Abstract
3′-End processing of pre-mRNAs prior to packaging and export to the cytoplasm of the mature transcript is a highly regulated process executed by several tens of protein factors that recognize poorly conserved RNA signals. Among them is Pcf11, a highly conserved, multidomain protein that links transcriptional elongation, 3′-end processing, and transcription termination. Here we report the structure and biochemical function of Pcf11's C-terminal domain, which is conserved from yeast to humans. We identify a novel zinc-finger fold, resembling a trillium flower. Structural, biochemical, and genetic analyses reveal a highly conserved surface that plays a critical role in both cleavage and polyadenylation. These findings provide further insight into this important protein and its multiple functional roles during cotranscriptional RNA processing.
Keywords: Pcf11, zinc finger, NMR, structure, mRNA 3′-end processing
INTRODUCTION
Prior to nucleocytoplasmic export, nascent mRNA precursors (pre-mRNAs) must undergo 3′-end processing, consisting of an endonucleolytic cleavage step followed by polyadenylation (Millevoi and Vagner 2010; Chan et al. 2011; Darmon and Lutz 2012). These reactions occur cotranscriptionally, are coupled with termination of transcription, and are highly regulated (Perales and Bentley 2009; Hsin and Manley 2012).
In all eukaryotes, 3′-end processing is orchestrated by a very complex protein machine composed of several tens of core essential and more regulatory factors, organized into stable subcomplexes with defined biochemical activity (Takagaki et al. 1989; Mandel et al. 2008). In yeast, these factors are organized into three complexes called cleavage factor IA (CF IA), cleavage factor II (CF II), and polyadenylation factor I (PF I). The structural basis for the activities of many individual proteins has been characterized (Noble et al. 2005, 2007; Paulson and Tong 2012; Xiang et al. 2014), but how these activities are integrated into the complete processing apparatus and how different steps are orchestrated and coordinated remain unclear at the molecular level.
Pcf11 is particularly interesting in this regard. While most proteins participate in defined steps in processing (e.g., cleavage or recognition of RNA-specific signals), Pcf11, one of the four subunits of CF IA, is a highly conserved protein that plays critical roles in both 3′-end processing and transcription termination, and has also been reported to have a conserved role in linking termination of transcription to the transfer of the mRNA to the export machinery (Amrani et al. 1997; Gross and Moore 2001; Johnson et al. 2009). Saccharomyces cerevisiae Pcf11 (ScPcf11) contains a well-characterized and highly conserved RNA polymerase II CTD interacting domain (CID) at its N terminus followed by a stretch of 20 glutamines, a domain interacting with the Rna14–Rna15 dimer (which in turn binds to RNA), and two conserved putative zinc fingers near its C terminus that flank a short sequence that interacts with the Clp1 subunit of CF IA (Fig. 1A). Yeast genetic studies have shown that both the N- and C-terminal domains of ScPcf11 are essential for cell viability (Sadowski et al. 2003). The N-terminal and Clp1 interaction regions have been characterized both structurally and functionally. Namely, mutations disrupting the Pcf11/Clp1 interaction cause defects in growth and 3′-end processing (Noble et al. 2007; Ghazy et al. 2012). This region might also maintain interactions with Yra1, an mRNA export factor (Johnson et al. 2009, 2011). However, the function of the conserved zinc fingers in Pcf11 is not known. Keller and colleagues indirectly demonstrated an essential role in cell viability for the C-terminal zinc finger by showing that cross-complementation of a Pcf11 protein missing its C-terminal 200 amino acids allele failed to rescue the growth defect of the pcf11-2 strain that included three mutations spread along this putative zinc finger (Sadowski et al. 2003). However, no structural, biochemical, or functional data have yet been reported for this region of Pcf11.
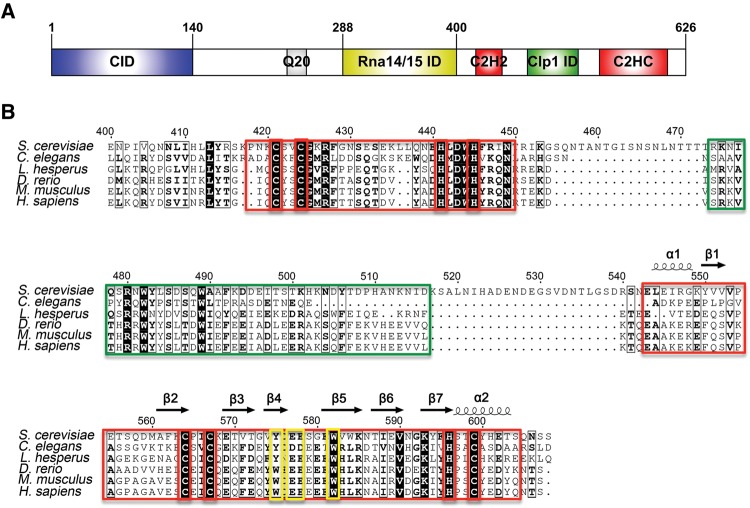
FIGURE 1.
(A) Domain organization of S. cerevisiae Pcf11, drawn with IBS (Liu et al. 2015). (B) Sequence alignment versus secondary structure for the C terminus of Pcf11 from S. cerevisiae to H. sapiens, drawn with ESPript (Robert and Gouet 2014). Conserved residues are indicated in black or in bold, based on similarity levels. Two putative zinc fingers and Clp1 interacting regions are highlighted in red boxes and a green box, respectively. Zinc coordinating Cys and His residues are highlighted in red boxes. Mutated residues Y575, E577, E578, and W582 are highlighted in yellow boxes.
Here we describe a novel flower-like structure for the C-terminal CCHC zinc-finger domain of ScPcf11 that is only distantly related to other zinc coordinating domains. Using a combination of NMR spectroscopy, biochemistry, and yeast genetics, we identify a conserved interface on this domain for potential binding partners and demonstrate that mutation of key residues along this conserved surface-exposed region disrupts 3′-end processing, illustrating for the first time the functional role of Pcf11's highly conserved C-terminal zinc-finger domain.
RESULTS
The CCHC zinc finger is the only stable domain within the C terminus of ScPcf11
The C-terminal region of ScPcf11, corresponding to residues 400–626, is the most conserved region of the protein besides the N-terminal CID domain, with conservation extending between organisms as divergent as fungi and humans (Fig. 1B). A BLAST (http://blast.ncbi.nlm.nih.gov) search for similar protein sequences using this portion of ScPcf11 finds only the Clp1-interacting motif (PDB #2NPI), while the flanking zinc-finger domains do not relate to any other protein with known structure. In order to characterize the structure and function of the C-terminal region of ScPcf11, we first cloned ScPcf11 405–608, which consists of the Clp1-interacting motif flanked by the two putative zinc fingers on either end, and ScPcf11 538–608, which only includes the second zinc finger. However, even freshly purified ScPcf11 405–608 rapidly degraded into a smaller fragment of a size consistent with that of the second zinc-finger domain, presumably as a result of cleavage by endogenous proteases. In addition, when digested with subtilisin, purified ScPcf11 405–608 was also rapidly converted into a smaller protein with a similar size to ScPcf11 538–608 (Supplemental Fig. S1), suggesting that the protein has a “beads-on-a-string” structure consisting of smaller folded domains separated by unfolded regions.
Subsequent NMR analysis of that proteolytically stable fragment revealed that the degradation product corresponds to the putative C-terminal CCHC zinc finger of Pcf11. By comparing the 1H–15N HSQCs (Supplemental Fig. S1) of the individually purified CCHC zinc finger with that of the longer, freshly prepared construct, we observed that the two spectra overlay well, indicating that the CCHC zinc finger is an independent folded domain that does not interact with the preceding Clp1-interacting motif or the first zinc finger. In addition, all dispersed resonances in the 1H–15N HSQC of ScPcf11 405–608 can be observed in the 1H–15N HSQC of ScPcf11 538–608, demonstrating that the CCHC zinc finger is the only well-folded and stable section in the C-terminal region of Pcf11 when purified alone in vitro. Altogether, we conclude that the CCHC zinc finger folds into an independent compact structure that follows the Clp1-interacting domain.
ScPcf11 538–608 folds into a novel flower-like structure
The well-dispersed 1H–15N HSQC spectrum of ScPcf11 538–608 indicates a well-ordered structure. As a consequence, we were able to obtain a good number of experimental restraints for structure determination, including a total of 907 NOE-derived distances, 28 hydrogen bonds, and 68 dihedral angle restraints that were used for calculating the structure (Table 1). The superposition of the 20 lowest-energy conformers together with the ribbon diagram of the representative structure is shown in Figure 2. The root mean square deviation (RMSD) of the 20 conformers for the backbone of the ordered region is only 0.36 Å, indicating an ordered and rigid conformation.
TABLE 1.
NMR constraints and structural statistics
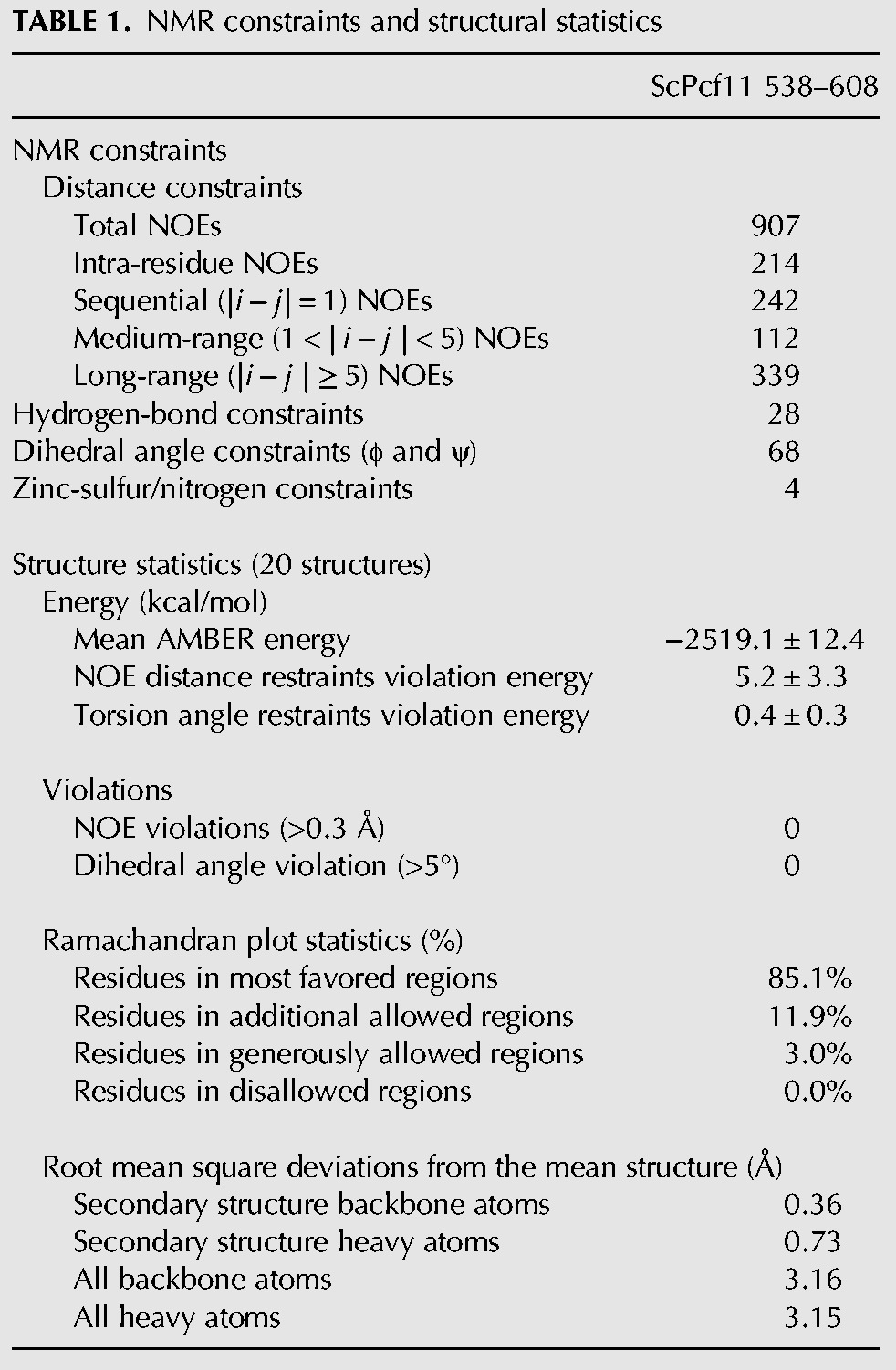
FIGURE 2.
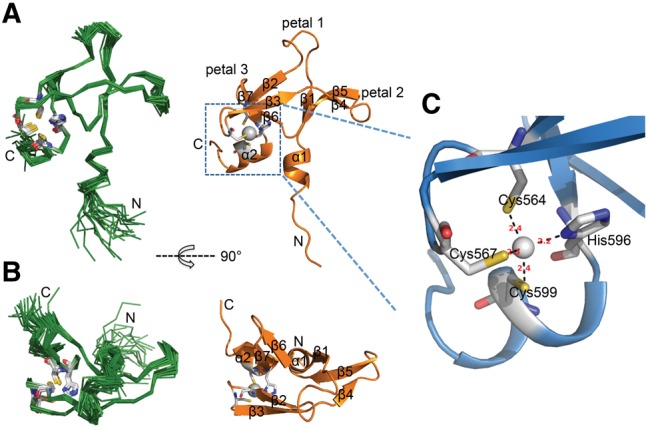
Overview of the solution structure of ScPcf11 538–608. (A) Superposition of the 20 lowest-energy structures on the left and the representative lowest energy structure, shown in ribbon on the right. Secondary structure elements are labeled and the side chains of zinc coordinating residues are drawn in sticks. (B) Same as A but rotated by 90° around the indicated axis. (C) Close-up view of the zinc-binding site. The distances between the zinc ion and its coordinating atoms are indicated in red (Å).
The CCHC zinc-finger domain of ScPcf11 adopts an atypical α1β1β2β3β4β5β6β7α2 fold that resembles a trillium flower, with one loop (petal) extending upward and two β-sheet petals extending to each side away from the helical stem stabilized by a treble-clef zinc finger (Fig. 2A,B). The N-terminal helix α1 is short and adopts a 310 helix conformation, rather than an α-helix structure, in several of the structures of the converged ensembles, while the C-terminal helix α2 is relatively long and stable enough to be present in all 20 conformers. The seven β strands correspond to residues Y550–V552 (β1), A561–C564 (β2), E569–T572 (β3), V574–D576 (β4), E581–K585 (β5), T587–V590 (β6), and K593–H596 (β7). Connecting β1 and β2, the 10-residue long loop forms the central petal 1. The β4β5 hairpin, together with β1, forms an antiparallel twisted β-sheet (petal 2), and β6 together with β7 forms β-hairpin petal 3. The β2β3 knuckle tilts inward to maintain the zinc finger with the C-terminal kinked helix α2. The tertiary structure of the zinc finger region consists of the β2β3-hairpin that positions the side chain thiolates of Cys564 and Cys567, and the kinked helix α2 that orients His596 and Cys599 side chains as ligands for zinc coordination (Fig. 2C). The Cα and Cβ chemical shift values for cysteines 564 (Cα 57.54 ppm and Cβ 30.65 ppm), 567 (Cα 58.80 ppm and Cβ 31.24 ppm), and 599 (Cα 63.60 ppm and Cβ 30.01 ppm), respectively, are consistent with sulfurs coordination of the zinc ion (Kornhaber et al. 2006). In order to determine the tautomeric state of the zinc coordinating nitrogen atoms (Nδ1 or Nε2) on the histidine ring, we calculated the chemical shift difference of Cε1 − Cδ2 to be ∼18.2 ppm, indicating Nδ1 coordination (Barraud et al. 2012). To further confirm the Zn–Nδ1 coordination, we performed the structure calculation in two ways: first with Zn–Nδ1 restrained; then with Zn–Nε2 restrained instead, and compared their compatibilities with the experimental NOEs. As expected, the structure calculated with Zn–Nδ1 coordination has fewer NOE violations, which further validates the presence of the Nε2–H tautomer, the most common configuration in CCHC-type zinc fingers (Barraud et al. 2012).
A highly conserved “petal” unique to Pcf11
Extensive efforts were made to detect homology between the CCHC zinc finger of Pcf11 with other proteins that could shed light on its function. However, a DALI server search (http://ekhidna.biocenter.helsinki.fi/dali_server/) returned no obvious structural neighbors. Among the four top hits with Z-scores larger than 2.0 (highest Z = 2.6), three are RING finger subunits in the crystal structure of the ubiquitin conjugating domain of a RING E3–E2–ubiquitin complex, and the fourth is RING finger protein 38. Only two equivalent residues are shared between Pcf11 and the four structural hits. On the other hand, a VAST (http://structure.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml) search identified 56 structural neighbors, mostly RING finger domains, as well as a considerable number of LIM domains, a structure composed of two contiguous zinc-finger domains, separated by a hydrophobic linker of two amino acids. Similar to the DALI search, the aligned residues between Pcf11 and its VAST structural neighbors are no more than 37 (∼50% coverage), indicative of a lack of significant similarity. In other words, the CCHC zinc-finger domain of ScPcf11 is a novel structure that cannot be classified as a member of any characterized zinc finger or β-stranded folds.
To further understand the uniqueness of this structure, we compared the topology arrangement of our structure with the RING finger and LIM domains, as shown in Figure 3. We selected RING finger protein 38 (the top hit in VAST, PDB #1X4J) and the C-terminal LIM domain of MLP/CRP3 (the highest ranking single LIM domain structure in VAST, PDB #2O13) as the representative of each family. Each of the three domains is a small protein composed of short antiparallel β sheets (Fig. 3A–C). Specifically, they share a common β hairpin (β6β7 in ScPcf11, β4β5 in RING finger, and β5β6 in LIM domain) and the subsequent α helix in the C terminus linked to the N-terminal β hairpin (β2β3 in ScPcf11 and RING finger, β3β4 in LIM domain) by a coordinated zinc ion. When alignment is performed with the complete structures, the RMSD values between ScPcf11, RING finger and LIM domains are poor, 2.7 Å and 9.3 Å, respectively. However, if we use the common structural elements to perform the alignment, the RMSD between ScPcf11 and the RING and LIM domains decreases to 1.4 Å and 1.7 Å, respectively, indicating their structural similarity near the zinc-binding region.
FIGURE 3.
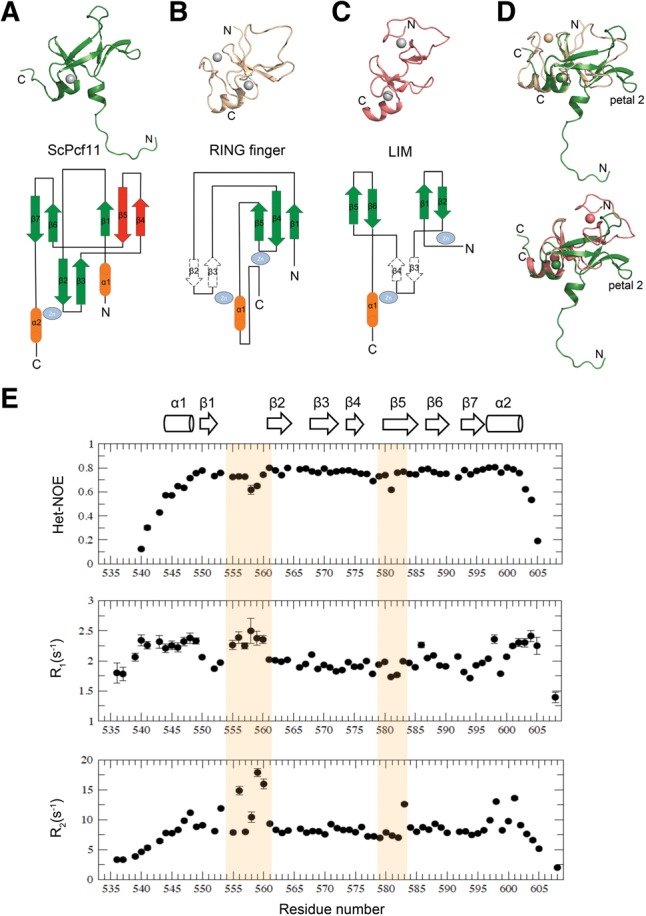
Structural comparison of ScPcf11 538–608 with RING finger and LIM domain proteins. (A) Representative structure of ScPcf11 538–608 (upper) and topology representation (lower). The uncommon “petal” is colored in red. (B) Structure (upper) and topology diagram of a RING finger protein (pdb #1x4j). (C) Structure (upper) and topology diagram (lower) of a LIM protein (pdb #2o13). (D) Alignment of ScPcf11 538–608 (green) with 1x4j (wheat) and with 2o13 (salmon) based on their common structural elements, as described in the main text. (E) Backbone relaxation data of ScPcf11 538–608. Heteronuclear {1H}–15N NOE values (top panel), 15N longitudinal relaxation rate constants R1 (middle panel), and 15N transverse relaxation rate constants R2 (bottom panel) plotted as a function of residue number. Corresponding secondary structures are presented on the top as a column for α helix and an arrow for β strand.
Nevertheless, ScPcf11 differs from the other two structures in several significant ways. First, with similar sequence length (∼70 amino acids), ScPcf11 has a higher percentage of β structure. For example, the stable zinc coordinating β hairpin in ScPcf11 is reduced to a simple turn loop in the other two domains. Secondly, due to its unusually long insertion (28 amino acids) following the zinc-finger β hairpin (β2β3 in ScPcf11 and RING finger, β3β4 in LIM domain), ScPcf11 has an additional β hairpin (β4β5) forming a twisted antiparallel β sheet with β1, designated petal 2, which is completely absent in the other two domains (Fig. 3D). Remarkably, most of the conserved residues of this region of ScPcf11 reside within this special β hairpin (Figs. 1B, 4A), including the absolutely conserved Trp582. In addition, the β4β5 hairpin region forms a negative surface charge patch with central petal 1, which could potentially serve as a binding interface for a basic protein instead of nucleic acids (Fig. 4A). By monitoring UV absorbance at 260 nm, we did not detect any residual nucleic acids binding to purified ScPcf11 538–608 or ScPcf11 405–608, as is instead often the case with RNA- or DNA-binding proteins. We also titrated RNA polyadenines A10 into the C-terminal zinc finger of Pcf11 and observed no interaction.
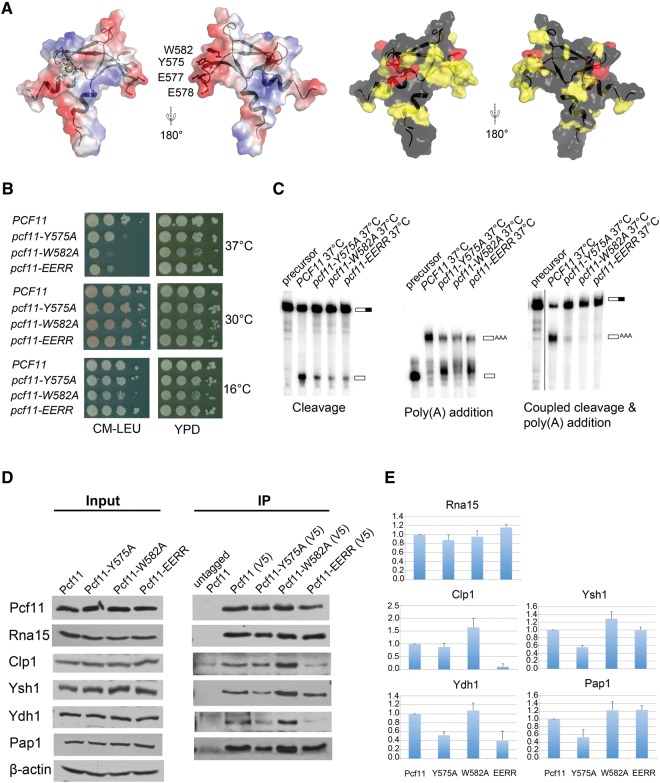
FIGURE 4.
Pcf11 is necessary for both cleavage and polyadenylation steps of mRNA 3′-end processing in yeast. (A) Highly conserved residues cluster in a negative-charged binding surface. The left panel corresponds to surface representations showing the electrostatic potential, with red indicating negative and blue indicating positive electrostatic potential, with the four mutated residues indicated. The right panel shows surface representations of conserved residues (completely conserved in red, less conserved in yellow). (B) Growth characteristics of pcf11-W582A, pcf11-Y575A, and pcf11-EERR mutant strains at 16°C, 30°C, and 37°C. (C) In vitro cleavage assay (left), in vitro polyadenylation assay (middle), and coupled cleavage and polyadenylation assay (right). The experiments were carried out at 37°C. The positions of precursor and product RNAs are indicated on the right side of each panel. (D) Coimmunoprecipitation with extracts prepared from cells expressing forms of Pcf11, Pcf11-Y575A, Pcf11-W582A, and Pcf11-EERR tagged with both V5 and Myc. Cells were grown at 30°C, then shifted to 37°C for 2 h. Coimmunoprecipitation was performed using anti-V5 antibody and the eluates were analyzed by Western blotting with antibodies against CF IA and CPF subunits. Pcf11 was detected with the Myc antibody. The control was done with extract from cells expressing only untagged Pcf11. The left panel shows the input used for each extract, with β-actin as the loading control. (E) Quantitation of co-IP analysis. The ratio of coimmunoprecipitated protein to that of Pcf11 was determined by scanning the Western blots using ImageJ for pulldowns from two different extract preparations. These ratios were then divided by the ratios observed for the wild-type protein and plotted to generate relative binding activities.
To further characterize this conserved β hairpin, we measured backbone dynamics of ScPcf11 538–608 using 15N relaxation methods, including steady-state 15N–[1H] heteronuclear Overhauser effect (Het-NOE) values, longitudinal relaxation rates R1, and the transverse relaxation rates R2. In total, 65 of 73 residues (including two N-terminal leftover residues from the cleavage site) were analyzed. The unanalyzed residues included two prolines that have no amide protons, two C-terminal residues that were unassigned, and four residues that were either overlapped or whose signal was too weak to be accurately analyzed. The values of R1, R2, and Het-NOE are plotted versus residue number in Figure 3E. Overall, the generally high Het-NOE values (>0.76) for residues in the secondary structures confirm that the domain is structurally rigid. Residues within the N- and C-terminal ends have Het-NOE values lower than 0.6, indicative of disorder, consistent with the large RMSD of structure ensembles for these regions (Fig. 2A). For the ordered part, residues 548–602, significant depression of the Het-NOE data between P554 to A561 corresponding to the center petal 1, as well as E578 and E581 from the β4β5 petal 2, are observed, indicating motional flexibility on the picosecond/nanosecond time scale for this region. Although the β6β7 petal 3 has a similar loop size as petal 2, only a mild depression was observed with the average Het-NOE value around 0.74. Plots of R1 and R2 values match the overall Het-NOE profile. Both the N- and C-terminal regions have a larger R1 and smaller R2, compared to average, and the decrease in R2 follows the pattern for Het-NOE values in these regions. The center petal 1 region with low Het-NOE is matched by larger R1 and R2 values, indicating fast motion on the picosecond/nanosecond time scale and slower motion on the microsecond/millisecond time scale occurring at the same time. Significantly larger R2 are observed for V583 within the β4β5 petal 2 region, in addition to lower Het-NOE values, without simultaneous variations of R1, indicating the presence of conformational exchange on a microsecond/millisecond time scale. Taken together, these data indicate that the β4β5 petal 2, as well as central petal 1, form the most dynamic site in this zinc finger with both picosecond/nanosecond and microsecond/millisecond motional flexibility.
The CCHC zinc finger of Pcf11 is necessary for mRNA 3′-end processing
The CCHC zinc finger of Pcf11 is essential for cell survival. Our yeast cell growth assay indicated that a pcf11 variant lacking the C-terminal CCHC zinc finger is not viable (Supplemental Fig. S2). To further investigate the functional significance of the β4β5 hairpin that is so highly conserved and unique to Pcf11, we generated three mutants, Pcf11-W582A, Pcf11-Y575A, and Pcf11-EERR (two consecutive conserved glutamic acids, E577 and E578, were mutated to arginine). The mutated residues, W582, Y575, and E577E578, are all conserved (Fig. 1B) and their side chains cluster into a negative-charged surface region likely mediating binding to other proteins or peptide fragments (Fig. 4A). The comparison of 1H–15N HSQCs of the mutants with wild-type Pcf11 indicated that the protein structure, stability, or solubility was not affected by the mutations (Supplemental Fig. S3).
We tested the growth phenotypes of these mutant strains at three different temperatures (16°C, 30°C, and 37°C). All three mutant strains of pcf11 grow normally at 30°C but exhibit growth defects at the less optimal temperatures of 16°C and 37°C (Fig. 4B). The pcf11-W582A mutant is defective for growth at 37°C and 16°C, and pcf11-Y575A and pcf11-EERR also display growth defects at 37°C on minimal medium, indicating that the putative protein–protein interaction surface of the CCHC zinc finger is essential to maintain the full cellular function of ScPcf11.
To further decipher how the pcf11 mutants exert the defective growth phenotype, we used extracts from the three pcf11 mutant strains grown at 37°C to perform in vitro cleavage and polyadenylation assays to test the role of the CCHC zinc finger in pre-mRNA 3′-end processing (Fig. 4C). In a coupled cleavage and polyadenylation assay performed using the GAL7 pre-mRNA, pre-mRNA processing in all three mutant extracts was nearly completely abolished. To dissect whether the mutants exerted their effects at either the cleavage or polyadenylation step, we performed decoupled cleavage and polyadenylation reactions using wild-type and mutant pcf11 extracts. In the cleavage reaction, all three mutants exhibited greatly diminished pre-mRNA cleavage ability relative to wild-type strain. Interestingly, in a polyadenylation reaction, mutant pcf11 extracts also exhibited diminished polyadenylation relative to wild type. Careful examination of the data shows that pcf11 mutants exhibit a buildup of a slightly larger species of RNA than the precursor substrate, suggesting that mutation of the β4β5 interface impairs interactions that promote processive polyadenylation.
The polyadenylation step of 3′-end pre-mRNA processing is directed by the cleavage and polyadenylation factor (CPF), and a few protein factors within CPF are reported to interact with Pcf11. In pull-down assays with in vitro translated proteins, Pcf11 interacts with the CPF subunit Ydh1 (Kyburz et al. 2003), suggesting that a direct interaction might occur between these proteins in vivo as well. Ysh1, another subunit of the CPF complex that forms a heterodimer with Ydh1, is also critical for both steps of 3′-end processing (Dominski et al. 2005; Xiang et al. 2014). To investigate how mutations of the β4β5 interface of Pcf11 affect its interactions with the CPF subunits Ydh1 and Ysh1, we used various V5-tagged constructs of Pcf11 to execute coimmunoprecipitation (co-IP) experiments. We also investigated pull-down levels of Clp1 and Rna15 (as a control) from CF IA and poly(A) polymerase Pap1 in the same experiments (Fig. 4D,E). As expected, binding to Rna15 was not affected by any of the mutations, a result consistent with the previous observation that the Rna15 binding site within Pcf11 is independent and located away from the C-terminal zinc finger (Amrani et al. 1997). Interestingly, binding to Clp1 was significantly enhanced by the W582A mutant but impaired by the EERR mutant. The binding capacity for Ysh1 was also improved by the W582A mutant but weakened by the Y575A mutant. Finally, the interaction with Ydh1 was disrupted by both mutants Y575A and EERR and the interaction with Pap1 was also disrupted by the Y575A mutant. Taken together, binding of Pcf11 to Clp1 and several tested CPF subunits were all affected by at least one mutation within the β4β5 interface of Pcf11.
DISCUSSION
Pcf11 coordinates transcription and its termination with pre-mRNA 3′-end processing and mRNA export (Mapendano et al. 2010). The highly conserved CID domain at the N terminus of Pcf11 has been well studied structurally as well as with regard to function, but its C-terminal domain has yet to be studied in depth, despite equivalent levels of conservation and requirement for viability. Within this highly conserved region of Pcf11, there is only one proteolytically stable fragment. We report here the solution structure of this domain, which contains the C-terminal CCHC zinc finger of yeast Pcf11. The C-terminal zinc-finger domain of Pcf11 forms a novel flower-like structure with no detectable similarity with other known proteins, which organizes a highly conserved exposed petal-like lobe. Its sequence conservation, structural comparison, and backbone dynamic, as well as the yeast growth phenotype and in vitro mRNA cleavage and polyadenylation biochemical activity, identify this β-hairpin as a protein binding site that performs an essential functional role in mRNA 3′-end processing by affecting both cleavage and polyadenylation steps.
The CCHC zinc finger within Pcf11 forms a typical treble clef fold with the first two cysteines coming from the β2β3 knuckle and the histidine and last cysteine from the N terminus of α helix 2 (Fig. 2C). According to the structural classification of zinc fingers (Krishna et al. 2003), the treble clef fold group can be divided into 10 families. In most families, the finger core is augmented by a loop and a β hairpin between the zinc coordinating β hairpin and an α helix (like RING fingers), but no known structure forms an additional β hairpin, as seen here in the CCHC zinc finger of Pcf11. During our examination of zinc fingers with longer spacer sequence, one special RING finger, the N-terminal zinc-finger domain of PARP-1 (Poly[ADP-ribose] polymerase 1), drew our attention because it has the same number of spacer residues as Pcf11 (CX2CX28HX2C), but instead of forming an additional β hairpin like Pcf11, PARP-1 adopts the same topology as other RING fingers with a longer β4β5 hairpin linked by a 310 helix (Supplemental Fig. S4). Intriguingly, this longer β4β5 hairpin in PARP-1 is one of two important recognition sites for its DNA-break substrate (Langelier et al. 2011; Eustermann et al. 2015). This comparison suggests that evolution provides these conserved, structured long spacer sequences for different functional purposes by appending them to zinc-finger structures that organize them structurally.
Based on the above analysis, the C-terminal zinc finger of Pcf11 very likely acts as a platform and bridge with other protein factors involved in 3′-end processing. If we divide ScPcf11 into three regions (the N-terminal CID; the central CF IA interacting segment, including the Rna14/Rna15 interacting region followed by the Clp1 interacting sequence; and the C-terminal zinc finger) the functional role of each segment becomes clear. The CF I complex contributes to recognition of RNA sequences that specify the poly(A) site on mRNA precursors and to cross-factor interactions with CPF in assembly of an active processing complex (Mandel et al. 2008). It has also been implicated in export of mRNA from the nucleus to the cytoplasm (Brodsky and Silver 2000; Hammell et al. 2002; Nino et al. 2013). As a scaffold protein in the CF I complex, we hypothesize that Pcf11 functions as a central hub in the assembly of the poly(A) site selection machinery and uses the interaction of its N-terminal CID domain with the Ser2 phosphorylated CTD of Pol II to localize proteins to pre-mRNA 3′-ends. According to the BioGRID database, there are seven interacting partners for Pcf11 identified by at least two experimental approaches, although some of these interactions might be mediated by another factor: Clp1, Rna14, Rna15, Rpo21, Ysh1, Ydh1, and Yra1. The middle region of Pcf11 helps in the assembly and localization of Rna14/Rna15 and Clp1 to the 3′-UTR (Noble et al. 2007; Qu et al. 2007; Gordon et al. 2011). Yra1 putatively interacts with Pcf11 through a region N-terminal to the very C-terminal zinc finger to facilitate assembly of the pre-mRNA for recognition by the RNA export machinery (Johnson et al. 2009, 2011). It is tempting to suggest, based on our data, that the C-terminal zinc finger of Pcf11 binds the remaining two proteins, Ydh1 and Ysh1, two main subunits of the CPF complex that form a heterodimer and are critical for both steps of 3′-end processing (Dominski et al. 2005; Xiang et al. 2014).
In our coimmunoprecipitation experiments, we found that binding to Clp1 was significantly enhanced by the W582A mutant but impaired by the EERR mutant. The interactions of Pcf11 with Ydh1, Ysh1, and Pap1 were all disrupted by the Y575A mutant, and binding capacity for Ydh1 was also impaired by the EERR mutant. Overall, Clp1 and Ydh1 were affected most by the β4β5 hairpin mutants of Pcf11. These co-IP results suggest two possibilities: The C-terminal zinc finger of Pcf11 binds to Ydh1 in such a way that changes in their interaction indirectly influence pull-down levels of Ydh1's interacting partners Clp1, Ysh1 and Pap1. Alternatively, considering that the Clp1-interacting sequence K475-S499, as revealed by the crystal structure of Clp1–Pcf11 complex, is located N-terminal to the very C-terminal zinc finger of Pcf11, the interaction between Clp1 and Pcf11 might extend to the C-terminal zinc finger and mediates their interaction with the CPF subunits.
Increased binding by mutation is not unusual when protein interfaces are involved in multiple interactions with other proteins (Sharabi et al. 2014). In order to maintain the stability of the complex and specificities toward all interacting partners, protein sequences evolve to optimize multiple interactions but not necessarily confer best binding affinity for each binding partner. Indeed, Holbein et al. (2011) reported a similar phenomenon, in which mutations in Clp1 improved its interaction with Ysh1. The solvent-exposed W582 in Pcf11 potentially plays such a balancing role that prevents too strong an interaction with some protein partners by steric repulsion. Our result that the Pcf11 mutation W582A that causes the most striking growth defect actually increased its interaction with Clp1 is interesting. We suggest two possible explanations. First, “moderate” binding between Pcf11, Clp1, and the CPF complex is necessary to provide the flexibility of the complete 3′-end processing complex in positioning Ysh1 over the mRNA cleavage site, and the flexibility of reorganization of the CPF complex so that Pap1 can reach the growing poly(A) tail. The Pcf11 W582A mutant might decrease this flexibility by increasing the interaction affinity, leading to dysfunctional cleavage and polyadenylation; second, since Yra1 shares the same binding region of Pcf11 with Clp1, “moderate” binding might enable switching from a complex with Clp1 during 3′-end processing to an interaction with Yra1 for mRNA export. Since the interaction with Clp1 and Yra1 may extend to the C-terminal zinc-finger domain of Pcf11, the Pcf11 W582A mutant favors Clp1 binding, which in turn blocks Yra1 and affects the interaction with CPF subunits.
In summary, our results indicate a critical role for the very C-terminal zinc finger of Pcf11 in its interaction with Ydh1/Ysh1, either directly or indirectly mediated by Clp1. Disruption of these interactions by mutation of highly conserved residues lead to 3′-end processing defects in biochemical assays and disrupts the biological function of Pcf11 in yeast.
MATERIALS AND METHODS
Protein expression and purification
Two constructs of ScPcf11 were prepared: ScPcf11 405–608 and ScPcf11 538–608. The PCR-amplified genes were inserted into a modified pET-28a (Novagen) vector with a Protein G B1 domain (GB1) fused to the N terminus. The recombinant plasmids were transformed into Escherichia coli BL21 (DE3) cells. Uniformly 15N- and 15N-, 13C-labeled ScPcf11 proteins were obtained by growing the transformed cells at 37°C in M9 minimal medium containing 0.5 g/L 15NH4Cl, 2 g/L 13C-glucose supplemented with trace elements and 20 µM ZnSO4. Protein expression was induced with 0.5 mM IPTG when OD600 of cells reached 0.6. After 3–4 h, cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris, 500 mM NaCl, 20 mM imidazole, pH 8.0). Cell lysate was extracted by sonication and cleared by centrifugation. The supernatant was applied onto a HisTrap column (GE Healthcare) equilibrated in lysis buffer. By washing with elution buffer (20 mM Tris, 500 mM NaCl, 10–300 mM imidazole, pH 8.0; the linear gradient increased the concentration of imidazole), the protein was eluted at 240 mM imidazole. The pooled protein was incubated with in-house purified tobacco etch virus (TEV) protease overnight and loaded onto HisTrap again to remove the cleaved His-GB1 tag and dialyzed into NMR buffer (20 mM Tris, 120 mM NaCl, pH 7.4). The NMR samples were first treated with excess Zn2+ to saturate the metal binding site, then with excess EDTA to remove free and unspecific bound Zn2+. As a final cleaning step, the protein solution was loaded onto a Superdex 75 column (GE Healthcare) to remove chelated EDTA from the solution and exchange it into NMR buffer.
NMR spectroscopy
NMR spectra were collected at 25°C on Bruker Avance 500 MHz and 600 MHz spectrometers equipped with 1H/13C/15N triple-resonance probes and pulse-field gradients. NMR data were processed with NMRPipe (Delaglio et al. 1995) and analyzed within CcpNmr (Vranken et al. 2005). 2D 1H–15N/13C HSQCs and 3D HNCO, HNCA, HNCACB, CBCA(CO)NH, HBHA(CO)NH, HCCH-TOCSY, and (H)CCH-TOCSY were performed to obtain the chemical shift assignments of backbone and side chain atoms. Chemical shifts were referenced to internal 2, 2-dimethyl-2-silapentanesulfonic acid (DSS). 3D 13C-edited aliphatic and aromatic NOESY-HSQC (mixing time 100 msec) and 15N-edited NOESY-HSQC experiments (mixing time 100 msec) were collected to generate distance restraints for structural calculations. Hydrogen–deuterium exchange experiments were carried out to identify the protected amides and confirm the hydrogen bonds network in secondary structures.
Structure determination
A total of 200 starting structures of ScPcf11 538–608 were calculated using CYANA (Güntert et al. 1997), from which 100 structures with the lowest target functions were selected to be refined in AMBER (Case et al. 2005). Only distance restraints obtained from 3D 13C/15N-edited NOESY-HSQC experiments were used as initial inputs for the first several rounds of calculation in CYANA. In the following iterations, 68 torsion angle restraints for both φ and ψ predicted by TALOS+ (Shen et al. 2009) were included as well, based on the secondary structure identified from the early structure ensembles. In addition, 28 hydrogen bonds were added according to the hydrogen–deuterium experiments and NOE patterns characteristic of β sheets. The structures clearly show a metal binding site with three highly conserved cysteines and a histidine clustered together. Therefore, in the final round of iterative calculation of CYANA, one zinc ion was incorporated by adding four zinc-sulfur/zinc-nitrogen bond restraints (∼2.4 Å). The 100 structures with the lowest target functions were refined in AMBER 8 by using a 20-ps restrained simulated annealing protocol. The annealing was done with the ff99 force field and generalized Born solvation model, restrained by the same distances, torsion angles, and hydrogen bonds used in CYANA with force constants of 35 kcal mol−1 Å−1, 50 kcal mol−1 rad−2, and 50 kcal mol−1 Å−1, respectively. In addition, chirality restraints were applied along the main chain to prevent inversions of chirality at high temperature. The 20 conformers with the lowest AMBER energy were finally selected and bonded with zinc ion using the Metal Center Parameter Builder (MCPB) module of Amber 14.
Backbone 15N relaxation measurements
The backbone 15N relaxation parameters (longitudinal relaxation rates R1 [1/T1]; transverse relaxation rates R2 [1/T2] and steady-state heteronuclear NOEs) were measured using standard pulse sequences (Farrow et al. 1994). Experiments were performed on Bruker Avance 600 MHz NMR spectrometer at 25°C. Spectral widths of 8417.5 Hz and 1946.3 Hz were used for 1H and 15N dimensions, respectively. For the T1 and T2 measurements, 1024 (1H) and 128 (15N) complex data points were collected with 16 transients and a recycle delay of 3 sec. The delays used for the T1 experiments were 10 (×2), 40, 90, 150, 300, 500, 800, 1100, 1500, and 2000 msec, and those used for the T2 experiments were 10 (×2), 20, 40, 60, 90, 120, 150, 200, 250, and 300 msec. The relaxation rate constants were obtained by fitting the peak intensities to a single exponential function using the nonlinear least squares method. For the {1H}–15N Het-NOE measurements, two spectra were performed in the presence and absence of a 3-sec proton pre-saturation period prior to the 15N excitation pulse and using recycle delays of 2 and 5 sec, respectively. In total, 32 transients were used for each experiment. Het-NOE values were calculated from the ratio of peak intensities with and without proton saturation.
Yeast strains used and plasmid construction
The S. cerevisiae strains used in this study are as follows:
CM332 (MATa ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11-Δ::TRP1 [pFL38-PCF11])
SDL83 (MATa ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11-Δ::TRP1 [pRS315-Myc-PCF11-V5])
SDL84 (MATa ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11-Δ::TRP1 [pRS315-Myc-pcf11-Y575A-V5])
SDL85 (MATa ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11-Δ::TRP1 [pRS315-Myc-pcf11-W582A-V5])
SDL86 (MATa ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 pcf11-Δ::TRP1 [pRS315-Myc-pcf11-EERR-V5])
For pRS315-PCF11, the PCF11 sequence is flanked by a Myc epitope tag at the N terminus and V5 tag at the C terminus, and the protein is expressed under the control of the MPE1 promoter and terminator sequences (900-bp upstream and downstream of the coding region of the MPE1 gene [Ghazy et al. 2012]). Point mutants pcf11-Y575A, pcf11-W582A, and pcf11-EERR were generated in pRS315-PCF11 using the QuikChange kit (Stratagene). All mutants were verified by sequencing.
Growth assays
To determine the effect of pcf11 mutations on yeast cell growth, the plasmid shuffle complementation assay was used (Boeke et al. 1987). The pRS315 plasmids containing pcf11 mutations were analyzed in the PCF11 plasmid shuffle strain CM332. Growth properties were analyzed by growing the strains in liquid YPD at room temperature to an optical density at 600 nm of 1.0, spotting 5 µL of 10-fold serial dilutions on plates containing rich medium (YPD) or plates with minimal medium lacking leucine and incubating the plates for 3–7 d at 16°C, 30°C, or 37°C.
In vitro RNA processing assays
Preparation of yeast cell extracts, transcription of [α-32P] UTP-labeled GAL7-1 RNA or precleaved GAL7-9, and processing assays were performed as described previously (Zhao et al. 1999).
Coimmunoprecipitation assays
Yeast processing extracts (1 mg) prepared as described previously (Zhao et al. 1999) were incubated with anti-V5 antibody (Invitrogen) overnight at 4°C in 500 µL of buffer IP-150 (10 mM Tris–HCl at pH 7.9, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM PMSF, 2 µM pepstatin-A, 0.6 µM leupeptin, 0.1% NP-40), followed by 1 h incubation with Protein A agarose beads (Santa Cruz). The beads were washed five times with 1 mL of buffer IP-150, resuspended in 100 µL of 3× SDS sample buffer, and incubated for 15 min at room temperature. Eluates and inputs were analyzed by SDS-8% polyacrylamide gel. Western blotting was performed according to standard procedures. Polyclonal antibodies against Rna15, Ydh1, and Ysh1 were a gift from H. Domdey. Antibodies against β-actin were purchased from Abcam (#ab8224) and the anti-myc antibody was the 9E10 monoclonal. Clp1 polyclonal antibody was prepared in the Moore laboratory.
DATA DEPOSITION
Atomic coordinates and related NMR restraints of ScPcf11 538–608 have been deposited in the Protein Data Bank as 2nax.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant RO1-GM064440 from the National Institutes of Health–National Institute of General Medical Sciences (NIH-NIGMS) to Gabriele Varani and grant MCB-1244043 from the National Science Foundation (NSF) to Claire Moore. We thank Dr. David Bentley for providing the plasmid corresponding to full-length ScPcf11.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.058354.116.
REFERENCES
- Amrani N, Minet M, Wyers F, Dufour ME, Aggerbeck LP, Lacroute F. 1997. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol 17: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Schubert M, Allain FH-T. 2012. A strong 13C chemical shift signature provides the coordination mode of histidines in zinc-binding proteins. J Biomol NMR 53: 93–101. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175. [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. 2005. The Amber biomolecular simulation programs. J Comput Chem 26: 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Choi E-A, Shi Y. 2011. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA 2: 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon SK, Lutz CS. 2012. mRNA 3′ end processing factors: a phylogenetic comparison. Comp Funct Genomics 2012: 876893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang X, Purdy M, Wagner EJ, Marzluff WF. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25: 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Wu W-F, Langelier M-F, Yang J-C, Easton LE, Riccio AA, Pascal JM, Neuhaus D. 2015. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell 60: 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. [DOI] [PubMed] [Google Scholar]
- Ghazy MA, Gordon JMB, Lee SD, Singh BN, Bohm A, Hampsey M, Moore C. 2012. The interaction of Pcf11 and Clp1 is needed for mRNA 3′-end formation and is modulated by amino acids in the ATP-binding site. Nucleic Acids Res 40: 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JMB, Shikov S, Kuehner JN, Liriano M, Lee E, Stafford W, Poulsen MB, Harrison C, Moore C, Bohm A. 2011. Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry 50: 10203–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moore C. 2001. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci 98: 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P, Mumenthaler C, Wüthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol 22: 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein S, Scola S, Loll B, Dichtl BS, Hübner W, Meinhart A, Dichtl B. 2011. The P-loop domain of yeast Clp1 mediates interactions between CF IA and CPF factors in pre-mRNA 3′ end formation. PLoS One 6: e29139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26: 2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Kim H, Erickson B, Bentley DL. 2011. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 18: 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhaber GJ, Snyder D, Moseley HNB, Montelione GT. 2006. Identification of zinc-ligated cysteine residues based on 13Cα and 13Cβ chemical shift data. J Biomol NMR 34: 259–269. [DOI] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV. 2003. Structural classification of zinc fingers. Nucleic Acids Res 31: 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Sadowski M, Dichtl B, Keller W. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res 31: 3936–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M-F, Planck JL, Roy S, Pascal JM. 2011. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA. J Biol Chem 286: 10690–10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, et al. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31: 3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino CA, Herissant L, Babour A, Dargemont C. 2013. mRNA nuclear export in yeast. Chem Rev 113: 8523–8545. [DOI] [PubMed] [Google Scholar]
- Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol 12: 144–151. [DOI] [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA. 2007. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res 35: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson AR, Tong L. 2012. Crystal structure of the Rna14-Rna15 complex. RNA 18: 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Perez-Canadillas J-M, Agrawal S, De Baecke J, Cheng H, Varani G, Moore C. 2007. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem 282: 2101–2115. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hübner W, Keller W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J 22: 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi O, Shirian J, Grossman M, Lebendiker M, Sagi I, Shifman J. 2014. Affinity- and specificity-enhancing mutations are frequent in multispecific interactions between TIMP2 and MMPs. PLoS One 9: e93712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. 2009. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev 3: 1711–1724. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696. [DOI] [PubMed] [Google Scholar]
- Xiang K, Tong L, Manley JL. 2014. Delineating the structural blueprint of the pre-mRNA 3′-end processing machinery. Mol Cell Biol 34: 1894–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kessler M, Helmling S, O'Connor JP, Moore C. 1999. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol 19: 7733–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.