Abstract
H3N3 and H1N1 influenza A viruses were isolated from Canadian pigs in 2001 and 2002. These viruses are phylogenetically related to waterfowl viruses and antigenically distinct from reference swine influenza viruses. The isolation of these viruses reemphasizes the potential for interspecies transmission of influenza viruses from waterfowl to pigs in North America.
Influenza A virus infections in animals are important contributors to the evolution of human influenza viruses (41, 65, 66, 68). In particular, waterfowl and seabirds constitute a global reservoir of all 15 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes of influenza A viruses (15, 65, 66, 68). Viruses from waterfowl reassorted with existing human influenza viruses to generate the 1957 (HA, NA, and PB1 polymerase genes were derived from an avian virus) and 1968 (HA and PB1 polymerase genes were derived from an avian virus) pandemic human influenza viruses (24, 54, 66) and may play a similar role in the creation of future pandemic viruses.
The specific host that supported the reassortment events leading to the 1957 and 1968 pandemic viruses remains unclear. Human infections with avian H5N1 and H9N2 influenza viruses in southeast Asia since 1997 (7, 11, 32, 46, 59), avian H7N7 viruses in Europe in 2003 (27a), and previous evidence of localized ocular or subclinical human infections with avian influenza viruses (28, 62, 67) all demonstrate that viruses can be directly transmitted from birds to humans. However, avian influenza viruses typically replicate poorly in humans and nonhuman primates, and vice versa, human influenza viruses generally do not replicate efficiently in birds (2, 14, 37, 58, 66, 68). The basis for species specificity of influenza viruses appears to be polygenic (27, 53, 58, 63, 65, 66, 68), but the HA is hypothesized to be a major factor because of its role as the receptor-binding protein (29, 68). Avian influenza viruses bind preferentially to sialyloligosaccharides with an α2,3 linkage of N-acetylneuraminic acid to galactose (NAcα2,3Gal), whereas human influenza viruses prefer NAcα2,6Gal receptors, consistent with the fact that NAcα2,3Gal is the major receptor found on duck intestinal cells and that human tracheal epithelial cells express predominantly NAcα2,6Gal molecules (8, 9, 16-18, 36, 49, 50, 68). Pigs, however, express both NAcα2,3Gal and NAcα2,6Gal receptors as well as α2,3-linked N-glycolneuraminic acid (NGc) receptors that are also expressed by duck intestinal cells (16, 17, 19, 60). Thus, pigs have been suggested to be the intermediary host in which avian influenza viruses adapt to replication in mammals and in which avian-human influenza virus reassortment may occur (3, 16, 53, 55, 66). As such, detection and characterization of avian influenza viruses in pigs are important components of influenza virus surveillance.
Kida and colleagues demonstrated that pigs can be infected experimentally with avian viruses of nearly all subtypes (H4 to H13) (25). In addition, avian H1N1, H3N2, and H9N2 viruses have been recovered from pigs in Asia (12, 26, 45), and avian H1N1 viruses spread widely among pigs in northern Europe after their introduction in 1979 (10, 13, 47, 52, 56, 66). In North America, reassortant H3N2, H1N2, and H1N1 viruses containing avian genes have been isolated since 1998 (5, 6, 20, 22, 23, 39, 40, 64, 69), and there has been limited serologic evidence for avian virus infection of pigs (4, 42). However, there has previously been only a single report of isolation of wholly avian viruses from North American pigs. These were H4N6 viruses that we isolated from pigs in Ontario in 1999 (21). We now report the genetic and antigenic characteristics of additional wholly avian viruses of H3N3 and H1N1 subtypes isolated from Canadian pigs.
The first virus, A/Swine/Ontario/42729A/01 (Sw/ON/42729), was isolated in October 2001 from one of a group of approximately 16-week-old pigs in eastern Ontario suffering from weight loss and coughing. (Two additional influenza viruses, 42729B and 42729C, were isolated at the same time from additional pigs involved in this outbreak. However, since complete genome sequencing ultimately demonstrated that the A, B, and C isolates were virtually identical genetically, only isolate 42729A is considered hereafter.) Interestingly, this is the same farm from which the H4N6 viruses were isolated previously (21). The source of the H4N6 viruses for pigs was thought to be raw lake water that was pumped into a barn from an adjacent lake, and this is a practice that continues on the farm. Bronchoalveolar pneumonia was confirmed upon histopathologic examination of the pig from which Sw/ON/42729 was isolated. Both influenza A virus and Mycoplasma hyopneumoniae antigens were detected in the lungs by immunohistochemistry, whereas immunohistochemistry assays for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 antigens were negative. Secondary bacterial pathogens (Pasteurella multocida and Streptococcus suis) were also recovered from the lungs. Influenza A viruses were isolated from lung tissue in both embryonated chicken eggs and Madin-Darby canine kidney (MDCK) cells, but all antigenic and genetic analyses were conducted on the MDCK cell isolate. The Sw/ON/42729 virus was initially subtyped by reverse transcription-PCR (RT-PCR) (20) as an H3 virus, but the NA subtype could not be determined by using N1- or N2-specific primer sets. RT-PCR amplification with global NA primers (70) and subsequent cycle sequencing (ABI Big Dye; PE Applied Biosystems, Foster City, Calif.) and BLAST analyses (1) demonstrated that this was an H3N3 virus.
A second virus, A/Swine/Ontario/K01477/01 (Sw/ON/K01477),was similarly shown to be an H3N3 virus. It was also isolated in October 2001 from pigs in eastern Ontario but on a farm that is located approximately 30 km from the farm of origin of Sw/ON/42729. The second farm uses only well water and stringently prevents pig and pig feed contact with birds. There is also no movement of animals between these two farms.
The third virus described in this report, A/Swine/Saskatchewan/18789/02 (Sw/SK), was isolated from pigs on a 1,200-sow, farrow-to-finish farm in Saskatchewan in May 2002. Influenza-like illness had been occurring on the farm since December 2000 involving pigs of all ages but most severely affecting the nursery pigs. Bacterial pneumonia agents (S. suis, Haemophilus parasuis, and Actinobacillus suis) were also endemic problems on the farm. RT-PCR, cycle sequencing, and BLAST analyses demonstrated that this was an H1N1 virus.
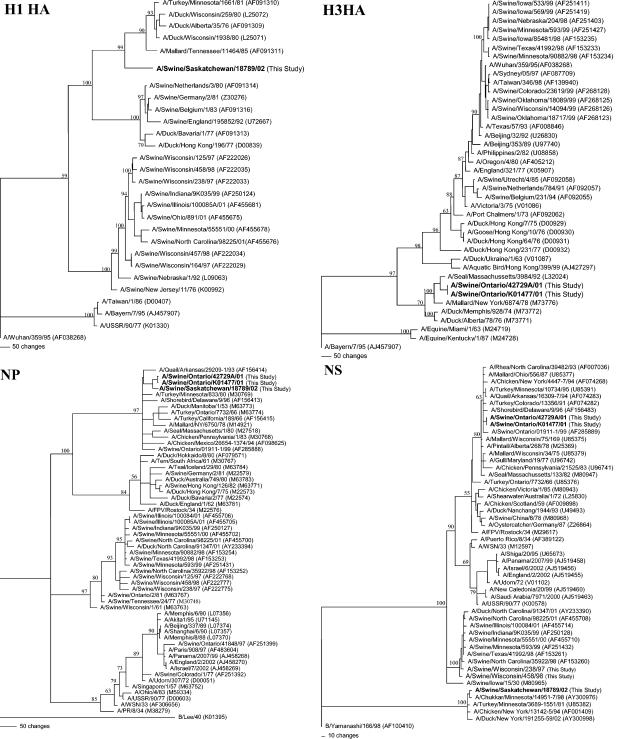
Following initial sequencing of the HA and NA genes of Sw/ON/42729, Sw/ON/K01477, and Sw/SK for subtype determination, the sequences of the full-length protein coding regions of all eight RNA segments for each virus were determined by cycle sequencing with influenza virus universal primers (70) and subsequently designed sequence-specific primers. (The sequences of the primers used for characterization of these viruses are provided in Table 1). BLAST analyses were conducted on each sequence to identify related reference viruses. Sequence comparisons to selected reference viruses were conducted by using DNASTAR software (version 5.0 for Win32), and phylogenetic relationships among Sw/ON/42729, Sw/ON/K01477, Sw/SK, and the reference viruses were estimated by the method of maximum parsimony (PAUP software, version 4.0b6; David Swofford, Smithsonian Institution, and Sinauer Associates, Sunderland, Mass.) using a bootstrap resampling method (500 replications) with a fast heuristic search algorithm. These analyses demonstrated that Sw/ON/42729, Sw/ON/K01477, and Sw/SK are all wholly avian viruses and that each of the RNA segments are phylogenetically related to viruses of the North American lineage of avian influenza viruses. Interestingly, these three viruses are also topologically closely related to one another on the nucleoprotein (NP), the matrix protein, and PB1, PB2, and PA polymerase gene phylograms. However, the nonstructural (NS) gene of Sw/SK was derived from a clade distinct from that of the Sw/ON viruses. Finally, the NA genes of the Sw/SK and Sw/ON viruses, though all clearly of North American avian origin, are phylogenetically distinct from one another since they represent different NA subtypes. By way of example of the phylogenetic analyses conducted, the H1 HA, H3 HA, NP, and NS phylogenetic trees are shown in Fig. 1.
TABLE 1.
Nucleotide sequences of primers used for RT-PCR and/or sequencing in this study
| Primer | 5′-3′ nucleotide sequence | Usea | Reference or source |
|---|---|---|---|
| SZAHA+ | CTCGAGAGCAAAAGCAGGGG | P, S | 70 |
| SZAHA− | AGTAGAAACAAGGGTGTTTTT | P, S | 70 |
| H1HA1095Rb | AGGCATGATAGATGGGTGG | S | This study |
| H3HA1169R | AAGCACTCAAGCCGCCAT | S | This study |
| SZANA+ | AGCAAAAGCAGGAGTTTAAAATG | P, S | 70 |
| SZANA− | AGTAGAAACAAGGAGTTTTTT | P, S | 70 |
| N1NA408F | AGGGGCCTTGTTGAATGA | S | This study |
| N3NA522F | GATATGTATTGCTTGGTCT | S | This study |
| SZANP+ | CTCGAGAGCAAAAGCAGGGT | P, S | 70 |
| SZANP− | AGTAGAAACAAGGGTATTTTTC | P, S | 70 |
| NP484F | AGGATGTGTTCTCTGATGC | S | This study |
| SZAM+ | CTCGAGCAAAAGCAGGTAGAT | P, S | 70 |
| SZAM− | AGTAGAAACAAGGTAGTTTTTT | P, S | 70 |
| SZANS+ | AGCAAAAGCAGGGTGACAAA | P, S | 70 |
| SZANS− | AGTAGAAACAAGGGTGTTTTTT | P, S | 70 |
| SZAPB1+ | AGCAAAAGCAGGCAAACCAT | P, S | 70 |
| SZAPB1− | AGTAGAAACAAGGCATTTTTTCAT | P, S | 70 |
| PB1589F | AAGAAAATGGTCACACAA | S | This study |
| PB11548R | AGTTTTGGAGTGTCTGGA | S | This study |
| SZAPB2+ | CTCGAGCAAAAGCAGGTCAA | P, S | 70 |
| SZAPB2− | AGTAGAAACAAGGTCGTTTTTAAAC | P, S | 70 |
| PB2657F | AGTGGCTGGTGGAACAAG | S | This study |
| PB21750R | AGCCATTCCAGTCTCTGG | S | This study |
| SZAPA+ | CTCGAGCAAAAGCAGGTACTGAT | P, S | 70 |
| SZAPA− | AGTAGAAACAAGGTACTTTTTTGGAC | P, S | 70 |
| PA691F | GCCTATGTGGATGGATTC | S | This study |
| PA1467R | CCAATGATAAGCAAATGT | S | This study |
P, PCR; S, sequencing.
R, reverse primer; F, forward primer.
FIG. 1.
Nucleotide phylograms for the H3 HA, H1 HA, NP, and NS genes of Sw/ON/42729 (H3N3), Sw/ON/K01477 (H3N3), Sw/SK (H1N1), and related reference viruses. The evolutionary relationships among these viruses were estimated by the method of maximum parsimony (PAUP software version 4.0b6; David Swofford, Smithsonian Institution, and Sinauer Associates) by using a bootstrap resampling method (500 replications) with a fast heuristic search algorithm. The numbers at the nodes of the phylograms indicate the bootstrap confidence levels. Horizontal line distances are proportional to the minimum numbers of nucleotide changes needed to join nodes and gene sequences. The vertical lines are present simply to space the branches and labels. The GenBank accession numbers for the sequences of all of the reference viruses are provided in parentheses following the virus names.
The replication of avian influenza viruses in pigs raises concerns for adaptation to mammalian cells and ultimately enhanced infectivity for humans. Therefore, the deduced amino acid sequences of the HA genes of Sw/ON/42729, Sw/ON/K01477, and Sw/SK were examined for signatures related to avian versus mammalian (human and swine) receptor preferences and/or evolutionary lineages as defined in previous studies (8, 17, 34-36, 38, 48, 49, 51, 61). As shown in Table 2, the Sw/ON/42729 and Sw/ON/K01477 H3 viruses maintain the glutamine typical of avian viruses and NAcα2,3Gal receptor use at residue 226 (38, 51). At residue 228, these viruses have an alanine; avian viruses that bind to NAcα2,3Gal receptors typically have a glycine at amino acid 228, while human viruses that bind to NAcα2,6Gal receptors typically have a serine (8, 38). The Sw/SK H1 virus maintains amino acids typical of avian viruses at residues 77, 138, 145, 155, 159, 186, 225, and 228 (using the H3 numbering scheme). However, at residues 190 and 194, this virus has amino acids typical of human H1 influenza viruses (17, 35, 36, 48). As regards NGc receptor signature sequences, the Sw/ON/42729 and Sw/ON/K01477 H3 viruses have amino acids typical of NGc receptor use at residues 143 and 158, but they have threonine at residue 155 in place of the tyrosine typical of NGc receptor binding (34). The Sw/SK H1 virus lacks the isoleucine or valine at residue 155 that is typical of H1 swine viruses and postulated to enhance binding to NGc receptors (35). Finally, note that the sequences defined for these viruses are very unlikely to have originated from cross-contamination or laboratory errors. The lab where the RT-PCR and sequencing were conducted had never worked with avian H3 or N3 viruses in the past and had not worked with avian H1 or N1 viruses for many months prior to the arrival of the Sw/ON and Sw/SK isolates. Furthermore, each of the PCR amplifications included negative control reactions containing all reagents except the template.
TABLE 2.
Analysis of the sequences of Sw/ON/42729 (H3N3), Sw/ON/K01477 (H3N3), and Sw/SK (H1N1) HA proteins at residues that define avian versus mammalian evolutionary lineages and/or receptor preferences
| HA analysis and amino acid residue positiona | Amino acid found in:
|
Amino acid(s) typical ofb:
|
|||
|---|---|---|---|---|---|
| Sw/ON/42729 and Sw/ON/K01477 | Sw/SK | Avian-lineage viruses and/or NAcα2,3Gal receptor use | Mammalian-lineage viruses and/or NAcα2,6Gal receptor use | NGcα2,3Gal receptor use | |
| H3 | |||||
| 143 | P | NAc | NAe | NAe | P/S |
| 155 | T | NAc | NAe | NAe | Y |
| 158 | G | NAc | NAe | NAe | G |
| 226 | Q | NAc | Q | L | NAf |
| 228 | A | NAc | G | S | NAf |
| H1 | |||||
| 77 | NAd | D | D | E | NAf |
| 138 | NAd | A | A | S | NAf |
| 145 | NAd | S | S | L | NAf |
| 155 | NAd | T | T | V/T | I/V |
| 159 | NAd | T | T | N/G | NAf |
| 186 | NAd | P | P | S | NAf |
| 190 | NAd | D | E | D | NAf |
| 194 | NAd | I | L | I | NAf |
| 225 | NAd | E/G | G | E | NAf |
All numbers are based on the H3 numbering scheme.
Lineage- and receptor-specific amino acids are based on previously published sequence analyses (8, 17, 34-36, 38, 48, 49, 51, 61).
NA, analysis applicable only to H3 HA analysis.
NA, analysis applicable only to H1 HA analysis.
NA, residue analyzed only in reference to NGc receptor analysis.
NA, residue not applicable to NGc receptor analysis.
Beyond these genetic comparisons, the antigenic characteristics of the HA proteins of Sw/ON/42729 and Sw/SK were also assessed in hemagglutination inhibition (HI) assays by using either monoclonal antibodies (MAbs) specific for H1 HA proteins or polyclonal anti-H1 or anti-H3 swine serum. The anti-H1 MAbs (ascites) had been produced against A/Swine/Indiana/1726/88 (H1N1) and A/Swine/Wisconsin/27/86 (H1N1) as previously described (33, 57). The anti-H1 polyclonal swine serum was collected from pigs 14 days following the administration of two doses of classical H1N1 swine influenza virus vaccine (MaxiVac-FLU; SyntroVet, Inc.) (31). The anti-H3 polyclonal swine serum was collected from pigs 7 days following intranasal infection with A/Swine/Minnesota/593/99 (H3N2) (30). The HI assays were conducted as previously described (42, 44) with serial twofold dilutions of MAb ascites (1:100 to 1:6,400) or polyclonal swine serum (1:10 to 1:640). HI titers were defined as the reciprocal of the highest dilution of MAb ascites or serum that completely inhibited virus-induced agglutination of chicken red blood cells. As shown in Table 3, the Sw/SK H1 virus lacked reactivity with two of the four MAbs that recognize classical swine influenza viruses. This pattern is consistent with previous results from a North American waterfowl H1N1 virus, A/Duck/Alberta/35/76 (33, 43, 57). The Sw/SK virus also reacted to an eightfold-lower titer with anti-H1 swine serum compared to that of a reference classical H1N1 swine virus. The Sw/ON/42729 H3 virus was completely nonreactive with serum recovered from a pig previously infected with a triple-reassortant H3N2 virus (30) that is typical of those that emerged among American pigs in 1998.
TABLE 3.
Reactivity of Sw/ON/42729 and Sw/SK compared to reference swine viruses in HI assaysb
| Virusa | HI titer in:
|
|||||
|---|---|---|---|---|---|---|
| Anti-H1 MAbs
|
Anti-H1 polyclonal swine serum | Anti-H3 polyclonal swine serum | ||||
| 1-6B2 | 3F2C | 2-15F1 | 7B1B | |||
| Sw/ON/42729A (H3N3) | −b | − | − | − | − | − |
| A/Swine/Minnesota/593/99 (H3N2) | − | − | − | − | − | 320 |
| Sw/SK (H1N1) | >6,400 | − | − | 800 | 10 | − |
| A/Swine/Indiana/1726/88 (H1N1) | >6,400 | >6,400 | 1,600 | >6,400 | 80 | − |
Sw/ON/42729 and Sw/SK are the wholly avian viruses described in this report. A/Swine/Minnesota/593/99 is a triple-reassortant H3N2 reference virus, and A/Swine/Indiana/1726/88 is a classical swine H1N1 reference virus.
−, HI titers <100 (MAb ascites) or <10 (serum).
In summary, this work documents the isolation of wholly avian H3N3 and H1N1 influenza viruses from pigs in Canada during 2001 and 2002. In one case, the likely source of virus for the pigs was lake water contaminated with waterfowl feces, while the source of virus in the other two instances is unclear. However, the phylogenetic data for all three viruses clearly reveal a North American waterfowl origin. We do not have data to address the extent of transmission of these viruses among the swine population. Nonetheless, it is important for diagnostic virologists to know that H1 and H3 viruses that are genetically and antigenically distinct from the H1 and H3 viruses currently circulating among pigs in North America may be present within the swine population. Finally, the isolation of these viruses reemphasizes the fact that bird-to-pig interspecies transmission of influenza viruses may occur not just in Asia but also in North America.
Nucleotide sequence accession numbers.
The GenBank accession numbers assigned to the gene sequences determined for this report are as follows: AY619970 to AY619977 for Sw/ON/42729, AY619962 to AY619969 for Sw/ON/K01477, AY619954 to AY619961 for Sw/SK, AY619979 for A/Swine/Wisconsin/238/97, and AY619978 for A/Swine/Wisconsin/458/98.
Acknowledgments
This work was supported in part by a grant to C.W.O. from the USDA National Research Initiative Competitive Grants Program.
We thank Gabriele Landolt for technical assistance and many helpful discussions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beare, A. S., and R. G. Webster. 1991. Replication of avian influenza viruses in humans. Arch. Virol. 119:37-42. [DOI] [PubMed] [Google Scholar]
- 3.Brown, I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74:29-46. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, T. M., V. S. Hinshaw, Y. Kawaoka, B. C. Easterday, and R. G. Webster. 1991. Influenza viral infection of swine in the United States 1988-1989. Arch. Virol. 116:261-265. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 87:173-179. [DOI] [PubMed] [Google Scholar]
- 6.Choi, Y. K., S. M. Goyal, and H. S. Joo. 2002. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch. Virol. 147:1209-1220. [DOI] [PubMed] [Google Scholar]
- 7.Claas, E. C. J., A. D. M. E. Osterhaus, R. Van Beek, J. C. de Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 8.Connor, R. J., Y. Kawaoka, R. G. Webster, and J. C. Paulson. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17-23. [DOI] [PubMed] [Google Scholar]
- 9.Couceiro, J. N. S. S., J. C. Paulson, and L. G. Baum. 1993. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29:155-165. [DOI] [PubMed] [Google Scholar]
- 10.Donatelli, I., L. Campitelli, M. R. Castrucci, A. Ruggieri, L. Sidoli, and J. S. Oxford. 1991. Detection of two antigenic subpopulations of A (H1N1) influenza viruses from pigs: antigenic drift or interspecies transmission. J. Med. Virol. 34:248-257. [DOI] [PubMed] [Google Scholar]
- 11.Gao, P., S. Watanabe, T. Ito, H. Goto, K. Wells, M. McGregor, A. J. Cooley, and Y. Kawaoka. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J. Virol. 73:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, Y., K. F. Shortridge, S. Krauss, P. H. Li, Y. Kawaoka, and R. G. Webster. 1996. Emergence of avian H1N1 influenza viruses in pigs in China. J. Virol. 70:8041-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinshaw, V. S., D. J. Alexander, M. Aymard, P. A. Bachman, B. C. Easterday, C. Hannoun, H. Kida, M. Lipkind, J. S. Mackenzie, K. Nerome, G. C. Schild, C. Scholtissek, D. A. Senne, K. F. Shortridge, J. J. Skehel, and R. G. Webster. 1984. Antigenic comparisons of swine-influenza-like H1N1 isolates from pigs, birds and humans: an international collaborative study. Bull. W. H. O. 62:871-878. [PMC free article] [PubMed] [Google Scholar]
- 14.Hinshaw, V. S., R. G. Webster, C. W. Naeve, and B. R. Murphy. 1983. Altered tissue tropism of human-avian reassortant influenza viruses. Virology 128:260-263. [DOI] [PubMed] [Google Scholar]
- 15.Hinshaw, V. S., R. G. Webster, and B. Turner. 1980. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 26:622-629. [DOI] [PubMed] [Google Scholar]
- 16.Ito, T. 2000. Interspecies transmission and receptor recognition of influenza A viruses. Microbiol. Immunol. 44:423-430. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., J. N. S. S. Couceiro, S. Kelm, L. G. Baum, S. Krauss, M. R. Castrucci, I. Donatelli, H. Kida, J. C. Paulson, R. G. Webster, and Y. Kawaoka. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 72:7367-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Suzuki, L. Mitnaul, A. Vines, H. Kida, and Y. Kawaoka. 1997. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology 227:493-499. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Suzuki, T. Suzuki, A. Takada, T. Horimoto, K. Wells, H. Kida, K. Otsuki, M. Kiso, H. Ishida, and Y. Kawaoka. 2000. Recognition of N-glycolylneuraminic acid linked to galactose by the α2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J. Virol. 74:9300-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karasin, A. I., G. A. Anderson, and C. W. Olsen. 2000. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J. Clin. Microbiol. 38:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karasin, A. I., I. H. Brown, S. Carman, and C. W. Olsen. 2000. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J. Virol. 74:9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karasin, A. I., J. G. Landgraf, S. L. Swenson, G. Erickson, S. M. Goyal, M. Woodruff, G. Scherba, G. A. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 24.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kida, H., T. Ito, J. Yasuda, Y. Shimizu, C. Itakura, K. F. Shortridge, Y. Kawaoka, and R. G. Webster. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75:2183-2188. [DOI] [PubMed] [Google Scholar]
- 26.Kida, H., K. F. Shortridge, and R. G. Webster. 1988. Origin of the hemagglutinin gene of H3N2 influenza viruses from pigs in China. Virology. 162:160-166. [DOI] [PubMed] [Google Scholar]
- 27.Klenk, H. D., and R. Rott. 1988. The molecular biology of influenza virus pathogenicity. Adv. Virus Res. 34:247-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Koopmans, M., B. Wilbrink, M. Conyn, G. Natrop, H. van der Nat, H. Vennema, A. Meijer, J. van Steenbergen, R. Fouchier, A. Osterhaus, and A. Bosman. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in The Netherlands. Lancet 363:587-593. [DOI] [PubMed] [Google Scholar]
- 28.Kurtz, J., R. J. Manvell, and J. Banks. 1996. Avian influenza virus isolated from a woman with conjunctivitis. Lancet 348:901-902. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 30.Landolt, G. A., A. I. Karasin, L. Phillips, and C. W. Olsen. 2003. Comparison of the pathogenesis of two genetically different H3N2 influenza A viruses in pigs. J. Clin. Microbiol. 41:1936-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen, D. L., A. I. Karasin, F. Zuckermann, and C. W. Olsen. 2000. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet. Microbiol. 74:117-131. [DOI] [PubMed] [Google Scholar]
- 32.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luoh, S. M., M. W. McGregor, and V. S. Hinshaw. 1992. Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J. Virol. 66:1066-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda, H., T. Suzuki, Y. Sugiyama, G. Horiike, K. Murakami, D. Miyamoto, K. I. P. J. Hidari, T. Ito, H. Kida, M. Kiso, K. Fukunaga, M. Ohuchi, T. Toyoda, A. Ishihama, Y. Kawaoka, and Y. Suzuki. 1999. Substitution of amino acid residue in influenza A virus hemagglutinin affects recognition of sialyl-oligosaccharides containing N-glycolylneuraminic acid. FEBS Lett. 464:71-74. [DOI] [PubMed] [Google Scholar]
- 35.Matrosovich, M., A. Tuzikov, N. Bovin, A. Gambaryan, A. Klimov, M. R. Castrucci, I. Donatelli, and Y. Kawaoka. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502-8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matrosovich, M. N., A. S. Gambaryan, S. Teneberg, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, and K. A. Karlsson. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224-234. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, B. R., D. L. Sly, E. L. Tierney, N. T. Hosier, J. G. Massicot, W. T. London, R. M. Chanock, R. G. Webster, and V. S. Hinshaw. 1982. Reassortant virus derived from avian and human influenza A viruses is attenuated and immunogenic in monkeys. Science 218:1330-1332. [DOI] [PubMed] [Google Scholar]
- 38.Naeve, C. W., V. S. Hinshaw, and R. G. Webster. 1984. Mutations in the hemagglutinin receptor-binding site can change the biological properties of an influenza virus. J. Virol. 51:567-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen, C. W. 2002. Emergence of novel strains of swine influenza virus in North America, p. 37-43. In A. Morilla, K.-J. Yoon, and J. J. Zimmerman (ed.), Trends in emerging viral infections of swine. Iowa State University Press, Ames, Iowa.
- 40.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 41.Olsen, C. W. 2004. Influenza in pigs and their role as the intermediate host. In K. G. Nicholson, R. G. Webster, A. Hay, and N. J. Cox (ed.), Textbook of influenza, in press. Blackwell Science, Oxford, United Kingdom.
- 42.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen, C. W., M. W. McGregor, A. J. Cooley, B. Schantz, B. Hotze, and V. S. Hinshaw. 1993. Antigenic and genetic analysis of a recently isolated H1N1 swine influenza virus. Am. J. Vet. Res. 54:1630-1636. [PubMed] [Google Scholar]
- 44.Palmer, D. F., W. R. Dowdle, M. T. Coleman, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis. U.S. Department of Health, Education and Welfare Immunology Series. U.S. Department of Health, Education and Welfare, Washington, D.C.
- 45.Peiris, J. S. M., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. S. Ip, R. W. M. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 47.Pensaert, M., K. Ottis, J. Vandeputte, M. M. Kaplan, and P. A. Bachmann. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull. W. H. O. 59:75-78. [PMC free article] [PubMed] [Google Scholar]
- 48.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers, G. N., and B. L. D'Souza. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317-322. [DOI] [PubMed] [Google Scholar]
- 50.Rogers, G. N., and J. C. Paulson. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361-373. [DOI] [PubMed] [Google Scholar]
- 51.Rogers, G. N., J. C. Paulson, R. S. Daniels, J. J. Skehel, I. A. Wilson, and D. C. Wiley. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76-78. [DOI] [PubMed] [Google Scholar]
- 52.Scholtissek, C., H. Burger, P. A. Bachmann, and C. Hannoun. 1983. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 129:521-523. [DOI] [PubMed] [Google Scholar]
- 53.Scholtissek, C., H. Burger, O. Kistner, and K. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287-294. [DOI] [PubMed] [Google Scholar]
- 54.Scholtissek, C., W. Rohde, V. Von Hoyningen, and R. Rott. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13-20. [DOI] [PubMed] [Google Scholar]
- 55.Scholtissek, C., and E. Naylor. 1988. Fish farming and influenza pandemics. Nature 331:215. [DOI] [PubMed] [Google Scholar]
- 56.Schultz, U., W. M. Fitch, S. Ludwig, J. Mandler, and C. Scholtissek. 1991. Evolution of pig influenza virus. Virology 183:61-73. [DOI] [PubMed] [Google Scholar]
- 57.Sheerar, M. G., B. C. Easterday, and V. S. Hinshaw. 1989. Antigenic conservation of H1N1 swine influenza viruses. J. Gen. Virol. 70:3297-3303. [DOI] [PubMed] [Google Scholar]
- 58.Snyder, M. H., A. J. Buckler-White, W. T. London, E. L. Tierney, and B. R. Murphy. 1987. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J. Virol. 61:2857-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Y. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki, T., G. Horiike, Y. Yamazaki, K. Kawabe, H. Masuda, D. Miyamoto, M. Matsuda, S. I. Nishimura, T. Yamagata, T. Ito, H. Kida, Y. Kawaoka, and Y. Suzuki. 1997. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 404:192-196. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki, Y., T. Ito, T. Suzuki, R. E. Holland, Jr., T. M. Chambers, M. Kiso, H. Ishida, and Y. Kawaoka. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825-11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, H. R., and A. J. Turner. 1977. A case report of fowl plague keratoconjunctivitis. Br. J. Ophthalmol. 61:86-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian, S.-F., A. J. Buckler-White, W. T. London, L. J. Reck, R. M. Chanock, and B. R. Murphy. 1985. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/N.Y./78 virus and its reassortants in squirrel monkey respiratory tract. J. Virol. 53:771-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webby, R. J., and R. G. Webster. 2001. Emergence of influenza A viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Webster, R. G., K. F. Shortridge, and Y. Kawaoka. 1997. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol. Med. Microbiol. 18:275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 69.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K.-J. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou, S. 1997. A practical approach to genetic screening for influenza virus variants. J. Clin. Microbiol. 35:2623-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]