Abstract
Human population genomic studies have repeatedly observed a decrease in heterozygosity and an increase in linkage disequilibrium with geographic distance from Africa. While multiple demographic models can generate these patterns, many studies invoke the serial founder effect model, in which populations expand from a single origin and each new population’s founders represent a subset of genetic variation in the previous population. The model assumes no admixture with archaic hominins, however, recent studies have identified loci in Homo sapiens bearing signatures of archaic introgression. These results appear to contradict the validity of analyses invoking the serial founder effect model, but we show these two perspectives are compatible. We also propose using the serial founder effect model as a null model for determining the signature of archaic admixture in modern human genomes at different geographic and genomic scales.
Introduction
Research in human evolution relies on multiple fields — such as archaeology, genetics, and linguistics — to give a history of Homo sapiens during the last hundred thousand years. In the last decade, investigations of worldwide human genomic variation based on multiple genetic marker types have observed three robust trends in summary statistics as a function of increasing geographic distance from Africa (Figure 1; see also DeGiorgio et al. [1]): a decrease in heterozygosity, an increase in linkage disequilibrium (LD), and an increase in the frequency of derived alleles [2–8]. Analyses of genomic datasets from human pathogens and parasites have also observed a reduction in heterozygosity as a function of distance from Africa [9–11]. Further, studies of anthropometric [12, 13], economic [14], cultural [15], and linguistic data [16] have reported that population-level diversity in various traits follows this same pattern.
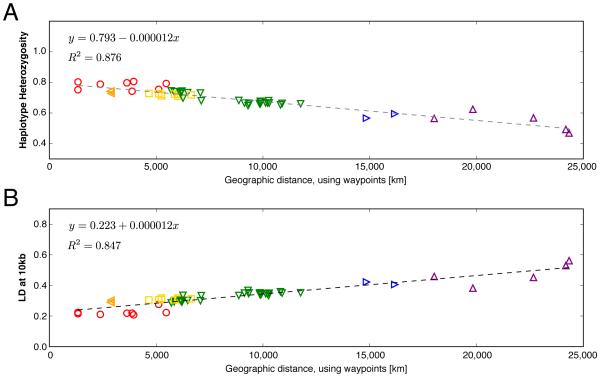
Figure 1. Observed genome-wide patterns of heterozygosity and linkage disequilibrium (LD) among worldwide human populations as functions of geographic distance from Addis Ababa, Africa (9 N, 38 E).
Haplotype heterozygosity (panel A, calculated as in Conrad et al. [95]) and average LD at 10kb (panel B, measured by r2) were calculated across 640,034 SNPs genotyped in the Human Genome Diversity Panel [7]. Note error bars are smaller than symbols. Equations for fitted lines and each linear model’s coefficient of determination are displayed within each panel. Symbols indicate geographic regions: red circles = Africa, orange left-pointing triangles = Middle East, yellow squares = Europe, green down-pointing triangles = Central/South and East Asia, blue right-pointing triangles = Oceania, purple up-pointing triangles = Americas; populations are assigned to regions as in Rosenberg et al. [38].
These studies each interpreted the observed reduction in trait diversity out of Africa as reflecting the population and geographic expansion of modern humans in the last 45k-60k years by invoking a model for the great human expansion known as the “serial founder effect model” [2, 3, 17–20]). In a serial founder effect model, a series of successive bottlenecks from a single origin of expansion produces a stepwise increase in genetic drift — and consequent decrease in genetic diversity — as a function of geographic distance from the origin. Although the ability to identify a unique model for human evolutionary history from genome-wide diversity patterns has generated debate across a range of disciplines (for example, [1, 21–32]), the serial founder effect model has proven to be a useful framework for both understanding the dynamic nonequilibrium history of human populations and identifying origins of large-scale human population expansions [6, 19, 33–35]. However, emerging high-quality archaic genomes and genomic datasets sampling increasing numbers of individuals from diverse human populations have shed new light on the complex demographic events that characterize human evolutionary history. Thus, the time is ripe to examine where the assumptions of the serial founder effect model falter, and what we can learn from genomic regions and populations in which genomic diversity patterns diverge from those predicted by the model.
We focus here on one particular assumption: the serial founder effect model, as invoked by Prugnolle et al. [2] and Ramachandran et al. [3], implicitly assumes that admixture with archaic hominins did not play a substantial role in the evolutionary history of Homo sapiens (see also DeGiorgio et al. [1], Pickrell and Reich [29]). Here, we review observed patterns of human genomic variation from large-scale studies in the last five years, examine this assumption closely in light of these empirical results, and propose a framework for disentangling the signatures of archaic introgression from those of modern human population interactions in worldwide human genomic datasets.
Human genomic variation reveals intercontinental clusters and intracontinental admixture
Analyses of human genomic data from globally distributed populations repeatedly find that multilocus genotypes from human populations produce genetic clusters largely corresponding to major geographic regions [36–41]. Within continents, inferred genetic clusters may identify geographically or culturally isolated populations, distinguish among various subsistence strategies, or reveal signatures of gene flow [8, 42, 43].
The existence of genetic clusters among worldwide human populations has been challenged for multiple reasons [44–46]. One point of contention regarding the “clusteredness” of humans is particularly relevant to this review: Serre and Pääbo [47] questioned whether the identification of clusters from human multilocus genotype data is an artefact due to the sampling design of datasets such as the Human Genome Diversity Panel [4, 5, 37, 48]. In response, Rosenberg et al. [38] showed that inferred clusters arise not from the geographic dispersion of sampled individuals, but rather are generated by small discontinuous jumps in genetic distance for population pairs on opposite sides of geographic barriers (Figure 2 and[44]).
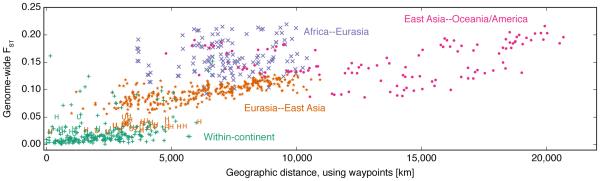
Figure 2. Genetic differentiation among worldwide human populations reflects intercontinental clusters and intracontinental clines.
Pairwise genome-wide FST was calculated across various population pairs in the HGDP. Different colors represent comparisons within and between continental groups defined by major representation in the same cluster in the K = 5 plot of Figure 2 in Rosenberg et al. [38]. Green +’s indicate comparisons within each of continental group (Africa, Eurasia, East Asia, Oceania, and America). “Eurasia” here combines Europe, the Middle East, and Central/South Asia. Orange ∗’s indicate pairs with one population from Eurasia and one from East Asia; purple ×’s indicate pairs with one population from Africa and the other from Eurasia; and pink circles indicate pairs with one population from East Asia and the other from either Oceania or America. Comparisons with the Hazara or Uygur are indicated with an “H” or “U”, respectively; these admixed populations are less differentiated from Eurasian populations than predicted by geographic distance alone. A linear regression of FST onto distance and geological barriers estimates FST = 0.0012+0.0072×D +0.0703×B, where D is great circle distance measured in thousands of km, and B is a boolean variable indicating whether at least one barrier such as oceans or the Himalayas occur between a given population pair [38]. Both regression coefficients are significantly different from 0 (p < 0.001); R2 = 0.773. Regression analyses reflecting the same qualitative pattern were generated by Rosenberg et al. [38] using microsatellite data in the HGDP.
The serial founder effect model assumes no gene flow between neighboring populations, and generates a positive correlation between genetic distance and geographic distance among population pairs [3]. As was observed by Rosenberg et al. [38] using microsatellite data, we find that the effect of a barrier that delineates a continental region — such as a large mountain range or a continental shelf — is to add to pairwise genetic distance between populations beyond the value predicted by geographic distance alone. In the case of HGDP SNP data analyzed in Figure 2, crossing a barrier adds 0.0703 to pairwise FST, the equivalent to traveling approximately 9,764 km within a continental region.
Thus, Figure 2 strongly suggests that qualitative features of the serial founder effect model hold true for human evolutionary history: modern human genetic diversity generally reflects a stepwise accumulation of genetic drift across geographic barriers, and intercontinental migration was not the norm in human evolutionary history. This is borne out by analyses of diverse human populations in conjunction with the HGDP [6–8, 34, 35, 49, 50]. Further, the bottlenecks in the serial founder effect model, which increase drift and short-range linkage disequilibrium [51], is supported by studies that infer parameters for human demography, all of which identify bottlenecks during the peopling of new continental regions [52–55]. Indeed, models that attempt to recreate genomic patterns of LD and heterozygosity without bottlenecks must invoke either very carefully constructed migration rates [29] or an instantaneous divergence model with monotonically decreasing population sizes [1].
In contrast to observed intercontinental clusters, analyses of intracontinental human genomic variation indicate limited population structure within continents (see Novembre et al. [56]), with counterexamples corresponding to unique population histories and waves of migration (for example, in sub-Saharan Africa [8], the Americas [43, 49], and India [57]). However, two populations which notably diverge from the expected increase of genetic distance with geographic distance shown in Figure 2 are the Hazara and Uygur, admixed populations deriving equal ancestry from Eurasian and East Asian genetic clusters inferred by Rosenberg et al. [38]. This deviation reflects the genomic signature of modern human admixture across geographic barriers, one of the situations in which the assumptions of the serial founder effect model are violated. Next we review empirical genomic patterns of archaic admixture with modern humans, which also deviate from patterns predicted by the model.
Genomic signatures of archaic admixture in Homo sapiens
After years of debate regarding the contribution of archaic hominids to the modern human gene pool, studies taking advantage of recent advances in the amplification of DNA from ancient human remains have produced compelling evidence that archaic admixture occurred with ancestral modern humans. Further studies have shown that some of these introgression events were important for adaptation within modern human populations (see Racimo et al. [58] for a review). Although archaic hominin mitochondrial and Y-chromosomal haplotypes are not present among any modern humans [59, 60], several studies of modern human populations outside of Africa infer genome-wide levels of archaic admixture ranging from 1% to 8% originating from more than one admixture pulse [61–65]. When combined with a serial founder effect in simulation, these inferred levels of archaic admixture produce genome-wide patterns in summary statistics like those observed in modern human data and unique patterns of population structure at a small fraction of loci in the human genome. In contrast, archaic admixture of 10% or greater both inflates measures of linkage disequilibrium and leads to a discontinuous relationship between heterozygosity and geographic distance [1].
Three genomic signatures are used to localize regions of archaic introgression in modern human genomes: identifying (i) loci where non-African modern humans carry a derived single-nucleotide variant seen in archaic hominins but not in Africans [61, 66]; (ii) non-African haplotypes with low sequence divergence to an archaic haplotype but high sequence divergence to African haplotypes [67]; and (iii) candidate haplotypes bearing the two previous signatures with lengths consistent with interbreeding between 40-90 kya [68, 69]. Loci meeting these criteria can then be tested for signatures of adaptive or balancing selection; to date, candidate genes for adaptive archaic introgression belong to skin pigmentation, immune, and metabolic pathways [58, 61, 67, 70–77].
While adaptive introgression has been shown for select genomic regions, individual maps of archaic ancestry in modern humans suggest that introgression also was associated with deleterious fitness consequences for modern humans [78]. For example, Sankararaman et al. [75] find that genic regions exhibit reduced Neandertal ancestry, implying that modern human genetic lineages bearing archaic sequences have been removed by purifying selection over time.
Our understanding of the relationship between ancestral modern humans and archaic hominins is limited by the small number of high-quality genomes available from archaic individuals. Although technology for extracting whole genomes from ancient hominin and human remains is improving at a fast pace, there are regions of the world that may never be represented in genomic studies of archaic hominin or ancestral modern human remains, due to unfavorable climates for DNA preservation in fossils. The recent development of methods for constructing individual maps of archaic ancestry from modern human population-genomic data and reference archaic genomes is thus a promising approach to gain insight into hominin evolutionary history, and one with power to exhume population-genomic information from extinct hominins without a large collection of archaic sequences [64, 65, 75, 79].
Leveraging the serial founder effect model in scans for archaic introgression
While the genome-wide patterns in Figure 1 are well-predicted by the serial founder effect model, population genomic data are shaped by a plethora of evolutionary events beyond bottlenecks: including, but not limited to, population expansions, admixture (with archaic hominins and among modern humans alone), slavery, isolation, and adaptation [80]. Each of these events leaves its own genomic footprint, which might be either distributed genome-wide (e.g., an excess of rare variants genome-wide due to post-agricultural population expansions [81]) or more confined to a local genomic region (e.g., support from X-chromosomal variation for male-biased European ancestry in African Americans [42]). Here we suggest an integrative framework for reconciling the coexistence of genomic signatures of archaic admixture with genomic signatures of modern human demographic processes: by using the robust genome-wide patterns present in intercontinental human population genomic datasets and predicted by the serial founder effect model as a null model, we can identify local genomic regions deviating from those patterns in predictable ways that indicate gene flow from archaic hominins (Figure 3).
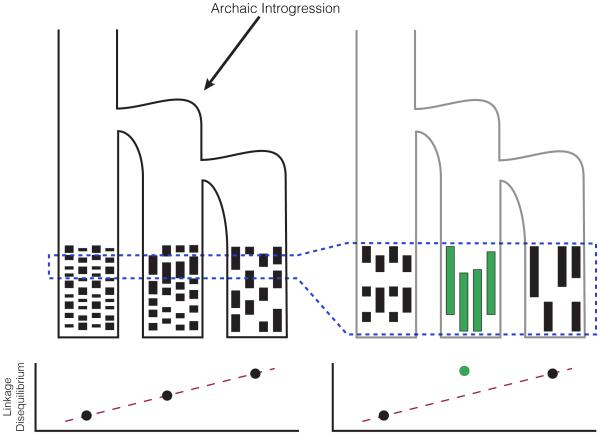
Figure 3. A new framework for identifying genomic targets of archaic introgression, by scanning for a localized inflation of linkage disequilibrium relative to genome-wide levels.
Genome-wide linkage disequilibrium (LD) increases linearly with distance from the source population, matching expectations from the serial founder effect model (bottom left). This pattern is disrupted at specific genomic regions with traces of archaic introgression (area outlined in dashed blue), which inflates LD in specific populations or geographic regions relative to genome-wide expectations (bottom right).
Archaic introgression at a given locus leads to an increase in LD at that locus relative to genome-wide levels of LD predicted by the serial founder effect model (Figures 1B, 4A-B, and [58]). In Figure 4A, the East Asian populations in the HGDP show a clear inflation in LD at the OAS gene cluster involved in immune response to viral infection; Mendez et al. [72] identified ~185 kb introgressed from Neandertals into present-day non-Africans (particularly in Oceanic individuals and Indonesians) in this gene cluster. Arguably the most celebrated example of archaic introgression occurred from the Denisovan lineage into Tibetans at the gene EPAS1, which encodes a transcription factor affecting response to hypoxia at high altitudes (Figure 4B). Analyses of a single genome from the southern Chinese Dai population did not reveal either the Denisovan EPAS1 haplotype or excess allele sharing with the Denisovan genome [63, 74]; Huerta-Sanchez et al. [74] further note that three single-nucleotide variants in EPAS1 are private alleles shared only between Tibetans and the archaic Denisovan genome, and that the putatively beneficial EPAS1 haplotype is not observed in Melanesian modern humans or in the Altai Neandertal from Siberia [82]. However, analysis of population genomic data has found higher Denisovan ancestry in southern Chinese populations – such as the Dai, highlighted in Figure 4B – than in northern Chinese populations [83].
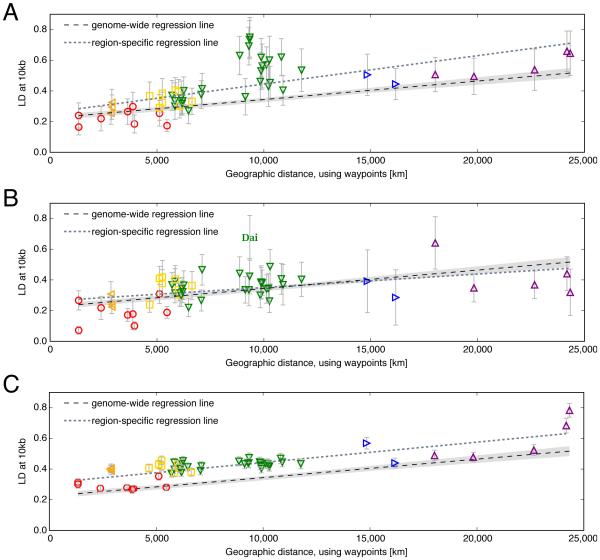
Figure 4. Linkage disequilibrium (LD) in loci undergoing archaic introgression and various forms of selection deviate from the patterns predicted by a serial founder effect model.
LD is calculated among SNP pairs 10kb apart as in [4], and error bars are standard errors of mean LD across all SNP pairs within each analyzed genomic region. Symbols match those in Figure 1; the regression line fitted in Figure 1B is shown as a dashed black line in each panel with 99% confidence bands shaded in gray. A: The OAS gene cluster (chr12:113300000-113600000 (hg19)), shown to have Neanderthal gene-flow in non-Africans [72], shows a deviation from genome-wide patterns, particularly in East Asia (green down-pointing triangles). B: EPAS1 (chr2:46524563-46613836), a gene crucial for high-altitude adaptation in Tibetans [96] and known to bear an introgressed Neanderthal haplotype [74], deviates from genome-wide patterns in the Dai population, a southern Chinese ethnic minority group. There are no Tibetan samples in the HGDP. C: 11Mb region surrounding FOXP2 (chr7:113600000-124700000), previously identified as an introgression desert [65]. This genomic region has elevated LD worldwide relative to genome-wide patterns (grey dashed regression line), and all populations seem to be equally affected, which may be a signature of background selection/conservation.
Other evolutionary forces, notably selection in the modern human lineage, can also lead to a local genomic increase in LD relative to the genome-wide pattern. Strong positive selection, for example, should lead to an increase in LD for the population or populations with the adaptive allele. In Figure 4C, we observe inflation in LD worldwide in the highly conserved gene FOXP2, a transcription factor crucial for language development [84] and part of an 11Mb introgression desert [65]. In fact, Kuhlwilm et al. [85] classify a 150kb segment within FOXP2 as exhibiting signatures of gene flow from modern humans into the Altai Neandertal (see also Coop et al. [86]). Conservation through strong purifying selection is expected to inflate measures of LD [87], but will produce a different genomic signature than introgression: many human populations, including those in Africa, should be affected to a similar degree (Figure 4C), in contrast to the local genomic signatures expected under positive selection confined to a particular geographic region (Figures 4A and B).
In Figure 4, we illustrate the utility of comparing patterns predicted by the serial founder effect model to local genomic patterns at known targets of archaic introgression and conservation by a simple comparison to a linear model generated by genome-wide data. Similar comparison of local genomic patterns to genome-wide patterns are often explored to determine the relative roles of hard adaptive sweeps versus demographic processes in shaping modern human genomic variation [88–90]. As more diverse human populations and archaic samples are sequenced at higher coverage, identifying loci undergoing archaic admixture will be done with greater precision. New methods will undoubtedly be developed that combine multiple statistics and multivariate outlier detection for this purpose (as was done to detect positive and balancing selection by Hunter-Zinck and Clark [91]). We suggest that the framework in Figures 3 and 4 outlines a fruitful avenue for future research, allowing investigators to disentangle the genome-wide footprints of modern human demographic history from evolutionary forces such as archaic admixture, selection, and conservation that act on local genomic targets.
Conclusions and future directions
DNA from human and archaic hominin remains offers unique snapshots into the evolutionary history of our species. However, our ability to integrate this data type into analyses of present-day human population genomic data is hampered by — and may always be hampered by — the limited number of archaic hominin samples with high-quality genomic data. More work needs to be done to evaluate how the systematic dearth of archaic samples in certain parts of the world biases downstream inferences. Additionally, the interaction between genetic drift in modern humans and pulses of archaic admixture has been underexplored (but see Skoglund and Jakobsson [83]). Therefore, approaches to identify the genomic signatures of archaic admixture from present-day human genomes, such as those developed by Vernot and Akey [64], Vernot et al. [65], Sankararaman et al. [75, 79], will be crucial to inferring hominin evolutionary history. Here we propose one approach for identifying targets of archaic introgression in modern human genomes that leverages the robust genome-wide patterns predicted by the serial founder effect model, and scans for regions that diverge from these genome-wide patterns. Further evaluation and development of our framework – particularly in the form of methods for determining the genome-wide significance of local genomic patterns of population-level heterozygosities – will be necessary before this framework can yield statistically sound inferences; still, we believe that the proposed framework will ultimately be a versatile and fruitful one. For example, future studies could use simulations to learn expected patterns of genome-wide versus local genomic diversity under a range of models that violate the assumptions of the serial founder effect: adaptive selection, super-exponential growth, and various pulses of archaic admixture (as studied by [65, 72, 79, 92], who aim to differentiate between signatures of Neandertal versus Denisovan admixture).
The framework we propose also underscores the fact that the relative role of archaic admixture in shaping present-day human genetic variation is small when compared to the genomic footprint of modern human population dynamics over the last ~75k years. Indeed, DeGiorgio et al. [1] reported that estimates of worldwide levels of archaic admixture (≤ 8%) are not sufficient to qualitatively change the patterns predicted by the serial founder effect model; also, non-trivial levels of Denisovan ancestry (~2-4%) are currently only observed in Oceanic populations [63, 65]. The minimal parameters in the serial founder effect model capture the dominant signal of drift that pervades human population-genomic data, yet oversimplify the complex nature of modern human demographic history. We must account for the strong genomic signatures this history has left on human genomes as we work to identify disease-associated mutations and adaptive mutations [89, 93].
Ultimately, the serial founder effect model has been a useful, parsimonious model for modern human evolutionary history in studies over the last decade. Moving forward, our focus should shift to what we can learn about local genomic regions that differ from the robust genome-wide patterns in summary statistics observed in Figures 1 and 2. As Cavalli-Sforza [94] noted in the preface to Genes, Peoples, and Languages in reference to the interdisciplinary nature of research in human evolution, “Singly, each approach [archaeology, genetics, and linguistics] has many lacunae, but hopefully their synthesis can help to fill the gaps.” Our hope is that integrating serial bottlenecks and pulses of archaic admixture into analyses of human population genomic data will offer new insight those traits that uniquely define humans.
Acknowledgements
We thank Trevor Pemberton, Sriram Sankararaman, and Carina Schlebusch for helpful discussions, Arthur Sugden for help generating Figure 3, and John Novembre for comments on an earlier draft of this manuscript. This work was supported by the Pew Charitable Trusts (SR is a Pew Scholar in the Biomedical Sciences), and US National Institutes of Health (NIH) grant R01GM118652 (to SR) and COBRE award P20GM109035. SR also acknowledges support from NSF CAREER Award DBI-1452622.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].DeGiorgio M, Jakobsson M, Rosenberg NA. Explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc Natl Acad Sci U S A. 2009;106:16057–16062. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Current Biology. 2005;15:R159–160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- [5].Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- [6].Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodríguez-Botigué L, Ramachandran S, Hon L, Brisbin A, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. American Journal of Human Genetics. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, Ranford-Cartwright L, Sawai H, Sakihama N, Ohmae H, et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- [11].Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–348. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hanihara T. Morphological variation of major human populations based on nonmetric dental traits. Am J Phys Anthropol. 2008;136:169–182. doi: 10.1002/ajpa.20792. [DOI] [PubMed] [Google Scholar]
- [14].Ashraf Q, Galor O. The ”Out of Africa” hypothesis, human genetic diversity, and comparative economic developmen. The American Economic Review. 2013;103:1–46. doi: 10.1257/aer.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rogers DS, Feldman MW, Ehrlich PR. Inferring population histories using cultural data. Proceedings of the Royal Society B. 2009;276:3835–3843. doi: 10.1098/rspb.2009.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Atkinson QD. Phonemic diversity supports a serial founder effect model of language expansion from Africa. Science. 2011;332:346–9. doi: 10.1126/science.1199295. [DOI] [PubMed] [Google Scholar]
- [17].Harpending H, Rogers A. Genetic perspectives on human origins and differentiatio. Annual Review of Genomics and Human Genetics. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- [18].Harpending HC, Eller E. Human diversity and its history. In: Kato M, editor. The Biology of Biodiversity. Springer-Verlag; Tokyo: 2000. pp. 301–314. chapter 20. [Google Scholar]
- [19].Deshpande O, Batzoglou S, Feldman MW, Cavalli-Sforza LL. A serial founder effect model for human settlement out of Afric. Proceedings of the Royal Society B. 2009;276:291–300. doi: 10.1098/rspb.2008.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci U S A. 2012;109:17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bowern C. Out of africa? the logic of phoneme inventories and founder effects. Linguistic Typology. 2011;15:207–216. [Google Scholar]
- [22].Dahl Ö . Are small languages more or less complex than big ones? Linguistic Typology. 2011;15:171–175. [Google Scholar]
- [23].Maddieson I, Bhattacharya T, Smith DE, Croft W. Geographical distribution of phonological complexit. Linguistic Typology. 2011;15:267–279. [Google Scholar]
- [24].Cysouw M, Dediu D, Moran S. Comment on “Phonemic diversity supports a serial founder effect model of language expansion from Africa”. Science. 2011;335:657–b. doi: 10.1126/science.1208841. [DOI] [PubMed] [Google Scholar]
- [25].Hunley K, Bowern C, Healy M. Rejection of a serial founder effects model of genetic and linguistic coevolutio. Proceedings of the Royal Society B. 2012;279:2281–2288. doi: 10.1098/rspb.2011.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moran S, McCloy D, Wright R. Revisiting population size vs. phoneme inventory siz. Language. 2012;88:877–893. [Google Scholar]
- [27].Wang C-C, Ding Q-L, Tao H, Li H. Comment on “Phonemic Diversity Supports a Serial Founder Effect Model”. Science. 2012;335:657–c. doi: 10.1126/science.1207846. [DOI] [PubMed] [Google Scholar]
- [28].d’Alpoim Guedes J, Bestor TC, Carrasco D, Flad R, Fosse E, Herzfeld M, Lamberg-Karlovsky CC, Lewis CM, Liebmann M, et al. Meadow R. Is poverty in our genes? a critique of ashraf and galor, “the ‘out of africa’ hypothesis, human genetic diversity, and comparative economic development,” american economic review. Curr Anthropol. 2013;54:71–79. doi: 10.1257/aer.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends in Genetics. 2014;30:377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30•].Creanza N, Ruhlen M, Pemberton TJ, Rosenberg NA, Feldman MW, Ramachandran S. A comparison of worldwide phonemic and genetic variation in human population. Proc Natl Acad Sci U S A. 2015;112:1265–1272. doi: 10.1073/pnas.1424033112. This study provides new conceptual and methodological approaches for studies of genetic and linguistic coevolution, rejects the serial founder effect model for sound units, and clarifies the different population processes that affect genes and languages at the global versus regional geographic levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hunley K. >Reassessment of global gene-language coevolution. Proc Natl Acad Sci U S A. 2015;112:1919–1920. doi: 10.1073/pnas.1425000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32•].Rosenberg NA, Kang JTL. Genetic diversity and societally important disparitie. Genetics. 2015;201:1–12. doi: 10.1534/genetics.115.176750. Through analysis of an expanded genetic dataset, the authors reject the claim made by Ashraf and Galor [14] that genetic diversity has driven comparative economic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement histor. Am J Hum Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, et al. Genetic variation and population structure in Native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- [38].Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design consideration. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- [40].Patterson N, Price AL, Reich D. Population structure and eigenanalysi. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo J-M, Wambebe C, Tishkoff SA, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, Contreras AV, Acuña-Alonzo V, Sandoval K, Eng C, Romero-Hidalgo S, et al. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344:1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Edwards AWF. Human genetic diversity: Lewontin’s fallacy. BioEssays. 2003;25:798–801. doi: 10.1002/bies.10315. [DOI] [PubMed] [Google Scholar]
- [45].Bamshad M, Wooding S, Salisbury BA, Stephens JC. Deconstructing the relationship between genetics and race. Nat Rev Genet. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- [46].Weiss KM, Fullerton SM. Racing around, getting nowher. Evolutionary Anthropology. 2005;14:165–169. [Google Scholar]
- [47].Serre D, Pääbo S. Evidence for gradients of human genetic diversity within and among continent. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cann HM, de Toma C, Cazes L, Legrand M-F, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Chen Z, Chu J, et al. A human genome diversity cell line panel. Scienc. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- [49].Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pemberton TJ, DeGiorgio M, Rosenberg NA. Population structure in a comprehensive genomic data set on human microsatellite variation. G3. 2013;3:891–907. doi: 10.1534/g3.113.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Slatkin M. Linkage disequilibrium–understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variatio. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Amos W, Hoffman JI. Evidence that two main bottleneck events shaped modern human genetic diversity. Proceedings of the Royal Society B. 2010;277:131–137. doi: 10.1098/rspb.2009.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Bustamante CD. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Pääbo S. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60•].Mendez FL, Poznik GD, Castellano S, Bustamante CD. The divergence of Neandertal and modern human Y chromosome. American Journal of Human Genetics. 2016;98:728–734. doi: 10.1016/j.ajhg.2016.02.023. This study analyses a Neandertal Y chromosome from Spain and places the lineage as a distant outgroup to modern human Y chromosomes; the estimated time to the most recent common ancestor between this Neandertal lineage and modern human Y chromosomes is over 580k years ago. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Meyer M, Kircher M, Gansauge M-T, Li H, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, et al. A high coverage genome sequence From an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64••].Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genome. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. The authors apply a two-stage computational approach to modern human genomes to identify population-level DNA sequence information from Neandertals, and infer a per-individual average of approximately 19.6 Mb of Neandertal sequences in over 600 European and East Asian individuals. This study also confirms that Neandertal admixture occurred multiple times with ancestral modern humans outside of Africa (see also [65, 79, 92]) [DOI] [PubMed] [Google Scholar]
- [65•].Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. This study focuses on identifying archaic introgression in Island Melanesian genomes, which bear signatures of archaic admixture with both Neandertals and the Denisovan genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human histor. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Mendez FL, Watkins JC, Hammer MF. A haplotype at STAT2 introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guine. American Journal of Human Genetics. 2012;91:265–274. doi: 10.1016/j.ajhg.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern human. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yotova V, Lefebvre JF, Moreau C, Gbeha E, Hovhannesyan K, Bourgeois S, Bédarida S, Azevedo L, Amorim A, Sarkisian T, et al. An X-linked haplotype of neandertal origin is present among all non-African populations. Mol Biol Evo. 2011;28:1957–1962. doi: 10.1093/molbev/msr024. [DOI] [PubMed] [Google Scholar]
- [71].Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo J-M, Lema G, Fu W, Nyambo TB, Rebbeck TR, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mendez FL, Watkins JC, Hammer MF. Global genetic variation at OAS1 provides evidence of archaic admixture in Melanesian population. Mol Biol Evol. 2012;29:1513–1520. doi: 10.1093/molbev/msr301. [DOI] [PubMed] [Google Scholar]
- [73].Wall JD, Yang MA, Jay F, Kim SK, Durand EY, Stevison LS, Gignoux C, Woerner A, Hammer MF, Slatkin M. Higher levels of Neanderthal ancestry in East Asians than in European. Genetics. 2013;194:199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74••].Huerta-Sanchez E, Jin X, Asan, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. The authors show that the adaptation of Tibetans to the hypoxic environment of the Tibetan plateau can only be explained by the introgression of a Denisovan haplotype around the gene EPAS1. This study suggests archaic admixture may have helped ancestral modern humans adapt to new environments during the modern human expansion out of Africa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75••].Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. This study identifies introgression deserts and illuminates the role purifying selection has played in removing genetic material from Neandertals in modern human genomes; findings suggest that the X chromosome played an important role in reproductive isolation of modern humans and archaic hominins. The authors also report multiple Neandertalderived alleles that increase disease risk in modern humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76•].Williams AL, Jacobs SBR, Moreno-Macías H, Huerta-Chagoya A, Churchhouse C, Márquez-Luna C, García-Ortíz H, José Gómez-Vázquez M, Burtt NP, Aguilar-Salinas CA, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. This study by the SIGMA Type 2 Diabetes Consortium identifies a new risk haplotype for type 2 diabetes spanning two solute carriers (SLC16A11 and SLC16A13) that introgressed into modern humans from Neandertals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova J-L, Patin E, Quintana-Murci L. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. The American Journal of Human Genetics. 2016;98:5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Currat M, Excoffier L. Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. Proc Natl Acad Sci U S A. 2011;108:15129–15134. doi: 10.1073/pnas.1107450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79•].Sankararaman S, Mallick S, Patterson N, Reich D. The combined landscape of denisovan and neanderthal ancestry in present-day humans. Curr Biol. 2016;26:1241–1247. doi: 10.1016/j.cub.2016.03.037. The authors unravel segments of Denisovan and Neandertal ancestry in 120 diverse modern human populations, and report more Denisovan ancestry in South Asians than is predicted by current models of archaic admixture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hunley KL, Healy ME, Long JC. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: Implications for biological race. Am J Phys Anthropol. 2009;139:35–46. doi: 10.1002/ajpa.20932. [DOI] [PubMed] [Google Scholar]
- [81].Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1 doi: 10.1038/ncomms1130. doi:10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82•].Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. This study reports a high-quality genome sequenced from a Neandertal woman found in Siberia, and infers several gene flow events in the Late Pleistocene among archaic hominins and between archaic hominins and modern humans. One of these gene flow events indicates the Denisovan lineage may derive ancestry from a currently unknown archaic group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc Natl Acad Sci U S A. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- [85••].Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. This study jointly analyses whole genomes from archaic and modern humans, and is the first to characterize gene flow from modern humans into Neandertals, in this case into Neandertals from the Altai Mountains of Siberia. The authors do not find evidence for gene flow from modern humans into the Denisovan or European Neandertal genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Coop G, Bullaughey K, Luca F, Przeworski M. The timing of selection at the human FOXP2 gene. Mol Biol Evol. 2008;25:1257–1259. doi: 10.1093/molbev/msn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zeng K, Charlesworth B. The joint effects of background selection and genetic recombination on local gene genealogies. Genetics. 2011;189:251–266. doi: 10.1534/genetics.111.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu Rev Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- [91].Hunter-Zinck H, Clark AG. Aberrant time to most recent common ancestor as a signature of natural selection. Mol Biol Evol. 2015;32:2784–2797. doi: 10.1093/molbev/msv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92•].Kim BY, Lohmueller KE. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am J Hum Genet. 2015;96:454–461. doi: 10.1016/j.ajhg.2014.12.029. The authors show that genetic drift cannot explain the observed higher proportion of Neandertal ancestry in East Asians compared to Europeans. Using simulations, this study find strong support for multiple pulses of Neandertal admixture in modern human genetic lineages (as observed by [64]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Cavalli-Sforza LL. Genes, Peoples, and Languages. University of California Press; 2000. [Google Scholar]
- [95].Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- [96].Jeong C, Alkorta-Aranburu G, Basnyat B, Neupane M, Witonsky DB, Pritchard JK, Beall CM, Di Rienzo A. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun. 2014;5 doi: 10.1038/ncomms4281. doi:10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]