Abstract
Enhancer regions and transcription start sites of estrogen-target regulated genes are connected by means of Estrogen Receptor long-range chromatin interactions. Yet, the complete molecular mechanisms controlling the transcriptional output of engaged enhancers and subsequent activation of coding genes remain elusive. Here, we report that CTCF binding to enhancer RNAs is enriched when breast cancer cells are stimulated with estrogen. CTCF binding to enhancer regions results in modulation of estrogen-induced gene transcription by preventing Estrogen Receptor chromatin binding and by hindering the formation of additional enhancer-promoter ER looping. Furthermore, the depletion of CTCF facilitates the expression of target genes associated with cell division and increases the rate of breast cancer cell proliferation. We have also uncovered a genomic network connecting loci enriched in cell cycle regulator genes to nuclear lamina that mediates the CTCF function. The nuclear lamina and chromatin interactions are regulated by estrogen-ER. We have observed that the chromatin loops formed when cells are treated with estrogen establish contacts with the nuclear lamina. Once there, the portion of CTCF associated with the nuclear lamina interacts with enhancer regions, limiting the formation of ER loops and the induction of genes present in the loop. Collectively, our results reveal an important, unanticipated interplay between CTCF and nuclear lamina to control the transcription of ER target genes, which has great implications in the rate of growth of breast cancer cells.
INTRODUCTION
Estrogen-induced gene transcription is regulated by cis-regulatory regions containing sets of DNA-binding proteins (1). The interaction of these proteins with DNA is time-specific and usually is orchestrated by estrogen. Moreover, these regulatory regions are located hundreds of kilobases away from transcription start sites (TSS) of the target gene (2). Cohesin and condensin have recently been shown to positively regulate transcription by modulating enhancer function and enhancer–promoter looping (3,4). Yet, the role of other architectural proteins involved in nuclear organization, such as the transcriptional regulator CCCTC-binding factor (CTCF), is unexplored. CTCF is an ubiquitous multivalent zinc finger protein that regulates higher-order chromatin organization (5). A large number of studies have focused on the mechanism underlying the different functions achieved by CTCF in genome biology. Most of the research supports the idea that CTCF is able to participate in insulation and various aspects of transcriptional regulation by mediating long-range interactions between two or more genomic loci. Other functions such as RNA pausing regulation and alternative splicing have also have been proposed (6,7). Interestingly, CTCF is frequently altered in ER positive breast cancers (8,9). Yet, the molecular role of CTCF in the regulation of estrogen-induced transcription and the consequences of its alterations are poorly understood.
Previous advances in sequencing technology have facilitated the genome-wide identification of regulatory elements based on chromatin accessibility, posttranslational histone modifications and the binding of regulatory factors. More recently, the incorporation of novel methodologies such as global run-on sequencing (GRO-seq) has allowed mapping of the position and the orientation of transcriptionally enhancer RNAs (eRNAs) at genome-wide level (1). These eRNAs are likely to have important functions in many regulated programs of gene transcription (10). Furthermore, the identification of estrogen induced eRNAs has aided the study of mechanisms underlying estrogen-ER regulation of breast cancer gene expression (1).
In this study, we focused on the role of the chromatin-binding protein CTCF in orchestrating estrogen-specific gene transcription. We observed that CTCF performs as a transcriptional repressor by interacting to chromatin regions of eRNAs and promoter to control transcriptional regulation induced by estrogen. CTCF regulates target gene expression by hindering the ER binding and the formation of ER chromatin loops induced by estrogen, thereby modulating a subset of estrogen-specific gene transcripts. Remarkably, the repressive function of CTCF is associated with its ability to impede the proliferation mediated by ER. Finally, our data showed that CTCF binding to eRNAs and promoters is facilitated by estrogen when chromatin establishes contacts with nuclear lamina.
MATERIALS AND METHODS
Cell culture
MCF-7 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were grown in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum.
Chromatin Immunoprecipitation (ChIP)
CTCF, ER and LaminB genomic regions were identified by using the cross-linking (X)-ChIP protocol as described previously (11). MCF-7 cells were grown in hormone-stripped media with 5% of charcoal-stripped serum and treated with vehicle (control) or estrogen for 0 h, 45 min or 3 h. Cells were fixed with 1% formaldehyde for 10 min and then quenched with 125 mM of glycine. Cells pellets were washed with phosphate buffered saline (PBS) and lysed with lysis buffer. DNA fragmentation was done by using Bioruptor sonicator (Diagenode) (17–20 cycles on and off 30 s). We inspected the fragmentation size of DNA after sonication tested resolution of the ChIP-Seq. Fragment length was typically 200–500 bp, visualized on a 1.5% agarose gel. Chromatin was incubated overnight at 4°C with 5 µg specific antibodies CTCF (Millipore 07–729; Lot: 2453497), ER (Santa Cruz sc543; Lot: F1215), FOXA1 (Abcam ab5089; lot GK122110-17, FOXA1 ab23738; lotGR176970-1), LaminB (Santa Cruzc-20; sc-6216; Lot F1715) and equal amounts of Protein A&G Dynal Beads (Life technologies). Western blot was performed visualizing specific bands of each antibody used for the ChIP-Seq (Additional file 1). The beads were washed and DNA was eluted by reverse crosslink overnight at 65°C. For the Re-ChIP similar protocol was used, however, after the first IP DNA was eluted in 100 μl Re-ChIP elution buffer (10 mM DTT 1% SDS) and incubated 30 min at 37° with shaking. The elution was removed from the beads and diluted 1:70 in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.0) before the second immunoprecipation (IP) was performed as a standard ChIP procedure.
The libraries for next-generation DNA sequencing (NGS) were made with Illumina TruSeq ChIP Sample Preparation Kit (IP-202-1012) where DNA of desired size was cut from gel after specific indexes had been added or Diagenode MicroPlex Library Preparation kit (C05010010 (AB-004-0012)). Each sample was run on bioanalyser before submission to NGS.
Transfection
Cells were seeded in 6 well plate to be 50% confluent upon transfection. Cells were transfected with siRNA targeting CTCF (ON-TARGET J-010319-05-0005, Thermo Fisher Scientific) and siControl Non-targeting (siNT) (AllStars, Qiagen) using Lipfectamine RNAiMax (Life technologies) to a final concentration of 40 nM.
ChIP sequencing data analyses
ChIP DNA was amplified as previously described (11). Reads generated by the genome analyzer were aligned against the human genome using Bowtie 2 software (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) with default parameters. Samtools was used to convert the SAM file into BAM and subsequently sort the BAM file. Duplicate reads were removed with samtools and the resulting file was converted into a BED file containing the aligned unique reads. Peaks were called with MACS2 with default parameters (P-value < 0.01) or MACS2 1.4.2 (P-value < 1e-4) was used to call the peaks over the MCF7 input DNA.
Moreover, sequencing data, CTCF and ER binding sites are available with GSE8510 accession number.
To determine the number of ER loops that presented CTCF signal, we considered all the ER start sites involved in loop formation and we measured the signal of CTCF (normalized by signal input) in a window of 10 Kb from the center of the ER start site. We considered ER loops with CTCF binding when in CTCF signal was positive (higher than 0). For the analysis, we considered only ER loops with a minimal distance of 20 Kb from the ER start to the ER end sites (2151 ER loops).
Real-time PCR
DNA eluted from ChIP, from 3C experiments and from cDNA were analyzed by real-time PCR (RT-PCR) using SYBR Green and specific primers. All the selected primers had annealing temperature 60° and are listed in the Supplementary Table S1. Primer-specific PCR mix were obtained by diluting Power SYBRGreen MasterMix (Applied Biosystems, cat. 4367659) 1:1 and primers to a final concentration of 100 nM. Typically, 2 μl of ChIP or 3C eluted DNA and of 1:100 cDNA dilution (1 μg of RNA retrotranscribed) were added to each well. For each experiment, samples were run in triplicates on MicroAmp plates (cat. 4346906, Applied Biosystems) for 40 reaction cycles in Applied Biosystem 7900 HT Fast RT-PCR System. The curves obtained were quantified with the software SDS 2.4 (Applied Biosystems). Inverse logarithms of ChIP Ct values were normalized on DNA content and on input from non-ChIPped material. For 3C experiments, 3C values were normalized over unligated samples and on DNA content. For expression analysis, gene-specific values were normalized over UBC control for each condition. A MIQE checklist table with details for RT-PCR is included in the Supplementary Data (Supplementary Table S2).
Motif analyses
To assess the presence of binding motifs of in the CTCF bound regions, we looked for overrepresented motifs using ‘findMotifsGenome.pl’ part of the HOMER package (v4.7) (12).
RNA isolation, quality control and RNA-seq analysis
After siCTCF transfection, cells were isolated and the culture medium was removed after the centrifugation of the cell suspension. Total RNA was isolated with the total RNA isolation kit according to the manufacturer's protocol (Qiagen RNeasy kit). RNA yield was assessed spectrophotometrically (NanoDrop 2000). Total RNA was isolated with the total RNA isolation kit (RNeasy, Qiagen) according to the manufacturer's protocol. RNA yield was assessed spectrophotometrically (NanoDrop 2000). A total of 500 ng of RNA were used for cDNA transcription with SuperScript III (Life technologies) and random primers. cDNA was analyzed by RT-PCR with gene-specific primers and SYBR Green. Quantification and quality control for RNA-seq libraries were performed by Agilent Bioanalyzer (RNA nano 6000, Agilent). RNA libraries for RNA-seq were prepared according to Illumina protocol and paired-end sequencing was performed at Norwegian Sequencing Centre.
The paired-end RNAseq data were aligned by using tophat. The gene expression values (FPKM) were calculated by using cufflinks. The differential expression values and plots were generated by using Cuffmerge and CummeRbund. RNA sequencing is available with GSE85108 accession number.
GRO-seq analysis
The GRO-seq data for respective time points (0 min, 40 min and 150 min) were downloaded from Hah et al. (1). With the help of liftover tool we converted the genomic coordinates from Hg18 to Hg19. For this analysis, first we categorized CTCF binding sites in to three different categories based on their genomic location: Promoter (±5 kb gene start), gene body (gene start +5 kb to gene end, including 3′) and rest as intergenic. The percentage of GRO-seq reads overlapping with genomic distribution (promoter, gene body, intergenic) of CTCF binding sites were calculated for respective time points. The R and python scripts used in the analysis are available at bit bucket (https://bitbucket.org/yogita_sharma/ctcf).
Metacore pathway analysis
Significantly differentially regulated identified genes were uploaded to MetaCore from GeneGo Inc for process (www.genego.com), network and pathway map analysis.
Cell proliferation assay
Cells were seeded at equal confluence and the effect of CTCF depletion on cell proliferation were measured by live cell imaging Incucyte (Essen BioScience).
Chromatin fractionation
MCF-7 cells were grown in the same manner as described in the previous sections. The medium was removed and the cells were scraped in cold PBS containing proteinase and phosphatase inhibitors (Thermo Scientific) and subsequently pelleted by centrifugation. Each cell pellet was incubated for 10 min in 200 μl of cold buffer A (10 mM Hepes [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease and phosphatase inhibitors (Thermo Scientific) supplemented with 0,1% Triton X-100. The samples were subjected to low-speed centrifugation (4 min, 1300 g, 4°C) to collect the nuclei, which were subsequently washed in 200 μl of buffer A. The pelleted nuclei were lysed in 200 μl of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease and phosphatase inhibitors (Thermo Scientific)) and kept on ice 30 min. After three washes in buffer B, the chromatin pellets were resuspended in 200 μl of buffer B and sonicated for 30 s (Bioruptor, Diagenode) to shear DNA. Chromatin was pelleted by centrifugation (5 min, 16 000 g, 4°C), and the pellets were resuspended in buffer B supplemented with 4x loading dye (Life Technologies GmbH).
Western blots and IP
Cells were lysed (50 mM Tris-HCl, pH7.4; 100 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1 mM NaF; 0.1% SDS; 0.5% Sodium Deoxycholate; 1% Triton-X-100; 2 mM Sodium Orthovanadate; Protease Inhibitor Cocktail (Thermofisher)). Protein levels were quantified by BSA assay (BSA Assay, Thermo fisher). Protein lysate was resolved using precast SDS-PAGE gels, and transferred to PVDF membrane. Blots were blocked and incubated overnight at 4°C with primary antibodies. Blots were incubated with an HRP-conjugated secondary antibody and developed using ECL Western blotting substrate. Antibodies used were: ERα (sc-543) from Santa Cruz Biotechnologies, β-Actin (4970S) from Cell signaling, CTCF (07-729 Millipore) and histone H3 (ab1791) from Abcam.
Mass spectrometry and data analysis
Proteins were dissolved in 50 μl 6 M urea and 100 mM ammonium bicarbonate, pH 7.8. For reduction and alkylation of cysteines, 2.5 μl of 200 mM DTT in 100 mM Tris-HCl, pH 8 was added and the samples were incubated at 37°C for 1 h followed by addition of 7.5 μl 200 mM iodoacetamide for 1 h at room temperature in the dark. The alkylation reaction was quenched by adding 10 μl 200 mM DTT at 37°C for 1 h. Subsequently, the proteins were digested with 2 μg trypsin for 16 h at 37°C. The digestion was stopped by adding 5 μl 50% formic acid and the generated peptides were purified using a Strata C18-E SPE column (Phenomenex, Værløse, Denmark), and dried using a Speed Vac concentrator (Savant, Holbrook, NY, USA). The dried peptides were dissolved in 10 μl of aqueous 2% acetonitrile containing 1% formic acid. Five microliters of sample was injected into a Dionex Ultimate 3000 nano-UHPLC system (Sunnyvale, CA, USA) coupled online to an LTQ-Orbitrap XL mass spectrometer (ThermoScientific, Bremen, Germany) equipped with a nano electrospray ion source. For liquid chromatography, aqueous 2% acetonitrile containing 0.1% formic acid was used a solvent A and solvent B contained 0.1% formic acid in 90% acetonitrile. Peptides were separated on an Acclaim PepMap 100 column (C18, 3 μm particle diameter, 100 Å pore size, 75 μm internal diameter, 50 cm length) at a flow rate of 0.3 μl/min using solvent gradients of 7% B to 35% B in 40 min, to 35% B to 50% B (3 min) and finally to 80% B in 2 min. An ion spray voltage of 1.7 kV, tube lens voltage of 120 V and transfer capillary temperature of 200°C were used. The mass spectrometer was operated in data-dependent acquisition mode and precursor ion survey scans (400 m/z to 2000 m/z) were acquired in the orbitrap analyzer with resolution R = 60 000 at 400 m/z (after accumulation to a target of 500 000 charges in the LTQ). The fragmentation spectra were acquired in the linear ion trap using collision induced dissociation at a target value of 10 000 charges (normalized collision energy 35%, activation time 30 ms, activation Q 0.25, isolation width 2) for the top seven most abundant ions with a dynamic exclusion time of 60 s.
The raw data were searched against the SwissProt human database (11/2013, 20 279 entries) using Mascot search engine (Matrix Science, London, UK, version: 2.4.0). The following search parameters were used: enzyme: trypsin, maximum missed cleavage sites: 1, precursor ion mass tolerance: 10 ppm, fragment ion tolerance: 0.6 Da, variable modifications: oxidation on methionine and acetylation at N-terminus. Peptide and protein identifications were further validated with Scaffold (version 4.4.4, Proteome Software Inc, Portland, OR, USA). Peptide identifications were accepted if they could be established at greater than 95% probability by the Peptide Prophet algorithm with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99% probability and contained at least two identified peptides. The Protein Prophet algorithm assigned protein probabilities.
Chromosome Conformation Capture (3C)
Job Dekker's protocol was followed with minor modifications (13). For each condition, one 15 cm dish of MCF-7 cells was transfected with siNT and siCTCF. After 72 h of hormone depletion, when they reached 70–80% confluence, cells were treated with Estrogen 100 nM for 0 h, 45 min or 3 h and crosslinked for 10 min in DMEM 1% formaldehyde. After quenching with glycine 125 mM, cells were scraped as previously described and each cell pellet was resuspended in 5 ml of lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, Complete proteinase inhibitor Roche). Nuclei were isolated by centrifugation at 500 g for 5 min and subsequently incubated for 1 h at 37° in 0.5 ml of 1.2x restriction enzyme buffer with 0.3% SDS. Triton-X-100 was added to the samples at a final concentration of 2% and samples were incubated for one more hour at 37°. After this, each sample was digested overnight with 400 U of the specific restriction enzyme (BglII for P2RY2 region, New England Biolabs). After restriction enzyme inactivation at 65°, the digested nuclei were resuspended in 7 ml of ligation buffer supplemented with 1% triton-X-100. After 1-h incubation at 37°, 800 U of T4 ligase (New England Biolabs) were added to each sample and samples were ligated for 4 h at 16° followed by 30 min at room temperature. Samples were reverse crosslinked and proteins were digested by Proteinase K (Life Technologies) overnight at 65°. After RNA digestion by RNAse A (Life Technologies), DNA was isolated by Phenol Chloroform extraction followed by Ethanol precipitation. DNA was quantified by Qubit and analyzed by RT-PCR Syber Green using primers specific for each genomic region.
RESULTS
Estrogen influences CTCF chromatin interactions in a time specific fashion
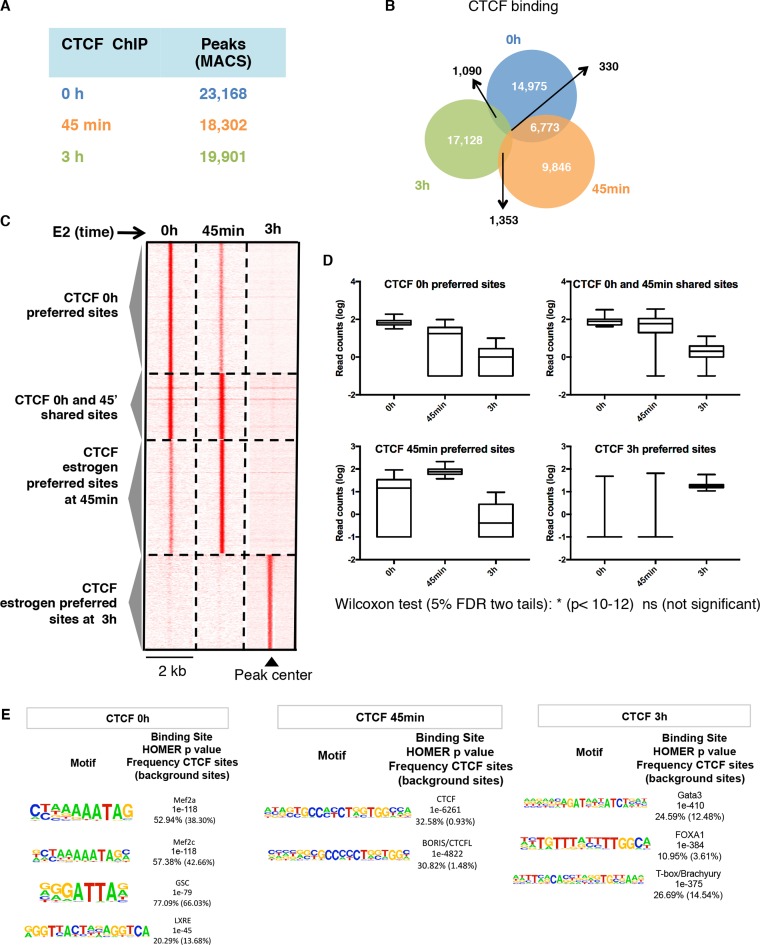
In order to determine whether estrogen might influence CTCF chromatin interactions we performed CTCF ChIP-sequencing in breast cancer cells responsive to Estrogen (MCF-7) upon different stimulation time points (0 h, 45 min and 3 h). CTCF chromatin binding sites were called using MACS (14) by considering unique sequencing reads. Differences in CTCF chromatin-binding regions were identified in cells treated with estrogen (18,302 in 45 min and 19,901 in 3 h) compared to non-treated cells (23,168 in 0 h) (Figure 1A; Additional file 2A). The comparison of CTCF binding between treated and non-treated cells revealed that estrogen influences CTCF by facilitating time-specific chromatin interactions (Figure 1B–D). In order to characterize time-specific CTCF interacting proteins, we performed de novo motif analysis of the regions bound by CTCF upon time-specific estrogen stimulations. We found a significant enrichment of CTCF and BORIS motifs at 45 min, whilst FOXA1 and GATA3 motifs were enriched at CTCF bound regions induced at 3 h (Figure 1E). The chromatin immunoprecipitation of FOXA1 followed by CTCF confirmed that FOXA1 mediates the CTCF chromatin interaction at 3 h of estrogen stimulation (Additional file 2B). All together, these results suggested that estrogen regulated CTCF binding occurs by direct contacts with chromatin or through ER-associated proteins such FOXA1.
Figure 1.
Estrogen influences CTCF chromatin binding interactions. (A) ChIP-sequencing of CTCF in MCF-7 cells treated with vehicle (0 h) or estrogen for 45 min or 3 h. (B) Overlap of CTCF binding events among different time of estrogen treatment. (C) Heatmap showing the signal intensity of CTCF chromatin regions with preferred binding at 0 h, 45 min and 3 h of estrogen stimulation. The heatmap of the overlapping regions is also shown. (D) Boxplots showing changes in CTCF signals for sites identified with estrogen treatment. (E) Top-enriched transcription factor motifs identified by de novo motif discovery at CTCF preferred binding at 0 h, 45 min or 3 h of estrogen stimulation.
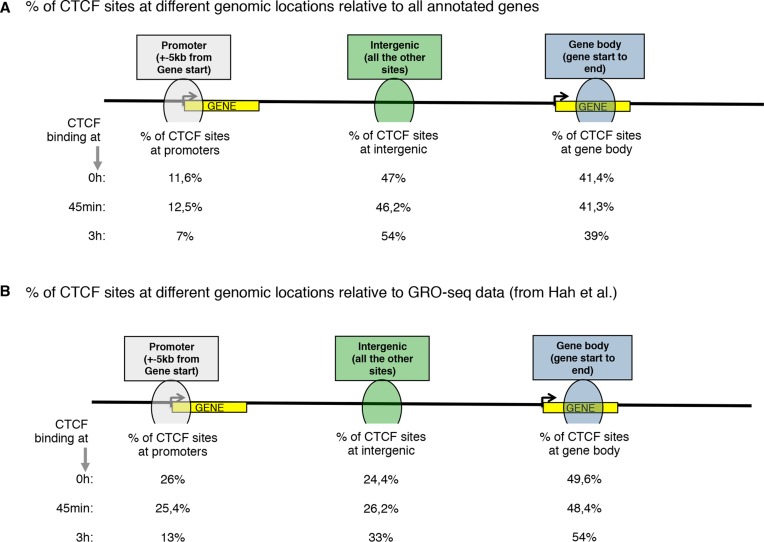
Next, we determined the distribution of estrogen-regulated CTCF chromatin interactions at genome-wide level. To facilitate this analysis we first classified CTCF binding sites according to their distance from annotated gene transcripts (i.e. those at promoter, gene body and intergenic). We observed that around 50% of the binding sites were localized at intergenic regions (Figure 2A). To get a better understanding of the mechanism of estrogen-influenced CTCF binding, we compared global run-on experiment followed by High throughput sequencing (GRO-seq) from MCF7 cells stimulated with estrogen at different time points. For that analysis we retrieved an existing GRO-seq data set in MCF-7 cells stimulated with estrogen for different time points (1). Then, we analyzed the binding of CTCF at eRNAs, promoter and gene transcript detected from GRO-seq analyses. We found that around 30% of CTCF binding sites overlapped with estrogen-induced transcripts in any of the genomic regions analyzed (data not shown). We focused on these overlapping regions and we observed a higher enrichment for CTCF binding at 45 min of estrogen stimulation to the promoter regions of early estrogen up-regulated genes (25.4%; Figure 2B) compared to non-estrogen regulated genes (12.5%; Figure 2A). We also detected enrichment of CTCF binding to the promoter of estrogen down-regulated genes or up-regulated at later time points (Figure 2A and B). Finally, we also observed a gain of CTCF binding at gene bodies of genes that were regulated at both early and late time points, although this enrichment was less clear than that for the early-regulated set of genes (Figure 2A and B).
Figure 2.
Estrogen-regulated CTCF binding correlates with estrogen-regulated gene transcripts and enhancer RNAs. Relative overlap of CTCF within (A) the total of transcripts (defined from UCSD genes) or (B) estrogen regulated transcripts (defined from GRO-seq (10)). The genomic regions were defined as: gene promoter (defined as ±5 kilobases from gene start), gene body (defined from +5 kilobases from gene start to gene end) and intergenic regions (defined from −5 kilobases from gene start to gene end).
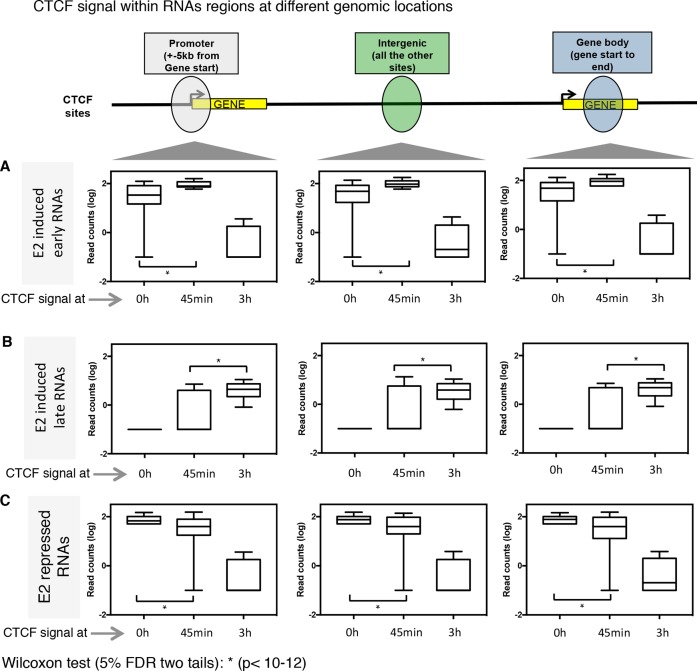
Next, we quantified the CTCF binding intensity at the overlapping regions with estrogen regulated eRNAs, promoter regions and gene transcripts across the different time points. The analysis showed a positive correlation between CTCF signal upon 0 h and 45 min of estrogen stimulation at both eRNAs and promoters/gene bodies of estrogen-induced genes and their expression at the same time points (Figure 3A). Interestingly, a significant increase of CTCF binding was detected at 45 min of estrogen stimulation at all the GRO-seq regions analyzed (Figure 3A). Consistent with these findings, we also observed that the gain of CTCF binding at 3 h resulted in reciprocal increase in estrogen-dependent transcription at a similar time point (Figure 3B). Finally, we could identify a similar correlation for estrogen down-regulated transcripts with CTCF binding sites at 0 h (Figure 3C).
Figure 3.
Time specific estrogen-induced CTCF binding correlates positively with time specific estrogen-induced transcripts. Box plots showing changes in CTCF signals from genomic regions as defined in Fig. 2 of (A) early estrogen-induced transcripts (40 min), (B) late estrogen-induced transcripts (150 min) or (C) estrogen-repressed transcripts (0 h).
To better understand how CTCF chromatin interactions may relate to estrogen receptor (ER) function, we analyzed the binding of ER in MCF-7 cells stimulated with estrogen at different time points (0 h, 45 min and 3 h) in GRO-seq regions containing CTCF binding and compared with GRO-seq regions without CTCF binding. Previously, we reported that FOXA1 is a transcription factor determinant for ER functionality (15). Hence, we also examined the binding of FOXA1 at these GRO-seq regions (i.e. with or without CTCF binding). Upon estrogen treatment for 45 min, we observed an increase of ER binding at both GRO-seq regions. However, ER binding was significantly increased at GRO-seq non-overlapping with CTCF. Interestingly, the binding of FOXA1 was significantly reduced CTCF and GRO-seq overlapping regions when cells were stimulated with estrogen at 45 min, whereas it was significantly increased at GRO-seq non-CTCF overlapping (Additional file 3A). Then, we investigated the enrichment of histone mark associated with active transcription (H3K4me1 and H3K36me3) at these GRO-seq genomic regions (i.e. with or without CTCF binding)(Additional file 3B). The binding of CTCF correlated negatively with the levels of histone modifications at GRO-seq and CTCF overlapping regions. By contrast, we detected a positive correlation for non-CTCF regions. All together, these results suggested that CTCF binding induced by 45 min of estrogen stimulation to chromatin regions associated with repression and that CTCF might play an important role in the control of estrogen-induced gene transcription.
CTCF binding prevents ER chromatin interaction and loop formation
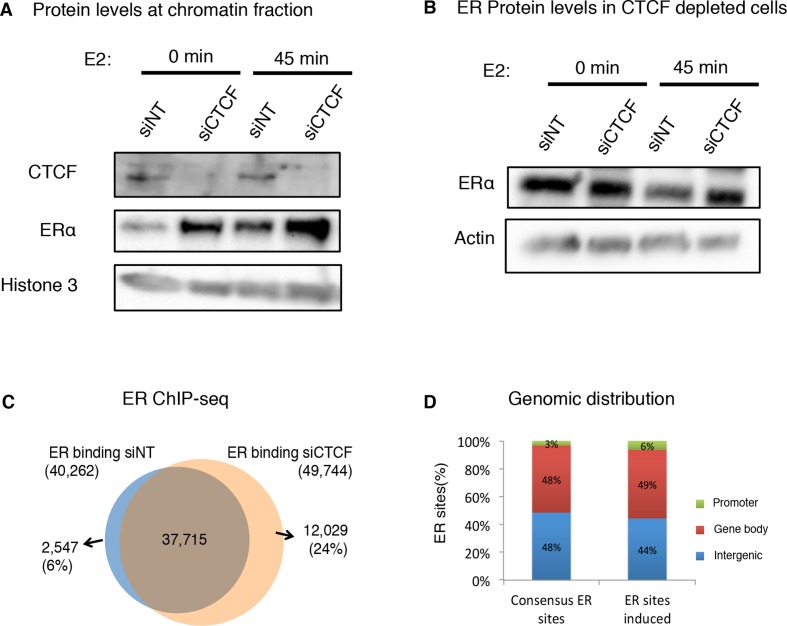
Previously, Hah et al. reported that about 50% of ER binding regions were in fact overlapping with eRNAs identified at intergenic regions (1). Interestingly, our results suggest that ER binding is significantly reduced at GRO-seq (including eRNAs) sites where CTCF binds. To investigate the effect of CTCF on ER binding to chromatin, we transfected MCF-7 cells with control small inhibitory RNA (siControl) or a siRNA against CTCF (siCTCF). After transfection, cells were hormone-deprived and consequently treated or non-treated with estrogen for 45 min. Subsequently, protein from the chromatin fraction was blotted against ER. The global binding of ER to chromatin was increased in CTCF depleted cells, while CTCF depletion did not make any significant effect on ER protein expression (Figure 4A and B). We validated this finding by silencing CTCF and analyzing the ER chromatin interaction by ChIP-sequencing (Figure 4C). Our results revealed that CTCF depletion resulted in a gain of 24% of novel ER binding regions, whereas almost all the ER interactions identified in CTCF expressing cells were still conserved (Figure 4C). The genomic distribution of these novel ER sites revealed a gain at promoter and introns compared to non-CTCF affected ER binding sites (Figure 4D).
Figure 4.
CTCF constrains global ER chromatin binding and ER chromatin looping. (A) MCF-7 cells were transfected with siControl or siCTCF, treated with vehicle (0 h) or estrogen for 45 min (E2) and were fractionated to enrich the chromatin fraction, which was protein blotted with CTCF and ER antibodies. Histone H3 was used as a control. (B) ER levels were tested from protein extracts of MCF-7 cells transfected with siControl or siCTCF, treated with vehicle (0 h) or estrogen for 45 min (E2). Actin was used as a loading control. (C) Overlap of ER binding events induced by estrogen (45 min) and analyzed by ChIP-sequencing of ER in MCF-7 cells transfected with siNT or siCTCF. (D) Genomic distribution of ER consensus or ER induced sites.
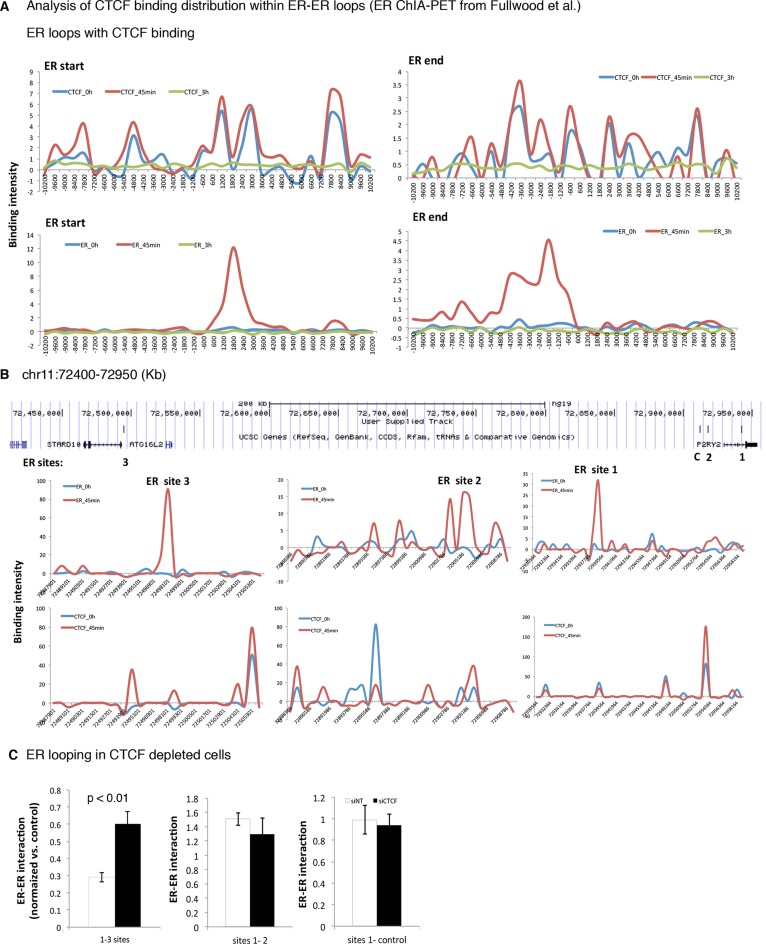
ER plays an important role in the establishment of intra-chromosomal interactions. In fact, ER chromatin loops induced by estrogen at 45 min correlate positively with estrogen-dependent target gene activation (16). To find out whether CTCF could be involved in estrogen-induced ER loops, we retrieved an existing ER ChIA-PET data set (16), which maps all the ER–ER chromatin loops induced by estrogen at 45 min. We assayed for the ER loops induced by estrogen and analyzed the fraction of ER loops containing CTCF binding. We mapped ER and CTCF binding in MCF-7 cells non-treated or treated with estrogen at 45 min and 3 h. The analysis showed that around 52% of the ER chromatin loops were associated with CTCF binding (Figure 5A, Additional file 4A). CTCF binding was detected around the estrogen-ER induced loops at 45 min. Interestingly, similar to ER, CTCF binding correlated with estrogen treatment, which suggested a positive correlation between CTCF and ER binding to chromatin (Figure 5A). Therefore, we hypothesized that CTCF could influence ER loop formation. To test the hypothesis, we investigated the effect of CTCF depletion on ER binding and ER-loop formation on the estrogen-induced gene P2RY2, which has been previously well characterized (16). A representative genome browser image of aforementioned ChIP-seq results is shown for P2RY2 locus (Figure 5A). Two ER–ER loops have been described to establish contact with P2RY2 gene: ER loop between ER site 1 and ER site2 (∼37 Kb) and ER loop between ER site 1 and ER site 3 (∼400 Kb). The CTCF map at this genomic region reveal the presence of at least two CTCF sites enriched with estrogen at 45 min, which were flanking the ER site 3. CTCF regions nearby ER sites 1 and 2 were also identified but these CTCF sites were not flanking ER binding sites. To appreciate the effect of CTCF depletion, we transfected MCF-7 cells with Control or CTCF targeting siRNA and stimulated with estrogen for 45 min. Next, we tested ER binding and ER–ER loop formation at the previously characterized sites. Chromosome conformation capture (3C)-PCR showed that ER site1-ER site 3 chromatin loop was significantly induced in CTCF depleted MCF-7 cells. On the contrary, the interactions between ER site1 and ER site 2 and a negative control region were not influenced (Figure 5C). Interestingly, an increase of ER binding at the ER looping site 3 was also observed upon CTCF silencing (Additional file 4B), suggesting that CTCF hinders the formation of precise ER–ER loops by reducing ER chromatin binding. Hence, these results indicate that CTCF limits gene loop formation and possibly the transcription of these genes.
Figure 5.
Estrogen-induced CTCF binding with chromatin at 45 min influences ER chromatin loop assembly. (A) Average signal intensity of ER and CTCF binding events of ER chromatin loops with CTCF overlapping binding regions. (B) CTCF binding intensity in non-stimulated cells (0 h, blue) or stimulated with estrogen (45 min, red) is plotted at ER sites 1 and 2 involved in ER looping formation of P2RY2 gene. RNA polymerase II binding intensity in stimulated cells with estrogen (45 min, green) is plotted at ER site 1. (C) MCF-7 cells were transfected with siControl or siCTCF, treated with vehicle (0 h) or estrogen for 45 min (E2) and ER ChIP and 3C was performed followed by RT-PCR of ER binding events already identified to be involved in ER loop formation for P2RY2 gene. The data are the mean of three independent replicates ± s.d.
CTCF modulates transcription of cell cycle associated genes and buffers cell growth
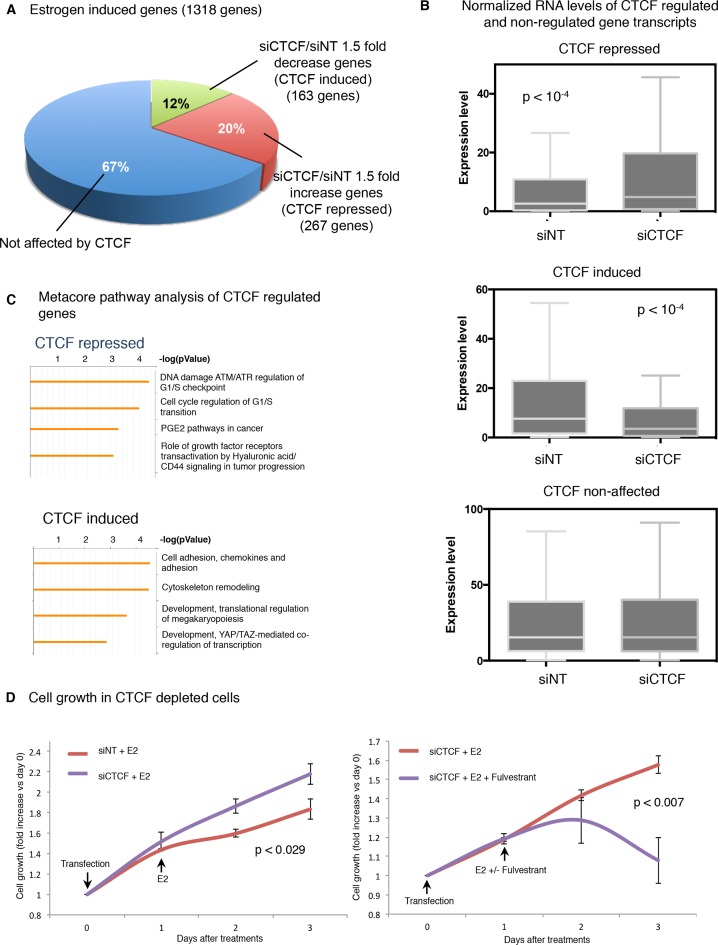
We hypothesized that CTCF recruitment to ER-regulated genomic regions upon estrogen treatment might reflect a buffering mechanism to dampen the expression of target genes. To confirm this hypothesis we assessed the effects of CTCF depletion on global estrogen regulated gene transcription. We transfected hormone-deprived MCF-7 cells with siControl or siCTCF and treated them with vehicle or estrogen. We performed RNA-sequencing (RNA-seq) and examined the impact of CTCF on estrogen-regulated genes. Strikingly, our analysis showed that around 32% of estrogen-induced gene transcripts were influenced by CTCF depletion. The major fraction of these transcripts was down regulated by at least 1.5-fold change, whereas the other smaller fraction of genes was up regulated (Figure 6A and B; Additional file 5A and B). These results suggested that CTCF represses the expression of a substantial number of estrogen-induced genes. We validated these findings by increasing CTCF protein in MCF-7 cells and determining the expression by RT-PCR of a subset of estrogen-induced genes that we identified to be up regulated in CTCF depleted cells. The overexpression of CTCF inhibited the transcription in all the genes tested (Additional file 6A and B). To determine the functional significance of CTCF regulated-gene transcription, we performed Metacore Pathway analysis of CTCF-regulated estrogen-induced genes. The top significant pathways associated with CTCF down regulated-genes included G1/S checkpoint and cell cycle regulation of G1/S transition (Figure 6C; Additional file 6C). Furthermore, CTCF silencing resulted in a significant increase of cell proliferation mediated by ER (Figure 6D)(P < 0.03). All together, these findings suggested that the primary function of CTCF in the control of ER-mediated proliferation is to modulate the expression of gene transcripts key for cell division.
Figure 6.
CTCF modulates the expression of a subset of estrogen-induced genes. (A) MCF-7 cells were transfected with siControl or siCTCF, treated with estrogen and RNA was sequenced. We used 1318 estrogen-induced genes at 40 min and published previously (1) to calculate expression levels for each gene induced by estrogen in cells depleted or expressing CTCF (RKPM). Then we measured the fold change (FC) expression (siCTCF/siNT) and determined the fraction of genes with an 1.5 FC increase or decrease. (B) Box plot of normalized RNA levels of CTCF regulated genes (with at least 1.5 FC) or non-regulated genes. (C) Metacore pathway analysis of CTCF repressed or CTCF induced genes. (D) In siControl or siCTCF transfected MCF-7 cells, cell growth was measured when cells were stimulated with E2 (100 nM) and/or with Fulvestrant (1 μM). The data are the mean of six independent replicates ± s.d.
Nuclear Lamina mediates CTCF repressive function
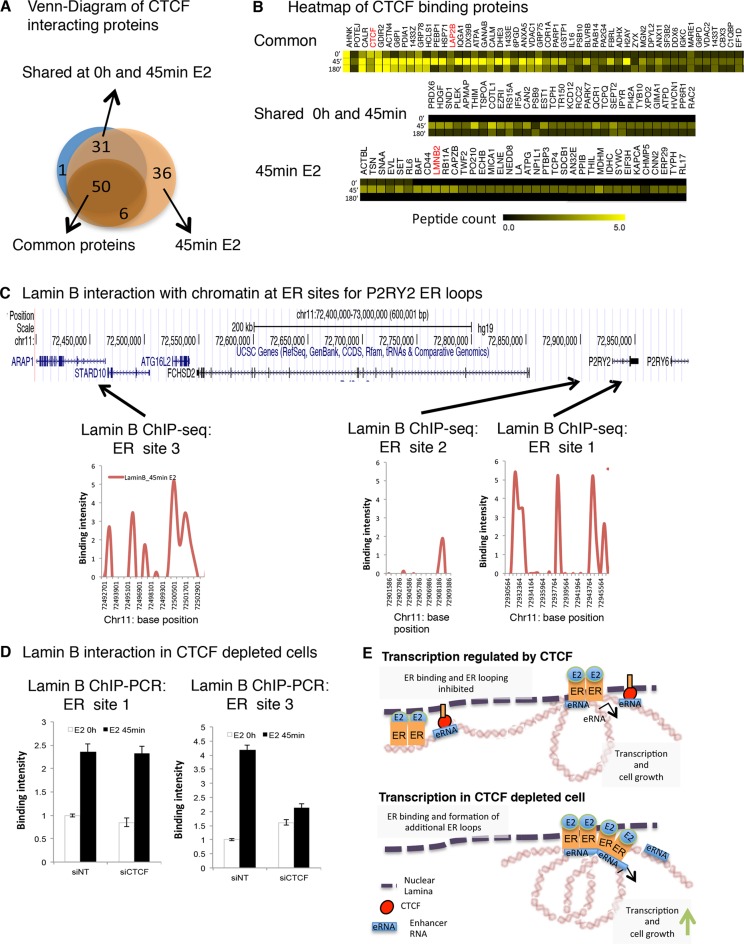
Next, we investigated the mechanism by which CTCF interacts with the chromatin upon estrogen treatment. We used proteomic approaches to identify regulatory targets of CTCF specific for breast cancer cells. For that, we performed CTCF immunoprecipitation of chromatin enriched protein fraction coupled with mass spectrometry as previously described (17) by means of Rapid Immunoprecipitation of Endogenous proteins method (RIME). This method uses formaldehyde cross-linked cell extracts, which maintain transient protein-protein interactions. Then, CTCF protein is immunoprecipitated and pulled down proteins are identified by mass spectrophotometry. We performed CTCF RIME in MCF-7 cells hormone depleted and treated with vehicle (0 h) or estrogen (45 min and 3 h) (Figure 7A). Proteins associated with nuclear lamina were identified among the CTCF–binding targets (Figure 7B). These partners were validated by CTCF immunoprecipitation and Western blot (Additional file 7A). Previously, nuclear lamina has been linked to transcription regulation by organizing chromatin into active and inactive domains (18,19). Moreover, nuclear lamina is known to interact with CTCF to tether chromatin regions into sub-nuclear periphery to modulate transcription (20). To get insight into the role of nuclear lamina in the control of CTCF binding to the chromatin, we characterized the Lamin B genomic interactions within the defined ER–ER chromatin loops associated with CTCF binding. For that, we mapped Lamin B interaction to the chromatin by ChIP-seq in MCF-7 cells treated with estrogen at 0 h, 45 min and 3 h. Then, we quantified Lamin B interaction within ER-chromatin loops containing CTCF binding (Additional file 7B). This analysis showed that the nuclear lamina established contacts with the ER chromatin loops in cells stimulated with estrogen for 45 min. Interestingly, we found a positive correlation between nuclear lamina and CTCF binding at these chromatin interaction. To appreciate the role of CTCF in nuclear lamina contacts with ER chromatin loops, we tested Lamin B chromatin interactions at the ER sites involved in the loop formation around P2RY2 gene upon CTCF silencing. We observed that ER site 1 and 3 were associated with nuclear lamina when cells were stimulated with estrogen at 45 min (Figure 7C). The depletion of CTCF resulted in the loss of contact between nuclear lamina and the ER binding site 3, which is a ER distal site involved in the formation of looping between ER site1 and ER site 3. On the contrary, the contact between nuclear lamina and the ER binding site 1 was unaffected (Figure 7D). These results, together with the finding that ER-chromatin looping between ER sites 1 and 3 was facilitated in CTCF depleted cells (Figure 5c), indicate that the positioning of ER loops to the nuclear lamina might be a mechanism by which CTCF might control the transcription of ER target genes. Interestingly, our results also show that ER site 2 is not associated with nuclear lamina (Figure 7C), which supports the hypothesis that the formation of ER loop between site 1 and 2 is not impeded by CTCF.
Figure 7.
Nuclear lamina facilitates CTCF chromatin interaction. (A) CTCF RIME was conducted in MCF-7 cells. Venn Diagram of CTCF interacting proteins in cells stimulated with vehicle (0 h) or estrogen at 45 min or 3 h. (B) Nuclear lamina associated proteins and Lamin B were identified as CTCF binding protein. The results include proteins that were identified in our CTCF RIME samples but not in in any of the IgG control RIME. (C) Signal intensity of Lamin B at ER sites for P2RY2 ER loops. (D) Lamin B interaction by ChIP-PCR in cells transfected with siCTCF or siNT oligonucleotides. ER sites 3 and 1 for P2RY2 ER loop were analyzed. (E) model showing plausible mechanism of interaction of CTCF with eRNAs to modulate the expression of estrogen induced genes.
All together, our data suggest that following the chromatin–nuclear lamina contact, CTCF associated with nuclear lamina binds to selective genomic regions around estrogen-induced genes as a mechanism to hinder the formation of additional ER loops and therefore modulate gene transcription. By contrast, in CTCF depleted cells, the binding of ER to chromatin and ER chromatin loop is facilitated and consequently gene transcription and cell growth is amplified (Figure 7E).
DISCUSSION
Our study reveals an important, unanticipated interplay between CTCF and nuclear lamina that control the transcription of ER target genes. Here, we show that CTCF plays an active role moderating gene transcription induced by estrogen through its direct interaction with transcriptionally active chromatin-regions. Importantly, we demonstrate that estrogen-ER establishes contacts with chromatin and nuclear lamina, which might facilitate the mRNA transport of cell cycle regulated genes to the cytoplasm and their subsequent translation. However, as this happens, the fraction of CTCF associated with the nuclear lamina establishes contacts with transcriptionally active chromatin domains, limiting the formation of ER loops. By doing so, the expression of estrogen-induced genes is modulated and this results in the moderation of breast cancer cell growth.
CTCF establishes contacts to enhancer chromatin regions to disturb ER function
Genome-wide analyses of ER chromatin-interactions have pointed out that most of the ER binding occurs at regions distal from ER target genes (21). More recent reports have shown that long-range chromatin interactions are induced by estrogen treatment and are essential for the tight regulation of ER-mediated transcription (16). Yet, the mechanisms contributing to modulate ER chromatin loop formation and its control of transcription remained unknown. Now, we have demonstrated that CTCF contributes to control the estrogen-induced transcription by establishing direct interaction with transcriptionally active chromatin regions (Figures 1–3, Additional file 2–3). Importantly, these chromatin interactions occur simultaneously to ER binding with the chromatin (Figures 4 and 5A). The results of this study also demonstrate that the depletion of CTCF is able to increase ER-chromatin interaction at the start and end of the loop containing the P2RY2 gene locus (Figure 5C). Therefore, CTCF impinges in the three-dimensional loop conformation that is needed to induce the transcription of P2RY2. Interestingly, our study has revealed that CTCF binds to half of the ER chromatin loops, which suggests that CTCF might function as a common negative regulator of ER looping formation. Other factors involved in chromatin organization, such as Cohesin, play a central role in regulating long-range interactions and transcription. While Cohesin seems to be generally involved in transcriptional activation (3), CTCF seems to block enhancer function when placed between enhancers and promoters. Recently, Sanborn et al. (22) have proposed the existence of chromatin domains, which are flanked by CTCF and cohesin. However, Sanborn's study is not in disagreement with the conclusions of our study. We propose that ER loops hindered by CTCF might occur within these chromatin domain described in Sanborn's study. In other words, ER loops might be formed inside larger CTCF loops. Then, one might hypothesize that CTCF plays different roles in the control of transcription by: (i) maintaining the structure of chromatin domains and (ii) regulating the formation of ER loops and therefore modulating the expression of ER target genes. Interestingly, considering that the average length of these chromatin domains might be of 185 kb and that ER loops with CTCF binding span around 68 kb (data not shown), both events might be occurring at the same time and do not need to exclude each other.
Previously, it has been reported that a substantial fraction of long-range chromatin interactions between enhancers and promoters are not impeded by the presence of CTCF binding sites (5). Indeed, we did also observe that the expression of a subset of estrogen-induced genes is induced by CTCF that suggests that in some instances CTCF may also favor enhancer–promoter interactions of a subset of estrogen-induced genes.
Other previous reports also showed a correlation between CTCF and ER regions and those regions are not strongly influenced by estrogen (15,23). Our Pearson correlation (Additional file 2) analysis revealed that CTCF chromatin interactions are stable but also demonstrate that selective CTCF chromatin interactions might be facilitated by estrogen (Figures 1–3). In addition, we also plotted the CTCF data set of that study toward that subset of ER chromatin loop. Importantly, we also detected a substantial enrichment of CTCF signal with estrogen when compared with vehicle (data not shown).
CTCF associated with nuclear matrix buffers cell growth by controlling estrogen induced gene transcription
Our study has also elucidated the mechanism underlying CTCF repressive effects on ER-induced genes. Here, we demonstrate that the fraction of CTCF associated with nuclear lamina, establishes contacts with enhancer chromatin regions and TSS when ER-loops establish contacts with nuclear lamina (Figure 7, Additional file 7). These findings led us to establish the hypothesis that CTCF-nuclear lamina complex play an active role in the control of transcription of ER target genes.
Interestingly, CTCF has been shown to physically interact with RNA polymerase (Pol) II and to recruit it to specific CTCF binding sites genome wide (24). This interaction between CTCF and RNA Pol II has been associated with transcriptional pausing (7). Our proteomic experiments reveal that CTCF binds with RNA pol2-associated proteins (Figure 7B), which is in agreement with previous publications. Together, these results suggest that CTCF binds to chromatin with different proteins, such as lamina components or RNA Pol2, to control estrogen induced gene transcription.
In this study we show that CTCF repressed genes are involved in cell cycle progression and that CTCF moderates the cell growth of ER positive cells (Figures 4–6, Additional file 4–6), suggesting that CTCF behaves as a tumor suppressor in ER positive tumors. There are several evidences that support this statement. First, it has been reported that a subset of luminal breast cancers carry mutations in the sequence of CTCF (25) and that most of these mutations detected in breast tumors are affecting the structure of the zinc-finger domain and therefore the binding to the DNA (26). CTCF mutations were mainly identified in Luminal A, B and HER2 enriched tumors, positive for ER. Moreover, in sporadic breast cancer, 43% of tumors exhibited cytoplasmic CTCF expression (27). Importantly, CTCF gene is located at the 16q chromosome arm and the loss of the chromosomal material at 16q is the most frequent genetic event in invasive and in situ (LCIS) lobular carcinoma of the breast (28). Finally, Ctcf hemizygous knockout mice are markedly susceptible to spontaneous, radiation- and chemically-induced cancer in a broad range of tissues. Ctcf± tumors are characterized by increased aggressiveness, including invasion, metastatic dissemination and mixed epithelial/mesenchymal differentiation (29). These evidences indicate that CTCF is a frequent target that breast cancer cells mutate or inactivate in order to prevail and outgrow normal cells. Notably, in this study, we highlight its importance for the definition of the chromatin landscape of breast cancer cells and we provide mechanistic cues to explain how any genomic alteration impacting CTCF function can affect proliferation of breast cancer cells. Therefore, CTCF emerges as a new player in the intricate network of transcription factors regulating gene expression during breast cancer progression.
Supplementary Material
Acknowledgments
The authors acknowledge support from University of Oslo, Research Council of Norway and Helse Sør-Øst authority. S.S was supported by Sciencia Fellow EU program and S.V.S. is supported by Norwegian Cancer Society. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the ‘Functional Genomics’ and ‘Infrastructure’ programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Author's contributions: E.F., Y.S. and A.H. designed all the experiments. E.F., S.G., S.W., S.V.S. and M.R.K. performed the experiments. The bioinformatics analyses were performed by Y.S., E.F. and S.S.S. A.U. and I.G.M. performed the motif analyses. The manuscript design and writing was performed by A.H. with assistance of E.F., Y.S. and S.G. The rest of authors have read the manuscript and agreed with the content of it.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Sciencia Fellow EU program [to S.S]; Norwegian Cancer Society [to S.V.S.]. Funding for open access charge: Norwegian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin C.Y., Vega V.B., Thomsen J.S., Zhang T., Kong S.L., Xie M., Chiu K.P., Lipovich L., Barnett D.H., Stossi F., et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt D., Schwalie P.C., Ross-Innes C.S., Hurtado A., Brown G.D., Carroll J.S., Flicek P., Odom D.T. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W., Hu Y., Oh S., Ma Q., Merkurjev D., Song X., Zhou X., Liu Z., Tanasa B., He X., et al. Condensin I and II complexes license full estrogen receptor alpha-dependent enhancer activation. Mol. Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong C.T., Corces V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong C.T., Corces V.G. Insulators as mediators of intra- and inter-chromosomal interactions: a common evolutionary theme. J. Biol. 2009;8:73. doi: 10.1186/jbiol165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D., Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Kandoth C., Reimand J., Lawrence M.S., Getz G., Bader G.D., Ding L., Lopez-Bigas N. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilfillan S., Fiorito E., Hurtado A. Functional genomic methods to study estrogen receptor activity. J. Mammary Gland Biol. Neoplasia. 2012;17:147–153. doi: 10.1007/s10911-012-9254-4. [DOI] [PubMed] [Google Scholar]
- 12.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker J. The three ‘C’ s of chromosome conformation capture: Controls, controls, controls. Nat. Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed H., D'Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L., et al. Endogenous purification reveals G.R.EB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H., Sifakis E.G., Sumida N., Millan-Arino L., Scholz B.A., Svensson J.P., Chen X., Ronnegren A.L., Mallet de Lima C.D., Varnoosfaderani F.S., et al. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol. Cell. 2015;59:984–997. doi: 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F., et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papantonis A., Cook P.R. Genome architecture and the role of transcription. Curr. Opin. Cell Biol. 2010;22:271–276. doi: 10.1016/j.ceb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J., et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross-Innes C.S., Brown G.D., Carroll J.S. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics. 2011;12:593. doi: 10.1186/1471-2164-12-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernukhin I., Shamsuddin S., Kang S.Y., Bergstrom R., Kwon Y.W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong X., Yang H., Zhao S., Shyr Y., Li B. Network-based stratification analysis of 13 major cancer types using mutations in panels of cancer genes. BMC Genomics. 2015;16(Suppl. 7):S7. doi: 10.1186/1471-2164-16-S7-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippova G.N., Qi C.F., Ulmer J.E., Moore J.M., Ward M.D., Hu Y.J., Loukinov D.I., Pugacheva E.M., Klenova E.M., Grundy P.E., et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 27.Butcher D.T., Rodenhiser D.I. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur. J. Cancer. 2007;43:210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Green A.R., Krivinskas S., Young P., Rakha E.A., Paish E.C., Powe D.G., Ellis I.O. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res. Treat. 2009;113:59–66. doi: 10.1007/s10549-008-9905-8. [DOI] [PubMed] [Google Scholar]
- 29.Kemp C.J., Moore J.M., Moser R., Bernard B., Teater M., Smith L.E., Rabaia N.A., Gurley K.E., Guinney J., Busch S.E., et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.