Abstract
In most animals, female meiosis completes only after fertilization. Sperm entry has been implicated in providing a signal for the initiation of the final meiotic processes; however, a maternal component required for this process has not been previously identified. We report the characterization of a novel family of three highly similar paralogs (memi-1, memi-2, memi-3) that encode oocyte-specific proteins. A hyper-morphic mutation memi-1(sb41) results in failure to exit female meiosis II properly; however, loss of all three paralogs results in a “skipped meiosis II” phenotype. Mutations that prevent fertilization, such as fer-1(hc1), also cause a skipped meiosis II phenotype, suggesting that the MEMI proteins represent a maternal component of a postfertilization signal that specifies the meiosis II program. MEMI proteins are degraded before mitosis and sensitive to ZYG-11, a substrate-specific adapter for cullin-based ubiquitin ligase activity, and the memi-1(sb41) mutation results in inappropriate persistence of the MEMI-1 protein into mitosis. Using an RNAi screen for suppressors of memi-1(sb41), we identified a sperm-specific PP1 phosphatase, GSP-3/4, as a putative sperm component of the MEMI pathway. We also found that MEMI and GSP-3/4 proteins can physically interact via co-immunoprecipitation. These results suggest that sperm-specific PP1 and maternal MEMI proteins act in the same pathway after fertilization to facilitate proper meiosis II and the transition into embryonic mitosis.
Keywords: Caenorhabditis elegans, PP1 phosphatase, female meiosis, fertilization, mitosis
FERTILIZATION represents a unique stage of development in all sexually reproducing animals. As the oocyte matures, it must synchronize the completion of the meiotic divisions with the timing of fertilization. During female meiosis, homologous chromosomes and sister chromatids separate in two successive asymmetric meiotic divisions, giving rise to tiny polar bodies and a single large cell. However, different animals use different strategies to coordinate these events with fertilization. In most species, oocytes initially arrest at prophase I. Upon hormonal or developmental stimulation, the oocyte undergoes meiotic maturation, whereby it exits this arrest. Depending on the species, the oocyte can then exhibit a second arrest at meiotic metaphase I [e.g., ascidians (McDougall et al. 2012) and insects (Von Stetina and Orr-Weaver 2011)], meiotic metaphase II [e.g., vertebrates (Masui and Markert 1971) and cephalochordates (Holland and Onai 2012)], or during the first mitosis [e.g., cnidarians and starfishes (Kishimoto 2004; Costache et al. 2014)]. Fertilization releases the cell from the second arrest, thus, in many species, the final stages of female meiosis are completed only after sperm entry.
Caenorhabditis elegans has become a powerful model to identify molecular pathways involved in gametogenesis (Hansen and Schedl 2013), fertilization (Marcello et al. 2013), and the transition to zygotic development (Kim et al. 2013; Robertson and Lin 2015). In C. elegans hermaphrodites, oocytes form in a single file and slowly migrate toward the spermatheca at the proximal end of the gonad arm. The oocytes arrest at diakinesis of prophase I. The most proximal oocyte, situated next to the spermatheca, responds to a hormone secreted by the sperm, termed the major sperm protein (MSP) (Miller et al. 2001). MSP performs at least two diverse functions: it forms cytoskeletal polymers for actin-independent motility of nematode spermatozoa (Bottino et al. 2002), and it can act as a hormone to stimulate oocyte meiotic maturation and gonadal sheath cell contraction (Miller et al. 2001). During maturation, the oocyte nuclear envelope breaks down, the cortical cytoskeleton rearranges, and a meiosis I spindle forms (Harris et al. 2006; Kim et al. 2013), similar to the cellular changes associated with meiotic maturation in other systems (Masui and Clarke 1979).
Gonadal sheath cells that surround the oocytes inhibit meiotic maturation when sperm are absent, at least in part, through the regulation of gap junction-based communication between sheath cells and oocytes (Govindan et al. 2006; Whitten and Miller 2007; Starich et al. 2014). The gonadal sheath also promotes meiotic maturation when sperm are present, likely via the Gαs-adenylate cyclase protein kinase A pathway (Govindan et al. 2006, 2009). Although the MSP receptor(s) on gonadal sheath cells remain unknown, MSP can bind to an Eph-related receptor tyrosine kinase (VAB-1), which is expressed in the oocyte and contributes to oocyte maturation (Miller et al. 2003; Corrigan et al. 2005; Govindan et al. 2006; Cheng et al. 2008). Upon encountering MSP, the somatic gonadal sheath contracts and pushes the oocyte through the spermatheca for fertilization (Samuel et al. 2001). The fertilized cell then enters the uterus, where it typically completes the meiosis I and meiosis II divisions in rapid succession (Yang et al. 2003). The one-cell embryo then enters a specialized mitosis involving migration of maternal and paternal pronuclei, followed by mitotic spindle assembly and the first mitotic cell division (reviewed in Muller-Reichert et al. 2010).
Unlike the oocytes of most species studied, C. elegans oocytes do not exhibit a second arrest after meiotic maturation. However, proper completion of the meiotic program nonetheless requires sperm entry (reviewed in Marcello et al. 2013). This was inferred by observing oocytes in mutant worms that have fertilization-defective sperm (Ward and Carrel 1979; McNally and McNally 2005). These oocytes still exhibit signs of maturation by progressing into meiotic metaphase I, and they initiate anaphase; however, they do not extrude the first polar body. Instead, they abort anaphase I, skip meiosis II entirely, and enter mitosis. The mitotic cells exhibit signs of cell-cycle progression, such as nuclear envelope breakdown and reformation, but they do not divide, possibly because they lack the centrioles normally contributed by the sperm (Albertson 1984; Mikeladze-Dvali et al. 2012). These results suggest that, upon fertilization, sperm normally contribute a second signal that stimulates the egg to extrude the first polar body and initiate meiosis II.
One sperm-contributed protein that is required for proper zygotic development is SPE-11. spe-11 mutants exhibit defects in eggshell formation, mitotic spindle positioning, and cytokinesis (Hill et al. 1989; Browning and Strome 1996), as well as an increased risk of polyspermy (Johnston et al. 2010). Live fluorescence imaging revealed that spe-11 embryos undergo both meiosis I and meiosis II, but they fail to extrude polar bodies at the end of each meiosis (McNally and McNally 2005). Although the paternal factor SPE-11 is required for proper female meiotic divisions, the spe-11 phenotype is distinct from unfertilized embryos, which completely skip meiosis II, suggesting the existence of additional sperm factor(s) involved in specifying the meiosis II program (McNally and McNally 2005). Presumably, the oocyte also contains a specific factor(s) that responds to this putative sperm signal.
Herein, we describe the identification and characterization of the memi gene family and its role in female meiosis II. MEMI proteins are maternally expressed and loss of all members results in a skipped female meiosis II. In contrast, a hyper-morphic mutation in memi-1 results in a failure to exit meiosis II properly, indicating that the activity of MEMI proteins must be strictly regulated during the meiosis-to-mitosis transition. MEMI protein levels are sensitive to CUL-2 E3 ligase activity, suggesting that MEMIs are targeted by this Cullin system for degradation prior to mitosis. Finally, through a genome-wide RNAi screen for suppression of the hyper-morphic memi-1 mutant, we identified a conserved sperm-specific PP1 phosphatase, GSP-3/4. We propose that MEMIs represent a maternal component of a signal that specifies meiosis II upon sperm entry.
Materials and Methods
Worm strains and culture conditions
C. elegans (var. Bristol) was cultured as described (Brenner 1974). The following strains were used: DR1786 dpy-13(e184) unc-24(e138) IV; mDp4[unc-17(e245)] IV, MAS182 memi-1(sb41) dpy-20(e1282) IV, MAS137 memi-1(sb41) dpy-20(e1282) ruIs57[pie-1::GFP-tbb-2], IV, MAS91 unc-119(ed3) III; itIs37[pie-1::mCherry-HIS58]; ruIs57[pie-1::GFP-tbb-2], MAS138 unc-119(ed3) III; memi-1(sb41) itIs37[pie-1::mCherry-HIS58] IV; ruIs57[pie-1::GFP-tbb-2], MAS123 Y17G9B.9(tm3099), Y62E10A.14(tm2638) IV, MAS131 Y17G9B.9(tm3099) IV, MAS132 Y62E10A.14(tm2638) IV, MAS133 H02I12.5(tm3158) IV, MAS134 Y17G9B.9(tm3099) H02I12.5(tm3158) IV, MAS135 Y62E10A.14(tm2638) H02I12.5(tm3158) IV, HR592 memi-1(sb41) dpy-20(e1282)/nT1[unc(n754dm) let] IV; +/nT1 V, ET113 unc-119(ed3) III; ekIs2[pie-1p::GFP::cyb-1 unc-119(+)]; MAS188 memi-1(sb41) dpy-20(e1282)/nT1[unc(n754dm) let] IV; +/nT1 V; ekIs2[pie-1p::GFP::cyb-1 unc-119(+)], MAS186 unc-119(ed3) III; itIs37[pie-1::mCherry-HIS58] unc-119(+); ruIs57[pie-1::GFP-tbb-2]; fer-1(hc1). memi-1 was previously referred to as mel-43 in the original study that identified the sb41 mutation (Mitenko et al. 1997); the gene name was changed after consultation with P. Mains (University of Calgary). Further details on strains and molecular lesions are available on Wormbase (http://www.wormbase.org).
Duplication analysis
mDp4 was predicted to cover the memi-1 region (LGIV: −27.0 to 4.7; Figure 3). To determine the effect of mDp4 on the phenotype, memi-1(sb41) dpy-20(e1282)/+ males were crossed to DR1786 (dpy-13(e184) unc-24(e138) IV; mDp4[unc-17(e245)] IV). Fifty F1 progeny were plated and allowed to self-fertilize at 15°. F1 worms were genotyped by scoring Dpy-20 or Dpy-13 progeny. Ten plates exhibited Dpy-13 and Dpy-20 phenotypes in the F2 generation. In order to confirm that Dpy-20 worms still had the memi-1(sb41) mutation, three Dpy-20 worms from each plate were transferred to 25° and scored for lethality. At permissive temperature (15°) the memi-1(sb41) dpy-20/+ heterozygotes exhibited 23.5% lethality and mDp4/+/+ exhibited 11% embryonic lethality. If memi-1(sb41) and mDp4 act independently, the expected embryonic lethality would be 32% [1 − (0.89 × 0.765) × 100]. Five wild-type worms, memi-1(sb41) dpy-20(e1282)/(dpy-13(e184) unc-24(e138) IV; plus or minus mDp4[unc-17(e245)] IV), were picked from five different plates and allowed to self-fertilize at 15°. All progeny were scored and the percentage of Dpy-13 progeny was used to distinguish between parents having mDp4 or not. memi-1(sb41) dpy-20(e1282)/dpy-13(e184) unc-24(e138) are expected to give 25% Dpy-13 Unc-24 progeny. However, mDp4 would decrease this frequency because it spans the Dpy-13 region. Using this method, it was possible to separate all 25 hermaphrodites into two classes. Two out of 25 plates exhibited 9% Dpy-13; these worms carried the mDp4 duplication. From each of these two plates, 7/17 wild-type progeny exhibited <13% Dpy-13 self-progeny. Worms that had mDp4 exhibited 58.7 ± 4.3% dead eggs, compared with sibling controls that lacked the duplication (7.6 ± 2.4% dead eggs). These observations indicated that the duplication mDp4 was not able to rescue the memi-1(sb41) mutation, rather it increased the severity of the phenotype. Combined with the fact that memi-1(RNAi) rescued memi-1(sb41) (see Results), this analysis revealed that memi-1(sb41) is likely hyper-morphic.
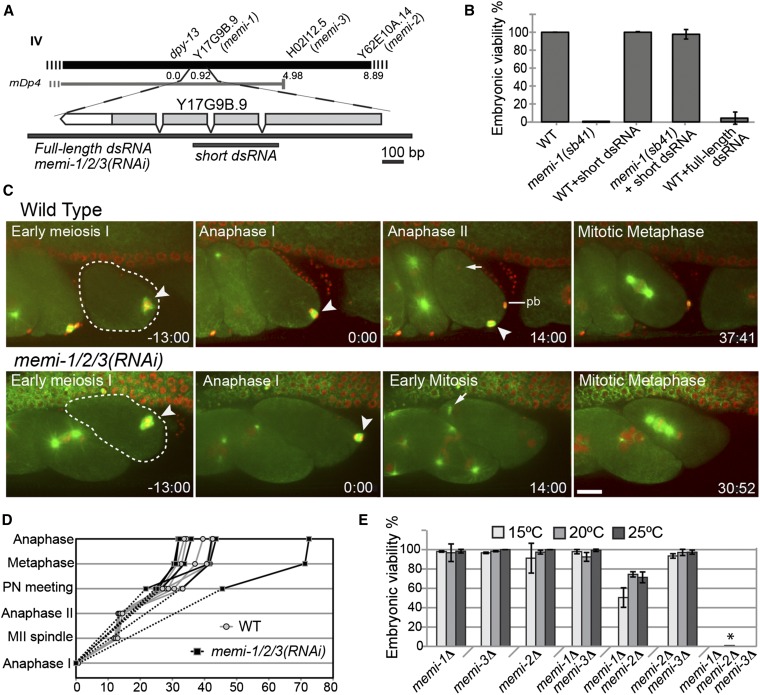
Figure 3.
Identification and characterization of memi-1 and two closely related paralogs. (A) The map position of memi-1 and the physical location of Y17G9B.9 and paralogs H02I12.5 and Y62E10A.14 on chromosome IV are shown. Although its physical end-point is unknown, the mDp4 duplication is estimated to end at position 4.7 (http://www.wormbase.org). The Y17G9B.9 dsRNA that is maximally divergent from H02I12.5 and Y62E10A.14 is shown below the gene (short dsRNA). Full-length dsRNA is 87% identical (overall) and is expected to efficiently target all three genes (referred to as memi-1/2/3(RNAi) in the text). (B) The viability (% of total progeny) for each worm strain at 25° is shown. memi-1(sb41) is rescued by the short Y17G9B.9 dsRNA. Viabilities were calculated from at least three hermaphrodites and >600 progeny. Bars show SD. (C) RNAi knock-down of memi-1, memi-2, and memi-3 together [memi-1/2/3(RNAi)] results in skipped meiosis II. Selected frames from an in utero time-lapse movie of wild-type and memi-1/2/3(RNAi) embryos in a strain expressing GFP-tubulin (microtubules; green) and mCherry-histone (DNA; red) are shown. Meiotic spindles are indicated with an arrowhead; anaphase I is time = 0. In memi-1/2/3(RNAi), sperm-derived centrosomes nucleate MTs in early mitosis immediately after meiosis I (compare arrows). Note the extra nuclei within a single cell in the adjacent embryo, illustrating the cytokinesis defect also present in memi-1/2/3(RNAi) embryos. pb = polar body. Bar, 10 µm. (D) Examples of the timing of events in wild-type (gray; n = 5) and memi-1/2/3(RNAi) (black; n = 6) embryos, from anaphase I (bottom) until anaphase of first mitosis (top). “Anaphase I” (t = 0) and “Anaphase II” refer to the time at which chromosomes separated and a clear microtubule midzone was first visible. “MII spindle” refers to the time when the spindle compressed to a length of ∼4 µm (only applies to WT). In memi-1/2/3(RNAi) embryos, anaphase I initiated but did not produce a polar body. (E) The viability (% of total progeny) for each homozygous deletion mutant strain at different temperatures is shown. Viabilities were calculated from at least three hermaphrodites and >200 progeny. Bars show SD. The triple deletion homozygotes produced only dead eggs (7 hermaphrodites, 1029 progeny) at 20° (asterisk).
Antibodies, immunostaining, and microscopy
Anti-MEMI rabbit antibodies were generated against full-length MEMI-1 fused to glutathione S-transferase. MEMI-1 antisera were affinity-purified using full-length MEMI-1 fused to maltose-binding protein. Bacterial acetone powder was added to the final sera to competitively bind remaining anti-bacterial antibodies prior to Western blot and immunostaining experiments. Rabbit anti-AIR-1 serum was generated against a mixture of GST-AIR-1 protein (full length) and AIR-1 C-terminal peptide (H2N-LTKSSRNNSTANQ-COOH) coupled to BSA; mouse anti-REC-8 (Abcam) was used at 1/100. Anti-GSP-3/4 antibodies were generated against a GST-fusion of a 28 a.a. C-terminal peptide, cloned using primers: GSP-3F_BamHI pGEX 5′-GGA TCC TCG GCT GCA ACA ATG-3′ and GSP-3R_NotI pGEX 5′-GCG GCC GCT TAT CCT CGA CGC ATG G-3′ and affinity-purified against an MBP-fusion protein using primers: GSP-3F_NotI pMAL 5′-GCG GCC GCT CGG CTG CAA CAA TGA AG-3′ and GSP-3R_BamHI pMAL 5′-GGA TCC TTA TCC TCG ACG CAT GGA C-3′.
Tubulin was visualized with mouse anti-tubulin antibodies (DM1A; 1:100; Sigma, St. Louis, MO). DAPI (5 µg/ml) was used to stain chromatin. Alexa647 goat anti-mouse, Alexa488 goat anti-rabbit, or Alexa546 goat anti-rabbit secondary antibodies (Invitrogen, Carlsbad, CA) were used at 1:100. Immunostained samples were mounted in 0.5% p-phenylenediamine, 20 mM Tris-Cl, pH 8.8, 90% glycerol.
To determine eggshell permeability, hermaphrodites were dissected in egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 25 mM Hepes, pH 7.4) with 0.25 µg/ml DAPI and incubated for ∼5 min, washed once, and mounted on agarose pads for imaging.
Confocal images of fixed and living embryos were obtained with a Hamamatsu Orca R2 camera on an inverted Olympus IX81 microscope with a Yokogawa CSU-10 spinning disc confocal head modified with a condenser lens in the optical path (Quorum Technologies). Images were acquired using a 60× oil (NA 1.42) or 60× Silicon (NA 1.3; for in utero imaging) objective lens and captured with a Hamamatsu Orca R2 camera controlled by MetaMorph software. Image files were analyzed using MetaMorph software. For quantifying MEMI cytoplasmic fluorescence levels, the average of integrated intensity of three circles drawn within the cytoplasm of each embryo was calculated. The intensity of each embryo was compared to N2 controls and displayed as relative fluorescence levels for the graphs.
Co-immunoprecipitation
Worm extracts were obtained by sonicating 3 g frozen gravid adult worms in lysis buffer [final concentration: 1 mM EGTA, 1 mM MgCl2, 50 mM HEPES pH 7.8, 100 mM KCl, 10% glycerol, 0.05% NP-40, and Complete Mini EDTA-free protease inhibitor (Roche)]. Antibodies (rabbit IgG, rabbit anti-MEMI, or rabbit anti-GSP-3/4) were coupled to Affiprep Protein A beads (Bio-Rad, Hercules, CA) as described by Moritz et al. (1998). Fifty micrograms of coupled random IgG, or anti-MEMI, or anti-GSP-3/4 antibodies were incubated with the extract at 4° overnight. Beads were washed 3× with lysis buffer + 0.05% NP-40 and 2× with lysis buffer. Bound proteins were eluted in sample loading buffer and separated via SDS-PAGE and analyzed by Western blotting.
Western blotting
Western blotting of MEMI-1 was performed on both whole-worm and collected embryo lysates. For each lane, embryos were prepared as in Gusnowski and Srayko (2011) from 100 worms. Protein samples were resolved via SDS-PAGE (10%). For whole-worm lysates, worms were first washed with 3× 400 µl H2O, retained in a final volume of 10 μl, to which loading buffer was added prior to loading. A prestained protein marker (7−175 kDa; New England Biolabs, Beverly, MA) was used to estimate the relative mass of the proteins. Proteins were transferred to nitrocellulose membrane (Hybond-N, GE Healthcare) at 100 V for 2 hr. The membrane was blocked in 8% skim milk in TBST for 1 hr (Tris buffer saline Tween 20; 20 mM Tris-HCl, pH 7.4, 500 mM NaCl, and 0.05% Tween 20). The anti-MEMI-1 and anti-tubulin antibodies were used at 1:200 and 1:400, respectively, in TBST + 4% skim milk and incubated for 1 hr at RT. Goat anti-rabbit and goat anti-mouse HRP-bound secondary antibodies (Bio-Rad) were used at 1:5000 in TBST + 4% skim milk and incubated with the membrane for 1 hr at RT. The secondary antibodies were detected via SuperSignal West Pico ECL (Thermo Fisher Scientific).
RNAi by feeding and injection
dsRNA was introduced to worms either by microinjection (Fire et al. 1998) or feeding dsRNA-expressing bacteria to L3-L4 hermaphrodites (Kamath et al. 2003). Control RNAi was the L4440 RNAi feeding vector (Addgene; A. Fire, Stanford University School of Medicine, Stanford, CA) lacking an insert. To produce dsRNA for microinjection, genomic DNA was amplified via PCR using forward and reverse primers that contained the T3 or T7 bacterial polymerase promoter sequences, respectively. dsRNA targeting all memi genes used a PCR product from forward 5′-T3-CTG ACA GCT GAC ACT CAC AAA AAC TG-3′ and reverse 5′T7p-TTG CGG GTT GCG GTG GGA AAA TAAC-3′ primers. For memi-1-specific dsRNA, forward 5′-T3p-GTC GAG CAC GTG TTT CTT CA-3′ and reverse 5′T7p-CAG TGT GGT TCT CAG GA-3′ primers were used. For in vitro transcription, templates were PCR fragments purified by the MEGAscript RNAi kit (Invitrogen, Carlsbad, CA). dsRNA was purified with QIAGEN RNeasy columns. 400 ng/μl of purified dsRNA was injected into the gonads of L4 C. elegans larvae. The injected worms were incubated at 25° overnight prior to use in further experiments. For RNA feeding experiments, PCR products were cloned into the L4440 feeding vector using primers 5′-ATC CCG GGA TGT CAG CTC CAT CTG GC-3′ and 5′-CGA CTA GTC TCG TCC TCT TCA TCC AG-3′ (for full-length dsRNA cloned into XmaI/SpeI) and forward 5′-ATC CCG GGT CGA CGT GTT TCT TCA-3′ and reverse 5′-CGACTAGTGTGTGGTTCTCAGGAGACG-3′ primers (for short dsRNA cloned into XmaI/SpeI).
Whole-genome sequencing
For sequencing memi-1(sb41) worms, strain MAS137 was cultured on 10 nematode growth media (NGM)-agarose plates with OP50 at 15° until plates were cleared of bacteria. Mixed stage worms were washed off with M9 (22 mM KH2PO4, 42 mM Na2HPO4, 85 mM NaCl, 1 mM MgSO4) followed by two centrifugation and washing steps. The pellet was resuspended in 0.5 ml of lysis buffer (10 mM Tris-Cl, 0.1 M EDTA, 0.5% SDS, 20 μg/ml DNase-free RNase) followed by 25 μl of 10% SDS and 2.5 μl of proteinase K (20 mg/ml) and incubated at 50° for 1 hr. A second 2.5 μl of 20 mg/ml of proteinase K was added and incubated at 50° for one more hour. Nucleic acid was isolated by standard phenol/chloroform and ethanol precipitation methods. Sequencing was performed by the BC Cancer Agency Genomic Sciences Center and sequence data analysis was kindly provided by the UBC C. elegans Gene Knockout Laboratory.
Ex utero and in utero confocal imaging
For ex utero imaging of fragile early meiotic embryos, three to five worms were dissected in 5 μl of egg buffer (see above) on poly-L-lysine-coated coverslips. A hand-made cover was used to prevent evaporation during imaging. Time-lapse imaging was performed on an Olympus IX81 spinning disk confocal microscope described above.
For in utero imaging, worms were picked into 7 μl of egg buffer containing 5 mM tetramisole hydrochloride to immobilize the worms prior to imaging. When the worms stopped moving, the cover slip was inverted onto a 2% agarose pad.
For imaging MAS138 (memi-1(sb41); GFP::tubulin; mCherry::histone), five planes (2 μm spacing) were taken every 20 sec from the time that the oocyte entered the spermatheca until first mitosis. Movies presented are made from stack projections. DIC imaging at the end of mitosis was used to assess cytokinesis. To measure cell-cycle events, the following landmarks were used: anaphase I, meiosis II spindle (at 4 µm length), anaphase II, meeting of pronuclei, mitotic metaphase, and mitotic anaphase. In memi-1(sb41) embryos, the nuclei were often not discernible and the sperm DNA remained condensed, therefore, pronuclear meeting for these embryos was defined as the stage when the sperm-derived chromatin migrated to meet the meiotic spindle/meiotic chromatin. For memi-1/2/3(RNAi) worms, imaging was performed 24 hr after the dsRNA injection. For line-scan quantification of mCherry-histone and GFP-tubulin fluorescence in meiosis II, a 9.6 μm line (width of 15 pixels) was drawn. The line was oriented along the spindle pole-to-pole axis for wild type. In most cases, memi-1(sb41) meiotic spindles appeared rounded, making spindle poles difficult to discern; however, multiple line orientations provided similar results. When possible, a line perpendicular to a “metaphase plate” was used to determine the meiosis II line orientation.
Genome-wide RNAi screen for suppression of memi-1(sb41)
A bacterial library in 384-well format was used to feed worms C. elegans dsRNA (Kamath et al. 2003). Disposable 96-pin replicators were used to inoculate 200 µl of LB ampicillin in 4 × 96-well plates from each 384-well stock plate, and the cultures were grown overnight at 37°, with gentle shaking. For each trial, a separate 96-well plate contained controls: L4440 + memi-1/2/3(RNAi) long fragment and L4440 + memi-1(RNAi) short fragment (Figure 3A). Control inoculations were from streaked LB Amp plates kept at 4°. RNAi-NGM agar for worm screening was in 48-well plates (1.3 ml agar per well, with 1 µM IPTG, 25 ng/ml carbenicillin, 100 units/liter nystatin). Plates were allowed to solidify, inverted, and left overnight at RT to prevent desiccation. Five microliters of each overnight culture of RNAi bacteria was transferred from the 96-well plate to the agar surfaces of the 48-well plate using an adjustable 8-tip multi-channel pipette (Rainin Pipet-lite XLS). L2-L4 worms (MAS137 memi-1(sb41) dpy-20; ruIs57[pie-1::GFP::tbb-2] or N2 wild-type controls) were washed off of NGM plates seeded with OP50 bacteria, washed four times with M9 in a microfuge tube, and aliquoted in minimal volume onto a large unseeded NGM RNAi plate and incubated at 25° for 1 hr to shed bacteria. Prior to transferring worms to the RNAi plates, 8 µl of sterile HPLC water was pipetted onto each well of the 48-well RNAi plate. The worms were picked individually and transferred into the liquid droplet to allow ease of transfer and to prevent scoring of the agar surface. Plates were incubated at 25° for 3–4 days and scored for suppression. The entire library was tested for suppression of memi-1(sb41) in at least three independent trials. Bacteria from suppressed wells (positives) were cultured and the plasmids were sequenced to confirm insert identity.
Data availability
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
memi-1(sb41) mutants exhibit cytoskeletal defects and cell-cycle delays at the meiosis-to-mitosis transition
sb41 is a dominant temperature-sensitive maternal-effect lethal mutation that was originally identified in a genetic screen for redundant genes affecting embryonic development (Mitenko et al. 1997). In the initial characterization using DIC microscopy, embryos from homozygous mutant sb41 mothers displayed excessive membrane blebbing, failure in polar body II extrusion, abnormal intracellular cytoskeletal structures at the end of meiosis II, sporadic cytokinesis failure in the first mitosis, and synchronous cell divisions in two-cell embryos (Mitenko et al. 1997).
To further examine the cellular defects associated with this mutation, we created an sb41 strain that expressed GFP-tubulin (microtubules) and mCherry-histone (chromosomes). sb41 embryos exhibited normal meiosis I spindle assembly, chromosome segregation, and extrusion of the first polar body. Meiosis II spindle assembly also appeared normal; however, meiosis II chromatid segregation and polar body II extrusion usually did not occur (16/20; Figure 1A).
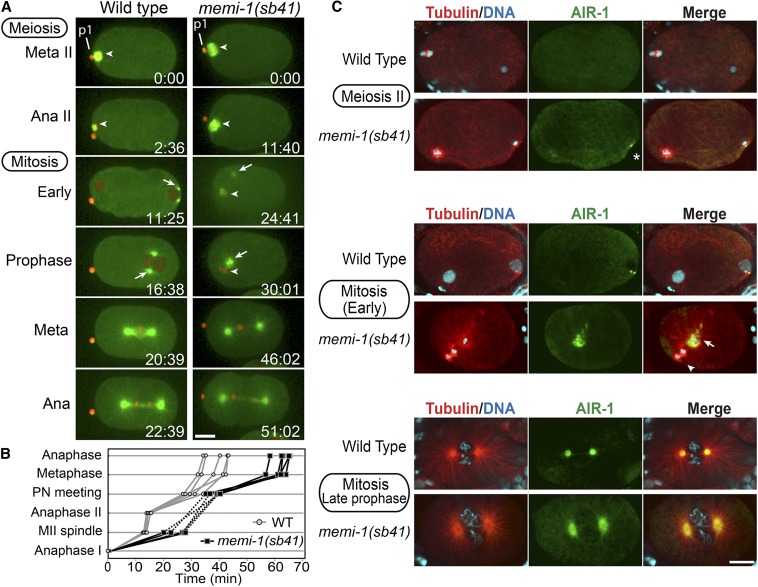
Figure 1.
memi-1(sb41) embryos exhibit defects at the meiosis II-to-mitosis transition. (A) Time-lapse images of wild-type and memi-1(sb41) embryos expressing mCherry-histone and GFP-tubulin. Panels start during the second meiotic metaphase (t = 0). The previous meiosis I division was successful, as evidenced by the polar body (p1). In memi-1(sb41), meiosis II chromatin does not segregate, centrosomal microtubles appear (t = 24.41) and the centrosomes migrate to the female chromatin (t = 30.01). A mitotic spindle-like structure eventually forms in memi-1(sb41), and cytokinesis is initiated. Meiotic spindle and centrosomes are shown with arrowheads and arrows, respectively. Bar, 10 µm. (B) Timing of events in wild-type (gray; n = 5) and memi-1(sb41) (black; n = 5) embryos, from anaphase I (bottom) until anaphase of first mitosis (top). “Anaphase I” (t = 0) and “Anaphase II” refer to the time at which chromosomes separated and a clear microtubule midzone was first visible. In these five memi-1(sb41) examples, meiosis II chromosomes did not appear to separate prior to dissolution of meiotic spindle microtubules (dotted line). “MII spindle” refers to the time when the bipolar spindle compressed to a length of ∼4 µm. Nuclear envelopes were not always observable during the MII-to-mitosis transition in memi-1(sb41), so pronuclei (PN) meeting in memi-1(sb41) embryos was estimated as the time when centrosome asters and meiotic structures neared each other (∼3 μm). (C) Immunofluorescence images of microtubules (red), DNA (blue), and the centrosomal Aurora kinase AIR-1 (green). In contrast to wild type, memi-1(sb41) mutant centrosomes recruit AIR-1 during metaphase of meiosis (asterisk). AIR-1 foci were observed near sperm DNA during meiosis II in memi-1(sb41) (10/12), but not in WT (0/10). Centrosome fragmentation (arrow) occurs in memi-1(sb41) embryos. Arrowhead indicates the meiosis II spindle with two masses of chromatin, which likely represent separated meiotic chromatids within the cytoplasm. In late prophase, two centrosomes are visible (bottom panels). Bar, 10 µm.
Using specific cellular events as landmarks (see Materials and Methods), we determined that cell-cycle delays occurred in sb41 embryos after meiosis I. This included a 10–15 min delay in reaching late metaphase II, the time at which the spindle shortens to ∼4 µm (Albertson and Thomson 1993; Yang et al. 2003) (Figure 1B). The transition from early mitosis to metaphase was also delayed in mutant embryos, but the subsequent mitotic metaphase-to-anaphase transition occurred at a rate similar to wild type (Figure 1B).
In wild-type embryos, the sperm-derived centrosomal microtubules appear only after the completion of meiosis II (Figure 1A; Supplemental Material, File S1 and McNally et al. 2012). In sb41 embryos, the meiosis II spindle persisted into mitosis, often moving in an unpredictable manner around the cell. Furthermore, centrosomal microtubule asters formed and migrated toward meiotic chromatin while the meiosis II spindle was still present (14/14; Figure 1A and File S2). Despite the coexistence of these structures in the sb41 embryos, a mitotic spindle eventually formed and the cell progressed into mitotic anaphase (File S2 and File S3). Some sb41 embryos displayed a failed mitotic cytokinesis (2/14; File S4), whereas others exhibited anaphase-like separation of microtubule asters and pseudocleavage, followed by reformation of two centrosomes (2/14; File S5). The sb41 embryos often exhibited synchronous cell divisions in two-cell embryos (Mitenko et al. 1997, and end of File S5), indicative of defects in proper polarity establishment (Figure S1). Based on the phenotypes of the dominant sb41 mutation, as well as additional phenotypic evidence presented below, we termed this gene memi-1 (meiosis-to-mitosis transition defect).
memi-1(sb41) centrosomes mature in the presence of late-stage meiosis II spindles
Because memi-1(sb41) embryos displayed meiotic and mitotic structures within the same cell, we used antibodies against Aurora A kinase, AIR-1, to assess the maturation state of the centrosomes (Figure 1C). AIR-1 is required for many steps leading to the full maturation of the centrosomes, including the recruitment of γ-tubulin, and is implicated in promoting microtubule growth (Mitenko et al. 1998; Hannak et al. 2001; Srayko et al. 2005). In wild-type embryos, AIR-1 was detected on the centrosomes as very small foci only after the completion of meiotic anaphase II, and AIR-1 levels at centrosomes increased in intensity with the cell cycle. In contrast, in memi-1(sb41) embryos, AIR-1 was detected on centrosomes in embryos that still displayed a meiosis II spindle. In addition, early mitotic memi-1(sb41) embryos often exhibited an abnormally fragmented pattern of AIR-1, suggesting an underlying defect in centrosome integrity in these embryos (Figure 1C, middle row). Based on live GFP-tubulin imaging, sections of astral microtubules separated away from the centrosome after the centrosomes started to migrate toward the meiotic chromatin (e.g., File S4). Therefore, memi-1(sb41) interferes with the completion of meiosis II but it does not prevent the initiation of mitotic events, giving rise to abnormal embryos with features of both meiosis II and mitosis. The fragmentation of AIR-1 and microtubule asters indicated that the centrosomes were also defective during the abnormal meiosis-to-mitosis transition in memi-1(sb41) embryos.
memi-1(sb41) mutants exhibit abnormal anaphase II during female meiosis
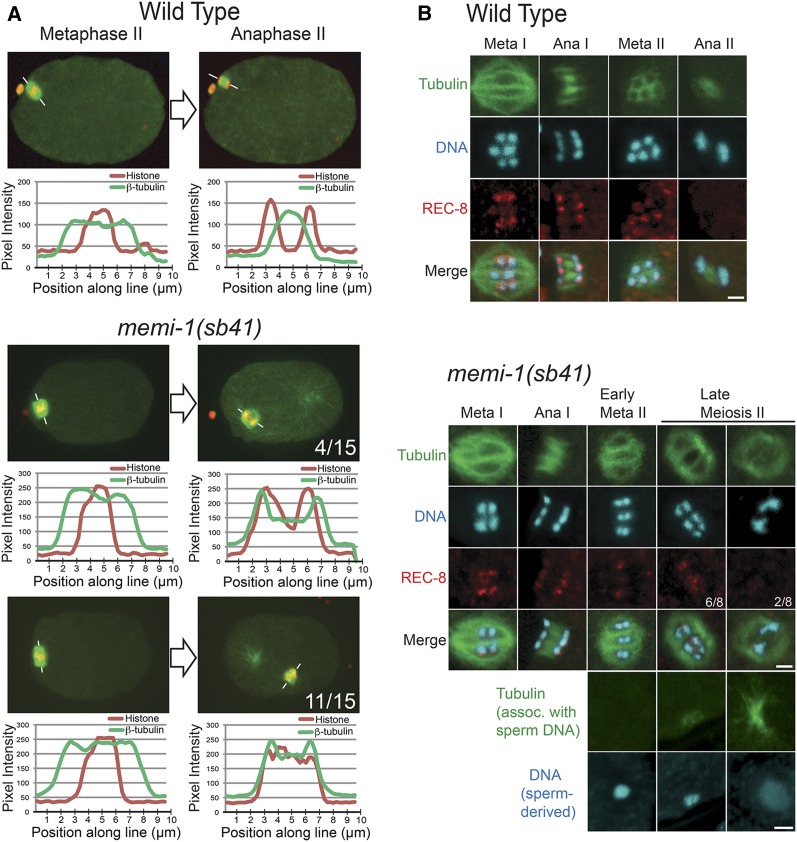
To assess chromatid congression and segregation in meiosis II, we performed line scans of fluorescence intensity for mCherry-histone and GFP-β-tubulin in wild-type and memi-1(sb41) mutant embryos. In metaphase II, both memi-1(sb41) and wild-type embryos exhibited a similar distribution of tubulin and chromatin along the spindle axis. Tubulin was concentrated at the spindle poles, and the chromatin was aligned at the spindle equator in metaphase, indicating that chromosomes can congress in the mutants (Figure 2A). In wild type, anaphase II is accompanied by a change in the distribution of tubulin, with microtubules appearing between the separating masses of chromatin (Albertson and Thomson 1993; Dumont et al. 2010; Muscat et al. 2015). In the memi-1(sb41) embryos, the meiosis II spindles did not exhibit a peak of tubulin fluorescence between chromatin masses (11/15; Figure 2A). In a few embryos, two masses of chromatin were separated (4/15), but tubulin fluorescence levels were more uniformly distributed along the pole–pole axis (Figure 2A). Based on these observations, we concluded that memi-1(sb41) embryos exhibited congression of chromatids in meiosis II, but that chromatid segregation was impaired, and spindle microtubule rearrangements typical of anaphase II did not occur.
Figure 2.
memi-1(sb41) results in defects in meiosis II chromatid segregation and spindle morphology. (A) Single plane confocal images of metaphase II and anaphase II from a wild-type embryo and two memi-1(sb41) embryos. Fluorescence intensities of histone (red) and tubulin (green) along the marked line are displayed as line-scan plots. The number of memi-1(sb41) embryos displaying the represented phenotypes are shown. (B) Immunofluorescence staining with anti-REC-8 antibody on wild-type and memi-1(sb41) embryos. In wild-type embryos, REC-8 locates between homologs and sister chromatids in metaphase I and between sister chromatids in meiosis II. In memi-1(sb41), the majority of late-stage metaphase II embryos exhibit REC-8 between chromatids and unsegregated chromatin (6/8). Some late-stage meiosis II embryos with two separated masses of chromatin exhibited very weak or no REC-8 (2/8). Meiotic spindles depicted in memi-1(sb41) were identified as early or late-stage meiosis II by the absence or presence, respectively, of microtubules near the sperm DNA, shown at the bottom. Bar, 2 µm.
We next tested whether memi-1(sb41) interfered with the release of sister chromatids. During anaphase I, REC-8/kleisin is specifically removed from homologous chromosomes but it remains associated with the sister chromatids until anaphase II, at which time it is removed to allow sister chromatid separation (Pasierbek et al. 2001; Severson et al. 2009). We examined the pattern of REC-8 between sister chromatids in memi-1(sb41) embryos. Late-stage meiosis II spindles in memi-1(sb41) embryos stained positive for REC-8 between chromatids, but REC-8 was not visible in embryos where chromatids had separated (Figure 2B). These observations suggested that the persistence of the REC-8/kleisin on sister chromatids correlated with a failure of anaphase II in memi-1(sb41) mutants. Based on live-cell imaging, embryos that exhibited sister chromatid separation nevertheless displayed various other defects such as abnormal polar body extrusion and cell cycle delays. Therefore, memi-1(sb41) perturbs multiple processes at the end of meiosis II, including timely REC-8 removal, proper spindle microtubule reorganization, and polar body formation.
memi-1 and two closely related paralogs are required for embryonic viability
memi-1(sb41) was mapped to the center of chromosome IV, <1 MU from dpy-13. Using mDp4, a chromosomal duplication that spans the sb41 genetic interval, we found that memi-1(sb41)/+/+ exhibited a more severe phenotype than memi-1(sb41)/+ at 15° (Figure 3A and Materials and Methods). This suggested that sb41 was hyper-morphic. Whole-genome sequencing of memi-1(sb41) worms revealed one mutation in a predicted gene, Y17G9B.9, near dpy-13. The Y17G9B.9 protein has no recognizable domains and no obvious homolog outside of nematodes. The memi-1(sb41) mutation resulted in a P74S substitution that is within a putative target region for a number of proline-directed kinases, such as cyclin-dependent kinase (Figure S2).
Y17G9B.9(tm3099) worms have a 195-bp deletion removing most of the first intron and 151 bp of the second exon of the gene (National Bioresource Project, Tokyo, Japan), but they displayed no obvious phenotypes, suggesting that Y17G9B.9 was nonessential. Database searches revealed the existence of two highly similar paralogs in C. elegans, Y62E10A.14 and H02I12.5 (Figure S2), each having 87% DNA (85% aa) sequence identity to Y17G9B.9, raising the possibility that this gene family was functionally redundant.
To knock-down all three paralogs, a 1717-bp dsRNA spanning the entire Y17G9B.9 gene was made, with the idea that 87% identity among the paralogs should permit RNAi-based knock-down of all three genes (Figure 3A). The full-length dsRNA resulted in nearly complete embryonic lethality at 25° (Figure 3B). In contrast, a short 420-bp dsRNA predicted to be more specific to Y17G9B.9 (80% identity with the paralogs) injected into wild type (WT) did not cause obvious defects (Figure 3B). Given that memi-1(sb41) exhibited hyper-morphic behavior but short Y17G9B.9(RNAi) caused no obvious phenotypes, we reasoned that short Y17G9B.9(RNAi) should rescue memi-1(sb41). Indeed, when short Y17G9B.9 dsRNA was injected into memi-1(sb41) mutants, the maternal-effect lethality of memi-1(sb41) at 25° was completely suppressed (Figure 3B). This confirmed the identity of memi-1 and indicated that sb41 was not a loss-of-function allele. Furthermore, the RNAi experiments suggested that one or both of the paralogs were functionally redundant, hence, Y62E10A.14 and H02I12.5 were termed memi-2 and memi-3, respectively.
MEMIs are required for the female meiosis II program
The embryonic lethality associated with memi-1(sb41) at 25° was likely caused by chromosomal aneuploidy due to a failure to complete meiosis II. Because full-length Y17G9B.9 dsRNA, hereafter referred to as memi-1/2/3(RNAi), also resulted in lethality (Figure 3B), we examined wild-type (File S6) and memi-1/2/3(RNAi) (File S7) embryos using in utero fluorescence microscopy. memi-1/2/3(RNAi) embryos exhibited normal meiosis I spindle assembly, metaphase I, as well as separation of chromosomes in anaphase I. However, cytokinesis did not occur at the end of anaphase I, resulting in a failure to extrude the first polar body (Figure 3C). Surprisingly, memi-1/2/3(RNAi) embryos did not assemble a meiosis II spindle (20/20; File S7). Instead, nuclear envelopes formed around decondensing DNA, and astral microtubules emanated from centrosomes, all of which indicated entry into mitosis (Figure 3C). Chromosome segregation occurred in mitosis (Figure 3C) and cytokinesis was initiated but failed, resulting in a multinucleated cell (e.g., adjacent cell in Figure 3C). The time to complete the meiosis-to-mitosis transition in memi-1/2/3(RNAi) embryos was variable, and not always shorter than wild type, despite the absence of meiosis II events (Figure 3D). The skipped meiosis II phenotype was similar to what has been reported for fertilization-defective mutants (Ward and Carrel 1979; McNally and McNally 2005). fer-1(hc1) spermatozoa are motility-defective and they are incapable of fertilizing oocytes (Argon and Ward 1980). Using live imaging of GFP-tubulin and mCherry-histone, unfertilized fer-1 oocytes exhibited a prolonged anaphase I, followed by nuclear envelope formation and mitotic cycling without centrosomes (File S8).
We next sought to determine the contributions of individual memi paralogs to the meiosis-to-mitosis transition. Viable strains for each single deletion were obtained (National Bioresource Project, Tokyo, Japan) and double-deletion strains were constructed. All single-deletion and double-deletion worms were scored for lethality at three temperatures, 15, 20, and 25°. Single deletions of memi-1, memi-2, or memi-3 appeared wild type; however, different double-deletion combinations exhibited some maternal-effect lethality, depending on the genes deleted (Figure 3E). The memi-1(Δ) memi-2(Δ) combination resulted in the most severe double-mutant phenotype (as low as 50% hatching; Figure 3E). DIC microscopy revealed defects similar to memi-1/2/3(RNAi) phenotypes, such as extra female pronuclei (5/20) likely resulting from meiotic failure and/or polar body extrusion defects (File S9) and cytokinesis failure (1/20; File S10), which were not observed in wild type (File S11). The double-deletion mutants indicated that each single gene provided partial function during meiosis II. A heterozygous recombinant was also recovered with the genotype memi-1(Δ)memi-3(+)memi-2(Δ)/memi-1(Δ)memi-3(Δ)memi-2(Δ), which produced 46% viable offspring (306/666). As expected, one-quarter of the surviving progeny from this recombinant grew to adulthood, but produced 100% dead eggs (Figure 3E). Diagnostic PCR confirmed that these worms were triple-deletion homozygotes, and DIC imaging of embryos from maternal-effect lethal sibling worms revealed one-cell phenotypes that were similar to the memi-1/2/3(RNAi) embryos (i.e., 10/11 embryos failed to make a polar body, 1/11 produced an abnormally large meiosis I polar body), and cytokinesis often failed (5/10) (File S12).
C. elegans embryos are encased in a protective trilaminar eggshell composed of an outer vitelline layer, a middle chitin-containing layer, and an inner layer that contains proteoglycans (Wharton 1980; Rappleye et al. 1999; Bembenek et al. 2007; Benenati et al. 2009; Olson et al. 2012). The vitelline layer forms initially, and is present on the oocyte before fertilization (Olson et al. 2012). The middle chitin layer is assembled after fertilization (Maruyama et al. 2007), and it provides mechanical strength and a block to polyspermy (Johnston et al. 2010). The inner proteoglycan layer is deposited during anaphase I, and then a distinct permeability barrier that involves fatty acid biosynthesis and modification enzymes (Tagawa et al. 2001; Rappleye et al. 2003; Benenati et al. 2009; Carvalho et al. 2011) forms during anaphase II (Olson et al. 2012). Because loss of memi-1/2/3 function resulted in a skipped meiosis II, we reasoned that these embryos might exhibit defects in eggshell permeability. Indeed, using the DNA stain DAPI as a tracer, we observed chromosome staining throughout the memi-1/2/3 triple-deletion embryos (28/28), but not the WT embryos (0/21; Figure 4). In contrast to the memi-1/2/3 deletion embryos, memi-1(sb41) embryos were not permeable to DAPI (0/24), suggesting that these embryos completed the final stages of egg shell synthesis.
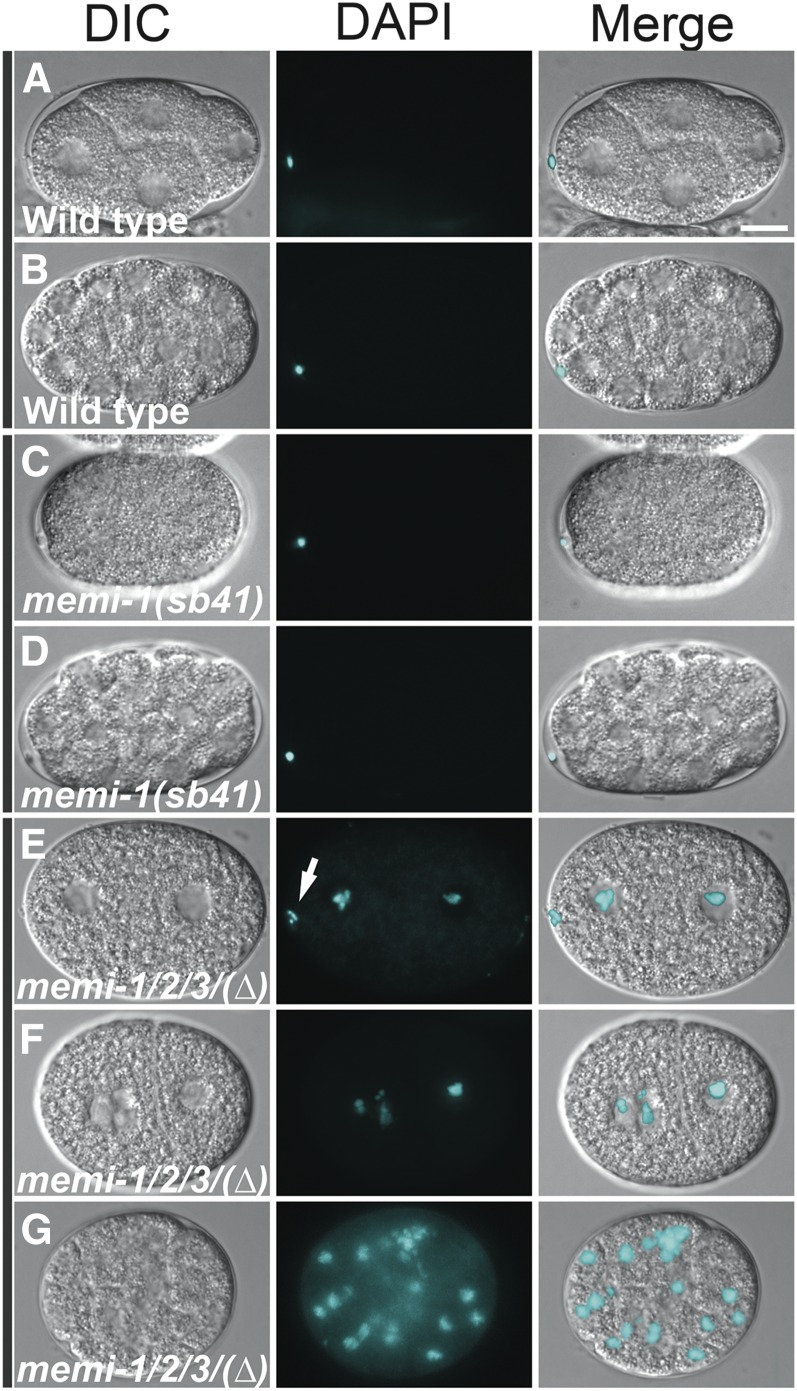
Figure 4.
Loss of MEMI function results in a defective eggshell permeability barrier. WT embryos and embryos from homozygous mutant mothers were incubated with DAPI DNA stain to assay permeability of the eggshell. (A and B) In WT, polar bodies from meiosis I stained with DAPI (21/21 embryos), but no other DNA was visible (0/21 embryos) due to a permeability barrier in the eggshell. (C and D) In memi-1(sb41), polar bodies from meiosis I stained with DAPI (24/24 embryos), but no other DNA was visible (0/24 embryos). (E–G) The memi-1/2/3(∆) triple-deletion mutants exhibited DNA fluorescence throughout the embryo (28/28). Segregated chromosomes from meiosis I were often detected (17/28 embryos). The embryo in E showed a single grouping of condensed DNA clustered near the inner edge of the cell (determined by viewing other focal planes), suggesting that chromosomes were not extruded (arrow). This embryo exhibited no other polar body. Nuclear envelopes formed around the chromatin, indicating entry into mitosis, consistent with a skipped meiosis II. F and G did not exhibit any polar bodies. Bar, 10 µm.
We also examined the location of DAPI-stained chromosomes in the memi-1/2/3 deletion embryos by fluorescence microscopy (Figure 4). Of the memi-1/2/3 deletion embryos, 17/28 displayed a grouping of condensed DNA close to the inner cortex of the cell, possibly indicative of an incomplete/failed extrusion of chromosomes during meiosis I. The remaining embryos (11/28) were in various stages of mitosis and did not display any signs of meiotic chromatin separation or polar bodies, consistent with a failure to complete both meiotic divisions. None of the embryos displayed any signs of having completed meiosis II (0/28), consistent with the phenotypes of memi-1/2/3(RNAi).
MEMI proteins are enriched in meiotic embryos
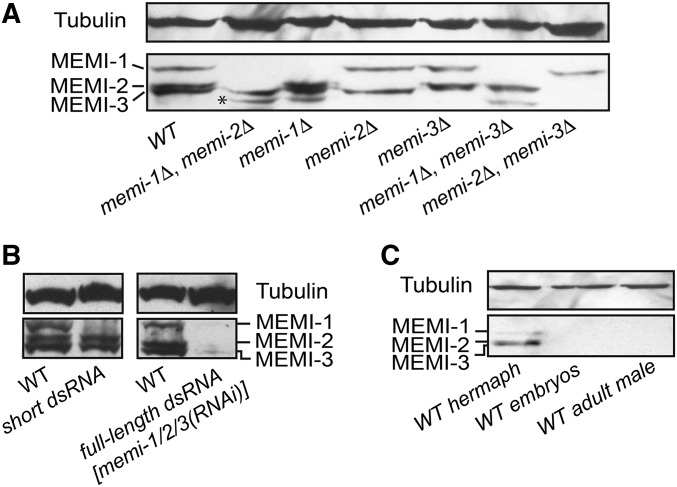
The MEMI proteins each have a predicted molecular weight of ∼40 kDa. Affinity-purified pan-specific α-MEMI antibodies detected three proteins in whole-worm hermaphrodite lysates, ranging in size from 45 to 52 kDa. Probed lysates from the deletion mutants and dsRNA-treated worms revealed the expected changes (Figure 5, A and B). The three proteins were not detected in lysates derived from adult males, consistent with MEMI-1 having a maternal function (Figure 5C). Furthermore, the MEMI proteins were not detected in lysates prepared from purified eggs (composed of mitotic embryos), suggesting that MEMI proteins are degraded early in development (Figure 5C).
Figure 5.
Pan-specific anti-MEMI antibodies detect maternally expressed proteins. (A) Western blots of gravid adult hermaphrodite worms homozygous for different memi deletions (Δ) indicate the identity of each MEMI protein. The asterisk indicates a truncated species observed in all lanes that contain memi-1(Δ). (B) Western blots of lysates from wild-type and RNAi-treated worms probed with anti-MEMI. A short dsRNA specifically targets MEMI-1, while the full-length dsRNA targets all three genes (refer to Figure 3 for RNA description). (C) MEMI-1, MEMI-2, and MEMI-3 proteins are enriched in WT hermaphrodite lysates, but not visible in embryo lysates, which are enriched for late-stage mitotic embryos. MEMI proteins are not detected in male whole-worm lysates. Tubulin was a loading control for all blots.
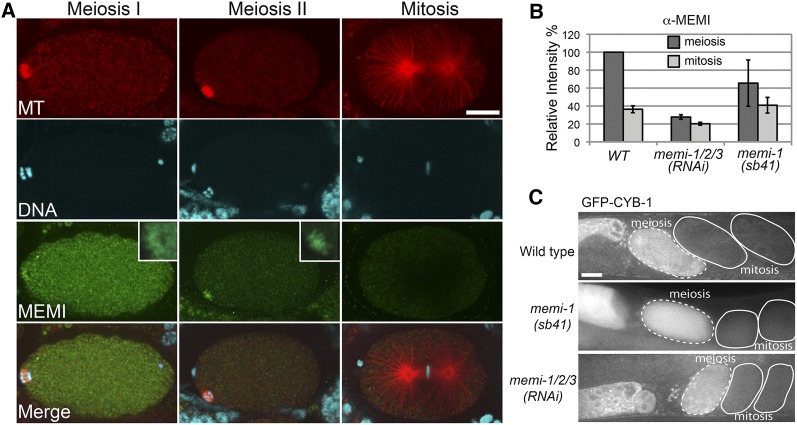
Indirect immunofluorescence of MEMI proteins in wild type revealed a strong cytoplasmic signal in oocytes and embryos in meiosis I. In anaphase I embryos, a distinct signal was also observed at the midzone of the spindle (Figure 6A). In meiosis II embryos, the cytoplasmic signal was reduced compared to meiosis I, but staining was often visible at the midzone of the anaphase II spindle (Figure 6A). Immunofluorescence was reduced in memi-1/2/3(RNAi) embryos (Figure 6B and Figure S3). Quantification of fluorescence intensities revealed that the cytoplasmic signal was considerably lower in one-cell mitotic embryos, compared with meiotic embryos (Figure 6B). memi-1(sb41) embryos similarly displayed reduced MEMI immunofluorescence in mitotic embryos, however, with more variability (Figure 6B and Figure S3). Because the antibodies were pan-specific, any changes in the levels of MEMI-1(sb41) specifically might be difficult to detect via immunostaining.
Figure 6.
Immunofluorescence staining with anti-MEMI antibodies. (A) Wild-type embryos exhibit a cytoplasmic distribution of MEMI in meiotic embryos. A fibrous staining pattern was also observed at the midzone of anaphase meiotic spindles, most prominently during meiosis II (insets). Microtubules in red, DNA in blue, and MEMI in green. Bar, 10 µm. (B) Quantification of cytoplasmic levels indicated MEMI levels in the first mitosis were lower than in meiotic embryos, and greatly diminished in the RNAi samples. MEMI levels in memi-1(sb41) showed a similar trend to wild type, but with more variability. Values were based on meiotic embryos (WT, n = 3; memi-1/2/3(RNAi), n = 11; memi-1(sb41), n = 14) and mitotic embryos (WT, n = 5; memi-1(RNAi), n = 7; memi-1(sb41), n = 3). Bars show SD. (C) Selected frames from a time-lapse movie of wild-type and memi-1/2/3(RNAi) embryos expressing GFP-CYB-1. GFP fluorescence was strong in oocytes and meiotic embryos (dashed line), but reduced in mitotic embryos (solid line). Bar, 10 µm.
Postmeiotic degradation of MEMI proteins requires a Cullin-ring ubiquitin ligase
In C. elegans, two different E3 ubiquitin ligase complexes are required for the progression through meiosis I and meiosis II. The anaphase-promoting complex (APC) is required for exit from female meiosis I in C. elegans (Furuta et al. 2000; Golden 2000). Knockout of any subunits of this complex results in an arrest in metaphase of meiosis I. Another E3 ubiquitin ligase, CUL-2 (cullin-ring ubiquitin ligase), and its substrate-specific adaptor, ZYG-11, is essential for the meiosis II metaphase-to-anaphase transition, the establishment of anterior–posterior polarity, and chromosome condensation (Feng et al. 1999; DeRenzo et al. 2003; Liu et al. 2004; Sonneville and Gonczy 2004; Vasudevan et al. 2007). Loss of CUL-2 or ZYG-11 results in cyclin B persistence and a ∼40 min delay in metaphase of meiosis II (Sonneville and Gonczy 2004; Vasudevan et al. 2007). These embryos eventually go through anaphase II but they fail to extrude the second polar body. The cul-2(RNAi) embryos also exhibit delays in forming the eggshell permeability barrier (Olson et al. 2012). memi-1(sb41) exhibits many of the same phenotypes, including severe delays in the meiosis-to-mitosis transition and polarity defects, but we did not observe obvious defects in eggshell permeability when compared to WT (Figure 4).
The zyg-11(RNAi) phenotype resembles memi-1(sb41) embryos, at least in part, thus we hypothesized that MEMI-1 might regulate the CUL-2 complex or be downstream of the complex. In order to test whether memi-1 affects the function of the CUL-2 complex, we monitored cyclin B levels in a GFP-CYB-1 strain in both memi-1(sb41) and memi-1/2/3(RNAi). In wild type, oocytes have high GFP-Cyclin B fluorescence levels and by fertilization, levels begin to decrease (Liu et al. 2004; Sonneville and Gonczy 2004). In both memi-1(sb41) and memi-1/2/3(RNAi) embryos, CYB-1 levels declined after the embryo exited the spermatheca, similar to wild type (Figure 6C and Figure S4). This indicated that MEMIs probably do not alter general APC or CUL-2/ZYG-11 functions.
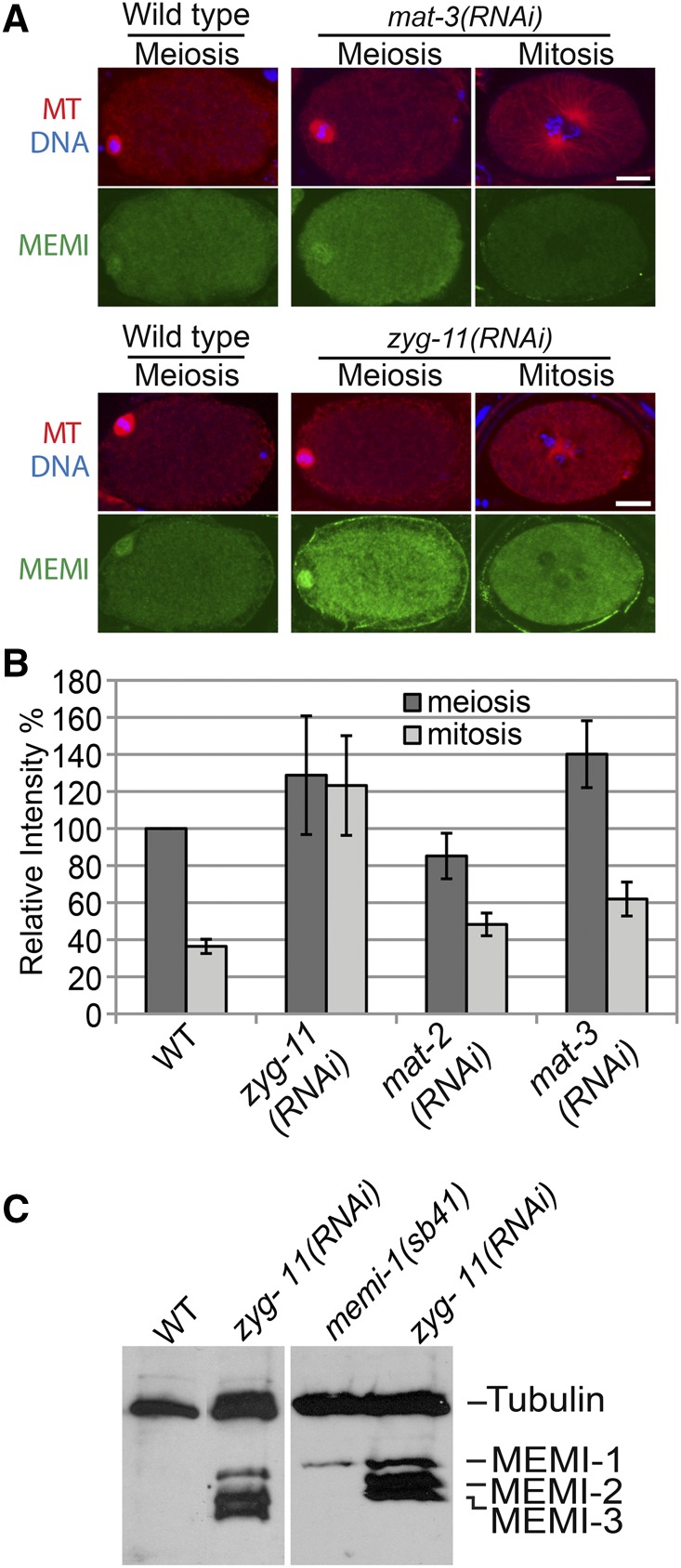
Since MEMI protein levels are decreased after meiosis, their degradation might be dependent on ubiquitin-mediated proteolysis. In order to test this, we used RNAi by feeding to inactivate the APC or CUL-2 complexes separately, and we measured MEMI levels by indirect immunofluorescence. mat-2 and mat-3 encode subunits of the APC and both of these genes are required for targeting proteins for degradation during the meiosis I metaphase-to-anaphase transition (Golden et al. 2000; Stein et al. 2007). Although RNAi of these genes caused a severe delay in meiosis I, the embryos eventually progressed. In both mat-2(RNAi) and mat-3(RNAi), the MEMI fluorescence signal was high in early meiotic embryos but reduced in embryos that progressed past meiosis II, indicating that MEMI proteins are eventually degraded in these defective embryos (Figure 7, A and B).
Figure 7.
CUL-2 E3 ligase substrate-specific adaptor ZYG-11 is required for degradation of MEMI. (A) Immunofluorescence staining of wild-type, mat-3(RNAi), and zyg-11(RNAi) embryos with anti-MEMI. Decreased cytoplasmic signal of MEMI in mitosis was observed in both wild-type and mat-3(RNAi) embryos, but levels remained high in mitotic zyg-11(RNAi) embryos. Tubulin (red), DNA (blue), and MEMI-1 (green). Bar, 10 µm. (B) Quantification of cytoplasmic levels of MEMI-1 in wild type, zyg-11(RNAi), mat-2(RNAi), and mat-3(RNAi) indicated that ZYG-11 was required for reduction of MEMI-1 levels. Values were based on meiotic embryos (WT, n = 3; zyg-11(RNAi), n = 26; mat-2(RNAi), n = 14; and mat-3(RNAi), n = 14) and mitotic embryos (WT, n = 5; zyg-11(RNAi), n = 12; mat-2(RNAi), n = 9; and mat-3(RNAi), n = 12). Bars show SD. (C) Western blot of WT and zyg-11(RNAi) mitotic embryo-prep lysates, probed with anti-MEMI and anti-tubulin. zyg-11(RNAi) causes MEMI-1, MEMI-2, and MEMI-3 proteins to persist in mitotic embryos. memi-1(sb41) mutants retain MEMI-1 protein in mitotic embryos, despite complete degradation of MEMI-2 and MEMI-3.
We next tested zyg-11(RNAi) embryos in order to determine if MEMI protein levels are altered by the loss of this substrate-specific adaptor of the CUL-2 E3 ligase complex (Liu et al. 2004; Sonneville and Gonczy 2004; Vasudevan et al. 2007). Using immunostaining, zyg-11(RNAi) revealed a strong signal for MEMI proteins even in embryos that exhibited mitotic characteristics (e.g., pronuclei), indicating a failure to degrade the MEMI proteins (Figure 7, A and B). Furthermore, Western blot analysis showed that zyg-11(RNAi) mitotic embryos contained all three MEMI proteins (Figure 7C). Thus, the CUL-2 E3 ligase complex is required for the degradation of MEMI proteins during the meiosis-to-mitosis transition. Western blots of memi-1(sb41) mitotic embryo lysates revealed that this hyper-morphic mutation resulted in the persistence of MEMI-1 protein in mitosis, without interfering with MEMI-2 or MEMI-3 degradation (Figure 7C).
An RNAi screen for suppressors of memi-1(sb41) identifies the sperm PP1 phosphatase GSP-3/4
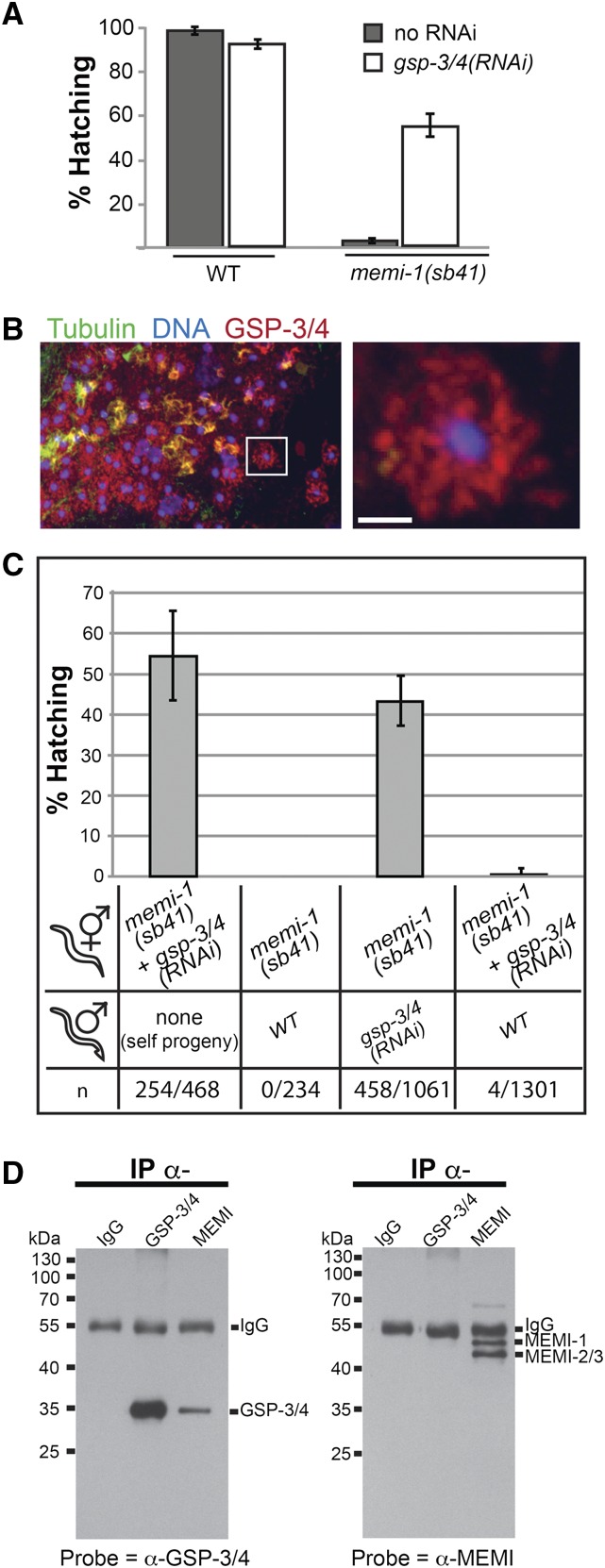
The skipped-meiosis II phenotype of memi-1/2/3(RNAi) is similar to what has been observed in fertilization-defective mutants, whereby oocytes are activated by MSP but are not fertilized (McNally and McNally 2005; File S8). Therefore, MEMI might trigger meiosis II in response to sperm entry, via an unidentified sperm component. In order to identify the putative sperm factor, we exploited the memi-1(sb41) mutation to identify genetic interactors. Given that memi-1(RNAi) rescued memi-1(sb41) lethality, loss of any other gene required for memi activity could potentially suppress the memi-1(sb41) mutation. Using a dsRNA-feeding approach (Kamath et al. 2003), we conducted a genome-wide RNAi suppressor screen of memi-1(sb41) worms. From ∼16,000 genes tested, two suppressors were identified, gsp-3 and gsp-4 (Figure 8A). Both genes encode a sperm-specific PP1 phosphatase related to GLC seven protein phosphatases (Wu et al. 2012). These two genes are highly similar (98% identity) and dsRNA directed against either gene is expected to target both. GSP-3/4 is required for spermatid activation, possibly by altering the polymerization dynamics of cytoskeletal MSP within the sperm cytoplasm (Wu et al. 2012). We raised antibodies to a C-terminal region of GSP-3/4 and confirmed the previously reported localization pattern in the sperm (Figure 8B and Figure S5).
Figure 8.
gsp-3/4 and memi exhibit genetic and physical interactions. (A) memi-1(sb41) maternal-effect lethality was suppressed when worms were treated with gsp-3/4(RNAi) (feeding method). Under the same conditions, wild-type worms exhibited a slight decrease in embryo survival. Bars are SEM. (B) Immunofluorescence of a section of male gonad, stained with DAPI to visualize DNA (blue), anti-tubulin to visualize microtubules (green), and anti-GSP-3/4 (red). The boxed region is enlarged (right) to show a single spermatid that exhibits a fibrous staining pattern for GSP-3/4. Bar, 2 µm. (C) gsp-3/4(RNAi) male-mating can suppress memi-1(sb41) hermaphrodites. memi-1(sb41) hermaphrodites [untreated or treated with gsp-3/4(RNAi)], were mated to WT or gsp-3/4(RNAi) males and the hatching rates were compared to unmated controls. Outcross (male) sperm is preferred to hermaphrodite sperm, hence, the RNAi treatment of the males determines the suppression outcome. Number of progeny scored (n) is shown. (D) Anti-MEMI antibodies co-immunoprecipitate GSP-3/4 proteins. Western blots of proteins immunoprecipitated by incubation with rabbit IgG, anti-GSP-3/4, or anti-MEMI antibodies. Blot on left was probed with anti-GSP; blot on right was probed with anti-MEMI.
Because the RNAi screen was performed on memi-1(sb41) hermaphrodites, it was formally possible that, despite the appearance of GSP-3/4 in the sperm, reduction of some maternally expressed fraction of GSP-3/4 in the oocyte was responsible for the suppression. To test this, we fed gsp-3/4(RNAi) bacteria to wild-type males, and then mated them to memi-1(sb41) hermaphrodites that were never exposed to gsp-34(RNAi) bacteria. Mated hermaphrodites store a mixture of outcross and self-cross sperm in their spermatheca, but the outcross sperm are preferentially used (Ward and Carrel 1979; L’Hernault 2006). After the mating, we observed suppression of the memi-1(sb41) hermaphrodites (13/17), suggesting that outcross sperm were responsible for the suppression (Figure 8C). In a similar experiment, we raised males to adulthood on normal bacteria (i.e., not exposed to gsp-3/4 dsRNA) and mated them to memi-1(sb41) hermaphrodites that were cultured on gsp-3/4(RNAi) bacteria. In this case, we observed two classes of results. In one class, suppression was observed but no male progeny were observed, suggesting that mating did not occur (2/17). In the other class, we observed no surviving progeny, despite the hermaphrodites feeding on gsp-3/4(RNAi) bacteria (15/17). We interpret this latter class as hermaphrodites that successfully mated, but that the outcross male sperm could not suppress the memi-1(sb41) phenotype. Because the spermatheca of mated hermaphrodites contains a mixture of male and self sperm, this result suggested that the suppression of memi-1(sb41) required fertilization by a gsp-34(RNAi) sperm, rather than contact with a diffusible factor released by gsp-3/4(RNAi) sperm.
The identification of GSP-3/4 in our screen suggested that it could be an activator of memi-1(sb41), but other explanations are possible. For example, reduction of GSP-3/4 activity could alleviate some poisonous effect of the sb41 mutation, without being involved in normal MEMI functions during the meiosis I-to-meiosis II transition. In order to test for a possible molecular relationship between GSP-3/4 and MEMI, we asked whether the wild-type MEMI proteins can physically associate with GSP-3/4. We found that α-MEMI antibodies were able to co-immunoprecipitate GSP-3/4 (Figure 8D); however, we did not observe immunoprecipitation of MEMIs with α-GSP-3/4 antibodies. Despite the lack of reciprocity, these results nonetheless indicated that MEMI and GSP-3/4 proteins can physically associate, which lends support for a model whereby PP1 phosphatase interacts with MEMI within the fertilized embryo. Because reduction of gsp-3/4 activity suppresses the overactive memi-1(sb41) mutation, we favor an idea whereby GSP-3/4 acts in the same pathway as MEMI. This could involve GSP-3/4 promoting or sustaining MEMI activity, or MEMI might modulate the function of GSP-3/4 after it enters the fertilized egg.
Discussion
Animals use various strategies to coordinate maturation of the oocyte with fertilization. In many cases, the final stages of female meiosis are controlled by external stimuli. Examples of such stimuli include environmental determinants, sex hormones, or sperm entry. C. elegans likely have two sperm-derived signals, a diffusible signal (MSP) that triggers the oocyte to exit MI prophase (McCarter et al. 1999; Miller et al. 2001), and a second hypothetical signal that is required for the meiotic MI-to-MII transition. Evidence for this latter signal comes from experiments involving fertilization-defective sperm (Ward and Carrel 1979; McNally and McNally 2005). These mutant sperm still produce MSP (Miller et al. 2001); however, because fer-1 sperm are unable to fertilize the oocyte, meiosis I is aborted in anaphase I, the cell fails to extrude the first polar body, and it progresses to mitosis instead of meiosis II. Although the fer-1 “embryos” lack all sperm-derived structures, the cell-cycle phenotypes are strikingly similar to what we observed with memi-1/2/3(RNAi), including an aborted meiosis at the end of anaphase I, and a skipped meiosis II. Cytokinesis defects observed in fer-1 mutants and memi-1/2/3(RNAi) could be due, at least in part, to egg shell defects and osmotic sensitivity, which has been reported for sep-1(RNAi) (Siomos et al. 2001).
Loss of memi results in a skipped meiosis II and egg shell permeability defects
The skipped meiosis II phenotype in C. elegans has previously only been attributed to fertilization-defective mutants like fer-1, and as a rare phenotype of weak, loss-of-function mutations in mat-1, which encodes the CDC-27 subunit of the APC E3 ligase complex (Shakes et al. 2003). Because the APC is also required for sperm function (Sadler and Shakes 2000), its involvement in female meiosis II could be related to an uncharacterized sperm-specific role in the fertilized embryo. Another possibility is that MEMI requires the APC for its function, and the skipped meiosis II phenotype in mat-1 mutants could be due to reduced MEMI activity. If so, we would expect that mat-1 loss-of-function should suppress memi-1(sb41). However, we did not recover any APC subunit genes in our RNAi suppressor screen, and subsequent tests with mat-1(RNAi) did not reveal any suppression of memi-1(sb41) (E. Sykes, unpublished results). In addition, such a role for the APC would likely not involve MEMI protein stability, since we did not observe a reduction in early meiotic MEMI levels after disruption of APC function. Thus, it is unclear if the reported skipped meiosis II phenotype in mat-1(lf) embryos is related to a loss of memi activity.
Although MEMI is required to initiate female meiosis II, its timely removal prior to mitosis is also critical. In C. elegans, the MII-to-mitosis transition requires the E3 CUL-2 and the substrate-recognition subunit ZYG-11 (Liu et al. 2004; Sonneville and Gonczy 2004; Vasudevan et al. 2007). memi-1(sb41) phenotypes resemble zyg-11(RNAi) (Liu et al. 2004; Sonneville and Gonczy 2004), for example, they both result in cell-cycle delays during meiosis II, and polarity defects. ZYG-11/CUL-2 are required for cyclin B degradation (Liu et al. 2004; Sonneville and Gonczy 2004) and MEMI degradation at the end of meiosis II. However, because cyclin B degradation was not obviously affected by memi-1(sb41), this suggests that MEMI persistence alone is responsible for some of the phenotypes observed in zyg-11(RNAi) embryos. The MEMI proteins could be substrates of ubiquitin-mediated proteolysis; however, further work is required to ascertain the molecular relationship between these two pathways.
Assembly of the trilaminar (vitelline, chitin, proteoglycan) egg shell is followed by the synthesis of a permeability barrier during anaphase II (Olson et al. 2012). Previous work determined that the middle chitin layer provides mechanical strength and is responsible for the block to polyspermy (Johnston et al. 2010). We did not observe polyspermy in the memi loss-of-function embryos, suggesting that the early stages of eggshell assembly occurred. However, the observation that memi-triple mutants were permeable to the DNA stain DAPI (Figure 4) is consistent with the idea that passage through anaphase II is required for production of the permeability barrier (Olson et al. 2012); memi-1/2/3(RNAi) embryos skip meiosis II, and would not be expected to complete this step. Interestingly, memi-1(sb41) embryos were not permeable to DAPI, suggesting that, despite the defective meiosis-to-mitosis transition in these embryos, a functional eggshell was synthesized. Previous work showed that CUL-2 activity was required for timely synthesis of the permeability barrier during anaphase of meiosis II (Olson et al. 2012). Therefore, the persistence of MEMI-1 alone did not seem to interfere with this process, despite other phenotypic similarities to zyg-11/cul-2. It is likely that a failure to degrade MEMI-1 interferes with a subset of processes that are regulated by CUL-2/ZYG-11. Examination of the eggshells at high resolution in memi mutants could help determine the precise nature of any structural defects.
memi-1(sb41) interferes with the meiosis-to-mitosis transition
Chromosomal duplication analysis, RNAi knock-down, and Western blotting revealed that memi-1(sb41) is likely hyper-morphic, resulting in persistent MEMI-1 activity that interferes with meiosis II exit. The memi-1(sb41) P74S alteration is within a predicted proline-directed kinase phosphorylation site. Further work is needed to confirm a role for phosphorylation in regulating MEMI activity; however, we speculate that phosphorylation could trigger the proteolytic demise of MEMIs at the end of meiosis II, with sb41 interfering with the timely removal of MEMI-1.
memi-1(sb41) interferes with the completion of meiosis II but it does not prevent the initiation of mitotic events; the mutation gives rise to embryos with features of both meiosis II and mitosis. Thus, the unusual memi-1(sb41) phenotype revealed that the presence of the meiotic spindle alone is not sufficient to block centrosome maturation. The centrosomes in late-stage meiosis II memi-1(sb41) embryos initiated microtubule nucleation; however, the centrosome fragmentation we observed suggested that the microtubule-centrosome attachments and/or the internal structure of the centrosomes were abnormally weak during this defective transition to mitosis.
Reduction of sperm PP1 phosphatase activity rescues memi-1(sb41)
The genome-wide RNAi screen for suppressors of memi-1(sb41) revealed two genes that encode PP1 phosphatase catalytic subunits, termed GSP-3 and GSP-4 (GLC seven-like phosphatase). The yeast PP1 phosphatase GLC7 is involved in glycogen metabolism, meiosis, sporulation, and mitosis (Feng et al. 1991; Peggie et al. 2002; Tan et al. 2003; Bharucha et al. 2008). GSP-3/4 are required for multiple C. elegans sperm functions, including the development and motility of sperm, as well as chromosome segregation in sperm meiosis (Chu et al. 2006; Wu et al. 2012). PP1 phosphatases are also necessary for sperm development and fertility in mice (Varmuza et al. 1999; Oppedisano et al. 2002; Chakrabarti et al. 2007). Interestingly, GSP-3/4 co-localizes with MSP (major sperm protein) (Wu et al. 2012), the cytoskeletal protein required for both the ameboid crawling of C. elegans sperm as well as the diffusible signal for oocyte maturation (Burke and Ward 1983; Sepsenwol et al. 1989; Miller et al. 2001; Kosinski et al. 2005).
Although GSP-3/4 has an established role in spermatid activation, our results suggest a new postfertilization function for this conserved PP1 phosphatase that is also sperm-specific. In order for the suppression of memi-1(sb41) to occur, GSP-3/4 activity must be reduced within the sperm. As previously reported, strong loss-of-function phenotypes for gsp-3/4 include sterility (Wu et al. 2012), which we also observed when we maintained worms on gsp-3/4(RNAi) feeding plates for two generations (J. Tegha-Dunghu, unpublished results). However, under the conditions that suppress memi-1(sb41) (i.e., a single generation, fed from the L2-L3 larval stage) we observed only a slight decrease in embryo viability (91% hatching, Figure 8A). Furthermore, we did not observe suppression when memi-1(sb41); gsp-3/4(RNAi) hermaphrodites were mated to untreated wild-type males. This suggested that sperm entry (and not a secreted sperm factor) was required for the suppression.
Another possibility is that the suppression of memi-1(sb41) by gsp-3/4(RNAi) was due to off-target effects of two other GLC7-like PP1 phosphatases with similarity to GSP-3/4, called GSP-1 and GSP-2. These PP1 enzymes have been shown to oppose Aurora kinase functions during female meiosis (Kaitna et al. 2002; Rogers et al. 2002; de Carvalho et al. 2008). However, we did not observe suppression of memi-1(sb41) with either gsp-1(RNAi) or gsp-2(RNAi) (E. Sykes, unpublished data), indicating that the genetic interaction was specific for gsp-3/4.
A model for a sperm-derived signal that initiates meiosis II
The nature of the sperm-derived signal that specifies postfertilization events in the one-cell zygote is still unclear, but our work presents strong evidence that PP1 phosphatase, in combination with MEMI, represents part of the sperm-to-oocyte signal for female meiosis II (Figure 9). MSP triggers oocyte maturation (McCarter et al. 1999; Miller et al. 2001), and, in our model, MEMI initiates meiosis II after sperm entry. This interpretation predicts that gsp-3/4 loss-of-function should also result in a skipped meiosis II phenotype, but this has not been reported. This could be because GSP-3/4 has an essential function in spermatid activation; thus, it might not be possible to identify a postfertilization role for GSP-3/4 without using a sensitive assay such as the suppression of memi-1(sb41). We contend that our approach of using the sensitized memi-1(sb41) background has revealed a postfertilization role for this PP1 enzyme, but the molecular relationship could be either direct or indirect. The Co-IP results suggest that PP1 could directly affect MEMI-dependent processes in the fertilized embryo, but it is also possible that PP1 acts on an unidentified substrate within the sperm, which, in turn, directly participates in MEMI functions after fertilization. Furthermore, it is unclear whether GSP-3/4 regulates MEMI or vice versa. The physical interaction between GSP-3/4 and MEMI raises the possibility that MEMIs might regulate the GSP-3/4 phosphatases. If MEMIs are regulatory subunits, they would likely activate GSP-3/4 in the fertilized egg, because the genetics suppression data suggest that they act in the same direction. However, expected docking motifs, such as the “RVxF” and “SILK” motifs present in many PP1 regulatory subunits (Hendrickx et al. 2009), are not apparent in MEMI-1 or its paralogs. Future work will focus on elucidating the molecular function of this sperm-delivered PP1 phosphatase and the MEMI proteins.
Figure 9.
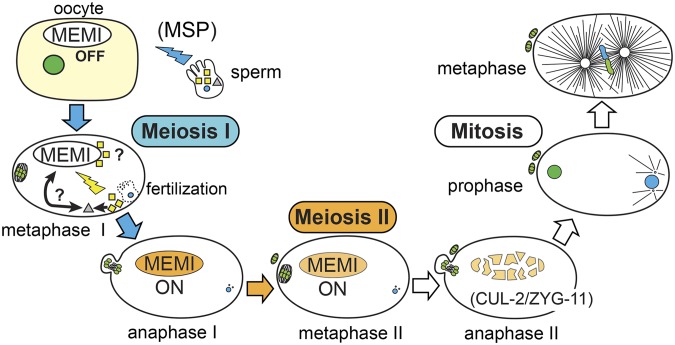
A model for meiosis II activation in the C. elegans embryo. A model for the fertilization-dependent signal that specifies meiosis II is shown. Sperm provide a signal involving secreted MSP protein (blue thunderbolt), which, via interactions with gonadal sheath cells and the oocyte, triggers oocyte maturation (nuclear envelope breakdown, cytoskeletal rearrangement, assembly of MI spindle). This cell will progress to anaphase I (blue arrows), but if sperm do not enter the oocyte, the cell will abort MI and skip MII. A second signal from the sperm (yellow thunderbolt) is delivered upon fertilization. MEMI is likely a maternal component of this signal, in part because loss of MEMI also results in a skipped MII phenotype. At least one additional component of the signal is the sperm-specific PP1 phosphatase, GSP-3/4 (yellow squares), which we identified as a suppressor of hyper-morphic memi-1(sb41). The positive influence on the MEMI pathway could occur either directly via GSP-3/4 interaction, or indirectly, via another unidentified sperm factor (gray triangle). Evidence from mating experiments supports a mechanism that requires GSP-3/4 (or intermediate factor) to enter the oocyte. MEMI is required for meiosis II (orange arrow), but must be inactivated prior to mitosis (white arrows). CUL-2/ZYG-11-dependent ubiquitin-mediated degradation inactivates MEMI before mitosis. The memi-1(sb41) mutation specifically interferes with MEMI-1 protein degradation, resulting in defects during the transition to mitosis.
Acknowledgments
We thank Kelly Adames for technical support and the Srayko laboratory for helpful discussions, and D. Moerman for help with genome sequence analysis. We thank the Caenorhabditis Genetics Center [funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440)] and the National Bioresource Project (Tokyo, Japan) for providing strains. This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (341474). M.S. was supported by a scholar award from the now defunct Alberta Heritage Foundation for Medical Research.
Footnotes
Communicating editor: D. I. Greenstein
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192997/-/DC1.
Literature Cited
- Albertson D. G., 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101: 61–72. [DOI] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. [DOI] [PubMed] [Google Scholar]
- Argon Y., Ward S., 1980. Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics 96: 413–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek J. N., Richie C. T., Squirrell J. M., Campbell J. M., Eliceiri K. W., et al. , 2007. Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase. Development 134: 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenati G., Penkov S., Muller-Reichert T., Entchev E. V., Kurzchalia T. V., 2009. Two cytochrome P450s in Caenorhabditis elegans are essential for the organization of eggshell, correct execution of meiosis and the polarization of embryo. Mech. Dev. 126: 382–393. [DOI] [PubMed] [Google Scholar]
- Bharucha J. P., Larson J. R., Konopka J. B., Tatchell K., 2008. Saccharomyces cerevisiae Afr1 protein is a protein phosphatase 1/Glc7-targeting subunit that regulates the septin cytoskeleton during mating. Eukaryot. Cell 7: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino D., Mogilner A., Roberts T., Stewart M., Oster G., 2002. How nematode sperm crawl. J. Cell Sci. 115: 367–384. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Strome S., 1996. A sperm-supplied factor required for embryogenesis in C. elegans. Development 122: 391–404. [DOI] [PubMed] [Google Scholar]
- Burke D. J., Ward S., 1983. Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. J. Mol. Biol. 171: 1–29. [DOI] [PubMed] [Google Scholar]
- Carvalho A., Olson S. K., Gutierrez E., Zhang K., Noble L. B., et al. , 2011. Acute drug treatment in the early C. elegans embryo. PLoS One 6: e24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Cheng L., Puri P., Soler D., Vijayaraghavan S., 2007. Protein phosphatase PP1 gamma 2 in sperm morphogenesis and epididymal initiation of sperm motility. Asian J. Androl. 9: 445–452. [DOI] [PubMed] [Google Scholar]
- Cheng H., Govindan J. A., Greenstein D., 2008. Regulated trafficking of the MSP/Eph receptor during oocyte meiotic maturation in C. elegans. Curr. Biol. 18: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. S., Liu H., Nix P., Wu T. F., Ralston E. J., et al. , 2006. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan C., Subramanian R., Miller M. A., 2005. Eph and NMDA receptors control Ca2+/calmodulin-dependent protein kinase II activation during C. elegans oocyte meiotic maturation. Development 132: 5225–5237. [DOI] [PubMed] [Google Scholar]
- Costache V., McDougall A., Dumollard R., 2014. Cell cycle arrest and activation of development in marine invertebrate deuterostomes. Biochem. Biophys. Res. Commun. 450: 1175–1181. [DOI] [PubMed] [Google Scholar]
- de Carvalho C. E., Zaaijer S., Smolikov S., Gu Y., Schumacher J. M., et al. , 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22: 2869–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C., Reese K. J., Seydoux G., 2003. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., Desai A., 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., et al. , 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1: 486–492. [DOI] [PubMed] [Google Scholar]
- Feng Z. H., Wilson S. E., Peng Z. Y., Schlender K. K., Reimann E. M., et al. , 1991. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J. Biol. Chem. 266: 23796–23801. [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Furuta T., Tuck S., Kirchner J., Koch B., Auty R., et al. , 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A., 2000. Cytoplasmic flow and the establishment of polarity in C. elegans 1-cell embryos. Curr. Opin. Genet. Dev. 10: 414–420. [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J. A., Cheng H., Harris J. E., Greenstein D., 2006. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr. Biol. 16: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Govindan J. A., Nadarajan S., Kim S., Starich T. A., Greenstein D., 2009. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development 136: 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnowski E. M., Srayko M., 2011. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J. Cell Biol. 194: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., Kirkham M., Hyman A. A., Oegema K., 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155: 1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. E., Govindan J. A., Yamamoto I., Schwartz J., Kaverina I., et al. , 2006. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev. Biol. 299: 105–121. [DOI] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., et al. , 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16: 365–371. [DOI] [PubMed] [Google Scholar]
- Hill D. P., Shakes D. C., Ward S., Strome S., 1989. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Dev. Biol. 136: 154–166. [DOI] [PubMed] [Google Scholar]
- Holland L. Z., Onai T., 2012. Early development of cephalochordates (amphioxus). Wiley Interdiscip. Rev. Dev. Biol. 1: 167–183. [DOI] [PubMed] [Google Scholar]
- Johnston W. L., Krizus A., Dennis J. W., 2010. Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr. Biol. 20: 1932–1937. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., Glotzer M., 2002. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12: 798–812. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., 2004. More than G1 or G2 arrest: useful starfish oocyte system for investigating skillful MAP kinase. Biol. Cell 96: 241–244. [DOI] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I., Greenstein D., 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132: 3357–3369. [DOI] [PubMed] [Google Scholar]
- L’Hernault, S. W., 2005 Spermatogenesis (February 20, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.85.1, http://www.wormbook.org
- Liu J., Vasudevan S., Kipreos E. T., 2004. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development 131: 3513–3525. [DOI] [PubMed] [Google Scholar]
- Marcello M. R., Singaravelu G., Singson A., 2013. Fertilization. Adv. Exp. Med. Biol. 757: 321–350. [DOI] [PubMed] [Google Scholar]
- Maruyama R., Velarde N. V., Klancer R., Gordon S., Kadandale P., et al. , 2007. EGG-3 regulates cell-surface and cortex rearrangements during egg activation in Caenorhabditis elegans. Curr. Biol. 17: 1555–1560. [DOI] [PubMed] [Google Scholar]
- Masui Y., Clarke H. J., 1979. Oocyte maturation. Int. Rev. Cytol. 57: 185–282. [DOI] [PubMed] [Google Scholar]
- Masui Y., Markert C. L., 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 177: 129–145. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McDougall A., Chenevert J., Dumollard R., 2012. Cell-cycle control in oocytes and during early embryonic cleavage cycles in ascidians. Int. Rev. Cell Mol. Biol. 297: 235–264. [DOI] [PubMed] [Google Scholar]
- McNally K. L., McNally F. J., 2005. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev. Biol. 282: 218–230. [DOI] [PubMed] [Google Scholar]
- McNally K. L., Fabritius A. S., Ellefson M. L., Flynn J. R., Milan J. A., et al. , 2012. Kinesin-1 prevents capture of the oocyte meiotic spindle by the sperm aster. Dev. Cell 22: 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., von Tobel L., Strnad P., Knott G., Leonhardt H., et al. , 2012. Analysis of centriole elimination during C. elegans oogenesis. Development 139: 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Ruest P. J., Kosinski M., Hanks S. K., Greenstein D., 2003. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 17: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitenko N. L., Eisner J. R., Swiston J. R., Mains P. E., 1997. A limited number of Caenorhabditis elegans genes are readily mutable to dominant, temperature-sensitive maternal-effect embryonic lethality. Genetics 147: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Zheng Y., Alberts B. M., Oegema K., 1998. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T., Greenan G., O’Toole E., Srayko M., 2010. The elegans of spindle assembly. Cell. Mol. Life Sci. 67: 2195–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat C. C., Torre-Santiago K. M., Tran M. V., Powers J. A., Wignall S. M., 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife 4: e06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S. K., Greenan G., Desai A., Muller-Reichert T., Oegema K., 2012. Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J. Cell Biol. 198: 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppedisano L., Haines G., Hrabchak C., Fimia G., Elliott R., et al. , 2002. The rate of aneuploidy is altered in spermatids from infertile mice. Hum. Reprod. 17: 710–717. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peggie M. W., MacKelvie S. H., Bloecher A., Knatko E. V., Tatchell K., et al. , 2002. Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J. Cell Sci. 115: 195–206. [DOI] [PubMed] [Google Scholar]
- Rappleye C. A., Paredez A. R., Smith C. W., McDonald K. L., Aroian R. V., 1999. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 13: 2838–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C. A., Tagawa A., Le Bot N., Ahringer J., Aroian R. V., 2003. Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev. Biol. 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S., Lin R., 2015. The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113: 1–42. [DOI] [PubMed] [Google Scholar]
- Rogers E., Bishop J. D., Waddle J. A., Schumacher J. M., Lin R., 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler P. L., Shakes D. C., 2000. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development 127: 355–366. [DOI] [PubMed] [Google Scholar]
- Samuel A. D., Murthy V. N., Hengartner M. O., 2001. Calcium dynamics during fertilization in C. elegans. BMC Dev. Biol. 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J. M., Ashcroft N., Donovan P. J., Golden A., 1998. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development 125: 4391–4402. [DOI] [PubMed] [Google Scholar]
- Sepsenwol S., Ris H., Roberts T. M., 1989. A unique cytoskeleton associated with crawling in the amoeboid sperm of the nematode, Ascaris suum. J. Cell Biol. 108: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Ling L., van Zuylen V., Meyer B. J., 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23: 1763–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Sadler P. L., Schumacher J. M., Abdolrasulnia M., Golden A., 2003. Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130: 1605–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomos M. F., Badrinath A., Pasierbek P., Livingstone D., White J., et al. , 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11: 1825–1835. [DOI] [PubMed] [Google Scholar]
- Sonneville R., Gonczy P., 2004. Zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131: 3527–3543. [DOI] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., Hyman A. A., 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell 9: 223–236. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198: 1127–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K. K., Davis E. S., Hays T., Golden A., 2007. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics 175: 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa A., Rappleye C. A., Aroian R. V., 2001. Pod-2, along with pod-1, defines a new class of genes required for polarity in the early Caenorhabditis elegans embryo. Dev. Biol. 233: 412–424. [DOI] [PubMed] [Google Scholar]
- Tan Y. S., Morcos P. A., Cannon J. F., 2003. Pho85 phosphorylates the Glc7 protein phosphatase regulator Glc8 in vivo. J. Biol. Chem. 278: 147–153. [DOI] [PubMed] [Google Scholar]
- Varmuza S., Jurisicova A., Okano K., Hudson J., Boekelheide K., et al. , 1999. Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cgamma gene. Dev. Biol. 205: 98–110. [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Starostina N. G., Kipreos E. T., 2007. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina J. R., Orr-Weaver T. L., 2011. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 3: a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Carrel J. S., 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73: 304–321. [DOI] [PubMed] [Google Scholar]
- Wharton D., 1980. Nematode egg-shells. Parasitology 81: 447–463. [DOI] [PubMed] [Google Scholar]
- Whitten S. J., Miller M. A., 2007. The role of gap junctions in Caenorhabditis elegans oocyte maturation and fertilization. Dev. Biol. 301: 432–446. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Go A. C., Samson M., Cintra T., Mirsoian S., et al. , 2012. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics 190: 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. Y., McNally K., McNally F. J., 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans. Dev. Biol. 260: 245–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.