Abstract
Centromeres consist of DNA repeats in many eukaryotes. Non-allelic homologous recombination (HR) between them can result in gross chromosomal rearrangements (GCRs). In fission yeast, Rad51 suppresses isochromosome formation that occurs between inverted repeats in the centromere. However, how the HR enzyme prevents homology-mediated GCRs remains unclear. Here, we provide evidence that Rad51 with the aid of the Swi/Snf-type motor protein Rad54 promotes non-crossover recombination between centromere repeats to prevent isochromosome formation. Mutations in Rad51 and Rad54 epistatically increased the rates of isochromosome formation and chromosome loss. In sharp contrast, these mutations decreased gene conversion between inverted repeats in the centromere. Remarkably, analysis of recombinant DNAs revealed that rad51 and rad54 increase the proportion of crossovers. In the absence of Rad51, deletion of the structure-specific endonuclease Mus81 decreased both crossovers and isochromosomes, while the cdc27/pol32-D1 mutation, which impairs break-induced replication, did not. We propose that Rad51 and Rad54 promote non-crossover recombination between centromere repeats on the same chromatid, thereby suppressing crossover between non-allelic repeats on sister chromatids that leads to chromosomal rearrangements. Furthermore, we found that Rad51 and Rad54 are required for gene silencing in centromeres, suggesting that HR also plays a role in the structure and function of centromeres.
INTRODUCTION
Repetitive DNA elements are prevalent in eukaryote genomes. In humans, repetitive elements including LINEs, SINEs, microsatellites, minisatellites, rDNA, telomere and centromere repeats account for half of the genome (1,2). Homologous recombination (HR) is considered to be a pathway that accurately repairs DNA damage such as DNA double-strand breaks (DSBs) because it uses intact DNA strands as templates. However, non-allelic HR between repetitive elements at different chromosomal loci gives rise to a risk of gross chromosome rearrangements (GCRs) such as deletions, duplications, inversions and translocations (3,4). An increasing body of evidence indicates that GCRs mediated by non-allelic HR lead to numerous types of genetic disease in humans (5,6).
Centromeres consist of specific DNA repeats in many eukaryotes (7). Centromeres provide a platform for the assembly of kinetochores, to which spindle microtubules attach during mitosis and meiosis. Despite their important role in the accurate segregation of chromosomes, DNA sequences of centromeres are highly variable between species (8,9). Furthermore, spontaneous GCRs occur in centromeres. Deletion of centromere repeats was observed when dicentric chromosomes were changed to monocentric chromosomes (10,11). Exchange of entire short arms between acrocentric chromosomes, known as Robertsonian translocations, are the most common translocations observed in humans (1/1000 individuals) (12). Such translocations between the same chromosomes produce isochromosomes, whose arms are mirror images of one another. Copy number variations of genes by isochromosomes can lead to genetic disorders such as Down syndrome (13,14). In the fission yeast S. pombe and in the pathogenic fungus C. albicans, isochromosomes are formed by recombination between inverted repeats in the centromere region (15–18).
There are several HR pathways, although the pathway choice mechanism is not fully understood (19). During the initial step of HR, stretches of single-stranded DNA (ssDNA) are produced at DSB ends or stalled replication forks. ssDNA invades homologous double-stranded DNA to form displacement loops (D-loops). D-loop migration accompanied by extensive leading strand synthesis occurs during break-induced replication (BIR), resulting in nonreciprocal transfer of large regions of chromosomes. A subunit of DNA polymerase δ (Polδ): budding yeast Pol32, fission yeast Cdc27 and human POLD3 is required for BIR (17,20–22). Disruption of D-loops after DNA synthesis from the 3′ end of invading strands leads to synthesis-dependent strand annealing (SDSA), which generates noncrossover (NCO) but not crossover (CO) products. The second end capture by D-loops creates double Holliday junctions (HJs). Dissolution of double HJs by helicases and topoisomerases forms only NCO products, whereas resolution of joint molecules (JMs) including D-loops and HJs by structure-specific endonucleases generates either CO or NCO products depending on the manner in which JMs are cleaved. Among the endonucleases that cleave JMs, the Mus81-Eme1 complex mainly generates CO products (23–26). CO and BIR at non-allelic genomic loci give rise to chromosomal rearrangements.
Rad51 forms nucleoprotein filaments on ssDNA and catalyzes strand exchange to produce D-loops. Loading of Rad51 onto RPA-coated ssDNA is mediated by Rad52 in yeast and by BRCA2 in mammals (27–29). Rad54 is a Swi/Snf-type motor protein (30,31) that functions before and after D-loop formation. Rad54 binds and stabilizes Rad51 nucleoprotein filaments even in the absence of ATP (32,33), and facilitates Rad51-mediated D-loop formation in the presence of ATP (34). After D-loop formation, the Rad54 motor protein dissociates Rad51 from heteroduplex DNA, allowing DNA Polδ to bind and extend D-loops; Rad54 also has the ability to disrupt D-loops, which may lead to SDSA (35–38). In vivo analyses of mating type switching in budding yeast have shown that Rad54 is dispensable for the recruitment of Rad51 to a donor locus (39), suggesting that Rad54 plays an essential role only after D-loop formation. Interestingly, deletion of Rad51 in fission yeast increases the spontaneous formation of isochromosomes, which are produced by recombination between inverted repeats in the centromere region (16). It appears paradoxical that Rad51 suppresses homology-mediated GCRs as it promotes HR. It remains unclear how Rad51 suppresses isochromosome formation in centromeres. The rad51 mutation also increases chromosome loss and sensitivity to a microtubule-destabilizing drug (16,18,40), suggesting a role in centromere function.
Here, we found that deletions of Rad51 and Rad54 in fission yeast epistatically increase the formation of isochromosomes whose breakpoints are located in centromere repeats. Mutations in the Rad54 ATPase domain, rad54KA and rad54KR, did not impair the ability to bind Rad51 but accumulated spontaneous Rad54 foci and increased isochromosome formation, suggesting that Rad54 is important after D-loop formation for suppressing isochromosome formation. In contrast to their high GCR rates, rad51 and rad54 mutations decreased gene conversion between inverted repeats in the centromere. Physical analyses of the recombinants revealed that rad51 and rad54 mutations increase the ratio of COs to NCOs. Deletion of Mus81 endonuclease decreased both COs and GCRs in the rad51Δ mutant. In contrast, the cdc27-D1 mutation that impairs BIR (17) did not decrease GCRs. These data suggest that Rad51 and Rad54 promote NCO recombination between centromere repeats on the same chromatid, thereby preventing non-allelic crossing over between sister chromatids that results in isochromosome formation. We also found that rad51 and rad54 mutations increase chromosome loss and sensitivity to a microtubule-destabilizing drug as well as impair transcriptional gene silencing in centromeres. HR mediated by Rad51 and Rad54 may also play a role in the chromatin structure of centromeres.
MATERIALS AND METHODS
Genetic procedures
The fission yeast strains used in this study are listed in Supplementary Table S1. Standard genetic procedures were used as described previously (16,41). Each amino acid was added to the medium at a final concentration of 225 μg/ml, or as indicated. YNB media contained 1.7 g/l of yeast nitrogen base (Difco 233520, BD Biosciences), 5 g/l of ammonium sulfate, 2% glucose. 5-FOA media are YNB media supplemented with 1 mg/ml of 5-fluoroorotic acid (Wako) and 56 μg/ml of uracil. rad54KA and rad54KR mutants were created by the pop-in/pop-out gene replacement procedure: pTN993 and pTN992 ura4+ plasmids containing rad54KA and rad54KR, respectively, were treated with HpaI and introduced into yeast cells, and Ura+ transformants were selected on Edinburgh minimal medium (EMM) plates. The ura4+ pop-outs were selected on 5-FOA plates. Correct integration was confirmed by PCR and DNA sequencing.
Western blotting
Yeast extracts were prepared in the presence of trichloroacetic acid as described previously (42), separated by 7.0% SDS-PAGE (59:1) and transferred onto Immobilon-P transfer membranes (Millipore). To detect GFP-tagged proteins, full-length Aequorea victoria GFP polyclonal antibody (Clontech) (1:2000) and peroxidase AffiniPure goat anti-rabbit IgG (heavy+light) (Jackson ImmunoResearch Laboratories) (1:10 000) were used as the primary and secondary antibodies, respectively. The blots were developed using SuperSignal West Femto substrate (ThermoScientific) and exposed to RX-U films (Fujifilm), after which they were stained with Coomassie Brilliant Blue.
Northern blotting
From log-phase cells in EMM+U, total RNAs were extracted by heating and freezing cells in the presence of phenol and SDS (43). Six micrograms of RNAs were separated by 1.0% agarose gel electrophoresis in MOPS/formaldehyde buffer (44), and transferred onto Nytran nylon membranes (Schleicher & Schuell). A 32P-labeled DNA probe was prepared using a 1.8 kb HindIII fragment containing the full-length ura4+ gene, and used for Northern hybridization. Radioactive signals were detected using a BAS2500 phosphorimager (Fujifilm) and quantified with Image Gauge Software (Fujifilm).
Yeast two-hybrid assay
The yeast two-hybrid assay was carried out using Matchmaker Two-Hybrid System 3 (Clontech) as described previously (42). Full-length Rad51 and Rad54 open-reading frames were cloned into expression vectors.
Fluorescence microscopy
Log-phase cells were treated with 1 μg/ml of Hoechst 33342 (Dojindo) for 5 min at room temperature. Signals were detected using a fluorescence microscope (BX51, Olympus) with a 100x objective (UPLSAPO 100XO, Olympus). Images were obtained using a charge-coupled device camera (DP72, Olympus) and processed using Adobe Photoshop CS6 software.
GCRs and chromosome loss
The rates of spontaneous GCRs and chromosome loss were determined essentially as described previously (16), with some modifications. A single colony formed on YE+LUA plates (YE media supplemented with leucine, uracil and adenine) was suspended in distilled water, and cells were plated onto YE plates to visualize Ade– red colonies. At 4–6 days after plating, the number of total colonies and that of red colonies were counted. Red colonies were transferred onto EMM+UA to inspect leucine prototrophs. The number of Leu– Ade– indicative of ChL loss was obtained by subtracting Leu+ Ade– from Ade–. Leu+ Ade– clones grown on EMM+UA were replicated onto EMM+U and EMM+A to confirm Ade– and inspect uracil prototrophs, respectively. The number of Leu+ Ura– Ade– indicative of GCRs was obtained by subtracting Leu+ Ura+ Ade– from Leu+ Ade–. Cells were incubated at 30°C. The rates of GCRs and chromosome loss per generation were determined by a fluctuation test using the method of medians (45).
To measure the GCR rate using ChLC, yeast cells grown in EMM+UA were plated onto YNB+UA (YNB media supplemented with uracil and adenine) and 5-FOA+UA plates (5-FOA media supplemented with adenine). At 5–9 days after plating, colonies were counted to determine the number of Leu+ and Leu+ Ura–. Leu+ Ura– colonies were transferred from 5-FOA+UA to EMM+A and EMM+U plates to confirm Ura– and inspect adenine prototrophs, respectively. The number of Leu+ Ura– Ade– cells was obtained by subtracting Leu+ Ura– Ade+ from Leu+ Ura–.
Pulse field gel electrophoresis (PFGE) of chromosome DNA
Chromosomal DNA was prepared in agarose plugs. GCR products were analyzed by PFGE, Southern hybridization and PCR, essentially as previously described (16). PFGE was carried out using CHEF-DRII (Bio-Rad) under the following conditions. Broad-range PFGE: switching time 1800–1000 s, 2 V/cm for 45 h and then 70 s for 3 h at 10°C in 1x TAE buffer using 0.55% Certified Megabase agarose gel (Bio-Rad). Short-range PFGE: 40–70 s, 4.5 V/cm for 24 h at 10°C in 0.5x TBE buffer using 0.6% Certified Megabase agarose gel. DNA probes used for Southern hybridization were prepared using a Random primer DNA labeling kit ver.2 (Takara) and [α-32P]dCTP (PerkinElmer Life Sciences). Radioactive signals were detected using a BAS2500 phosphorimager (Fujifilm).
Recombination between ade6B and ade6X heteroalleles located in cen1
The ade6B and ade6X mutations were constructed by digestion and fill-in of the BamHI and XhoI sites, respectively. ade6B and ade6X heteroalleles were introduced into cen1 by a series of transformations (Supplementary Figure S11). After introduction of a 1.5-kb SspI fragment containing ura4+ at the HpaI site of imr1L, a 1.9-kb DraI fragment containing ade6B was introduced into the same site to replace ura4+. Another round of transformation was carried out for imr1R, using ade6X instead of ade6B. ura4+ and ade6B/X transformants were selected on EMM and 5-FOA plates, respectively. Replacement of ura4+ by ade6B/X mutant genes was carried out in the clr4Δ strain background to exclude false-positive clones that may have grown on 5-FOA plates because of gene silencing.
Yeast strains containing ade6B/X heteroalleles were grown on YE+A plates for 3–5 days. Ten milliliters of EMM+A was inoculated with a single colony formed on YE+A plate. After 1–3 days of incubation, cells were plated onto EMM+A and EMM+G (EMM supplemented with 50 μg/ml of guanine to prevent adenine uptake). After 3–5 days of incubation, colonies were counted to determine the number of colony-forming units and Ade+ recombinants. Rates of Ade+ formation were determined by a fluctuation test as described above.
To prepare yeast DNA, ≥4.0×108 cells were collected, washed with 5 ml of TE10:25 (10 mM Tris-HCl (pH 8.0), 25 mM EDTA) and resuspended in 1 ml of SP1 (20 mM sodium citrate, 20 mM di-sodium hydrogen phosphate, 40 mM EDTA (pH 5.6)). After addition of 10 μl of β-mercaptoethanol, the cell suspension was incubated at 30°C for 20 min. After centrifugation, the cells were resuspended with 0.5 ml of SP1 containing 350 μg/ml lyticase (Sigma) and incubated at 37°C for 30–60 min. Spheroplasts were collected by centrifugation and suspended with 300 μl of TE 50:20 (50 mM Tris-HCl (pH 8.0), 20 mM EDTA). After addition of 100 μl of 10% SDS, tubes were incubated at 65°C for 20 min. A total of 300 μl of 5 M KAc was added and the tubes were kept on ice for 10 min, followed by centrifugation at 17900 × g for 10 min. The supernatant was recovered, and DNA was precipitated by addition of 750 μl isopropanol. After treatment with AfeI or XhoI, DNA fragments were separated by PFGE using CHEF-DRII (Bio-Rad). The agarose gel was irradiated with 300 J UV light, treated with 1.2 M NaCl and 0.4 M NaOH buffer for 40 min, and DNA was transferred to a Nytran nylon membrane followed by Southern hybridization. A cnt1 probe was prepared using the 2.8-kb EcoRI-HindIII fragment from pKT110 (46).
Statistical analysis
The Fisher's exact test and Mann–Whitney test were performed using GraphPad Prism version 6.0g for Mac (GraphPad Software, La Jolla, CA, USA).
RESULTS
Rad51 and Rad54 suppress gross chromosomal rearrangements
Taking advantage of an extra minichromosome ChL derived from chromosome 3 (chr3), GCRs can be detected by monitoring three genetic markers (Figure 1A). Previously, we plated yeast cells onto minimum medium containing 5-fluoroorotic acid (5-FOA) to isolate Ura– clones as the first step in identifying GCR clones of Leu+ Ura– Ade– (16). Here, we used YE media and isolated Ade– red colonies as the first step to identify GCR clones (see Materials and Methods). Fluctuation analysis showed that a rad51 deletion (rad51Δ) increases the rate of spontaneous GCRs (Figure 1B), confirming the importance of Rad51 in GCR suppression.
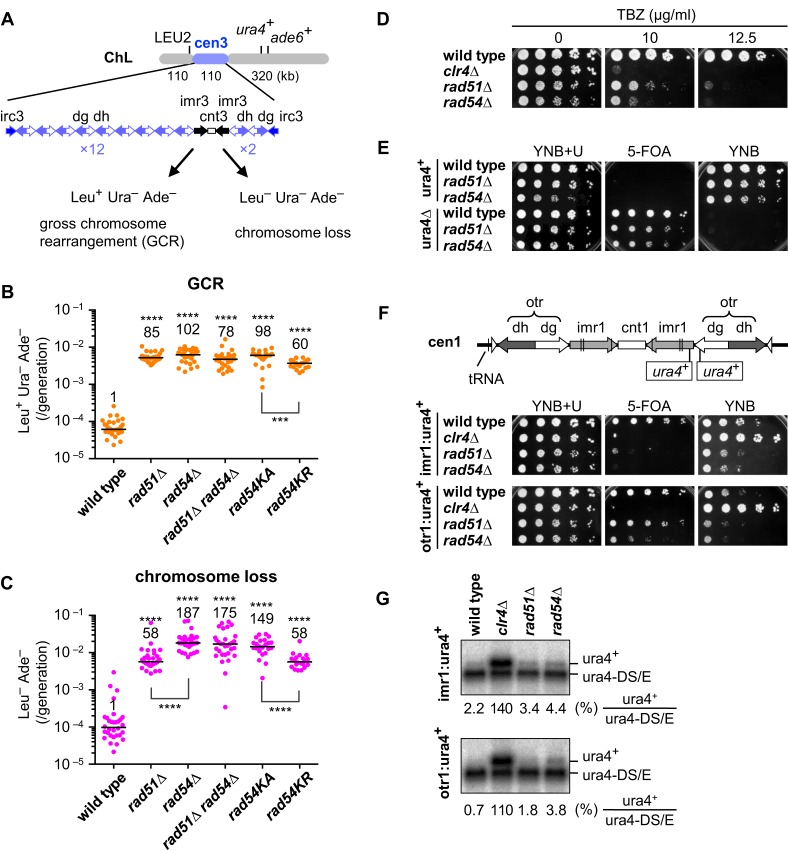
Figure 1.
Rad51 and Rad54 suppress gross chromosome rearrangements and are important for the structure and function of centromeres. (A) Illustrated are DNA repeats in centromere 3 (cen3) and three genetic markers introduced into ChL (16). GCRs associated with the loss of ura4+ and ade6+ result in Leu+ Ura– Ade–, whereas complete loss of ChL results in Leu– Ura– Ade–. Spontaneous rates of (B) GCRs and (C) chromosome loss were determined in wild-type, rad51Δ, rad54Δ, rad51Δ rad54Δ, rad54KA and rad54KR strains (TNF3896, 4034, 4048, 4942, 4599 and 4563, respectively). Independent experimental values are shown in scatter plots. Lines indicate medians. Rates relative to wild-type value are indicated at the top of each column. P-values were determined by the two-tailed Mann–Whitney test. ***P < 0.001; ****P < 0.0001. (D) Exponentially growing cells of wild-type, clr4Δ, rad51Δ and rad54 strains (TNF2399, 2803, 2621 and 3284, respectively) in EMM+U were 5-fold serially diluted with distilled water and spotted onto YE+U supplemented with thiabendazole (TBZ) at the indicated concentrations. (E) Wild-type, rad51Δ and rad54Δ cells of ura4+ (TNF35, 2610 and 3719, respectively), and those of ura4Δ (TNF2347, 2404 and 5015, respectively) were grown in EMM+U and spotted onto the indicated plates. (F) Illustrated are the integration sites of ura4+ in imr1 and otr1 (96). Wild-type, clr4Δ, rad51Δ and rad54Δ cells that contain imr1:ura4+ (TNF2399, 2803, 2621 and 3284, respectively), and those containing otr1:ura4+ (TNF2648, 2900, 2848 and 3273, respectively) grown in EMM+U were spotted onto the indicated plates. Cells were grown at 30°C. (G) Northern blot analysis of ura4+ and ura4DS/E RNAs in wild-type, clr4Δ, rad51Δ and rad54Δ cells. % of ura4+ RNAs compared to ura4-DS/E RNAs is shown below each panel.
To test whether another HR enzyme, Rad54, is also required to suppress GCRs, we deleted the rad54 gene and found that rad54Δ increases the GCR rate similarly to rad51Δ (Figure 1B). A rad51Δ rad54Δ double mutant exhibited a GCR rate similar to each single mutant, showing that Rad51 and Rad54 function in the same pathway to suppress GCRs. Substitution of a conserved lysine residue in the ATPase domain of Rad54 to alanine (rad54K300A) or arginine (rad54K300R) also increased the GCR rate, suggesting that the ATP-dependent function of Rad54 is involved in GCR suppression. rad54KR caused a mild defect compared to rad54KA. This may be because Rad54KR but not Rad54KA has residual ATPase activity (47). These results suggest that Rad51 suppresses GCRs through homologous recombination with the aid of the Rad54 motor protein.
Rad51 and Rad54 are required for the structure and function of centromeres
As observed previously (16), rad51Δ increased the rate of ChL chromosome loss (Figure 1C). We found that rad54Δ increases the loss rate to a greater extent than rad51Δ (Figure 1C). Rad54-specific functions such as chromatin remodeling (48–50), rather than the ineffective attempts of recombination by Rad51 in rad54Δ cells (51), may account for the high rate of chromosome loss, as rad51Δ rad54Δ increased the loss rate to the same level as rad54Δ. Histone H3K9 methyltransferase Clr4 is required for the heterochromatin structure in centromeres (52,53), and clr4Δ cells are hypersensitive to a microtubule-destabilizing drug, thiabendazole (TBZ) (54). Consistent with chromosome loss, a serial dilution assay showed that rad54Δ as well as rad51Δ cells are hypersensitive to TBZ, although they are less sensitive than clr4Δ cells (Figure 1D), suggesting that Rad51 and Rad54 play a role in centromere function.
Higher-order chromatin structures in centromeres repress transcription and are required for the faithful segregation of chromosomes (7,55). To determine whether Rad51 and Rad54 affect chromatin structure in centromeres, we examined transcriptional gene silencing of ura4+ integrated into centromere 1 (cen1). Before evaluating gene silencing, we confirmed that rad51Δ and rad54Δ cells form colonies on 5-FOA plates only when ura4+ was absent (Figure 1E, ura4+ and ura4Δ). Even when ura4+ was present in the inner repeat (imr1), wild-type cells formed colonies on 5-FOA because of gene silencing (Figure 1F, imr1:ura4+). As expected, the clr4Δ mutation that destroys the heterochromatin structure impaired colony formation on 5-FOA and accelerated that on YNB plates. rad51Δ and rad54Δ mutations impaired colony formation on 5-FOA, although no effects were observed on YNB plates. Because the cells that are defective in the silencing and express ura4+ are sensitive to 5-FOA, this result suggests that Rad51 and Rad54 are required for maintaining the silencing status of ura4+ in imr1. When ura4+ was present in the outer repeat (otr1), rad51Δ had no obvious effects, while rad54Δ resulted in a slight defect in gene silencing (Figure 1F, otr1:ura4+). To detect ura4+ transcripts, we prepared total RNAs from yeast cells and carried out Northern blotting (Figure 1G). In addition to ura4+ RNAs, ura4-DS/E mini RNAs transcribed from the arm region were detected as a control. rad51Δ and rad54Δ slightly increased the level of ura4+ RNAs compared to wild-type strains. Similar results were obtained in both imr1:ura4+ and otr1:ura4+ strains. Although rad54Δ exhibited slightly more severe phenotypes than rad51Δ, these data show that Rad51 and Rad54 are involved in transcriptional gene silencing in centromeres. Collectively, our data suggest that Rad51 and Rad54 are required for the structure and function of centromeres.
Rad51 and Rad54 suppress isochromosome formation in the centromere
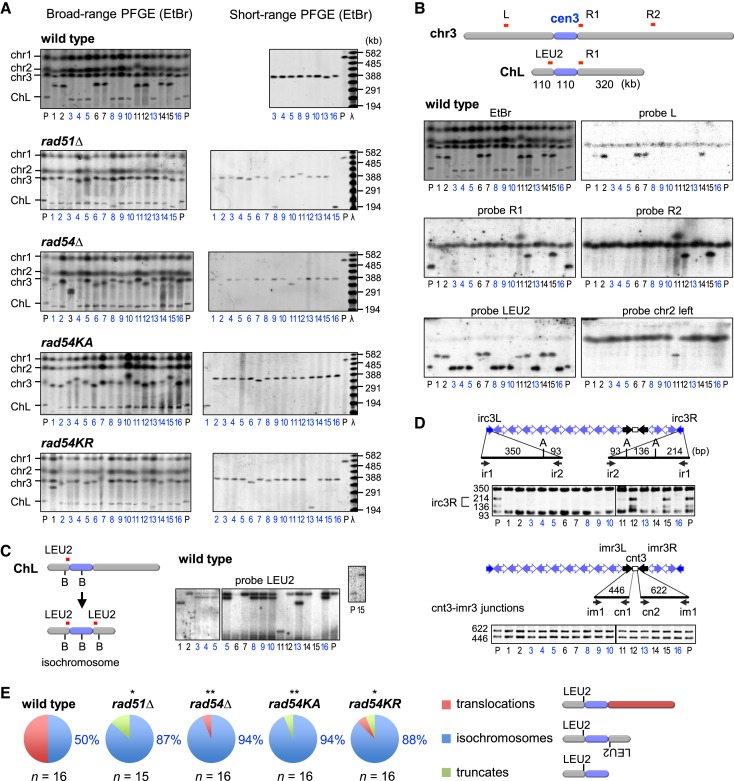
To characterize GCR products, chromosomal DNA was prepared from parental and independent GCR clones, separated by PFGE and stained with ethidium bromide (EtBr) (Figure 2A). In wild-type cells, approximately half of the GCR products were larger than the parental ChL (Figure 2A, Broad-range PFGE, clone 1, 2, 6, 7, 11, 12, 14 and 15) and were detected using probes specific to chr3 or chr2 by Southern hybridization (Figure 2B, probe L, R2 and chr2 left), indicating that they are translocations. Although all GCR products were detected using the LEU2 probe, GCR products smaller than the parental ChL do not retain the right side of cen3 (Figure 2B, probe R1) and their sizes matched those of isochromosomes (300–400 kb) (Figure 2A, Short-range PFGE). Two different sizes of restriction fragments were detected using the LEU2 probe (Figure 2C, clone 3, 4, 5, 8, 9, 10, 13 and 16), demonstrating that they are isochromosomes. PCR analysis of the GCR products recovered from agarose gel revealed that irc3R was lost while cnt3-imr3 junctions were retained (Figure 2D), indicating that breakpoints of isochromosomes are present in centromere repeats. In contrast to wild type, most GCR products formed in rad51Δ, rad54Δ, rad54KA and rad54KR mutants were isochromosomes (Figure 2A and Supplementary Figures S1 and S2), whose breakpoints are present in centromere repeats (Supplementary Figure S3). A small number of truncates of ∼200 kb and translocations of ∼2 Mb were also observed in the mutant strains. Figure 2E summarizes the distributions of the three types of GCRs detected: translocations, isochromosomes and truncates. There were no significant differences between the GCR types of rad51 and rad54 mutants. Together with the GCR rate (Figure 1B), these data demonstrate that Rad51 and Rad54 suppress the formation of isochromosomes whose breakpoints are present in centromere repeats.
Figure 2.
Rad51 and Rad54 suppress isochromosome formation in the centromere. (A) Pulse field gel electrophoresis (PFGE) analysis of gross chromosome rearrangements (GCR) products. Chromosomal DNA were separated by broad-range PFGE (left panel) and short-range PFGE (right panel) (see Materials and Methods). Sizes of λ ladders (ProMega-Markers) are indicated on the right. GCR products smaller than the parental ChL are labeled in blue. P, parental. (B) Southern blotting of wild-type GCR products. DNA was transferred to a nylon membrane and hybridized with the indicated probes. (C) Chromosomal DNA was digested with BmgBI, loaded onto a 0.6% agarose gel, separated by PFGE (switching time 1 to 6 s, 6 V/cm for 22 h) in 0.5x TBE buffer, and subjected to Southern hybridization using LEU2 probe. B, BmgBI. (D) PCR analysis of GCR products. ChLs recovered from agarose gel were used as templates for PCR. irc3L and irc3R regions were amplified, treated with ApoI and separated by standard agarose gel electrophoresis (upper panels). cnt3-imr3 junctions were also amplified (lower panels). Primer sequences are shown in Supplementary Table S2. A, ApoI. (E) Pie charts show the distribution of translocations, isochromosomes and truncates in each strain. The proportion of isochromosomes is indicated. Numbers of isochromosomes and others were compared between wild-type and mutant strains with the two-tailed Fisher's exact test. *P < 0.05; **P < 0.01.
ATPase activity of Rad54 is essential at a late step of recombination
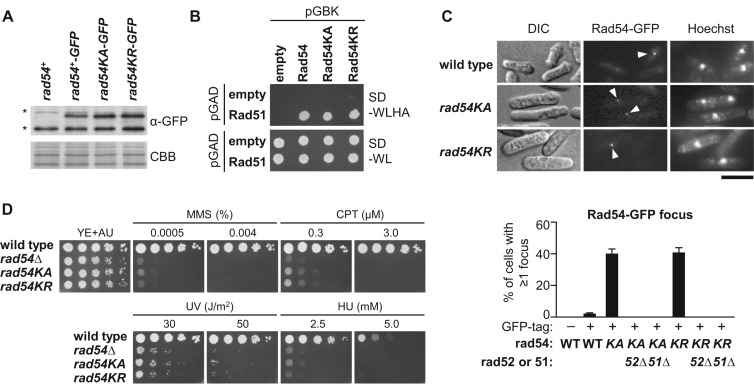
To understand the role of Rad54 ATPase activity in fission yeast, we further examined the rad54KA and rad54KR mutants. Detection of Rad54-GFP proteins by Western blotting using anti-GFP antibodies showed that neither rad54KA nor rad54KR changes the protein expression level (Figure 3A). Neither mutation affected the Rad51–Rad54 interaction detected in the yeast two-hybrid assay (Figure 3B). However, microscopic analyses revealed that rad54KA and rad54KR increase the proportion of cells exhibiting spontaneous Rad54-GFP foci (Figure 3C). Rad52 facilitates Rad51 loading onto ssDNA and remains at recombination foci even after strand invasion (27,28,56). Deletion of rad52 or rad51 eliminated Rad54KA and Rad54KR foci, despite the similar levels of Rad54-GFP expression (Figure 3C and Supplementary Figure S4), showing that focus formation is dependent on Rad52 and Rad51. rad54KA and rad54KR also increased the proportion of cells containing Rad52-GFP foci (Supplementary Figure S5A and B), and >70% of Rad54-GFP and Rad52-mCherry foci colocalized when they were co-expressed (Supplementary Figure S5C), suggesting that the Rad54 mutant proteins localize to spontaneous HR sites. rad54KA and rad54KR cells were hypersensitive to the following DNA damaging agents: methyl methanesulfonate, camptothecin and ultraviolet (UV) irradiation, and to replication stress caused by hydroxyurea (HU) treatment (Figure 3D). These data are consistent with a previous study of mouse ES cells expressing Rad54 ATPase mutant proteins (33), and suggest that the ATP-dependent activity of Rad54 at a late step of recombination is important for suppressing isochromosome formation.
Figure 3.
rad54KA and rad54KR accumulate cells containing Rad54 foci and cause hypersensitivity to DNA damage and replication stress. (A) Immunostaining of Rad54-GFP. Extracts were prepared from rad54+, rad54+-GFP, rad54KA-GFP and rad54KR-GFP cells (TNF3864, 3945, 4465 and 4489, respectively). CBB, Coomassie Brilliant Blue. Asterisks indicate non-specific bands. (B) Yeast two-hybrid interaction between Rad51 and Rad54. Budding yeast AH109 was transformed with pGBKT7 or its derivatives expressing Rad54, Rad54KA or Rad54KR (pTN997, 1005 or 1004, respectively), in combination with pGADT7 or its derivative expressing Rad51 (pTN998). Transformants grown in SD-WL media were spotted onto SD-WLHA and SD-WL plates, and incubated at 30°C for 3 days. (C) Rad54-GFP focus formation. Arrowheads indicate Rad54-GFP foci. Scale bar indicates 5 μm. Percentages of the cells containing at least one Rad54-GFP focus in rad54+, rad54+-GFP, rad54KA-GFP, rad54KA-GFP rad52Δ, rad54KA-GFP rad51Δ, rad54KR-GFP, rad54KR-GFP rad52Δ and rad54KR-GFP rad51Δ strains (TNF3864, 3945, 4465, 4410, 4409, 4489, 4413 and 4412, respectively) are shown. More than 200 cells were examined by fluorescence microscopy in each measurement. DIC, differential interference contrast. WT, wild-type. n = 3, mean ± s.d. (D) Sensitivity to DNA damage and replication stress. Exponentially growing cells of wild-type, rad54Δ, rad54KA and rad54KR strains (TNF672, 4491, 4144 and 4143, respectively) were 5-fold serially diluted with distilled water and spotted onto YE+UA supplemented with methanesulfonate (MMS), camptothecin (CPT) or hydroxyurea (HU) at the indicated concentrations. The plates were UV-irradiated with indicated doses and incubated at 30°C.
Rad51 and Rad54 promote gene conversion between DNA repeats in the centromere
Isochromosome breakpoints were present in centromere repeats (Figure 2D and Supplementary Figure S3), suggesting that recombination between centromere repeats produces isochromosomes. However, mutations in Rad51 and Rad54 recombinases greatly increased the formation of such isochromosomes. It is possible that Rad51 and Rad54 promote gene conversion between centromere repeats, a conservative mechanism of recombination, thus preventing chromosomal rearrangements. To detect gene conversion in the centromere region, we introduced the ade6B and ade6X mutant genes into the HpaI site in imr1L and imr1R repeats of cen1, respectively (Figure 4A). We chose imr1 because most of the breakpoints in rad51Δ cells were located in imr (16), and ade6+ was not transcriptionally repressed in the kinetochore region of centromeres (Supplementary Figure S6A). Gene conversion between the ade6B/X heteroalleles within the same chromatid or on sister chromatids can produce Ade+ prototrophs, but isochromosomes of chr1 are not detected because it is lethal in haploid cells. Remarkably, fluctuation analysis revealed that rad51 and rad54 mutations severely decrease the rate of spontaneous Ade+ formation (Figure 4B), showing that Rad51 and Rad54 are required for gene conversion between inverted repeats in the centromere. It should be noticed that rad54KR only partially decreased the gene conversion rate as compared to rad54KA, likely because of residual ATPase activity of Rad54KR (47). The strong inverse correlation between gene conversion (Figure 4) and GCR rates (Figure 1B) suggests that Rad51 and Rad54 promote gene conversion between centromere repeats to prevent isochromosome formation.
Figure 4.
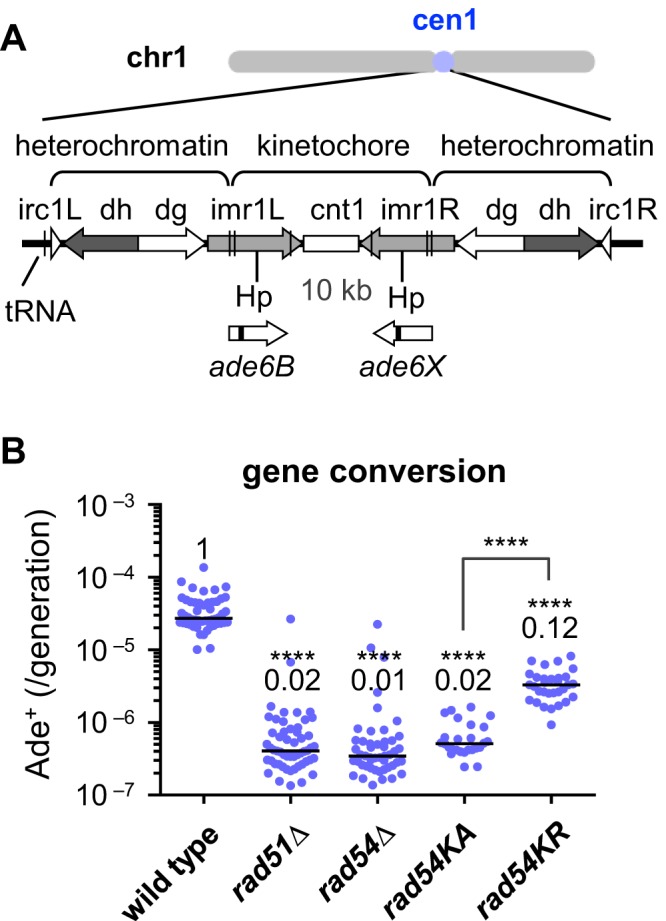
Gene conversion between inverted repeats in the centromere occurs in a Rad51- and Rad54-dependent manner. (A) ade6B and ade6X heteroalleles introduced into cen1 are illustrated. The central region (cnt1) is surrounded by a set of inverted repeats (imr1, dg, dh and irc1). Hp, HpaI. The introduction of ade6B/X heteroalleles did not change the sensitivity to a microtubule destabilizing drug, TBZ (Supplementary Figure S6B), suggesting that it does not interfere with centromere function. (B) Spontaneous rate of gene conversion between inverted repeats in the centromere region. Wild-type, rad51Δ, rad54Δ, rad54KA and rad54KR strains (TNF3144, 3257, 3286, 4299 and 4311, respectively) were grown in the presence of adenine, and subsequently Ade+ recombinants were identified on EMM plates. Cells were grown at 33°C.
Rad51 and Rad54 promote non-crossover recombination between centromere repeats
Crossing over between centromere inverted repeats on the same chromatid results in inversion of the central region, cnt1 (Figure 5A), whereas crossing over between sister chromatids may generate isochromosomes. To determine whether Rad51 and Rad54 are required for crossing over, chromosomal DNA was prepared from independent Ade+ recombinants, digested with AfeI and separated by PFGE; the fragments containing cnt1 were detected by Southern hybridization (Figure 5B). In wild-type samples, only 1 of 75 Ade+ recombinants was CO (∼1%). rad51Δ and rad54Δ strikingly increased the proportion of COs to 24% and 35%, respectively (Figure 5C and Supplementary Figure S7), indicating that Rad51 and Rad54 preferentially promote NCO recombination between centromere repeats. The increased incidence of CO by rad54KA and rad54KR (Figure 5C) shows the importance of the ATP-dependent function of Rad54 in NCO recombination.
Figure 5.
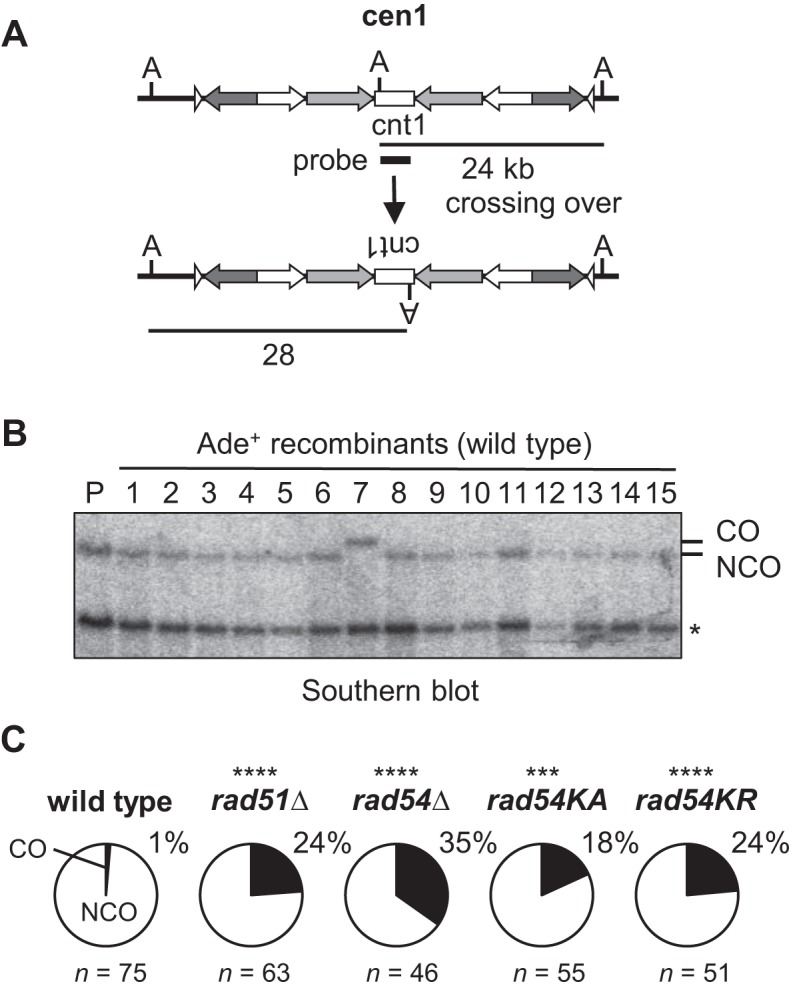
Rad51 and Rad54 suppress crossing over between centromere repeats. (A) Crossing over between inverted repeats in cen1. ade6B/X heteroalleles are omitted in the illustration for simplicity. A, AfeI. (B) Chromosomal DNA was prepared from parental and Ade+ recombinants, treated with AfeI, and applied to 0.6% agarose gel PFGE (switching time 1 to 6 s, 6 V/cm for 14 h, 10°C) in 0.5x TBE buffer. Asterisk indicates the band derived from cen3. (C) Pie charts show the proportion of COs among Ade+ recombinants examined. P-values were determined by the two-tailed Fisher's exact test.
Mus81 but not Cdc27/Pol32 is required for isochromosome formation in rad51Δ cells
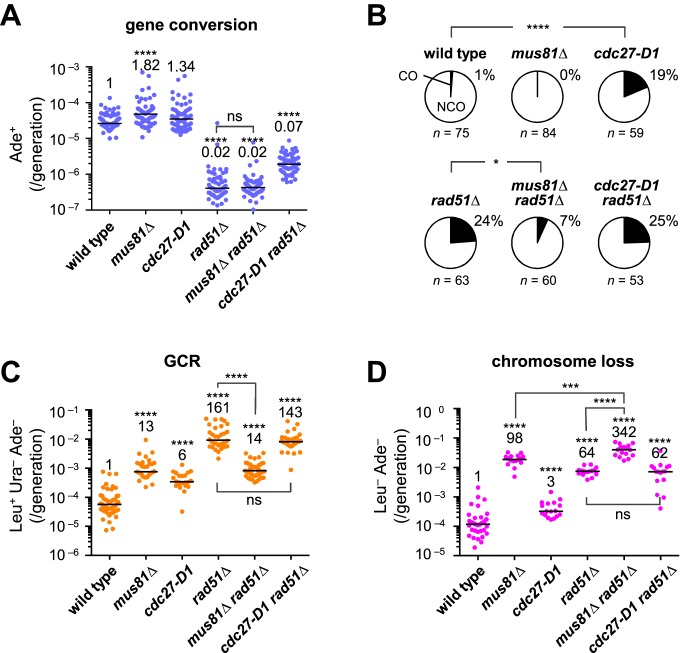
Specific resolution of JMs such as D-loops and Holliday junctions results in COs. The Mus81-Eme1 complex is a structure-specific endonuclease that generates COs (23–26). To see whether Mus81-Eme1 is required for crossing over between inverted repeats in the centromere, we disrupted Mus81, the catalytic subunit of Mus81-Eme1, measured the recombination rate (Figure 6A), and determined the proportion of COs (Figure 6B and Supplementary Figure S8) among the recombinants. In the wild-type background, it was difficult to detect the effect of mus81Δ on the proportion of COs (Figure 6B). In the rad51Δ background, however, mus81Δ did not change the recombination rate (Figure 6A), but decreased the proportion of COs from 24% to 7% (Figure 6B), demonstrating that Mus81 is required for crossing over between centromere repeats in the rad51Δ mutant.
Figure 6.
Mus81 is required for COs and GCRs in the rad51Δ mutant. (A) Rates of spontaneous gene conversion in the centromere and (B) proportion of COs among Ade+ recombinants were determined using wild-type, mus81Δ, cdc27-D1, rad51Δ, mus81Δ rad51Δ and cdc27-D1 rad51Δ strains (TNF3144, 6035, 6312, 3257, 6234 and 6335, respectively). Spontaneous rates of (C) GCRs and (D) chromosome loss were determined using wild-type, mus81Δ, cdc27-D1, rad51Δ, mus81Δ rad51Δ, and cdc27-D1 rad51Δ strains (TNF5369, 5669, 5402, 5411, 5974 and 5671, respectively) that contain ChLC minichromosome. Only the position of the ura4+ marker differs between ChLC and ChL: 10 and 170 kb from irc3R, respectively. Because the rate of chromosome loss was too high to obtain sufficient GCR clones to determine GCR rates in the mus81Δ rad51Δ mutant, GCR rates were determined using cells grown in EMM+UA media to pre-select the cells that retained the minichromosome (see Materials and Methods). Pre-selection can, at least in part, explain why the GCR rate of rad51Δ was relatively higher (161-fold increase) than that shown in Figure 1B (85-fold increase), where cells were grown in YE+LUA rather than EMM+UA. In Figure 1B, the GCR rate of the mutants that lost minichromosomes at a high rate such as rad51Δ may be slightly underestimated, as the cells that did not retain the minichromosome were included in the total count. ns, P > 0.05.
Because rad51 and rad54 mutations increase both isochromosome formation and the CO ratio, isochromosomes formed in these mutants might be produced by crossing over between centromere inverted repeats on sister chromatids. To test this possibility, we examined the effect of mus81Δ on GCRs. In the wild-type background, mus81Δ increased the GCR rate (Figure 6C), as expected from previous studies (3,57). In the rad51Δ background, however, mus81Δ reduced the GCR rate by ∼10-fold, indicating that Mus81 is required for GCRs that occur in the absence of Rad51. A mus81Δ rad51Δ double mutant exhibited an increased rate of chromosome loss compared to individual single mutants (Figure 6D), showing that Rad51 and Mus81 have non-overlapping functions to maintain chromosomes.
When GCRs are induced by DSB formation outside the centromere, isochromosomes are produced by BIR, as the cdc27-D1 mutation that eliminates a C-terminal half containing PCNA-binding motif decreased isochromosome formation (17,58). In our system, however, cdc27-D1 increased the GCR rate by 6-fold in the rad51+ background and did not change the rate in the rad51Δ background (Figure 6C). PFGE analysis showed that most GCR products formed in cdc27-D1 and cdc27-D1 rad51Δ mutants are isochromosomes (Supplementary Figure S9). Thus, BIR does not appear to be the major pathway of spontaneous isochromosome formation. It should be noted that cdc27-D1 and rad51Δ increased the proportion of COs in an epistatic manner (Figure 6B), suggesting that Cdc27 (or Polδ) and Rad51 act in the same pathway to facilitate NCO recombination (see Discussion).
DISCUSSION
Here, we found that not only Rad51 but also Rad54 suppresses GCRs in fission yeast. Detailed analyses of the GCR products revealed that rad51Δ, rad54Δ, rad54KA and rad54KR mutations increase the formation of isochromosomes with breakpoints located in centromere repeats. rad54KA and rad54KR mutants accumulated Rad54 foci, which are dependent on Rad51 and Rad52, suggesting defects in a late step of HR. In sharp contrast to GCRs, rad51 and rad54 decreased gene conversion between ade6B/X heteroalleles introduced into cen1. Strikingly, rad51 and rad54 increased the ratio of COs among the gene conversion products. Deletion of the Mus81 endonuclease in a rad51Δ background decreased both COs and GCRs, while the cdc27-D1 mutation of Cdc27/Pol32 did not. These data suggest that Rad51 and Rad54 promote NCO recombination between centromere repeats, thereby preventing crossing over of inverted repeats on sister chromatids resulting in isochromosome formation. We also found that rad51Δ and rad54Δ increase chromosome loss and TBZ sensitivity, as well as partially impair gene silencing in centromeres, suggesting that HR mediated by Rad51 and Rad54 is also required for maintaining the integrity of centromere chromatin.
It appears paradoxical that Rad51 suppresses spontaneous GCRs produced by recombination between centromere repeats (16). We found that rad51Δ and rad54Δ similarly increase the rate of isochromosome formation and that their effects are epistatic. Mutations in the ATPase domain of Rad54: rad54KA and rad54KR also increased isochromosome formation. Although rad54KA and rad54KR accumulated Rad54 foci, introduction of wild-type rad54+ into the mutant strains decreased Rad54 foci to the wild-type level and completely suppressed their defects in GCRs, chromosome loss and gene conversion (Supplementary Figure S10), demonstrating that rad54KA and rad54KR are recessive loss-of-function alleles. These data suggest that HR catalyzed by Rad51 and Rad54 is important for suppressing centromere GCRs.
How do Rad51 and Rad54 suppress isochromosome formation in the centromere? Fission yeast centromeres consist of a series of inverted repeats (imr, dg, dh and irc) surrounding the central region (cnt). We found that Rad51 and Rad54 are specifically required for gene conversion between inverted repeats in the centromere. rad51Δ and rad54Δ decreased the rate of gene conversion, but increased that of isochromosome formation. While rad54KA and rad54KR caused hypersensitivity to DNA damage to the same extent, rad54KR only partially affected gene conversion and GCR rates as compared to rad54KA, further supporting a tight correlation between gene conversion and isochromosome formation. Gene conversion and isochromosome formation are likely two alternative outcomes initiating from the same event such as pausing of the replication fork (59–61). Our data suggest that Rad51 and Rad54 suppress isochromosome formation by promoting gene conversion between centromere repeats.
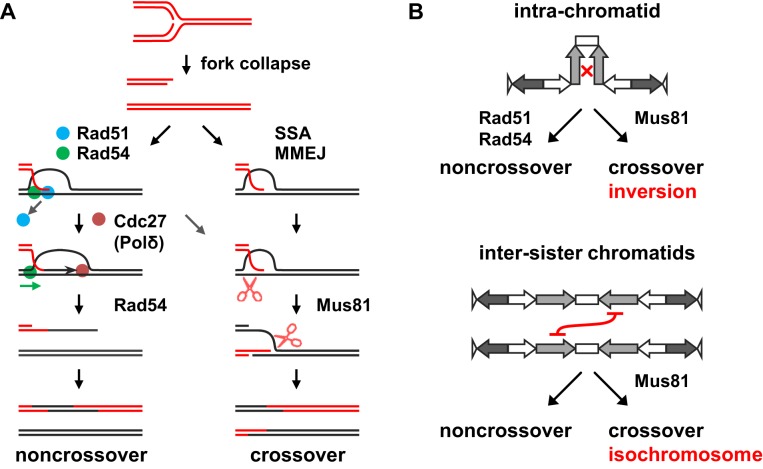
To better explain our findings, we propose models of centromere repeat recombination (Figure 7). One-end DSBs formed by replication fork collapse may initiate spontaneous recombination in centromeres (Figure 7A). After processing of DNA ends to have single-stranded tails, Rad51 with the aid of Rad54 forms D-loop structures between homologous centromere repeats (34,36). Following D-loop formation, the Rad54 motor protein releases Rad51 from heteroduplex DNA, allowing Cdc27 (or Polδ) to bind and extend the 3′ end of invading strands. After DNA synthesis, Rad54 dissociates D-loop structures to promote SDSA, which results in NCOs (Figure 7A). Crossing over between inverted repeats on the same chromatid results in ‘inversion’ of the central region (Figure 7B, intra-chromatid). Such COs were very rare and represented ∼1% of total Ade+ recombinants in the wild-type strain (Figure 5). However, rad51Δ and rad54Δ strikingly increased the proportion of COs to 20–30%, indicating that Rad51 and Rad54 preferentially promote NCO recombination (Figure 7A). The preference for NCOs appears to stem from the nature of HR mediated by Rad51 and Rad54, as an increase in the CO ratio has also been observed for rad51, rad54 and rad52 mutations in the arm regions of budding yeast (62–65). However, further comparison between the centromere and arm regions is required. In vitro analyses have shown that, in an ATP-dependent manner, Rad54 releases Rad51 from heteroduplex DNA in D-loop structures to facilitate DNA synthesis by Polδ (37,38) and Rad54 dissociate D-loop structures (33,35,36,66). These Rad54 functions on D-loop structures may be important for NCO recombination in centromeres, as we found that mutations in the ATPase domain of Rad54: rad54KA and rad54KR increase COs in the centromere (Figure 5). Cdc27/Pol32/POLD3 facilitates the DNA synthesis activity of Polδ (67–69). It is likely that Polδ with the aid of Cdc27 extends the 3′ ends of invading strands in D-loop structures to facilitate the SDSA reaction, as the cdc27-D1 mutation increased the CO ratio in an epistatic manner with rad51Δ (Figure 6). It was also observed in budding yeast that both rad51 and pol32 mutations increase half-crossovers in the repair of one-end DSBs (70,71). cdc27-D1 did not alter the total rate of recombination but changed the balance between NCOs and COs (Figure 6A and B), indicating that recombination intermediates in cdc27-D1 cells are channeled from the NCO pathway to the CO pathway (Figure 7A). It remains unclear how JMs are formed in the absence of Rad51 or Rad54 in centromeres. However, homology-mediated recombination that occurs independently of Rad51 and Rad54 has been reported, such as single-strand annealing (SSA) and microhomology-mediated end joining (MMEJ) (for review, see (72,73)) (Figure 7A). A set of proteins including Rad59, Rdh54 and Rad50 have been implicated in the Rad51-independent pathway of DSB repair in budding yeast (74). Not only Rad51 but also TLS/FUS, HLTF/Rad5, Rad52 and Rad51 paralogs: Rad51C-XRCC3 and Rad51D-XRCC2 complexes have the ability to form D-loop structures by themselves (75–80). Remarkably, we found that, in the absence of Rad51, Mus81 is required for CO recombination (Figure 6B and C), indicating that JMs formed independently of Rad51 are likely D-loop structures or nicked Holliday junctions that are good substrates for Mus81-Eme1 resolvase in vitro (23,24,81,82). It appears that Rad51-Rad54 and Mus81-Eme1 function in distinct pathways: NCO and CO recombination, respectively (Figure 7A). Consistent with this, rad51Δ and mus81Δ synergistically increased chromosome loss (Figure 6D). A synergistic increase in DNA damage sensitivity was also observed in the mus81Δ rad51Δ and the mus81Δ rad54Δ double mutants (80,83,84).
Figure 7.
Models in which Rad51 and Rad54 prevent isochromosome formation in the centromere region. (A) Recombination between centromere repeats occurs in the presence or absence of Rad51 and Rad54. Replication fork collapse creates one-end DSBs. Displacement-loop (D-loop) is produced between a single-stranded tail of a DSB and its homologous repeat sequence. In the presence of Rad51 and Rad54, synthesis-dependent strand annealing (SDSA) that results in noncrossover products may occur as follows. After the formation of D-loops, Rad54 releases Rad51 from heteroduplex DNA and recruits Cdc27 (Polδ) to catalyze DNA synthesis from the 3′ end of invading strands. After DNA synthesis, Rad54 dissociates D-loop structures, providing displaced strands that will be annealed to a single-stranded tail of a DSB formed by a converging fork. In the absence of Rad51 or Rad54, recombination intermediates between centromere repeats may be formed by single-strand annealing (SSA), microhomology-mediated end joining (MMEJ) or some other mechanisms. The intermediates are cleaved by Mus81-Eme1 endonuclease (shown as scissors), resulting crossover or half-crossover products. (B) Non-allelic HR between inverted repeats in the centromere can occur either on the same chromatid or between sister chromatids. Crossing over between inverted repeats on the same chromatid results in ‘inversion’ of the intervening region (top panel), whereas that between sister chromatids results in isochromosome formation (bottom panel). We propose that via the SDSA mechanism shown in (A), Rad51 and Rad54 promote non-crossover recombination between centromere repeats on the same chromatid, thereby preventing crossover recombination between sister chromatids that results in isochromosome formation.
In theory, in contrast to intra-chromatid recombination, crossing over between inverted repeats on sister chromatids results in isochromosome formation (Figure 7B, inter-sister chromatids). We propose that Rad51 and Rad54 promote intra-chromatid NCO recombination between centromere repeats and, in the absence of Rad51 or Rad54, intermediates are channeled to CO recombination between non-allelic repeats on sister chromatids that results in isochromosome formation. As predicted from our model, deletion of Mus81 endonuclease decreased both COs (i.e. inversions) and isochromosomes in the rad51Δ mutant, and the mus81Δ rad51Δ double mutant exhibited higher rates of chromosome loss than the single mutants (Figure 6). These data establish that Mus81-dependent crossing over is responsible for isochromosome formation in the rad51Δ background. A high incidence of GCRs and chromosome loss observed in the mus81Δ single mutant may be related to its role in replication fork progression and in faithful chromosome segregation (85,86). In contrast to isochromosomes induced by HO-endonuclease mediated DSBs outside centromeres (17), BIR does not appear to be the major pathway of spontaneous isochromosome formation. The cdc27-D1 mutation that impairs BIR (17) did not decrease spontaneous isochromosomes either in the presence or absence of Rad51 (Figure 6C). Recently, it was shown that Mus81 and converging forks reduce the likelihood of BIR occurring in S phase (87). The difference may be related to the different stages of the cell cycle: HO-induced and spontaneous GCRs occur in the G2 and S phases, respectively. Future studies should address whether residual levels of COs and GCRs observed in mus81Δ rad51Δ cells depend on BIR or on endonucleolytic cleavage by other nucleases such as Rad1-Rad10 complexes (88,89). Our study clearly shows that, rather than BIR, crossing over is the major pathway producing spontaneous isochromosomes in the absence of Rad51 (Figure 7). It appears that the high incidence of isochromosome formation in rad51 and rad54 mutants results from a combination of crossing over and inter-sister recombination. rad51 and rad54 mutations increased the GCR rate to much higher levels than cdc27-D1, while cdc27-D1 increased the CO ratio to the same level as rad51 and rad54 (Figures 1, 5 and 6), indicating that the increase in COs only partly explains the high GCR rates of rad51 and rad54 mutants. Remarkably, rad51 and rad54 but not cdc27-D1 greatly decreased gene conversion between inverted repeats in the centromere (Figures 4 and 6). Although it is unclear whether gene conversion occurs on the same chromatid or between sister chromatids in our current system, only recombination between sister chromatids can produce isochromosomes through crossing over. We propose that Rad51 and Rad54 promote intra-chromatid rather than inter-sister recombination between centromere repeats (Figure 7B).
It has been proposed that recombination intermediates formed between centromere repeats on the same chromatid affect the geometry of centromeres, which is important for centromere function (40). Not only rad51 but also rad54 mutations increased chromosome loss and sensitivity to a microtubule-destabilizing drug, TBZ (Figure 1C and D). Furthermore, rad51 and rad54 partially impaired transcriptional gene silencing in centromeres (Figure 1F and G). The silencing defect was more prominent in inner repeats (imr) compared to outer repeats (otr). Breakpoints of isochromosomes also accumulated in the imr region in the rad51Δ mutant (16). These observations are consistent with the idea that stem-loop structures are produced in the central region of the centromere where recombination intermediates provide covalent linkage of the stem (40) (Figure 7B). It has been reported that, in Candida albicans, Rad51 and Rad52 are required for the stable localization of centromere-specific histone H3 variant CENP-A at centromeres (90). Together, these data suggest that HR mediated by Rad51 and Rad54 plays a role in the structure and function of centromeres. Unexpectedly, we found that Rad54 has Rad51-independent function, as rad54Δ caused more severe defects than rad51Δ in chromosome loss, TBZ sensitivity and gene silencing in centromeres (Figure 1). Interestingly, a Rad54 paralog in budding yeast, Rdh54, localizes to kinetochores even in the absence of DNA damage. Rad54 can also localize to kinetochores in the absence of Rdh54 (50). Chromatin remodeling activity may be important for the Rad51-independent function of Rad54, as rad54KA exhibited a higher rate of chromosome loss than rad54KR and rad51Δ (Figure 1C).
Repetitive elements are prevalent in higher eukaryote genomes and used as templates for non-allelic HR (87,91). The breast cancer susceptibility gene BRCA2 involved in Rad51 loading onto ssDNA is required for gene conversion and suppresses homology-mediated GCRs (92,93). RAD54 knockdown in human cells causes isochromosome formation (94). In budding yeast, rad51Δ induces the fusion of nearby inverted repeats, resulting in dicentric and acentric chromosomes (95). Thus, the function of Rad51 and Rad54 to promote NCO recombination appears to be conserved throughout evolution and is important for safeguarding the integrity of both centromere and non-centromere regions. Our finding that Mus81 is required for chromosomal rearrangements in the mutant strain of Rad51 makes it a potential target of chemical therapy for the treatment of HR-deficient tumors.
Supplementary Material
Acknowledgments
The authors are grateful to Robin C. Allshire (The University of Edinburgh, UK), Stuart A. MacNeill (The University of St Andrews, UK), Yoshinori Watanabe (University of Tokyo) and Takeshi Sakuno (University of Tokyo) for yeast strains and plasmids, and Dayalini Weerasekara for critical comments on this manuscript. The authors also thank Yoshiki Okihara for technical assistance.
Footnotes
Present address: Rei Matsuyama, Department of Oncogene Research, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita, Osaka 565-0871, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [JP21114513, JP23570212 and JP26114711 to T.N.]. Funding for open access charge: JSPS KAKENHI [JP21114513, JP23570212 and JP26114711 to T.N.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putnam C.D., Hayes T.K., Kolodner R.D. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki M., Lange J., Keeney S. Genome destabilization by homologous recombination in the germ line. Nat. Rev. Mol. Cell Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell I.M., Gambin T., Dittwald P., Beck C.R., Shuvarikov A., Hixson P., Patel A., Gambin A., Shaw C.A., Rosenfeld J.A., et al. Human endogenous retroviral elements promote genome instability via non-allelic homologous recombination. BMC Biol. 2014;12:74. doi: 10.1186/s12915-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 7.Allshire R.C., Karpen G.H. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks. Nat. Rev. Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S., Ahmad K., Malik H.S. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 9.Plohl M., Mestrovic N., Mravinac B. Centromere identity from the DNA point of view. Chromosoma. 2014;123:313–325. doi: 10.1007/s00412-014-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stimpson K.M., Song I.Y., Jauch A., Holtgreve-Grez H., Hayden K.E., Bridger J.M., Sullivan B.A. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010;6:e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato H., Masuda F., Takayama Y., Takahashi K., Saitoh S. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr. Biol. 2012;22:658–667. doi: 10.1016/j.cub.2012.02.062. [DOI] [PubMed] [Google Scholar]
- 12.Page S.L., Shin J.C., Han J.Y., Choo K.H., Shaffer L.G. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum. Mol. Genet. 1996;5:1279–1288. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez D., Fisher E.M. Down syndrome genetics: unravelling a multifactorial disorder. Hum. Mol. Genet. 1996;5(Suppl. 1):1411–1416. doi: 10.1093/hmg/5.supplement_1.1411. [DOI] [PubMed] [Google Scholar]
- 14.Kurabayashi N., Sanada K. Increased dosage of DYRK1A and DSCR1 delays neuronal differentiation in neocortical progenitor cells. Genes Dev. 2013;27:2708–2721. doi: 10.1101/gad.226381.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmecki A., Forche A., Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura K., Okamoto A., Katou Y., Yadani C., Shitanda T., Kaweeteerawat C., Takahashi T.S., Itoh T., Shirahige K., Masukata H., et al. Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J. 2008;27:3036–3046. doi: 10.1038/emboj.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinline-Purvis H., Savory A.P., Cullen J.K., Dave A., Moss J., Bridge W.L., Marguerat S., Bahler J., Ragoussis J., Mott R., et al. Failed gene conversion leads to extensive end processing and chromosomal rearrangements in fission yeast. EMBO J. 2009;28:3400–3412. doi: 10.1038/emboj.2009.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P.C., Petreaca R.C., Jensen A., Yuan J.P., Green M.D., Forsburg S.L. Replication fork stability is essential for the maintenance of centromere integrity in the absence of heterochromatin. Cell Rep. 2013;3:638–645. doi: 10.1016/j.celrep.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Symington L.S., Rothstein R., Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lydeard J.R., Jain S., Yamaguchi M., Haber J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 21.Malkova A., Ira G. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., Halazonetis T.D. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 24.Osman F., Dixon J., Doe C.L., Whitby M.C. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 25.Ho C.K., Mazon G., Lam A.F., Symington L.S. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith G.R., Boddy M.N., Shanahan P., Russell P. Fission yeast Mus81·Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinohara A., Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 28.New J.H., Sugiyama T., Zaitseva E., Kowalczykowski S.C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 29.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazin A.V., Mazina O.M., Bugreev D.V., Rossi M.J. Rad54, the motor of homologous recombination. DNA Repair (Amst) 2010;9:286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceballos S.J., Heyer W.D. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazin A.V., Alexeev A.A., Kowalczykowski S.C. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal S., van Cappellen W.A., Guenole A., Eppink B., Linsen S.E., Meijering E., Houtsmuller A., Kanaar R., Essers J. ATP-dependent and independent functions of Rad54 in genome maintenance. J. Cell. Biol. 2011;192:735–750. doi: 10.1083/jcb.201011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petukhova G., Stratton S., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 35.Bugreev D.V., Mazina O.M., Mazin A.V. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 36.Wright W.D., Heyer W.D. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell. 2014;53:420–432. doi: 10.1016/j.molcel.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Heyer W.D. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Stith C.M., Burgers P.M., Heyer W.D. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugawara N., Wang X., Haber J.E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 40.McFarlane R.J., Humphrey T.C. A role for recombination in centromere function. Trends Genet. 2010;26:209–213. doi: 10.1016/j.tig.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Alfa C., Fantes P., Hyams J., Mcleod M., Warbrick E. Experiments with Fission Yeast. NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 42.Maki K., Inoue T., Onaka A., Hashizume H., Somete N., Kobayashi Y., Murakami S., Shigaki C., Takahashi T.S., Masukata H., et al. Abundance of prereplicative complexes (Pre-RCs) facilitates recombinational repair under replication stress in fission yeast. J. Biol. Chem. 2011;286:41701–41710. doi: 10.1074/jbc.M111.285619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Lin M., Chang C.J., Green N.S. A new method for estimating high mutation rates in cultured cells. Mutat. Res. 1996;351:105–116. doi: 10.1016/0027-5107(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petukhova G., Van Komen S., Vergano S., Klein H., Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 48.Jaskelioff M., Van Komen S., Krebs J.E., Sung P., Peterson C.L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 49.Alexeev A., Mazin A., Kowalczykowski S.C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 50.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 51.Mason J.M., Dusad K., Wright W.D., Grubb J., Budke B., Heyer W.D., Connell P.P., Weichselbaum R.R., Bishop D.K. RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells. Nucleic Acids Res. 2015;43:3180–3196. doi: 10.1093/nar/gkv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rea S., Eisenhaber F., O'Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 53.Ekwall K., Javerzat J.P., Lorentz A., Schmidt H., Cranston G., Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 54.Ekwall K., Nimmo E.R., Javerzat J.P., Borgstrom B., Egel R., Cranston G., Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell. Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 55.McKinley K.L., Cheeseman I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016;17:16–29. doi: 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyazaki T., Bressan D.A., Shinohara M., Haber J.E., Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dendouga N., Gao H., Moechars D., Janicot M., Vialard J., McGowan C.H. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka H., Ryu G.H., Seo Y.S., MacNeill S.A. Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2-Cdc24 complex to DNA polymerase δ. Nucleic Acids Res. 2004;32:6367–6377. doi: 10.1093/nar/gkh963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenfeder S.A., Newlon C.S. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 1992;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaco I., Canela A., Vera E., Blasco M.A. Centromere mitotic recombination in mammalian cells. J. Cell. Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rozenzhak S., Mejia-Ramirez E., Williams J.S., Schaffer L., Hammond J.A., Head S.R., Russell P. Rad3ATM decorates critical chromosomal domains with γH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 2010;6:e1001032. doi: 10.1371/journal.pgen.1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai Y., Symington L.S. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 63.Rattray A.J., Symington L.S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson J.A., Fink G.R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- 65.Haber J.E., Hearn M. RAD52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bugreev D.V., Hanaoka F., Mazin A.V. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 67.Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 68.Zuo S., Bermudez V., Zhang G., Kelman Z., Hurwitz J. Structure and activity associated with multiple forms of Schizosaccharomyces pombe DNA polymerase δ. J. Biol. Chem. 2000;275:5153–5162. doi: 10.1074/jbc.275.7.5153. [DOI] [PubMed] [Google Scholar]
- 69.Masuda Y., Suzuki M., Piao J., Gu Y., Tsurimoto T., Kamiya K. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35:6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith C.E., Lam A.F., Symington L.S. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase δ complex. Mol. Cell. Biol. 2009;29:1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deem A., Barker K., Vanhulle K., Downing B., Vayl A., Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179:1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sfeir A., Symington L.S. Microhomology-mediated end joining: A back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verma P., Greenberg R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016;30:1138–1154. doi: 10.1101/gad.280545.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ira G., Haber J.E. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baechtold H., Kuroda M., Sok J., Ron D., Lopez B.S., Akhmedov A.T. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 76.Kagawa W., Kurumizaka H., Ikawa S., Yokoyama S., Shibata T. Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem. 2001;276:35201–35208. doi: 10.1074/jbc.M104938200. [DOI] [PubMed] [Google Scholar]
- 77.Kurumizaka H., Ikawa S., Nakada M., Eda K., Kagawa W., Takata M., Takeda S., Yokoyama S., Shibata T. Homologous-pairing activity of the human DNA-repair proteins Xrcc3·Rad51C. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurumizaka H., Ikawa S., Nakada M., Enomoto R., Kagawa W., Kinebuchi T., Yamazoe M., Yokoyama S., Shibata T. Homologous pairing and ring and filament structure formation activities of the human Xrcc2·Rad51D complex. J. Biol. Chem. 2002;277:14315–14320. doi: 10.1074/jbc.M105719200. [DOI] [PubMed] [Google Scholar]
- 79.Burkovics P., Sebesta M., Balogh D., Haracska L., Krejci L. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 2014;42:1711–1720. doi: 10.1093/nar/gkt1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Doe C.L., Osman F., Dixon J., Whitby M.C. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 2004;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cromie G.A., Hyppa R.W., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doe C.L., Ahn J.S., Dixon J., Whitby M.C. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 83.Ii M., Brill S.J. Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr. Genet. 2005;48:213–225. doi: 10.1007/s00294-005-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghamrasni S.E., Cardoso R., Li L., Guturi K.K., Bjerregaard V.A., Liu Y., Venkatesan S., Hande M.P., Henderson J.T., Sanchez O., et al. Rad54 and Mus81 cooperation promotes DNA damage repair and restrains chromosome missegregation. Oncogene. 2016;35:4836–4845. doi: 10.1038/onc.2016.16. [DOI] [PubMed] [Google Scholar]
- 85.Fu H., Martin M.M., Regairaz M., Huang L., You Y., Lin C.M., Ryan M., Kim R., Shimura T., Pommier Y., et al. The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat. Commun. 2015;6:6746. doi: 10.1038/ncomms7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Minocherhomji S., Ying S., Bjerregaard V.A., Bursomanno S., Aleliunaite A., Wu W., Mankouri H.W., Shen H., Liu Y., Hickson I.D. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 87.Mayle R., Campbell I.M., Beck C.R., Yu Y., Wilson M., Shaw C.A., Bjergbaek L., Lupski J.R., Ira G. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazon G., Lam A.F., Ho C.K., Kupiec M., Symington L.S. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat. Struct. Mol. Biol. 2012;19:964–971. doi: 10.1038/nsmb.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazon G., Symington L.S. Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol. Cell. 2013;52:63–74. doi: 10.1016/j.molcel.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mitra S., Gomez-Raja J., Larriba G., Dubey D.D., Sanyal K. Rad51-Rad52 mediated maintenance of centromeric chromatin in Candida albicans. PLoS Genet. 2014;10:e1004344. doi: 10.1371/journal.pgen.1004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tutt A., Bertwistle D., Valentine J., Gabriel A., Swift S., Ross G., Griffin C., Thacker J., Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu V.P., Koehler M., Steinlein C., Schmid M., Hanakahi L.A., van Gool A.J., West S.C., Venkitaraman A.R. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 94.Ruiz-Herrera A., Smirnova A., Khouriauli L., Nergadze S.G., Mondello C., Giulotto E. Gene amplification in human cells knocked down for RAD54. Genome Integr. 2011;2:5. doi: 10.1186/2041-9414-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paek A.L., Kaochar S., Jones H., Elezaby A., Shanks L., Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.