Abstract
Nature utilizes two groups of enzymes to catalyze methane conversions, methyl-coenzyme M reductases (MCRs) and methane monooxygenases (MMOs). These enzymes have been difficult to incorporate into industrial processes due to their complexity, poor stability, and lack of recombinant tractability. Despite these issues, new ways of preparing and stabilizing these enzymes have recently been discovered, and new mechanistic insight into how MCRs and MMOs break the C-H bond in nature’s most inert hydrocarbon haved been obtained. This review focuses on recent findings in the methane biocatalysis field, and discusses the impact of these finding on designing MMO and MCR-based biotechnologies.
Graphical Abstract
Introduction
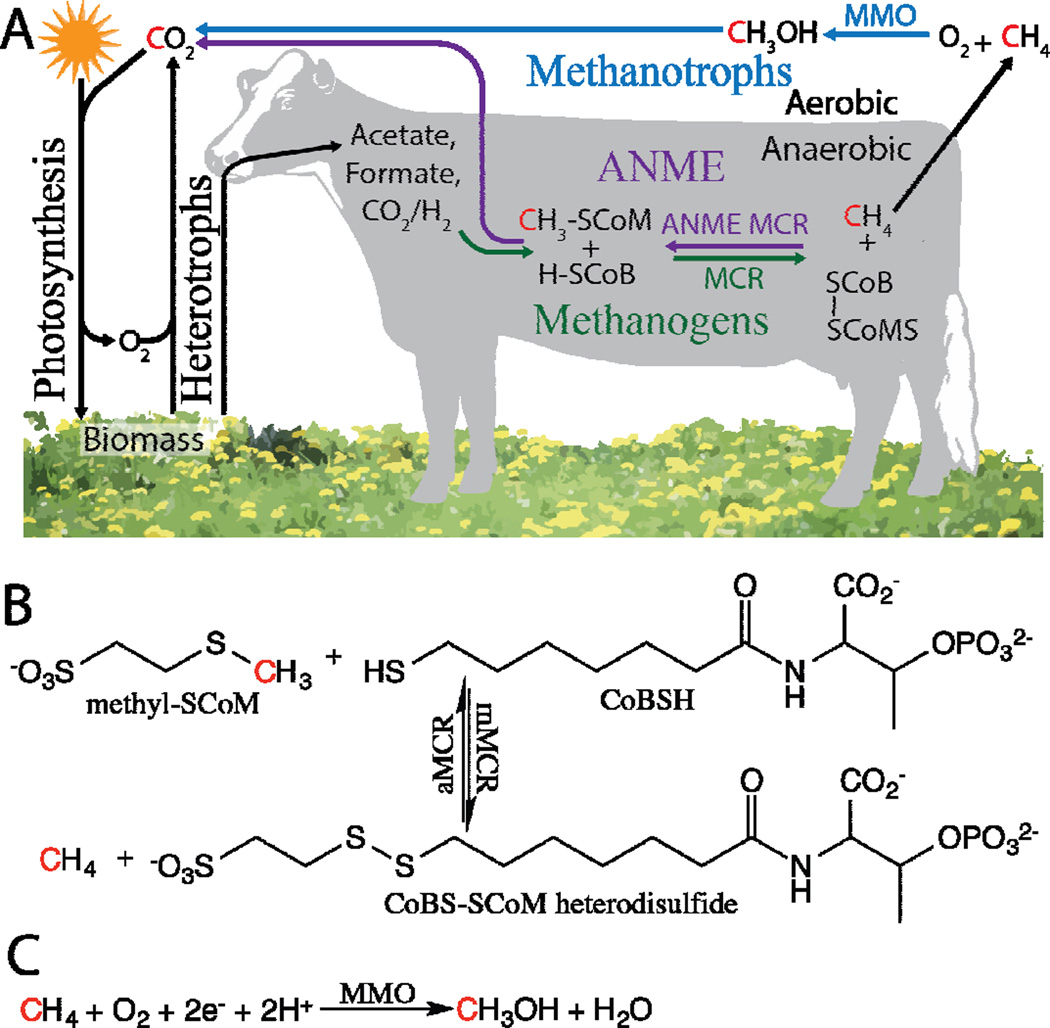
Methane is a potent greenhouse gas formed geologically in anaerobic environments from the thermal decomposition of kerogen (fossil fuel), from the reduction of oxidized carbon compounds by archaeal methanogens, and possibly by abiotic synthesis [1,2]. Globally, 500–600 million metric tons of methane are emitted to the atmosphere per year with 69% of emissions derived from microbial sources (Figure 1A) [1]. As the most reduced carbon molecule in the cycle, methane is an excellent carbon and energy source, and is utilized by anaerobic methane oxidizing archaea (ANME) and aerobic methane oxidizing bacteria (methanotrophs) (Figure 1A), which have been reported to mitigate 10–60% and 20% of methane emissions in some environments, respectively [1,3–5]. The recent availability of cheap natural gas has sparked renewed interest in biological methane oxidation from the biotechnology sector, and recent advances in understanding the microbiology, genetics, and metabolism of methane oxidation now suggest that biological conversion of methane gas to liquid fuels (Bio-GTL) is scientifically possible and economically viable [6–12]. Moreover, since methane can be produced from waste feedstocks via methanogenesis, Bio-GTL has the potential to be a renewable energy technology [13]. The only enzymes known to catalyze biological methane conversions are methyl-coenzyme M reductases (MCRs) and methane monooxygenases (MMOs) [14–16].
Figure 1.
(A) The role of methane in the biological carbon cycle. The cow is representative of anaerobic environments, not just the bovine ruminant. No indication of how cows release methane is implied. (B) Reversible anaerobic chemistry catalyzed by MCRs. (C) Aerobic chemistry catalyzed by MMOs.
MCRs catalyze anaerobic methane conversions whereas MMOs catalyze aerobic methane oxidation [14–16]. Methanogenic MCR (mMCR) catalyzes the final step of methane synthesis in methanogens using a nickel tetrapyrrole active site (F430) to couple reductive demethylation of methyl-coenzyme M (methyl-SCoM) to the oxidation of coenzyme B (CoBSH), resulting in the formation of the CoBS-SCoM heterodisulfide and methane (Figures 1B, 2A) [17]. In ANME, an MCR homolog (aMCR) is believed to catalyze the reverse reaction, the oxidation of methane and the reduction of the CoBS-SCoM heterodisulfide (Figure 1B) [18]. While nickel cofactors are used exclusively in anaerobic methane conversions, the soluble and particulate MMOs (sMMO and pMMO), use iron and copper, respectively, to aerobically oxidize methane to methanol with the production of one water molecule and the input of two electrons (Figures 1C, 2B, 2C) [15,16]. Of the two distinct MMOs, most methanotrophs express pMMO [8,19], a few only possess the genes for sMMO [20], and a subset can express either sMMO or pMMO, depending on the availability of copper [21].
Figure 2.
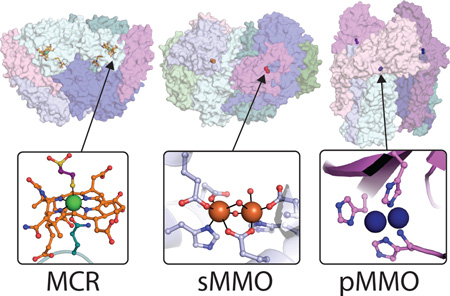
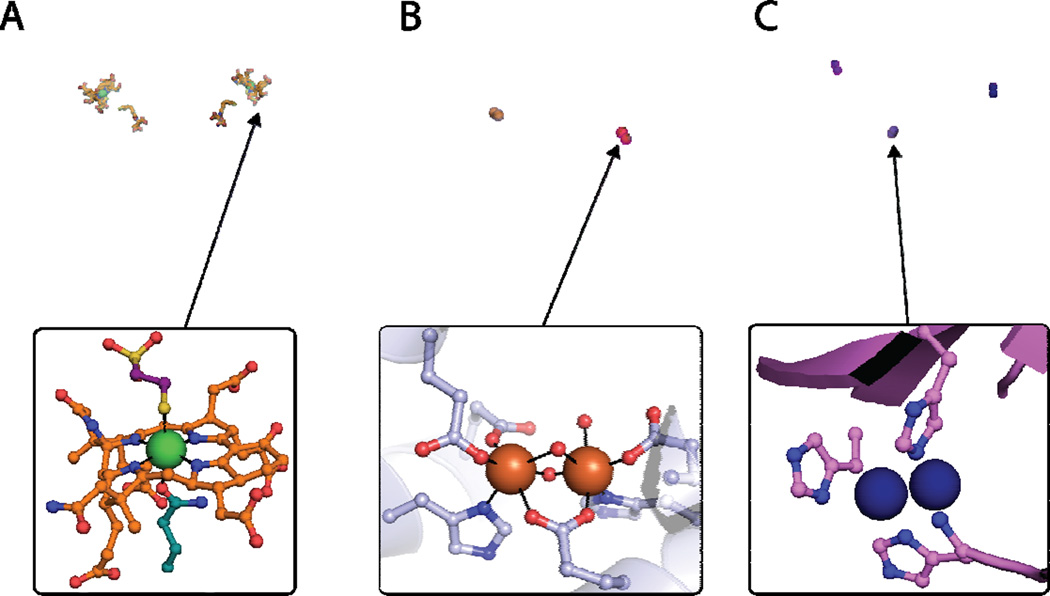
(A) Crystal structure of the MCR dimer from Methanothermobacter marburgenesis (PDB code 1MRO) with cofactors shown as orange sticks, α subunits shown in light cyan and teal, β subunits shown in light blue and blue, and γ subunits shown in pink and magenta (top). The MCRox1-silent active site is shown with F430 in orange, the Ni ion in green, and methyl-SCoM in purple. (B) Crystal structure of the MMOH-MMOB complex from Methylococcus capsulatus (Bath) (PDB code 4GAM) with iron ions shown in orange, α subunits shown in shades of blue, β subunits shown in teal and light cyan, γ subunits shown in green, and MMOB shown in pink and magenta (top). The oxidized form of the MMOH diiron site from M. capsulatus (Bath) (PDB code 1MTY). (C) Crystal structure of the pMMO trimer from Methylococcus capsulatus (Bath) (PDB code 3RGB) with copper ions shown as blue spheres, PmoB subunits shown in shades of purple and pink, PmoA subunits shown in shades of blue, and PmoC subunits shown in shades of teal and cyan. The active site of pMMO is shown modeled as a dicopper center (bottom).
Studies of MMOs and MCRs were initiated in the 1950s and 1970s, respectively [22,23]. Major accomplishments in the field include identification of their active sites, determination of their crystal structures, and elucidation of key catalytic intermediates [14,15,24–29]. Their suitability for Bio-GTL applications has been recently reviewed in depth [6,30–34]. Likewise, much is known about the genetics and regulation of these enzymes, as well as the enzymes involved in the up/downstream portions of the methane metabolic pathways [4,8,19,21,22,35]. As interest in exploiting methane-oxidizing enzymes for biotechnology applications intensifies, it is important to identify new ways to utilize these enzymes and to study the basic factors that affect their performance. In this Current Opinion, we highlight major findings from the last three years, emphasizing their relevance to Bio-GTL applications.
MCR: Advances in recombinant expression and mechanism
MCRs are the slowest methane-oxidizing enzymes and biosynthesis of their cofactors is more challenging than the biosynthesis of cofactors involved in aerobic methane oxidation [34]. However, the anaerobic C-C bond forming pathways found in ANME are expected to outperform methanotroph pathways in Bio-GTL processes [6,30], rendering MCRs an important area of research. MCRs have been difficult to study due to the challenges in obtaining active enzyme. mMCR, which is capable of catalyzing the reverse reaction [36], must be isolated under strict anaerobic conditions to retain the active Ni(I) state referred to as MCRred1 [37]. aMCR has never been isolated in an active form. The crystal structures of both enzymes have been determined, and reveal an (αβγ)2 homodimer with the F430 cofactor noncovalently bound near the interface of the three subunits, buried deep within the protein [27,38] (Figure 2A). While aMCR remains mechanistically uncharacterized, the mechanism of mMCR has been pursued extensively, leading to the characterization of 16 spectroscopically distinct states and two major mechanistic proposals (Figure 3) [25,29].
Figure 3.
Possible Ni intermediates in the MCR mechanism.
Ideally, anaerobic Bio-GTL technology would employ aMCR, but very little is known about how this enzyme works. In general, expressing MCRs in traditional recombinant hosts is not feasible, mainly owing to the complex biosynthetic machinery required to generate the F430 cofactor [39,40]. ANME organisms grow syntropically in consortia with bacterial reducers that transfer reducing equivalents from methane to terminal electron acceptors, including sulfur, oxidized metals, and nitrate [41–44]. As a result, ANME are extremely difficult to grow, and purification of large quantities of aMCR from native organisms grown in the laboratory is not feasible [5]. In addition, the choice of a terminal electron acceptor is a critical parameter in designing anaerobic Bio-GTL systems [34]. Crystals for the aMCR structure determination, which represents the only available in vitro data for aMCR, were obtained from a heterogeneous sample; this material came from microbial mats retrieved from the Black Sea floor [27]. However, expression of aMCR in the genetically tractable methanogen Methanosarcina acetovorans has been demonstrated recently [45]. The presence of aMCR was confirmed via Western blotting, and the engineered organism was capable of consuming methane at a rate twice that of the control strain when coupled to iron reduction; growth of methane was verified using isotopically labeled methane and bicarbonate. Whether functional aMCR can be isolated from this system remains to be seen, but this is clearly the most practical method for producing the enzyme since it does not involve the use of a submersible. Similarly, reversing the methanogen carbon assimilation pathway is an encouraging first step toward designing anaerobic Bio-GTL technology.
Another way to use MCR in methane oxidation pathways is to engineer mMCR to favor the reverse reaction. There are some structural differences between mMCR and aMCR [27,38], but whether these features relate to the direction of the reaction remains unclear. Understanding the mechanisms of mMCR and aMCR is critical to realizing this possibility. The mMCR catalytic cycle begins with F430 in the reduced Ni(I) form, MCRred1, and it is known that methyl-SCoM, the methyl donor, binds first followed by CoBSH, the electron donor, yielding the CoBS-SCoM heterodisulfide and methane [25,46]. Following binding of methyl-SCoM to the electron paramagnetic resonance (EPR)-active MCRred1 state, two possible Ni-bound intermediates have been proposed, an EPR-active methyl-Ni(III) species (mechanism I) and an EPR-silent Ni(II)-thiolate (mechanism II) (Figure 3) [14,25,29]. Using a truncated CoBSH analog (CoB6SH) that decreases the reaction rate, this intermediate has been trapped and characterized [29]. In this work, stopped-flow experiments and EPR analysis of rapid freeze quench samples in conjunction with rapid chemical quench activity assays indicate that MCRred1 decays to an EPR-silent state with concomitant release of methane. Not only is the disappearance of the EPR signal consistent with mechanism II, but if the reaction followed mechanism I, simultaneous decay of MCRred1 and release of methane would not be expected due to the comparatively more stable methyl-Ni(III) intermediate. The EPR-silent intermediate was identified by comparing the magnetic circular dichroism (MCD) spectra of RFQ samples with those of known mMCR states: the intermediate clearly resembles the previously characterized MCRox1-silent state in which Ni is coordinated by the thiolate group of SCoM (Figure 2A) [38,47]. While these findings represent a major leap forward in understanding the MCR mechanism, observations that support mechanism I still need to be reconciled with the new data [48,49], and it remains unclear whether the proposed radical intermediates can be detected [50]. Perhaps most important for Bio-GTL is knowing what factors affect the direction of the reaction and whether aMCR simply proceeds via the reverse mechanism.
sMMO: The structure of Q and new insight into protein-protein interactions
As the fastest and the most well understood methane-oxidizing enzyme [16,24,28], sMMO is arguably the most viable target for Bio-GTL [34]. Overall, it is the most tractable methane-oxidizing enzyme for functional recombinant expression, but attempts to do so have been minimally successful [34,51]. sMMO comprises three components: a regulatory protein (MMOB) required for efficient methane oxidation, a reductase (MMOR), and the active site-containing component (MMOH) [15,16]. MMOH is an (αβγ)2 homodimer, of which the α subunit contains a diiron center coordinated by four glutamates, two histidines, and several water molecules (Figure 2B) [52]. Electrons for methane oxidation are transferred from NADH to this active site via FAD and [2Fe-2S] clusters in MMOR [15,16]. Extensive kinetic, spectroscopic, and computational studies have established the key intermediates in the sMMO catalytic cycle (Figure 4), and provide constraints on their possible structures [16,24]. Recent work has resulted in structural characterization of the key reactive intermediate [53] as well as molecular level insight into how all three sMMO components work in concert to drive methane oxidation [54–56].
Figure 4.
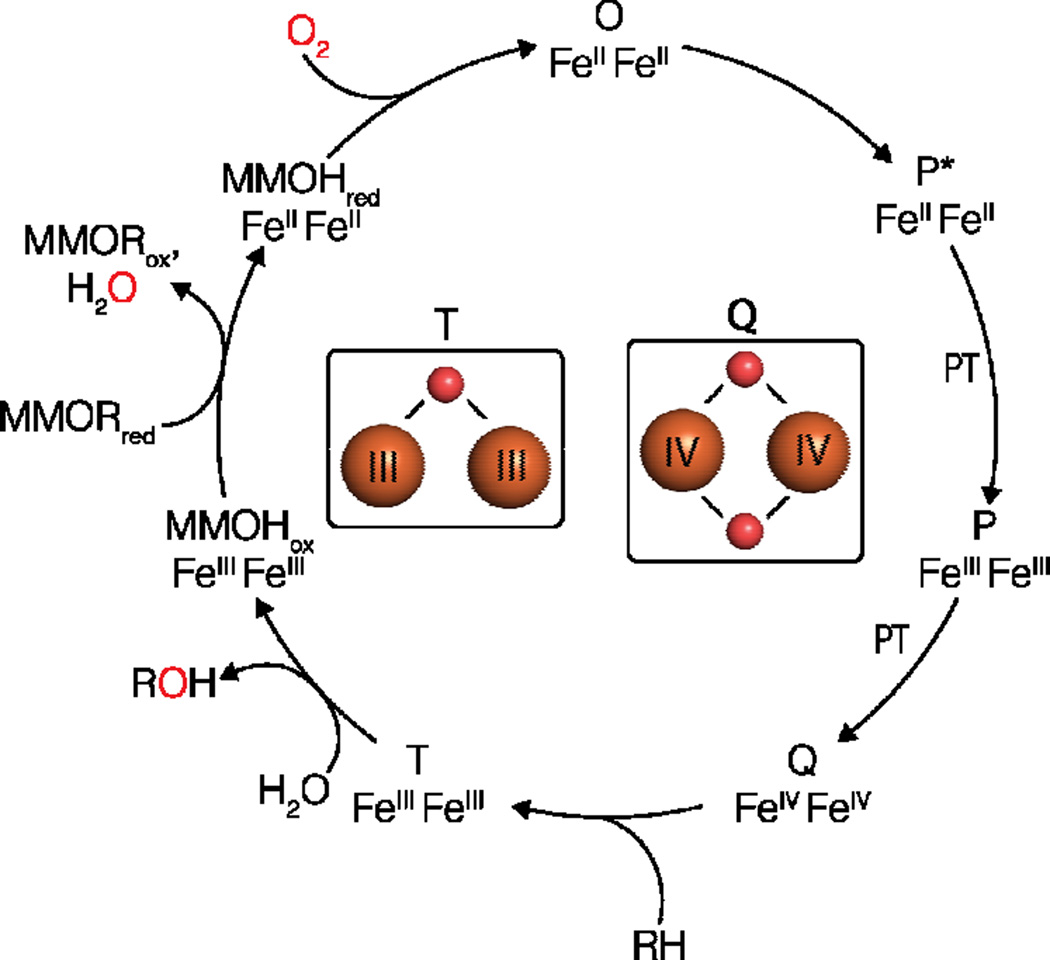
The catalytic cycle of sMMO with the structures of intermediates Q and T recently assigned by resonance Raman spectroscopic studies [52]. Iron ions are shown as orange spheres and oxygen atoms are shown as red spheres.
The mechanism of sMMO has been reviewed in depth recently [16,24]. Briefly, the diiron(III) site of MMOH (MMOHox) is reduced by MMOR in two sequential electron transfer events to the diiron(II) state (MMOHred) [28]. Oxygen reacts with MMOHred to form intermediate O, followed by the peroxo intermediates P*, a diiron(II) species [57], and P [57–59], a peroxo-bridged diiron(III) species that is converted to the diiron(IV) intermediate Q, which is defined by its characteristic absorption feature at 420 nm [16,59,60]. Q is believed to react with methane resulting in the formation of the product complex, T [16]. The structures of MMOHox and MMOHred have been determined by X-ray crystallography (Figure 2B) [52,61], but molecular details of the intermediates have been elusive due to their transient nature. As the methane-oxidizing species, Q has been of special interest, and a number of open core and closed core structures have been proposed [16,24]. This issue has been addressed recently by time-resolved resonance Raman spectroscopy. In this study, the 16O2 (Δ18O2) vibration frequency of Q was identified at 690 (−36) cm−1, and assigned as a diamond di-(μ-oxo) diferryl core based on comparison to model compound data [53]. The data also suggest that Q forms via homolytic cleavage of the O-O bond. The trick to finally detecting Q’s vibration was to react MMOHred-MMOB alternately with buffer containing either 16O2 or 18O2 and collect difference spectra, which exhibit only vibrations originating from dioxygen. In addition, the structure of T was assigned as a mono-(μ-oxo) diferric core. These findings set the stage for further spectroscopic and computational investigation of Q.
Other recent studies have focused on how MMOR and MMOB together control the delivery of electrons, protons, and methane to the diiron site. It has long been established that MMOB increases the reaction rate of sMMO with dioxygen by 2–3 orders of magnitude, and that binding of MMOB to MMOH alters the electronic structure and reduces the reduction potential of the diiron center [15]. The crystal structure of the MMOB-MMOH complex reveals that MMOB binds in the so-called canyon region of MMOH (Figure 2B), with its N-terminal 35 residues forming a ringlike structure. Complexation closes a hydrophilic pore suggested to play a role in proton transfer and product release and opens a channel between the diiron site and several proposed substrate-binding cavities [54]. MMOB also induces structural changes at the active site, but this could also be attributed to photoreduction of the diiron site during data collection. Double electron-electron resonance (DEER) experiments indicate that the MMOB N-terminus is more rigid when MMOH is reduced and fluorescence anistropy measurements suggest that MMOB has a higher affinity for MMOHred than MMOHox [55]. It is unclear how these data relate to the lowered reduction potential of MMOHox in the presence of MMOB, which suggests that MMOB stabilizes the diiron(III) state. Regardless, these results strongly suggest that MMOB gates access to the diiron site. Multiple lines of new evidence indicate that MMOR also binds in the canyon region. These data includes crosslinking studies, hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS), computational docking, and the crystal structure of the MMOH homolog toluene 4-monooxygenase hydroxylase with its reductase component bound in the same location as its regulatory protein binds [56,62]. In addition, MMOB inhibits MMOR binding and electron transfer [56,62].
These findings have led to a proposed model in which MMOR reduces MMOHox to MMOHred followed by binding of MMOB to open the substrate channel [28]. Upon return to the MMOHox state, MMOR is proposed to replace MMOB, which would allow product release and another cycle of catalysis. This model is consistent with the observations that suggest MMOB and MMOR occupy the same site, but replacement of MMOB by MMOR has not been established experimentally. It is also not clear how to reconcile previously reported complex formation between MMOR and MMOB with them having the same binding site on MMOH [63]. Regardless, the emerging picture of sMMO’s mechanism is one in which MMOB and MMOR interact with MMOH in a highly choreographed three-way dance to trigger specific events within the catalytic site. Such a mechanism avoids futile turnover and prevents oxidative damage to the protein. It also presents special challenges for using sMMO in BioGTL applications. Not only must industrial hosts functionally express the sMMO subunits and incorporate the iron cofactor, but the relative expression levels of the individual components must be tuned to ensure maximal activity.
pMMO: The active site, native reductant, and printable bioreactors
pMMO presents several unique challenges for characterization and for use in Bio-GTL applications [34]. First, pMMO is an integral membrane protein that is natively expressed in extensive intracytoplasmic membranes (ICMs) [15,16]. The crystal structure of pMMO reveals an (αβγ)3 homotrimer comprising the subunits PmoB, PmoA, and PmoC (Figure 2C) [64]; the N-terminus of PmoB is located in the periplasm whereas the N-termini of PmoA and PmoC are cytoplasmic. Recombinant expression of intact pMMO has been minimally successful, likely due in part to not understanding the role of the ICMs as well as to difficulties assembling the three subunits correctly [65]. The active site is believed to be a copper center coordinated by three histidine residues in the N-terminal periplasmic region of PmoB [66], which is also problematic for recombinant expression since excess copper is effluxed in most recombinant hosts [67]. pMMO is difficult to maintain in an active form, with 10-fold loss in methane oxidation activity occurring after lysis, and an additional 10-fold loss observed upon solubilization and purification from the native membranes [15]. Thus, spectroscopic and kinetic characterization of active pMMO has been hindered, and spectroscopic analysis of the active site is further complicated by copper binding at multiple locations within the pMMO structure [15,34].
Given these issues, recent investigations of pMMO have continued to address the nature of the copper active site as well as the roles of other observed metal binding sites. The active site was identified using a recombinant, soluble fragment of the PmoB subunit denoted spmoB [66], and recent mutagenesis studies of this protein support the assignment [68]. While the active site is suggested to consist of two copper ions on the basis of X-ray absorption and optical spectroscopic data [15], a single copper ion was modeled in recent structures of pMMO from Methylocystis sp. strain Rockwell [69] and of a soluble fragment of a related ammonia monooxygenase subunit [70]. However, the lack of or low activity of these samples precludes drawing any conclusions regarding the nuclearity. Some attention has also focused on a second metal binding site present in the PmoC subunit that can be occupied with zinc if crystallization is performed in zinc-containing buffers. Mutagenesis studies of a related hydrocarbon monooxygenase suggest that this site is functionally relevant, and recent data show that zinc partially inhibits pMMO, likely by binding at this site [69,71].
Whereas reduction of MMOH by NADH via MMOR is well understood, the native reductant of pMMO remains controversial. In one model, ubiquinol generated by a type 2 NADH:quinone oxidoreductase is used, consistent with the empirical use of duroquinol for in vitro reduction of pMMO [15,34]. Another possibility, first suggested 30 years ago [72], is that methanol oxidation by methanol dehydrogenase (MDH) is coupled to methane oxidation, providing electrons via the electron acceptor of MDH, cytochrome cL. Interestingly, recent metabolic modeling studies of the methanotroph Methylomicrobium buryatense 5G are most consistent with this pathway or with a related model in which methanol oxidation partially supports methane oxidation [10]. Also in support of this model, biolayer interferometry studies reveal a protein-protein interaction between pMMO and MDH. The interaction is also observed for the spmoB protein, consistent with the localization of MDH in the periplasm [73].
One way to circumvent issues with expression in an industrial host is to use isolated enzymes instead. This strategy has recently been pursued for pMMO. As the major component of methanotroph membranes, reasonably pure pMMO can be isolated by simple cell lysis and centrifugation, but as a membrane protein, it is not amenable to typical enzyme immobilization approaches. A biocatalytic material was created by embedding membrane-bound pMMO in polyethylene glycol diacrylate (PEGDA) hydrogels. Remarkably, this PEG-pMMO hydrogel exhibits similar activity to membranes containing pMMO, and can be washed and reused without significant loss in activity. The PEG-pMMO material was also printed into a 3D silicone lattice and tested in a miniature continuous bioreactor, which was capable of stable methanol production for over two hours [74]. Although there are still many limitations to this approach (e.g. using NADH as a reductant, improving pMMO rates, conversion of methanol to a high value product), unique reactor designs like this have the potential to overcome mass transfer issues associated with gas fermentation processes. It is also encouraging that these materials can stabilize pMMO for long periods of time. Overall, these materials are promising, and open up interesting possibilities for Bio-GTL applications.
Conclusion
What a successful Bio-GTL process will look like remains unclear, but it will inevitably be dependent on how the methane-oxidizing enzyme is produced. It is possible that recent technological developments will lead to successful recombinant expression of a methane-oxidizing enzyme in a traditional industrial host or that genetic manipulation of the native organisms will advance enough to enable stable incorporation of foreign biosynthetic pathways. Alternatively, a hybrid cell-free approach could utilize enzymes produced in separate organisms. These uncertainties in the future design of Bio-GTL applications underscore the importance of studying MMOs and MCRs.
Many of the discoveries highlighted here have the potential to impact future Bio-GTL applications. The work on MCRs and pMMO opens up whole new possibilities within the realm of biotechnology. Recombinant expression of aMCR and reversal of the methanogen biosynthetic pathways suggests that synthetic biology based on anaerobic methane oxidation is possible, and understanding the mechanism of mMCR may lead to rational approaches to improving the reverse reaction. Similarly, work on printable pMMO polymers could lead to new types of methane-oxidizing materials. In addition, investigations into reductant utilization of methanotrophs expressing pMMO now set an upper limit on the efficiency of aerobic methane oxidation. At the same time, recent studies of sMMO emphasize the complexities of methane biocatalysis. Precisely timed protein-protein interactions are required to trigger electron transfer, oxygen binding, and finally proton transfer to form the methane reactive intermediate, Q, the structure of which is now known. The mechanisms of MCR and pMMO are likely just as complicated, and we are just starting to scratch the surface in our understanding of these enzymes. The last three years have indeed seen big progress toward breaking the small methane substrate, and the next three are likely to be just as exciting.
Highlights.
The MCR reaction proceeds via a Ni(II)-thiolate intermediate.
A recombinant expression system for ANME MCR has been developed.
The structure of a key intermediate in the sMMO catalytic cycle has been elucidated.
The crystal structure of an sMMO protein-protein complex has been determined.
Catalytically active pMMO has been incorporated into a printable polymer.
Acknowledgments
This work was supported by the Advanced Research Projects Agency-Energy (ARPA-E) REMOTE program (DE-AR0000435) and the National Institutes of Health (GM118035).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 2009;1:285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 2.Sephton MA, Hazen RM. On the origins of deep hydrocarbons. Rev Mineral Geochem. 2013;75:449–465. [Google Scholar]
- 3.Conrad R. Advances in Agronomy. Elsevier; 2007. Microbial ecology of methanogens and methanotrophs; pp. 1–63. [Google Scholar]
- 4.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knittel K, Boetius A. Anaerobic oxidation of methane: Progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 6.Haynes CA, Gonzalez R. Rethinking biological activation of methane and conversion to liquid fuels. Nat Chem Biol. 2014;10:331–339. doi: 10.1038/nchembio.1509. [DOI] [PubMed] [Google Scholar]
- 7.Kalyuzhnaya MG, Puri AW, Lidstrom ME. Metabolic engineering in methanotrophic bacteria. Metab Eng. 2015;29:142–152. doi: 10.1016/j.ymben.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova L, Lidstrom PME. Aerobic methylotrophic prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin Heidelberg: Springer; 2013. pp. 267–285. [Google Scholar]
- 9.Puri AW, Owen S, Chu F, Chavkin T, Beck DAC, Kalyuzhnaya MG, Lidstrom ME. Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol. 2015;81:1775–1781. doi: 10.1128/AEM.03795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torre A, Metivier A, Chu F, Laurens LML, Beck DAC, Pienkos PT, Lidstrom ME, Kalyuzhnaya MG. Genome-scale metabolic reconstructions and theoretical investigation of methane conversion in Methylomicrobium buryatense strain 5G(B1) Microb Cell Fac. 2015;14:188. doi: 10.1186/s12934-015-0377-3. • This study provides insight into the physiological reductant for pMMO.
- 11.Gilman A, Laurens LM, Puri AW, Chu F, Pienkos PT, Lidstrom ME. Bioreactor performance parameters for an industrially-promising methanotroph Methylomicrobium buryatense 5GB1. Microb Cell Fac. 2015;14:182. doi: 10.1186/s12934-015-0372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A, Gowda GAN, Raftery D, Fu Y, Bringel F, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4 doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- 13.Weiland P. Biogas production: Current state and perspectives. Appl Microbiol Biotechnol. 2010;85:849–860. doi: 10.1007/s00253-009-2246-7. [DOI] [PubMed] [Google Scholar]
- 14.Ragsdale SW. Metal Ions in Life Sciences. Netherlands: Springer; 2014. Biochemistry of methyl-coenzyme M reductase: The nickel metalloenzyme that catalyzes the final step in synthesis and the first step in anaerobic oxidation of the greenhouse gas methane; pp. 125–145. [DOI] [PubMed] [Google Scholar]
- 15.Sirajuddin S, Rosenzweig AC. Enzymatic oxidation of methane. Biochemistry. 2015;54:2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sazinsky MH, Lippard SJ. Methane monooxygenase: Functionalizing methane at iron and copper. In: Kroneck PMH, Sosa Torres ME, editors. Metal Ions in Life Sciences. Springer International Publishing; 2015. pp. 205–256. [DOI] [PubMed] [Google Scholar]
- 17.Ferry JG. How to make a living by exhaling methane. Annu Rev Microbiol. 2010;64:453–473. doi: 10.1146/annurev.micro.112408.134051. [DOI] [PubMed] [Google Scholar]
- 18.Thauer RK. Anaerobic oxidation of methane with sulfate: On the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Opin Microbiol. 2011;14:292–299. doi: 10.1016/j.mib.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Trotsenko YA, Murrell JC. Advances in Applied Microbiology. Elsevier; 2008. Metabolic aspects of aerobic obligate methanotrophy; pp. 183–229. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, Stott MB, Alam M, Theisen AR, Murrell JC, Dunfield PF. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J Bacteriol. 2010;192:3840–3841. doi: 10.1128/JB.00506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 22.Anthony C. The biochemistry of methylotrophs. Academic Press; 1982. [Google Scholar]
- 23.Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 24.Tinberg CE, Lippard SJ. Dioxygen activation in soluble methane monooxygenase. Acc Chem Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ermler U. On the mechanism of methyl-coenzyme M reductase. Dalton Trans. 2005;0:3451–3458. doi: 10.1039/b506697b. [DOI] [PubMed] [Google Scholar]
- 26.Culpepper MA, Rosenzweig AC. Architecture and active site of particulate methane monooxygenase. Crit Rev Biochem Mol. 2012;47:483–492. doi: 10.3109/10409238.2012.697865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shima S, Krueger M, Weinert T, Demmer U, Kahnt J, Thauer RK, Ermler U. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature. 2012;481:98–101. doi: 10.1038/nature10663. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Liang AD, Lippard SJ. Coupling oxygen consumption with hydrocarbon oxidation in bacterial multicomponent monooxygenases. Acc Chem Res. 2015;48:2632–2639. doi: 10.1021/acs.accounts.5b00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wongnate T, Sliwa D, Ginovska B, Smith D, Wolf MW, Lehnert N, Raugei S, Ragsdale SW. The radical mechanism of biological methane synthesis by methyl-coenzyme M reductase. Science. 2016;352:953–958. doi: 10.1126/science.aaf0616. •• This study definitively establishes a Ni(II)-thiolate as a key intermediate in the mechanism of MCR.
- 30.Nazem-Bokaee H, Gopalakrishnan S, Ferry JG, Wood TK, Maranas CD. Assessing methanotrophy and carbon fixation for biofuel production by Methanosarcina acetivorans. Microb Cell Fac. 2016;15:1. doi: 10.1186/s12934-015-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez R, Conrado RJ. Envisioning bioconversion of methane to liquids. Science. 2014;343:621–623. doi: 10.1126/science.1246929. [DOI] [PubMed] [Google Scholar]
- 32.Fei Q, Guarnieri MT, Tao L, Laurens LML, Dowe N, Pienkos PT. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol Adv. 2014;32:596–614. doi: 10.1016/j.biotechadv.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Strong PJ, Xie S, Clarke WP. Methane as a resource: Can the methanotrophs add value? Environ Sci Technol. 2015;49:4001–4018. doi: 10.1021/es504242n. [DOI] [PubMed] [Google Scholar]
- 34. Lawton TJ, Rosenzweig AC. Methane-oxidizing enzymes: An upstream problem in biological gas-to-liquids conversion. J Am Chem Soc. 2016 doi: 10.1021/jacs.6b04568. • This review quantitatively assesses the potential of each methane-oxidizing enzyme for Bio-GTL.
- 35.Semrau JD, DiSpirito AA, Vuilleumier S. Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett. 2011;323:1–12. doi: 10.1111/j.1574-6968.2011.02315.x. [DOI] [PubMed] [Google Scholar]
- 36.Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature. 2010;465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- 37.Goubeaud M, Schreiner G, Thauer RK. Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F430 is reduced to the nickel(I) oxidation state by titanium(III) citrate. Eur J Biochem. 1997;243:110–114. doi: 10.1111/j.1432-1033.1997.00110.x. [DOI] [PubMed] [Google Scholar]
- 38.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Crystal structure of methyl-coenzyme M reductase: The key enzyme of biological methane formation. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 39.Graham DE, White RH. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat Prod Rep. 2002;19:133–147. doi: 10.1039/b103714p. [DOI] [PubMed] [Google Scholar]
- 40.Warren MJ, Deery E, Rose R-S. Tetrapyrroles. New York: Springer; 2009. Biosynthesis of siroheme and coenzyme F430; pp. 344–351. [Google Scholar]
- 41.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damsté JSS, Camp den HJMO, Jetten MSM, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 42.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 43.Beal EJ, House CH, Orphan VJ. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 44.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 45. Soo VWC, McAnulty MJ, Tripathi A, Zhu F, Zhang L, Hatzakis E, Smith PB, Agrawal S, Nazem-Bokaee H, Gopalakrishnan S, et al. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb Cell Fac. 2016;15:1. doi: 10.1186/s12934-015-0397-z. • This study describes the first recombinant system for aMCR.
- 46.Wongnate T, Ragsdale SW. The reaction mechanism of methyl-coenzyme M reductase: how an enzyme enforces strict binding order. J Biol Chem. 2015;290:9322–9334. doi: 10.1074/jbc.M115.636761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Craft JL, Horng Y-C, Ragsdale SW, Brunold TC. Nickel oxidation states of F430 cofactor in methyl-coenzyme M reductase. J Am Chem Soc. 2004;126:4068–4069. doi: 10.1021/ja038082p. [DOI] [PubMed] [Google Scholar]
- 48.Dey M, Li X, Kunz RC, Ragsdale SW. Detection of organometallic and radical intermediates in the catalytic mechanism of methyl-coenzyme M reductase using the natural substrate methyl-coenzyme M and a coenzyme B substrate analogue. Biochemistry. 2010;49:10902–10911. doi: 10.1021/bi101562m. [DOI] [PubMed] [Google Scholar]
- 49.Scheller S, Goenrich M, Mayr S, Thauer RK, Jaun B. Intermediates in the Catalytic Cycle of Methyl Coenzyme M Reductase: Isotope Exchange is Consistent with Formation of a σ–Alkane–Nickel Complex. Angew Chem Int Edit. 2010;49:8112–8115. doi: 10.1002/anie.201003214. [DOI] [PubMed] [Google Scholar]
- 50.Lawton TJ, Rosenzweig AC. Methane—make it or break it. Science. 2016;352:892–893. doi: 10.1126/science.aaf7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jahng D, Wood TK. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1994;60:2473–2482. doi: 10.1128/aem.60.7.2473-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 53. Banerjee R, Proshlyakov Y, Lipscomb JD, Proshlyakov DA. Structure of the key species in the enzymatic oxidation of methane to methanol. Nature. 2015;518:431–434. doi: 10.1038/nature14160. •• This study assigns intermediate Q of sMMO as a diamond di-(μ-oxo) diferryl core.
- 54. Lee SJ, McCormick MS, Lippard SJ, Cho U-S. Control of substrate access to the active site in methane monooxygenase. Nature. 2013;494:380–384. doi: 10.1038/nature11880. •• This study reports the first crystal structure of MMOH in complex with MMOB.
- 55.Wang W, Lippard SJ. Diiron oxidation state control of substrate access to the active site of soluble methane monooxygenase mediated by the regulatory component. J Am Chem Soc. 2014;136:2244–2247. doi: 10.1021/ja412351b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Iacob RE, Luoh RP, Engen JR, Lippard SJ. Electron transfer control in soluble methane monooxygenase. J Am Chem Soc. 2014;136:9754–9762. doi: 10.1021/ja504688z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerjee R, Meier KK, Münck E, Lipscomb JD. Intermediate P* from soluble methane monooxygenase contains a diferrous cluster. Biochemistry. 2013;52:4331–4342. doi: 10.1021/bi400182y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Nesheim JC, Lee SK, Lipscomb JD. Gating effects of component B on oxygen activation by the methane monooxygenase hydroxylase component. J Biol Chem. 1995;270:24662–24665. doi: 10.1074/jbc.270.42.24662. [DOI] [PubMed] [Google Scholar]
- 59.Tinberg CE, Lippard SJ. Revisiting the mechanism of dioxygen activation in soluble methane monooxygenase from M. capsulatus (Bath): Evidence for a multi-step, proton-dependent reaction pathway. Biochemistry. 2009;48:12145–12158. doi: 10.1021/bi901672n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S-K, Lipscomb JD. Oxygen activation catalyzed by methane monooxygenase hydroxylase component: Proton delivery during the O–O bond cleavage steps. Biochemistry. 1999;38:4423–4432. doi: 10.1021/bi982712w. [DOI] [PubMed] [Google Scholar]
- 61.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem Biol. 1995;2:409–418. [PubMed] [Google Scholar]
- 62.Acheson JF, Bailey LJ, Elsen NL, Fox BG. Structural basis for biomolecular recognition in overlapping binding sites in a diiron enzyme system. Nat Commun. 2014;5:5009. doi: 10.1038/ncomms6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox BG, Liu Y, Dege JE, Lipscomb JD. Complex formation between the protein components of methane monooxygenase from Methylosinus trichosporium OB3b. J Biol Chem. 1991;266:540–550. [PubMed] [Google Scholar]
- 64.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 65.Gou Z, Xing X-H, Luo M, Jiang H, Han B, Wu H, Wang L, Zhang F. Functional expression of the particulate methane mono-oxygenase gene in recombinant Rhodococcus erythropolis. FEMS Microbiol Lett. 2006;263:136–141. doi: 10.1111/j.1574-6968.2006.00363.x. [DOI] [PubMed] [Google Scholar]
- 66.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Festa RA, Thiele DJ. Copper: An essential metal in biology. Curr Biol. 2011;21:877–883. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Culpepper MA, Cutsail GE, III, Gunderson WA, Hoffman BM, Rosenzweig AC. Identification of the valence and coordination environment of the particulate methane monooxygenase copper centers by advanced EPR characterization. J Am Chem Soc. 2014;136:11767–11775. doi: 10.1021/ja5053126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sirajuddin S, Barupala D, Helling S, Marcus K, Stemmler TL, Rosenzweig AC. Effects of zinc on particulate methane monooxygenase activity and structure. J Biol Chem. 2014;289:21782–21794. doi: 10.1074/jbc.M114.581363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawton TJ, Ham J, Sun T, Rosenzweig AC. Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins. 2014;82:2263–2267. doi: 10.1002/prot.24535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL, Holmes AJ. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. The ISME Journal. 2011;6:171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leak DJ, Dalton H. Growth yields of methanotrophs 2. A theoretical analysis. Appl Microbiol Biotechnol. 1986;23:477–481. [Google Scholar]
- 73.Culpepper MA, Rosenzweig AC. Structure and protein–protein interactions of methanol dehydrogenase from Methylococcus capsulatus (Bath) Biochemistry. 2014;53:6211–6219. doi: 10.1021/bi500850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Blanchette CD, Knipe JM, Stolaroff JK, DeOtte JR, Oakdale JS, Maiti A, Lenhardt JM, Sirajuddin S, Rosenzweig AC, Baker SE. Printable enzyme-embedded materials for methane to methanol conversion. Nat Commun. 2016;7:11900. doi: 10.1038/ncomms11900. • This report describes the first example of embedding a methane-oxidizing enzyme in a polymer.