb in vivo (15), we used LXRα, a ligand-regulated nuclear receptor whose expression is robustly induced during infection, as a model TF to validate the enhancer function of Alu repeats. Several complementary approaches revealed that LXR binding sites in Alu repeats are engaged and function as bona fide enhancers during infection. Moreover, they are associated with genes implicated in early innate immune and metabolic responses to infection, including those controlling metabolism of cholesterol, a critical carbon source for Mtb replication. These findings are supported by analysis of independent H3K4me1 datasets derived from ChIPseq studies of Mtb-infected primary human dendritic cells (16) and macrophage differentiation and stress responses (17). These studies define macrophage transcriptional responses to Mtb infection, and underline the importance of Alu element transposition as a platform for shaping human/primate transcription programs in innate immunity.
MATERIALS AND METHODS
Cell and bacteria culture
THP-1 cells (ATCC® TIB-202™) were cultured in RMPI-1640 with l-glutamine and 25mM HEPES (Wisent®) with 10% FBS. H37Rv (ATCC® 25618™) and H37Ra (ATCC® 25177™) were cultured in Middlebrook 7H9 (Difco®) with 0.05% Tween-80, 0.1% glycerol and 10% ADC enrichment (BD Biosciences).
Macrophage infections
1 × 106 THP-1 cells were differentiated by 20 nM PMA for 24 h in RPMI with 10% charcoal-stripped FBS. H37Rv or H37Ra (OD between 0.2 and 0.8) were resuspended in RPMI with 10% charcoal-stripped FBS by 10 repeated passages through a 27 G needle. THP-1 cells were infected with Mtb at the multiplicity of infection (MOI) of 5 for 4 h. Cells were washed three times with RPMI, followed by incubation in RPMI with 10% charcoal-stripped FBS containing either vehicle DMSO or TO901319 as indicated.
ChIP Assays, ChIPseq and bioinformatics analysis
Biological duplicates of PMA-differentiated infected and uninfected THP-1 cells (20 × 106 cells) were collected after 1 and 24 h of infection. Cells were fixed by adding formaldehyde directly to the medium to a final concentration of 1% followed by incubation for 20 min at room temperature on a rocking platform. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating at room temperature for 5 min on a rocking platform. The cells (20 × 106 cells per condition) were collected by centrifugation (1200 rpm) and washed twice with ice cold PBS. Cell lysis was performed by adding 1 ml of cell-lysis buffer (5 mM PIPES-pH 8.5, 85 mM KCl, 1% (v/v) IGEPAL CA-630, 50 mM NaF, 1 mM PMSF, 1 mM phenylarsine oxide, 5 mM sodium orthovanadate and additional inhibitors) incubating for 30 min on ice and removing cytoplasmic components by centrifugation. Nuclear pellets were dissolved in 500 μl nuclei-lysis buffer (50 mM Tris–HCl pH8.0, 10mM EDTA, 1% (w/v) SDS, 50 mM NaF, 1 mM PMSF, 1 mM phenylarsine oxide, 5 mM sodium orthovanadate, and additional inhibitors) and incubated 15 min on ice. Lysates were sonicated on a Bioruptor to a repeated cycle of 10 s ON and 20 s OFF for 15 cycles, five times to result in DNA fragments of 200–600 bp. Cellular debris were removed by centrifugation. 50 μl of the lysate were diluted 1:10 in ChIP dilution buffer H from Diagenode kit and 6 μg of specific antibody and non-specific anti-IgG rabbit were added and the samples were incubated overnight at 4°C on a rotating platform. The immunocomplexes were collected using 40 μl of protein A-beads for 2 h at 4°C with rotation. The beads were washed then for 4 min in rotating platform with 1 ml of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1), high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris–HCl, pH 8.1) and LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1). Finally, the beads were washed twice with 1 ml TE buffer (1 mM EDTA, 10 mM Tris–HCl, pH 8.0) and the immune complexes were eluted twice using 200 μl elution buffer (1% SDS, 100 mM NaHCO3). The supernatants were combined and the immune complexes were reverse cross-linked for at least 6 h at 65°C in the presence of proteinase K (20 mM). DNA was purified using MinElute PCR Purification Kit from Qiagen. The libraries were then prepared using the Trueseq library kit from Illumina according to the manufacture's recommendations. DNA libraries for each IP and mock IP (a.k.a. input) were sequenced on an Illumina HiSeq sequencer. All experiments were performed in biological duplicates (see each corresponding BED file at http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3192/) obtaining an average of 60 M reads per sample using 100 based long reads and ∼300 bases insert size. Reads were mapped to the human genome (hg19) with BWA 0.5.9-r16 (18) using default settings and converted to BAM files with SAMtools (19). For each experiment, Chip-Seq peaks were obtained by comparing the read from the IP to those from mock IP and calling peaks using MACS 1.4.2 (20), with a fold of at least 2 and 5% FDR. BED files were annotated and filtered using SnpEff. In a few cases where input samples were not available, a pooled input strategy was used. For histone marks H3K4me1, peaks obtained in Mtb-infected cells 1h and 24 h but which did not overlap those found in uninfected cells were assigned to the set of VnotN. Peak set operations were performed using BedTools (21). De novo motif discovery and enrichment was performed using Homer (software v4.7.2 and annotation v5.4) with custom nuclear receptor motif matrices based on RGGTCA or RGKTCA half-sites arranged as direct, inverted, and everted repeats (22). The parameter mset was used to limit the analysis to vertebrate motifs. The analysis was performed with and without TEs/repeat sequence masking, since these elements can be a source of transcription factor binding sites (TFBS) (22–24). Homer motif analysis used only 200 bp around the peaks to limit the number of false positive that could come from compositional bias or low-complexity regions in the >2 kb regions identified by MACS. Background sequences were generated from the histone marks sequences with uShuffle using parameters n = 5 and k = 3 (25). GREAT (26) was used to identify genes near the histone marks regions and GO analysis. Gene set enrichment analysis was performed using a greedy algorithm based on Fisher exact test and MSigDb database (27). Venn diagrams were generated with the Vennerable R package. Homer and in-house programs were used to screen the human genome (hg19) for putative DR2, DR4, MEF2 and ATF sites (28). The ChIPseq data are available on ArrayExpress accession: E-MTAB-3192 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-3192/). The distribution of regions with ATF1/DR2/DR4/MEF2C sites within 100 kb of TSS of T09 target genes were calculated from the distance in GREAT region-gene association tables with a custom Perl program. The intersectBed bedtools program was used to identify sites within Alu from the hg19 RepeatMasker annotation track of the UCSC Genome Browser. The position of the sites within Alu were calculated from the genomic coordinates of sites and Alu in the intersectBed results. Primers used for region amplification for qPCR validation are listed in Supplemental Table S11. H3K4me1 regions from Mtb-infected and non-infected dendritic cells were identified with MACS2 v2.1.0.20150731 using the bdgpeakcall command with default parameters from the supplementary files available in the NCBI GEO database (accession GSE64175) (Supplementary Tables S4 and S5). The published epigenomic clusters of monocytes and macrophages for the analysis done for Supplementary Tables S6 and S7 were taken from the Supplementary Table S4 from (17). For statistical analysis of the association of H3K4me1 regions with TEs, the Genomic Association Tester (GAT) tool was used to estimate with 10 000 simulations the probability to obtain a similar overlap by chance between H3K4me1 regions and TEs (Supplementary Table S2) (29).
RNA extraction, reverse transcription and qPCR/microarray
RNA extraction was performed with Trizol (Invitrogen®) as per manufacturer's instructions. The samples were then treated with DNase (RQ1 from Promega) for 30 min at 37°C. RT was performed with iScript cDNA Synthesis Kit (Bio-Rad®) from 500 ng pure RNA and qPCR was performed with Fast Universal mix (Roche®), on a 1/50 dilution, normalized to 18S, EEIF1A1, ACTB (beta-actin) and/or GAPDH RNA levels as required by the MIQE guidelines. Primers used are listed in Supplementary Table S11. qPCR were performed on a LC96 machine from Roche and analyzed on the associated software. Microarrays were performed using Human Gene 2.0 ST arrays (Affymetrix®) at the Innovation Center of Genomics at McGill University. Flexarray v1.6.3 software (URL http://genomequebec.mcgill.ca/FlexArray) was used to normalize chip signal using the Affymetrix Power Tools Robust Multi-Array Average algorithm. The EB (Wright and Simon) algorithm (30) was used for statistical analysis to calculate fold transcript. The networks and pathway analysis were generated through the use of QIAGEN's Ingenuity Pathway Analysis (IPA® QIAGEN Redwood City, www.qiagen.com/ingenuity). The microarrays results were validated by performing RT-qPCR following the MIQE guideline requirements.
eRNA detection and 3C assays
For detection of eRNAs, experiments were performed as described above. Briefly, THP-1 cells were infected for 4h with H37Ra at a MOI of 5, followed by 24 h of treatment with vehicle (DMSO) or TO901317 (20 nM) and RNA extraction. Reverse transcription (Roche kit) was performed using random hexamers only, and cDNAs were used directly without dilution for qPCR detection of putative eRNAs in the SCD intergenic region, using multiple set of primers (Supplementary Table S11). For ChIP 3C experiments, 20 million THP-1 were seed and differentiated per condition, for 24 h with 10−8 M of PMA. Then, cells were infected with H37Ra at a MOI of 5, for 4 h and washed three times prior to treatment with vehicle or TO901317 as indicated for 24 h. Then, as for a regular ChIP, cells were crosslinked with 1% formaldehyde for 20 min, and quenched with 125 mM glycine. Cells were gently lysed with a 3C buffer (100 mM Tris, 100 mM NaCl, 0.5% NP40, protease inhibitor) for 60 min with occasional swirling. Lysates were sonicated in the restriction enzyme buffer (cutsmart from New England Biolabs; NEB) containing 0.3% SDS in order to obtain fragments enrichment with a size below 5 kb. IP with anti-LXR antibody or IgG were used as indicated and as described previously for ChIP overnight. Then, the beads were washed prior to 24 h digestion at 25°C with ApaI (NEB), followed by 24 h digestion at 37°C with PsiI (NEB). The beads were washed and resuspended in ligation buffer with or without ligase (NEB) and incubated overnight at 16°C. Finally, the samples were decrosslinked with protease K and incubated overnight at 45°C, and DNA were extracted. RNAse (Ambion) treatment were performed for 30 min at 37°C. qPCR were performed with specific primers around the digestion sites (verification of digestion) and the ligation site (3C product), and ChIP efficiency were determined with the primers already used and validated for ChIP in DR4 within Alu repeats (Supplementary Table S11).
Western blots
Protein extracts from THP-1 cells were processed for Western blotting (Lysis with RIPA buffer, cycles of freezing −80°C/defreezing, and sonication three times 15 s ON, 15 s OFF at high power on the bioruptor (Diagenode)) and separated on Tris–SDS 5–20% gradient protein gels (Biorad®) using standard transfer protocol and blotting in TBS–5% milk for 1 h at RT before incubating O/N at 4C with the primary antibody. Then classic protocols of washing were used, Luminata Crescendo or Luminata Forte (Millipore®) ECL or Femto ECL (Pierce) were used to reveal the WB depending on the signal.
Antibodies and reagents
Anti-LXRα (ab41902 Abcam), anti-ATF1 (sc270, Santa Cruz), anti-ATF3 (sc188, Santa Cruz), anti-ATF5 (sc377168, Santa Cruz), anti-RXRA (sc553, Santa Cruz), anti-RARA (sc551, Santa Cruz), anti-MEF2A (ab7606. Abcam), anti-MEF2C (ab64644, Abcam), anti-LC3 (NB100-2220, Novus), anti-β-actin (sc-1616, Santa Cruz), anti-H3K27ac (ab4729),anti-NCoR (ab24552), anti-KAT3B/p300 (ab14984), anti-KAT13A/SRC1/NCoA1 (ab84), anti-H3K4me1 (ab8895, abcam), anti-PPARγ (ab41928, Abcam), anti-Mtb-FITC (ab20962, Abcam), anti-rabbit Alexa647 (A-21245, Life Technologies), Prolong Gold with Dapi (Life Technologies), anti-rabbit-Alexa488 (Life Technologies), Bodipy 493 (Life Technologies). TO901317, ATRA and BAX Inhibiting Peptide V5 were bought from Sigma and resupended in DMSO. Nelfinavir (Nel) and Ritonavir (Rit) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases. Auto Histone ChIP-seq Kit (Protein A) was bought from Diagenode.
CFU assay and antimicrobial quantification
At indicated time points, tissue culture plates were scrapped directly with the medium, then the medium was centrifuged to pellet any liberated mycobacteria at 3220×g for 20 min. Medium was aspirated and macrophages were lysed with water for 5 min, after which an equal volume of 2 × 7H9 with 0.1% Tween-80 was added. Samples were vigorously resuspended by repeated passages into 27 G needle and plated on Middlebrook 7H10 (Difco) with 10% OADC enrichment (BD Biosciences). To assess Mtb viability from infected monocytes, we used the real-time PCR–based method, comparing 16S rRNA to genomic DNA for assessing the viability of bacteria, as previously described (31). For experiments with cholesterol efflux inhibitors, Ritonavir (30 μM) or Nelfinavir (10 μM) were added to medium 20 min prior to addition of TO901317 (20 nM), as described (31). Infected cells were harvested for mycobacterial DNA and RNA content 3 days later.
Human macrophage isolation and treatment
Following informed written consent, blood was drawn from healthy adult males. Peripheral blood mononuclear cells (PBMCs) were isolated as previously described (1). Macrophages were treated with GM-CSF in order to allow the differentiation in macrophages for 3days. Protocols for the collection of blood for research purposes were approved by New England Institutional Review Board.
Microscopy and quantification
Following 4% paraformaldehyde fixation during 15 min, cells were washed with PBS twice. Cells were then permeabilized with 0.1% Triton X-100 for 5 min and incubated in PBS–0.2% BSA for an additional 5min. Primary antibody at the right concentration is incubated at 37°C for 1 h in a humidified chamber in phosphate-buffered saline with the addition of 1% BSA. Following three washes, cells are incubated with the secondary antibody coupled to a specific fluorophore during 45 min at RT in the dark. Slides were mounted in ProlongGold containing DAPI (Life technologies) and observed with a Zeiss Axiovert X100 bright field microscope or a Zeiss LSM750 × 100 confocal microscope. Images were acquired with Zen or Axiovision software. Quantification of size, intensity and number of spots corresponding to LD has been done with Metamorph5.0 and the figures mounted with Photoshop (Adobe) and ImageJ. Images processing and analysis for this manuscript was performed in the McGill University Life Sciences Complex Advanced BioImaging Facility (ABIF).
Apoptosis and necrosis assays
The Cell death Elisa Plus detection kit from Roche was used and performed at 24 or 72 h after infection, as indicated, on the cells for apoptosis detection or on the supernatants for necrosis detection. The experiment has been done with the attenuated and the virulent strain of Mtb due to their difference in host cell death induction.
Statistical analysis
Student's t-test or one-way ANOVA were performed where indicated using GraphPad software.
RESULTS
Regions of de novo H3K4 monomethylation in Mtb-infected cells are enriched for Alu repeats of the J and S subtypes
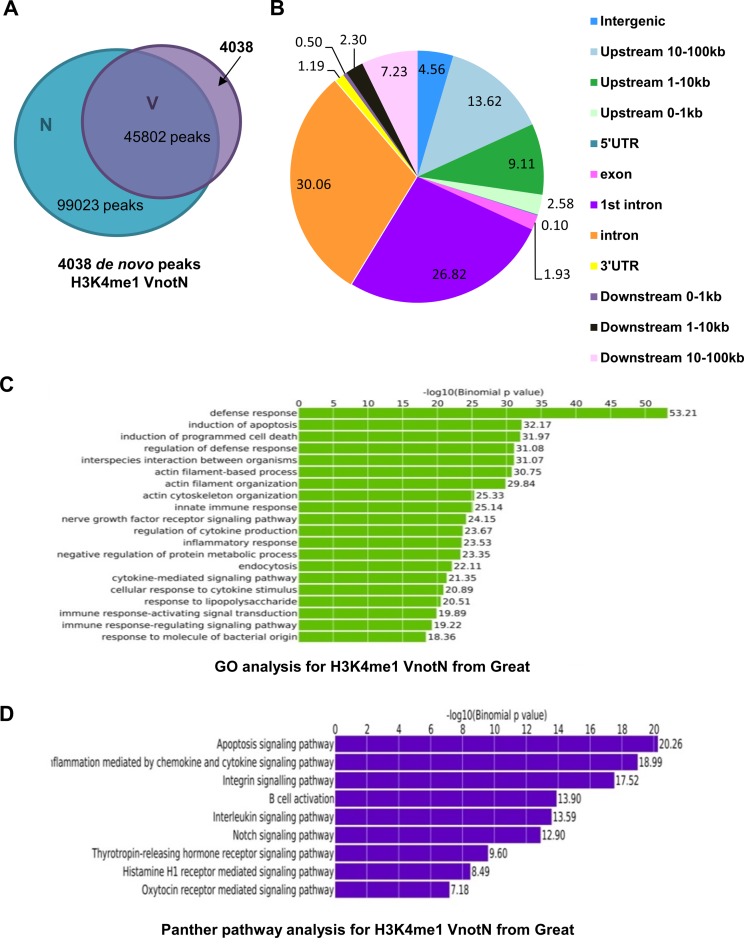
To decipher the transcriptomic responses of human macrophages within the first 24 h of H37Rv infection, we analyzed by ChIPseq the genome-wide profiles of the enhancer marker H3K4me1. 99 023 H3K4me1 regions were mapped in uninfected differentiated THP-1 macrophages, with a mean peak size of ∼2.7 kb, whereas in infected cells, 45 802 regions were identified, with an average peak size of 3.6 kb. The reduced number of peaks in infected cells occurred in spite of the fact that the number of sequence reads was not significantly different between the two conditions (Supplementary Table S1), and is thus consistent with transcriptional reprogramming in response to infection. Of the 45 802 peaks in infected cells, 4038 represented regions of de novo monomethylation, in agreement with an ∼10% ratio of dynamic to stable H3K4me1 regions seen in studies of macrophage priming (17).
While genes associated with any of the 45 802 H3K4me1-enriched sites in infected cells (V) may be of physiological significance, we focused initially on the 4038 de novo regions (VnotN) observed in infected cells but not in non-infected cells (N) (Figure 1A), as they should contain enhancers engaged shortly after Mtb infection. In this regard, induction of toll-like receptor (TLR) signaling prompted macrophage transcriptional responses through a combination of existing enhancers and de novo regions of H3K4 monomethylation (32). De novo peaks in our dataset were most concentrated at proximal promoter sequences (<10 kb upstream) or in first or middle introns, (Figure 1B), in agreement with their implication in transcriptional regulation. Moreover, genes proximal to de novo peaks identified by GREAT (26) were enriched for loci implicated in human immune defense responses and cell death pathways (Figure 1C and D), consistent with their roles in innate immune responses to H37Rv.
Figure 1.
Analysis of H3K4me1 peaks in uninfected and H37Rv-infected THP-1 cells. (A) Venn diagram representing the number of H3K4me1 peaks in cells infected with H37Rv (V) and uninfected controls (N). (B) Representation of the genomic distribution of the H3K4me1 peaks present in infected but not uninfected cells (VnotN). (C) Gene Ontology analysis of H3K4me1 peaks in VnotN performed with GREAT software. (D) Panther pathway analysis for H3K4me1 VnotN peaks performed with GREAT software.
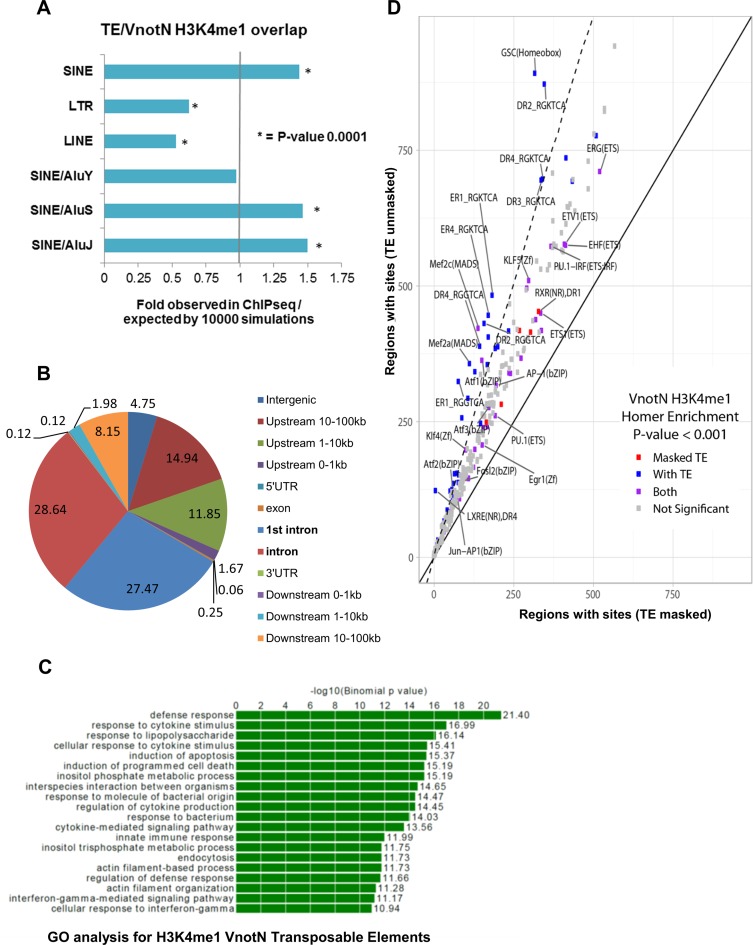
Analysis of the 4038 de novo H3K4me1 peaks using the motif discovery algorithm HOMER revealed that 40% (1620/4038) contained human/primate specific TEs. Of these 1620 regions, repetitive elements corresponded to least 50% of the 200 pb around the center of H3K4me1 peaks. As repetitive elements are abundant in the human genome (e.g. Alu repeats represent >10% of human genomic DNA), we performed a statistical analysis of the association of de novo H3K4me1 regions with TEs using GAT (Genome Association Tester; (29) and Materials and Methods) (Supplementary Table S2). This revealed that Alu repeats of the older J and S subtypes showed statistically significant enrichment of 1.5- and 1.46-fold, respectively, in H3K4me1 VnotN peaks compared to chance occurrence if the same number of genomic regions were chosen randomly. In contrast, younger AluY elements were not enriched, and LTR and LINE elements were depleted (Figure 2A, Supplementary Table S2; cf. p- and q-values). The distributions of the 1620 H3K4me1 peaks containing repetitive elements and all 4038 de novo regions were essentially identical (compare Figures 1B and 2B), with an enrichment in introns and gene-proximal locations relative to intergenic DNA. Moreover, the associated loci were heavily enriched in genes implicated in host immune responses (Figure 2C), suggesting that the regions containing repetitive elements contribute to transcriptional responses to infection.
Figure 2.
Enrichment for SINEs (AluJ and AluS) in H3K4me1 peaks. (A) GAT simulation showing 1.4-fold more AluJ and AluS in H3K4me1 regions with P-value of 0.0001. (B) Genomic distribution of the H3K4me1 peaks containing Alu repeats present in infected but not uninfected cells (VnotN). (C) Gene ontology analysis on genes associated with VnotN peaks that contain a TE. (D) Scatter Plot of the Homer Analysis showing that the analysis including transposable elements (TEs) (y-axis) highlighted new TF (blue) not present in the mask analysis only (red) (x-axis), TF present in both analysis are represented in purple.
H3K4me1 Alu repeats contain TFBS for multiple factors implicated in macrophage biology
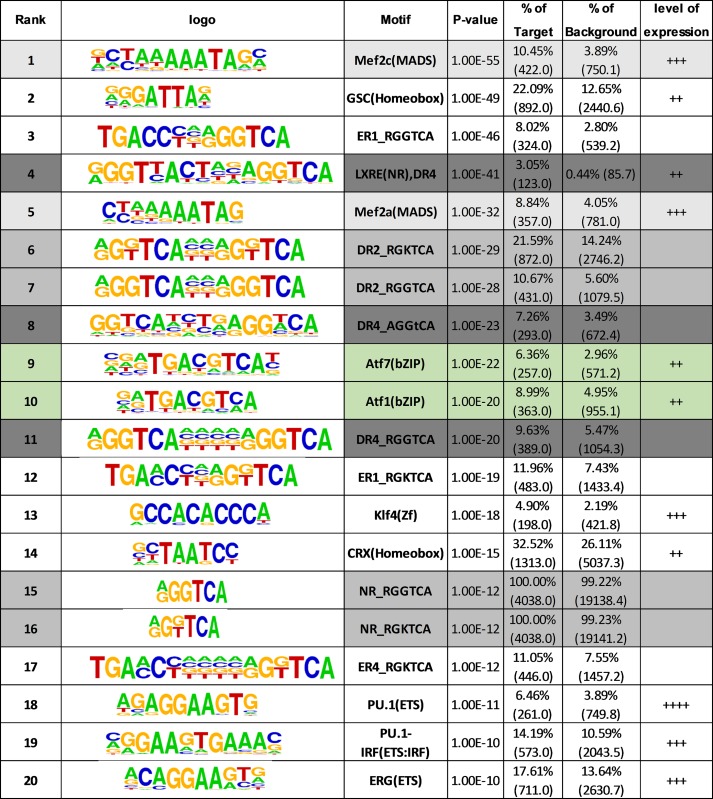
To determine whether repetitive elements within H3K4me1 regions contain TFBS, we performed HOMER analysis with (Supplementary Table S3) and without (Table 1) the parameter ‘Mask’ to compare sequence data with and without repetitive elements. Note that repetitive element masking is the default setting generally used in HOMER analysis of ChIPseq datasets. The scatter-plot representation showed that multiple TFBS were significantly enriched only when the TEs were included in the analysis (Figure 2D). Moreover, there was an enrichment of binding sites for several TFs implicated in macrophage biology in the unmasked sequence. These included homeobox proteins and MEF2 TFs (Figure 2D; Table 1), which are implicated in macrophage differentiation and survival (5). Microarray analysis, RT/qPCR and western blotting confirmed the expression of multiple MEF2 TFs in infected cells (Supplementary Table S4; Figure S1A and B). Also enriched were motifs recognized by ATFs, which are regulated downstream of numerous (patho)physiological stimuli, including TLR signaling and responses to infection (6,7). Several ATFs are expressed in infected cells, and ATFs 3 and 5 are induced by infection (Supplementary Table S4; Figure S1C and D), suggesting that ATF motifs in Alu repeats may be bound by several members of the CREB/ATF family.
Table 1. Motif search in unmasked VnotN H3K4me1 peaks from Homer analysis. Results of TF motif search in VnotN H3K4me1 peaks from Homer analysis. Analysis was performed without repeat sequence/TE masking (see Materials and Methods for details). +: weakly expressed, ++: well expressed, +++: strongly expressed in uninfected THP-1 cells (see (1)). Motifs shaded in pale grey are recognized by various nuclear receptors, whereas those shaded in darker grey are high affinity sites for LXRs. ATF motifs are shaded in green and MEF2 motifs in white/grey.
|
Enriched motifs also featured one or more RGGTCA or RGKTCA (RGG/TTCA) motifs, which correspond to response elements bound by nuclear receptors. These included DR4 motifs (LXREs), high-affinity response elements for liver X receptor/retinoid X receptor (LXR/RXR) heterodimers (see below), and DR2 motifs, which are bound by retinoic acid receptor (RAR)/RXRs (Figure 2D; Table 1). RXRs α and β are expressed in THP-1 cells, as are all three RARs (Supplementary Figure S1E–H). Everted repeats separated by a single nucleotide (ER1) were also present; however, it is not clear whether this configuration is recognized with moderate or high affinity by nuclear receptors (8).
In addition, several sites recognized by ETS family members were enriched in de novo H3K4me1 peaks in either filtered or unfiltered sequences (Table 1 and Supplementary Table S3). ETS factors control myeloid and lymphoid cell differentiation and are associated with responses to pathogen stimulation (33). Finally, filtered sequences were also enriched in DR1-type response elements, which are bound by peroxisomal proliferator-activated receptors (PPARs) with high affinity and by LXRs with moderate affinity (Supplementary Table S3). PPARγ was the most highly expressed, but its expression was flat or declined during infection (Supplementary Figure S1I and J).
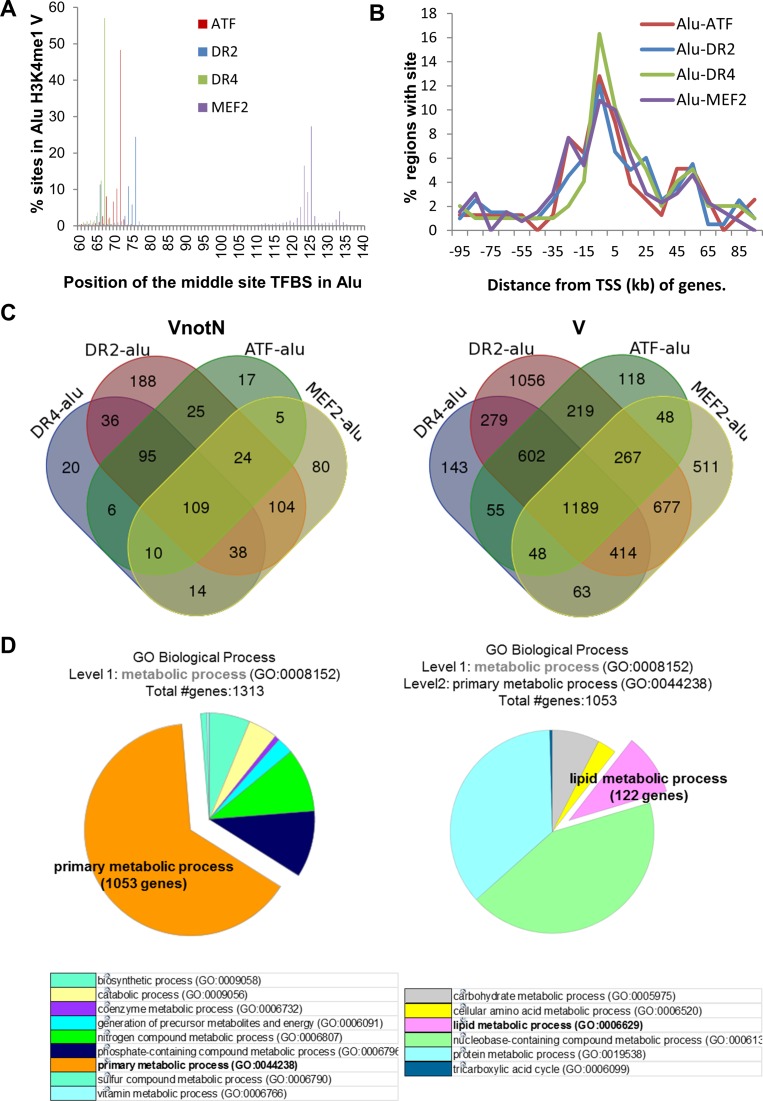
We analyzed further the distribution of ATF, MEF2, RAR (DR2) and LXR (DR4) binding sites present in H3K4me1 regions in our data, and the loci associated these elements in VnotN and V datasets. All four motifs were present at conserved positions in AluJ and AluS elements (Figure 3A), and were similarly distributed with respect to transcription start sites (TSS; Figure 3B). The majority of Alu repeats within H3K4me1 regions contained more than one TFBS (476/781, 61% and 3861/5689, 67% in VnotN and V, respectively; Figure 3C), suggesting that Alu repeats integrate multiple signaling pathways. There were variations; whereas the large majority (184/202, 91%) of DR4 motifs were in Alu repeats containing multiple TF motifs, 138/395 (35%) of elements contained isolated DR2 motifs (VnotN; Figure 3C).
Figure 3.
De novo H3K4 monomethylation regions within Alu repeats are enriched for specific transcription factor motifs in Mtb-infected cells. (A) Positions of the centers of ATF, -DR2, –DR4 and MEF2 motifs in Alu repeats located in H3K4me1 V regions. The positions of the sites within Alu were calculated from the genomic coordinates of sites and Alu in the intersectBed results. (B) Distribution of distances of ATF, -DR2, -DR4 and MEF2 motifs located Alu repeats in VnotN peaks relative to TSS (kb). The distribution of regions with ATF/DR2/DR4/MEF2 sites within 100 kb of TSS of T09 target genes were calculated from the distance in GREAT region-gene association tables with a custom Perl program. (C) Venn Diagrams representing the number of Alu repeats containing DR1, DR4, MEF2 and/or ATF motifs located in VnotN (left) or V (right) H3K4me1 regions. (D) In depth Panther analysis on Metabolic Processes (GO:0008152) enriched in Alu/DR4, /DR2, /MEF2, /ATF motifs found in H3K4me1 V peaks (Cf. Supplementary Figure S2). The left-hand pie chart corresponds to level 1 analysis of Metabolic Processes and its distribution in different pathways. The pie chart at right corresponds to level 2 analysis of primary metabolic processes from the left-hand panel and its detailed distribution into different pathways, notably in lipid metabolism.
To further validate these findings, we analyzed available ChIPseq datasets from primary human dendritic cells infected with Mtb (16) (GEO dataset GSE64175, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64175) and from macrophage differentiation and priming (17) (GEO dataset GSE58310, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58310). We used HOMER with and without filtering for repetitive elements, as above, which revealed a selective enrichment of TFBS for LXRs, RARs, MEF2, and ATFs in unmasked datasets, in agreement with our data (compare Supplementary Tables S5 and S6 for infected dendritic cells, and S7 and S8 for macrophage differentiation). The enrichment of DR4 elements, MEF2 and ATF motifs was also retained in analysis of unmasked subsets of data from the macrophage study (17), including macrophage differentiation and lipopolysaccharide (LPS)-induced tolerance (Supplementary Table S9). Collectively, these data provide strong evidence for the enrichment of Alu repeats containing TFBS in enhancers associated with multiple myeloid cell processes, in particular, responses to infection. The results also validate the importance of analysis of ChIPseq datasets including repetitive elements, and suggest that reanalysis of available datasets without filtering will identify new TFBS.
LXRα binding to cognate motifs in Alu repeats is enhanced by Mtb infection
We used the GREAT algorithm to identify loci associated with Alu repeats containing MEF2, ATF, LXR (DR4) or RAR (DR2) binding sites. PANTHER analysis of these loci revealed that 50% of the genes were enriched in metabolic processes (Supplementary Figure S2). Of the TFs identified in Alu repeats, nuclear receptors are of particular interest because they are ligand-regulated, which facilitates analysis of enhancer function. They also represent potential targets of therapeutic development. DR4 elements are high-affinity sites for LXR nuclear receptors, whose signaling controls cholesterol and lipid homeostasis. 1053 of 1313 of the DR4-Alu associated loci were linked to primary metabolic processes, including carbohydrate, protein, nucleic acid metabolism (Figure 3D, left). The list also included 122 genes associated with lipid metabolism (Figure 3D, right). This is intriguing, as intracellular cholesterol stores in macrophages provide a critical source of carbon units for Mtb proliferation (34,35). Moreover, animal studies have shown that LXRα depletion in mice renders them more susceptible to Mtb infection (15). DR4 elements thus represent excellent models for validation of the role of TFBS present in Alu repeats in response to infection.
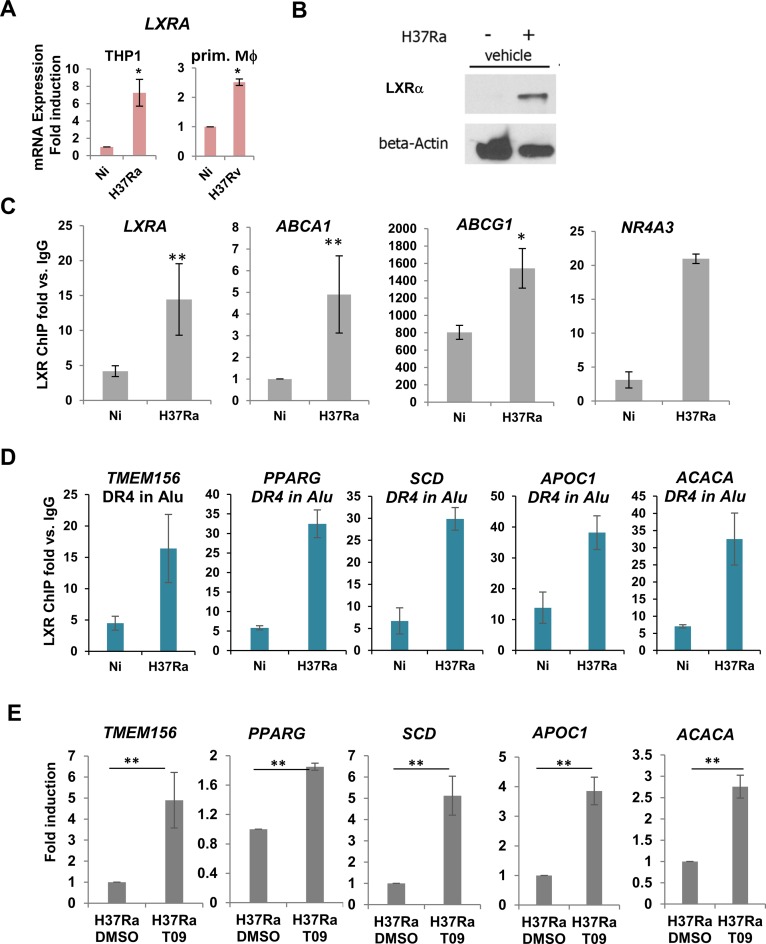
We probed further the potential role of Alu repeats as putative binding sites for LXRs, and how LXR signaling within Alu repeats controls Mtb burden. Expression profiling showed that expression of LXRα, but not related LXRβ, is induced in THP-1 cells infected with either H37Rv or H37Ra [Supplementary Table S4, and (1)]. LXRα induction during infection was confirmed by RT/qPCR in infected THP-1 cells and primary human macrophages (Figure 4A) and by western blotting in THP-1 cells (Figure 4B). LXR DNA binding to previously characterized DR4 response elements in the LXRA, ABCG1, ABCA1 and NR4A3 genes, which lie within H3K4me 1 peaks, increased in H37Ra-infected cells (Figure 4C). Importantly, we also found that LXRα binds in infected cells to Alu/DR4 elements associated with multiple genes implicated in lipid metabolism (Figure 4D), whose expression is induced by LXR agonist treatment (Supplementary Figure S3 and Figure 4E, and see below). Note that for all of the Alu/DR4 elements tested, the motifs represented only the consensus LXR binding site in the vicinity of gene. These findings confirm the capacity of LXRs to recognize DR4 elements within the context of Alu repeats in Mtb infected cells.
Figure 4.
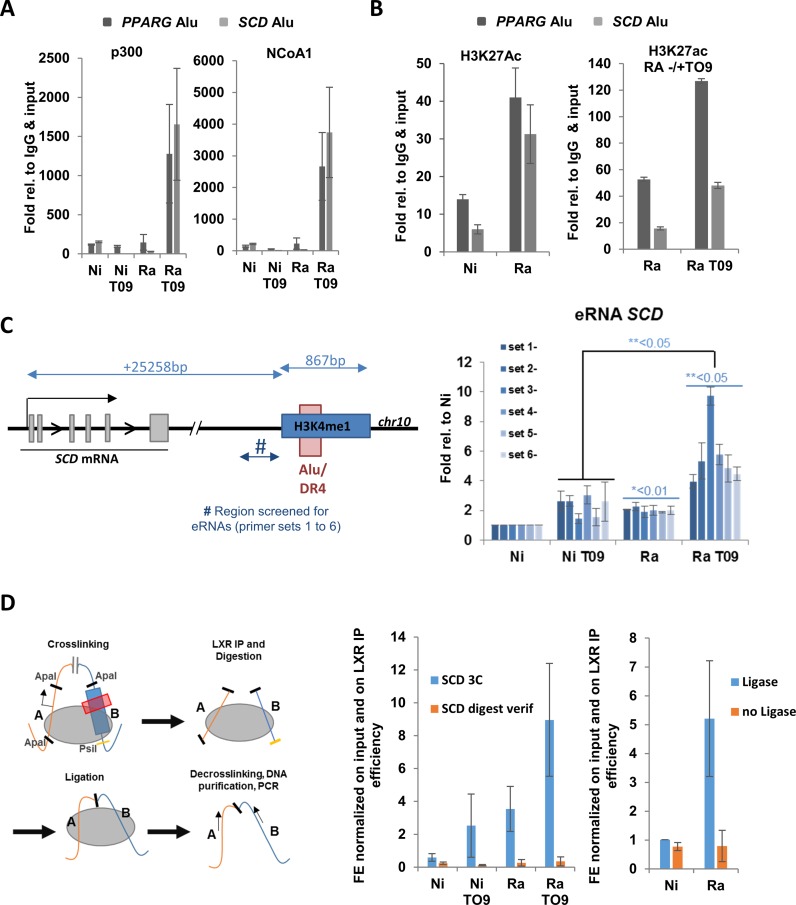
LXRα binding to cognate motifs in Alu repeats is enhanced by Mtb infection. (A) RT-qPCR of LXRA in THP-1 cells or primary human macrophages uninfected (Ni) and infected with H37Ra or H37Rv, as indicated. Expression was normalized to reference genes (18S, GAPDH, b-actin; n is a minimum of 3). (B) Western blots showing LXRα expression in uninfected and H37Ra-infected THP-1 cells. (C) ChIP-qPCR analysis of the effect of H37Ra infection of LXRα binding to previously characterized DR4 elements in the LXRA, ABCA1, ABCG1, NR4A3 genes. (D) ChIP-qPCR analysis of LXRα binding to newly identified DR4 elements in Alu repeats in VnotN peaks corresponding to genes TMEM156, PPARG, APOC1, SCD and ACACA. (E) RT/qPCR analysis of regulation by LXR agonist TO901317 of genes analyzed by ChIP-qPCR in (D). Results are average of 3 experimental replicates performed in biological triplicates, ***P-values < 0.005.
LXRα/RXR heterodimers bind DNA in the absence of agonist and recruit corepressors NCoR and SMRT (36,37), whereas agonist binding induces coactivator recruitment and activation of target gene expression. To analyze the effects of Mtb infection on the transcriptional status of LXRs, we performed ChIP analyses for association of NCoR and the coactivators p300 and NCoA1 with LXREs in uninfected and infected cells. Under conditions where total LXRα binding to DR4 elements in the LXR, ABCA1, and ABCG1 genes was elevated in infected cells (Supplementary Figure S4A), there were no substantial increases in levels of NCoA1 or p300 associated with the same sites (Supplementary Figure S4A). In contrast, association of NCoR with the same elements increased to essentially the same degree as LXR binding (Supplementary Figure S4A). These results suggest that infection under these conditions is not accompanied by an increased production of endogenous LXR agonists. Indeed, we did not observe any substantial regulation of genes encoding the enzymes CYP27A1, CYP46A1, CH25H and DHCR24 (Supplementary Figure S4B), which catalyze the formation of endogenous LXR ligands (38,39), a result confirmed by RT-qPCR (Supplementary Figure S4C). While the catalytic activities or expression levels of these enzymes could be regulated post-transcriptionally, the data taken together indicate that the enhanced expression of LXRα in infected cells is not accompanied by an increase in the proportion of agonist-bound receptor under these conditions.
Function of Alu/DR4 elements as enhancers in response to infection
Several potent LXRα-selective agonists have been developed, including TO901317, which specifically activates LXRα at low nanomolar concentrations (40). We used 20nM TO901317, a concentration that does not activate other receptors such as FXRs (41). Recruitment of NCoA1 and p300 to previously characterized DR4 elements was substantially increased in TO901317-treated infected cells (Supplementary Figure S5) in the absence of an increase in LXRα binding. A similar TO901317-induced recruitment of coactivators was observed to the DR4 elements located in Alu repeats within H3K4me1 regions adjacent to the PPARG and SCD genes (Figure 5A). We also observed that H3K27 acetylation, a marker of active transcription and active enhancers, in the same regions was increased by infection and further enhanced by TO901317 treatment (Figure 5B). As an additional probe for TO910317-dependent enhancer function, we screened for production of enhancer RNAs (eRNAs; (42)) using primer sets centered 500–800 bp upstream of the DR4 adjacent to the SCD gene. This element was chosen because it lies in an intergenic region downstream of the SCD gene. This revealed that levels of eRNAs were significantly elevated in infected cells, and further enhanced by treatment with TO901317, consistent with the function of the Alu-DR4 as an LXR-dependent enhancer. Finally, we performed ChIP-3C (Chromatin Conformation Capture) assays to test for interaction of the Alu repeat downstream of the SCD gene with the TSS of the gene. This revealed an infection- and TO901317-dependent increase in the association of the Alu repeat region with the SCD TSS (Figure 5D). Taken together, these results show that Alu repeats containing DR4 motifs can function as bona fide enhancers. Moreover, they suggest that TO901317 treatment should alter macrophage transcriptional and biochemical responses to infection.
Figure 5.
Function of DR4 motifs in Alu repeats as enhancer elements. (A) LXR agonist TO901317 induces recruitment of coactivators NCoA1 and p300 to Alu repeats containing DR4 elements in the regions of the PPARG and SCD genes, as assessed by ChIP assay. (B) ChIP assays reveal that H3K27 acetylation is enhanced in regions of Alu repeats of the PPARG and SCD genes by H37Ra (Ra) infection and TO901317 (TO9). (C) Left: Schematic representation of the SCD gene and its downstream region including sequences upstream of the Alu repeat screened for eRNAs. Right: Results of screening for eRNAs in non-infected (Ni) or H37Ra-infected (Ra) THP-1 cells in the absence or presence of TO901317. (D) Analysis of association of the downstream Alu repeat with the TSS of the SCD gene by ChIP-chromatin conformation capture assay. Left: Schematic representation of ChIP-3C assay. Center: ChIP-3C signal (Blue) obtained by amplification across the junction of the ApaI 3C ligation product in non-infected (Ni) or H37Ra (Ra)-infected THP-1 cells treated with vehicle or TO901317, as indicated. An amplification (orange) across the ApaI site in the region of the TSS of the SCD gene is also shown to control for digestion (see Supplementary Table S11 for primers). Right: A control experiment showing the amount of ChIP-3C product generated in non-infected or H37Ra-infected cells in the presence or absence of ligase.
LXR agonists reduce Mtb viability through altered cholesterol metabolism
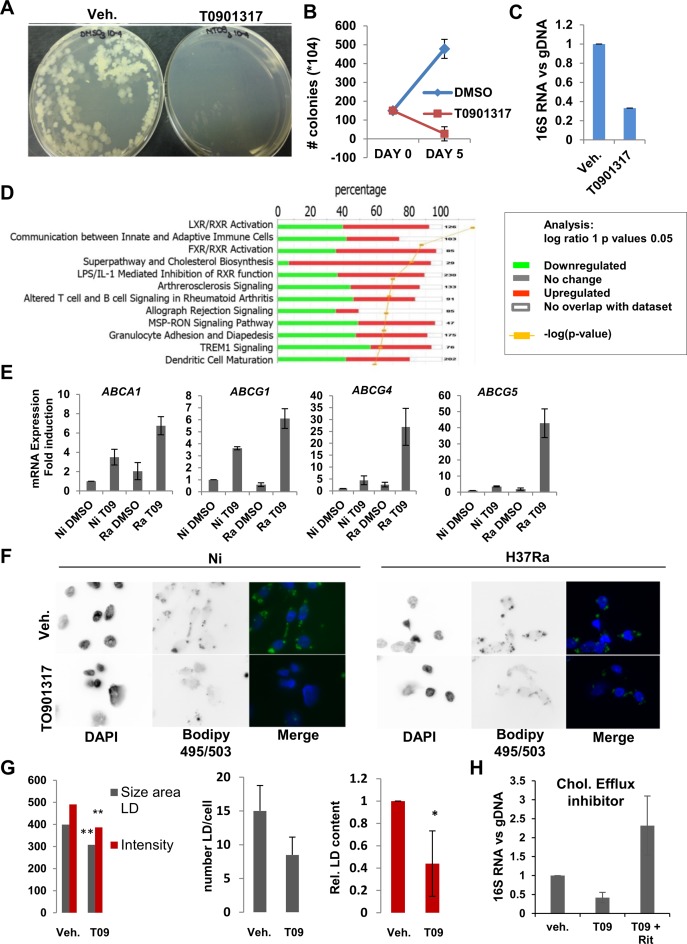
To confirm the importance of LXR signaling in responses to infection we tested the effect of TO901317 on Mtb viability. In control experiments, TO901317 added to Mtb cultured in broth 7H9 media had no effect on mycobacterial growth (Supplementary Figure S6). In contrast, TO901317 treatment of macrophages led to an 18-fold reduction in mycobacterial burden after 5 days of infection as measured by colony-forming units (Figure 6A and B), results supported by quantification of the ratio of mycobacterial 16S RNA to total genomic DNA 3 days post-infection (Figure 6C; (43)).
Figure 6.
Analysis of the effects of LXR agonist TO901317 on LXRα target gene regulation in infected cells. (A) Pictures of representative results of CFU analysis of DMSO- or TO901317- (20 nM) treated THP-1 cells infected with H37Ra for 5 days: dilution 10−5 of H37Ra spread on 7H10 plates and incubated for 3 weeks. (B) Quantification of CFU assay results at day 0 and day 5 after infection and treatment, n = 3. (C) Assessment of Mtb replication after DNA and RNA extraction was performed by (RT-)qPCR analysis of the ratio of 16SRNA mycobacterial RNA to genomic DNA. (D) Results of IPA (Ingenuity®) pathway enrichment analysis for genes regulated at least 2-fold by treatment with LXR agonist TO901317. Green is for downregulated genes, and red is for upregulated genes. (E) RT-qPCR analysis of expression of genes encoding cholesterol transporters ABCA1, ABCG1, ABCG4 and ABCG5 in uninfected (Ni) or H37Ra (Ra)-infected THP-1 cells treated 24 h with DMSO- or TO901317 (20nM). (F) Imaging of Lipid Droplets (LD) in infected (H37Ra) vs uninfected (NI) THP-1 cells treated with DMSO (veh.) or TO901317, and stained with Bodipy 495/503 and DAPI (Axiovert microscope, X100 objective). (G) Quantification of LD, by size and intensity, number of LD per cell and the total lipid droplet content in H37Ra-infected THP-1 cells treated with DMSO or TO901317. Measurement was done on 20 different images for each condition and on at least 10 cells per image. Measurement and quantification were done using Imaris. **P < 0.05, *P < 0.005. (H) Cholesterol efflux inhibitors Ritonavir (30μM) or Nelfinavir (10 μM) reverse the antimycobacterial effects of TO901317. Quantification of Mtb replication after DNA and RNA extraction was performed by qPCR. Results are represented as expression of 16SRNA normalized to genomic DNA.
LXR target gene regulation in the presence of TO901317 in THP-1 cells has been characterized (44), but not in the context of infection. Therefore, we performed Affymetrix Gene2ST microarrays in THP-1 cells infected with H37Ra for 24h in the absence or presence of 20nM TO901317. A total of 254 and 288 genes were up- or downregulated, respectively, at least 1.5-fold, (Supplementary Table S10, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64335). In addition to the genes tested above containing Alu/DR4 elements (Figure 4D), we validated the regulation of several TO901317 target genes by RT-qPCR (Supplementary Figure S7A). Pathways analysis (IPA) of TO901317-regulated genes revealed enrichment for loci controlling LXR signaling and cholesterol biosynthesis (Figure 6D; Supplementary Figure S7B), consistent with previously published work (40,45), as well as pathways of signaling between innate and adaptive immune cells. This is significant, as the enrichment of lipid metabolic pathways in TO901317-regulated genes links LXR signaling in infected cells to Alu/DR4 elements associated with loci implicated in lipid metabolism (see above and Supplementary Figure S2). There was very limited overlap between our results (Supplementary Figure S8) and TO901317-regulated gene expression profiles already published in differentiated THP-1 cells (44), which may arise from the lack of Mtb infection and differences in ligand concentrations (20nM vs 10μM Cf. (44)) between the two studies.
Previous studies have shown that ablation of LXR expression increases Mtb burden in vitro and in vivo (15,46). However, specific metabolic pathways regulated by LXR agonist signaling in infected cells have not been examined. Among the genes most highly induced by TO901317 were ABCA1 and ABCG1, which encode cholesterol transporters. Significantly, infection and TO901317 combined to synergistically induce expression of both genes (Figure 6E), as well as that of ABCG4 and ABCG5, which have been recently characterized and implicated in cholesterol efflux (47). LXR-stimulated expression of cholesterol transporters can lead to elevated efflux of cholesterol (48), a critical carbon source for replicating Mtb, from macrophages (34). There was a reduction in lipid droplet size and intensity of BODIPY staining for neutral lipids and other nonpolar lipids, a trend towards reduced numbers of droplets, and a significant reduction in the total lipid content of droplets in H37Ra-infected cells (Figure 6F and G). Importantly, the diminution in the ratio of 16S RNA to total genomic DNA was reversed by preincubation of TO901317-treated infected cells with cholesterol efflux inhibitors Ritonavir or Nelfinavir (31). This data provide first evidence that reduction of cholesterol stores induced by LXR signaling contribute to its anti-mycobacterial actions (Figure 6H).
Numerous studies have shown that autophagy plays a critical role in controlling the viability of intracellular pathogens such as Mtb (49). We probed the effect of TO901317 on autophagy in infected cells by analyzing the colocalization of Mtb and LC3II protein, a marker of autophagosomes, 3 days after infection and found no differences in colocalization or signal intensity (Supplementary Figure S9A). Macrophage necrosis increased in H37Rv-infected cells (Supplementary Figure S9B), as expected (50,51), but TO901317 treatment had no apparent effect on necrosis in either uninfected or infected cells. However, TO901317 selectively enhanced apoptosis in H37Rv and in H37Ra-infected cells, but not in uninfected cells (Supplementary Figure S9C). This finding is important: while necrosis may facilitate bacterial dissemination and transmission (77), macrophage apoptosis during intracellular infection is bactericidal and contributes to innate immune defenses against Mtb (52), thus providing a mechanism for the mycobactericidal effects of TO901317 treatment. Indeed, inhibiting apoptosis with a peptide that blocks BAX activity (53) completely eliminated the effect of TO901317 on Mtb viability (Supplementary Figure S9D). These studies reveal that the phenotypic effects of LXR signaling, driven in part by LXRα binding to high-affinity response elements present in Alu repeats, control Mtb viability through altering cholesterol metabolism and apoptosis.
DISCUSSION
Analysis of the epigenetic changes occurring in macrophages infected by Mtb has been understudied. However, such changes can provide a window to discover novel signaling pathways implicated in host immune responses, and to understand transcriptomic regulatory responses under specific pathophysiological conditions. In this regard, recent studies reported that changes in DNA methylation patterns can rapidly occur in response to certain environmental stimuli (54–56), and Pacis et al. suggested that changes in DNA methylation could contribute to short-term memory in innate immune cells, such as dendritic cells (16). Here, we characterized changes in the H3K4me1 profile in H37Rv-infected macrophages. Our systems biology approach combining bioinformatic prediction, genomic analysis, and molecular validation revealed the roles of AluJ and AluS repeats as platforms for TFBS in host transcriptomic responses to Mtb infection.
We found that, for 40% of de novo H3K4me1 in infected macrophages, at least 50% of the 200 bp around their center could be attributed to human/primate-specific TEs, predominantly Alu repeats. Several lines of evidence have suggested a potential role of Alu repeats as sources of several TFBS (13,23,57). Moreover, bioinformatic analyses have shown that genes proximal to Alu elements are selectively enriched for H3K4me1 marks and are associated with functional enhancers (14). Analysis of our ChiPSeq data showed that Alu repeats present in H3K4me1 peaks in Mtb-infected cells contain several putative motifs for TFs central to macrophage biology. Moreover, they were present in the vicinity of numerous genes associated with immunity and metabolic processes. Enriched TFBS included those recognized by members of the MEF2 and ATF families, which have been implicated in macrophage differentiation, survival and responses to stress (6,58), as well as binding sites for LXR and RAR nuclear receptors. These findings were largely reproduced by analyzing independent H3K4me1 datasets derived from Mtb-infected primary human dendritic cells, and macrophages induced to undergo differentiation or responses mimicking infection (16,17). This reinforces the robustness of our findings and the implication of these motifs in a broad range of myeloid cell responses.
Analysis of loci associated with Alu repeats in our dataset revealed that many of the genes are implicated in host defense and regulation of primary metabolic processes. Other studies have suggested that RAR signaling controls Mtb infection partly by regulating cholesterol metabolism (31). Similarly, LXRs are key regulators of lipid metabolism in several tissues, including macrophages (40). Previous work on the effects of LXRα ablation in THP-1 cells on Mtb burden (15,46) provided a correlation between elevated lipid body content and reduced apoptosis during H37Ra infection and loss of LXRα. We show for the first time that the anti-mycobacterial activity of LXRα agonist TO901317 could be ascribed to its capacity to alter cholesterol metabolism and selectively enhance apoptosis versus necrosis in infected cells. Similar beneficial effects of TO901317 may not be limited to Mtb infection; host cholesterol and fatty acids can also provide important carbon sources for other pathogens, including human Mycobacterium leprae (59), as well as viruses such as immunodeficiency virus type 1 (60). Our results thus suggest that, similar to vitamin A supplementation (61), which enhances RAR signaling, LXRs represent a target for potential therapeutic intervention in infectious diseases with the development of tissue-specific modulators that target their activities in macrophages.
Our findings also reinforce the importance of Alu element transposition in the evolution of primate transcriptional programs. The Alu repeats enriched in H3K4me1 regions in infected cells relative to random genomic sequence were of the more ancient AluJ and AluS subtypes, unlike younger AluY elements. The expansion of J and S subtypes occurred relatively early in primate evolution, approximately 35–55 million years ago. A very specific but nonetheless telling example of an acquisition of enhancer function in innate immunity is the well-conserved, promoter-proximal insertion of an AluS repeat containing a vitamin D response element (VDRE) in the gene encoding the antimicrobial peptide cathelicidin (62,63). This insertion has been retained in both old- and new-world primates (63). This event was a key element in the evolution of human/primate-specific mechanisms of innate immune regulation by vitamin D signaling (64).
Other work has also emphasized the importance of TE transposition as sources of TFBS expansion. Reanalysis of ChIPSeq datasets for seven different TFs showed that binding sites for 5 of were prevalent several types of retrotransposons (23). It is unclear why TEs provided vehicles for dispersion of some TFBS and not others apart from the obvious possibility that TEs may lack progenitor sequences for specific classes of TFs. The authors of the above study (23) speculated that TE transposition provided a mechanism for expansion of TFBS with more complex sequence compositions, which would be less likely to evolve by mutation than simpler motifs. In support of this notion, previous work has shown that 7SL genes and ancient Alu elements contain multiple hexamers that resemble the consensus RGGTCA half-sites recognized by several nuclear receptors (65). LXREs and RAREs, like VDREs (DR3 elements), represent complex, bipartite binding sites with relatively constrained sequence specificities. It is thus likely that Alu element transposition was a powerful tool for their dispersion throughout the genome.
In conclusion, this study provides extensive evidence that TF binding sites present in Alu repeats contribute to macrophage transcriptional responses to Mtb. Our results also suggest that Alu repeats may play a larger role in the regulatory genome in general, and that specific TF binding sites within will be selectively conserved if beneficial to the organism. Alu repeats are present in numerous genes associated with host defenses and metabolic responses to infection, and a combination of bioinformatic and biochemical approaches connected loci associated with Alu repeats to macrophage metabolic responses to Mtb. In particular, Alu repeats serve as platforms for binding of RAR and LXR nuclear receptors, whose signaling greatly reduces mycobacterial burden at least partly through altering macrophage lipid metabolism. These results provide compelling evidence that expansion of repetitive elements in the human genome has contributed on a large scale to the evolution of transcriptional responses to (patho)physiological signals, and that the potential contribution of these elements must be taken into account in genomic analyses of transcription.
Supplementary Material
Acknowledgments
We thank the McGill University and Genome Quebec Innovation Centre for performing gene expression analysis and ChIP sequencing. We also thank the Advanced BioImaging Facility of McGill University for the images taken with their microscope and the software used for the imaging analysis. We would like to thank Dr. John Lambourne for his help for the SOP of the ChIP sequencing and his precious advice for the library preparation. We would like to thank Vassil Dimitrov for his help in planning and trouble-shooting in the ChIP-3C experiment.
Author contributions: M.B. and J.H.W. designed the study. M.B. designed and performed experiments, analysed data and wrote the paper with J.H.W; D.L. developed analytical tools and performed the bioinformatics analysis. M.A.B. contributed to studies with H37Rv. B.M, J.M., M.V., A.F. and E.M. designed and performed some experiments; S.M., R.S. and L.B.B. contributed to genomic studies and critically read the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Quebec and the Canadian Institutes of Health Research [CIHR MOP-106439 to J.H.W.]; Fonds de Recherche en Santé, Québec (M.B.). Funding for open access charge: CIHR [MOP-106439].
Conflict of interest statement. None declared.
REFERENCES
- 1.Verway M., Bouttier M., Wang T.-T., Carrier M., Calderon M., An B.-S., Devemy E., McIntosh F., Divangahi M., Behr M.A., et al. Vitamin D induces interleukin-1β expression: paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013;9:e1003407. doi: 10.1371/journal.ppat.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobat A., Gallant C.J., Simkin L., Black G.F., Stanley K., Hughes J., Doherty T.M., Hanekom W.A., Eley B., Jaïs J.-P., et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J. Exp. Med. 2009;206:2583–2591. doi: 10.1084/jem.20090892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 4.Hedges D.J., Batzer M.A. From the margins of the genome: mobile elements shape primate evolution. BioEssays News Rev. Mol. Cell. Dev. Biol. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 5.Canté-Barrett K., Pieters R., Meijerink J.P.P. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33:403–410. doi: 10.1038/onc.2013.56. [DOI] [PubMed] [Google Scholar]
- 6.De Nardo D., Labzin L.I., Kono H., Seki R., Schmidt S.V., Beyer M., Xu D., Zimmer S., Lahrmann C., Schildberg F.A., et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 2014;15:152–160. doi: 10.1038/ni.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tattoli I., Sorbara M.T., Vuckovic D., Ling A., Soares F., Carneiro L.A.M., Yang C., Emili A., Philpott D.J., Girardin S.E. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Rőszer T., Menéndez-Gutiérrez M.P., Cedenilla M., Ricote M. Retinoid X receptors in macrophage biology. Trends Endocrinol. Metab. TEM. 2013;24:460–468. doi: 10.1016/j.tem.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 11.Cui F., Sirotin M.V., Zhurkin V.B. Impact of Alu repeats on the evolution of human p53 binding sites. Biol. Direct. 2011;6:2. doi: 10.1186/1745-6150-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu T.J., Yi X., Zhao X.W., Zhao Y., Yin J.Q. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonaki A., Demetriades C., Polyzos A., Banos A., Vatsellas G., Lavigne M.D., Apostolou E., Mantouvalou E., Papadopoulou D., Mosialos G., et al. Genomic analysis reveals a novel nuclear factor-κB (NF-κB)-binding site in Alu-repetitive elements. J. Biol. Chem. 2011;286:38768–38782. doi: 10.1074/jbc.M111.234161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su M., Han D., Boyd-Kirkup J., Yu X., Han J.-D.J. Evolution of Alu elements toward enhancers. Cell Rep. 2014;7:376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Korf H., Vander Beken S., Romano M., Steffensen K.R., Stijlemans B., Gustafsson J.-A., Grooten J., Huygen K. Liver X receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice. J. Clin. Invest. 2009;119:1626–1637. doi: 10.1172/JCI35288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacis A., Tailleux L., Morin A.M., Lambourne J., Maclsaac J.L., Yotova V., Dumaine A., Danckaert A., Luca F., Grenier J.-C., et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015 doi: 10.1101/gr.192005.115. doi:10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed S., Quintin J., Kerstens H.H.D., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.-C., Ratter J., Berentsen K., van der Ent M.A., et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatem A., Bozdağ D., Toland A.E., Çatalyürek Ü.V. Benchmarking short sequence mapping tools. BMC Bioinformatics. 2013;14:184. doi: 10.1186/1471-2105-14-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez-Gonzalez R.H., Bonnal R., Caccamo M., Maclean D. Bio-samtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol. Med. 2012;7:6. doi: 10.1186/1751-0473-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma. Oxf. Engl. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laperriere D., Wang T.-T., White J.H., Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.-L., Ruan Y., Wei C.-L., Ng H.H., et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie M., Hong C., Zhang B., Lowdon R.F., Xing X., Li D., Zhou X., Lee H.J., Maire C.L., Ligon K.L., et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang M., Anderson J., Gillespie J., Mayne M. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinformatics. 2008;9:192. doi: 10.1186/1471-2105-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol. Biol. Clifton NJ. 2014;1150:153–160. doi: 10.1007/978-1-4939-0512-6_9. [DOI] [PubMed] [Google Scholar]
- 28.Bourdeau V., Deschênes J., Laperrière D., Aid M., White J.H., Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008;36:76–93. doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heger A., Webber C., Goodson M., Ponting C.P., Lunter G. GAT: a simulation framework for testing the association of genomic intervals. Bioinforma. Oxf. Engl. 2013;29:2046–2048. doi: 10.1093/bioinformatics/btt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright G.W., Simon R.M. A random variance model for detection of differential gene expression in small microarray experiments. Bioinforma. Oxf. Engl. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 31.Wheelwright M., Kim E.W., Inkeles M.S., De Leon A., Pellegrini M., Krutzik S.R., Liu P.T. All-trans retinoic acid-triggered antimicrobial activity against Mycobacterium tuberculosis is dependent on NPC2. J. Immunol. Baltim. MD. 2014;192:2280–2290. doi: 10.4049/jimmunol.1301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C., et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carotta S., Wu L., Nutt S.L. Surprising new roles for PU.1 in the adaptive immune response. Immunol. Rev. 2010;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 34.Pandey A.K., Sassetti C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee W., Vander Ven B.C., Fahey R.J., Russell D.G. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghisletti S., Huang W., Jepsen K., Benner C., Hardiman G., Rosenfeld M.G., Glass C.K. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astapova I., Lee L.J., Morales C., Tauber S., Bilban M., Hollenberg A.N. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19544–19549. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cyster J.G., Dang E.V., Reboldi A., Yi T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat. Rev. Immunol. 2014;14:731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- 39.Ignatova I.D., Angdisen J., Moran E., Schulman I.G. Differential regulation of gene expression by LXRs in response to macrophage cholesterol loading. Mol. Endocrinol. Baltim. MD. 2013;27:1036–1047. doi: 10.1210/me.2013-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz J.R., Tu H., Luk A., Repa J.J., Medina J.C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D.J., et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houck K.A., Borchert K.M., Hepler C.D., Thomas J.S., Bramlett K.S., Michael L.F., Burris T.P. T0901317 is a dual LXR/FXR agonist. Mol. Genet. Metab. 2004;83:184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y., Wong C.-H., Birnbaum R.Y., Li G., Favaro R., Ngan C.Y., Lim J., Tai E., Poh H.M., Wong E., et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez A.N., Lahiri R., Pittman T.L., Scollard D., Truman R., Moraes M.O., Williams D.L. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J. Clin. Microbiol. 2009;47:2124–2130. doi: 10.1128/JCM.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldmann R., Geikowski A., Weidner C., Witzke A., Kodelja V., Schwarz T., Gabriel M., Erker T., Sauer S. Foam cell specific LXRα ligand. PLoS One. 2013;8:e57311. doi: 10.1371/journal.pone.0057311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitro N., Vargas L., Romeo R., Koder A., Saez E. T0901317 is a potent PXR ligand: Implications for the biology ascribed to LXR. FEBS Lett. 2007;581:1721–1726. doi: 10.1016/j.febslet.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 46.Mahajan S., Dkhar H.K., Chandra V., Dave S., Nanduri R., Janmeja A.K., Agrewala J.N., Gupta P. Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPARγ and TR4 for survival. J. Immunol. Baltim. Md 1950. 2012;188:5593–5603. doi: 10.4049/jimmunol.1103038. [DOI] [PubMed] [Google Scholar]
- 47.Wang N., Lan D., Chen W., Matsuura F., Tall A.R. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9774–9779. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy M.A., Barrera G.C., Nakamura K., Baldán A., Tarr P., Fishbein M.C., Frank J., Francone O.L., Edwards P.A. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Sanjurjo L., Amézaga N., Vilaplana C., Cáceres N., Marzo E., Valeri M., Cardona P.-J., Sarrias M.-R. The scavenger protein apoptosis inhibitor of macrophages (AIM) potentiates the antimicrobial response against Mycobacterium tuberculosis by enhancing autophagy. PLoS One. 2013;8:e79670. doi: 10.1371/journal.pone.0079670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M., Gan H., Remold H.G. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J. Immunol. Baltim. MD. 2006;176:3707–3716. doi: 10.4049/jimmunol.176.6.3707. [DOI] [PubMed] [Google Scholar]
- 51.Arcila M.L., Sánchez M.D., Ortiz B., Barrera L.F., García L.F., Rojas M. Activation of apoptosis, but not necrosis, during Mycobacterium tuberculosis infection correlated with decreased bacterial growth: role of TNF-alpha, IL-10, caspases and phospholipase A2. Cell. Immunol. 2007;249:80–93. doi: 10.1016/j.cellimm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Keane J., Remold H.G., Kornfeld H. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. Baltim. MD. 2000;164:2016–2020. doi: 10.4049/jimmunol.164.4.2016. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida T., Tomioka I., Nagahara T., Holyst T., Sawada M., Hayes P., Gama V., Okuno M., Chen Y., Abe Y., et al. Bax-inhibiting peptide derived from mouse and rat Ku70. Biochem. Biophys. Res. Commun. 2004;321:961–966. doi: 10.1016/j.bbrc.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 54.Klug M., Heinz S., Gebhard C., Schwarzfischer L., Krause S.W., Andreesen R., Rehli M. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2183–E2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marr A.K., MacIsaac J.L., Jiang R., Airo A.M., Kobor M.S., McMaster W.R. Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. 2014;10:e1004419. doi: 10.1371/journal.ppat.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Huang S. Nuclear function of Alus. Nucl. Austin TX. 2014;5:131–137. doi: 10.4161/nucl.28005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stehling-Sun S., Dade J., Nutt S.L., DeKoter R.P., Camargo F.D. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat. Immunol. 2009;10:289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- 59.Mattos K.A., Oliveira V.C.G., Berrêdo-Pinho M., Amaral J.J., Antunes L.C.M., Melo R.C.N., Acosta C.C.D., Moura D.F., Olmo R., Han J., et al. Mycobacterium leprae intracellular survival relies on cholesterol accumulation in infected macrophages: a potential target for new drugs for leprosy treatment. Cell. Microbiol. 2014;16:797–815. doi: 10.1111/cmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H., Badralmaa Y., Yang J., Lempicki R., Hazen A., Natarajan V. Retinoic acid and liver X receptor agonist synergistically inhibit HIV infection in CD4+ T cells by up-regulating ABCA1-mediated cholesterol efflux. Lipids Health Dis. 2012;11:69. doi: 10.1186/1476-511X-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reduced Mortality among Children in Southern India Receiving a Small Weekly Dose of Vitamin A — NEJM. doi: 10.1056/NEJM199010043231401. [DOI] [PubMed] [Google Scholar]
- 62.Wang T.-T., Nestel F.P., Bourdeau V., Nagai Y., Wang Q., Liao J., Tavera-Mendoza L., Lin R., Hanrahan J.W., Mader S., et al. Cutting edge: 1, 25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. Baltim. MD. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 63.Gombart A.F., Saito T., Koeffler H.P. Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics. 2009;10:321. doi: 10.1186/1471-2164-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dimitrov V., White J.H. Species-specific regulation of innate immunity by vitamin D signaling. J. Steroid Biochem. Mol. Biol. 2015 doi: 10.1016/j.jsbmb.2015.09.016. doi:10.1016/j.jsbmb.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 65.Vansant G., Reynolds W.F. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8229–8233. doi: 10.1073/pnas.92.18.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.