Abstract
The virus/host interaction is a complex interplay between pro- and anti-viral factors that ultimately determines the spread or halt of virus infections in tissues. This interplay develops over multiple rounds of infection. The purpose of this study was to determine how cellular-level processes combine to impact the spatial spread of infection. We measured the kinetics of virus replication (VSV), antiviral paracrine signal upregulation and secretion, spatial spread of virus and paracrine antiviral signaling, and inhibition of virus production in antiviral-exposed A549 human lung epithelial cells. We found that initially infected cells released antiviral signals 4-to-7 hours following production of virus. However, the subsequent rapid dissemination of signal and fast induction of a robust and persistent antiviral state ultimately led to a suppression of infection spread. This work shows how cellular responses to infection and activation of antiviral responses can integrate to ultimately control infection spread across host cell populations.
Keywords: antiviral state, virus spread, infection spread, interferon, paracrine signaling, A549 cells
Background and Motivation
The spread of viruses from cell to cell within tissues is determined by the dynamics of the virus-host interaction over multiple rounds of infection. Early in viral infections, virus spread is governed by the interaction between pro-viral factors and cellular antiviral innate immune signaling responses (Dobrovolny et al., 2013; McLaren and Butchko, 1978; Miao et al., 2010). In the first 1–6 days postinfection, the predominant antiviral responses are the localized auto- and paracrine innate antiviral signaling networks involving the interferon (IFN) responses, among others (McLaren and Butchko, 1978; Miao et al., 2010). These responses consist of intracellular receptors that recognize viral factors and trigger synthesis of a set of signaling molecules including type I and III interferons (IFNs), which are secreted and signal cells, sometimes synergistically (Voigt and Yin, 2015), to enter into an antiviral state that inhibits viral replication. Antiviral states are stimulated not only in infected cells, but also in cells proximal to infection through diffusion or transport of paracrine-signaling antiviral molecules away from the infection site. Often described as a “race,” the early spread of virus infection through tissues is not determined by a single round of virus/host interaction, but through multiple rounds of infection and innate immune responses during which antiviral signals and infectious virus spread through tissues.
Many studies of virus/host interactions, for example in lung epithelial cells during respiratory virus infection (Liu et al., 2008; Vester et al., 2009; Yoshikawa et al., 2010), have been conducted on uniformly infected cells, using mRNA microarrays or mass spectrometry for measurement of cellular responses to viral infection. Such results can be useful for developing mathematical or computational models of infection, which seek to predict how rates of spread depend on the coupled biological reactions that describe virus growth, immune responses, and the physical processes such as diffusion that govern how virus and cytokines may spread [e.g., (Baccam et al., 2006; Bocharov and Romanyukha, 1994; Getto et al., 2008; Haseltine et al., 2008; Howat et al., 2006)]. While viral infection spread has been modeled, experimental studies of the corresponding intracellular viral and immune processes that drive infection spread that can inform these models are rare (Lam et al., 2005; Timm and Yin, 2012). Most previous modeling efforts of intracellular virus/host interactions are either conducted on an in vivo basis, or these efforts assume well-mixed conditions in an in vitro system, reflecting available experimental data. A lack of experimental data designed to inform understanding of spatial infection spread has been specifically noted, particularly the need to test the effects of specific cellular immune components on virus replication in in vitro infections (Dobrovolny et al., 2013).
To understand factors involved in multi-round spread and arrest of infection, a cell/virus system is required that reflects the observed pattern of virus replication in vivo, where an initial phase of virus replication is eventually limited by the host antiviral response. The A549 human lung epithelial cell line is a suitable host system, as it retains an intact innate immune system despite immortalization and is widely used to study antiviral immunity. However, lab-adapted strains of classic human respiratory viruses such as influenza and human rhinovirus (HRV) do not replicate well on A549 cells, potentially reflecting these robust immune responses. For this study, we used a well-characterized variant of vesicular stomatitis virus, which carries an M51R mutation in its matrix protein. Its properties are useful in this model system because: 1) it replicates well in A549 cells to high titers, 2) it strongly induces measurable antiviral responses, 3) this virus strain does not significantly block cellular antiviral responses, allowing one to study the full and intact antiviral cytokine response network, and 4) VSV is amenable to reverse-genetics engineering for creation of reporter virus strains (Ahmed et al., 2003; Lawson et al., 1995).
This work elucidated how different viral and cellular processes combine to influence infection spread during early phases of acute viral infections in human lung cells. We identified and measured rate-limiting kinetic processes that occur during virus replication and activation of host responses, which may in turn be used to predict the infection outcome. We found that the generation of antiviral cytokines by initially infected cells lags significantly behind the production of infectious virus, creating a temporal handicap for the antiviral responses, a handicap that must be overcome in later rounds of infection. An estimated 10-fold faster diffusion of antiviral cytokines over virus combined with the rapid activation and long-term persistence of cellular antiviral responses to antiviral cytokines eventually halted infection spread.
Methods and Materials
Cell culture
Human lung epithelial carcinoma (A549, ATCC CCL-185) cells were obtained from the American Type Culture Collection and grown in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals). Baby hamster kidney cells (BHK-21) for plaque assays were originally obtained from Isabel Novella (University of Toledo) and grown on minimal essential medium (MEM, Corning) with 10% FBS and 2 mM Glutamax I (Gibco). All cell lines were cultured in a humidified incubator at 37°C in a 5% CO2 atmosphere.
IFIT2 immune activation reporter A549 human lung epithelial cells (plFIT2:GFP cells) were created using the Duet011 lentiviral gene integration system (Addgene) as described previously (Swick et al., 2013). These cells have been engineered to have an enhanced promoter region for the interferon stimulated immune gene IFIT2, with a 5× repeat of the interferon-stimulated response element (ISRE) region, and to produce ZsGreen1 fluorescent protein upon IFIT2 gene upregulation (Swick, 2013). IFN reporter A549 cells (plFN:GFP), with eGFP under the control of the IFNβ promoter, were obtained from Richard Randall (University of St. Andrews) and cultured as normal A549 cells (Chen et al., 2010).
Virus strains
Fluorescent reporter virus strains incorporating DsRed2 or DsRed-Express fluorescent genes into the fifth genomic position of VSV-lndiana were created previously using reverse genetics techniques (Swick et al., 2013; Voigt et al., 2013). The M51R reporter DsRed-Express virus strain used in this work was originally created by introducing the methionine-to-arginine substitution in the 51st position of the M protein via oligonucleotide-directed mutagenesis using the same viral genomic backbone as the other viral strains (Swick et al., 2013). This point mutation abolishes the binding to the host protein Rae1 and relieves suppression of innate immune responses during VSV infection (Ahmed et al., 2003; Rajani et al., 2012).
Plaque spread
Ninety-five percent confluent monolayers of A549 cells (plFN:GFP, plFIT2:GFP, and normal) were plated in 12-well tissue culture plates at a density of 1 × 105 cells/well and cultured overnight (12 hrs) in RPMI media with 10% FBS. The cell layers were infected with DsRed-Express M51R vesicular stomatitis virus at a multiplicity of approx. 10 pfu/well and 1 pfu/well in separate wells in 100 µl of infection media, RPMI supplemented with 2% FBS. Virus was allowed to adsorb for one hour with periodic rocking of the plate to fully distribute infectious virus over the monolayer. The infection media was then removed, the cells rinsed with 1 ml PBS per well, and finally 1 ml of infection medium with 0.6% agar was added.
Cells were incubated on the microscope stage at 37°C in a 5% CO2 and 75% relative humidity atmosphere in a warming chamber (InVivo Scientific, Saint Louis, MO, USA) and a stage-top incubator chamber (Pathology Devices, Westminster, MD, USA). Infection was visualized every 8 hours over three days by a Nikon TE Eclipse 300 microscope equipped with a Chroma PhotoFluor fluorescent light source. Stage automation allowed for creation of large-scale composite images for each infection well, and was produced by a Prior ProScanll automated microscope stage controlled by metaMorph v.7.7.8 imaging software (Molecular Devices, Sunnyvale, CA, USA). Resulting images were processed using JEX, a customized JAVA-based batch-processing image analysis platform incorporating much of the functionality of ImageJ (Rasband, 1997–2012) that can be found as shareware at <http://sourceforge.net/projects/jextools> (Warrick et al., 2016). Image processing steps included correction for nonuniform fluorescent illumination across the image range, subtraction of background and autofluorescence (rolling ball radius of 50 pixels), and stitching of 7×7 image arrays to create larger composite images.
Individual spreading plaques that developed independently were identified, in which neither virus reporter nor immune activation reporter signals collided with other plaques’ virus or immune signals over the 48 hours of infection. Around these independent plaques, identically-sized regions of interest that spanned the maximum virus and antiviral signal spread areas were identified for each plaque and cropped out of composite images at each timepoint for further analysis. Numbers of virus reporter (RFP) or immune-reporter (GFP) positive cells were calculated for each plaque and timepoint using ImageJ software on the corresponding fluorescent images as follows. Images were thresholded into fluorescent positive and negative areas and particles with a minimum area of 50 pixels2 (virus RFP), 30 pixels2 (IFN GFP) or 10 pixels2 (IFIT2 GFP) were counted using the ImageJ “Analyze Particles” function. To account for particle areas consisting of multiple overlapping cells, the area of each particle was divided by the average size of virus-RFP-positive, IFN-GFP-positive, or IFIT2-GFP-positive single-cells and rounded to produce # positive cells for that particle. Cell counts were summed over the entire image to produce # positive cells/plaque. Radius of virus- and immune-signal spread was drawn at the radius from plaque center that included approx. 95% of total positive cells. To best display plaque-spread images scaled appropriately for print, cropped fluorescent images from representative plaques were filtered and adjusted for optimum contrast and brightness.
One-step virus infection
Medium was removed from 95% confluent monolayers of A549 cells in 12-well tissue culture plates (approximately 4 × 105 cells), and 80 µl of virus solution was added. After 1 hour of adsorption at 37°C and 5% CO2with gentle rocking every 20 minutes, the inoculum was removed and the cells rinsed with 1 ml PBS. One ml of infection medium, RPMI supplemented with 2% FBS, was then added. Supernates were then harvested at times indicated, and frozen for later assays for virus titer and secreted antiviral signaling molecules.
Plaque assays
For quantification of infectious virus particles in infection supernates, samples were serially diluted in 1:10 dilutions in MEM supplemented with 2% FBS and 2 mM Glutamax. BHK cells were plated 18 hours prior to assay at a concentration of 5×l05 cells/well in 6-well tissue culture plates and allowed to form monolayers. Cell monolayers were infected with 200 µl virus dilution and incubated for one hour with gentle rocking every 20 minutes. The virus-containing sample solution was then removed, cells rinsed with PBS, and then cell monolayers were overlaid with 2 ml of MEM supplemented with 2% FBS, 2 mM Glutamax, and 0.6% melted agar. The plates were cooled until agar solidified, and incubated at 37°C, 5% CO2 for approximately 24 hours, until plaques appeared. Agar layers were then removed; cells were fixed for 20 minutes with a 4% paraformaldehyde (PFA) solution containing 5% sucrose in PBS. The PFA solution was then removed and cell layers were stained with 0.1% crystal violet in 20% ethanol to visualize plaques. Virus titers were calculated as plaque-forming unit (PFU) per average cell in the monolayer.
Antiviral secretion assay
Supernates collected from one-step virus infections were thawed and irradiated with 7000 J/m2 of UVC irradiation over 20 minutes to inactivate infectious virus. The supernates were then assayed for paracrine antiviral signaling molecules as previously described (Voigt et al., 2013). Briefly, serial 1:2 dilutions were incubated over A549 cells in 96-well plates for 24 hours, and the antiviral state of the cells was then challenged by infection with VSV-DsRed2 virus at MOI 5. After 24 hours incubation, successful infection as indicated by DsRed2 fluorescence was read using a GE Typhoon FLA 9000 Biomolecular Imager at 555/580 nm under BSL2 conditions. Inhibition of virus replication indicated presence of antiviral signaling molecules, the concentration of which was determined by calculating the IC50 dilution and presented in units (U) per ml in the original one-step virus infection. Here, one unit of antiviral activity per ml is the amount that inhibits 50% of VSV replication in A549 cells using the assay described above.
ELISA
Cellular supernates were also tested for IFNβ levels using a VeraKine Human Interferon Beta ELISA kit (PBL Assay Science, Piscataway, NJ, USA, lot K2247). Supernates were diluted in cell culture media prior to assay to keep IFNβ concentrations within assay operating range, as per the manufacturer’s recommendations. Sample dilutions were as follows: t=0, 2, 4, 6 hr undiluted, the t=8 hr sample diluted 1:2, the t=10 hr sample diluted 1:5, and the t=12,18, 24 hr samples diluted 1:10. The manufacturer’s protocol was followed exactly.
Preparation of conditioned media with secreted antivirals
A549 human lung epithelial cells were seeded into T-225 cell culture flasks and grown to 90% confluency. Cells were then infected by VSV-M51R at MOI 5 in infection media to stimulate production of cell-derived paracrine-signaling antiviral cytokines. After 24 hours of infection, antiviral-containing supernates were harvested and (JVC irradiated by 7000 J/m2 over 20 minutes to inactivate virus, and cellular debris removed using a 0.2 um syringe filter. Solutions were then titered for antiviral activity and stored at 4°C for future use. Antiviral activity of solutions was regularly re-titered to account for decline in activity.
Antiviral state assay
The kinetics of cellular antiviral state development after treatment with antiviral signaling molecules was measured by pretreatment of 96-well plates containing A549 cell monolayers with a solution containing antiviral signaling molecules. After a pretreatment period, antiviral states were probed by infection with DsRed2-VSV at MOI 5. At 24 hours postinfection, plates were scanned for RFP signal on a GE Typhoon FLA 9000 Biomolecular Imager. Inhibition of RFP signal reflected on the reduction in virus replication within cells as a result of antiviral pretreatment.
Results
Virus infection spread is controlled by cellular antiviral responses
How does the innate immune response dynamically affect infection spread? Does the process occur over one or several rounds of infection? To address these questions we employed a model spreading-plaque infection and tracked over time the relative spread of cytokine-mediated cellular responses versus VSV infections of a human lung epithelial cell line. Low-multiplicity (MOI=1 × 10−5) infections were conducted on monolayers of plFN:GFP A549 cells (Chen et al., 2010), and plFIT2:GFP cells (Swick et al., 2013). We used VSV-M51R-dsRedEx reporter virus, which triggers cellular antiviral responses and lacks the ability to suppress these responses (Ahmed et al., 2003). Spread of fluorescent foci resulting from point infections was allowed to progress under agar overlays for 48 hours, imaged every 4 hours at 4× magnification.
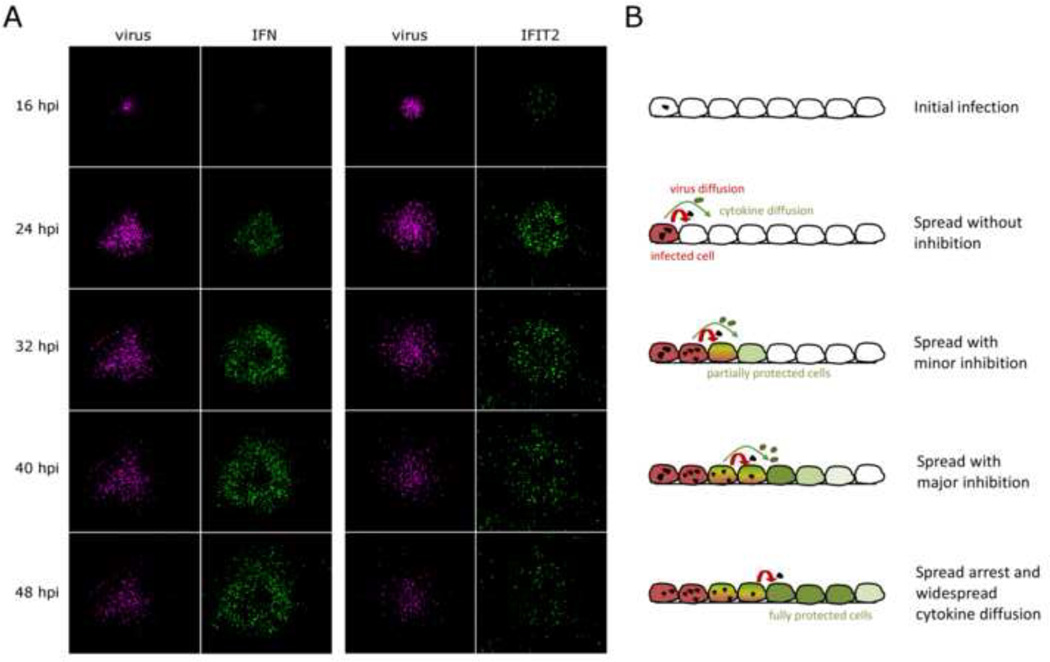
Fluorescent reporter of virus replication (red) became apparent 8–16 hours postinfection (hpi) (Fig. 1A, red). Spread of virus slowed and halted by 32 hpi, soon after the induction and spread of antiviral responses. Fluorescence resulting from antiviral gene activation appeared 16–24 hours postinfection (Fig. 1A, green), initially in cells that were directly infected, and then later spreading radially to uninfected cells proximal to foci of viral infection. The plaques all followed a pattern of initial infection and uninhibited spread, immune activation and continued spread, and finally spread arrest and widespread cytokine diffusion far beyond the site of infection initiation (Fig 1B).
Figure 1. Infection spread through A549 cell monolayers is governed by virus/host interactions over multiple infection rounds.
Panel A: Monolayers of reporter A549 human lung epithelial cells, which express GFP upon stimulation of either the IFN promoter (left) or the IFIT2 antiviral protein promoter (right), were infected with VSV-M51R-dsRedExpress reporter virus at a low MOI. Cell monolayers were imaged every 8 hours under 4× magnification to visualized virus spread and propagation of immune responses through the cellular monolayers. Panel B: Schematic representing corresponding stages of multi-round infection spread over the 48 hour experimental period.
In infections with wild-type VSV-dsRedExpress virus, which suppresses host immunity, we saw continuous virus spread for the duration of the experiment, with little cellular immune activation (data not shown), consistent with previous observations of wild-type VSV spread (Swick et al., 2013).
Image quantification reveals that virus infection directly triggers IFNs, which in turn diffuse beyond infected cells and halt infection spread
Time-lapse microscopy images were analyzed to quantify spread of virus, IFNβ upregulation, and IFIT2 upregulation for multiple spreading-plaque infections. Five VSV-M51R-dsRedEx plaques on plFN:GFP A549 cell monolayers and six VSV-M51R-dsRedEx plaques on plFIT2:GFP cell monolayers were analyzed by measuring over time the number of positive RFP and GFP cells, reflecting gene expression by virus infection and host-cell immune activation, respectively. Further, the extent of radial spread for each reporter was quantified using ImageJ image processing and analysis software (version 1.46r, imagej.nih.gov).
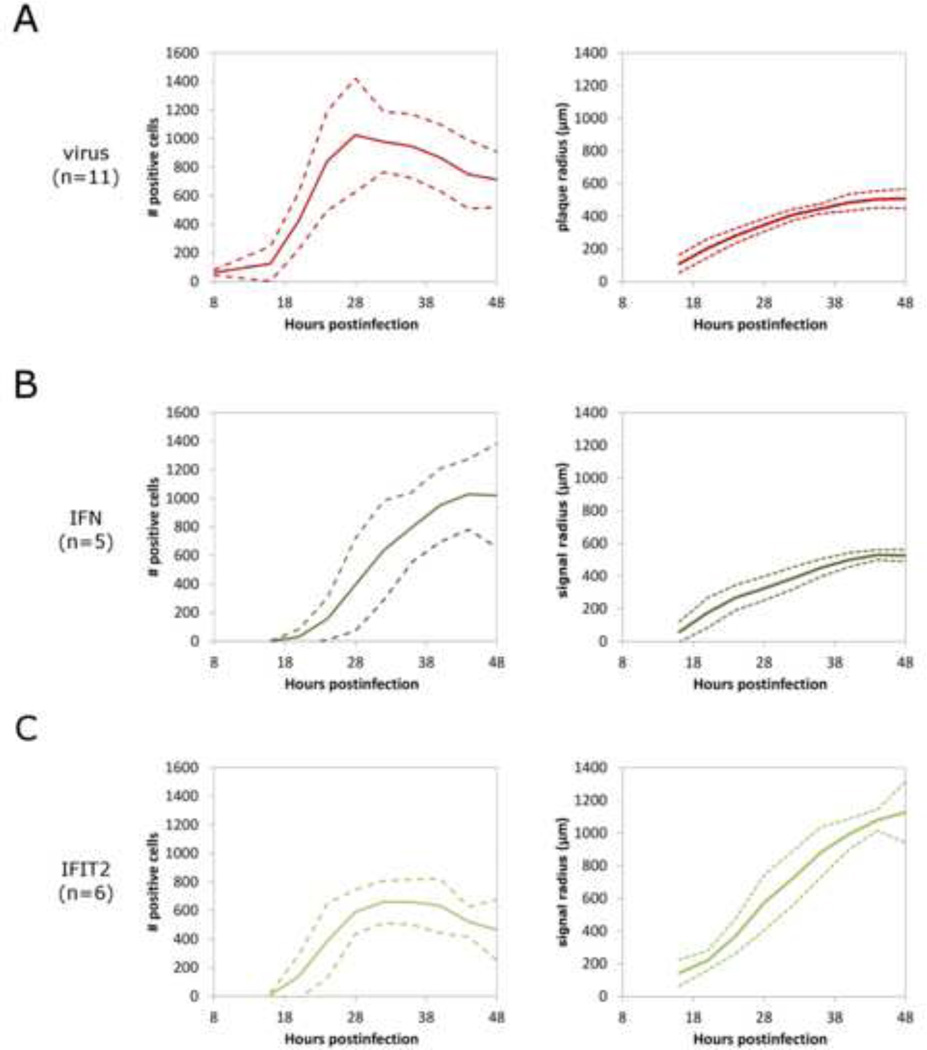
Average numbers of virally-infected cells for each plaque reached a maximum after 28 hours of infection (Fig. 2A). Radial spread of virus away from the site of infection slowed and ceased by 48 hpi. Fluorescence resulting from IFN activation was apparent at 16 hpi at the center of point infections (Fig. 2B). IFN activation spread radially, initially spatially lagging behind the virus infection front and approaching a maximum number of active cells towards the end of the 48 hour infection period. IFN activation appeared to be spatially restricted to areas with active virus infection, not spreading beyond the borders of infection. IFIT2 activation was also apparent starting 16 hours after infection, and, similar to IFN activation, spread radially outwards following the spread of virus (Fig. 2C). However, unlike the IFN signal, which remained within the borders of active virus infection, IFIT2 activation spread beyond the plaque borders, showing extensive activation with lower signal by 40 hpi. IFIT2 activation also appears nonuniform and stochastic in nature, consistent with previous reports of stochastic, IFN-dependent ISG activation by interferons (Rand et al., 2012).
Figure 2. Quantification of virus and immune response spread in plaque-spread infections of A549 cells.
Time-lapse images of plaque-spread infections were analyzed for # of cells that show positive signal and spread radius of reporter signal for A) VSV-M51R-DsRedExpress virus replication, B) cellular IFNβ induction, and C) interferon-stimulated antiviral effector protein IFIT2 induction. Quantification of plaque spread in terms of # of positive reporter cells is given per plaque, +/− standard deviations over the given number of plaques. Quantification of plaque spread radius is calculated by area of positive signal spread from the origin of infection per plaque, +/− standard deviations over the given number of plaques.
These results provide evidence that IFN production requires stimulation by active viral infection in cells, and not by secreted IFN or other antiviral signals alone. In contrast, IFIT2 is known to be stimulated both directly by virus and indirectly by IFN signaling (Elco et al., 2005), allowing cells beyond the border of infection to receive IFN signal as it diffuses away from sites of infection, and enter an antiviral state that inhibits subsequent infection. Tracking the spatial spread of virus, IFN induction, and IFIT2 effector protein responses reveal that virus initially replicates and spreads readily with little inhibition. However, the subsequent activation of cellular antiviral responses beyond the immediate area of infection slows virus replication and ultimately stops infection spread over further rounds of replication.
Virus replication precedes release of antiviral cytokines in initially infected cells
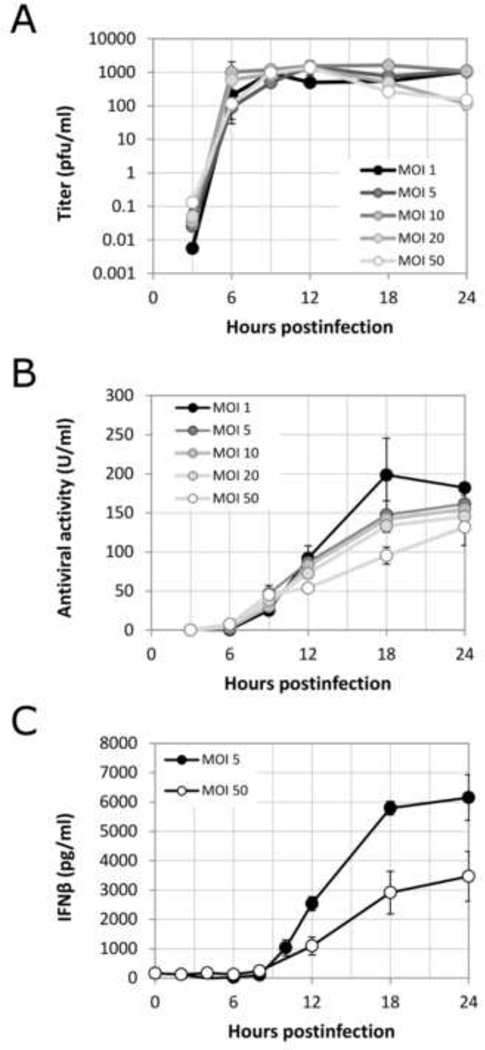
The results in Figure 1 suggest that there is an initial lag between replication/spread of virus and production/spread of interferon, but that the spread of antiviral responses eventually overtakes and halts virus spread. To better understand the early events that precede the spread of infection beyond the first initial cell, we measured the kinetics of virus production and secreted antiviral signaling molecules in a single round of virus infection. Cells were uniformly infected with different multiplicities of VSV-M51R-dsRedExpress reporter virus ranging from 1 to 50 pfu/cell. Infection supernates were collected at various times during infection and assayed for progeny virus by plaque assay (Fig. 3A), antiviral cytokines by a functional bioassay (Fig. 3B), and interferon beta (IFNβ) by ELISA (Fig. 3C).
Figure 3. Viral and antiviral dynamics during single-round viral infection of A549 cells.
Panel A: infectious progeny virions produced and released from a population of A549 cells uniformly infected with VSV-M51R. Panel B: Antiviral secretions released from A549 cells uniformly infected at different MOI, measured by functional titer-reduction antiviral activity assay. Panel C: lnterferon-3 protein secreted from uniformly infected A549 cells at different MOI, measured by ELISA.
Infectious virus progeny from this single-round infection were first detectable at 3 hpi, reaching half-maximal levels at 7.4±1.6 hpi. The levels of virus at 3 hpi depended on the initial multiplicity, but peak viral yields of ~1000 infectious virions/ml were independent of MOI. Antiviral cytokines and IFNβ secreted from cells were first detected at 6 hpi at all MOI, reaching half-maximum secretion levels only at 12.2±0.9 hpi (functional antivirals) and 13.8±0.4 hpi (IFNβ protein), lagging virus replication by 4.8 and 6.4 hours, respectively. Initially, higher antiviral activities were detected for higher MOI infections (for 6 to 9 hpi), however, this trend then reversed by later timepoints, and ultimately the cells infected with lowest initial MOI produced the highest levels of antiviral secretions. Similar results were found by analysis of IFNβ levels, where cells in MOI 5 infections activated release of about two-fold higher IFNβ levels than cells from MOI 50 infections (Fig. 3C). These results indicate that VSV infections in the absence of immunosuppressive function start with a 4–7 hour advantage over host antiviral responses.
Cells respond rapidly to paracrine signals
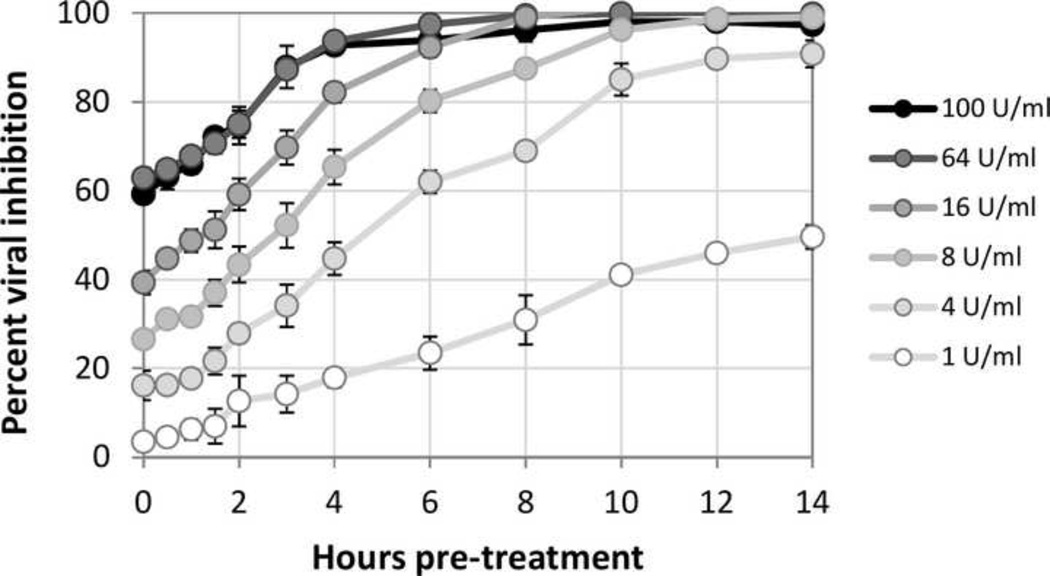
The ability of defensive cytokine signaling to contain infection spread will depend not only on how rapidly cells produce cytokines, but also how rapidly they respond to such signals. To characterize how rapidly and effectively an antiviral state can develop, A549 cells were treated with cellular supernatants containing antiviral secretions from previously uniformly-infected cells, and the susceptibility of these cells to infection was then quantified by challenge with the VSV-dsRed2 reporter virus. These measures were carried out for different durations of antiviral pretreatment.
The development of cellular antiviral states in response to antiviral paracrine signals was rapid and dose-dependent, as shown in Figure 4. Cells could significantly inhibit virus replication, even when they were simultaneously stimulated and infected (0 hour pretreatment). At the most potent levels tested (64 and 100 U/ml), virus production was reduced by 60 percent with no pretreatment and fully blocked following 6 hours pretreatment. At a 100-fold lower dose, 1 U/ml, secreted cytokines were unable to initially inhibit virus growth. Instead cells slowly developed an antiviral state, requiring 14 hours to reach 50% virus inhibition. These data show that responses of cells to cytokine signals can be rapid (0-to-10 hours) compared to the 10-to-24 hours cells need to make cytokines in response to infection.
Figure 4. Upregulation of the antiviral state in response to cytokine treatment.
Experimentally measured development of the antiviral state in cells in response to treatment with antiviral molecules, for six separate treatment doses. Naive A549 cells were treated with conditioned media containing antiviral cytokines, at six different doses of measured antiviral activity. Antiviral cellular states were probed by infection with the VSV-DsRed2 viral replication reporter over time post-treatment, viral RFP signal was measured 24 hpi, and signal was normalized to untreated infected and uninfected controls.
Cellular antiviral states can persist over multiple days
The persistence of cellular antiviral states in lung epithelial cells may also contribute to the ability of tissues to inhibit viral infection spread over multiple rounds of virus infection. To characterize the persistence of the antiviral state, A549 cells were stimulated for 16 hours by cell-derived antiviral secretions to induce cellular antiviral states. These secretions were then removed, the cells were rinsed, and the subsequent decline of cellular resistance to infection was measured (Figure 5).
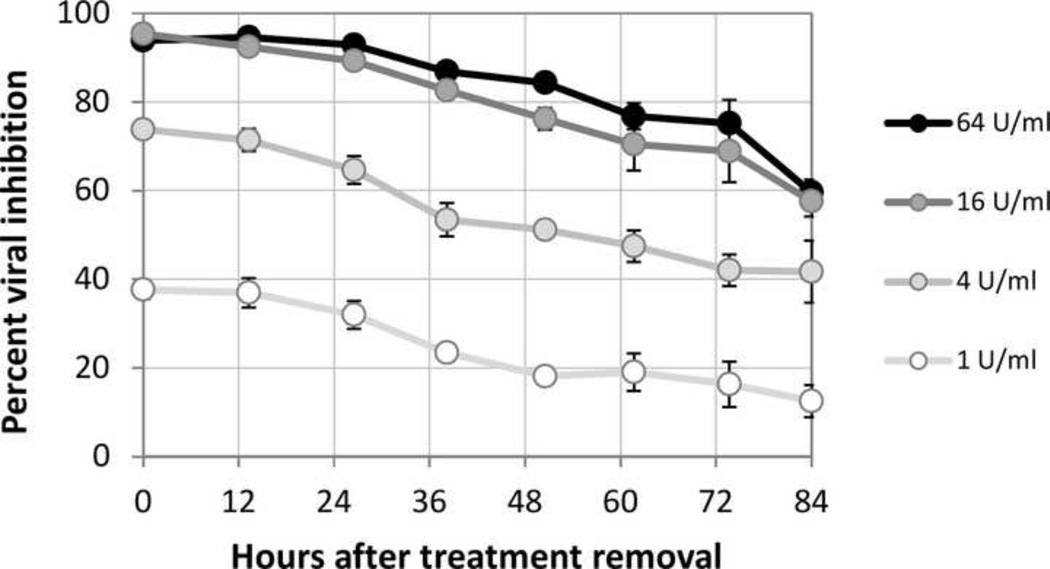
Figure 5. Persistence of the antiviral state after removal of cytokine treatments.
A549 cells were treated for 16 hours with secreted antiviral cytokines at four different doses, in order to induce cellular antiviral states. Cytokine was then removed and thoroughly rinsed away. Antiviral cellular states were probed by infection with the VSV-DsRed2 viral replication reporter at various times. Fluorescent signal of virus replication was used to calculate % remaining cellular viral inhibitory level.
Cellular antiviral states persisted long after the removal of activating cytokines, with 60 percent viral inhibition present even after 3.5 days in cells that were initially fully resistant to infection. Degradation of the antiviral state (or return to an infection-susceptible state) proceeded in a linear fashion at all original cytokine treatment doses, decreasing for each dose by approximately 30% over the course of 3.5 days. These results indicate that once induced, antiviral states in A549 cells persist for a duration equivalent to multiple virus replication cycles, protecting signaled cells from infection and reducing the probability that the infection will continue to spread to susceptible host cells.
Discussion
Spread of virus through a population of susceptible host cells early in infection is a spatial process where the outcome is determined by the virus/host interaction over multiple rounds of infection. In vitro plaque-spread experiments in the presence of cellular immunity highlight a critical role for cytokine-mediated paracrine signaling in arresting intercellular viral infection spread.
We visualized, over several rounds of VSV infection in A549 cells, the initial rapid spread of the infection with subsequent slowing and arrest. With the aid of IFNβ and IFIT2 reporter cells we also visualized the activation of these innate immune markers in infected cells, followed by the spread of antiviral activation beyond the borders of the infected region. Activation of cellular antiviral immunity was likely due to a mixture of secreted paracrine-signaling molecules, demonstrated in this experimental system to include IFNα, IFNβ, and IFNλ1/2/3 using ELISA (Voigt and Yin, 2015). This functional antiviral cocktail may also contain other secreted paracrine-signaling antiviral factors such as ISG15, TNFct, and IP-10 (D’Cunha et al., 1996; O’Neill and Bowie, 2010; Randall and Goodbourn, 2008; Sadler and Williams, 2008), which would function together with type I and III interferons to induce the development of cellular antiviral states.
These results are consistent with VSV spreading infections on PC3 prostate cancer cells in a similar dual-reporter system (Swick et al., 2013), however here we have elucidated the relative rates of virus and cytokine production in response to initial infection, as well as the development and persistence of cellular antiviral states in response to secreted cytokines. While the detailed dynamics of infection spread will depend on specific features of the virus and its interaction with its host cell, the role of not only innate response activation, but also the extent of its persistence are likely to be key factors in halting infection spread in diverse tissues. We note here that these innate cytokine responses and their persistence also inform the development of later cellular and humoral immune responses, which this model system of the earliest, localized immune events does not address.
Here we found that for a rapidly replicating RNA virus such as VSV, viral replication and release from cells was earlier and faster than the secretion of antiviral cytokines by A549 cells, even when employing a VSV variant (M51R) that is impaired in its ability to inhibit activation of an innate immune response. The inability of antiviral responses in early-infected cells to measurably affect initial virus replication in these same cells is perhaps not surprising, though to the best of our knowledge this has not been previously reported. While the initially infected cells gain no benefit from the auto- and paracrine antiviral signals that they expend energy and cellular resources to synthesize and secrete, proximal cells can gain substantial benefit from these physically smaller, faster-diffusing antiviral signaling molecules in later rounds of infection, allowing for infection arrest.
During single-round infection of resting cells, which may serve as a model for the initial round of infection in spreading plaques, VSV replication occurred quickly and initial infectious dose did not significantly affect final virus titers in A549 human lung epithelial cells. Previous studies in baby hamster kidney (BHK) cells also showed final titers were independent of virus inoculum dose (Stauffer-Thompson, 2008; Timm and Yin, 2012). This suggests that cells have a maximum capacity for virus production, defined by a cellular resource that is eventually exhausted by the replicating virus.
Different viral inoculum doses do, however, influence the kinetics and final titers of antiviral molecule secretion (Fig. 3) with interesting behaviors. While higher virus infection doses induce more potent early antiviral secretions, their later production is less potent, reflecting the negative effects of high virus load on cellular function.
The relatively low levels of antivirals secreted in this system indicate the cooperative nature of early antiviral responses, even between cells of a single type, to produce sufficient antiviral secretions to substantially inhibit future infection cycles. The antiviral titers observed in Fig. 3 (note that ~4 × 105 cells per well contributed to the measured antiviral secretion into 1 ml media) indicate that even under optimal conditions, an average of more than 1000 cells are needed to secrete each active unit (U) of antivirals. These phenomena emphasize that the ability of the innate immune response to stop infection spread is a result of a multi-round cooperative process between a large number of cells, in which both virus and antiviral signals are being created, diffusing, and propagating from cell to cell.
Secreted antiviral molecules such as interferons are more than ten-fold smaller in average diameter than virions, and thus diffuse faster through liquids and semi-solid matrices such as agar. For spreading infections, mathematical models suggest how velocities of spatial expansion can depend on the kinetics of virus production and the effective diffusivity of the virus particles (Yin and McCaskill, 1992; You and Yin, 1999). Based on estimated diffusivities for the virus and interferon, and measured time delays for the production of virus and interferon (Figure 3), estimated velocities of interferon spread (450–550 µm/h) are nearly ten-fold higher than velocities of infection spread (40–70 µm/h) (supplemental information). This provides one mechanism for antiviral cytokines to overcome the delay in their initial production relative to virus replication, and create an infection-resistant band of host cells ahead of the spreading infection.
A second mechanism to inhibit infection spread is provided by the rapid cellular responses to antivirals relative to the rates of cytokine induction and virus replication. The substantial decrease of viral titers even with concurrent antiviral treatment and viral infection suggests that downstream antiviral signaling occurs on similar time scales to the ~6 hour VSV replication cycle. Not only is induction of cellular antiviral states in epithelial cells after interferon treatment rapid, this effect remains potent for days to weeks after initial stimulation, independent of any other immune mechanisms. Other work suggests that these strong downstream effects may result from independent and/or synergistic interactions between multiple secreted antiviral cytokines, thus enhancing signaling effects (Voigt and Yin, 2015).
We have demonstrated that although the initial secretion of antiviral signaling molecules is slow compared to the replication, packaging, and release of initial progeny VSV particles, the relatively rapid activation of antiviral response during subsequent rounds of infection acts to counter the slow start. This may reflect the need for tight immunological control in organisms. Antiviral signaling needs to be slow and difficult to trigger in order to prevent tissue damage that results from to false alarms and hyperimmunity (Kim et al., 2008); however, when induced by veritable viral infection, downstream effector signaling must respond quickly and persist in order to prevent large-scale infection spread and minimize virus-induced tissue damage.
The study of infection spread in expanding plaques provides a framework for better understanding factors that contribute to the dynamics of infection spread in vivo. Typical studies characterize virus growth and innate immune activation over the time scale of a single infection cycle. In real infections, however, multiple cycles of infection are accompanied by multiple cycles of innate immune activation, which in turn affects infection spread and further signaling. In a spreading infection, every cell writes its own unique history, dependent on the changing local levels of virus and cytokines, which reflect the histories of near and distant infected and signaled cells. These facets of infection are seldom studied in well-controlled in vitro systems. Here, by studying how cells respond over time to a broad range of virus and cytokine doses, we have sampled a cross-section of the diverse changing environments that a cell may encounter in an expanding plaque. This work integrates this simple average-cell single-round kinetic data with spreading plaque infection data to explain spatially observed phenomena and reveals vital features of multidimensional spreading infection processes.
Future work will seek to integrate data from homogeneous culture measures with properties of virus and cytokine diffusional movement in mathematical models to predict rates of infection spread and containment. Moreover, experimental systems for infection spread will be developed to account for a broader array of structures or processes that impede or enhance the spread of virus and cytokines in vivo, for example, the extracellular matrix, cell-cell junctions, mucosal layers and cilial movements.
Conclusions
The virus-host interaction is a complex interplay between pro- and anti-viral forces that ultimately determines the spread or halt of virus infections in tissues. This interplay is worked out over multiple rounds of infection, with initially infected cells often unable to successfully defend themselves, but capable nonetheless of secreting antiviral compounds that signal proximal cells to enter into antiviral states that ultimately allow for arrest of viral replication and spread. The dynamic factors of this process suggest that while antiviral responses are triggered by viruses slowly relative to VSV replication, this handicap is overcome in later rounds of infection, predominantly by rapid downstream antiviral signaling of proximal cells that receive paracrine antiviral signals prior to or concurrent with infection. These induced cellular antiviral states are persistent, lasting for days even after removal of cytokine stimulants. These results are consistent with concepts of immunological control, where immune responses should be slow to trigger, to avoid autoimmunity, but act efficiently once triggered in order to successfully combat infection spread.
Supplementary Material
Acknowledgments
We thank Sri Ram for his valuable suggestions and input on data analysis and presentation, as well as Douglas Lyles for advice regarding experimental design and manuscript writing. We also thank Bahar Inankur and Ashley Baltes for help with time lapse microscopy setup and troubleshooting. Richard Randall (University of St. Andrews) generously provided the IFN-reporter A549 cells. E.V. and A.S. were supported by National Science Foundation Predoctoral Fellowships. We gratefully acknowledge support for this work by the National Institutes of Health (AI091646, AI104317).
References
- Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J Virol. 2003;77:4646–4657. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P, Beauchemin C, Macken CA, Hayden FG, Perelson AS. Kinetics of influenza A virus infection in humans. J Virol. 2006;80:7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov GA, Romanyukha AA. Mathematical model of antiviral immune response III - Influenza A virus infection. Journal of Theoretical Biology. 1994;167:323–360. doi: 10.1006/jtbi.1994.1074. [DOI] [PubMed] [Google Scholar]
- Chen S, Short JAL, Young DF, Killip MJ, Schneider M, Goodbourn S, Randall RE. Heterocellular induction of interferon by negative-sense RNA viruses. Virology. 2010;407:247–255. doi: 10.1016/j.virol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Cunha J, Knight E, Jr, Haas AL, Truitt RL, Borden EC. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U S A. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolny HM, Reddy MB, Kamal MA, Rayner CR, Beauchemin CAA. Assessing Mathematical Models of Influenza Infections Using Features of the Immune Response. PLoS One. 2013;8:20. doi: 10.1371/journal.pone.0057088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Williams BRG, Sen GC. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappa B, and interferon but not Toll-like receptor 3. J Virol. 2005;79:3920–3929. doi: 10.1128/JVI.79.7.3920-3929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getto P, Kimmel M, Marciniak-Czochra A. Modelling and analysis of dynamics of viral infection of cells and of interferon resistance. Journal of Mathematical Analysis and Applications. 2008;344:821–850. [Google Scholar]
- Haseltine EL, Lam V, Yin J, Rawlings JB. Image-guided modeling of virus growth and spread. Bulletin of Mathematical Biology. 2008;70:1730–1748. doi: 10.1007/s11538-008-9316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howat TJ, Barreca C, O'Hare P, Gog JR, Grenfell BT. Modelling dynamics of the type I interferon response to in vitro viral infection. Journal of the Royal Society Interface. 2006;3:699–709. doi: 10.1098/rsif.2006.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, Benoit LA, Byers DE, Alevy Y, Tucker J, Swanson S, Tidwell R, Tyner JW, Morton JD, Castro M, Polineni D, Patterson GA, Schwendener RA, Allard JD, Peltz G, Holtzman MJ. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nature Medicine. 2008;14:633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam V, Duca KA, Yin J. Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnology and Bioengineering. 2005;90:793–804. doi: 10.1002/bit.20467. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant Vesicular Stomatitis Virus from DNA. Proc Natl Acad Sci U S A. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Song WJ, Wang P, Lee KC, Chan W, Chen HL, Cai ZW. Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics. 2008;8:1851–1858. doi: 10.1002/pmic.200700757. [DOI] [PubMed] [Google Scholar]
- McLaren C, Butchko GM. Regional T-cell and B-cell responses in influenza-infected ferrets. Infect Immun. 1978;22:189–194. doi: 10.1128/iai.22.1.189-194.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao HY, Hollenbaugh JA, Zand MS, Holden-Wiltse J, Mosmann TR, Perelson AS, Wu HL, Topham DJ. Quantifying the Early Immune Response and Adaptive Immune Response Kinetics in Mice Infected with Influenza A Virus. J Virol. 2010;84:6687–6698. doi: 10.1128/JVI.00266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. Sensing and signaling in antiviral innate immunity. Curr Biol. 2010;20:R328–R333. doi: 10.1016/j.cub.2010.01.044. [DOI] [PubMed] [Google Scholar]
- Rajani KR, Kneller ELP, McKenzie MO, Horita DA, Chou JW, Lyles DS. Complexes of Vesicular Stomatitis Virus Matrix Protein with Host Rael and Nup98 Involved in Inhibition of Host Transcription. Plos Pathogens. 2012;8:16. doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand U, Rinas M, Schwerk J, Nohren G, Linnes M, Kroger A, Flossdorf M, Kaly-Kullai K, Hauser H, Hofer T, Koster M. Multi-layered stochasticity and paracrine signal propagation shape the type-l interferon response. Molecular Systems Biology. 2012;8:13. doi: 10.1038/msb.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. Journal of General Virology. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nature Reviews Immunology. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer-Thompson KA. Chemical and Biological Engineering. University of Wisconsin-Madison; 2008. Quantitative effects of defective interfering virus-like particles on the growth and population behavior of vesicular stomatitis virus. [Google Scholar]
- Swick A. Cell and Molecular Biology. University of Wisconsin-Madison; 2013. Visualing vesicular stomatitis virus (VSV) infection spread and resulting host immune responses in vitro with a dual color fluorescent reporter system. [Google Scholar]
- Swick A, Baltes A, Yin J. Visualizing infection spread: Dual-color fluorescent reporting of virus-host interactions. Biotechnology and Bioengineering. 2013:1–10. doi: 10.1002/bit.25170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm A, Yin J. Kinetics of virus production from single cells. Virology. 2012;424:11–17. doi: 10.1016/j.virol.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester D, Rapp E, Gade D, Genzel Y, Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics. 2009;9:3316–3327. doi: 10.1002/pmic.200800893. [DOI] [PubMed] [Google Scholar]
- Voigt E, Inankur B, Baltes A, Yin J. A quantitative infection assay for human type I, II, and III interferon antiviral activities. Virology Journal. 2013;10:10. doi: 10.1186/1743-422X-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt EA, Yin J. Kinetic Differences and Synergistic Antiviral Effects Between Type I and Type III Interferon Signaling Indicate Pathway Independence. J Interferon Cytokine Res. 2015;35:734–747. doi: 10.1089/jir.2015.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JW, Timm A, Swick A, Yin J. Tools for Single-Cell Kinetic Analysis of Virus-Host Interactions. PLoS One. 2016;11:e0145081. doi: 10.1371/journal.pone.0145081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, McCaskill JS. Replication of viruses in a growing plaque - a reaction-diffusion model. Biophysical Journal. 1992;61:1540–1549. doi: 10.1016/S0006-3495(92)81958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. Dynamic Innate Immune Responses of Human Bronchial Epithelial Cells to Severe Acute Respiratory Syndrome-Associated Coronavirus Infection. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LC, Yin J. Amplification and spread of viruses in a growing plaque. Journal of Theoretical Biology. 1999;200:365–373. doi: 10.1006/jtbi.1999.1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.