Abstract
Ca2+ antagonist drugs are widely used in therapy of cardiovascular disorders1,2. Three chemical classes of drugs bind to three separate, but allosterically interacting, receptor sites on CaV1.2 channels, the most prominent voltage-gated Ca2+ (CaV) channel type in myocytes in cardiac and vascular smooth muscle3–9. The 1,4-dihydropyridines are used primarily for treatment of hypertension and angina pectoris and are thought to act as allosteric modulators of voltage-dependent Ca2+ channel activation, whereas phenylalkylamines and benzothiazepines are used primarily for treatment of cardiac arrhythmias and are thought to physically block the pore1,2. The structural basis for the different binding, action, and therapeutic uses of these drugs remains unknown. Here we present crystallographic and functional analyses of drug binding to the bacterial homotetrameric model CaV channel CaVAb, which is inhibited by dihydropyridines and phenylalkylamines with nanomolar affinity in a state-dependent manner. The binding site for amlodipine and other dihydropyridines is located on the external, lipid-facing surface of the pore module, positioned at the interface of two subunits. Dihydropyridine binding allosterically induces an asymmetric conformation of the selectivity filter, in which partially dehydrated Ca2+ interacts directly with one subunit and blocks the pore. In contrast, the phenylalkylamine Br-verapamil binds in the central cavity of the pore on the intracellular side of the selectivity filter, physically blocking the ion-conducting pathway. Structure-based mutations of key amino-acid residues confirm drug binding at both sites. Our results define the structural basis for binding of dihydropyridines and phenylalkylamines at their distinct receptor sites on CaV channels and offer key insights into their fundamental mechanisms of action and differential therapeutic uses in cardiovascular diseases.
CaV1 channels are composed of a complex of a pore-forming α1 subunit associated with β, γ, and α2δ subunits1,10. The α1 subunits contain four homologous domains with six transmembrane segments in each11,12. Transmembrane segments S1–S4 form the voltage-sensing module, and S5, S6 and the intervening P-loop form the pore1. The overall architecture of the mammalian skeletal muscle CaV1.1 channel was recently elucidated at a resolution of ~4–6 Å by cryo- electron microscopy13. However, higher-resolution structural analysis of mammalian CaV channels has not yet been achieved. The bacterial voltage-gated Na+ channel NaChBac and its relatives are homote-trameric proteins composed of four identical subunits, each analogous to one domain of a mammalian voltage-gated Na+ or Ca2+ channel14,15. These bacterial channels probably represent the evolutionary ancestors of both mammalian channel families. The structures of bacterial Na+ channels have been determined at high resolution by X-ray crystallography in pre-open16 and inactivated17,18 states. Moreover, the structural basis for Ca2+ conductance and selectivity has been elucidated at atomic resolution through studies of CaVAb, a site-directed mutant of NaVAb with full Ca2+ channel function19. We have used derivatives of CaVAb (see Methods) to define receptor sites and mechanisms of action of Ca2+ antagonist drugs at atomic resolution.
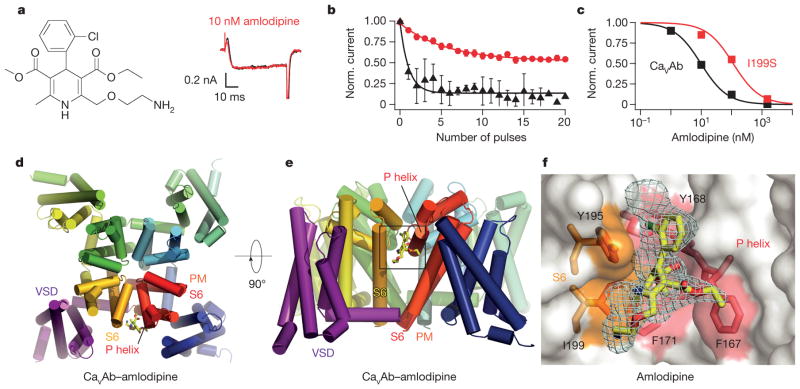
CaVAb was inhibited by amlodipine with high affinity (Fig. 1a–c). No inhibition was observed during single depolarizations, indicating that amlodipine does not enter the open pore and block it (Fig. 1a). However, inhibition increased progressively during trains of depolarizations, reflecting increased binding affinity for the activated and/or inactivated states of CaVAb (Fig. 1b). After a train of 20 depolarizing pulses, the half-maximum inhibitory concentration (IC50) for inhibition by amlodipine was 10 nM (Fig. 1c). This affinity was surprisingly high, considering the evolutionary distance between CaVAb and mammalian CaV1.2 channels, which have IC50 values from 0.3 nM to 1 μM for various dihydropyridines20.
Figure 1. Structural basis for inhibition of CaVAb by amlodipine.
a, Amlodipine structure. Ba2+ currents for 0 nM (black) and 10 nM (red) amlodipine during depolarization from −120 mV to 0 mV. b, State-dependent block by amlodipine after 50-ms pulses at 1 Hz from −120 mV to 0 mV (10 nM, circles; 100 nM, triangles; mean ± s.e.m.; n = 3–5. c, Inhibition by amlodipine. Data were fit by the Hill equation with nH = 1. CaVAb: IC50 = 10 ± 0.4 nM; CaVAb I199S: IC50 = 112 ± 10 nM; n = 3–5; mean ± s.e.m. d, Structure of CaVAb (top view in cylinders) binding amlodipine (yellow sticks). PM, pore module; VSD, voltage-sensing domain. e, CaVAb with bound amlodipine in side view. f, Dihydropyridine-binding pocket of CaVAb with the Fo–Fc electron density map (2.5σ, cyan) and amlodipine (yellow sticks). CaVAb residues contacted by amlodipine are highlighted in colours and labelled.
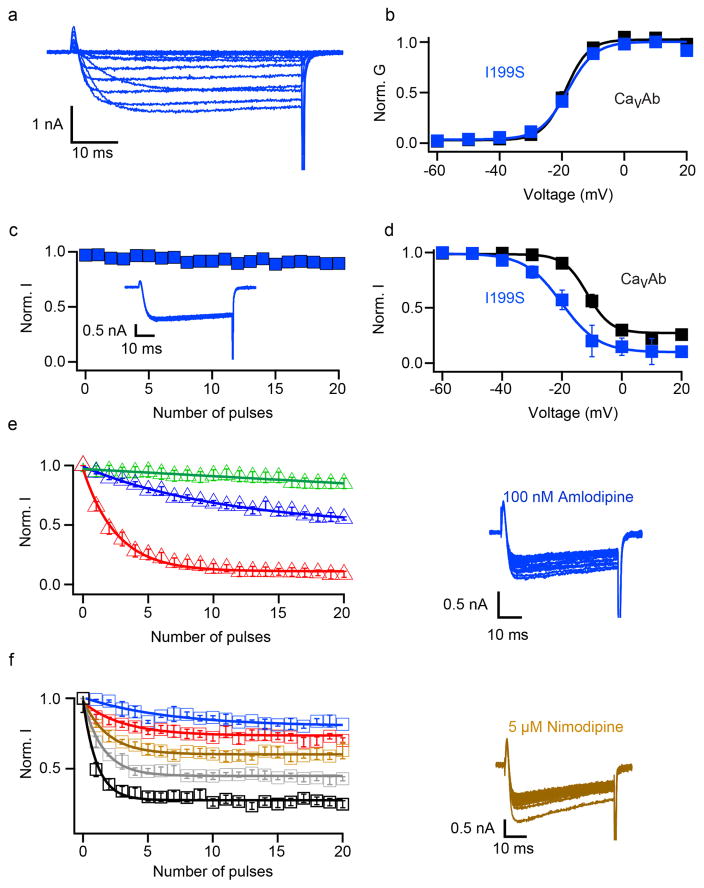
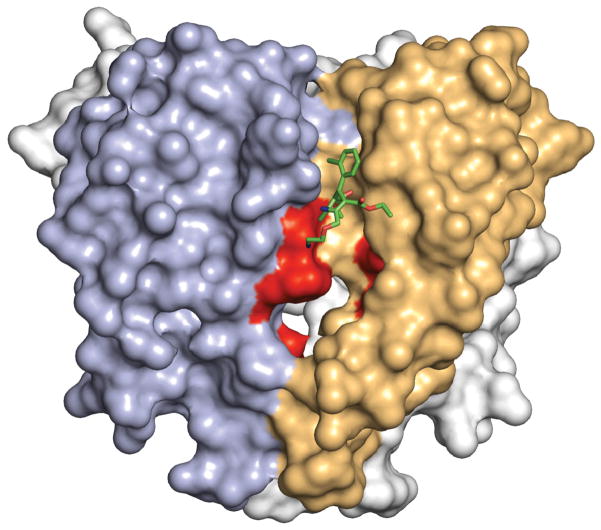
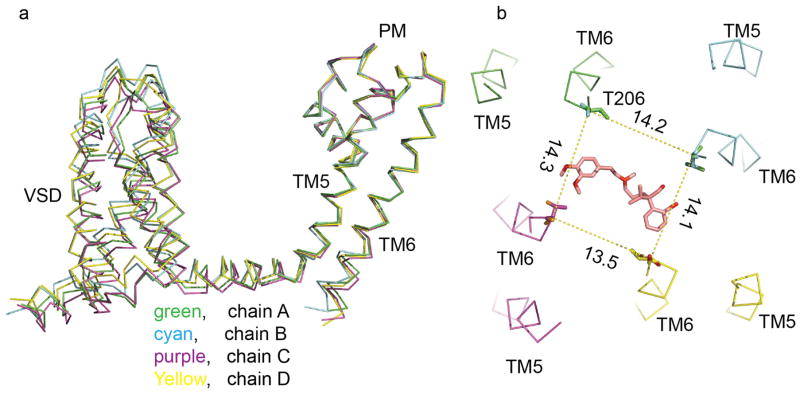
Photoaffinity labelling and site-directed mutagenesis suggest that dihydropyridines bind to a receptor site at the interface of homologous domains III and IV and the adjacent pore module in domain III in CaV1.2 channels4–7,21,22. In CaVAb, four identical subunits form a homotetramer (Fig. 1d)19. The structure of the amlodipine–CaVAb complex reveals the antagonist bound on the outer, lipid-facing surface of the pore module in the intersubunit crevice formed by neighbouring tilted S6 helices and the P-helix of the selectivity filter (Fig. 1d, e, yellow sticks). Despite the homotetrameric structure of CaVAb, only a single drug-binding site per tetramer is occupied, suggesting that drug-induced conformational changes prevent occupancy of more than one site. Amino-acid residues Y195, I199, F171, Y168 and F167 form a hydrophobic pocket for interaction with amlodipine (Fig. 1f). The dihydropyridine ring is sandwiched between Y195 of S6 and F167 of the P-loop. F171 and I199 of S6 form the bottom of the cleft that accommodates the bound drug. Mutations of I199 (for example, I199S) had minimal effects on CaVAb function (Extended Data Fig. 1), but markedly reduced the affinity for amlodipine (IC50 = 112 nM; Fig. 1c).
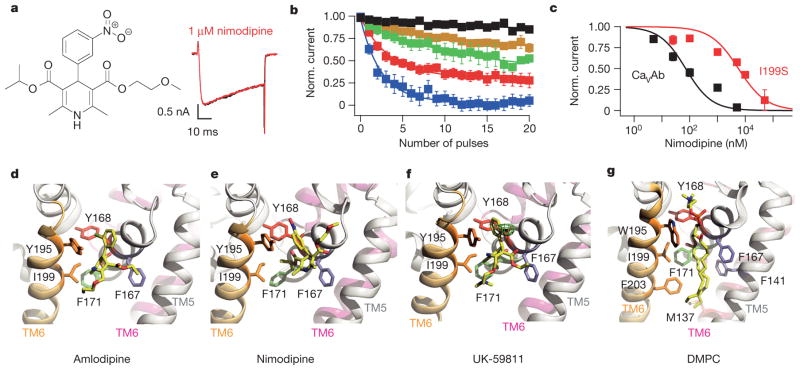
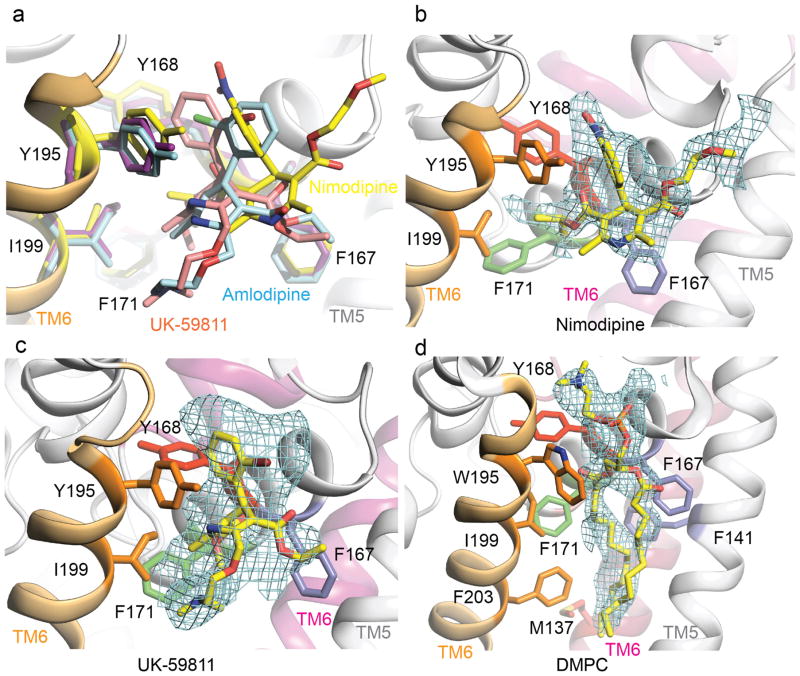
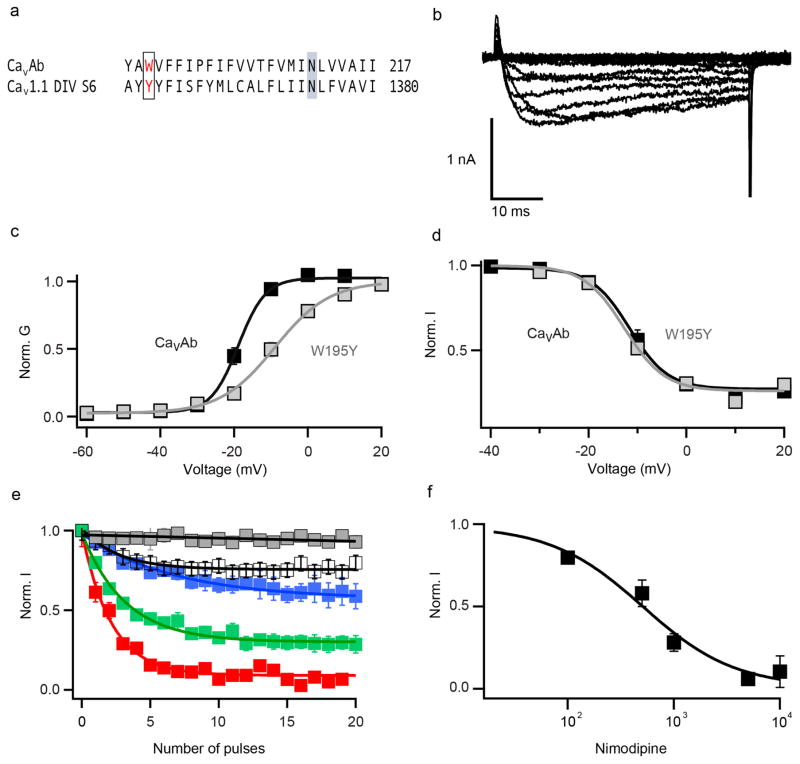
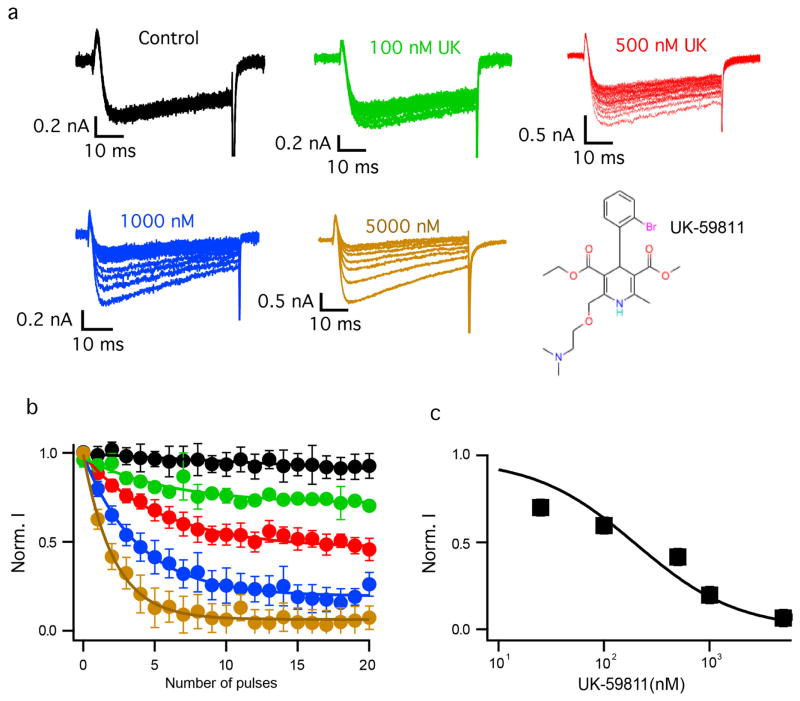
Nimodipine inhibited CaVAb like amlodipine, but its IC50 was 100 nM (Fig. 2a–c). Nimodipine binds to the same site as amlodipine (Fig. 2d, e and Extended Data Fig. 2a, b). The substitution I199S increased the IC50 for nimodipine from 100 nM to 5.7 μM (Fig. 2c), and W195Y increased it to 508 nM (Extended Data Fig. 3). The experimental Br-dihydropyridine derivative UK-59811 inhibited CaVAb with IC50 = 194 nM (Extended Data Fig. 4) and bound in a similar position (Fig. 2f and Extended Data Fig. 2a, c). Anomalous scattering density from its Br atom further confirmed the location of the dihydropyridine-binding site at the interface between the S6 segments of two adjacent subunits surrounded by Y195, F171, F167, and I199 (Fig. 2f, green mesh). High-resolution structures of CaVAb revealed 16 molecules of bound lipid per tetramer19. Without drugs, we found a single molecule of DMPC lipid aligned in the dihydropyridine-binding pocket with its polar headgroup facing the extracellular side and its long hydrocarbon tails projecting deep into the crevice formed by neighbouring S6 helices (Fig. 2g and Extended Data Fig. 2d). Thus, our structures reveal that dihydropyridine binding displaces an endogenous lipid molecule from their common binding site on CaVAb.
Figure 2. Inhibition of CaVAb by dihydropyridine binding at a lipid site.
a, Nimodipine structure. Current records as in Fig. 1a. b, State-dependent block by nimodipine as in Fig. 1b: 5 nM (black), 25 nM (brown), 100 nM (green), 1 μM (red), and 5 μM (blue); mean ± s.e.m.; n = 3–14. c, Inhibition by nimodipine as in Fig. 1c. CaVAb: IC50 = 100±9 nM; CaVAb I199S, IC50 = 5.7±0.6μM; n = 3–14; means.e.m. d, Amlodipine (yellow sticks) bound to CaVAb. S5 and S6 helices in ribbons; residues surrounding amlodipine in sticks. e, Nimodipine bound to CaVAb. f, UK-59811 bound to CaVAb. Anomalous scattering density (3σ, green mesh) for Br in UK-59811. g, DMPC lipid in the drug-free dihydropyridine-binding site in yellow sticks.
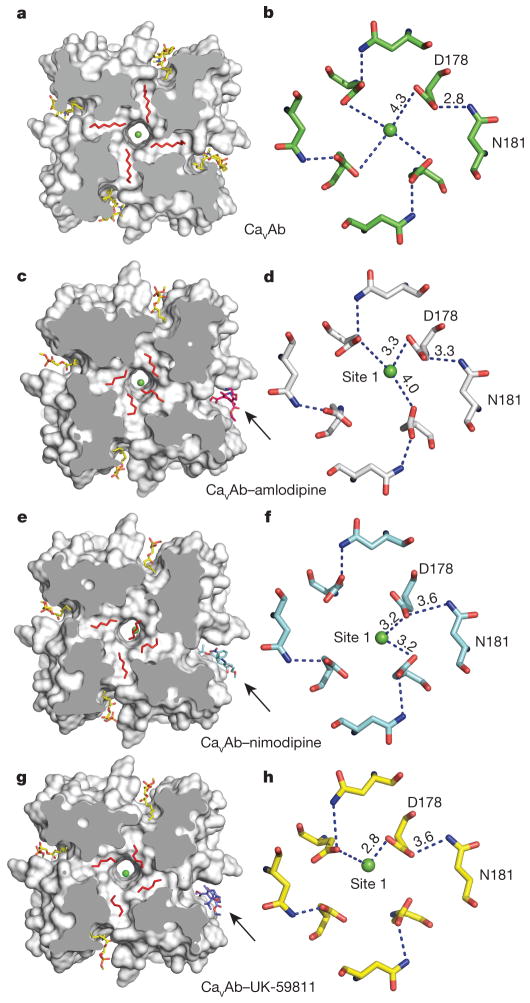
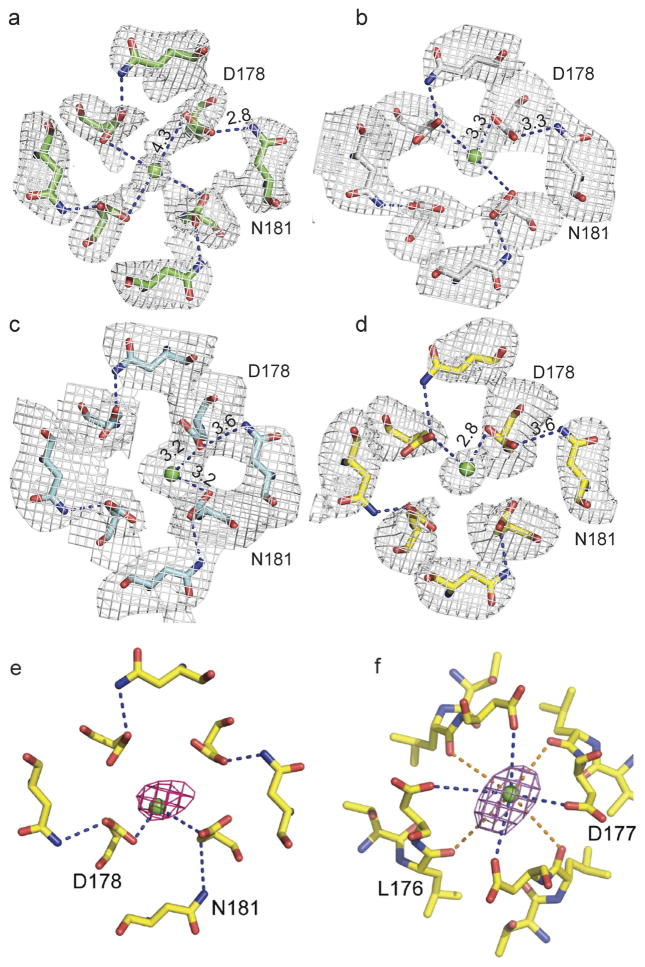
In the absence of dihydropyridines, the CaVAb structure has fourfold symmetry around the pore axis19. Four lipid molecules are found in the central cavity, occupying fenestrations that connect to the exterior of the channel (Fig. 3a). Binding of dihydropyridines to CaVAb rearranges the quaternary structure and breaks the fourfold symmetry (Fig. 3c, e, g, compare shaded cross-sections; see Supplementary Discussion of asymmetry induced by drug binding). With drug bound, the four lipid molecules in the central cavity lose their symmetric spatial organization, and the fenestration closest to the drug-binding site is no longer occupied by a lipid chain.
Figure 3. Dihydropyridine binding allosterically modifies Ca2+ binding in the selectivity filter.
a, Outward view. Four symmetrical lipids (red sticks) occupy fenestrations in CaVAb without dihydropyridine. Four additional lipids bind to the side of the pore module (yellow sticks). b, Top view. Site 1 with hydrated Ca2+ (green) coordinated directly by D178 and indirectly by N181 on extracellular end of the selectivity filter. c, Amlodipine binding (magenta sticks) induces asymmetry and causes rearrangement of lipids (red sticks). d, Top view. Site 1 with partially dehydrated Ca2+ and direct interaction with D178 due to binding of amlodipine. e, Binding of nimodipine (cyan sticks) induces asymmetry and reorganizes bound lipid. f, Partially dehydrated Ca2+ binds at site 1 with coordination distance of 3.2 Å to carboxylate side chains of D178. g, Binding of UK-59811 (blue sticks) to the dihydropyridine binding site induces asymmetry and reorganizes bound lipid. h, Ca2+ binds at Site 1 with coordination distance of 2.8 Å to a carboxylate side chain of D178.
By introducing asymmetry, dihydropyridine binding triggers allosteric changes at the selectivity filter of CaVAb and alters binding of the substrate ion. There are three Ca2+-binding sites in the CaVAb selectivity filter: two high-affinity sites (Sites 1 and 2) followed by one lower-affinity site (Site 3) arranged sequentially from its extracellular to intracellular end19. Without drug, Ca2+ binds near the central axis of the pore in a fully hydrated state, coordinated symmetrically by four D178 carboxylate side chains (Fig. 3b)19. With dihydropyridines bound, Ca2+ binds to Site 1 asymmetrically in a partially dehydrated state—significantly off the central axis of the pore and closer to one or two D178 carboxylate groups at a distance of 2.8–3.3 Å (Fig. 3d, f, h and Extended Data Fig. 5a–d). This binding distance suggests direct interaction of bound Ca2+ with the carboxylate side chain (Supplementary Discussion). In contrast, binding of Ca2+ at Site 2 is unchanged (data not shown). The anomalous scattering density of Ca2+ confirms its off-axis location in Site 1 and on-axis location in Site 2 (Extended Data Fig. 5e, f).
Studies with quaternary phenylalkylamine analogues revealed that these drugs inhibit CaV1.2 channels only after cytoplasmic application, and that drug binding is increased by repetitive depolarization to open the pore2,23. It was therefore concluded that tertiary phenylalkylamines such as verapamil penetrate the membrane in uncharged form, are re-protonated in the cytosol, and block the CaV1.2 channel by entering the intracellular mouth of the open pore in their protonated form and binding to their receptor site2,23. Photoaffinity labelling and site-directed mutagenesis revealed that the phenylalkylamine receptor site is formed by S6 segments in domains III and IV of CaV1.2 channels, consistent with drug binding in the pore4–7,24,25.
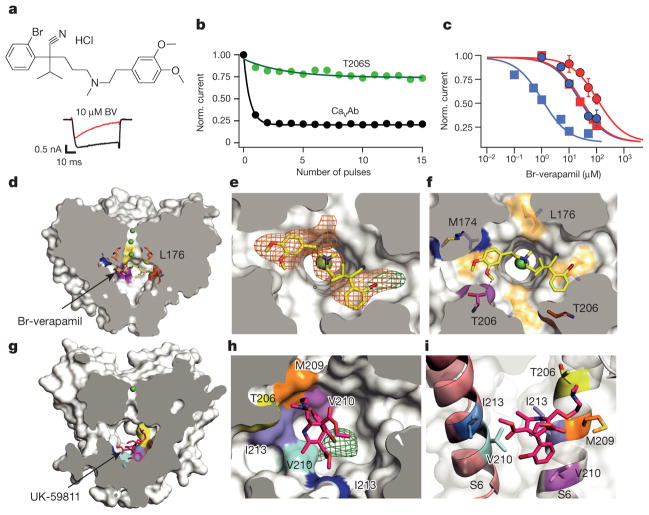
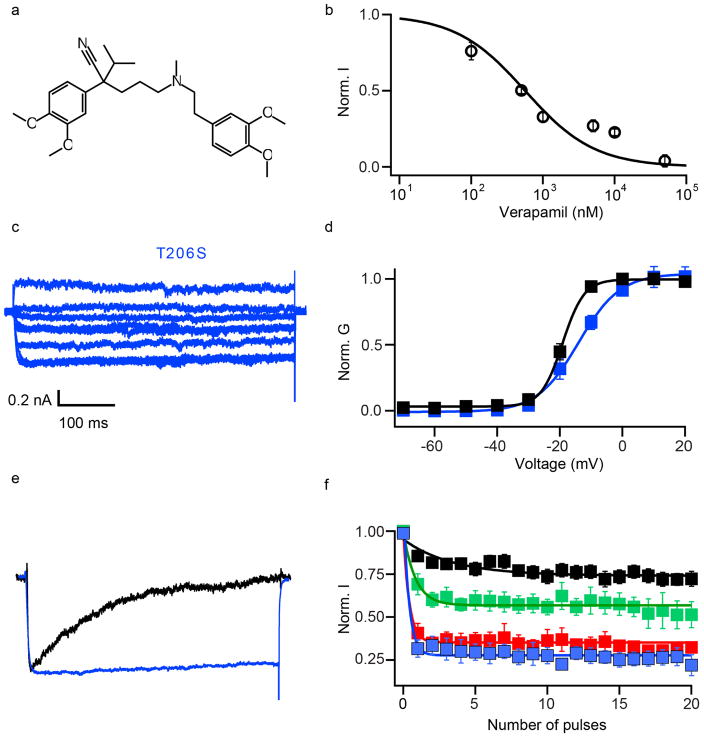
When Br-verapamil was perfused at −120 mV, the first depolarization to 0 mV showed progressive reduction of the current during the pulse (Fig. 4a). This profile supports a pore-blocking mechanism, in which the drug progressively enters and blocks the open pore. Repetitive depolarizing stimuli increased inhibition of CaVAb by Br-verapamil (Fig. 4b), yielding IC50 values of 810 nM for Br-verapamil (Fig. 4c, blue squares) and 475 nM for verapamil (Extended Data Fig. 6a, b) at steady state. The action of these drugs is strikingly state- dependent: the IC50 for Br-verapamil in the resting state is 24 μM, 30-fold higher than observed after a train of depolarizing stimuli (Fig. 4c, blue circles).
Figure 4. State-dependent inhibition by pore block with Br-verapamil and UK-59811.
a, Br-verapamil. Ba2+ current records for CaVAb with 0 μM (black) and 10 μM (red) during the depolarizing pulse. b, State-dependent block of CaVAb (n = 7) and CaVAb T206S (n = 3) at 10 μM during trains of depolarizations at 1 Hz from −120 mV to 0 mV. The error bars for all the data points on this graph are too small to be visible. c, Inhibition by Br-verapamil for CaVAb and CaVAb T206S at V = −120 mV and following trains of depolarizations as in b. CaVAb: resting state block, blue circles, IC50 = 24 ±1.6 μM; state-dependent block, blue squares, IC50 =810 ±80 nM. CaVAb T206S: resting state block, red circles, IC50 = 115±3.2μM; state-dependent block, red squares, IC50 =24±0.8μM; n =3–11; mean ± s.e.m. d, Side view of the pore module sectioned through the selectivity filter with Br-verapamil bound (yellow sticks). Ca2+, green spheres. e, Fo–Fc electron density (2.5σ, orange mesh) and anomalous scattering density (3σ, green mesh) for Br defines location of Br-verapamil. f, The two aromatic rings of verapamil are close to T206 of adjacent subunits. g, UK-59811 (red sticks) binds with its dihydropyridine ring deep in the central cavity. h, Anomalous scattering density (3.5σ, green mesh) of Br in UK-59811. i, S6 segments with residues surrounding UK-59811 in sticks.
Our crystal structures revealed a single molecule of Br-verapamil bound in the central cavity on the intracellular side of the ion selectivity filter (Fig. 4d, e; see Supplementary Discussion of asymmetry induced by drug binding). The bound drug is oriented with its characteristic positively charged tertiary amino group facing in the extracellular direction pointing towards Site 3 in the selectivity filter. In this position, the bound phenylalkylamine would physically block the pore. The distance between the tertiary amino group and Ca2+ coordinated by the carbonyls of L176 is 5 Å. The methoxy groups in the aromatic rings are located close to the inner end of the fenestrations, surrounded by T206, M209 of the neighbouring subunit and T175, M174, L176 of the selectivity filter (Fig. 4f). The two aromatic rings of Br-verapamil interact with T206 residues from two neighbouring S6 helices (Fig. 4f). A view from the intracellular side shows that Br-verapamil binds closer to two subunits on one side of the pore (Fig. 4f). The anomalous scattering from Br-verapamil further defines the position of the aromatic ring that is farther from the amino group and confirms its interaction with T206 (Fig. 4e, green mesh). Mutations in T206 impair inactivation of CaVAb (Extended Data Fig. 6c, e) and markedly reduce the affinity for Br-verapamil. For example, the conservative mutation T206S increases the IC50 for state-dependent inhibition from 810 nM to 24 μM (Fig. 4c, red squares) and the IC50 for resting state inhibition of CaVAb from 24 μM to 115 μM (Fig. 4c, red circles). The effects of these mutations on both resting and state-dependent block confirm that there is a direct interaction between the drug and T206. These results define the receptor site for pore block by phenylalkylamines at high resolution. Similar to the dihydropyridine-binding site, the phenylalkylamine-binding site is also occupied by lipid molecules in the absence of the drug.
At concentrations above 1 μM, dihydropyridines inhibit voltage-gated Na+ channels in a manner consistent with pore block26. At the high drug concentrations used in our crystallization studies, we found binding of UK-59811 (Fig. 4g–i) and other dihydropyridines in the pore of CaVAb. The anomalous scattering density of its Br places the dihydropyridine ring deep in the central cavity where it forms hydrophobic contacts with two neighbouring subunits (Fig. 4h, green mesh). Compared to Br-verapamil, UK-59811 bound more towards the intracellular base of the central cavity and was located closer to one subunit (Fig. 4g–i). Low-affinity block of Na+ channels by dihydropyridines bound in this site may contribute to cardiac arrhythmias caused by toxic overdoses of these drugs.
Overall, our results provide a structural basis for understanding how dihydropyridines and phenylalkylamines bind at two distinct, but allosterically coupled receptor sites on CaV1.2 channels and have different efficacy for treatment of hypertension and angina pectoris versus cardiac arrhythmias1–3,5–7. Consistent with photoaffinity-labelling and site-directed mutagenesis5–7, dihydropyridines bind on the outer, lipid-facing surface of the pore module at the interface between two subunits of CaVAb, in analogy with their proposed site of action between domains III and IV of CaV1.2 channels4,21. Their binding site is exposed to the extracellular side of the membrane, but not to the intracellular side. These structural results reveal why charged dihydropyridines are ineffective when applied intracellularly27, and they are consistent with location of the drug-binding site ~11–14 Å from the outer surface of the lipid bilayer as inferred from studies of charged derivatives of amlodipine with hydrophobic linkers of increasing length28. These comparisons reveal a close analogy between the site of dihydropyridine binding in our crystal structures of CaVAb and the expectations from studies of CaV1.2 channels, but the exact position of the drug-binding site in CaVAb is approximately one helical turn towards the extracellular side from the amino-acid residues implicated in dihydropyridine binding by studies of CaV1.2 channels (Extended Data Fig. 7). This difference may reflect the great evolutionary distance between CaVAb and mammalian CaV channels and/or indirect allosteric effects of mutations studied in CaV1 channels.
Binding of a single dihydropyridine to CaVAb induces a conformational change that alters the fourfold symmetry of the quaternary structure and induces changes in the three unoccupied dihydropyridine-binding sites that may prevent drug occupancy (Extended Data Fig. 8). Drug binding also disrupts the symmetry of the ion selectivity filter, allowing direct coordination of Ca2+ by carboxylate side chains. This conformational change is mediated in part by an altered pattern of hydrogen bonds formed by N181 in the subunit binding the dihydropyridine (Fig. 3). These structural results correlate closely with ligand-binding studies of CaV1.2 channels, which suggested that dihydropyridines induce high-affinity Ca2+ binding and block of the pore29,30. Our structural studies reveal exactly how dihydropyridines act as indirect allosteric blockers of the pore of Ca2+ channels. Dihydropyridine binding to CaV1.2 channels is voltage-dependent because of the high affinity for the inactivated state1,5–7. In a remarkable parallel, dihydropyridine binding causes a conformational change to an asymmetric pore structure in CaVAb, which is similar to the asymmetry induced in inactivated states of the parent NaVAb channel17 and its relative NaVRh18. Dihydropyridine binding may induce a similar asymmetric, Ca2+-blocked state of CaV1.2 channels and thereby enhance their inactivation, allowing selective inhibition in persistently depolarized cells. This mechanism underlies the use of dihydropyridines in treatment of hypertension and angina pectoris, in which vascular smooth muscle cells of resistance vessels are persistently depolarized, and their CaV1.2 channels are selectively inhibited by dihydropyridines.
The phenylalkylamine receptor site was localized to the S6 segments in domains III and IV of CaV1.2 channels by photoaffinity labelling and mutational analysis, and it was proposed that the amino-acid side chains involved in drug binding point towards the lumen of the pore4–6,24,25. Our structural results correlate precisely with this expectation and reveal the exact structure of the drug–receptor complex. Br-verapamil is stretched between two subunits of CaVAb, consistent with drug binding at the interface of domains III and IV in CaV1.2 channels4–6,24,25. As for dihydropyridines, phenylalkylamine binding at this site disrupts the fourfold symmetry of the pore (Extended Data Fig. 9). Location of the phenylalkylamine receptor site deep in the central cavity in the pore reveals why binding of these drugs is state-dependent. Access of phenylalkylamines to their receptor is greatly enhanced by opening the intracellular activation gate, which allows diffusion to the drug receptor site. Drug binding is therefore frequency-dependent, allowing selective block of CaV1.2 channels in rapidly firing cardiac myocytes2. This mechanism is the basis for use of verapamil for cardiac arrhythmias.
Overall, our structural studies illuminate the complex pharmacology and therapeutic uses of Ca2+ antagonist drugs in treatment of different cardiovascular disorders at the atomic level (see Supplementary Discussion). These structural models will be important for design and development of next-generation Ca2+ antagonist drugs to provide safer and more effective treatment of hypertension, angina pectoris, cardiac arrhythmia, and other medical conditions.
METHODS
CaVAb constructs and drugs
As originally defined, CaVAb was constructed by introducing the mutations E177D, S178D and M181D as a triple mutant into NaVAb19. This construct was used for all electrophysiological studies, except as noted in figure legends. In this work, we have also used CaVAb E177D S178D M181N, which has an identical structure and Ca2+-binding properties and has high Ca2+ selectivity19. It gives greater consistency of high-resolution crystal structures. We have also added the mutation W195Y, which substitutes the Y residue from the analogous position in mammalian CaV1.1 channels for W195 in CaVAb. This mutant gives better resolution of drugs bound at the dihydropyridine site. CaVAb E177D S178D M181N W195Y was used for all structural studies presented here, except as noted in the figure legends. Similar structural results were obtained for both versions of CaVAb. We found that amlodipine and other dihydropyridines (Figs. 1 and 2 and Extended Data Figs 1, 3 and 4) and verapamil and other phenylalkylamines (Fig. 4 and Extended Data Fig. 6) effectively blocked CaVAb and gave high-resolution crystal structures; however, we were unable to prepare crystals with bound diltiazem for structural biology so we have not addressed the structure of the benzothiazepine receptor site in this work.
Electrophysiology
All measurements were done in insect cells (Trichoplusia ni cells; High5). All CaVAb constructs used were made on the background of N49K mutation. Mutation N49K shifts the activation curve ~75 mV to more positive potentials compared to wild-type CaVAb and abolishes the use-dependent inactivation as described previously19,31. All constructs showed good expression, allowing measurement of ionic currents 24–48 h after infection. Whole-cell Ba2+ currents were recorded using an Axopatch 200 amplifier (Molecular Devices) with glass micropipettes (2–4 MΩ). Capacitance was subtracted and 80–90% of series resistance was compensated using internal amplifier circuitry. Extracellular solution contained in (mM) 10 BaCl2, 140 NMDG-methanesulphonate, 20 HEPES, (pH 7.4, adjusted with Ba(OH)2, [Ba2+]total = 13 mM). Intracellular solution contained in (mM) 105 CsF, 35 NaCl, 10 HEPES, 10 EGTA, (pH 7.4, adjusted with CsOH). Current–voltage (I–V) relationships were recorded in response to steps to voltages ranging from −120 to +50 mV in 10-mV increments from a holding potential of −120 mV. Conductance-voltage (G–V) curves were calculated from the corresponding (I–V) curves. Pulses were generated and currents were recorded using Pulse software controlling an Instrutech ITC18 interface (HEKA). Data were analysed using Igor Pro 6.2 (WaveMetrics). Sample sizes were chosen to give s.e.m. values of less than 10% of peak values based on prior experimental experience. Inhibition curves were fit with a Hill equation with nH = 1.0 unless indicated otherwise in the figure legends.
Protein expression and purification
The pFastBac-Flag-CaVAb was used as the construct for producing homotetrameric model voltage-gated Ca2+ channel19. I199S, W195Y, and T206S constructs were generated via site-directed mutagenesis using QuickChange (Stratagene). Recombinant baculovirus were produced using the Bac-to-Bac system (Invitrogen), and T. ni insect cells were infected for large-scale protein purification. Cells were harvested 72 h post-infection and re-suspended in buffer A (50 mM Tris-HCl, pH = 8.0, 200 mM NaCl) supplemented with protease inhibitors and DNase. After sonication, digitonin (EMD Biosciences) was added to 1%, and solubilization was carried out for 1–2 h at 4 °C. Clarified supernatant was then incubated with anti-Flag M2-agarose resin (Sigma) for 1–2 h at 4 °C with gentle mixing. Flag-resin was washed with ten column volumes of buffer B (buffer A supplemented with 0.12% digitonin) and eluted with buffer B supplemented with 0.1 mg ml−1 Flag peptide. The eluant was concentrated and then passed over a Superdex 200 column (GE Healthcare) in 10 mM Tris-HCl pH =8.0, 100 mM NaCl and 0.12% digitonin. The peak fractions were concentrated using a Vivaspin 30K centrifugal device.
Crystallization and data collection
CaVAb and the W195Y mutant were concentrated to ~20 mg ml−1 and reconstituted into DMPC:CHAPSO (Anatrace) bicelles according to standard protocols16,17,32,33. The protein-bicelle preparation and a well solution containing 1.8–2.0 M ammonium sulphate, 100 mM Na-citrate pH = 5.0 was mixed in 1:1 ratio and set up in a hanging-drop vapour-diffusion format. All the antagonist complex crystals were obtained through co-crystallization by incubating the protein-bicelle with 100 μM antagonist overnight before setting up crystallization trials. For the UK-59811 complex, both 100 μM and 200 μM antagonist were used for UK-59811/CaVAb crystals. Crystals were cryoprotected by soaking in 0.1 M Na-acetate, pH 5.0, 26% glucose, 2.0 M ammonium sulphate, and 5 mM Ca2+. Crystals were plunged into liquid nitrogen and maintained at 100 K during all data collection procedures.
The anomalous diffraction data sets for Br were collected at 0.9194 Å, and the anomalous data sets for Ca2+ were collected at 1.75 Å with the same synchrotron radiation source (Advanced Light Source, BL8.2.1). To optimize the anomalous scattering signal, the data sets were collected by using the ‘inverse beam strategy’ with the wedge size of 5°.
Structure determination, refinement, and analysis
X-ray diffraction data were integrated and scaled with the HKL2000 package34 and further processed with the CCP4 package35. The structure of CaVAb and its antagonist complex were solved by molecular replacement using an individual subunit of the CaVAb structure (PDB code 4MS2) as the search template. The data sets were processed in P21221 space group, in which there are four molecules in one asymmetric unit. Crystallography and NMR System software36 were used for refinement of coordinates and B-factors. Final models were obtained after several cycles of refinement with REFMAC37 and PHENIX38 plus manual re-building using COOT39. The geometries of the final structural models of CaVAb and its antagonist complexes were verified using PROCHECK40. Divalent cations were identified by anomalous difference Fourier maps calculated using data collected at wavelengths of 1.75 Å for Ca2+. The Br atoms of UK-59811 and Br-verapamil were identified by anomalous difference Fourier maps calculated using data collected at wavelengths of 0.9194 Å. Procedures accounting for merohedral twinning were performed during structural refinement of amlodipine, nimodipine, and Br-verapamil data sets. Detailed crystallographic data and refinement statistics for all constructs are shown in Extended Data Table 1. All structural figures were prepared with PyMol41.
Extended Data
Extended Data Figure 1. Biophysical characterization of CaVAb I199S.
a, Ba2+ currents recorded from a holding potential of −120 mV to test potentials from −60 mV to 20 mV in 10 mV steps for I199S. b, G–V curves of CaVAb and CaVAb I199S derived from peak I–V relationships. The voltages for half-maximal activation and slopes are: CaVAb: V1/2 = − 18.8±0.3, k =3.68±0.43, n = 7; CaVAb I199S: V1/2 = −18.8±0.3, k =3.88±0.47 (n = 5). c, Repetitive depolarization to 0 mV at 1 Hz from a holding potential of − 120mV (n 5). d, Steady-state inactivation of CaVAb and CaVAb I199S. Two pulses were applied: a 300-ms conditioning pulse to the indicated potentials followed by 50-ms test pulse to 0 mV (n =3). e, State-dependent block of CaVAb I199S by 10 nM (green), 100 nM (blue), or 1.5 μM (red) amlodipine during repetitive depolarizations to 0 mV (left, n = 3–5 cells). Ba2+ currents in 100 nM amlodipine for CaVAb I199S (right). f, Concentration-dependent block of CaVAb I199S by nimodipine at 100 nM (blue), 1 μM (red), 5 μM (brown), 10 μM (grey) and 50 μM (black) (left, n = 4–5 cells for each curve). Ba2+ currents in the presence of 5 μM nimodipine for CaVAb I199S (right).
Extended Data Figure 2. Structural comparison of the binding modes of amlodipine, nimodipine, and UK-59811.
a, Superposition of CaVAb in complexes with amlodipine (cyan), nimodipine (yellow), and UK-59811 (magenta) at the dihydropyridine binding site viewed from the side of the pore module. The side chains of dihydropyridine-interacting residues are shown in sticks. b, An Fo–Fc simulated annealing omit map contoured at 2.5σ for nimodipine. c, An Fo–Fc simulated annealing omit map contoured at 2.5σ for UK-59811. d, An Fo–Fc simulated annealing omit map contoured at 2.5σ for DMPC.
Extended Data Figure 3. Biophysical characterization and drug block of CaVAb W195 and CaVAb Y195.
a, Sequence alignment of CaVAb S6 segment and CaV1.1 DIV S6. W195 in CaVAb is equivalent to Y1358 in CaV1.1. b, Ba2+ currents recorded from a holding potential of −120 mV to test potentials from 60 mV to 20 mV in 10 mV steps for CaVAb W195Y. c, G–V curves for CaVAb W195 and CaVAb Y195 derived from peak I–V relationships. The voltages for half-maximal activation and slopes are: CaVAb W195 V1/2 = −18.8 ± 0.3, k = 3.7 ± 0.43, n = 7; CaVAb Y195, V1/2 = −9 ± 0.3, k = 7.4 ± 0.1, n = 5. d, Steady-state inactivation of CaVAb W195 and CaVAb Y195 (n = 3). Two pulses were applied: a 300-ms conditioning pulse followed by 50-ms test pulse to 0 mV. e, State-dependent block of CaVAb W195Y by nimodipine at 100 nM (white), 500 nM (blue), 1 μM (green), 5 μM (red), and control (grey). f, Concentration-dependent block of CaVAb W195Y by nimodipine. IC50 = 508 ± 93 nM (n = 4–5 cells for each point).
Extended Data Figure 4. Biophysical characterization of block by UK-59811.
a, Ba2+ currents for state-dependent block by different concentrations of UK-59811. b, State-dependent block of CaVAb by UK-59811 at 0 nM (black), 100 nM (green), 500 nM (red), 1 μM (blue), and 5 μM (brown). For each curve, n = 4–5 cells. c, Concentration-response curve for UK-59811. Data were fit with a Hill equation assuming a 1:1 binding. IC50 = 194 ± 22 nM, n = 4–5.
Extended Data Figure 5. Evidence for the partially dehydrated Ca2+ binding and carboxyl-carboxylate pairs at the selectivity filter entryway.
a, Top view of an Fo–Fc simulated annealing omit map contoured at 3σ for residues 178 and 181 for the wild-type channel without drug. b, Top view of an Fo–Fc simulated annealing omit map contoured at 3σ for residues 178 and 181 for CaVAb–amlodipine. c, Top view of an Fo–Fc simulated annealing omit map contoured at 2.5σ for residues 178 and 181 for CaVAb–nimodipine. d, Top view of an Fo–Fc simulated annealing omit map contoured at 3σ for residues 178 and 181 for CaVAb– UK-59811. e, Top view of Site 1 with the anomalous difference Fourier map density (red mesh, contoured at 3σ) calculated with diffraction data of crystals collected at 1.75 Å wavelength. Ca2+ is shown as a green sphere. Site 1 residues are shown in sticks. Hydrogen bonds are indicated with dashed lines. f, Top view of Site 2 with the anomalous difference Fourier map density (magenta mesh, contoured at 3σ).
Extended Data Figure 6. Biophysical characterization of verapamil block of CaVAb and functional properties of CaVAb T206S.
a, Chemical structure of verapamil. b, Concentration dependence of verapamil inhibition of CaVAb. The amplitude of the peak Ba2+ current was recorded after applying 20 pulses at a frequency of 1 Hz, where the block reaches steady state. The data were fit by a Hill equation assuming a 1:1 binding ratio. n = 4–7 cells. IC50 = 475 ± 25 nM. c, Ba2+ currents of CaVAb T206S. d, G–V curves. CaVAb (black): V1/2 = 18.8 ± 0.3 mV, k = 3.7 ± 0.43 ( n = 5); CaVAb T206S (blue): V1/2 = −15 ±1.8 mV, k = 6.6 ±0.4 ( n =5). e, Current traces of CaVAb (black) and CaVAb T206S (blue) during a 1-s depolarizing pulse from a holding potential of –120 mV to −10 mV. f, State-dependent inhibition of CaVAb T206S by Br-verapamil at 10 μM (black), 25 μM (green), 50 μM (red), and 100 μM (blue). For each curve, n = 4–5 cells.
Extended Data Figure 7. Comparison of dihydropyridine binding site in CaVAb and CaV1.2.
The pore domain of CaVAb is illustrated with two subunits in view, one in tan corresponding to domain III of CaV1.2 and one in blue corresponding to domain IV of CaV1.2. The amino acid residues in CaVAb corresponding to those that are important for dihydropyridine binding to CaV1.2 channels are highlighted in red. Bound amlodipine is illustrated with green sticks.
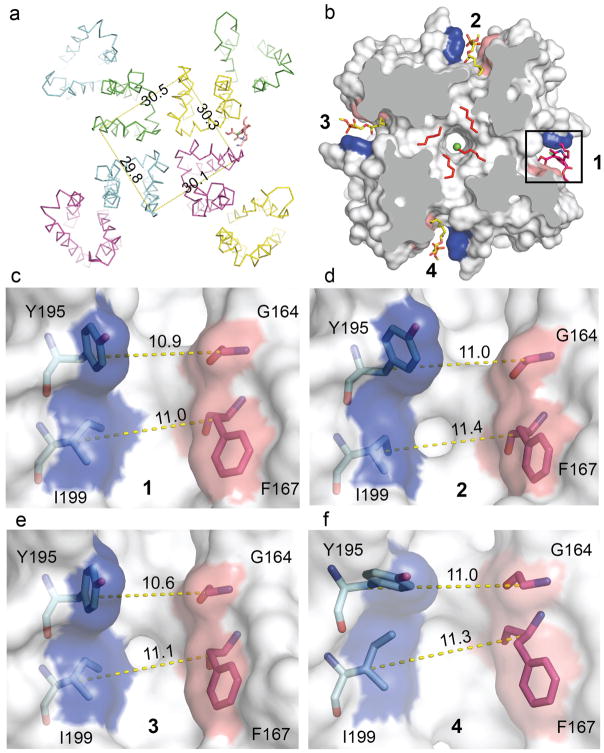
Extended Data Figure 8. Amlodipine binding breaks symmetry.
a, The overall structure of CaVAb in complex with amlodipine (shown in ribbon representation). Measuring the Cα distances of V196 (nearing the amlodipine binding pocket) from the 4 subunits shows the channel is asymmetrical. b, Binding of amlodipine (sticks in red) induces asymmetry and causes rearrangement of the lipid in the central cavity. c–f, The amlodipine binding pocket showing the Cα–Cα distance at two layers (Y195–G164 and I199–F167) horizontally. At layer 1 (Y195–G164), the Cα–Cα distance of its neighbouring sites (11.0 Å in d and 11.0 Å in f) matches the drug binding site (10.9 Å in c), but the diagonal site (e) is too narrow (10.6 Å). At layer 2 (I199–F167), the pocket width of the diagonal site (11.1 Å in e) matches the drug-binding site (11.0 Å in c), but the two diagonal sites are too wide (11.4 Å in d and 11.3 Å in f).
Extended Data Figure 9. Br-verapamil binding breaks symmetry.
a, Alignment of the 4 subunits of CaVAb in complex with Br-verapamil showing the voltage sensor module (VSD) and the ends of S6 are different. b, Measuring the Cα distances between T206 residues in adjacent subunits shows that the channel is indeed asymmetrical with Br-verapamil in the pore.
Extended Data Table 1.
Data collection, phasing and refinement statistics
| CavAb 5mM Ca2+ |
CavAb1 (W195Y) UK-59811 5mM Ca2+ |
CavAb2 UK-59811 5mM Ca2+ |
CavAb (W195Y) Amiodipine 5mM Ca2+ |
CavAb1 (W195Y) Nimodipine 5mM Ca2+ |
CavAb1 Br-verapamil 5mM Ca2+ |
|
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P21221 | P21221 | P21221 | P21221 | P21221 | P21221 |
| Cell dimensions | ||||||
| a, b, c (Å) | 124.9 125.7 191.5 |
125.9 126.0 192.1 |
125.5 125.9 191.7 |
125.6 125.3 191.7 |
125.3 125.4 191.6 |
125.6 125.6 192 |
| α,β,γ(°) | 90 90 90 |
90 90 90 |
90 90 90 |
90 90 90 |
90 90 90 |
90 90 90 |
| Resolution (Å) | 2.7 | 3.3 | 3.3 | 3.2 | 3.2 | 3.3 |
| Rsym or Rmerge | 11.4(98.4) | 12.6(74.2) | 11.7(60.1) | 12.1(49.1) | 11.2(68.6) | 18.8(86.1) |
| CC1/2(%) | 99.8(87.7) | 99.8(84.3) | 99.7(87.4) | 99.4(89.2) | 99.7(86.7) | 98.4(70.3) |
| //s/ | 13.4(2.4) | 13.4(3.3) | 13.1(3.1) | 10.2(2.8) | 15.2(3.5) | 6.0(1.7) |
| Completeness (%) | 92.5(97.8) | 95.0(100.0) | 92.1(82.0) | 92.7(94.5) | 99.8(99.8) | 97.7(98.8) |
| Redundancy | 10.1(9.8) | 9.5(9.9) | 9.4(9.2) | 5.1(5.2) | 9.3(9.0) | 4.9(5.0) |
| Refinement | ||||||
| Resolution (Å) | 30-2.7 | 30-3.3 | 30-3.3 | 30-3.2 | 30-3.2 | 30-3.2 |
| No. reflections | 76513 | 46606 | 42515 | 46657 | 50390 | 49327 |
| Rwork / Rfree | 22.1/26.2 | 28.0/30.5 | 27.5/30.0 | 23.3/27.7 | 21.7/25.6 | 25.1/29.4 |
| No. atoms | 9684 | 7400 | 7403 | 7380 | 7393 | 7366 |
| Protein | 8780 | 7192 | 7200 | 7192 | 7192 | 7200 |
| Ligand/ion | 887 | 205 | 199 | 187 | 189 | 166 |
| Water | 17 | 3 | 2 | 1 | 28 | |
| B-factors | ||||||
| Protein | 76.9 | 104.4 | 111.4 | 100.9 | 108.0 | 114.1 |
| Ligand/ion | 74.2 | 99.9 | 95.8 | 85.7 | 86.3 | 100.6 |
| Water | 46.6 | 57.9 | 63.2 | 48.8 | 52.8 | |
| R.m.s deviations | ||||||
| Bond lengths (Å) | 0.009 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 |
| Bond angles (°) | 1.15 | 1.74 | 1.75 | 1.73 | 1.54 | 1.74 |
| Ramachandran statistics | ||||||
| Favored | 96% | 96% | 96% | 95.0% | 96% | 96.0% |
| Allowed | 4.1% | 3.6% | 3.6% | 4.6% | 4.0% | 4.1% |
| Outliers | 0.28% | 0.23% | 0.23% | 0.23% | 0.12% | 0.12% |
This data set is collected at 0.9198 Å.
This data set is collected at 1.75 Å.
All other data sets are collected at 1.0 Å.
Acknowledgments
We are grateful to the beamline staff at the Advanced Light Source (BL8.2.1 and BL8.2.2) for their assistance during data collection. Research reported in this publication was supported by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health under award number R01 HL112808 (W.A.C. and N.Z.), and a National Research Service Award from training grant T32 GM008268 (T.M.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by research grants from the Howard Hughes Medical Institute (N.Z.) and by the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health under award number R01 NS26254 (W.A.C.). D.C.P. acknowledges support from Neusentis, Pfizer Inc., Cambridge, UK during the course of this work.
Footnotes
Author Information The coordinates and structure factors have been deposited in the Protein Data Bank with the following accession codes 5KLB (CavAb, 5 mM Ca2+ 2.7Å; 5KLG (CavAb-W195Y-UK-59811, 5 mM Ca2+); 5KLS (CavAb-UK-59811, 5 mM Ca2+); 5KMD (CavAb-W195Y-amlodipine, 5 mM Ca2+); 5KMF (CavAb-W195Y nimodipine, 5 mM Ca2+); and 5KMH (CavAb-Br-verapamil, 5 mM Ca2+).
Author Contributions L.T., T.M.G., T.M.S., T.S., D.C.P., N.Z., and W.A.C. designed the experiments. D.C.P. provided compound UK-59811. L.T. conducted protein purification, crystallization, and X-ray diffraction experiments for dihydropyridines. L.T. and T.M.S. conducted protein purification, crystallization, and X-ray diffraction experiments for Br-verapamil. L.T. and T.M.S. determined the structures and analysed the structural results with input from T.M.G. and N.Z. T.M.G. designed and analysed mutants that block drug binding and performed all of the electrophysiological studies. T.M.G., T.S., and W.A.C. analysed the electrophysiological results. All authors contributed to the interpretation of the structures in light of the physiological data. L.T., N.Z., and W.A.C. wrote the manuscript with input from all co-authors.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
References
- 1.Zamponi GW, Striessnig J, Koschak A, Dolphin AC. The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol Rev. 2015;67:821–870. doi: 10.1124/pr.114.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hondeghem LM, Katzung BG. Antiarrhythmic agents: the modulated receptor mechanism of action of sodium and calcium channel-blocking drugs. Annu Rev Pharmacol Toxicol. 1984;24:387–423. doi: 10.1146/annurev.pa.24.040184.002131. [DOI] [PubMed] [Google Scholar]
- 3.Murphy KM, Gould RJ, Largent BL, Snyder SH. A unitary mechanism of calcium antagonist drug action. Proc Natl Acad Sci USA. 1983;80:860–864. doi: 10.1073/pnas.80.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catterall WA, Striessnig J. Receptor sites for Ca2+ channel antagonists. Trends Pharmacol Sci. 1992;13:256–262. doi: 10.1016/0165-6147(92)90079-l. [DOI] [PubMed] [Google Scholar]
- 5.Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annu Rev Pharmacol Toxicol. 1997;37:361–396. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- 6.Striessnig J. Pharmacology, structure and function of cardiac L-type Ca2+ channels. Cell Physiol Biochem. 1999;9:242–269. doi: 10.1159/000016320. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann F, Lacinová L, Klugbauer N. Voltage-dependent calcium channels: from structure to function. Rev Physiol Biochem Pharmacol. 1999;139:33–87. doi: 10.1007/BFb0033648. [DOI] [PubMed] [Google Scholar]
- 8.Cheng RC, Tikhonov DB, Zhorov BS. Structural model for phenylalkylamine binding to L-type calcium channels. J Biol Chem. 2009;284:28332–28342. doi: 10.1074/jbc.M109.027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tikhonov DB, Zhorov BS. Structural model for dihydropyridine binding to L-type calcium channels. J Biol Chem. 2009;284:19006–19017. doi: 10.1074/jbc.M109.011296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi M, Seagar MJ, Jones JF, Reber BF, Catterall WA. Subunit structure of dihydropyridine-sensitive calcium channels from skeletal muscle. Proc Natl Acad Sci USA. 1987;84:5478–5482. doi: 10.1073/pnas.84.15.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanabe T, et al. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- 12.Mikami A, et al. Primary structure and functional expression of the cardiac dihydropyridine-sensitive calcium channel. Nature. 1989;340:230–233. doi: 10.1038/340230a0. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015;350:aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- 14.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 15.Catterall WA, Zheng N. Deciphering voltage-gated Na+ and Ca2+ channels by studying prokaryotic ancestors. Trends Biochem Sci. 2015;40:526–534. doi: 10.1016/j.tibs.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. Voltage-gated calcium channels: introduction. IUPHAR/BPS Guide to Pharmacology. 2011 ( http://guidetopharmacology.org/GRAC/FamilyIntroductionForward?familyId=80)
- 21.Striessnig J, Murphy BJ, Catterall WA. Dihydropyridine receptor of L-type Ca2+ channels: identification of binding domains for [3H](+)-PN200-110 and [3H]azidopine within the α1 subunit. Proc Natl Acad Sci USA. 1991;88:10769–10773. doi: 10.1073/pnas.88.23.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi S, et al. Key roles of Phe1112 and Ser1115 in the pore-forming IIIS5-S6 linker of L-type Ca2+ channel α1C subunit (CaV 1.2) in binding of dihydropyridines and action of Ca2+ channel agonists. Mol Pharmacol. 2003;64:235–248. doi: 10.1124/mol.64.2.235. [DOI] [PubMed] [Google Scholar]
- 23.Hescheler J, Pelzer D, Trube G, Trautwein W. Does the organic calcium channel blocker D600 act from inside or outside on the cardiac cell membrane? Pflügers Archiv. 1982;393:287–291. doi: 10.1007/BF00581411. [DOI] [PubMed] [Google Scholar]
- 24.Striessnig J, Glossmann H, Catterall WA. Identification of a phenylalkylamine binding region within the α1 subunit of skeletal muscle Ca2+ channels. Proc Natl Acad Sci USA. 1990;87:9108–9112. doi: 10.1073/pnas.87.23.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockerman GH, Johnson BD, Scheuer T, Catterall WA. Molecular determinants of high affinity phenylalkylamine block of L-type calcium channels. J Biol Chem. 1995;270:22119–22122. doi: 10.1074/jbc.270.38.22119. [DOI] [PubMed] [Google Scholar]
- 26.Yatani A, Brown AM. The calcium channel blocker nitrendipine blocks sodium channels in neonatal rat cardiac myocytes. Circ Res. 1985;56:868–875. doi: 10.1161/01.res.56.6.868. [DOI] [PubMed] [Google Scholar]
- 27.Kass RS, Arena JP, Chin S. Block of L-type calcium channels by charged dihydropyridines. Sensitivity to side of application and calcium. J Gen Physiol. 1991;98:63–75. doi: 10.1085/jgp.98.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bangalore R, Baindur N, Rutledge A, Triggle DJ, Kass RS. L-type calcium channels: asymmetrical intramembrane binding domain revealed by variable length, permanently charged 1,4-dihydropyridines. Mol Pharmacol. 1994;46:660–666. [PubMed] [Google Scholar]
- 29.Peterson BZ, Catterall WA. Calcium binding in the pore of L-type calcium channels modulates high affinity dihydropyridine binding. J Biol Chem. 1995;270:18201–18204. doi: 10.1074/jbc.270.31.18201. [DOI] [PubMed] [Google Scholar]
- 30.Glossmann H, Ferry DR, Goll A, Striessnig J, Zernig G. Calcium channels and calcium channel drugs: recent biochemical and biophysical findings. Arzneimittelforschung. 1985;35:1917–1935. [PubMed] [Google Scholar]
- 31.Gamal El-Din TM, Martinez GQ, Payandeh J, Scheuer T, Catterall WA. A gating charge interaction required for late slow inactivation of the bacterial sodium channel NavAb. J Gen Physiol. 2013;142:181–190. doi: 10.1085/jgp.201311012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 33.Faham S, et al. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. PyMol molecular viewer (V.1.2r3pre) 2002 ( http://www.pymol.org)