Abstract
Topoisomerase II alleviates DNA entanglements that are generated during mitotic DNA replication, transcription, and sister chromatid separation. In contrast to mitosis, meiosis has two rounds of chromosome segregation following one round of DNA replication. In meiosis II, sister chromatids segregate from each other, similar to mitosis. Meiosis I, on the other hand, segregates homologs, which requires pairing, synapsis, and recombination. The exact role that topoisomerase II plays during meiosis is unknown. In a screen reexamining Caenorhabditis elegans legacy mutants isolated 30 years ago, we identified a novel allele of the gene encoding topoisomerase II, top-2(it7). In this study, we demonstrate that top-2(it7) males produce dead embryos, even when fertilizing wild-type oocytes. Characterization of early embryonic events indicates that fertilization is successful and sperm components are transmitted to the embryo. However, sperm chromatin is not detected in these fertilized embryos. Examination of top-2(it7) spermatogenic germ lines reveals that the sperm DNA fails to segregate properly during anaphase I of meiosis, resulting in anucleate sperm. top-2(it7) chromosome-segregation defects observed during anaphase I are not due to residual entanglements incurred during meiotic DNA replication and are not dependent on SPO-11-induced double-strand DNA breaks. Finally, we show that TOP-2 associates with chromosomes in meiotic prophase and that chromosome association is disrupted in the germ lines of top-2(it7) mutants.
Keywords: top-2, Caenorhabditis elegans, meiosis, spermatogenesis, topoisomerase II
TYPE II DNA topoisomerases play a critical role in chromosome fidelity by alleviating topological stresses that arise within chromosomes. Eukaryotic type II topoisomerases (topoisomerase II) are structurally and functionally conserved (Holm et al. 1985; Drake et al. 1989; Ramos et al. 2011). Topoisomerase II is a large ATP-dependent, homodimeric enzyme. Each subunit breaks one DNA strand, passes a second unbroken strand through the break, and then reseals the break (reviewed in Nitiss 2009). Thus, during mitotic divisions, topoisomerase II enzymes solve topological problems that arise during replication, transcription, sister chromatid segregation, and recombination. Topoisomerase II is also crucial for the maintenance of mitotic chromosome structure. In mammals and yeast, it is found along the chromosome axes and knock down by either mutation or RNA interference (RNAi) in mammals, yeast, and Drosophila leads to chromosome condensation defects (Earnshaw et al. 1985; Gasser et al. 1986; Uemura et al. 1987; Xu and Manley 2007; Mengoli et al. 2014).
Although the actions of topoisomerase II in mitosis have been well characterized, studies investigating the roles of topoisomerase II during meiosis are emerging. Unlike mitosis, which has a single round of DNA replication followed by one round of chromosome segregation, meiosis consists of a single round of DNA replication followed by two rounds of chromosome segregation. During the first meiotic division, homologous chromosomes pair, synapse, and form physical connections through recombination that, in partnership with sister chromatid cohesion, aid in the segregation of the homolog pairs (reviewed in Hillers et al. 2015; Zickler and Kleckner 2015). The second meiotic division is similar to mitosis with respect to the segregation of sister chromatids. Due to the requirement for topoisomerase II during mitosis, most strong loss-of-function mutations are lethal, limiting studies during meiosis. Shifting temperature-sensitive mutants of diploid Saccharomyces cerevisiae and Schizosaccharomyces pombe top2 to a nonpermissive temperature leads to a cell-cycle arrest during meiosis I (Rose et al. 1990; Hartsuiker et al. 1998). Meiosis-specific Top2 RNAi in Drosophila results in sterility due to defects in homolog segregation (Hughes and Hawley 2014). Topoisomerase II inhibitors in male and female mouse meiosis lead to defects in chromosome condensation and segregation during the first meiotic division (Marchetti et al. 2001; Li et al. 2013; Gómez et al. 2014). Taken together, these studies demonstrate that topoisomerase II also plays a critical role in the segregation of homologs during meiosis I. Localization studies in yeast and mammals have demonstrated that, as in mitosis, topoisomerase II localizes along the chromosome axes of meiosis I chromosomes (Klein et al. 1992; Li et al. 2013; Gómez et al. 2014). Besides these data, the mechanism of topoisomerase II action and regulation during meiosis and its interaction with the vast and diverse molecular machinery needed to pair, synapse, link, and segregate homologous chromosomes have not been solved.
In Caenorhabditis elegans, a single gene encodes the topoisomerase II homolog, top-2. TOP-2 shares 52% amino acid sequence identity with human topoisomerase IIα. Within the C. elegans genome, a recent duplication event gave rise to a top-2 paralog called cin-4, however, this gene lacks the ATPase domain and is unlikely to have enzymatic function (Stanvitch and Moore 2008). Our genetic studies further suggest that cin-4 is not essential for embryonic or germ-line development. Previous studies found that RNAi depletion of top-2 results in chromosome nondisjunction during embryonic mitotic divisions (Bembenek et al. 2013). In another analysis, top-2 RNAi uncovered a role for TOP-2 in generating the DNA damage that occurs at the onset of zygotic genome activation in the primordial germ cells of embryos (Butuči et al. 2015). While these two studies have demonstrated that, similar to other organisms, C. elegans TOP-2 plays important roles in chromosome fidelity during mitotic divisions, its role in meiosis has not been examined.
In this article, we describe a newly identified allele of C. elegans topoisomerase II, top-2(it7). We show that top-2(it7) mutant males sire dead embryos and that this lethality is due to a failure to segregate DNA properly during the meiotic divisions of spermatogenesis. Instead, chromatin bridges form during anaphase I, and the chromatin becomes trapped in the residual body. We demonstrate that these chromosome-segregation defects result in anucleate sperm that are capable of fertilization. After fertilization, early embryonic events proceed normally, but all the resultant embryos arrest during later divisions. In addition, we have found that the chromosome-segregation defects observed during anaphase I in top-2(it7) males are not due to residual entanglements incurred during mitotic and meiotic DNA replication and are not dependent on SPO-11-induced double-strand DNA breaks (DSBs).
Materials and Methods
Strains
C. elegans strains (listed in Supplemental Material, Table S1) were cultured using standard conditions (Brenner 1973).
RNAi
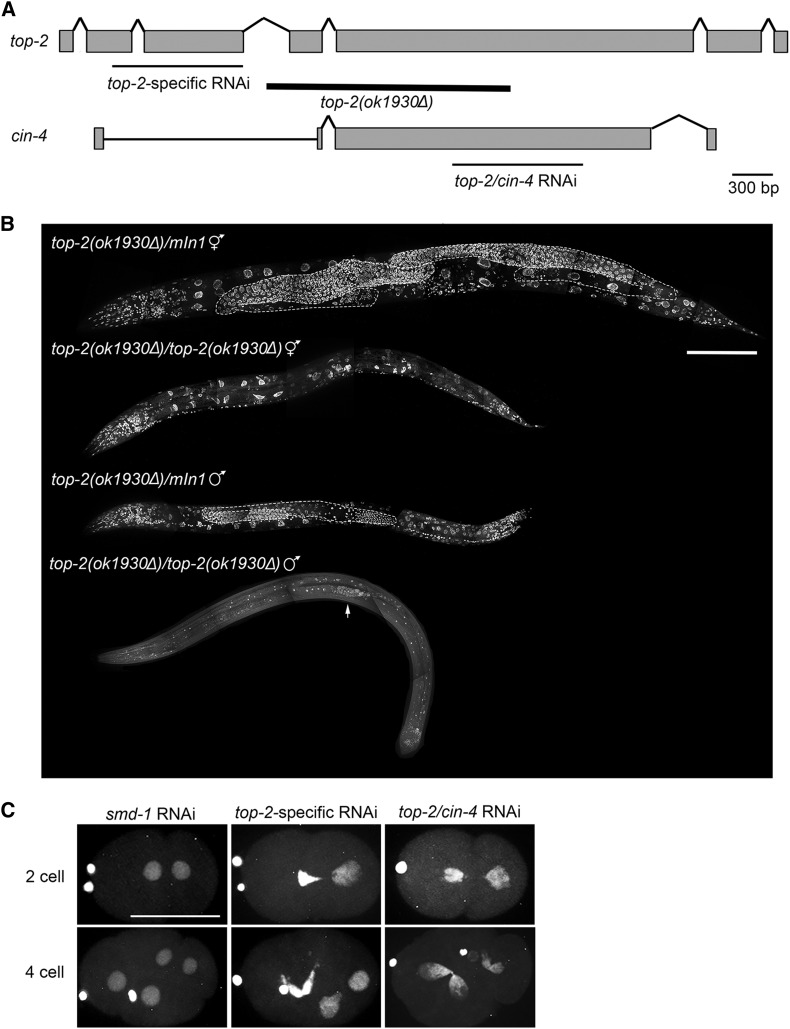
RNAi was performed via the feeding method of Timmons et al. (2001). L2/L3 worms were fed bacteria expressing double-stranded RNAi for 24 hr. Feeding clones were from the Ahringer feeding library (Kamath and Ahringer 2003) and included the following genes: smd-1 (negative control) and top-2/cin-4 (see Figure 5A). To obtain a top-2-specific RNAi feeding construct (pAJL17), a 1-kb fragment spanning exons two to three was amplified and cloned into a modified L4440 (pCR88) feeding vector by Gateway Cloning (see Figure 5A).
Figure 5.
top-2 is required for multiple cell cycles and developmental stages. (A) Schematic comparing the gene structure of top-2 and its paralog cin-4. Thick black line below the top-2 gene structure indicates the extent of the ok1930Δ deletion. Thin black lines below the gene structures indicate the sequences targeted by either top-2-specific RNAi or the RNAi construct that targets sequences present in both genes. (B) Whole-mount top-2(ok1930Δ) and control top-2(ok1930Δ)/mIn1 hermaphrodites and males were prepared for imaging by methanol fixation and DAPI staining. A minimum of three animals of each genotype were imaged. Germ lines in the heterozygous control animals are outlined in white. Arrow points to the few mitotic germ cells generated in the top-2(ok1930Δ) homozygous mutant male. Bar, 100 μm. (C) top-2 RNAi causes segregation defects in the early embryo. L3 hermaphrodites expressing mCherry::H2B were plated on RNAi plates at 24° for 24 hr. Hermaphrodites were dissected and embryos imaged with a confocal microscope. 0% of two-cell (n = 16) and four-cell (n = 16) smd-1 RNAi embryos scored had the cross-eyed phenotype. Of the top-2-specific RNAi embryos scored, 75% of two cell (n = 32) and 81.6% of four cell (n = 38) had the cross-eyed phenotype. 62.7% of two-cell (n = 51) and 81.8% (n = 33) of four-cell top-2/cin-4 RNAi embryos had the cross-eyed phenotype. Bar, 30 μm.
Embryonic viability assays
Single L3 or L4 hermaphrodite larvae were plated and allowed to lay embryos on individual plates at 15 or 24° for a 24 hr period. Adult worms were transferred to fresh plates every 24 hr until no additional embryos were produced. Percent hatching is the number of hatched larvae divided by the total number of embryos laid.
Genetic mapping and molecular identification of top-2(it7ts)
Genetic mapping:
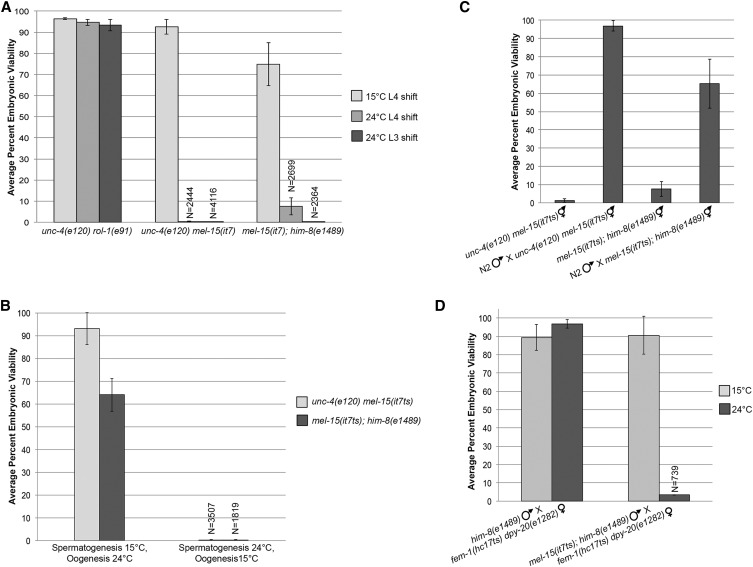
Classical three-factor genetic mapping was performed by mating mel-15(it7ts); him-8(e1489) males with unc-4(e120) rol-1(e91) hermaphrodites. From F1 cross-progeny, homozygous F2 Unc non-Rol (n = 21) and Rol non-Unc (n = 34) recombinant lines were assayed for the embryonic lethality phenotype of mel-15(it7ts) at 24°. A total of 6 of 21 Unc non-Rol hermaphrodites were embryonic lethal (28%), placing mel-15(it7ts) 1.43 cM to the right of unc-4. A total of 17 of 34 Rol non-Unc were embryonic lethal (50%), placing mel-15(it7ts) 4.20 cM to the left of rol-1. With this genetic data, mel-15 was placed at ∼4.3 cM on chromosome II.
Whole genome sequencing:
The genetic position of mel-15(it7ts) determined by three-factor mapping (above) was confirmed by Hawaiian single-nucleotide-polymorphism (SNP) mapping and whole genome sequencing (WGS) (Doitsidou et al. 2010). CB4856 Hawaiian strain crossing, F2 screening, library construction, and sequencing were performed as in Jaramillo-Lambert et al. (2015), yielding a 23-fold genome coverage of a single-end, 50-bp sequence. Variant calling with a pipeline of BFAST (Homer et al. 2009), SAMtools (Li et al. 2009), and ANNOVAR (Wang et al. 2010) failed to identify any novel protein-coding mutations within the map interval. However, an alternate pipeline of BBMAP (Bushnell 2015), FreeBayes (Garrison and Marth 2012), and ANNOVAR identified a single protein-coding variant in the top-2 gene. The it7ts mutation is a C→T missense allele at position 11,878,049 (genome version WS250, http://www.wormbase.org) that encodes an Arg828 to Cys substitution. We redesignated this allele as top-2(it7ts).
Complementation with deletion allele:
Balanced top-2(ok1930Δ)/mIn1-gfp; him-8(e1489) L4 males were mated with homozygous L4 unc-4(e120) mel-15(it7ts) hermaphrodites at 15°. L4 non-GFP, non-Unc F1 hermaphrodite cross-progeny were picked to individual plates and incubated at 24°. Embryonic viability was assessed as described above. Control complementation tests were performed as follows: Balanced mel-20(b317)/mnC1-gfp; him-8(e1489) L4 males were mated with homozygous L4 unc-4(e120) mel-15(it7ts) hermaphrodites at 15°. L4 non-GFP, non-Unc hermaphrodite cross-progeny were picked to individual plates and incubated at 24°, and embryonic viability assessed as described previously.
Testing for paternal-effect embryonic lethality
fem-1(hc17ts) is a temperature-sensitive allele that causes feminization of the germ line at 24° (Doniach and Hodgkin 1984). To produce “females” (no sperm produced), gravid fem-1(hc17ts) dpy-20(e1282) hermaphrodites were placed at 24° and their progeny were raised at the restrictive temperature for 48 hr until reaching the L4 larval stage. A single L4 fem-1(hc17ts) dpy-20(e1282) female was mated with five or six L4 males of indicated genotype at either 15 or 24°. Every 24 hr the adult worms were transferred to fresh plates until the female stopped laying embryos. The number of hatched larvae and dead embryos were scored for each 24 hr period and the percent embryonic viability was calculated.
Immunostaining
Immunostaining was essentially performed as in Martinez-Perez and Villeneuve (2005). Gonads were dissected in 30 μl 1× Egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2 · 2H2O, 2 mM MgCl2 · 6H2O, 25 mM Hepes, pH 7.4) and 0.1% Tween 20 on a coverslip. After dissection, 15 μl of the liquid was removed and replaced with 15 μl 2% paraformaldehyde solution (made from 16% EM Sciences in 1× Egg buffer, 0.1% Tween 20). The solution was carefully pipetted to mix and help extrusion of the gonads. Then, 15 μl of the liquid was removed and a Superfrost Plus microscope slide (Daigger Scientific, Vernon Hills, IL) was placed over the isolated gonads and the sample was allowed to fix for 5 min. The slide preparation was then placed in liquid nitrogen. After freezing, the coverslip was snapped off and the slide was immediately placed in −20° methanol for 1 min. Next, the slides were washed three times in PBS with Tween 20 (PBST) (1× PBS, 0.1% Tween 20) for 5 min each, followed by a 1-hr room temperature incubation in blocking solution (0.7% BSA in PBST). Primary antibody was diluted in blocking solution and slides were incubated for 16–18 hr at 4°, and secondary antibody for 2 hr in a humidified chamber. The slides were then stained with DAPI (2 μg/ml) for 5 min, followed by a final wash in PBST for 5 min, and then mounted with Vectashield. A slight modification to the wash steps of the above protocol was performed for immunostaining with anti-FLAG antibody: After freeze/cracking and incubation in −20° methanol for 1 min, slides were washed once in 1× PBS + 0.5% Triton X-100 for 10 min, then washed two times in PBST for 5 min.
The following primary antibodies were used: rabbit anti-phospho-Histone H3 (Ser10) (H3pS10) (1:100) (Upstate Biotechnology, Lake Placid, NY) and mouse anti-FLAG M2 (1:500) (Sigma Chemical, St. Louis, MO). The secondary antibodies Alexa Flour-561 and Alexa Fluor-488 (1:200) were purchased from Invitrogen (Thermo Fisher Scientific, Waltham, MA).
Methanol fixation protocol for whole-mount animals
Whole-mount worm fixations were prepared as follows: 5–10 animals were picked into 5 μl of M9 minimal medium (M9) on a regular glass slide (Daigger). All M9 was removed from the animals by wicking away the liquid with a Kimwipe, then 15 μl of 100% methanol was added to the animals on the slide. After the methanol evaporated (∼30 sec), 12 μl of DAPI solution (2 μg/ml in water) was placed on the fixed animals and covered with a coverslip for 30 min prior to imaging.
Hoechst staining to visualize DNA in live germ cells (male meiotic divisions)
Meiotic sperm spreads were generated by dissecting male gonads in sperm media (50 mM Hepes, pH 7, 1 mM MgSO4, 25 mM KCl, 45 mM NaCl, 5 mM CaCl2, 1 mg/ml BSA) (Nelson and Ward 1980) containing 2 μg/ml Hoechst dye 33342 (H-3570; Molecular Probes, Waltham, MA). A coverslip was placed over the isolated gonads and gentle pressure applied to flatten and create a single layer of spermatocytes and spermatids. The spreads were immediately imaged for red fluorescence under a 60× objective with DIC optics.
Live imaging of female meiosis
Adult hermaphrodites (OCF8 and AG260) were anesthetized with 2 mM levamisole (Sigma Chemical), mounted on an agarose pad, and covered with a coverslip. For time-lapse imaging, a single focal plane of red fluorescence images was captured at 10 sec intervals.
Image collection and processing
Collection of images was performed using a Nikon (Garden City, NY) Eclipse E800 spinning disk confocal microscope and MetaMorph imaging software. Images were processed and analyzed using Fiji Is Just ImageJ (Fiji) (Schindelin et al. 2012). All imaging experiments were repeated a minimum of three times.
CRISPR/Cas9-mediated genome editing
CRISPR-mediated gene editing was performed to recreate the top-2(it7ts) mutation in the N2 and top-2::3xflag backgrounds and to revert unc-4(e120) top-2(it7ts) to wild type via the clone-free method (Paix et al. 2015), using dpy-10 as a co-CRISPR marker (Arribere et al. 2014). All of the following injections were done using an injection mix of Cas9 protein (10 μg), dpy-10 CRISPR RNA (crRNA) (3.2 μg), dpy-10(cn64) repair oligonucleotide (0.2 μg), universal trans-activating crRNA (tracrRNA) (20 μg, Dharmacon, GE Life Sciences, Pittsburgh, PA), top-2 sequence targeting crRNA (CUUCUUCCAAUCGGACAGUUGUUUUAGAGCUAUGCUGUUUUG, 8 μg), and a gene-/allele-specific repair oligonucleotide (0.8 μg). To revert the cytosine 2977 to thymine (codon 828) in top-2(it7ts) back to wild type, unc-4(e120) top-2(it7ts) young adult hermaphrodites were injected with the above mix containing a repair oligonucleotide containing the sequence to edit the single nucleotide from T to a wild-type C (bold), additional changes to prevent further Cas9 cleavage (italics), and a PstI recognition site (underlined) to distinguish the reverted line from unc-4(e120) top-2(it7ts) (CTCGCTCAAGATTACGTTGGCTCCAACAACATCAACCTGCTTCTTCCAATAGGGCAATTCGGTACTCGTCTGCAGGGTGGAAAGGACAGTGCTTCAGCTCGTTACATCTTCACTCAACTGTCGCC). A single line was isolated that contained the T2977→C change, the PstI site, and the additional silent changes (confirmed by sequencing; Eurofins MWG Operon LLC, Louisville, KY). This line was given the allele designation av74.
To create a C-terminal 3xFLAG-tagged version of TOP-2 (top-2::3xflag), the C-terminal of the top-2 sequence was targeted for Cas9 cleavage using two single guide RNAs (sgRNAs) (sgRNA 1, CGCGTCGTCGACTCCGACT; and sgRNA 2, GGATCAGCCAAAGAAGAAG) and cloned into pDD162 (pAJL20 and pAJL21, respectively) (Dickinson et al. 2013). To integrate a 3xFLAG sequence, N2 young adults were injected with a dpy-10 target sequence in pDD162 (pSS4, 25 ng/μl), a dpy-10 repair oligonucleotide (for coconversion screening, 15 ng/μl), the two top-2 C-terminal sgRNAs, and a top-2::3xflag repair oligonucleotide (CTTACGACGTGGATTCAGGATCCGATTCGGATCAGCCAAAGAAGAAGAGGAGACGCGTCGTCGACTCCGACTCAGATGACTACAAAGACCATGACGGTGATTATAAAGATCATGATATCGATTACAAGGATGACGATGACAAGTAAATTAATTTGTTTCCCACCTTCCTTAAGTGTTTCTAATTTATTTTCTTTCC, 30 ng/μl). The top-2::3xflag repair oligonucleotide contained the 3xFLAG sequence (in bold), silent mutations to remove the protospacer adjacent motif (PAM) sites for both sgRNAs (italics), EcoRV and ClaI restriction sites (underlined, recognition sites overlap), and 50 nt of perfect homology flanking the 3xFLAG insertion. Three independent lines were generated and confirmed by sequencing. These lines were given the allele designation av64.
To recreate the single-nucleotide change of the it7ts allele (cytosine 2977 to thymine) in N2 and top-2::3xflag (see construction notes above), either N2, AG274, or AG275 young adult hermaphrodites were injected with the above mix containing a repair oligonucleotide with the sequences to edit the single nucleotide from a wild-type C to a T (bold), additional changes to prevent further Cas9 cleavage (italics), and a PstI recognition site (underlined) to distinguish the recreated line from N2 or top-2::3xflag (CTCGCTCAAGATTACGTTGGCTCCAACAACATCAACCTGCTTCTTCCAATAGGGCAATTCGGTACTTGTCTGCAGGGTGGAAAGGACAGTGCTTCAGCTCGTTACATCTTCACTCAACTGTCGCC, 0.8 μg). We isolated three independent lines of top-2(it7ts) in the N2 background and two independent lines of top-2(it7ts) in the top-2::3xflag background. Sequence changes were confirmed with sequencing. cin-4, a top-2 paralog, was also sequenced to ensure there was no off-target editing. These lines were given the allele designations av73 (recreation of it7ts) and av77ts (it7ts recreation in top-2::3xflag).
cin-4(av59) was generated by CRISPR/Cas9 genome editing to recreate the published cin-4(mr127ts) allele which changes Glu304 to Gly (Stanvitch and Moore 2008). The cin-4 sequence was targeted for Cas9 cleavage using two sgRNAs (sgRNA 1, CATTCAATTCCATGTCTTG; and sgRNA 2, TAGTTGAAGAAATCTTTTA) and cloned into pDD162 (pAG172 and pAG173, respectively). These sgRNAs target separate PAM sites that flank Glu304. N2 young adult hermaphrodites were injected with the two sgRNAs (25 ng/μl each) and a repair oligonucleotide (CAGGGGCTGGCTGAAGAGTGCCTCTACAACAAAGAAACTCGATTTGTCACTCTGAAAGATTTCTTCAACTACGGAATAGTTTGCTCATGGAATTTGCATTCAATTCCATGTCTTGTCGATGGTTTGAAACCAGGACAGCGGAAGGTTCTCTTCGCGTGCTTCAAAAGA, 30 ng/μl) that codes for the Glu304Gly alteration (in bold); the destruction of both target PAM sites (italics); and a novel, but silent, XmnI restriction site for screening purposes (underlined). In addition, coconversion screening with dpy-10 was performed as described above. A single line contained the Glu304Gly mutation, the XmnI site, and the altered PAM sites (confirmed by sequencing). Because there is high homology between cin-4 and top-2 in this region, additional sequencing of the relevant top-2 region was performed. Sequencing confirmed that no changes were made in the top-2 gene. This line was given the allele designation av59.
In addition to the recreation of mr127ts, a second cin-4 mutant line was generated by CRISPR/Cas9 genome editing to create two stop codons at Tyr303 and Glu304. Using the same coconversion injection strategy stated above, N2 young adults were injected with the same cin-4 sgRNAs (25 ng/μl) and a cin-4 repair oligonucleotide (CAGCAGGGGCTGGCTGAAGAGTGCCTCTACAACAAAGAAACTCGATTTGTCACACTAAAAGATTTCTTCAACTAATAAATAGTTTGCTCATGGAATTTGCATTCAATTCCATGTCTTGTTGATGGTTTGAAACCAGGACAGCGGAAGGTTCTCTTCGCGTGCTTCAAAAGA, 30 ng/μl). The cin-4 repair oligonucleotide coded for Tyr303Stop, Glu304Stop (in bold), the destruction of both PAM sites (italics), and the destruction of a FokI restriction site (for screening purposes, underlined). A single line was generated bearing the two consecutive stop codons (confirmed by sequencing). Sequencing confirmed that the homologous top-2 sequence was unchanged. This line was given the allele designation av61.
Data availability
All strains, reagents, and imaging data sets generated in this study are available upon request.
Results
Identification of mel-15(it7) as a temperature-sensitive, paternal-effect lethal allele
To better understand paternal contributions (i.e., sperm-provided components) required for early embryogenesis, we reexamined embryonic lethal mutants identified in early genetic screens (molecular identity unknown) whose embryonic phenotypes could be rescued by male mating. One such allele, it7, fit this description, but was previously characterized as a maternal-effect lethal (Mel) temperature-sensitive (ts) mutant (Kemphues et al. 1988). The original mel-15(it7) mutation is linked to unc-4(e120), which causes an abnormal movement phenotype. We generated a second strain to separate mel-15(it7) from unc-4(e120) and to introduce the him-8(e1489) allele (Phillips et al. 2005), which produces ∼30% male offspring due to chromosomal nondisjunction [mel-15(it7); him-8(e1489)]. To assess temperature sensitivity and embryonic lethality, progeny viability assays were performed on unc-4(e120) mel-15(it7) and mel-15(it7); him-8(e1489) hermaphrodites by counting the number of live progeny vs. dead embryos at both the permissive (15°) and restrictive (24°) temperatures. Both strains produce viable progeny at the permissive temperature of 15° (Figure 1A). mel-15(it7); him-8(e1489) had a lower percentage of viable progeny at 15° than unc-4(e120) mel-15(it7) (74.8 vs. 92.6%), but we believe this enhancement is due to an interaction with a second modifier mutation that was introduced during the him-8(e1489) cross. At the restrictive temperature (24°), progeny viability is reduced in both mel-15(it7) strains compared to the control strain unc-4(e120) rol-1(e91) (0.3 and 7.5%, respectively, Figure 1A). Because the strains were shifted to the restrictive temperature at the L4 stage when hermaphrodite spermatogenesis is in progress, we reasoned that the viable progeny could be from embryos fertilized by functional sperm that had completed spermatogenesis prior to the temperature shift. Progeny viability assays were repeated on hermaphrodites shifted to 24° at the L3 stage while the germ cells are actively undergoing spermatogenesis. Progeny viability was further reduced when hermaphrodites were shifted to the restrictive temperature at this earlier larval stage (Figure 1A). These results demonstrate that it7 is a temperature-sensitive allele and from here forward will be referred to as it7ts.
Figure 1.
mel-15(it7) is a temperature-sensitive, paternal-effect embryonic lethal allele. (A) Average percent embryonic viability from unc-4(e120) rol-1(e91), unc-4(e120) mel-15(it7), and mel-15(it7); him-8(e1489) at 15 and 24°. The progeny of at least 22 hermaphrodites were scored for each strain at 15 and 24°. Error bars represent SD of the average percentage of three individual replicate experiments. N, total number of embryos and hatched larvae scored. For bars in which N is not stated, N > 2400. (B) Average percent embryonic viability from unc-4(e120) mel-15(it7ts) and mel-15(it7ts); him-8(e1489) hermaphrodites shifted to 24° during either spermatogenesis (L2/L3 to adult) or oogenesis (young adult). The progeny of at least 18 hermaphrodites were scored for each strain at each temperature shift. Error bars represent SD of the average percentage of three individual replicate experiments. (C) Embryonic lethality is rescued by mating with wild-type males. unc-4(e120) mel-15(it7ts) and mel-15(it7ts); him-8(e1489) hermaphrodites were mated with N2 males at 24°. Percent embryonic viability was calculated from the cross-progeny. (D) mel-15(it7ts) is a paternal-effect embryonic lethal allele. L4 males of each mutant were crossed with a single L4 fem-1(hc17ts) dpy-20(e1282) female at 15 or 24°. Percent embryonic viability was calculated from the cross-progeny.
To determine whether the mutant phenotype was due to a defect in oocytes, a defect in spermatocytes, or a combination of both, hermaphrodites were shifted to 24° only during oogenesis (after the L4 to adult transition) or only during spermatogenesis (L3 until young adult) and embryonic lethality determined by progeny viability assays. Both unc-4(e120) mel-15(it7ts) and mel-15(it7ts); him-8(e1489) had a high percentage of viable progeny when they underwent spermatogenesis at 15° and oogenesis at 24° (Figure 1B). However, embryonic viability was dramatically reduced when the hermaphrodite germ lines underwent spermatogenesis at 24° and oogenesis at 15° (0.3 and 0.2%, respectively, Figure 1B).
To further test if the embryonic lethality of mel-15(it7ts) is due to a defect in oogenesis or a defect in spermatogenesis, reciprocal mutant crosses were performed. First, both unc-4(e120) mel-15(it7ts) and mel-15(it7ts);him-8(e1489) hermaphrodites were mated with N2 (wild type) males at the restrictive temperature (24°) and embryonic viability was determined by progeny viability assays. Embryonic viability was restored to 96.9 and 65.2% upon mating to N2 males compared to mutant hermaphrodite self-mating (Figure 1C). These results suggest that the defect originates with the mel-15(it7ts) sperm. As an additional test to determine whether mel-15(it7ts) causes a sperm-specific defect, mel-15(it7ts); him-8(e1489) males were mated to fem-1(hc17ts) dpy-20(e1282) females; fem-1(hc17ts) animals only produce oocytes at 24°. Control him-8(e1489) males crossed with fem-1(hc17ts) dpy-20(e1282) females produce a high percentage of viable progeny at both 15 and 24° (89.4 and 96.8%, Figure 1D). At 15°, mel-15(it7ts); him-8(e1489) males produced viable cross-progeny (90.6%); however, at 24°, embryonic viability of cross-progeny was reduced (3.4%, Figure 1D). These data demonstrate that mel-15(it7ts) is not a maternal-effect lethal mutant as originally characterized but a paternal-effect embryonic lethal allele.
Identification of mel-15(it7ts) as an allele of top-2
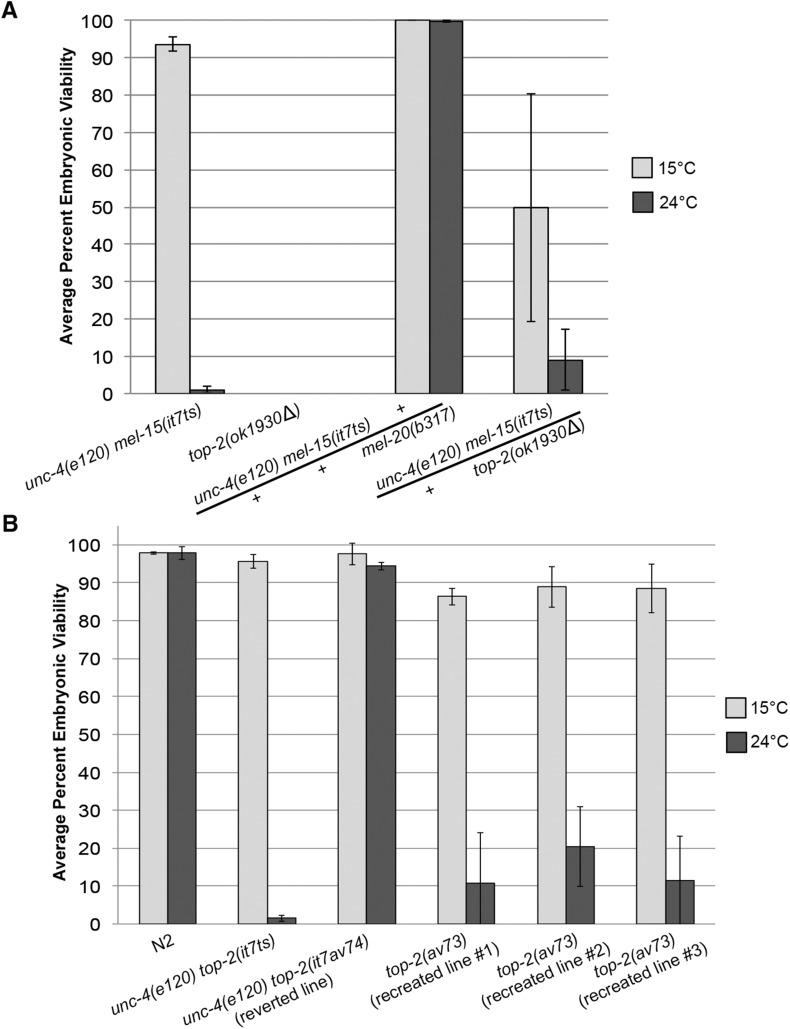
Genetic mapping placed mel-15(it7ts) at 4.3 cM between unc-4 and rol-1 on chromosome II. Using a combination of Hawaiian SNP mapping and WGS (WGS/SNP mapping) (Jaramillo-Lambert et al. 2015), a single nonsynonymous protein-coding candidate mutation was identified in the interval containing the mel-15(it7ts) allele. The protein-coding variant changed a cytosine (nucleotide 2977 of the coding sequence) to a thymine in the top-2 gene which results in arginine 828 being changed to a cysteine (Arg828Cys). To determine if the it7ts lesion is indeed an allele of the top-2 gene, complementation tests were performed between mel-15(it7ts) and top-2(ok1930Δ). Homozygous top-2(ok1930Δ) hermaphrodites are sterile and uncoordinated (Figure 5B) and do not produce any self-progeny at either 15 or 24° (Figure 2A). Individual trans-heterozygous top-2(ok1930Δ)/mel-15(it7ts) hermaphrodites were scored for embryonic viability at the restrictive temperature. The two alleles failed to complement, producing only 9.0% viable progeny at 24°; whereas a control cross of unc-4(e120) mel-15(it7ts) with an unrelated Mel mutant [mel-20(b317)] also located on chromosome II did complement, producing 99.8% viable progeny at 24° (Figure 2A). In addition, F1 mel-15(it7ts)/top-2(ok1930Δ) trans-heterozygotes exhibited a reduced percentage of viable progeny at the permissive temperature compared to the mel-15(it7ts) homozygotes (49.7 vs. 93.7%, Figure 2A). This observation suggests that the deletion allele combined with the missense allele produces insufficient active gene product at the permissive temperature.
Figure 2.
mel-15(it7ts) paternal-effect embryonic lethality is caused by a point mutation in top-2. (A) Complementation tests were performed by crossing unc-4(e120) mel-15(it7ts) hermaphrodites with either top-2(ok1930Δ)/mIn1-gfp; him-8(e1489) or mel-20(b317)/mnC1-gfp; him-8(e1489) males. The average percent embryonic viability at 15 or 24° was calculated by scoring the progeny of F1 trans-heterozygous hermaphrodites. The number of hermaphrodites scored for each cross at 15° were: unc-4(e120) mel-15(it7ts)/mel-20(b317), n = 30; and unc-4(e120) mel-15(it7ts)/top-2(ok1930Δ), n = 42. The number of hermaphrodites scored for each cross at 24° were unc-4(e120) mel-15(it7ts)/mel-20(b317), n = 30; and unc-4(e120) mel-15(it7ts)/top-2(ok1930Δ), n = 40. The average percent embryonic viability for self-mating unc-4(e120) mel-15(it7ts) and top-2(ok1930Δ) hermaphrodites is also indicated (15°, n = 36 and 16, respectively; 24°, n = 21 and 20). Note: top-2(ok1930Δ) homozygotes are sterile Uncs and do not produce any progeny. Error bars indicate SD. (B) Average percent embryonic viability from N2 (wild type), unc-4(e120) top-2(it7ts), and the CRISPR/Cas9-mediated reverted line of unc-4(e120) top-2(it7ts) to wild type [unc-4(e120) top-2(av74)], and the three it7ts recreated lines in N2 [top-2(av73)] at 15 or 24°. The progeny of at least 28 hermaphrodites were scored for each strain at 15 and 24°. Error bars represent SD of the average percentage of three individual replicate experiments.
To further validate that the molecular lesion of it7ts is in the top-2 gene, we used CRISPR/Cas9 genome editing and oligonucleotide-mediated repair in two ways. First, we reverted mel-15(it7ts) by changing the it7ts mutation (cytosine 2977→thymine) back to wild type (cytosine). A single edited line, designated it7av74, was recovered. Embryonic viability of unc-4(e120) top-2(it7av74) (reverted line) was restored to 94.4% at 24° while the embryonic viability of the nonreverted strain, unc-4(e120) top-2(it7ts), was only 1.5% (Figure 2B). Second, to confirm that the single-nucleotide change found in top-2 by WGS/SNP mapping is the cause of the mel-15(it7ts) temperature-sensitive phenotype, we edited the genome of N2 wild-type animals to recreate the cytosine 2977→thymine missense allele of mel-15(it7ts). While 97.8% of N2 progeny hatched at 24°, the percentage of hatching of three independent lines of top-2(av73) (recreated) was decreased (10.8, 20.4, and 11.4%). The decreases in embryonic viability, although significant, are not as severe as observed in the original unc-4(e120) mel-15(it7ts) (1.5%, Figure 2B). Because the unc-4(e120) mel-15(it7ts) mutation was generated through an EMS mutagenesis, this line may have other mutations in the strain background that act as enhancers of embryonic lethality. Taken together, the complementation tests, the recovery of viability by the mel-15(it7av74) reversion, and recreation of the temperature-sensitive embryonic lethality by a single-nucleotide change confirms that mel-15(it7ts) is an allele of top-2; this allele will henceforth be referred to as top-2(it7ts).
top-2 codes for the C. elegans homolog of topoisomerase II, an enzyme that alleviates topological stresses that arise in chromosomes. The top-2(it7ts) cytosine 2977 to thymine creates an Arg828 to Cys change in the central catalytic domain of the protein. This domain has high nucleotide sequence identity (92%) with cin-4, a top-2 paralog that arose by a recent duplication in C. elegans. cin-4(mr127ts) was reported as a temperature-sensitive embryonic lethal allele identified in a screen for chromosome-instability mutants (Stanvitch and Moore 2008). To determine the genetic relationship between these two paralogs, we sought to make double mutants with these two apparently essential genes. Because the strain bearing the cin-4(mr127ts) allele is no longer available from the original authors, we used CRISPR/Cas9 genome editing to recreate the Glu304Gly mutation of cin-4(mr127ts) in N2. A single independent line was generated, cin-4(av59) dpy-10(av60) (recreate). Embryonic lethality of the recreated strain was determined by counting the number of dead embryos vs. live progeny at 15, 24, and 25°. cin-4(av59) dpy-10(av60) (recreate) had unexpectedly high levels of embryonic viability at all three temperatures, similar to controls [dpy-10(cn64) and dpy-10(e128)] (Figure S1A); the temperature-sensitive embryonic lethality previously attributed to this specific mutation was not reproduced. An additional potential null allele was created using CRISPR/Cas9 genome editing to change both Tyr303 and Glu304 to stop codons. A single line bearing the Tyr303Stop Glu304Stop changes was isolated, cin-4(av61). Progeny viability assays revealed that this line had a high percentage of embryonic viability similar to controls (Figure S1B). Because neither cin-4(av59) dpy-10(av60) (recreate) nor cin-4(av61) dpy-10(av60) caused embryonic lethality, no further tests were performed with cin-4.
top-2(it7ts) embryos lack paternal DNA
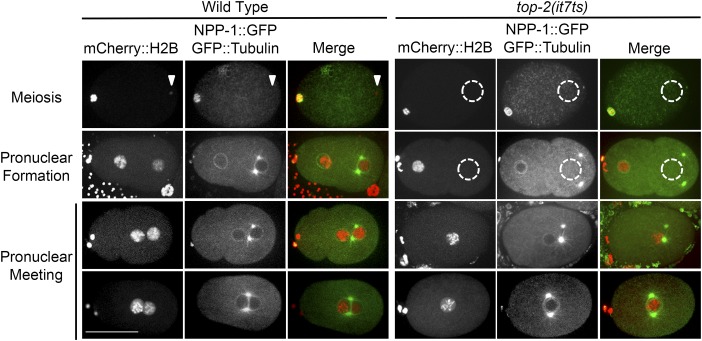
We began characterizing top-2(it7ts) using a combination of live imaging and fluorescence microscopy. In wild-type embryos, soon after fertilization, the maternal DNA completes two rounds of meiotic divisions while the ultracondensed paternal (sperm) DNA remains near its entry point in the posterior of the embryo. After the maternal meiotic divisions have completed, both the maternal and paternal DNA decondense and form pronuclei (48 of 48 observed embryos had both maternal and paternal DNA). Next, the two pronuclei migrate toward each other, come into close proximity, progress to metaphase, and set up a bipolar spindle for the first mitotic division (Figure 3). In newly fertilized embryos from homozygous top-2(it7ts) hermaphrodites shifted to 24° as L3 larvae, the maternal DNA completes the meiotic divisions but, strikingly, these embryos lack paternal DNA (47 of 47 observed embryos) (Figure 3). Other paternally provided components (e.g., centrosomes) are transferred to the embryo during fertilization (Figure 3). In the absence of the paternal pronucleus, the maternal pronucleus migrates posteriorly, associates with the paternally provided centrosomes, assembles the first mitotic spindle, and segregates the haploid complement of DNA (Figure S2); similar to observations of these events in other mutants that produce anucleate sperm (Sadler and Shakes 2000). These haploid embryos eventually arrest around the 200-cell stage (data not shown).
Figure 3.
top-2(it7ts) embryos lack paternal DNA. Wild type (OCF8) and top-2(it7ts) hermaphrodites were shifted to 24° as L4s. After 24 hr, hermaphrodites were dissected and the embryos imaged. Red, chromatin (mCherry::H2B); Green, tubulin (GFP::TBA-2) and nuclear envelope (NPP-1::GFP). White triangle indicates paternal DNA. Dashed-line circle demarks area of missing paternal DNA. Two example images of pronuclear meeting are given. The number of wild-type embryos observed was 48. The number of top-2(it7ts) embryos observed was 47. Bar, 30 μm.
top-2(it7ts) is required for meiotic chromosome segregation in the male germ line
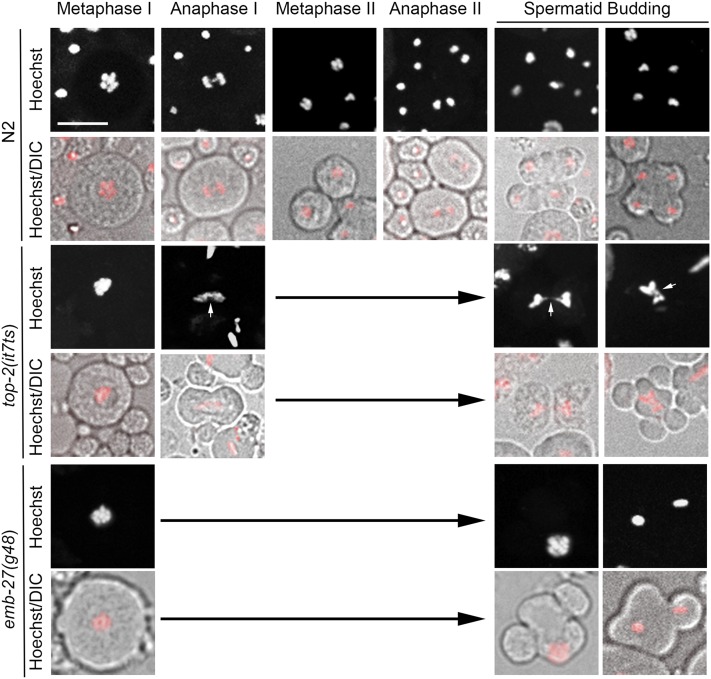
Due to the observation that top-2(it7ts) embryos lack paternal DNA, we examined the germ lines of both top-2(it7ts) hermaphrodite and male adults by DAPI staining after a 16 hr incubation at 24° (shift at L3 larval stage). The events of meiotic prophase were similar to N2 in both hermaphrodites and males (Figure S3). However, abnormal chromosome morphology was observed in the spermathecae of top-2(it7ts) hermaphrodites and after the highly condensed karyosome stage of spermatocytes in top-2(it7ts) males in the region of the meiotic divisions (Figure S3). Abnormal chromosome structures were also observed in the it7ts recreated line, top-2(av73) (data not shown). To determine where these abnormal chromosome structures arise, we examined meiotic divisions in the male germ line through Hoechst staining and live imaging. In N2 spermatocyte meiosis, homologous chromosomes first separate (primary spermatocytes) followed by the separation of sister chromatids (secondary spermatocytes) to generate four haploid spermatids that bud off a central residual body (Figure 4, top panels); a special compartment of the terminal differentiation of spermatids where cytoplasmic contents not needed for fertilization and embryogenesis are segregated (Ward 1986; Machaca et al. 1996). In top-2(it7ts) germ lines at the restrictive temperature (L3 shift, 24° for 16 hr), chromatin bridges form during anaphase of the first meiotic division. No secondary spermatocytes are observed, but spermatocytes progress to spermatid budding. The incompletely separated chromosomes become trapped in the residual body resulting in anucleate sperm (Figure 4, middle panels, and Table S2). Of the 953 spermatids examined, all were anucleate, while in N2 all 733 spermatids observed contained chromatin. This chromosome-morphology phenotype is distinct from other mutations that produce anucleate sperm [e.g. the anaphase promoting complex mutant emb-27(g48) arrests at metaphase I and no chromatin bridging is observed, Figure 4, bottom panels, and Table S2].
Figure 4.
top-2(it7ts) male germ lines have chromosome-segregation defects. Images from live analysis of N2, top-2(it7ts); him-8(e1489), and emb-27(g48) males. Males were shifted to 24° as L3s. After 16 hr, male gonads were dissected and stained with Hoechst dye (red in merged panels) and visualized by DIC optics. White arrows indicate examples of chromatin bridges and atypical chromosome segregation. The number of germ lines scored for each genotype were: N2, n = 32; top-2(it7ts), n = 25; and emb-27(g48), n=21. Bar, 5 μm.
Although the genetic data suggests that top-2(it7ts) affects only the male germ line, we examined the meiotic divisions of oocyte DNA in the one-cell embryo. To examine the effect of mutant top-2(it7ts) strictly during oocyte meiosis, hermaphrodites were shifted to the restrictive temperature as young adults (∼24 hr post-L4), after hermaphrodite spermatogenesis has completed. In addition, for these experiments we used a temperature shift of 26°, a temperature at which most temperature-sensitive alleles are more penetrant. In wild-type embryos, the oocyte DNA completes two rounds of meiosis, extruding two sets of chromosomes into polar bodies (six of seven, Figure S4A). In top-2(it7ts), six of seven one-cell embryos completed meiosis similar to wild type (example 1, Figure S4B). In one embryo, we observed what appeared to be a lagging chromosome during the first meiotic division. The lagging chromosome eventually segregates, the first polar body is formed, and the chromosomes appear to separate successfully in the second meiotic division (example 2, Figure S4B). This lagging-chromosome phenotype was also observed at low frequency in wild-type embryos (one of seven wild-type embryos) and may be a consequence of one of the transgenes that these strains harbor. From these data we conclude that TOP-2 is required for segregating chromosomes in spermatocyte meiosis, but top-2(it7ts) does not affect oocyte meiosis.
The top-2 gene product functions in multiple cell cycles
Type II topoisomerases have well-established roles in the mitotic cell cycle, including DNA replication and chromosome segregation. To determine whether this specific allele of C. elegans top-2 also disrupts mitosis, we examined the proliferative zone of top-2(it7ts) males and hermaphrodites where germ cells undergo mitotic proliferation prior to entering meiosis. When shifted to the restrictive temperature of 24°, both top-2(it7ts) male and hermaphrodite germ lines had enlarged mitotic nuclei (Figure S5, A and B), indicative of an S-phase arrest (MacQueen and Villeneuve 2001). This phenotype was not 100% penetrant (≤61.5% of gonads examined had enlarged nuclei, Figure S5C) and not every nucleus in the proliferative zone was enlarged (Figure S5, A and B). However, enlarged nuclei were observed in both hermaphrodite and male germ lines even after a very short temperature shift (T = 2 hr). In addition, germ lines shifted to 24° for 16 hr or longer had a statistically significant decrease in the number of phospho-Histone H3 (Ser10) positive germ cells (H3pS10) (Figure S5D), a histone modification associated with metaphase; indicating that the number of cells undergoing mitosis is decreased. Next we examined a nonconditional deletion allele of top-2. The deletion allele, ok1930Δ, removes exon four and part of exon five (Figure 5A), causing a predicted frameshift and a premature stop at amino acid 414. Germ-line proliferation fails in both top-2(ok1930Δ) hermaphrodites and males, suggesting that top-2 has a mitotic function in the proliferative region of the germ line (Figure 5B).
To examine the effect of mutant top-2 on embryonic mitotic divisions, we performed RNAi-mediated knockdown of top-2. The available top-2 RNAi feeding vector targets the central region of exon five (Kamath and Ahringer 2003), which has the greatest homology to its paralog, cin-4 (Figure 5A). Because this feeding clone likely depletes both top-2 and cin-4 RNA, a second, top-2-specific RNAi feeding clone was generated; targeting exons two through three which are not present in cin-4 (Figure 5A). In embryos from control RNAi mothers (smd-1, a nonlethal house-keeping gene), oocyte meiosis proceeded normally followed by two successful rounds of mitotic cell divisions (two- and four-cell embryos, Figure 5C). RNAi depletion of top-2 using both the top-2-specific and top-2/cin-4 RNAi clones resulted in the cross-eyed phenotype (Figure 5C), where cytokinesis proceeds despite a failure in chromosome segregation (Bembenek et al. 2013). These data demonstrate that TOP-2 functions in multiple cell cycles (mitosis and meiosis) and stages of development (adult germ line and embryogenesis). For the remainder of this article the focus will be on the functions of TOP-2 during meiosis.
top-2(it7ts) meiotic chromosome-segregation defects are not due to S-phase defects
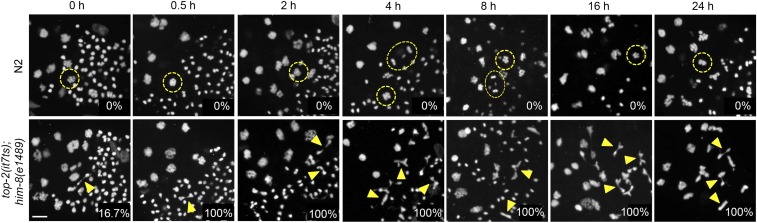
Because type II topoisomerases have known roles in DNA replication, the chromosome-segregation defects observed in meiotically dividing germ cells of top-2(it7ts) males could stem from defects that occurred earlier during either mitotic S phase or premeiotic S phase in the distal germ line. To test this hypothesis, we performed a time course of temperature shifts. It takes ∼20–24 hr at 20° for germ cells to progress from premeiotic S phase to the meiotic division zone (Jaramillo-Lambert et al. 2007). N2 and top-2(it7ts) males were shifted to 24° for varying lengths of time (0–24 hr), the gonads dissected and DAPI stained, and the meiotic division zone analyzed for chromosome-segregation defects. Meiotic chromosome-segregation abnormalities in germ cells from animals undergoing short temperature shifts (0.5–8 hr) cannot be due to mitotic or premeiotic S-phase defects (in the proliferative zone), because these germ cells were not at the restrictive temperature during premeiotic S phase (completed S phase at the permissive temperature of 15°). A small percentage (16.7%) of top-2(it7ts) germ lines have germ cells with chromosome-segregation defects at 15° (Figure 6). However, in contrast to top-2(it7ts) at 15° and to N2 germ lines at 24°, 100% of germ lines from top-2(it7ts) males had at least one example of a meiotically dividing germ cell with abnormal chromosome morphology (i.e., chromatin bridges) for all lengths of temperature shifts (Figure 6). Germ lines exposed to longer temperature shifts (24° for T = 16 and 24 hr) had a greater number of germ cells with abnormal chromosome morphology and no postmeiotic sperm with the compact, round DNA morphology observed in N2 germ lines. Only zero to two nuclei transition from the karyosome stage to the meiotic divisions at a time (Shakes et al. 2009). The accumulation of germ cells with abnormal chromosome morphology is likely due to more germ cells entering the meiotic divisions, along with a delay in the divisions as the germ cells attempt to partition the incompletely separated chromosomes into the residual bodies. Shorter temperature shifts resulted in a mix of normal (wild type) postmeiotic sperm DNA and chromatin with abnormal morphology; suggesting that some germ cells had completed the meiotic divisions prior to top-2(it7ts) inactivation. However, abnormal chromosome structures were observed as little as 30 min after the temperature shift (Figure 6), indicating that the mutant phenotype can be elicited relatively rapidly. These data demonstrate that TOP-2 plays a role in chromosome segregation during the meiotic divisions of sperm, and that this defect is not the result of mitotic or premeiotic S-phase errors.
Figure 6.
top-2(it7ts) meiotic chromosome-segregation defects are not incurred during mitotic or premeiotic DNA replication. Young adult N2 or top-2(it7ts) males (16 hr post-L3) were shifted to 24° for the indicated time intervals, the gonads dissected, fixed, and stained with DAPI. Chromosome segregation was monitored by fluorescence microscopy. Images are from the spermatogenesis condensation and meiotic division zones. Examples of germ cells in metaphase (circles) and anaphase (ovals) are encircled in yellow. Yellow triangles indicate examples of chromatin bridges and incompletely separated chromosomes. The percentage of germ lines with at least one germ cell with chromosome-segregation defects is indicated in the bottom right corner for each time point. The number of N2 germ lines examined were: 0 hr, 15°, n = 9; 0.5 hr, n = 9; 2 hr, n = 8; 4 hr, n = 10; 8 hr, n = 9; 16 hr, n = 13; 24 hr, n = 15. The number of top-2(it7ts); him-8(e1489) germ lines examined were: 0 hr, 15°, n = 15; 0.5 hr, n = 15; 2 hr, n = 22; 4 hr, n = 14; 8 hr, n = 10; 16 hr, n = 12; 24 hr, n = 18. Bar, 5 μm.
top-2(it7ts) chromosome-segregation defects are not dependent on SPO-11-induced DSBs
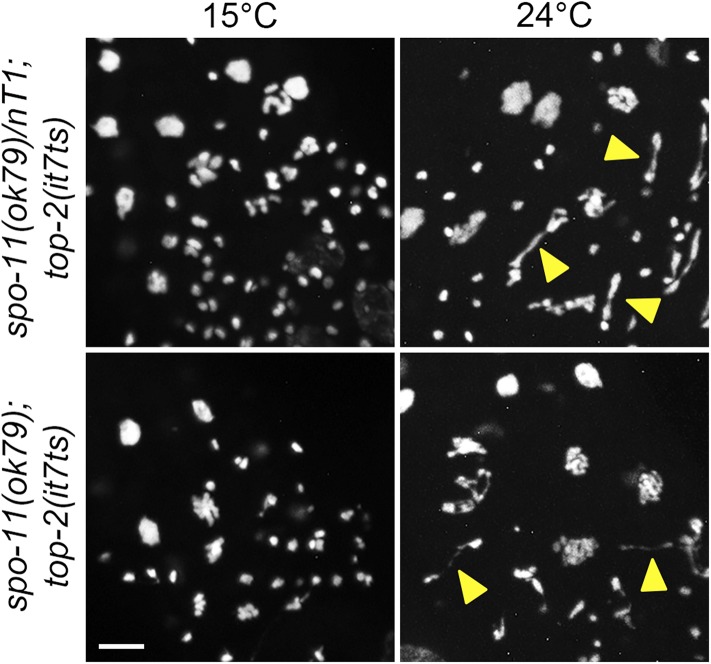
Homologous chromosome segregation during the first meiotic division requires the creation of physical connections formed through crossover (CO) recombination. COs in partnership with sister chromatid cohesion tether homologs together and promote bipolar attachment of homologs to the first meiotic spindle (reviewed in Hillers et al. 2015; Zickler and Kleckner 2015). Given the known enzymatic role of topoisomerase II in disentangling topological problems that arise in double-stranded DNA, we reasoned that TOP-2 might solve topological problems that arise during meiotic CO recombination. To test this hypothesis, we analyzed spo-11(ok79); top-2(it7ts) double mutants for chromosome-segregation defects in male germ lines. SPO-11 is the enzyme that creates the DSBs which initiate meiotic recombination (Dernburg et al. 1998). Germ lines from control spo-11(ok79)/nT1; top-2(it7ts) males raised at 15° did not have abnormal chromosome structures in the meiotic division zone while 100% of male germ lines in spo-11(ok79)/nT1; top-2(it7ts) shifted to 24° had chromosome bridges (Figure 7). In the double mutant, spo-11(ok79); top-2(it7ts), 50% of germ lines at 15° had meiotic chromosome-segregation defects; suggesting a partial synthetic interaction between spo-11 and top-2. However, in spo-11(ok79); top-2(it7ts) male germ lines shifted to 24°, we found that chromatin bridges and chromosome-segregation defects were still present in every germ line examined (arrowheads, Figure 7). No chromosome bridges were observed in the germ lines of spo-11(ok79) single mutants (0 of 13 at 24°, data not shown). These data suggest that the preclusion of meiotic recombination events alone cannot suppress the defects of top-2(it7ts) in male meiotic chromosome segregation.
Figure 7.
top-2(it7ts) chromosome-segregation defects are not dependent on SPO-11-induced DSBs. Young adult spo-11(ok79)/nT1; top-2(it7ts) or spo-11(ok79); top-2(it7ts) males (16 hr post-L3) were incubated at either 15 or 24° for 4 hr, the germ lines dissected, fixed, and stained with DAPI. Chromosome segregation was monitored by fluorescence microscopy. Images are from the spermatogenesis condensation and meiotic division zones. Yellow triangles indicate examples of chromatin bridges and incompletely separated chromosomes. The number of germ lines with chromosome-segregation defects at 15°: spo-11(ok79)/nT1; top-2(it7ts), 0 of 7; spo-11(ok79); top-2(it7ts), 5 of 10. The number of germ lines with chromosome-segregation defects at 24°: spo-11(ok79)/nT1; top-2(it7ts), 16 of 17; spo-11(ok79); top-2(it7ts), 14 of 14. Bar, 5 μm.
TOP-2 localizes to meiotic prophase chromosomes
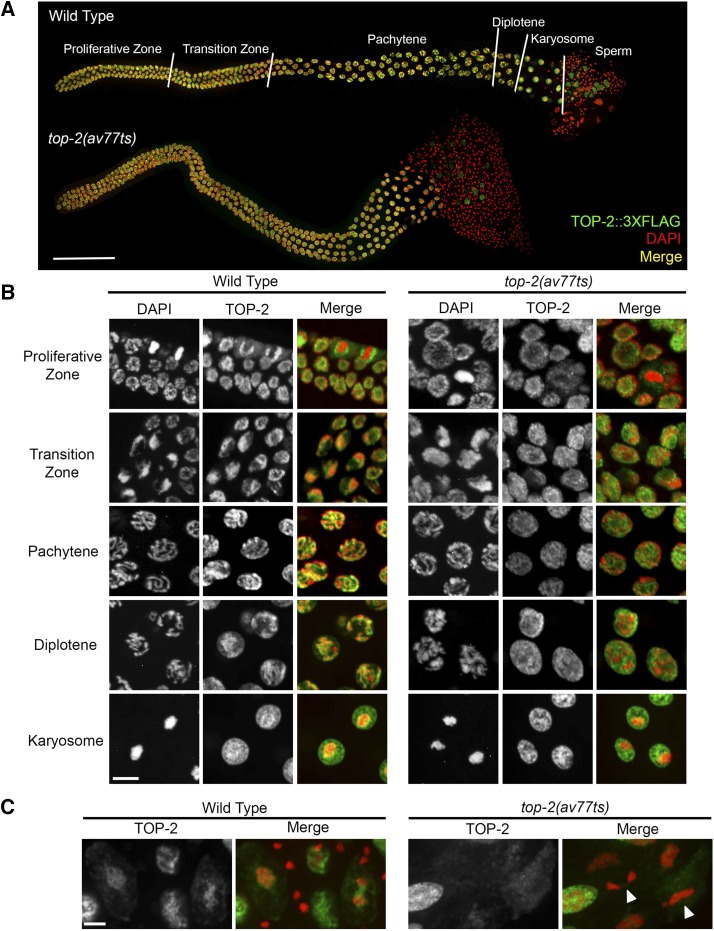
To determine TOP-2 localization in the germ lines of hermaphrodites and males, we used CRISPR/Cas9 to C-terminally tag TOP-2 with a FLAG polypeptide (top-2::3xflag) and examined the staining pattern during the different stages of germ cell development using an anti-FLAG antibody. Within the C. elegans gonad, germ cells are arranged in a spatiotemporal order with a population of mitotically dividing germ cells with dispersed chromosomes at the distal-most end. As germ cells progress through the gonad tube, they enter meiosis in the transition zone, where chromosomes cluster in a crescent-shaped pattern on one side of the nucleus (MacQueen and Villeneuve 2001). This region is followed by an extended meiotic prophase I where distinct substages can be visualized by chromosome morphology. In the pachytene stage of meiotic prophase I, the homologs are fully synapsed and redistributed around the nuclear periphery. After pachytene, chromatin morphology differs between oogenesis and spermatogenesis. In oogenic germ lines pachytene is followed by diplotene, where chromosomes desynapse but homologs remain associated by chiasmata as they condense. In the proximal-most end of the gonad, germ cell nuclei have entered diakinesis where chromosomes detach from the nuclear envelope and continue to condense (Goldstein and Slaton 1982). The oocyte meiotic divisions occur in the one-cell embryo after fertilization. In spermatogenic germ lines, proximal germ cells enter a “condensation zone” where the paired homologous chromosomes aggregate into a single mass called the karyosome. After karyosome formation, spermatocytes enter diakinesis and proceed through two rounds of meiotic divisions in the proximal germ line (Shakes et al. 2009).
In hermaphrodite germ lines shifted to 24° for 4 hr (and in 15° germ lines), TOP-2 was detected in germ cell nuclei throughout the gonad (green, Figure S6A). In the proliferative zone, TOP-2 is nucleoplasmic but appears to concentrate on the mitotic spindle of cells in metaphase. In the transition zone, TOP-2 associates with the clustered chromosomes and then localizes along the lengths of DAPI-stained chromatin of pachytene nuclei (colocalization shown in yellow, Figure S6B). As germ cell nuclei start to condense in diplotene, some TOP-2 remains associated with the chromosomes, but a pool of TOP-2 redistributes to the nucleoplasm. TOP-2 in diakinetic nuclei has completely disassociated from the chromosomes (Figure S6B). Similar to hermaphrodite germ lines, TOP-2 was detected in all germ cell nuclei in the male germ line (15 and 24° for 4 hr, Figure 8A). In male germ lines, TOP-2 is nucleoplasmic in most proliferative zone germ cells, except in metaphase cells where it is concentrated on mitotic spindles (Figure 8B, top panels). In the transition zone, TOP-2 starts to associate with the clustered chromosomes and then tracks along DAPI-stained chromatin in pachytene nuclei (colocalization in yellow, Figure 8B). In diplotene nuclei, TOP-2 begins to dissociate from chromosomes and is mostly nucleoplasmic in karyosome nuclei (Figure 8B). However, TOP-2 concentrates around the periphery of karyosome DNA (Figure 8B). During the meiotic divisions, TOP-2 encircles metaphase chromosomes and staining detects a weaker cytoplasmic pool of TOP-2. During anaphase, TOP-2 encircles chromosomes and becomes concentrated between the segregating chromosomes. No TOP-2 was detected in postmeiotic spermatids (Figure 8C).
Figure 8.
TOP-2 localization in the male germ line. (A) Immunolocalization of TOP-2::3xFLAG (green) in wild type [top-2(av64)] and top-2(av77ts) (it7ts recreate) in entire dissected male germ lines counterstained with DAPI (red; colocalization, yellow). Young adult animals were shifted to 24° for 4 hr prior to dissection and staining. Stages of meiotic prophase are delineated by white lines on the wild-type germ line. Bar, 50 μm. (B) Representative images of Z-projected confocal sections through the proliferative zone, transition zone, pachytene nuclei, diplotene nuclei, and karyosome nuclei (24° for 4 hr) stained with anti-FLAG antibody (green) and counterstained with DAPI (red; colocalization, yellow). Bar, 5 μm. (C) Images of TOP-2::3xFLAG (green) immunolocalization in the meiotic division zone in wild-type and top-2(av77ts) male germ lines (24° for 4 hr) counterstained with DAPI (red). White triangles point to examples of chromatin bridges and incompletely separated chromosomes in the top-2(av77ts) mutant. Experiments were repeated a minimum of three times and a minimum of eight germ lines were examined for each condition. Bar, 5 μm.
Similar to wild type, mutant TOP-2 [top-2(av77ts)::3xflag, CRISPR recreation of the it7ts allele in top-2::3xflag] at 15° colocalizes with chromosomes in the transition zone and pachytene, although to a lower extent than in wild type with more nucleoplasmic TOP-2 (data not shown). In contrast to wild-type animals at 24°, mutant TOP-2 [top-2(av77ts)::3xflag] at 24° (4 hr) fails to localize to chromosome axes in pachytene nuclei, remaining nucleoplasmic throughout meiotic prophase I in both male (Figure 8B) and hermaphrodite (Figure S6) germ lines. In addition, little or no TOP-2 staining was observed in nuclei undergoing meiotic divisions in male mutant germ lines (Figure 8C). Expression of the mutant protein is similar to wild type in germ cells throughout meiotic prophase, except at the karyosome stage where expression is reduced (Figure S7). These data suggest that the it7ts mutation disrupts the association of TOP-2 with chromosomes.
Discussion
In this study, we have shown that a novel mutation in top-2 affects chromosome segregation, specifically in the male germ line. During spermatogenesis, top-2(it7ts) mutants display chromosome-segregation defects during anaphase of meiosis I. Chromatin bridges form between the segregating chromatin masses, which fail to segregate to the budding spermatids; creating anucleate sperm. Oogenic germ lines appear unaffected by the top-2(it7ts) mutation. We have shown that the chromosome-segregation defects of top-2(it7ts) do not stem from a lack of top-2 function during mitotic and premeiotic DNA replication nor does removal of SPO-11-dependent DSBs suppress top-2(it7ts) chromosome-segregation defects. In addition, we show changes in TOP-2 localization in both male and hermaphrodite mutant germ lines. However, only germ lines undergoing spermatogenesis appear to be sensitive to this disruption.
top-2(it7ts) has a mutation in the tower domain
WGS identified a single missense mutation in top-2(it7ts) that changes arginine 828 to a cysteine. This arginine is highly conserved and is located within the catalytic domain of the protein. Interestingly, this amino acid residue lies at the base of a subdomain called the “tower” and interacts with another highly conserved residue (Glu960, human Glu924, S. cerevisiae Glu902; J. Berger, personal communication). Consistent with our TOP-2::3xFLAG localization data (Figure 8 and Figure S6), the tower domain is believed to be involved in DNA binding (Dong and Berger 2007). However, the purpose of this subdomain is still unclear as, up until this study, mutagenesis experiments have failed to show a critical function for the tower. How the arginine 828 to cysteine change of top-2(it7ts) is affecting the overall conformation of the TOP-2 protein and whether this mutation affects TOP-2 enzymatic activity remains to be determined. We do not believe that top-2(it7ts) disrupts protein stability because the mutant protein was detected in our immunolocalization experiments (Figure 8 and Figure S6), although expression of the mutant protein appears to be lower in karyosome-stage germ cells (Figure S7). Future structure-function analyses will help elucidate the role of the tower domain in TOP-2 activity.
TOP-2 role in spermatocyte vs. oocyte meiosis
We have shown that the top-2(it7ts) mutation affects the meiotic divisions during spermatogenesis but not oogenesis (Figure 1, Figure 3, Figure 4, and Figure S4). We provide evidence that top-2 itself is not specific to spermatogenesis as the protein is expressed in both hermaphrodite and male germ lines (Figure 8 and Figure S6). In addition, a null mutation results in a failure in germ-line proliferation in both males and hermaphrodites (Figure 5), although this is most likely due to defects in TOP-2 function during mitotic DNA replication. Why is the male germ line more sensitive to loss of top-2 function? Although meiosis in both oogenesis and spermatogenesis result in haploid germ cells, there are distinct differences in the two meiotic programs. For example, the length of meiotic prophase, germ-cell regulation by checkpoints, and germ-line apoptosis differ between spermatogenesis and oogenesis (Hunt and Hassold 2002; Cohen et al. 2006; Jaramillo-Lambert et al. 2007, 2010; Chu and Shakes 2013). Differences are also observed during the meiotic divisions where spermatocyte chromosomes are equally partitioned via a centriole-based meiotic spindle, while oocyte chromosomes are asymmetrically segregated on acentriolar spindles (Gard 1992; Albertson and Thomson 1993; Brunet et al. 1999). Sex-specific differences in chromosome condensation also exist. Late in meiosis, sperm chromatin is remodeled by a systematic replacement of somatic histones with sperm-specific histone variants and sperm nuclear basic proteins (SNBPs) that result in the hyper-compaction of DNA for sperm transit and to establish paternal epigenetic information (Braun 2001; Chu and Shakes 2013). Intriguingly for our current study, meiotic prophase ends with an aggregation of paired homologous chromosomes into a single mass called a karyosome in several organisms (Gruzova and Parfenov 1993; Orr-Weaver 1995; Shakes et al. 2009). During C. elegans spermatogenesis, karyosomes form after chromosomes desynapse. This chromosome aggregation does not occur in C. elegans oogenesis; oocyte homolog pairs condense to single entities as they prepare for the meiotic divisions. We propose that this is the meiotic stage that is sensitive to the loss of TOP-2 function (Figure 9). Several lines of evidence support this hypothesis. First, our data indicate that TOP-2 functions late in meiosis, as very short temperature shifts result in chromosome-segregation defects (Figure 6). Second, unlike in some other organisms where top-2 plays a role in recombination or in the resolution of recombined chromosomes (Rose et al. 1990; Hartsuiker et al. 1998), top-2(it7ts) chromosome-segregation defects are not ameliorated by the preclusion of meiotic COs in spo-11(ok79); top-2(it7ts) (Figure 7). Third, TOP-2 localizes along the lengths of pachytene chromosomes and surrounds the chromatin of karyosomes (Figure 8), positioning the protein for action during chromosome condensation. Furthermore, the karyosome stage is where mutant TOP-2::3xFLAG expression is at its lowest (Figure S7). TOP-2 localization is also consistent with the incorporation of C. elegans SNBPs at pachytene exit. Topoisomerase II has been implicated in chromosome condensation in both somatic cells and meiotic cells in several organisms including Drosophila, yeast, mouse, and human tissue culture cells (Hartsuiker et al. 1998; Maeshima and Laemmli 2003; Xu and Manley 2007; Li et al. 2013; Hughes and Hawley 2014; Mengoli et al. 2014). Furthermore, studies in mitosis reveal that type II topoisomerases colocalize with and, in some cases, directly interact with structural maintenance of chromosome proteins (Bhat et al. 1996; Vos et al. 2011). The function of topoisomerases II in chromosome condensation remains unclear and further studies are needed to uncover this role. An additional unknown is the function of karyosome formation. Although it has been proposed to facilitate chromosome segregation, this structure is poorly understood in all organisms. As analysis of the C. elegans top-2(it7ts) mutant appears to indicate that TOP-2 functions during the time of both chromosome condensation and karyosome formation in the male germ line, perhaps this unique, conditional mutant can elucidate the physiological role of these processes.
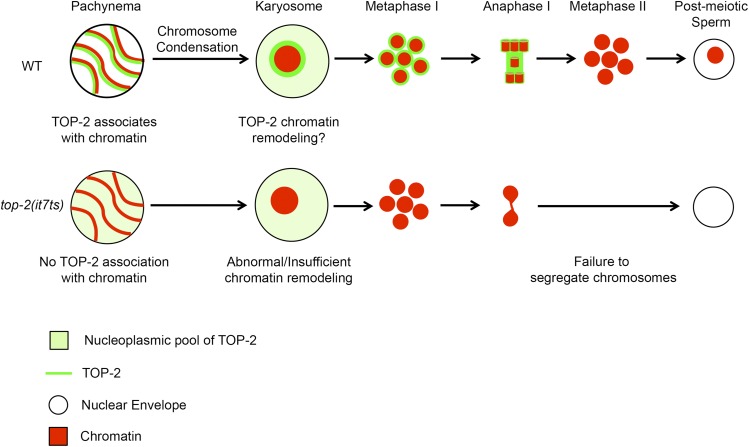
Figure 9.
Model for TOP-2 function in male meiosis. In wild-type germ lines TOP-2 first localizes along chromosome axes of germ-line nuclei during pachytene and then envelops the highly compact chromosome structure of karyosome nuclei, positioning TOP-2 to interact with DNA during chromosome condensation. TOP-2-mediated chromatin remodeling prepares the chromosomes for segregation during the meiotic divisions. In the top-2(it7ts) mutant, TOP-2 fails to localize along the chromosome tracks of pachytene nuclei and around the karyosome DNA. This results in insufficient chromatin remodeling leading to chromosome-segregation defects. WT, wild type.
In summary, we have identified a novel, conditional allele of the C. elegans topoisomerase II homolog, top-2(it7ts), where sperm DNA fails to segregate properly during the first meiotic division. We propose that TOP-2 localization during late pachytene positions the protein to function in chromosome condensation/karyosome formation prior to the meiotic divisions. When TOP-2 localization is disrupted in the top-2(it7ts) mutant, either abnormal or insufficient chromatin remodeling occurs during late prophase resulting in aberrant chromosome segregation (Figure 9). This is only one of many possible models. Further studies will help elucidate how the enzymatic activity of TOP-2 relates to the late stages of meiotic prophase or whether TOP-2 has a novel, nonenzymatic role in meiosis.
Acknowledgments
We thank K. Stein for genetic mapping of mel-15(it7ts); A. Chandra for helping with the mel-15(it7ts) complementation tests; James Berger for analysis of TOP-2 protein structure; N. Devaul, R. Meseroll, and A. Walters for comments on the manuscript; and members of the Baltimore Worm Club for helpful discussions. We are grateful to Ken Kemphues for the original isolation and characterization of the mel-15(it7ts) allele, which inspired this project. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD-010440). Some deletion allele strains were generated by the C. elegans Gene Knockout Consortium at the University of British Columbia led by D. Moerman. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Communicating editor: M. P. Colaiácovo
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.195099/-/DC1.
Literature Cited
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek J. N., Verbrugghe K. J. C., Khanikar J., Csankovszki G., Chan R. C., 2013. Condensin and the spindle midzone prevent cytokinesis failure induced by chromatin bridges in C. elegans embryos. Curr. Biol. 23: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M. A., Philp A. V., Glover D. M., Bellen H. J., 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell 87: 1103–1114. [DOI] [PubMed] [Google Scholar]
- Braun R. E., 2001. Packaging paternal chromosomes with protamine. Nat. Genet. 28: 10–12. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1973. The genetics of behaviour. Br. Med. Bull. 29: 269–271. [DOI] [PubMed] [Google Scholar]
- Brunet S., Maria A. S., Guillaud P., Dujardin D., Kubiak J. Z., et al. , 1999. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J. Cell Biol. 146: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, B., BBMap Short-Read Aligner, and Other Bioinformatics Tools. Available at: http://sourceforge.net/projects/bbmap. Accessed: July 10, 2015.
- Butuči M., Williams A. B., Wong M. M., Kramer B., Michael W. M., 2015. Zygotic genome activation triggers chromosome damage and checkpoint signaling in C. elegans primordial germ cells. Dev. Cell 34: 85–95. [DOI] [PubMed] [Google Scholar]
- Chu D. S., Shakes D. C., 2013. Spermatogenesis. Adv. Exp. Med. Biol. 757: 171–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. E., Pollack S. E., Pollard J. W., 2006. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27: 398–426. [DOI] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP Mapping Strategy. PLoS One 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K. C., Berger J. M., 2007. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450: 1201–1205. [DOI] [PubMed] [Google Scholar]
- Doniach T., Hodgkin J., 1984. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 106: 223–235. [DOI] [PubMed] [Google Scholar]
- Drake F. H., Hofmann G. A., Bartus H. F., Mattern M. R., Crooke S. T., et al. , 1989. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry 28: 8154–8160. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Halligan B., Cooke C. A., Heck M. M., Liu L. F., 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard D. L., 1992. Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev. Biol. 151: 516–530. [DOI] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907.
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K., 1986. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188: 613–629. [DOI] [PubMed] [Google Scholar]
- Goldstein P., Slaton D. E., 1982. The synaptonemal complexes of Caenorhabditis elegans: comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma 84: 585–597. [DOI] [PubMed] [Google Scholar]
- Gómez R., Viera A., Berenguer I., Llano E., Pendás A. M., et al. , 2014. Cohesin removal precedes topoisomerase IIα-dependent decatenation at centromeres in male mammalian meiosis II. Chromosoma 123: 129–146. [DOI] [PubMed] [Google Scholar]
- Gruzova M. N., Parfenov V. N., 1993. Karyosphere in oogenesis and intranuclear morphogenesis. Int. Rev. Cytol. 144: 1–52. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Bähler J., Kohli J., 1998. The role of topoisomerase II in meiotic chromosome condensation and segregation in Schizosaccharomyces pombe. Mol. Biol. Cell 9: 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J., V. Jantsch, E. Martinez-Perez, and J. L. Yanowitz, 2015 Meiosis (in press), WormBook, ed. The C. elegans Research Community Wormbook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Holm C., Goto T., Wang J. C., Botstein D., 1985. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41: 553–563. [DOI] [PubMed] [Google Scholar]
- Homer N., Merriman B., Nelson S. F., 2009. BFAST: an alignment tool for large scale genome resequencing. PLoS One 4: e7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Hawley R. S., 2014. Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet. 10: e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P. A., Hassold T. J., 2002. Sex matters in meiosis. Science 296: 2181–2183. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., Engebrecht J., 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20: 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Fuchsman A. S., Fabritius A. S., Smith H. E., Golden A., 2015. Rapid and efficient identification of Caenorhabditis elegans legacy mutations using Hawaiian SNP-based mapping and whole-genome sequencing. G3 (Bethesda) 5: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Kusch M., Wolf N., 1988. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics 120: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Laroche T., Cardenas M. E., Hofmann J. F., Schweizer D., et al. , 1992. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 117: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-M., Yu C., Wang Z.-W., Zhang Y.-L., Liu X.-M., et al. , 2013. DNA topoisomerase II is dispensable for oocyte meiotic resumption but is essential for meiotic chromosome condensation and separation in mice. Biol. Reprod. 89: 118. [DOI] [PubMed] [Google Scholar]
- Machaca K., DeFelice L. J., L'Hernault S. W., 1996. A novel chloride channel localizes to Caenorhabditis elegans spermatids and chloride channel blockers induce spermatid differentiation. Dev. Biol. 176: 1–16. [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Villeneuve A. M., 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K., Laemmli U. K., 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell 4: 467–480. [DOI] [PubMed] [Google Scholar]
- Marchetti F., Bishop J. B., Lowe X., Generoso W. M., Hozier J., et al. , 2001. Etoposide induces heritable chromosomal aberrations and aneuploidy during male meiosis in the mouse. Proc. Natl. Acad. Sci. USA 98: 3952–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A. M., 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 19: 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengoli V., Bucciarelli E., Lattao R., Piergentili R., Gatti M., et al. , 2014. The analysis of mutant alleles of different strength reveals multiple functions of topoisomerase 2 in regulation of Drosophila chromosome structure. PLoS Genet. 10: e1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G. A., Ward S., 1980. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell 19: 457–464. [DOI] [PubMed] [Google Scholar]
- Nitiss J. L., 2009. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., 1995. Meiosis in Drosophila: seeing is believing. Proc. Natl. Acad. Sci. USA 92: 10443–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E., Torre E. A., Bushey A. M., Gurudatta B. V., Corces V. G., 2011. DNA topoisomerase II modulates insulator function in Drosophila. PLoS One 6: e16562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Thomas W., Holm C., 1990. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell 60: 1009–1017. [DOI] [PubMed] [Google Scholar]
- Sadler P. L., Shakes D. C., 2000. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development 127: 355–366. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Wu J.-C., Sadler P. L., LaPrade K., Moore L. L., et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanvitch G., Moore L. L., 2008. cin-4, a gene with homology to topoisomerase II, is required for centromere resolution by cohesin removal from sister kinetochores during mitosis. Genetics 178: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., et al. , 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50: 917–925. [DOI] [PubMed] [Google Scholar]
- Vos S. M., Tretter E. M., Schmidt B. H., Berger J. M., 2011. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., 1986. The asymmetric localization of gene products during the development of Caenorhabditis elegans spermatozoa, pp. 55–75 in Gametogenesis and the Early Embryo, edited by Gall J. G. Alan R. Liss, New York. [Google Scholar]
- Xu Y.-X., Manley J. L., 2007. The prolyl isomerase Pin1 functions in mitotic chromosome condensation. Mol. Cell 26: 287–300. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7: a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains, reagents, and imaging data sets generated in this study are available upon request.