Abstract
Exogenous transfer RNAs (tRNAs) favor translation of bovine papillomavirus 1 wild-type (wt) L1 mRNA in in vitro translation systems (Zhou et al. 1999, J. Virol., 73, 4972–4982). We, therefore, investigated whether papillomavirus (PV) wt L1 protein expression could be enhanced in eukaryotic cells following exogenous tRNA supplementation. Both Chinese hamster ovary (CHO) and Cos1 cells, transfected with PV1 wt L1 genes, effectively transcribed the genes but did not translate them. However, L1 protein translation was demonstrated following co-transfection with the L1 gene and a gene expressing tRNASer(CGA). Cell lines, stably transfected with a bovine papillomavirus 1 (BPV1) wt L1 expression construct, produced L1 protein after the transfection of the tRNASer(CGA) gene, but not following the transfection with basal vectors, suggesting that tRNASer(CGA) gene enhanced wt L1 translation as a result of endogenous tRNA alterations and phosphorylation of translation initiation factors elF4E and elF2α in the tRNASer(CGA) transfected L1 cell lines. The tRNASer(CGA) gene expression significantly reduced translation of L1 proteins expressed from codon-modified (HB) PV L1 genes utilizing mammalian preferred codons, but had variable effects on translation of green fluorescent proteins (GFPs) expressed from six serine GFP variants. The changes of tRNA pools appear to match the codon composition of PV wt and HB L1 genes and serine GFP variants to regulate translation of their mRNAs. These findings demonstrate for the first time in eukaryotic cells that translation of the target genes can be differentially influenced by the provision of a single tRNA expression construct.
INTRODUCTION
Translation is a key facet of the central dogma of molecular biology (1), which uses the genetic information in mRNA to synthesize protein. Transfer RNAs (tRNAs) play a central role in translation, acting as the carrier of the monomeric units of proteins, the amino acids and the growing polypeptide chains (2). Both tRNA and mRNA eventually pair on the ribosome to determine which amino acid is inserted at a particular point in the nascent polypeptide chain. To ensure protein synthesis, correct base pairing between the three bases of the codons on the mRNAs and those of the anticodons of the cognate aminoacyl (aa)-tRNAs dictates the sequence of the polypeptide chain. In addition, the cellular content of tRNA isoaccepting species is clearly a determinant of the rates and amounts of protein synthesis (3). In unicellular organisms, a correlation between tRNA content constraint and gene expression level has been demonstrated using the concept of optimal codons (4). A shortage of critical tRNAs could result in slowed elongation of the nascent peptide or premature termination of translation (5,6). The tRNAs from uninfected cells can rescue the translation of global proteins in Autographa california nucleopolyhedrovirus-infected Ld652Y cells (7). Stimulated expression of GTase with tRNAAGA and tRNAAGG resulted in a 5-fold increase in GTase production in Escherichia coli, in which codon usage is highly biased and the tRNAs specific to codons AGAarg and AGGarg are extremely rare (8). Expression of a single tRNA gene has enhanced the expression of heterologous genes, especially for genes containing rare codons in bacterial expression systems (9,10). The data support the argument that the variable copy number of individual isoacceptor tRNAs is a primary determinant of translation (11). However, it is not clear whether provision of a single tRNA gene expression can mediate the expression of target genes at either the transcriptional or translational levels in eukaryotic cells.
The tRNA molecules and tRNA-like structures have been identified as crucial elements in the replication of a very diverse group of viruses including plant viruses (12,13) and mammalian retroviruses (14). Recently, a human tRNASer(CGA) gene has been cloned and expressed in mammalian cells (15,16). The cloned human tRNASer(CGA) gene containing 400 bp has previously been verified for its integrity and functionality (15,16), via its ability to act in reverse transcription of a retroviral vector harboring a complementary primer binding site sequence (16). The human tRNASer(CGA) molecule was originally identified via a screening of the human BOSC23 retroviral packaging cell line (17) for tRNA molecules functionable as primers for murine leukemia virus (MLV) replication (16). But it is not clear whether this tRNASer(CGA) molecule could be functional for protein translation of MLV and other viruses. More recently, tRNA-like structure of turnip yellow mosaic virus has been reported to be a 3′-translational enhancer, which enhanced expression of a firefly luciferase (LUC) reporter gene up to 25-fold (18). Therefore, we are interested to investigate whether and how the expression of the human tRNASer(CGA) gene would affect the expression of wild-type (wt) and codon-modified (HB) papillomavirus (PV) L1 genes in cultured mammalian cells.
Both wt and HB PV L1 genes were chosen as the target genes throughout the experiments; we initially considered the choice for three reasons. First, PVs are exclusively epitheliotropic viruses. The PV life cycle is tightly linked to epithelial cell differentiation. Production of viral capsid proteins (L1 and L2) expressed from the late gene (L1 and L2) is restricted to the most terminally differentiated keratinocytes in the upper layers of the epithelium. Expression of L1 and L2 proteins from wt PV L1 and L2 genes has not been observed in proliferating transfected keratinocytes (19) and other replicating mammalian cells (20). However, mRNAs encoding the capsid proteins can be detected in less-differentiated keratinocytes and other replicating mammalian cells, suggesting that the expression of the late genes is post-transcriptionally regulated (21). When the L1 and L2 genes are cloned under cytomegalovirus (CMV) or simian virus 40 (SV40) promoters in continuously growing transformed cell lines in transient transfection assays, large amounts of PV L1 and L2 mRNAs are made, but no proteins can be detected (20). Second, blockage to the translation of PV L1 mRNAs has been overcome by two means: point mutation (19) and codon modification (20,22–24). Introduction of point mutations in the 5′ end of the L1 gene which altered the RNA sequence without affecting the protein sequence resulted in the production of high levels of HPV 16 L1 mRNA and protein in HeLa cells (19). Codon modification of the PV L1 and L2 genes utilizing mammalian preferred codons without changing the protein sequence can express L1 and L2 proteins in large amounts in replicating mammalian cells (20,22–24). The argument proposed from the codon modification studies is that codon usage of the PV L1 and L2 genes may be a major determinant of PV capsid protein expression. The observation we made recently that bovine papillomavirus (BPV) 1 can undergo its life cycle in BPV1 virus-infected Saccharomyces cerevisiae (25,26) supports this argument because codon usage of BPV1 genes more closely resembles that of yeast genes than that of the mammalian consensus (20). The low expression efficiency of several viral proteins in HIV due to unfavorable codon usage has been reported by various groups (27–29). Third, provision of exogenous tRNAs either from bovine liver or from yeast could greatly assist the translation of the BPV1 L1 gene and human PV 16 (HPV16) E7 translation in different in vitro translation systems (20,30). These observations support the argument that codon usage of the mRNAs tends to be matched to the population of tRNAs in cells to regulate the translational efficiency. Previous studies have indicated that codon usage in highly expressed genes is biased toward ‘optimal’ codons corresponding to the tRNAs in the tissue or cell environments (31). Varenne et al. (31) reported that synthesis of a number of colicins is linked to the difference in tRNA availability for the various codons. However, it is not clear whether and how the tRNA abundance in cells matches the corresponding codons in mRNAs of the target genes to regulate their expression at the translational levels in specific mammalian cell lines.
In this report, we first examined whether the expression of the tRNASer(CGA) gene construct could enhance translation of wt PV L1 mRNAs in two replicating eukaryotic cell lines: Chinese hamster ovary (CHO) cell system that closely mimics living cells (32) and Cos1 cells that effectively transcribe wt PV L1 genes but do not produce L1 proteins (20). We then established two cell lines, CHO L1 and Cos1 L1, transfected with a wt BPV1 L1 expression construct, to determine whether the tRNASer(CGA) gene could assist translation of BPV1 L1 mRNA. At the same time, we examined whether the supplement of exogenous tRNAs would enhance translation of wt PV L1 mRNAs in cell culture systems. We then examined whether and how the expression of the tRNASer(CGA) gene affected the tRNA populations, phosphorylation of the translation initiation factors elF4E and elF2α, and expression of cellular proteins in the two L1 cell lines. We further studied whether the tRNASer(CGA) gene affected the expression of the L1 proteins encoded by two HB PV L1 genes (BPV1 and HPV6b L1 genes) in which CGA was a rarely used codon in the two cell lines. Finally, we investigated whether and how the tRNASer(CGA) gene affected expression of green fluorescent proteins (GFPs) expressed from hm gfp gene and six serine GFP variants, in which a leader sequence of six consecutive identical codons from one of the six serine codons (AGC, AGU, UCA, UCC, UCG and UCU) was introduced into the hm gfp gene downstream from the AUG codon in CHO cells.
MATERIALS AND METHODS
Plasmid constructions
The tRNASer(CGA) gene construction
The tRNASer(CGA) DNA (∼400 bp) was cut from pGEM-T tRNASer(CGA) [kindly provided by Lund et al. (15)] and ligated into a pSVneo vector at the EcoRI site to construct a pSV2tRNASer(CGA) plasmid [tRNASer(CGA) hereafter].
Construction of wild-type and codon-modified papillomavirus L1 genes
Plasmids pCDNA3BPV1 L1, pCDNA3BPV1 HB L1, pCDNA3HPV6b L1 and pCDNA3HPV6b HB L1 used in the experiments have been described previously (20). Briefly, both BPV1 and HPV6b wt L1 open-reading frames (ORFs) are ∼1.5 kb in length encoding 500 amino acids. The PV wt L1 genes show a strong codon usage bias, amongst degenerately encoded amino acids, toward 18 codons mainly with T at the third position that are rarely used by mammalian genes (20,33). We artificially modified BPV1 and HPV6b L1 genes, in which the L1 ORFs are substituted with codons preferentially used in the mammalian genome. We made about 250 base substitutions in 250 codons rarely used in mammalian cells to produce unmodified L1 proteins encoded from the L1 ORFs with consensus codon usage using the strategy previously reported (20). All the wt and codon-modified PV L1 sequences were sequenced and found to be error free; they were then cloned into the mammalian expression vector pCDNA3 containing SV40 ori (Invitrogen, Australia), giving four expression plasmids pCDNA3BPV1 L1, pCDNA3BPV1 HB L1, pCDNA3HPV6b L1 and pCDNA3HPV6b HB L1.
Construction of hm gfp gene and serine GFP variants
As described previously by Zhao et al. (35), a leader sequence of six consecutive identical codons from one of the six serine codons (AGC, AGU, UCA, UCC, UCG and UCU) was introduced into an hm gfp gene (Invitrogen, Australia) downstream from the AUG to produce six serine GFP variants. All the serine GFP variant and hm gfp cDNAs were produced from hm gfp by PCR using two flanking primers as described previously (35) and ligated into the EcoRI and KpnI sites of the pCDNA3 vector to construct seven GFP plasmids: hm gfp, AGC GFP, AGU GFP, UCA GFP, UCC GFP, UCG GFP and UCU GFP. All constructs were sequenced to ensure that no PCR-induced mutations are introduced. The correct orientations of all the plasmids used were confirmed by enzyme restriction analysis before being used for transfection.
Cell culture and DNA transfection
CHO cells, grown in Ham's F12 medium supplemented with 10% fetal bovine serum (FBS) (CSL, Australia), were brought up to 70% confluence in monolayer and then transfected with plasmid DNA using lipofectamine, following the instructions provided by the manufacturer (Invitrogen, Australia). After transfection, cells continued to grow in Ham's F12 medium supplemented with 10% FBS. Cos1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. Cells were transfected with plasmid DNA using Effectene reagent, following the instructions provided by the manufacturer (Qiagen, Australia). After transfection, cells continued to grow in DMEM medium supplemented with 10% FBS.
Establishment of BPV1 L1 expression cell lines
CHO or Cos1 cells were transfected with pCDNA3 BPV1 L1. G418 (Invitrogen, Australia) supplemented medium was used to select cells to establish two cell lines, CHO L1 and Cos1 L1. Expression of BPV1 L1 was confirmed by PCR after two or three passages; the cell lines were transfected with pSV tRNASer(CGA) or control expression constructs. Cells were collected 40 h after transfection and pelleted by centrifugation for Hirt and total genomic DNA preparation according to the methods described by Zhao et al. (34) and Thomas et al. (36). The DNA samples were digested overnight with DpnI/BamHI/EcoRI to remove any residual input DNA. Digested DNA was electrophoresed on 1% agarose and blotted on a nylon membrane. The membrane was hybridized with 32P-labeled mixed probes of BPV L1 and tRNASer(CGA) genes.
Protein sample preparation and western blotting
Both DNA-transfected CHO and Cos1 cells were collected for protein preparation 42 h post-transfection. Cell pellets were resuspended in phosphate-buffered 0.15 M sodium (PBS), pH 7.4, containing 2 mM phenylmethylsulpfnyl fluoride (PMSF) and sonicated for 40 s. An aliquot of 30 μg of total protein samples was resuspended in 40 μl of 1× Laemmli buffer (37), boiled for 8 min and loaded onto a 10% (w/v) polyacrylamide gel (SDS–PAGE) electrophoresis. After electrophoresis, the gel was electrotransferred onto a PVDF membrane (Bio-Rad). The blot was washed with PBS for 10 min, then blocked in PBS containing 5% skim milk for 1 h at room temperature and labeled with monoclonal antibody against BPV1 L1 protein (26) overnight at 4°C. The blot was incubated with secondary antibody goat anti-mouse immunoglobulin G (IgG) (Silenus, Australia), conjugated with horseradish peroxidase (HRP) at room temperature for 4 h and developed using an enhanced chemiluminescence (ECL) kit (Amersham, Australia). The L1-probed blots were stripped and relabeled with monoclonal antibody against β-tubulin (Sigma, Australia) to confirm equal loading of proteins samples.
Total RNA sample preparation and northern blot analysis
Total RNA was extracted from cells transfected with various plasmids, using the Machery-Nagel RNAII Kit (Integrated Sciences, Australia). An aliquot of 15 μg total RNA sample digested with DNase I was electrophoresed in 1.2% denatured agarose gels and blotted onto a Nylon N+ membrane (Amersham, Australia), then hybridized either with a 32P-labeled L1 gene or actin gene probe.
Immunofluorescence labeling
Cells were grown on 8-well chamber slides, transfected with the different plasmids, fixed and permeabilized with 85% ethanol for 42 h post-transfection. The fixed cells were then blocked with 5% skim milk–PBS and probed with monoclonal antibody against BPV1 L1 protein, followed by fluorescein-isothiocyanate-conjugated (FITC) anti-mouse IgG (Sigma, Australia). Nuclei were countstained by DAPI. Cells were examined by immunofluorescent microscopy.
tRNA preparation and northern blot hybridization
Total tRNAs were extracted and purified using a Qiagen kit as instructed by the supplier from CHO L1 or Cos1 L1 cell lines with or without transfection of the tRNASer(CGA) gene. An aliquot of 2 μg tRNA was fractioned by 10% denaturing PAGE containing 8 M Urea and 0.1 M sodium acetate (pH 5.2). Electrophoresis was carried out in 0.1 M sodium acetate (pH 5.2) at 80 V for ∼5 h. After electrophoresis, the tRNA gel was electroblotted onto Nytran Plus membrane in 40 mM Tris–acetate (pH 4.2) and 2 mM EDTA at 20 V for 90 min at 4°C. The membrane was crosslinked by 254 nm irradiation. The blot was prehybridized with hybridization buffer containing 6× SSC, 10× DEN, 0.2% SDS and 1 mM EDTA at 37°C for at least 4 h. The blot was then hybridized with DNA oligonucleotide probes complementary to the tRNASer(CGA) and three other tRNAs: tRNALys(AAA), tRNALys(AAG) and tRNAMet(initiator) in 4× SET buffer (1× SET = 0.15 M NaCl, 0.03 M Tris–HCl, 2 mM Na2EDTA, pH 8.0) at 37°C overnight. The DNA oligonucleotide probes complementary to the tRNASer(CGA) is 5′-CCTGAGCTTTAGGTTACC-3′; to tRNALys(AAA) is 5′-TCACTATGGAGATTTTA-3′, to tRNALys(AAG) is 5′-CGCCCAACGTGGGGCTC-3′ and to the tRNAMet(initiator) is 5′-TAGCAGAGGATGGTTTC-3′. The DNA oligomers were labeled with T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol, Amersham) at the first 5′ end. Specific activities of 108 to 109 c.p.m./μg were generally reached. Approximately 107 c.p.m. of oligomers was used per blot in hybridization reactions. Blots were washed with 1× SET buffer at 37°C and autoradiographed.
Aminoacyl-methionine-tRNA determination
The tRNA samples were used for direct analysis of aminoacyl-methionine-tRNA (aminoacyl-Met-tRNA). An aliquot of 2 μg tRNA sample was treated with Tris base to strip aminoacyl-tRNAs (0.5 M of Tris, pH 9.0, 37°C for 30 min), and the 2 μg tRNA sample was left untreated as control. Both Tris-treated and untreated tRNAs were fractioned by a denaturing PAGE and blotted onto Nytran Plus membrane described as above. The blot was then probed with DNA probes complementary to either tRNASer (CGA) or tRNAMet (initiator) as above.
Synthesis of cellular proteins
Synthesis of cellular proteins in CHO L1 or Cos1 L1 cell lines with or without transient transfection of the tRNASer(CGA) gene or pSVneo vector was studied by labeling with [35S]methionine. Cells at 32 h after transfection were incubated in 2 ml of medium supplemented with 10 μCi of l-[35S]Met (370 kBq) for 4 h. This extended labeling time is required to incorporate sufficient radioactivity into the newly synthesized proteins, a consequence of the relatively slow growth rates of these G418-treated cells. An aliquot of 10 μg protein sample was separated by 10% SDS–PAGE. Gels were stained with Coomassie blue or dried and autoradiographed.
RESULTS
Co-transfection of tRNASer(CGA) assists protein expression of PV wt L1 genes
Expression of the BPV1 L1 gene in cell-free translation systems is enhanced by the addition of exogenous tRNAs (20). To study whether provision of a single tRNA species similarly affected L1 expression, we examined BPV1 L1 transcription and translation in CHO and Cos1 cells transfected with BPV1 L1 gene constructs, either unmodified or codon modified to match eukaryotic consensus codon usage, and co-transfected with a tRNASer(CGA) expression construct. Northern blot hybridization revealed that both cell lines effectively transcribed BPV1 L1 whether or not codon modified (Figure 1A), confirming previous results obtained in our laboratory (20). Co-transfection of the tRNASer(CGA) expression construct did not affect the transcription of the BPV1 L1 gene in the two cell lines (Figure 1A). We then examined whether the L1 protein was expressed (Figure 1B). Neither cell line produced detectable L1 protein when transfected with the BPV1 wt L1 gene (Figure 1B), though, as previously shown (20), the codon-modified L1 gene was expressed. The L1 protein was expressed from the BPV1 wt L1 gene in cells co-transfected with the tRNASer (CGA) expression construct (Figure 1B). Immunofluorescent labeling confirmed that the L1 protein was expressed in both CHO and Cos1 cells following co-transfection of the BPV1 wt L1 and the tRNASer(CGA) expression constructs (Figure 1C). We also tested whether the tRNASer(CGA) gene could affect expression of the L1 gene from HPV 6b in the two cell lines, and similar results were obtained (data not shown). Thus, the tRNASer(CGA) gene product appeared to assist expression of L1 protein encoded by PV wt L1 genes in replicating mammalian cells.
Figure 1.
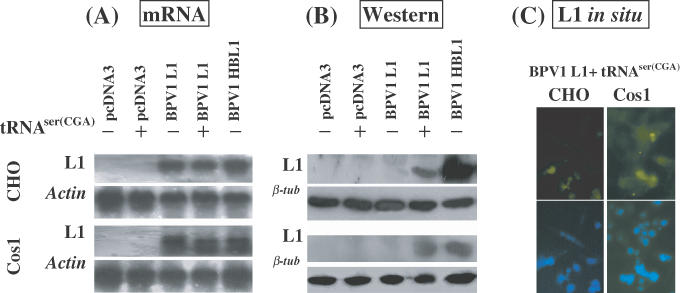
Translation of BPV1 L1 mRNA is enhanced by provision of tRNASer(CGA). CHO or Cos1 cells were transiently transfected with (i) pCDNA3, (ii) pCDNA3 + pSVtRNASer(CGA), (iii) pCDNA3BPV1 L1, (iv) pCDNA3BPV1 L1 + pSVtRNASer(CGA) and (v) pCDNA3BPV1 HBL1 (codon modified). (A). Northern blot hybridization of L1 mRNA transcription. RNA samples were prepared at 42 h post-transfection. Of each sample of DNase-I-digested total RNA, 10 μg was electrophoresed on a 1.2% denatured agarose gel and blotted onto a nylon membrane. The northern blots were hybridized with 32P-labeled wt and HB L1 probe mixture. As controls, the northern blots were hybridized with 32P-labeled actin gene probe. (B) Immunoblotting analysis of the L1 protein. Monoclonal antibody against BPV L1 protein was used to probe the blots. Upper panels show the results of L1 immunoblotting assay; lower panels show the results of β-tubulin immunoblotting assay indicating equal loading of the protein samples. (C) L1 expression in CHO and Cos1 cells co-transfected with pCDNA3BPV1 L1 and pSVtRNASer(CGA) was demonstrated by indirect immunofluorescence microscopy. Upper panels show the L1 labeling; lower panels show the nuclear staining by DAPI.
To confirm that enhanced translation of L1 mRNA was specifically related to the expression of the tRNASer(CGA) gene, we examined the effects of translation of cells with the pSVNeo basal vector or with exogenous supplementation of yeast tRNAs. L1 was effectively transcribed in BPV1 L1 transfected cells, whether or not the cells were co-transfected with the pSVNeo basal plasmid or exposed to exogenous yeast tRNAs (Figure 2A). However, the L1 protein was only detected in cell lines co-transfected with the BPV1 L1 and tRNASer(CGA) expression constructs (Figure 2B). Results further indicate that expression of the tRNASer(CGA) gene can thus assist translation of BPV1 L1 protein in non-differentiated mammalian cells.
Figure 2.
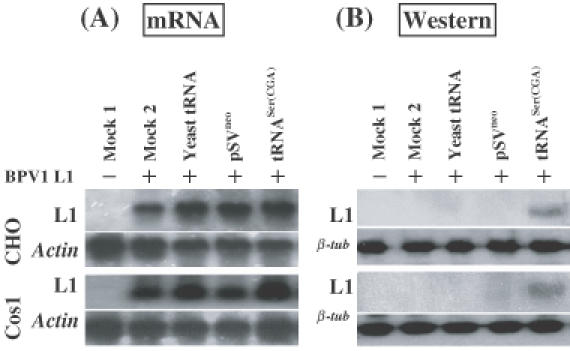
Enhancement of translation of BPV1 wt L1 mRNA by tRNASer(CGA) is specific. CHO or Cos1 cells were transiently transfected with (i) pCDNA3 (mock 1), (ii) pCDNA3BPV1 L1 (mock 2), (iii) pCDNA3BPV1 L1 plus exogenous supplement of yeast tRNAs in culture medium (commercial bake yeast tRNAs from Sigma, Australia, were added into CHO or Cos1 cell cultures at 1 μg/ml following transfection of BPV1 wt L1 expression construct), (iv) pCDNA3BPV1 L1 plus pSVneo basal vector, and (v) pCDNA3BPV1 L1 plus pSVtRNASer(CGA). (A). Northern blot hybridization of L1 mRNA transcription. RNA samples were prepared at 42 h post-transfection. Of each sample of DNase-I-digested total RNA, 10 μg was electrophoresed on a 1.2% denatured agarose gel and blotted onto a nylon membrane. Northern blots were hybridized with 32P-labeled BPV1 wt L1 probe (upper panel). As controls, the northern blots were also hybridized with 32P-labeled actin gene probe (lower panel). (B) Western blotting analysis of L1 protein. Monoclonal antibody against BPV L1 protein was used to probe on the blot. Upper panels show the results of L1 immunoblotting assay; lower panels show the results of β-tubulin immunoblotting assay indicating equal loading of the protein samples.
tRNASer(CGA) induces L1 protein expression in cell lines stably transfected with L1 gene
To examine whether enhanced translation of the BPV1 L1 gene, when co-transfected with the tRNASer(CGA) expression construct, is due to a consequence of their co-transfection or expression of the tRNASer(CGA) gene, we established cell lines, CHO L1 and Cos1 L1, stably transfected with a BPV1 L1 expression construct (Figure 3A). Multiple replication forms revealed by Southern blot hybridization using total genomic DNA samples confirmed the presence of integrated BPV1 L1 genes (data not shown). However, the episomal format of the BPV1 L1 gene was also observed in Hirt DNA samples, presumably because of passage number of the cell lines (Figure 3B). The tRNASer(CGA) gene was only detected in the two L1 cell lines following transfection with the tRNASer(CGA) expression construct (Figure 3B).
Figure 3.
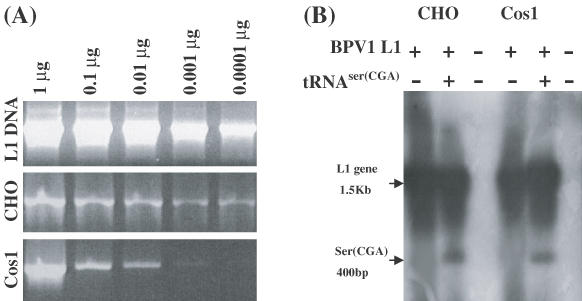
Replication of BPV1 L1 and tRNASer(CGA) genes in CHO L1 and Cos1 L1 cells. (A) PCR analysis of DNA prepared from CHO L1 or Cos1 L1 cells, confirming that the cell lines retained a BPV1 L1 expression construct. (B) Southern blot analysis of Hirt DNA for BPV1 L1 and tRNASer(CGA) gene replication in both CHO L1 and Cos1 L1 cells. 20 μg Hirt DNA samples prepared from CHO L1 or Cos1 L1 cells either untransfected or transfected with a tRNASer(CGA) expression plasmid were restricted with BamHI and EcoRI plus DpnI for at least 4 h and electrophoresed on a 1.0% agarose gel and blotted onto nylon membrane. The Southern blot was hybridized with a mixture of 32P-labeled BPV1 L1 and tRNASer(CGA) gene probes.
The two L1 cell lines transcribed the BPV1 L1 gene equally, whether they were transfected with the tRNASer(CGA) gene or a basal plasmid and also when exposed in culture to exogenous yeast tRNAs (Figure 4A). However, the L1 protein was produced only in cell lines transfected with the tRNASer(CGA) expression construct, but not with control plasmids, and/or supplemented with exogenous yeast tRNAs. Again, immunofluorescent labeling confirmed that L1 protein was produced in the two L1 cell lines after transfection of the tRNASer(CGA) gene (Figure 4C). The results confirm further that the L1 protein was produced due to the presence of the tRNASer(CGA) gene product.
Figure 4.
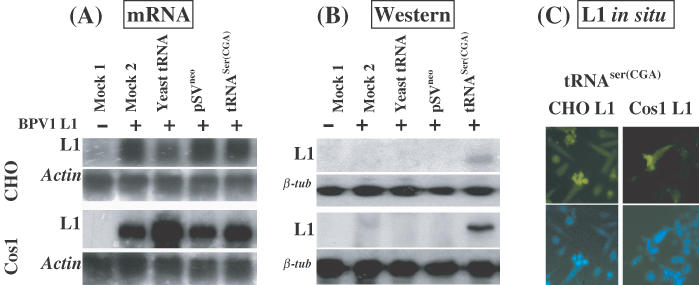
Expression of BPV1 L1 gene in cell lines carrying an L1 expression construct is influenced by transfection of pSV tRNASer(CGA). (i) CHO or cos1 cells only (mock 1). The CHO L1 and Cos1 L1 cell lines were transiently transfected with (ii) pCDNA3; (iii) no plasmid, but exogenous supplement of yeast tRNAs in culture medium as described in Figure 2; (iv) pSVneo basal vector; and (v) pSV tRNASer(CGA). (A) Northern blot hybridization of L1 mRNA transcription. RNA samples were prepared at 42 h post-transfection. Of each sample of DNase-I-digested total RNA, 10 μg was electrophoresed on a 1.2% denatured agarose gel and blotted onto a nylon membrane. The northern blots were hybridized with 32P-labeled BPV1 L1 probe (upper panel). As controls, the northern blots were hybridized with 32P-labeled actin gene probe (lower panel). (B) Western blotting analysis of L1 protein. Monoclonal antibody against BPV1 L1 protein was used to probe the blots. Upper panels show the results of L1 immunoblotting assay; lower panels show the results of β-tubulin immunoblotting assay indicating equal loading of the protein samples. (C) Immunofluorescence labeling of L1 expression in CHO and Cos1 L1 cells after transfection of the tRNASer(CGA) gene. Upper panels show the L1 labeling; lower panels show the nuclear staining by DAPI.
Action of tRNASer(CGA) on expression of BPV1 wt L1 protein is specific
To examine whether the transfection itself might induce expression of the BPV1 L1 protein, we transfected CHO L1 cell lines with not only the tRNASer(CGA) expression construct, but also several vectors including pIBT100, pUC19, pcDNA3 and pSVneo. No L1 protein expression was detected following the transfection of the control vectors in CHO L1 cells (Figure 5), suggesting further that expression of the BPV1 L1 protein was due to the tRNASer(CGA) gene product.
Figure 5.
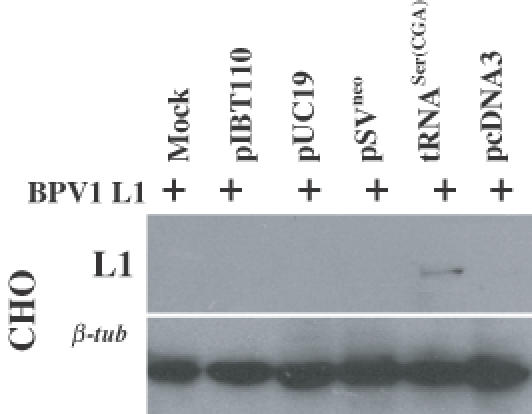
Expression of BPV1 L1 protein in CHO L1 cell line requires provision of tRNASer(CGA). Immunoblotting for L1 protein in CHO L1 cells transfected with various plasmids shown in upper panel using a monoclonal antibody against L1 protein and for β-tubulin shown in lower panel indicating equal loading of the protein samples.
tRNASer(CGA) results in enhanced expression of other tRNA species
It was unexpected that the expression of a single tRNA gene would induce translation of wt PV L1 mRNAs as the block to translation might be expected to be a consequence of shortages of many tRNA species critical to optimal translation of this gene. Thus, to determine why this might occur, we examined whether the transduced tRNASer(CGA) gene affected expression of tRNAs other than tRNASer(CGA) in the two L1 cell lines. We first used polyacrylamide gel electrophoresis to fractionate total tRNAs, prepared from CHO L1 and Cos1 L1 cell lines, either untransfected or following transfection of the tRNASer(CGA) expression construct. As shown in Figure 6A, total tRNA populations prepared from the two L1 cell lines differed according to whether they had been transfected with the tRNASer(CGA) gene (Figure 6A). Several extra tRNA bands in relative low molecular weights were observed in the L1 cell lines after transfection of the tRNASer(CGA) gene (Figure 6A), suggesting that expression of the tRNASer(CGA) gene may result in enhanced production of new tRNA species.
Figure 6.
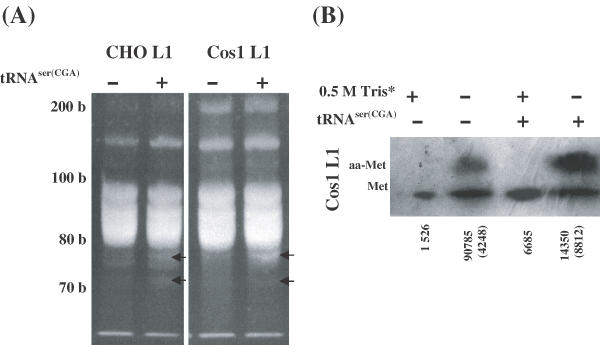
Fractionation of total tRNAs isolated from CHO L1 and Cos1 L1 cells with or without transfection of the tRNASer(CGA) gene and northern blot analysis for aminoacyl (aa) forms of methionine tRNA. (A) Total tRNAs on 10% denatured PAGE, extra tRNA bands induced by tRNASer(CGA) expression construct are marked by solid arrows on the right side. (B) Analysis of uncharged and aa-forms of methionine tRNA. Parts of the tRNA samples denoted by an asterisk was treated with 0.5 M Tris at pH 9.0 to strip aa-tRNAs before fractionated on the denaturing PAGE as described in Materials and methods. tRNA blots were probed with a 32P-lableled oligonucleotide complementary to specific regions of tRNAMet(ATG). Uncharged and aminoacyl forms of methionine tRNA are marked on the left position. The relative levels of uncharged and aminoacyl-methionine tRNA shown by arbitrary units (number) were measured from the blots by densitometry. The aa-methionine tRNA signals are shown by the arbitrary units in parentheses.
We arbitrarily chose four oligonucleotide probes that were complementary to specific regions of the four tRNA species, tRNASer(CGA), tRNALys(AAA), tRNALys(AAG) and tRNAMet(AUG) for northern blot analysis. The four probes mixed to detect the tRNA blot revealed that expression of the tRNASer(CGA) gene slightly increased the signals of the four tRNAs in the two L1 cell lines based on densitometric analysis (data not shown). To further examine whether the tRNASer(CGA) gene affected aminoacylation of the tRNA species in the two L1 cell lines, we detected the different forms (uncharged and aminoacyl-tRNAs) of the tRNASer(CGA) and initiator tRNAMet(AUG) by northern blot hybridization. The tRNASer(CGA) gene increased slightly the levels of aminoacylation of the tRNASer(CGA) in both L1 cell lines (data not shown). In contrast, two forms of uncharged and aminoacyl-tRNAMet(AUG) were well-separated in Cos1 L1 cells (Figure 6B), although not in CHO L1 cells. A high percentage of the tRNAMet(AUG) was aminoacylated (Figure 6B), and tRNASer(CGA) gene significantly increased the levels of aminoacylation of the tRNAMet(AUG) in Cos1 L1 cell line (Figure 6B), suggesting that expression of the tRNASer(CGA) gene may affect translation initiation.
tRNASer(CGA) induces phosphorylation of translation initiation factors
We then examined the phosphorylation status of two translation initiation factors, elF4E and elF2α, in the L1 cell lines with or without transfection of the tRNASer(CGA) gene to investigate whether the tRNASer(CGA) gene product affects phosphorylation of the translation initiation factors (Figure 7). By performing western blotting analysis with antibodies that specifically recognize the phosphorylation forms of elF4E and elF2α, no phosphorylation of both elF4E and elF2α was observed in CHO L1 and Cos1 L1 cells (Figure 7), in contrast, phosphorylated elF4E and elF2α were detected in CHO L1 and Cos1 L1 cells transfected with the tRNASer(CGA) gene (Figure 7). At the same time, the phosphorylation levels of both elF4E and elF2α are much higher in CHO L1 cells than in Cos1 L1 cells. Consistent with the L1 protein detected only in both CHO L1 and Cos1 L1 cells transfected with the tRNASer (CGA) gene (Figure 7), the results suggest that the phosphorylation of the two translation initiation factors elF4E and elF2α induced by the tRNASer(CGA) gene product may be involved in enhancing the translation of the L1 protein in the two cell lines examined.
Figure 7.
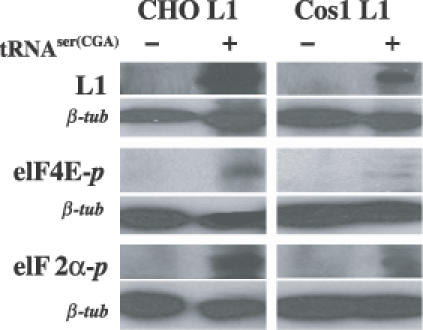
Effects of the tRNASer(CGA) gene product on phosphorylation of two translation initiation factors, elF4e and elF2α, in CHO L1 and Cos1 L1 cell lines. Protein samples of 40 μg prepared from the two L1 cell lines with or without transfection of the tRNASer(CGA) gene were resolved by SDS–PAGE, transferred to the PVDF membrane, and analyzed by western blotting with antibodies against L1 (top panel), eIF4E-P phosphorylated at serine 209 [eIF4E-P (Ser 209), middle panel] and eIF2α-P phosphorylated at serine 51[eIF2α-P (Ser 51), bottom panel].
tRNASer(CGA) increases yields of the total cellular proteins
As both the alteration of tRNA populations and the increased aminoacylation of initiator tRNAMet(ATG), together with the phosphorylation of both elF4e and elF2α, might impact on the synthesis of cellular proteins, we examined whether expression of the tRNASer(CGA) gene affected the production of cellular proteins in the two L1 cell lines. Cells (1 × 106/well) either untransfected or following transfection with the basal pSVneo vector and the tRNASer(CGA) gene were cultured for 48 h and collected for total protein analysis. Untransfected CHO L1 cells and the CHO L1 cells following transfection of the basal pSVneo vector produced significantly less proteins (0.547 ± 0.011 mg and 0.566 ± 0.014 mg, respectively) than CHO L1 cells following transfection of the tRNASer(CGA) gene (0.606 ± 0.015 mg) (Figure 8A). Cos1 L1 cells following transfection of the tRNASer(CGA) gene produced 0.690 ± 0.009 mg protein, slightly more than untransfected Cos1 L1 cells (0.665 ± 0.008 mg) and the pSVneo-transfected Cos1 L1 cells (0.653 ± 0.012 mg) (Figure 8A). SDS–PAGE was used to fractionate equal amounts of the cellular proteins from the two L1 cell lines either untransfected or following transfection with basal pSVneo vector and the tRNASer(CGA) gene, which was stained with Commasie blue (Figure 8B). The SDS–PAGE profiles of the total cellular proteins in the two L1 cell lines with or without transfection of the tRNASer(CGA) gene are different (Figure 8B). But transfection of the basal pSVneo vector appeared not to affect the SDS–PAGE profiles of the total cellular proteins in the two L1 cell lines (Figure 8B). The levels of some cellular proteins were significantly enhanced by the tRNASer(CGA) gene (Figure 8B). At the same time, a few proteins seemed to be reduced in their amounts in the L1 cells after transfection with the tRNASer(CGA) gene. These observations were confirmed by [35S]methionine labeling experiments (Figure 8B), suggesting that the tRNASer(CGA) gene product had negative effects on the synthesis of some cellular proteins, although it increased the yields of the total cellular proteins in the two L1 cell lines.
Figure 8.
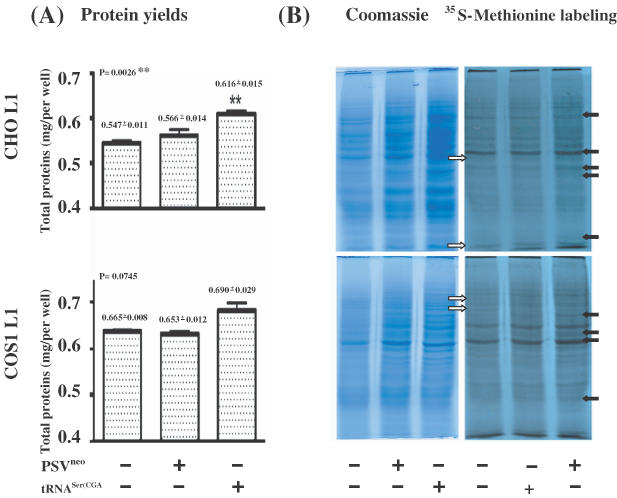
Effects of the tRNASer(CGA) gene product on cellular proteins in CHO L1 and Cos1 L1 cell lines. (A) Yields of total cellular proteins. CHO L1 and Cos1 L1 cells (1 × 106) were transfected with either the tRNASer(CGA) gene or vector pSVneo, or not transfected, and analyzed for total cellular proteins after 48 h. The yields of total cellular proteins shown in milligrams (mgs) over the histograms are the means of triplicates. Vertical bars indicate the standard errors (n = 3). P-values are shown on the top corner of the left side. Double asterisks indicate that the difference in total proteins in the CHO L1 cell lines without and with the tRNASer(CGA) gene transfection, and between the tRNASer(CGA) gene and vector pSVneo transfection is significant different (P < 0.01). (B) Synthesis of cellular proteins. An aliquot of 10 μg of the protein samples was separated by 10% SDS–PAGE in duplicates. Left panels show the PAGE gels stained with Coomassie blue. Right panels show the newly synthesized proteins labeled by [35S]methionine after the PAGE gels were dried on 3 M paper and exposed to X-ray film. Solid arrows on the right indicate that relative levels of the proteins are increased in both L1 cell lines after transfection of the tRNASer(CGA) gene, and empty arrows on the left indicate that relative levels of the proteins are reduced in both L1 cell lines after transfection of the tRNASer(CGA) gene.
tRNASer(CGA) reduces expression of the L1 proteins encoded by codon-modified PV L1 genes
To explain why the tRNASer(CGA) gene overexpression has variable effects on expression of different cellular proteins, we hypothesize that the matching between tRNAs and codon usage of the genes regulates expression of the cellular proteins in the cells. To test our hypothesis, we examined whether the tRNASer(CGA) gene could affect expression of L1 protein encoded by a codon-modified BPV1 L1 gene (BPV1 HB L1), which has been demonstrated to translate L1 protein in large amounts in replicating mammalian cells in previous studies (20,30) and in the current one (Figure 1B). Both CHO and Cos1 cells transfected solely with BPV1 HB L1 gene or co-transfected with BPV1 HB L1 and tRNASer(CGA) genes, were incubated for 42 h and collected for L1 mRNA and protein expression analysis (Figure 9). Northern blot hybridization confirmed that the BPV1 HB L1 gene is effectively transcribed (Figure 9A). No significant differences in the levels of L1 mRNA between transfection solely with BPV1 HB L1 gene and co-transfection with BPV1 HB L1 and tRNASer genes were observed (Figure 9A), which was confirmed by quantitative RT–PCR analysis using the L1 mRNAs (data not shown). However, western blot analysis shows that the tRNASer(CGA) gene resulted in a reduction in the levels of the L1 protein expressed from the BPV1 HB L1 gene by >50% (Figure 9B) in the two cell lines, whereas β-tubulin expression remained unaffected (Figure 9B). To confirm these observations, we examined whether expression of another codon-modified PV L1 gene, HPV6b HB L1, was also affected by expression of the tRNASer(CGA) gene (Figure 9B). The results (Figure 9) are in agreement with those obtained in the experiments using BPV1 HB L1 gene, supporting the idea that tRNAs may complement the codon composition of genes to regulate gene translation in mammalian cells.
Figure 9.
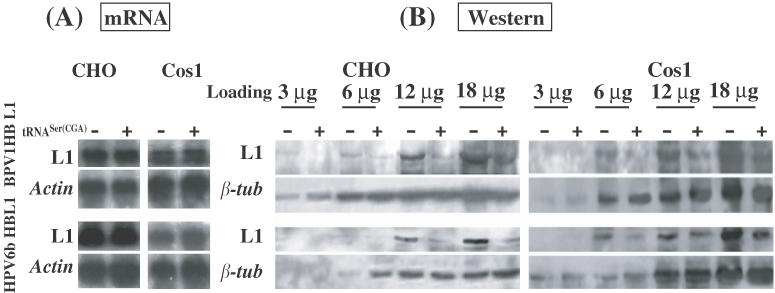
Effects of the tRNASer(CGA) expression construct on expression of L1 proteins encoded by codon-modified PV L1 genes (BPV1 HB L1 and HPV6b HB L1) in CHO and Cos1 cells. (A) L1 mRNA levels. CHO or Cos1 cells were transiently transfected with PV HB L1 gene construct only, or with PV HB L1 gene construct and the tRNASer(CGA) gene. (B) L1 protein expression. Four dilutions of protein samples from cells either transfected with PV HB L1 gene construct only, or co-transfected with PV HBL1 gene construct and the tRNASer(CGA) gene were loaded onto SDS-PAGE gel for protein fractionation and blotted on PVDF membranes for immunoblotting assay. Monoclonal antibody against BPV L1 protein was used to probe the blots. Upper panels show the results of L1 immunoblotting assay; lower panels show the results of β-tubulin immunoblotting assay indicating equal loading of the protein samples between two treatments.
tRNASer(CGA) differentially regulates expression of serine GFP variants
Based on the above results (Figure 9), we hypothesize further that the tRNASer(CGA) gene may favor the expression of the target genes in which UCG or other codons from the six serine codons are used more frequently. To test this hypothesis, we studied transcription and translation of the hm gfp gene and the six serine GFP variants in both CHO and Cos1 cells either transfected solely with hm gfp and six serine GFP variants or co-transfected with the GFP expression constructs and the tRNASer(CGA) gene. The serine GFP variants express the same protein sequences, but the six additional serines at the N-terminal of their GFP molecules following the AUG codon were encoded by one of the six serine codons (AGC, AGU, UCA, UCC, UCG and UCU), respectively. Northern blot hybridization revealed that the hm gfp gene and the six serine GFP variants were transcribed effectively in the two cell lines and co-transfection of the tRNASer(CGA) gene did not affect significantly the levels of their transcripts (data not shown). Western blot analysis first shows that all six serine GFP variants exhibited levels of immunoreactive GFPs significantly higher than hm gfp gene (Figure 10A), confirming our previous study that addition of serine codons to hm gfp gene downstream from the AUG increased significantly steady-state levels of GFP (35). Then, western blot analysis shows that co-transfection of the tRNASer(CGA) gene barely affected the levels of immunoreactive GFPs produced by hm gfp gene and two serine (AGC and UCU) GFP variants (Figure 10B). However, tRNASer(CGA) increased significantly the levels of immunoreactive GFPs produced by AGU and UCG GFP variants (Figure 10B), in contrast to the significant reduction of the level of immunoreactive GFPs produced by the UCA GFP variant followed the co-transfection of the tRNASer(CGA) gene (Figure 10B). Moreover, tRNASer(CGA) slightly increased the level of immunoreactive GFP produced by UCC GFP variants (Figure 10B). In the parallel experiments, we used flow cytometry analysis (FAC) to examine fluorescence intensity in both CHO and Cos1 cells either transfected solely with hm gfp and six serine GFP variants, or co-transfected with the GFP expression constructs and the tRNASer(CGA) gene. Results obtained from the FAC analysis (data not shown) confirmed the western blotting analysis for immunoreactive GFPs produced by the hm gfp gene and six serine GFP variants in the two cell lines, proving our hypothesis.
Figure 10.
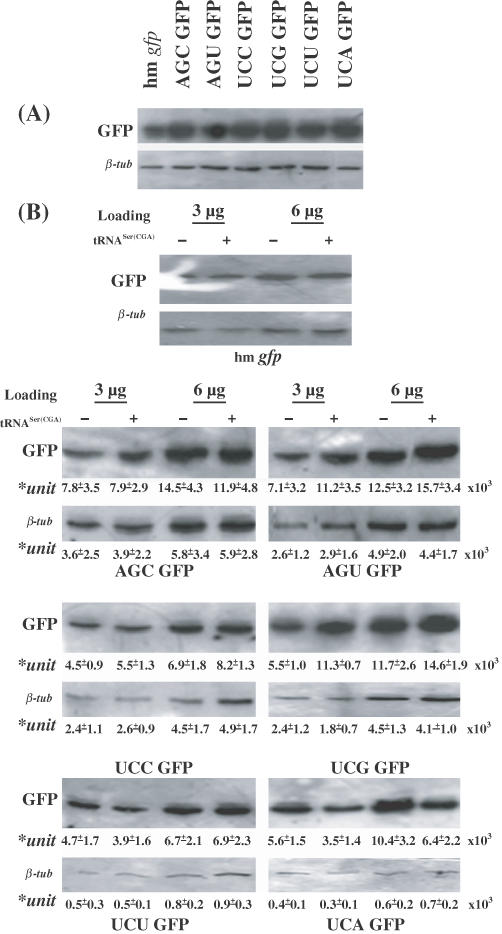
Variable effects of the tRNASer(CGA) expression construct on expression of GFPs encoded by hm gfp and six serine GFP variants in CHO cells. (A) Protein expression of hm gfp and six serine GFP variants in CHO cells transiently transfected solely with the seven GFP plasmid DNAs 42 h post-transfection. Protein sample 4 μg was separated by SDS–PAGE and blotted on PVDF membrane. Signals of both GFPs produced by the GFP expression constructs and β-tubulin were detected by monoclonal antibodies against both GFP and β-tubulin. (B) Effects of the tRNASer(CGA) expression construct on expression of GFPs produced by the hm gfp gene and six serine GFP variants in CHO cells. Two dilutions of protein samples from CHO cells either transfected solely with hm gfp and six Serine GFP variants, or co-transfected with GFP gene construct and the tRNASer(CGA) gene 42 h post-transfection were loaded onto SDS–PAGE gel for protein fractionation and blotted on PVDF membranes for immunoblotting assay. Upper panels show the results of GFP immunoblotting assay; lower panels show the results of β-tubulin immunoblotting assay indicating equal loading of the protein samples between two treatments. The relative levels of GFPs and tubulin produced by the six serine GFP variants with or without co-transfection of the tRNASer(CGA) gene are shown by arbitrary units (means + SD× 103), which were measured from the four blots by densitometry from two independent experiments.
DISCUSSION
In the present study, supplementation of exogenous tRNAs did not enhance the translation of PV wt L1 genes in two cell lines. The results cannot confirm our and other previous observations that supplementation of exogenous tRNAs enhanced the translation of PV L1 and E7 genes in in vitro translation systems (20,30), but they are consistent with the studies carried out by the other researchers that exogenous tRNAs have no effect on protein synthesis in permeabilized cells (32,38). The major reason is different translation machinery used in the in vitro translation systems and cell culture systems. Using CHO cell system, previous work has provided strong evidence that the translation apparatus is highly organized in cultured cells (32). Exogenous tRNAs cannot enter the translation machinery in cultured cells despite the fact that exogenous tRNA can rapidly distribute throughout the cells and can be aminoacylated by minor portion of the cellular aminoacyl-tRNA synthetases (aaRS) in permeabilized cells (38). Moreover, exogenous aminoacyl-tRNA in permeable cells is not sequestered and protected against RNase treatment, in contrast to endogenous tRNA molecules (39), because during translation endogenous tRNAs are never in a state in which they can mix with and be competed against by exogenous tRNA molecules (38). All of the above observations may explain why exogenous tRNAs did not play a role in enhancing the translation of PV L1 mRNA in both CHO and Cos1 cells examined in the present study if they were efficiently taken up by the two cell lines. It is also possible that exogenous tRNAs were not taken up by the cells so no functional roles could be identified.
Previous studies have indicated that changes of physiological factors resulted in alteration in the populations of specific or total tRNAs in the mammalian tissues or cells (40–44). For example, ovariectomy resulted in alterations in the population of uterine serine tRNA (40). Due to carcinogenesis, cellular tRNA distribution was shifted in the tRNA population with an increased level of initiator tRNAMet in the malignant tissues (44). Selenium, an essential trace element that functions in proteins (selenocysteine, Sec) critical for a variety of cellular processes, influences the level of the Sec tRNA[ser]sec population and the distribution of the Sec tRNA[ser]sec isoacceptors in both mammalian cells grown in culture (45) and mammalian tissues (46) and the turnover of Sec tRNA[ser]sec in CHO cells (43). The present data indicate further that the expression of the tRNASer(CGA) gene increased not only the relative levels of the tRNASer(CGA) and the initiator tRNAMet but also probably resulted in new tRNA species produced in the two cell lines. Consequently, expression of the tRNASer(CGA) gene altered the gel profiles of the total tRNAs in the two cell lines. Our results are in agreement with previous studies (47–52) in that genes for expressing tRNA species introduced into cells change tRNA contents in the targeted cells. Thus, all the observations suggest that mammalian cells may have a sensitive mechanism to sense and respond to both changes in physiological factors and introduction of foreign tRNA genes, by altering their tRNA populations.
Here, expression of the tRNASer(CGA) gene was identified to enhance the expression of L1 proteins encoded by the PV wt L1 genes and of GFPs encoded by two (UCG and AGU) serine variants in two cell lines examined. Previously, there have been examples in which synthesis of a specialized protein is associated with expression of a single tRNA gene due to modification of the tRNA population in different expression systems (9,10). Expression of the tRNASer(CGA) gene was then found to increases the synthesis of some cellular proteins in the present study. As a result, expression of 5–10% of total cellular proteins was increased in the L1 cells following transfection of the tRNASer(CGA) gene when compared to the control L1 cells. Our results support previous studies that tRNAs, even a single tRNA, affect protein expression encoded by heterologous genes in different expression systems (9,10,20,30,40,41). For example, tRNAs from oviducts of estrogen-stimulated chicks or from oviducts of laying hens produced an enhanced stimulation of ovalbumin synthesis in vitro (40). The use of tRNA (AGA/AGG) increased human interferon HUIFN-a2 and HCV core protein expression (9,10), but barely affected the human IFN-α8 expression in an E.coli expression system (9,10). However, expression of the tRNASer(CGA) gene reduced expression of L1 proteins encoded by the PV HB L1 genes and of GFP encoded by UCA serine variant and of some cellular proteins. The result was consistent with a few other studies. A tRNASer(UCN) gene flanked with the deafness-associated mitochondrial DNA (mtDNA) T7445C mutation at the 3′ end causes an average reduction of ∼70% in the levels of the tested proteins and a decrease of ∼45% in protein synthesis rate in the cell lines analyzed (53). Recently, Akama and Beier (54) reported that co-transfection of Arabidopsis hypocotyls with amber suppressor tRNASer(CGA) gene and the GUS reporter gene resulted in 10% of the GUS activity found in the same tissue transformed solely with the GUS gene. Therefore, we can speculate that contents and modifications of the endogenous tRNAs have effects on the expression of a large number of both foreign and host genes through tRNA function in translation in mammalian cells. Our findings are consistent with the conclusion that the host cell tRNAs can affect the synthesis of gene products quantitatively and qualitatively in translating heterologous genes (55,56).
The most interesting finding in the present study is that the tRNASer(CGA) gene had variable effects on expression of the PV wt and HB L1 genes and of hm gfp gene and serine GFP variants at protein translational levels in the two cell lines examined. Previous studies have reported that the expression of the PV capsid genes (L1 and L2) is controlled by several means: the late viral promoters depend on the differentiation status of the cells, polyadenylation signals terminate transcription before reaching the late regions (57–60) and mRNAs encoding the capsid proteins contain inhibitory elements that prevent nuclear export or destabilize the message (19,61–66). However, none of these studies produced the PV L1 and L2 proteins expressed from the wt L1 and L2 genes in replicating mammalian cells. In contrast, previous findings in our laboratory have indicated that a strong bias in codon usage is apparent between the papillomavirus and its host, which blocks BPV1 capsid genes from expressing their proteins in non-differentiated mammalian cells (20,33). On the basis of the hypothesis that the match between codon usage and tRNA contents regulates the gene expression at the protein levels (20), the limitation of PV capsid protein expression has been overcome by modifying the codon usage of these genes toward mammalian ‘consensus’ without changing the protein sequence, demonstrating that codon usage is one of the determinants of the rate of HPV gene expression in non-differentiated mammalian cells (20,22–24,67). In the present study, we take a different approach to test the hypothesis that the match between codon usage and tRNAs regulate PV L1 genes to express their proteins. We expressed a tRNASer(CGA) gene in the two non-differentiated cell lines. Expression of the tRNASer(CGA) gene resulted in alterations in tRNA patterns in the two cell lines examined, which enhanced translation of two PV wt L1 mRNAs that contain large numbers of the codons rarely used by mammalian genes (20,33). Meanwhile, the enhanced expression of the serine UCG GFP variant was probably corresponding to the expression of the tRNASer(CGA) gene according to a previous statement that correct translation of the UCG codon depends on the tRNASer(CGA) molecule (68). Although it has been reported that tRNA1Ser(G34) with the anticodon GGA can recognize not only UCC and UCU codons but also UCA and UCG codons (69), but the enhanced expression of the serine AGU GFP variant by the expression of the tRNASer(CGA) gene is presently not understood. Moreover, we observed that expression of the tRNASer(CGA) gene resulted in a dramatic reduction in the translation of PV HB L1 and UCA serine GFP variant mRNAs in the two cell lines. Thus, the phenomena observed in the present study may be well explained by two previous statements by Del Tito et al. (55) and Hatfield et al. (70). These statements have described how the tRNAs in cells are related to the gene expression at translation levels: (i) if the level of certain tRNAs was low, then one would predict translational difficulties in decoding mRNA species containing large numbers of the rare codons (55) and (ii) the cellular content of tRNA species often correlates positively with the codon bias of mRNAs for the major homologous protein products (70). Taken together with these statements (55) and our previous studies (20), current data strengthen support for the hypothesis that codon usage matches tRNAs to regulate expression of the target genes in both non-differentiated and differentiated epithelium.
Recently, Walsh and Mohr (71) provided the first evidence that phosphorylation of the translation initiation factor elF4E by the elF4E kinase mnk-1 is crucial for viral protein synthesis and replication of the herpes simplex virus-1 (HSV-1) in quiescent cells. Consistent with their report, phosphorylation of the two translation initiation factors elF4E and elF2α induced by the tRNASer(CGA) gene appears to be required for the translation of the PV wt L1 mRNAs in the two cell lines examined in the present study. In contrast to this expectation, however, the phosphorylation of elF4E and elF2α may have selective effects, even reducing the translation of the PV HB L1 and serine UCA GFP mRNAs. Thus, the expression of the tRNASer(CGA) gene was able to regulate the phosphorylation levels of both elF4E and elF2α, suggesting that another possible mechanism is involved by which the PV L1 genes and serine GFP variants may differentially regulate cap-dependent and cap-independent translation initiation.
Based on our observations, we can conclude that alterations in tRNAs and phosphorylation of elF4E and elF2α caused by the expression of the tRNASer(CGA) gene are important in modulating expression of the PV L1 genes with different codon usage and serine GFP variants at protein translational levels in the two cell lines. However, tRNAs must associate with their cognate aaRSs that catalyze the attachment of a particular amino acid to the 3′ end of an appropriate tRNAs containing the anticodon corresponding to that amino acid to generate aminoacyl-tRNAs necessary for protein synthesis. But, it is not clear whether the expression and activities of any aaRSs is affected by the tRNASer(CGA) gene. Detailed studies are therefore necessary to examine how expression of the tRNASer(CGA) gene affects the other components of the protein-synthesizing machinery, including the aaRSs and the elongator factors in the two cell lines.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rebecca Brown and Nigel McMillan for reading the manuscript. This work was funded in part by the Queensland Cancer Fund (Q52 to K.N.Z. and I.H.F., Q68 to K.N.Z.), the National Health and Medical Research Council of Australia Industry Research Fellowship (301256 to K.N.Z.).
REFERENCES
- 1.Crick F. (1970) Central dogma of molecular biology. Nature, 227, 561–563. [DOI] [PubMed] [Google Scholar]
- 2.Hershey J.W. (1991) Translational control in mammalian cells. Annu. Rev. Biochem., 60, 717–755. [DOI] [PubMed] [Google Scholar]
- 3.Smith D.W. (1975) Reticulocyte transfer RNA and hemoglobin synthesis. Science, 190, 529–535. [DOI] [PubMed] [Google Scholar]
- 4.Chen G.F. and Inouye,M. (1990) Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res., 18, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura Y. and Tabata,S. (1997) Codon-anticodon assignment and detection of codon usage trends in seven microbial genomes. Microb. Comp. Genomics, 2, 299–312. [DOI] [PubMed] [Google Scholar]
- 6.Oba T., Andachi,Y., Muto,A. and Osawa,S. (1991) Translation in vitro of codon UGA as tryptophan in Mycoplasma capricolum. Biochimie, 73, 1109–1112. [DOI] [PubMed] [Google Scholar]
- 7.Mazzacano C.A., Du,X. and Thiem,S.M. (1999) Global protein synthesis shutdown in Autographa californica nucleopolyhedrovirus-infected Ld652Y cells is rescued by tRNA from uninfected cells. Virology, 260, 222–231. [DOI] [PubMed] [Google Scholar]
- 8.Imamura H., Jeon,B., Wakagi,T. and Matsuzawa,H. (1999) High level expression of Thermococcus litoralis 4-alpha-glucanotransferase in a soluble form in Escherichia coli with a novel expression system involving minor arginine tRNAs and GroELS. FEBS Lett., 457, 393–396. [DOI] [PubMed] [Google Scholar]
- 9.Acosta-Rivero N., Sanchez,J.C. and Morales,J. (2002) Improvement of human interferon HUIFNalpha2 and HCV core protein expression levels in Escherichia coli but not of HUIFNalpha8 by using the tRNA(AGA/AGG). Biochem. Biophys. Res. Commun., 296, 1303–1309. [DOI] [PubMed] [Google Scholar]
- 10.Dieci G., Bottarelli,L., Ballabeni,A. and Ottonello,S. (2000) tRNA-assisted overproduction of eukaryotic ribosomal proteins. Protein Expr. Purif., 18, 346–354. [DOI] [PubMed] [Google Scholar]
- 11.Pavesi A. (1999) Relationships between transcriptional and translational control of gene expression in Saccharomyces cerevisiae: a multiple regression analysis. J. Mol. Evol., 48, 133–141. [DOI] [PubMed] [Google Scholar]
- 12.Joshi R.L., Joshi,S., Chapeville,F. and Haenni,A.L. (1983) tRNA-like structures of plant viral RNAs: conformational requirements for adenylation and aminoacylation. EMBO J., 2, 1123–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeiffer P. and Hohn,T. (1983) Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell, 33, 781–789. [DOI] [PubMed] [Google Scholar]
- 14.Coffin J.M., Hughes,S.H. and Varmus,H.E. (1997) (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 15.Lund A.H., Duch,M. and Pedersen,F.S. (2000) Selection of functional tRNA primers and primer binding site sequences from a retroviral combinatorial library: identification of new functional tRNA primers in murine leukemia virus replication. Nucleic Acids Res., 28, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lund A.H., Schmitz,A., Pedersen,F.S. and Duch,M. (2000) Identification of a novel human tRNA(Ser(CGA)) functional in murine leukemia virus replication. Biochim. Biophys. Acta, 1492, 264–268. [DOI] [PubMed] [Google Scholar]
- 17.Pear W.S., Nolan,G.P., Scott,M.L. and Baltimore,D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA, 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda D. and Dreher,T.W. (2004) The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology, 321, 36–46. [DOI] [PubMed] [Google Scholar]
- 19.Collier B., Oberg,D., Zhao,X. and Schwartz,S. (2002) Specific inactivation of inhibitory sequences in the 5′ end of the human papillomavirus type 16 L1 open reading frame results in production of high levels of L1 protein in human epithelial cells. J. Virol., 76, 2739–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., Liu,W.J., Peng,S.W., Sun,X.Y. and Frazer,I. (1999) Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol., 73, 4972–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cumming S.A., Repellin,C.E., McPhillips,M., Radford,J.C., Clements,J.B. and Graham,S.V. (2002) The human papillomavirus type 31 late 3′ untranslated region contains a complex bipartite negative regulatory element. J. Virol., 76, 5993–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leder C., Kleinschmidt,J.A., Wiethe,C. and Muller,M. (2001) Enhancement of capsid gene expression: preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J. Virol., 75, 9201–9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W.J., Gao,F., Zhao,K.N., Zhao,W., Fernando,G.J., Thomas,R. and Frazer,I.H. (2002) Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T-lymphocyte induction and anti-tumour activity. Virology, 301, 43–52. [DOI] [PubMed] [Google Scholar]
- 24.Liu W.J., Zhao,K.N., Gao,F.G., Leggatt,G.R., Fernando,G.J. and Frazer,I.H. (2001) Polynucleotide viral vaccines: codon optimisation and ubiquitin conjugation enhances prophylactic and therapeutic efficacy. Vaccine, 20, 862–869. [DOI] [PubMed] [Google Scholar]
- 25.Zhao K.N. and Frazer,I.H. (2002) Replication of bovine papillomavirus type 1 (BPV-1) DNA in Saccharomyces cerevisiae following infection with BPV-1 virions. J. Virol., 76, 3359–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K.N. and Frazer,I.H. (2002) Saccharomyces cerevisiae is permissive for replication of bovine papillomavirus type 1. J. Virol., 76, 12265–12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas J., Park,E.C. and Seed,B. (1996) Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol., 6, 315–324. [DOI] [PubMed] [Google Scholar]
- 28.zur Megede J., Chen,M.C., Doe,B., Schaefer,M., Greer,C.E., Selby,M., Otten,G.R. and Barnett,S.W. (2000) Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol., 74, 2628–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kofman A., Graf,M., Deml,L., Wolf,H. and Wagner,R. (2003) Codon usage-mediated inhibition of HIV-1 gag expression in mammalian cells occurs independently of translation. Tsitologiia, 45, 94–100. [PubMed] [Google Scholar]
- 30.De Pasquale C. and Kanduc,D. (1998) Modulation of HPV16 E7 translation by tRNAs in eukaryotic cell-free translation systems. Biochem. Mol. Biol. Int., 45, 1005–1009. [DOI] [PubMed] [Google Scholar]
- 31.Varenne S., Buc,J., Lloubes,R. and Lazdunski,C. (1984) Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol., 180, 549–576. [DOI] [PubMed] [Google Scholar]
- 32.Negrutskii B.S., Stapulionis,R. and Deutscher,M.P. (1994) Supramolecular organization of the mammalian translation system. Proc. Natl Acad. Sci. USA, 91, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K.N., Liu,W.J. and Frazer,I.H. (2003) Codon usage bias and A + T content variation in human papillomavirus genomes. Virus Res., 98, 95–104. [DOI] [PubMed] [Google Scholar]
- 34.Zhao K.N., Sun,X.Y., Frazer,I.H. and Zhou,J. (1998) DNA packaging by L1 and L2 capsid proteins of bovine papillomavirus type 1. Virology, 243, 482–491. [DOI] [PubMed] [Google Scholar]
- 35.Zhao K.N., Tomlinson,L., Liu,W.J., Gu,W. and Frazer,I.H. (2003) Effects of additional sequences directly downstream from the AUG on the expression of GFP gene. Biochim. Biophys. Acta, 1630, 84–95. [DOI] [PubMed] [Google Scholar]
- 36.Thomas J.T., Hubert,W.G., Ruesch,M.N. and Laimins,L.A. (1999) Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl Acad. Sci. USA, 96, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 38.Stapulionis R. and Deutscher,M.P. (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl Acad. Sci. USA, 92, 7158–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negrutskii B.S. and Deutscher,M.P. (1992) A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc. Natl Acad. Sci. USA, 89, 3601–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma O.K. and Borek,E. (1976) A mechanism of estrogen action on gene expression at the level of translation. Cancer Res., 36 (11 Pt. 2), 4320–4329. [PubMed] [Google Scholar]
- 41.Sharma O.K. and Kuchino,Y. (1977) Infidelity of translation of encephalomyocarditis viral RNA with tRNA from human malignant trophoblastic cells. Biochem. Biophys. Res. Commun., 78, 591–595. [DOI] [PubMed] [Google Scholar]
- 42.Kanduc D. (1997) Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch. Biochem. Biophys., 342, 1–5. [DOI] [PubMed] [Google Scholar]
- 43.Jameson R.R., Carlson,B.A., Butz,M., Esser,K., Hatfield,D.L. and Diamond,A.M. (2002) Selenium influences the turnover of selenocysteine tRNA([Ser]Sec) in Chinese hamster ovary cells. J. Nutr., 132, 1830–1835. [DOI] [PubMed] [Google Scholar]
- 44.Kanduc D., Grazia di Corcia,M., Lucchese,A. and Natale,C. (1997) Enhanced expression of initiator TRNA(Met) in human gastric and colorectal carcinoma. Biochem. Mol. Biol. Int., 43, 1323–1329. [DOI] [PubMed] [Google Scholar]
- 45.Hatfield D., Lee,B.J., Hampton,L. and Diamond,A.M. (1991) Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res., 19, 939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond A.M., Choi,I.S., Crain,P.F., Hashizume,T., Pomerantz,S.C., Cruz,R., Steer,C.J., Hill,K.E., Burk,R.F., McCloskey,J.A. et al. (1993) Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem., 268, 14215–14223. [PubMed] [Google Scholar]
- 47.Hoekema A., Kastelein,R.A., Vasser,M. and de Boer,H.A. (1987) Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol. Cell. Biol., 7, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pak M., Willis,I.M. and Schulman,L.H. (1994) Analysis of acceptor stem base pairing on tRNA(Trp) aminoacylation and function in vivo. J. Biol. Chem., 269, 2277–2282. [PubMed] [Google Scholar]
- 49.Borel F., Hartlein,M. and Leberman,R. (1993) In vivo overexpression and purification of Escherichia coli tRNA(ser). FEBS Lett., 324, 162–166. [DOI] [PubMed] [Google Scholar]
- 50.Rojiani M.V., Jakubowski,H. and Goldman,E. (1989) Effect of variation of charged and uncharged tRNA(Trp) levels on ppGpp synthesis in Escherichia coli. J. Bacteriol., 171, 6493–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojiani M.V., Jakubowski,H. and Goldman,E. (1990) Relationship between protein synthesis and concentrations of charged and uncharged tRNATrp in Escherichia coli. Proc. Natl Acad. Sci. USA, 87, 1511–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson C., Ragnini,A. and Fukuhara,H. (1989) Analysis of the regions coding for transfer RNAs in Kluyveromyces lactis mitochondrial DNA. Nucleic Acids Res., 17, 4485–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan M.X., Enriquez,J.A., Fischel-Ghodsian,N., Puranam,R.S., Lin,C.P., Maw,M.A. and Attardi,G. (1998) The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol. Cell. Biol., 18, 5868–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akama K. and Beier,H. (2003) Translational nonsense codon suppression as indicator for functional pre-tRNA splicing in transformed Arabidopsis hypocotyl-derived calli. Nucleic Acids Res., 31, 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Del Tito B.J. Jr, Ward,J.M., Hodgson,J., Gershater,C.J., Edwards,H., Wysocki,L.A., Watson,F.A., Sathe,G. and Kane,J.F. (1995) Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol., 177, 7086–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith D.W. (1996) Problems of translating heterologous genes in expression systems: the role of tRNA. Biotechnol. Prog., 12, 417–422. [DOI] [PubMed] [Google Scholar]
- 57.Grassmann K., Kratzer,F., Petry,K.U. and Iftner,T. (1996) Functional characterization of naturally occurring mutants of human papillomavirus type 16 with large deletions in the non-coding region. Int. J. Cancer, 68, 265–269. [DOI] [PubMed] [Google Scholar]
- 58.Grassmann K., Wilczynski,S.P., Cook,N., Rapp,B. and Iftner,T. (1996) HPV6 variants from malignant tumors with sequence alterations in the regulatory region do not reveal differences in the activities of the oncogene promoters but do contain amino acid exchanges in the E6 and E7 proteins. Virology, 223, 185–197. [DOI] [PubMed] [Google Scholar]
- 59.Baker C.C. and Howley,P.M. (1987) Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J., 6, 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker C.C., Phelps,W.C., Lindgren,V., Braun,M.J., Gonda,M.A. and Howley,P.M. (1987) Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol., 61, 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collier B., Goobar-Larsson,L., Sokolowski,M. and Schwartz,S. (1998) Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem., 273, 22648–22656. [DOI] [PubMed] [Google Scholar]
- 62.Tan T.M. and Ting,R.C. (1995) In vitro and in vivo inhibition of human papillomavirus type 16 E6 and E7 genes. Cancer Res., 55, 4599–4605. [PubMed] [Google Scholar]
- 63.Tan W., Felber,B.K., Zolotukhin,A.S., Pavlakis,G.N. and Schwartz,S. (1995) Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol., 69, 5607–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokolowski M., Tan,W., Jellne,M. and Schwartz,S. (1998) mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol., 72, 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kennedy I.M., Haddow,J.K. and Clements,J.B. (1990) Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol., 64, 1825–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy I.M., Haddow,J.K. and Clements,J.B. (1991) A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol., 65, 2093–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cid-Arregui A., Juarez,V. and zur Hausen,H. (2003) A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J. Virol., 77, 4928–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capone J.P., Sharp,P.A. and RajBhandary,U.L. (1985) Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO. J., 4, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamada Y., Matsugi,J. and Ishikura,H. (2003) tRNA1Ser(G34) with the anticodon GGA can recognize not only UCC and UCU codons but also UCA and UCG codons. Biochim. Biophys. Acta, 1626, 75–82. [DOI] [PubMed] [Google Scholar]
- 70.Hatfield D., Choi,I.S., Mischke,S. and Owens,L.D. (1992) Selenocysteyl-tRNAs recognize UGA in Beta vulgaris, a higher plant, and in Gliocladium virens, a filamentous fungus. Biochem. Biophys. Res. Commun., 184, 254–259. [DOI] [PubMed] [Google Scholar]
- 71.Walsh D. and Mohr,I. (2004) Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev., 18, 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]


