Abstract
Short interfering RNA (siRNA)-triggered gene knockdown via the RNA interference (RNAi) pathway is widely used to study gene function and siRNA-based therapeutics are in development. However, since the guide strand of an siRNA can function like a natural microRNA (miRNA), siRNAs often repress hundreds of off-target transcripts with complementary only to the seed region (nucleotides 2–8) of the guide strand. Here we describe novel guide strand 3′ end modifications derived from 1-ethynylribose and copper-catalyzed azide/alkyne cycloaddition reactions and evaluate their impact on target vs miRNA-like off-target knockdown. Surprisingly, when positioned at the guide strand 3′ end the parent 1-ethynylribose modification substantially reduced off-target knockdown while having no measurable effect on on-target knockdown potency. In addition, these modifications are shown to modulate siRNA affinity for the hAgo2 PAZ domain. However, the change in PAZ domain binding affinity is not sufficient to predict a modification’s effect on miRNA-like off targeting.
Introduction
Short interfering RNA (siRNA) is a commonly used tool in controlling gene expression in mammalian cells 1. In addition, advanced stage clinical trials of siRNA-based therapeutics are currently underway 2. A typical siRNA is composed of a passenger strand, and a complementary guide strand, typically 21 nucleotides (nt) in length with 3′ overhangs containing two nucleotides (nt) 1. Upon cellular entry, the guide strand is loaded into the protein Argonaute 2 (Ago2) 3. The guide strand directs binding to a sequence-complementary mRNA, which triggers cleavage by Ago2 resulting to gene silencing 4. Using siRNAs as therapeutics is promising, however, the development of these molecules as drugs is hindered by instability in serum, immune stimulation, and off-target effects. Chemical modification is a strategy to mitigate these effects and render the siRNAs viable for therapeutic purposes 5. A crucial drawback in using siRNAs both in therapeutics and as a gene silencing tool, is that any ~21 to 23 nt RNA binding to Ago2 can act as a miRNA, a form of endogenous noncoding RNA that also silences genes in an Ago-dependent manner 6,7. MiRNAs only require seed region (nt 2–8) binding to the target RNA to activate gene silencing 8. Therefore, siRNAs often have low target specificity, resulting in the knockdown of hundreds of off-target mRNA transcripts which can lead to unwanted phenotypes 9. Thus, development of chemical modifications that avoid miRNA-like off targeting is an active area of research.9–12
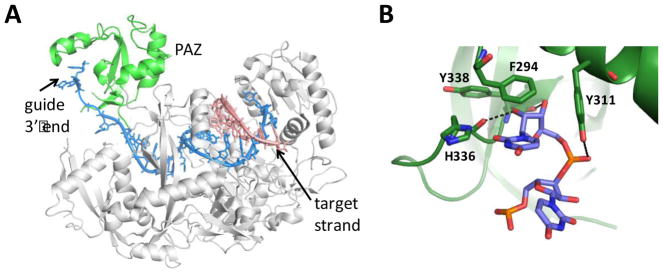
High resolution structures reveal that Ago2 has a conserved structure made up of four distinct domains – the N terminus, MID, PIWI, and the Piwi-Argonaute-Zwille (PAZ) (Figure 1) 13,14. The PAZ domain interacts with the 3′ end of the siRNA guide strand 15,16. Early structural work with bacterial Ago proteins indicated that extensive duplex formation with target beyond the guide strand seed region required the 3′ end to disengage from the PAZ domain 17. This is consistent with the “two-state” model of Zamore that suggests Ago2 initially binds target RNA by contacting the guide strand seed region while the 3′ end remains bound to the PAZ domain 18. Then, the 3′ end disengages from PAZ and the remaining duplex forms leading to the cleavage-competent complex. Indeed, a recently proposed minimal kinetic model for turnover in the human Ago2-catalyzed cleavage reaction involves consecutive guide 3′ end binding to, and release from, the PAZ domain during the catalytic cycle 19. However, since Ago2 catalytic turnover is not required for miRNA-like gene silencing and release of the 3′ end from the PAZ domain is apparently not necessary for seed-only binding, guide strands with altered PAZ domain binding properties may have reduced miRNA-like off target effects. Other groups have incorporated aromatic derivatives at the 3′ ends of siRNA guide strands and observed enhanced binding affinity to the Ago2 PAZ without compromising RNAi activity 20,21. In contrast, a computational analysis of a variety of 3′ end modifications suggested that weaker PAZ domain binding correlates with high RNAi activity 22. However, there have been no reports on how guide strand 3′ end modification can affect silencing of miRNA-like targets. Here, we describe the effects of modifications to the 3′ end of a siRNA guide strand on RNAi potency, miRNA-like off targeting and binding to the hAgo2 PAZ domain. We find that miRNA-like off targeting can indeed be reduced by modifying the guide strand 3′ end, but PAZ domain binding affinity alone is a poor predictor of effects on RNAi specificity.
Figure 1.
Recognition of the guide strand 3′ end by hAgo2. A) Structure of human Ago2 bound to guide strand (blue) and target strand (salmon) complementary to the seed region showing PAZ domain in green 23. B) Close up view of hAgo2 PAZ domain interactions with guide 3′ end nucleotides 23.
Results
Synthesis of 1-ethynyl ribose (1-ER) controlled pore glass and generation of modified guide strands
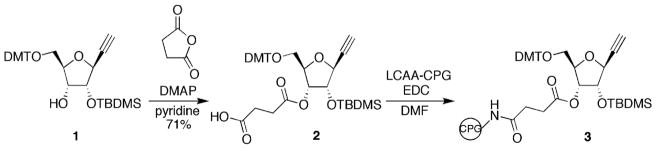
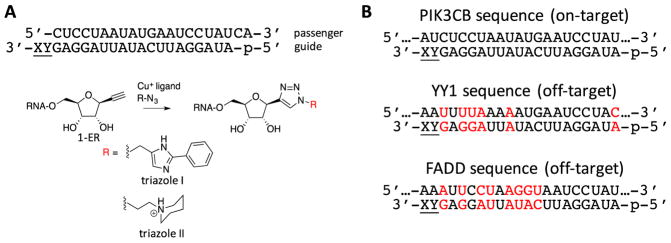
We previously described 1-ethynyl ribose (1-ER) modified RNA 24. The 1-ER nucleoside analog is a versatile precursor to nucleobase replacements in RNA via triazole-forming, copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions 12,24. In our earlier studies, 1-ER phosphoramidite was used to prepare modified RNA 12,24. However, modification of the 3′ terminal nucleotide of an oligoribonucleotide required preparation of a new solid phase synthesis support bearing 1-ER (Scheme 1). To this end, our previously reported DMT and TBDMS-protected 1-ER (1) 24 was allowed to react with succinic anhydride in pyridine in the presence of DMAP to give carboxylic acid (2) in 71% yield. Long chain alkyl amino controlled pore glass (LCAA CPG) was then treated with compound 2 and EDC in DMF to provide solid phase synthesis support 3 for incorporation of 1-ER at the 3′ end of oligoribonucleotides. Synthesis support 3 and the 1-ER phosphoramidite were used in the preparation of siRNA guide strands targeting the human PIK3CB message using automated RNA synthesis (Figure 2)24. Since PAZ domains have been reported to interact with the two nucleotides at the RNA 3′ end, we modified either the terminal (position 21, X) or penultimate (position 20, Y) position of the guide strand (Figure 2) 25. The crude oligonucleotides containing 1-ER were further functionalized using copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions and two different azides to produce 1-ER-derived triazoles I and II (Figure 2) 24. These triazoles had been shown in previous studies to modulate siRNA potency 24 and selectivity 12 at different guide strand positions. For instance, 1-ER triazole I was discovered in a computational screen for analogs that fit the hAgo2 guide strand 5′ end binding site24. A recent crystal structure of hAgo2 bound to a guide strand bearing 1-ER triazole I at the 5′ end revealed details of its interaction with the protein including a stacking interaction between the triazole ring and Tyr 529 in the hAgo2 MID domain 12. Since Phe 294 of the hAgo2 PAZ domain stacks onto the nucleobase of the guide strand 3′ end nucleotide, 1-ER-derived triazoles may maintain this favorable interaction while introducing new interactions via triazole substituents. 1-ER triazole II has been shown to maintain high levels of siRNA knockdown at position 12 of a guide strand 24. This effect was not due to changes in duplex RNA stability but rather likely arise from favorable interactions with hAgo2 and its N-ethylpiperidine group, although details of this interaction have not yet been determined 24.
Scheme 1.
Synthesis of 1-ER controlled pore glass (3) for 3′ end modification of RNA by solid phase synthesis.
Figure 2.
Sequences and modifications used in this study. (A) 1-ER and derivatives incorporated at position 21 (X) or 20 (Y) of the guide strand of an siRNA targeting PIK3CB. (B) PIK3CB on target sequence and two known off target sequences (YY1 and FADD). Red color indicates position of mismatches and lower case p indicates 5′ phosphate.
On-target versus off-target knockdown activities of 3′-end modified siRNA
We evaluated the knockdown efficiencies of the modified siRNAs in HeLa cells using a dual luciferase reporter assay. The PIK3CB siRNA has been shown to knockdown its target mRNA as well as other transcripts with complementarity to the guide strand seed region, including the YY1 and FADD mRNAs (Figure 2) 26. To facilitate our analysis, we used reporter plasmids with copies of the PIK3CB, YY1 or FADD target sequences inserted into the 3′ UTR of the Renilla luciferase gene 12. These plasmids also encode firefly luciferase as a transfection control.
We measured the half maximal inhibitory concentration (IC50) using knockdown data derived from titrations of unmodified and 3′-modified siRNAs for the PIK3CB on-target sequence, as well as the YY1 and FADD off targets (Table 1, Table 2) 12. In addition, as a measure of the effect on selectivity, we used the measured IC50 values to calculate the off-target/on-target ratio for each modification for both the YY1 and FADD off targets. With unmodified siRNA, this ratio is 1.0 for YY1 and 0.5 for FADD with higher values corresponding to increased specificity for the on-target sequence.
Table 1.
Potency and Selectivity of siRNAs with 3′ end modified guide strands (position 21)
| X | On-target IC50 pM, (PIK3CB) | Off-target IC50 pM, (YY1) | Off/On ratio (YY1/PIK3CB) | Off-target IC50 pM, (FADD) | Off/On ratio (FADD/PIK3C |
|---|---|---|---|---|---|
| U | 15 ± 5 | 15 ± 5 | 1.0 | 8 ± 3 | 0.5 |
| 1-ER | 16 ± 5 | 680 ± 30 | 42.5 | 127 ± 53 | 7.9 |
| Triazole I | 173 ± 68 | > 1200 | > 6 | 614 ± 37 | 3.5 |
| Triazole II | 23 ± 1 | 74 ± 3 | 3.2 | 20 ± 1 | 0.9 |
Table 2.
Potency and Selectivity of siRNAs with 3′ end modified guide strand (position 20)
| Y | On-target IC50 pM, (PIK3CB) | Off-target IC50 pM, (YY1) | Off/On ratio (YY1/PIK3CB) | Off-target IC50 pM, (FADD) | Off/On ratio (FADD/PIK3C |
|---|---|---|---|---|---|
| A | 15 ± 5 | 15 ± 5 | 1.0 | 8 ± 3 | 0.5 |
| 1-ER | 200 ± 62 | > 1200 | > 6 | 340 ± 65 | 1.7 |
| Triazole I | 238 ± 87 | 1035 ± 242 | 4.3 | > 1200 | > 5 |
| Triazole II | 71 ± 8 | 216 ± 88 | 3.0 | 170 ± 106 | 2.4 |
Interestingly, at the 3′ terminal nucleotide (position 21, X), the three different modified nucleosides have different effects on both knockdown and selectivity (Table 1). Whereas 1-ER and triazole II appear to be well tolerated for on-target knockdown, triazole I causes an approximate 10-fold reduction in potency (Table 1). However, these analogs alter off-target knockdown potency in a different manner. In this case, 1-ER and triazole I cause a substantial reduction in potency whereas triazole II has less of an effect (Table 1). Taken together, these results indicate that the 1-ER modification at position 21 has the greatest effect on the selectivity ratio (42.5 for the YY1/PIK3CB ratio and 7.9 for the FADD/PIK3CB ratio) while causing no change in on-target potency (IC50 = 16 ± 5 pM vs 15 ± 5 pM for unmodified siRNA (Table 1). This observation confirms our original hypothesis that guide strand 3′ end modifications can lead to reduced miRNA-like off targeting while maintaining on target potency.
Each of the three analogs tested at position 20 (Y) caused a reduction in on-target potency with 1-ER and triazole I leading to IC50 increases of 13-fold and 16-fold, respectively (Table 2). Triazole II is tolerated best at position 20, causing an increase in the measured IC50 of 4.7-fold. However, the effect on selectivity for this analog at this position indicates only modest improvement over unmodified siRNA (selectivity ratio = 3.0 for YY1/PIK3CB and 2.4 for FADD/PIK3CB) (Table 2). These results suggest that, while these analogs reduce off-target potency when placed at position 20, their effects on on-target potency make these modifications at position 20 unattractive for improving siRNA performance.
PAZ domain binding affinity of 3′-end modified siRNA
The RNAi potency and selectivity studies described above indicated that modification at position 21 with 1-ER improved on-target selectivity without compromising on-target potency. To determine if this effect was due to changes in binding affinity for the human Ago2 PAZ domain, we expressed and purified the fragment of Ago2 (residues 226-379) corresponding to the PAZ domain and used it in fluorescence polarization assays to measure the protein’s affinity for modified siRNAs (Figure 3) 25. For these experiments, we used a passenger strand for the PIK3CB siRNA bearing a 3′-end fluorescein hybridized to unmodified and modified PIK3CB guide strands and measured changes in fluorescence polarization upon protein titration 27. We found that these guide strand 3′ end modifications alter the measured dissociation constants (KD) for binding to the PAZ domain. For instance, triazole II at position 21 increased the measured KD from 75 ± 6 nM for unmodified siRNA to 194 ± 10 nM for modified siRNA. On the other hand, triazole II at position 20 decreased the measured KD to 32 ± 2 nM. However, while these analogs appear to alter binding affinity for the PAZ domain, the trends observed did not correlate with trends in siRNA potency or selectivity. For example, the 1-ER analog at position 21, the modification pattern displaying optimal potency and selectivity, bound the PAZ domain with a KD nearly identical to that of unmodified siRNA (Figure 3).
Figure 3.
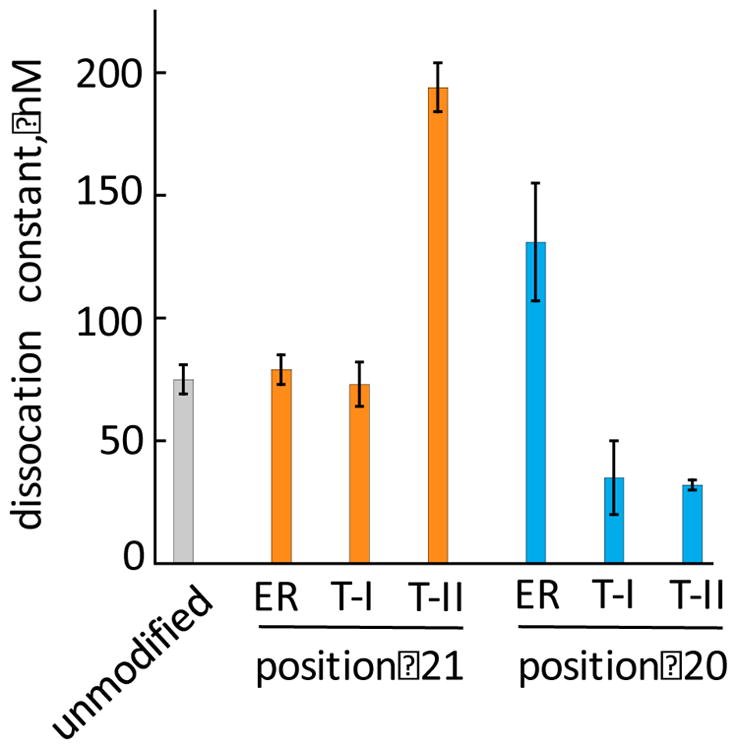
Comparison of dissociation constants (KD) measured for human Ago2 PAZ domain binding to modified siRNAs using a fluorescence polarization assay. ER = 1-ER modification, T-I = triazole I, T-II = triazole II, structures shown in Figure 2.
Discussion
MiRNA-like off target effects are difficult to avoid with siRNAs since they can arise from the same guide strand-hAgo2 complex required for target strand cleavage in the RNAi pathway 28. While a target RNA that is perfectly matched to the guide (i.e. siRNA target) will be cleaved by the nuclease activity of hAgo2, a target with seed only complementarity (i.e. miRNA target) forms a stable complex with hAgo2-guide that leads to repression of translation by mechanisms that do not involve hAgo2-catalyzed cleavage 29,30. The role of PAZ domain binding to the RNA 3′ end is apparently different in cleavage-dependent and cleavage-independent repression mechanisms. While miRNA-like target recognition appears to be compatible with anchoring the 3′ end in the Ago PAZ domain 23, siRNA-like recognition and cleavage, involving duplex formation to the target beyond the seed region, is believed to require release of the 3′ end from the PAZ domain and re-binding during catalytic turnover 19. These observations suggested to us that modification of an siRNA guide strand 3′ end may not only have effects on knockdown potency 20 but also on miRNA-like off target effects. Indeed, our studies show that modification of the 3′ terminal nucleotide of an siRNA guide strand with the 1-ER nucleoside analog reduces miRNA-like targeting without compromising on-target potency (Table 1). Modification with different analogs at the 3′ terminal nucleotide of the guide and modification at the 3′ penultimate nucleotide were less effective strategies at improving siRNA performance primarily due to reductions in on-target potency. Since the PAZ domain is known to bind the 3′ end of the guide, we measured the effect of our 3′ end modifications on the binding affinity of the isolated hAgo2 PAZ domain. However, the changes in affinity observed did not correlate with effects on siRNA potency or selectivity indicating that the reduction in miRNA-like off targeting imparted by the 1-ER modification is not simply the result of an increase or decrease in affinity for the PAZ domain. Indeed, the 1-ER modified siRNA bound the hAgo2 PAZ domain with the same affinity as does unmodified siRNA (Figure 3).
It is perhaps not surprising that the siRNA modified with 1-ER at position 21 and unmodified siRNA (with U at this position) have similar PAZ domain affinities. Examining the interactions between a 3′ terminal U and the hAgo2 PAZ domain in available structures suggests 1-ER could maintain hydrogen bonds between the main chain carbonyl of His 336 and the 2′-hydroxyl group of the ribose, the main chain carbonyl of Tyr 338 and the 3′-hydroxyl, and the side chain of Tyr 311 and the phosphodiester linking the terminal two nucleotides (Figure 1B) 23,31. These interactions are constant even when 3′ U is changed to a 3′ G, with additional hydrogen bonds formed by the phosphate group to His 316 and His 271, due to a slight tilting of the phosphate to accommodate the larger base 13. Both a 3′ U and 3′ G exhibit a stacking interaction to Phe 294. The 1-ER analog, bearing an ethyne substituent replacing the nucleobase, is likely to have a weaker stacking interaction with Phe 294. Past reports showed that the 3′ end binding pocket of the PAZ domain can accommodate a variety of large hydrophobic modifications such as pyrene, anthracene and naphthalene20, as well as nucleoside analogs containing phenyl groups 21. However, while docking studies predicted that pyrene would have the highest PAZ domain binding affinity, it resulted in low knockdown efficiency. The authors attributed this to the Ago cleavage mechanism that requires the 3′ end of the guide strand be released from the PAZ during the reaction suggesting that too much stabilization could result in a decrease in RNAi activity 20. It is possible that the 1-ER modification tested here facilitates the PAZ domain binding, release and re-binding cycle, but is less effective at anchoring the 3′ end in the PAZ domain. Such an effect could arise from changes in association and dissociation rates caused by the modification. Additional experiments directed at evaluating how various 3′ end modifications change PAZ domain association/dissociation kinetics will be necessary to test this idea. Furthermore, it is possible that nucleoside analogs that bind the PAZ domain in a primarily entropically-driven manner could behave differently than those whose binding is enthalpically driven. Further studies characterizing the enthalpic and entropic contributions to PAZ binding of different nucleoside analogs at the guide strand 3′ end could address this point. Also, guide strand loading into the different Ago family members may be altered by modification of the 3′ end. Indeed, hAgo1 plays a role in at least some of the known PIK3CB siRNA off targeting 32. Guide strand 3′ end modification with 1-ER could reduce loading for the cleavage-inactive Ago family members (e.g. hAgo1, hAgo3 and hAgo4) while maintaining efficient hAgo2 loading.
Regardless of the precise origin of the effect, 1-ER at the guide strand 3′ end reduced miRNA-like off-target knockdown for the PIK3CB siRNA while maintaining on-target potency whereas 1-ER triazole I and 1-ER triazole II did not. These results clearly show that miRNA-like off-target effects can be controlled by 3′ end modifications. We recently also reported that 1-ER triazole I at the guide strand 5′ end reduces miRNA-like off-target knockdown 12. A crystal structure of hAgo2 bound to the modified guide strand indicated that the phenyl-imidazole of 1-ER triazole I extends into the enzyme’s RNA-binding central cleft and likely inhibits formation of the miRNA-like recognition complex while allowing siRNA-like recognition to take place when the central cleft is more open 12. Thus, modification with nucleobase analogs at either end of an siRNA guide strand can reduce off-target effects. For 5′ end modifications, this appears to be possible because of the different conformational states adopted by hAgo2 and their relative importance in miRNA vs siRNA mechanisms.
Conclusion
The studies described here establish the siRNA guide strand 3′ terminal nucleotide as a site where modification can reduce miRNA-like off target activity and suggest the effects are not simply due to changes in PAZ domain binding affinity. In addition, the 1-ER solid phase synthesis support is shown to be a useful new tool for oligoribonucleotide 3′ end modification.
Materials and Methods
3,6-Anhydro-1,2-dideoxy-7-O-(4,4′-Dimethoxytrityl)-4-O-(tert-butyldimethylsilyl)-5-O-succinyl-D-allo-hept-1-ynitol (2)
Compound 1 (30 mg, 0.052 mmol) 24 was coevaporated with anhydrous acetonitrile, and the dried residue was dissolved in anhydrous pyridine (1 mL). To the solution were added succinic anhydride (16 mg, 0.16 mmol) and DMAP (6 mg, 0.05 mmol), and the mixture was stirred at room temperature for 20 h. The reaction mixture was diluted with EtOAc (20 mL), and washed with H2O (20 mL) and brine (20 mL). The organic phase was dried over Na2SO4, and the solvent was removed under reduced pressure. The residue was purified by column chromatography on a silica gel with hexane/EtOAc (3:1) containing 0.5% pyridine to give 5 (25 mg, 71%) as a pale yellow oil. 1H NMR (CD2Cl2, 300 MHz): δ (ppm) 7.48 (d, J = 7.5 Hz, 2H), 7.37-7.22 (m, 7H), 6.84 (d, J = 8.4 Hz, 4H), 5.22-5.19 (m, 1H), 4.58 (dd, J = 6.9, 5.4 Hz, 1H), 4.42 (dd, J = 6.9, 1.5 Hz, 1H), 4.06-4.04 (m, 1H), 3.78 (s, 6H), 3.34 (dd, J = 10, 3.0 Hz, 1H), 3.03 (dd, J = 10, 3.3 Hz, 1H), 2.63 (m, 5H), 0.90 (s, 9H), 0.16 (s, 3H), 0.08 (s, 3H). 13C NMR (CD2Cl2, 75 MHz): δ(ppm) 171.4, 159.1, 159.0, 145.4, 136.3, 136.1, 130.5, 128.5, 128.2, 127.1, 113.4, 86.7, 82.4, 81.9, 76.7, 75.1, 72.6, 64.3, 55.6, 29.2, 25.8, 18.3, −4.62, −4.87. ESIHRMS (m/z): calcd for C38H46O9Si (M+H)+ 697.2803,, obsd 697.2803.
3,6-Anhydro-1,2-dideoxy-7-O-(4,4′-Dimethoxytrityl)-4-O-(tert-butyldimethylsilyl)-D-allo-hept-1-ynitol LCAA-CPG (3)
EDC and LCAA-CPG were added to a solution of 2 in anhydrous DMF, and the mixture was kept at room temperature for 20 h. The CPG was filtered and washed with pyridine, MeOH and CH2Cl2. The remaining amino groups were capped by treatment with pyridine/Ac2O (9:1, 4 mL) and DMAP (49 mg). After 2 h, the resulting solid support was filtered and washed with MeOH, acetone and CH2Cl2, and dried under reduced pressure to give 3. The loading amount was estimated by DMTr cation assay to be 30 μmol/g.
Synthesis, purification and quantification of 1-ER, 1-ER triazole I and 1-ER triazole II containing siRNAs
The PIK3CB guide strand with 1-ER at position 21 was synthesized at the University of Utah DNA/Peptide Core Facility using the 1-ethynyl ribose controlled pore glass (3) at a 200 nmol scale on an ABI 394 Synthesizer. The PIK3CB siRNA guide strand with 1-ER at position 20 was synthesized using the 1-ethynyl ribose phosphoramidite on a 200 nmol scale 24. SiRNA guide strands were chemically 5′-phosphorylated during automated synthesis. Triazoles were formed using CuAAC reactions as previously described 24. MALDI-TOF MS (m/z) 1-ER position 20 [M−H]−: calcd 6717.0, obsd 6716.1; 1-ER triazole I position 20 [M−H]−: calcd 6915.1, obsd 6913.6; 1-ER triazole II position 20 [M−H]−: calcd 6868.0, obsd 6867.9; 1-ER position 21 [M−H]−: calcd 6739.0, obsd 6738.8; 1-ER triazole I position 21 [M−H]−: calcd 6938.1, obsd 6935.6; 1-ER triazole II position 21 [M−H]−: calcd 6894.2, obsd 6893.4
RNAi activity assay
HeLa cells (ATCC) were grown at 37 °C in humidified 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) and 1% antibiotic-antimycotic (100x Anti-Anti, GIBCO). The cells were maintained in exponential growth. For the dual-luciferase RNA interference assay, HeLa cells were grown to 80–90% confluence then detatched using Accutase (Innovative Cell Technologies, without calcium or magnesium) and diluted to a concentration of 1 × 105 cells/mL in fresh medium (DMEM, 10% FBS, 1x Anti-Anti). Reverse transfection of HeLa cells was accomplished using siPORT NeoFX (Ambion) in Opti-MEM media (GIBCO) to deliver the reporter plasmid and siRNA.26 For these experiments we used on-target and off-target activity reporter plasmids with the PIK3CB, YY1 and FADD target sequences inserted into the 3′ UTR of the Renilla luciferase gene 12,24. The two off target reporter plasmids contain four copies of either the YY1 or FADD off-target sequences inserted into the Renilla luciferase gene 3′ UTR12. Insertion of multiple copies of the off-target sequence into the mRNA allowed for evaluation of knockdown activity using low siRNA concentrations. Reverse transfection was performed in 96-well plates, where each well contained 0.5 μL siPORT, 20 ng reporter plasmid and varying concentrations of siRNA or buffer as a negative control. As an additional negative control we transfected a scrambled sequence siRNA (guide: 5′-p-gaauagguacguaauuauaug-3′, passenger: 3′-cacuuauccaugcauuaauau-5′) and compared its activity to knockdown observed with the unmodified PIK3CB siRNA (Supplementary Figure 1). After a 22–24h incubation (37 °C, 5% CO2), the Dual-Luciferase® Reporter (DLR™) Assay System (Promega) was used to determine the RNAi knockdown activities of the siRNAs. The dual luciferase assay is performed by sequentially measuring the firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferase activities of the same sample, and the results are expressed as the ratio of firefly to Renilla luciferase activity (Fluc/Rluc). Luminescence was measured using a CLARIOstar plate reader (BMG Labtech, Inc.). To calculate the IC50, the (Fluc/Rluc) ratios were plotted versus siRNA concentrations to create a dose-response curve. Kaleidagraph (Synergy Software) was used to estimate the concentration of siRNA that lead to RNAi knockdown by 50% as compared to the control cells. This is designated as the IC50 and is determined by curve fitting test data using the following four parameter square fit equation: Y = ((Lmax − Lmin)/(1 + (X/IC50)n)) + Lmin. where Lmax is the luminescence of the control cells (without siRNA), Lmin is the luminescence of the cells in the presence of the highest siRNA concentration, Y is the observed luminescence, X is the siRNA concentration, IC50 is the siRNA concentration that leads to 50% knockdown compared to the control cells, and n is the slope of the curve. For each IC50 value, three independent transfections were carried out and values determined were averaged to give the IC50 values and standard deviations reported in Tables 1 and 2.
Cloning, expression, and purification of Ago2 PAZ domain
The hAgo2 PAZ domain was cloned into the BamHI and EcoRI sites of pET28-c vector using Gibson cloning 33. To generate the PAZ domain, a fragment of hAgo2 (residues 226-379) was amplified from a plasmid expressing FLAG-tagged full-length hAgo2 (pcDNA5-FRT) 34 by using the following primers: forward (5′-TGGGTCGCGGATCCGCACAGCCAGTAATCGAGTTTGTTTG-3′) and reverse (5′-ACGGAGCTCGAATTCTTATAATCTCTTCTTGCCGATCGGGC-3′). Transformed E. coli cells were grown in LB medium containing 50 ug/ml kanamycin at 37°C to the mid-log phase (A600 = 0.6). IPTG (isopropyl β-D-1-thiogalactopyranoside) (0.5 mM) was added to induce the expression of protein, and the cells were incubated at 20°C with shaking for 8 h. Cells were lysed using sonication in Ni-NTA lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). The histidine-tagged PAZ domain was purified from the soluble cell extract using Ni-NTA agarose resin (Qiagen) followed by Amicon Ultrafree-MC centrifugal filter unit (50 kDa membrane cut off) to remove high molecular weight contaminants. A final purification step was carried out using Amicon Ultrafree-MC centrifugal filter unit (10 kDa membrane cut off) and dialysis (50 mM NaH2PO4, 100 mM NaCl, 20% glycerol, 70 uM β-mercaptoethanol) to remove excess imidazole. The purified PAZ was quantified using SDS-PAGE with bovine serum albumin (BSA) as standards.
Fluorescence polarization assay
A 3′-fluorescein-labeled PIK3CB passenger strand (Dharmacon, GE Life Sciences) was hybridized with either the unmodified and modified PIK3CB guide strand by annealing the two strands in hybridization buffer (10 mM Tris-HCl, 50 mM KCl, pH 7.6). The solution was heated to 95°C for 5 minutes and slow-cooled to room temperature. The PAZ domain protein was serially diluted from 15 uM to 0.73 nM in 25 uL of 50 mM NaH2PO4, 100 mM NaCl, pH 7.6) in Eppendorf tubes. The fluorescein labeled siRNA was added to each tube, yielding a final concentration of 25 nM. The mixture was incubated at room temperature for 30 minutes. The samples were transferred to a black 384-well plate and the fluorescence polarization was measured using a CLARIOstar plate reader (BMG Labtech, Inc.). To determine the KD, the data were fitted to the following equation: fraction bound = A * [protein]/([protein] + KD), where the KD is the fitted dissociation constant and A is the fitted maximum fraction of RNA bound.
Supplementary Material
Acknowledgments
PAB acknowledges support from the National Institutes of Health in the form of grant R01GM08784.
Footnotes
1H, 13C NMR spectra for compound 2, binding plots for hAgo2 PAZ-RNA complexes, comparison of knockdown activity for PIK3CB and scrambled sequence siRNAs
References
- 1.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Bobbin ML, Rossi JJ. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu Rev Pharmacol Taxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 3.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 4.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 5.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Disc. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HS, et al. Abasic pivot substitution harnesses target specificity of RNA interference. Nat Commun. 2015;6:10154. doi: 10.1038/ncomms10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. Nucleic Acids Res. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bramsen JB, et al. A screen for chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suter SR, et al. Structure-Guided Control of siRNA Off-target Effects. J Am Chem Soc. 2016;138:8667–8669. doi: 10.1021/jacs.6b06137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkayam E, et al. The Structure of Human Argonaute-2 in Complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirle NT, MacRae IJ. The Crystal Structure of Human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 16.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 19.Deerberg A, Willkomm S, Restle T. Minimal mechanistic model of siRNA-dependent target RNA slicing by recombinant human Argonaute 2 protein. Proc Natl Acad Sci U S A. 2013;110:17850–17855. doi: 10.1073/pnas.1217838110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somoza A, Terrazas M, Eritja R. Modified siRNAs for the study of the PAZ domain. Chem Comm. 2010;46:4270–4272. doi: 10.1039/c003221b. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, et al. Structure-Based Design of Novel Chemical Modification of the 3′-Overhang for Optimization of Short Interfering RNA Performance. Biochemistry. 2015;54:1268–1277. doi: 10.1021/bi500602z. [DOI] [PubMed] [Google Scholar]
- 22.Kandeel M, Kitade Y. Computational Analysis of siRNA Recognition by the Ago2 PAZ Domain and Identification of the Determinants of RNA-Induced Gene Silencing. PLOS One. 2013;8:e57140. doi: 10.1371/journal.pone.0057140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onizuka K, et al. Short Interfering RNA Guide Strand Modifiers from Computational Screening. J Am Chem Soc. 2013;135:17069–17077. doi: 10.1021/ja4079754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 26.Jackson AL, et al. Widespread siRNA “off target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson BJ, Larkin C, Guja K, Schildbach JF. Using Fluorophore-labeled Oligonucleotides to Measure Affinities of Protein-DNA Interactions. Methods Enymol. 2008;450:253–272. doi: 10.1016/S0076-6879(08)03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ipsaro JJ, Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol. 2015;22:20–28. doi: 10.1038/nsmb.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickers TA, et al. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 “Slicer” independent and can be mediated by Ago1. Nucl Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 34.Vaishnaw AK, et al. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.