Abstract
Somatic mutations in the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) gene are reportedly associated with sensitivity of lung cancers to gefitinib (Iressa), kinase inhibitor. In-frame deletions occur in exon 19, whereas point mutations occur frequently in codon 858 (exon 21). We found from sequencing the EGFR TK domain that 7 of 10 gefitinib-sensitive tumors had similar types of alterations; no mutations were found in eight gefitinib-refractory tumors (P = 0.004). Five of seven tumors sensitive to erlotinib (Tarceva), a related kinase inhibitor for which the clinically relevant target is undocumented, had analogous somatic mutations, as opposed to none of 10 erlotinib-refractory tumors (P = 0.003). Because most mutation-positive tumors were adenocarcinomas from patients who smoked <100 cigarettes in a lifetime (“never smokers”), we screened EGFR exons 2-28 in 15 adenocarcinomas resected from untreated never smokers. Seven tumors had TK domain mutations, in contrast to 4 of 81 non-small cell lung cancers resected from untreated former or current smokers (P = 0.0001). Immunoblotting of lysates from cells transiently transfected with various EGFR constructs demonstrated that, compared to wild-type protein, an exon 19 deletion mutant induced diminished levels of phosphotyrosine, whereas the phosphorylation at tyrosine 1092 of an exon 21 point mutant was inhibited at 10-fold lower concentrations of drug. Collectively, these data show that adenocarcinomas from never smokers comprise a distinct subset of lung cancers, frequently containing mutations within the TK domain of EGFR that are associated with gefitinib and erlotinib sensitivity.
Tyrosine kinases (TKs) regulate signaling pathways that control critical cellular activities (1). When overexpressed or activated by mutations, TKs can contribute to the development of cancers. If tumor cells depend on a mutant TK for survival, as illustrated by certain mouse models of cancer (2, 3), the mutated enzyme can fortuitously serve as an Achilles' heel for cancer therapy (4). Human examples include BCR-ABL-dependent chronic myelogenous and acute lymphoblastic leukemias (5), KIT- and PDGFRA-dependent gastrointestinal stromal tumors (6), and PDGFRA-dependent hypereosinophilic syndrome (7). In each disease, activated oncogenes encode TKs; inhibition by imatinib mesylate (Gleevec) leads to rapid and durable clinical responses.
EGFR is a TK of the ErbB family that is the presumptive target of the TK inhibitor (TKI) gefitinib. This drug is an anilinoquinazoline (Fig. 5, which is published as supporting information on the PNAS web site) that reversibly competes with ATP at a critical ATP-binding site (lysine 745; K745) within the epidermal growth factor (EGF) receptor (EGFR) protein (8, 9). In vitro, gefitinib selectively inhibits the kinase activity of EGFR versus a handful of other kinases (10). In two phase II trials, radiographic regressions of tumors were observed in 28% of patients treated in Japan and 10% of those studied in Europe and the U.S. (11, 12). Dramatic responses occurred within the first two weeks of initiating therapy (e.g., ref. 13), similar to those seen in the murine and human examples noted above. Assuming that the drug did affect a kinase, these kinds of responses suggested that at least some lung tumors depended on a specific genetic lesion for tumor survival. However, when gefitinib was approved as second- or third-line treatment for patients with non-small cell lung cancer (NSCLC), the clinically relevant target(s) of the drug in human tumors were unknown. Analyses of both preclinical xenograft models (14) and specimens from gefitinib-sensitive and -refractory tumors (15) did not reveal any obvious relationship between EGFR expression levels and tumor sensitivity. Retrospective epidemiologic analyses suggested that gefitinib is more likely to be effective in Japanese patients (11), individuals with adenocarcinomas of the bronchioloalveolar carcinoma (BAC) subtype, and “never smokers” (16).
Recently, two groups have shown that mutations in the TK domain of EGFR are associated with sensitivity of NSCLC to gefitinib (17, 18). In total, deletions or amino acid substitutions in exons 18, 19, and 21 of EGFR were found in 13 of 14 tumors sensitive to the drug, but in none of 11 tumors with no response. Lynch and colleagues (17) found mutations in another 2 of 25 primary NSCLCs, and Paez et al. (18) found EGFR mutations in 16 of 119 unselected tumors, with a striking predominance of mutations found in 15 of 58 (28%) specimens from Japan as compared to 1 of 61 from the U.S. (2%).
To confirm and extend data on gefitinib sensitivity, we examined the status of the TK domain of EGFR in tumors that were sensitive and refractory to the drug. To determine whether a related but distinct TKI, erlotinib (Fig. 5), “targets” a similar subset of NSCLCs, we also profiled erlotinib-sensitive and -refractory tumors. The clinically relevant target of erlotinib has not yet been documented. To examine whether smoking history is predictive of the likelihood of EGFR mutations, we determined the incidence of EGFR TK domain mutations in 96 resected NSCLCs from never smokers, as well as former and current smokers who had never received a TKI. Finally, in an effort to explain the selective advantage of cells with mutant EGFR and the drug sensitivity conferred upon mutant-bearing tumors, we began to characterize some biochemical properties of EGFR mutants in vitro.
Methods
Tissue Procurement. Tumor specimens were obtained on protocols approved by the Institutional Review Board and the Human Tissue Utilization Committee of Memorial Sloan-Kettering Cancer Center. Paraffin blocks of tumor material, obtained from patients before systemic treatment for lung cancer, were collected retrospectively for patients on gefitinib (n = 18) and prospectively for patients on erlotinib (n = 17). Frozen tumor specimens from untreated patients with Stage I-IIIA NSCLC (n = 96) were prospectively collected at the time of surgical resection (Supporting Text and Table 3, which are published as supporting information on the PNAS web site).
Mutational Analyses of EGFR in Lung Tumors. Genomic DNA was derived from either tumors embedded in paraffin blocks or from fresh frozen tumors (see Supporting Text and Tables 4 and 5, which are published as supporting information on the PNAS web site for PCR primers). All sequencing reactions were performed in both forward and reverse directions, and all mutations were confirmed by PCR amplification of an independent DNA isolate. For drug-sensitive tumors that did not have mutations, sequences from exons 19 and 21 were also determined from at least two independently derived PCR products. Fisher's exact test was used to calculate P values.
Functional Analyses of Mutant EGFRs. EGFR has two numbering systems. The first denotes the initiating methionine in the signal sequence as amino acid -24. The second, used here, denotes the methionine as amino acid +1. Commercial antibodies, such as the Y1068-specific anti-phospho-EGFR, use the first nomenclature. To be consistent, we consider Y1068 as Y1092.
Mutations were introduced into full-length EGFR by using a QuikChange Site-Directed Mutagenesis kit (Stratagene; see Supporting Text). All mutant clones were fully resequenced to ensure that no additional mutations were introduced. cDNAs were cloned into pcDNA3.1(-) expression vectors (Invitrogen). For transient transfections, 293T human embryonic kidney cells were transfected (2 × 105 cells per well in six-well plates) by using FuGENE (Roche Applied Science) and 0.8 μg of plasmid DNA. Cells were grown in DMEM with high glucose, 10% FCS, 2 mM l-glutamine, 10 units/ml penicillin, and 10 μg/ml streptomycin at 37°C and 5% CO2. Cells were serum-starved in media containing 0.1% serum. EGF (Cell Signaling Technology, Beverly, MA) was used at 100 ng/ml. Cells were treated with various concentrations of gefitinib or erlotinib, which were provided by AstraZeneca and Genentech, respectively. At least three independent experiments were performed for all analyses.
Immunoblotting. See Supporting Text for details on cell lysis and immunoblotting. Specific proteins were detected by using enhanced chemiluminescence (Amersham Pharmacia) and the following antibodies: horseradish peroxidase (HRP)-conjugated anti-phosphotyrosine (RC20) 1:3,333 and anti-total EGFR 1:2,500 (BD Transduction Laboratories), anti-actin 1:2,000 (Sigma), anti-phospho-EGFR (Tyr-1068; Y1092) 1:1,000 (Cell Signaling Technology), HRP-conjugated anti-rabbit Ig 1:5,000 (Amersham Pharmacia), and HRP-conjugated anti-mouse IgG 1:2,000 (Roche Applied Science). Blots were stripped with Restore Stripping Buffer (Pierce) at 37°C for 15 min; stripping was verified by reexposure to film before reprobing. Densitometry was performed by using imagequant v1.2 (Molecular Dynamics). At least three independent experiments were performed for all analyses.
Results
Mutations in EGFR Are Commonly Found in Lung Tumors Sensitive to Gefitinib. To ascertain whether mutations within the TK domain of EGFR are associated with sensitivity to the TKI, gefitinib (17, 18), we performed mutational profiling of exons 18-24 of EGFR in tumors from 10 patients who demonstrated a partial response or marked clinical improvement (defined in Supporting Text) when treated with the drug as a single-agent at the Memorial Sloan-Kettering Cancer Center. All patients were selected retrospectively. Seven tumors (70%) had mutations, six of which had in-frame nucleotide deletions in exon 19 that occur adjacent to K745 encoded by nucleotides 2233-2235. K745 has been shown to be critical for binding ATP (19). A seventh patient had a nonsynonymous mutation at nucleotide 2573 (T → G) in exon 21, resulting in a substitution of arginine for leucine at position 858 (L858R). This change occurs adjacent to the highly conserved DFG motif (amino acids 855-857) in the activation loop of the kinase (20) and was previously detected in NSCLCs (17, 18) (Table 1 and Fig. 6 A-D, which is published as supporting information on the PNAS web site). Mutations were not found in DNA from peripheral blood available from four patients (G2, G3, G5, and G6), implying that the lesions arose in somatic cells. Among the seven tumors with mutations, five arose in never smokers, six had adenocarcinoma histology with features of BAC, and none were from patients of East Asian origin.
Four of the exon 19 deletion mutations were the same as the previously described del E746-A750 (17, 18), and two were not previously described (del L747-S752 and del E746-T751insI). The del E746-T751insI mutation may have resulted from a double deletion of nucleotides 2229-2236 and 2245-2252 and an insertion of a single T nucleotide (Fig. 1 A and B). Remarkably, in this mutant, despite elimination of the codon for K745, the remaining nucleotide sequence reencoded a lysine but eliminated amino acids LREA, as in the other exon 19 deletions. The LREA motif is completely conserved among EGFRs in vertebrates (Fig. 1C).
Fig. 1.
Deletion mutations in exon 19 of EGFR from NSCLCs sensitive to TKIs, gefitinib (G) or erlotinib (E); all lack four amino acids, LREA, which are conserved among vertebrate species. (A) Nucleotide alignments. (B) Amino acid alignments. (C) Amino acid alignments of EGFR from various species. All alignments were generated by using Vector nti software. See Table 1 for characteristics of patients.
Most somatic EGFR mutations in NSCLCs have been reported to be heterozygous. However, in the analysis of two of seven tumors (G1 and G3), no wild-type sequence was detected in the region encompassing the deletion. This result implies that (i) the mutations in tumors from patients G1 and G3 were hemi- or homozygous, (ii) the mutant gene was selectively amplified in these tumors, or (iii) mutations in general may be homozygous, with wild-type sequence originating from contaminating “normal” DNA (Table 1 and Fig. 6A; see Discussion).
Table 1. Characteristics of patients sensitive to gefitinib (G) and erlotinib (E).
| Patient | Sex | Smoking | Histology | Mutation | Duration | OS |
|---|---|---|---|---|---|---|
| G1 | M | Former | BWFI | del E746-A750* | 5+ | 5+ |
| G2 | F | Never | AWBF | del E746-A750 | 28 | 30+ |
| G3 | F | Never | AWBF | del L747-S752* | 18 | 23+ |
| G4 | F | Never | AWBF | del E746-T751 insl | 9 | 14 |
| G5 | M | Never | AWBF | del E746-A750 | 8 | 15 |
| G6 | M | Never | AWBF | del E746-A750 | 5+ | 5+ |
| G7 | F | Former | ADENO | L858R | 5 | 8 |
| G8 | F | Never | AWBF | None | 7+ | 7+ |
| G9 | M | Former | BAC | None | 10+ | 10+ |
| G10 | F | Never | SQUAM | None | 9 | 16 |
| E1 | M | Never | AWBF | del L747-S752insQ | 8+ | 8+ |
| E2 | M | Never | BAC | del E746-A750 | 8.5 | 21+ |
| E3 | F | Never | AWBF | R776C and L858R | 13 | 22+ |
| E4 | M | Former | AWBF | L858R | 3 | 3.5 |
| E5 | F | Never | AWBF | L858R | 6 | 17+ |
| E6 | F | Former | AWBF | None | 11 | 11+ |
| E7 | F | Former | AWBF | None | 11 | 14+ |
M, male; F, female. Smoking indicates smoking history; never, smoked <100 cigarettes in a lifetime; former, smoked 100 or more cigarettes and quit > 1 year prior to diagnosis of lung cancer. BWFI, BAC with focal invasion; AWFB, adenocarcinoma with BAC features; ADENO, adenocarcinoma; SQUAM, squamous. Mutation indicates amino acids affected in EGFR; for all tumor specimens, exons 18-24 of EGFR, which encode the TK domain, were examined; asterisks denote homozygous deletions; none, no mutation observed. Duration indicates months of drug-induced response. OS indicates months of overall survival after starting therapy with gefitinib or erlotinib. +, still on drug and/or alive at the time of writing of this manuscript. No mutations were observed in exons 18-24 in 8 and 10 patients refractory to gefitinib and erlotinib, respectively.
None of eight gefitinib-refractory tumors contained mutations within exons 18-24 of EGFR (P = 0.004). Thus, EGFR mutations appear to be associated with sensitivity of NSCLCs to gefitinib.
Lung Tumors Sensitive to Erlotinib Also Harbor Mutations in EGFR. Erlotinib, like gefitinib, is an ATP-competitive inhibitor of the EGFR TK (Fig. 5). Erlotinib also appears to be particularly effective in lung cancers with BAC histology and in patients who are never smokers,‡‡ but the drug's clinically relevant target in tumors has not yet been documented. To see whether lung tumors sensitive to erlotinib also contained mutations within the TK domain of EGFR, we analyzed exons 18-24 in seven tumors from patients who demonstrated a partial response while on a Phase II trial of this agent in BAC.
Five tumors contained mutations, two of which were multinucleotide in-frame deletions within exon 19 near the ATP-binding site (K745), similar to those found in gefitinib-sensitive tumors. These deletions all eliminated amino acids LREA (amino acids 747-750) (Fig. 1 A and B). One mutation (del L747-S752insQ) has not been previously reported. Three tumors contained the L858R mutation at nucleotide 2573 that is also found in gefitinib-responsive tumors. One specimen also had a second mutation that was previously unreported: a C → T change at nucleotide 2326 in exon 20, substituting cysteine for arginine at position 776 (R776C), carboxy-terminal to the P-loop (Fig. 6E). Four of five mutation-positive tumors were obtained from never smokers, and none were from patients of East Asian origin.
Matched normal tissue available from four patients (E1-E3 and E5) showed only the wild-type sequence, indicating that mutations arose in somatic cells. No mutations were observed in exons 18-24 in tumors from 10 patients on the trial who did not have a partial response (P = 0.003). Thus, similar to results with gefitinib, mutations in the EGFR TK domain are also associated with sensitivity of NSCLCs to erlotinib.
EGFR Mutations Are Commonly Found in Lung Adenocarcinomas From Never Smokers. Nine of 12 (75%) mutation-positive tumors in this study had adenocarcinoma histology and were derived from never smokers (Table 1). Moreover, ≈10% of all cases of lung cancer arise in patients who have no history of tobacco use, most of which have adenocarcinoma histology (21, 22). To determine in a prospective manner the frequency of EGFR mutations in lung adenocarcinomas from never smokers, we sequenced exons 2-28 of EGFR in 15 tumor specimens that fit these clinical criteria. These samples were selected from our prospectively collected tissue bank of surgically resected NSCLCs derived from patients with stage I, II, and IIIA disease. At the time of surgery and tissue banking, none of the patients from whom the primary tumor was resected had received any treatment for lung cancer, including TKIs. In this “enriched” population of tumors from never smokers, 7 of 15 (47%) had mutations in the TK domain of EGFR. One tumor (specimen 230) had a deletion in exon 19 (del E746-A750), and five (specimens 3, 24, 25, 42, and 166) had the L858R amino acid substitution in exon 21 (Table 2). The seventh tumor (specimen 77) contained a previously unreported point mutation in exon 21 at nucleotide 2504 (A → T), resulting in substitution of leucine for histidine at position 835 (H835L) (Fig. 6F). This mutation is predicted to lie under or adjacent to the activation loop of the EGFR TK domain, in contrast to the L858R mutation, which is in the loop itself. No mutations were found in adjacent normal-appearing tissue from all seven patients.
Table 2. Somatic mutations in the TK domain of EGFR are common in surgically resected NSCLCs derived from never smokers but infrequent in former or current smokers.
| Specimen | Sex | Smoking | Histology | Mutation |
|---|---|---|---|---|
| 3 | F | Never | AWBF | L858R |
| 16 | F | Never | BWFI | None |
| 20 | F | Never | BWFI | None |
| 24 | M | Never | AWBF | L858R |
| 25 | F | Never | AWBF | L858R |
| 33 | F | Never | AWBF | None |
| 42 | M | Never | AWBF | L858R |
| 77 | F | Never | AWBF | H835L |
| 82 | F | Never | AWBF | None |
| 120 | F | Never | AWBF | None |
| 124 | F | Never | AWBF | None |
| 166 | F | Never | AWBF | L858R |
| 169 | F | Never | AWBF | None |
| 176 | F | Never | ADENO | None |
| 230 | M | Never | ADENO | E746-A750 |
| 5 | F | Former | ADENO | L858R |
| 65 | M | Former | AWBF | L858R |
| 98 | F | Former | AWBF | L858R |
| 134 | M | Former | ADENO | L858R |
Exons 2-28 from EGFR were examined for mutations in 96 surgically resected NSCLCs. Mutations in the TK domain were detected in 7 of 15 never smokers with adenocarcinomas and in 4 of 81 NSCLCs resected from former or current smokers. All abbreviations as per Table 1.
We also sequenced exons 2-28 of EGFR from an additional 81 primary NSCLCs randomly selected from the same tumor bank. All tumors in this cohort were derived from former or current smokers, and 24 tumors had squamous cell histology. Four of 81 (5%) had mutations in the EGFR TK domain; all were the previously observed L858R amino acid substitution within exon 21 (Table 2). Corresponding normal tissue, available for three of these four tumors (specimens 5, 65, and 134), had only wild-type sequence. Interestingly, among the four tumors with the L858R mutation, three (specimens 65, 98, and 134) arose in patients with a limited exposure to cigarette smoking: all three had smoked ≤1 pack per day for 9 years (9 “pack years”) and had quit at least 30 years before surgery. Specimen 5 was resected from an individual with a 14 pack-year history who quit 1 month before surgery. Taken together, these data demonstrate that EGFR mutations are commonly found in NSCLCs from never smokers as opposed to former or current smokers (7 of 15 vs. 4 of 81; P = 0.0001), and that tumors likely to be mutation-positive can be identified by using specific clinical characteristics.
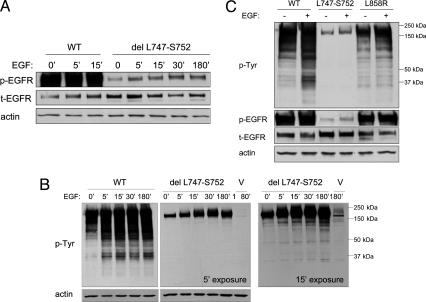
Biochemical Properties of EGFR Mutants. To gain further insight as to why cells containing mutant EGFRs are selected during the growth of certain NSCLCs and why mutations confer susceptibility to TKIs, we have begun to characterize the mutant proteins in cultured cells. Wild-type, del L747-S752, and L858R EGFR were produced by transient transfection with expression vectors in 293T cells, which have very low levels of endogenous EGFR (Fig. 2). Expression of total EGFR (t-EGFR) was assessed by immunoblotting using an anti-EGFR monoclonal antibody, and actin served as an indicator of relative levels of protein per sample. The size of mutant EGFRs was virtually indistinguishable from wild-type EGFR when assessed by immunoblotting. Interestingly, the amount of t-EGFR relative to actin was, on average, 3-fold higher for the del L747-S752 protein than that for wild-type EGFR (n = 5). The differences in t-EGFR were not likely caused by varying transfection efficiencies, as equal numbers of cells were used for each separate transfection and levels of t-EGFR for another EGFR mutant (L858R) were comparable to that of wild type (Fig. 2C).
Fig. 2.
The del L747-S752 mutant EGFR appears to have reduced kinase activity. 293T cells were transiently transfected with vector alone (V) or vector containing wild-type (WT) EGFR, del L747-S752, or L858R. Thirty-six hours later, cells were serum-starved for 24 h and then harvested for immunoblot analyses using anti-phosphotyrosine (p-Tyr), anti-phospho-EGFR (p-EGFR Y1092), anti-total EGFR (t-EGFR), and anti-actin antibodies as described in Methods.(A and B) Time course of ligand-induced activation of del L747-S752 mutant EGFR. Cells were treated with 100 ng/ml EGF for 0-180 min. (B Left) A 5-min exposure of an immunoblot assayed with an anti-phosphotyrosine antibody. (B Right) A 15-min exposure. (C) Comparison of WT EGFR with two EGFR mutants, as assessed by detection of p-Tyr and p-EGFR (Y1092).
We next used immunoblotting of extracts from cells expressing the various EGFRs to assess various aspects of protein activity and drug sensitivity. As a surrogate gauge of kinase activity, we measured the levels of “autophosphorylated” Tyr-1092 on EGFR (Y1092; the site of binding by adaptor molecules such as Grb2, which leads to activation of the mitogen-activated protein kinase/extracellular signal-related kinase cascade) by using an Y1092-specific antibody (i.e., phospho-EGFR or p-EGFR), in relation to levels of t-EGFR protein. We also assessed the pattern and levels of induced tyrosine-phosphorylation of cell proteins by using an anti-phosphotyrosine antibody (RC-20). In the absence of serum and EGF, extracts from cells transfected with wild-type EGFR demonstrated, on average, a 16-fold greater ability to autophosphorylate Y1092 than did the del L747-S752 mutant, even after addition of EGF (n = 5) (Fig. 2 A and C). Consistent with these results, the del L747-S752 mutant also induced markedly low levels of tyrosine-phosphorylated proteins compared to wild-type EGFR (Fig. 2 B and C). A longer exposure demonstrated that the relative intensities of such proteins appeared to be qualitatively different from that of wild type as well (Fig. 2B Right). In contrast to the deletion mutant, results with the Y1092-specific antibody for the L858R mutant were similar to those observed with wild-type EGFR (Figs. 2C and 3B). However, the pattern of phosphotyrosine staining of cell proteins was still distinct (Fig. 2C).
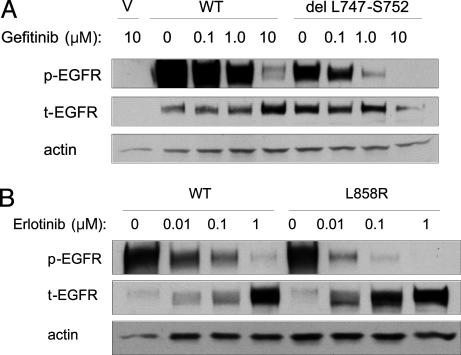
Fig. 3.
Compared to wild-type EGFR, the del L747-S752 mutant has similar sensitivity to TKIs, whereas the L858R mutant is inhibited at ≈10-fold lower concentrations of drug. (A) Dose-dependent inhibition by gefitinib of del L747-S752 mutant EGFR as compared to wild type. Cells were treated with gefitinib at various concentrations for 1 h before lysis. Results with erlotinib and in the presence of EGF were similar (data not shown). (B) Dose-dependent inhibition by erlotinib of L858R mutant as compared to wild-type EGFR. Dilution points at 0.0001 micromolar are not shown. Results with gefitinib were similar (data not shown). V, vector alone.
Finally, we assessed the sensitivity of the del L747-S752 and L858R EGFR mutants to TKIs, by measuring the ratio of p-EGFR/t-EGFR in lysates from transiently transfected cells that were serum-starved and pretreated with gefitinib or erlotinib. Wild-type EGFR and the del L747-S752 mutant appeared to have approximately the same sensitivity to both gefitinib (Fig. 3A) and erlotinib (data not shown). By contrast, the L858R mutant had an ≈10-fold greater sensitivity to both gefitinib (data not shown) and erlotinib (Fig. 3B and Fig. 7, which is published as supporting information on the PNAS web site).
Discussion
In this study, we confirm and extend recent work associating EGFR mutations with sensitivity to the TKI, gefitinib (17, 18). Furthermore, we establish that tumors sensitive to a related kinase inhibitor, erlotinib, contain similar types of EGFR mutations. When data from the two published reports and this study are used, 25 of 31 (81%) tumors from individuals experiencing partial responses or marked clinical improvement while taking gefitinib or erlotinib contain mutations in the EGFR TK domain. By contrast, none of 29 specimens from patients refractory to these agents had such mutations (P < 10-10). These findings demonstrate that mutations in the TK domain of EGFR are associated with sensitivity to these two drugs. Whether gefitinib and erlotinib target exactly the same or overlapping sets of NSCLC patients and whether distinct mutations confer greater sensitivity to specific EGFR-TK inhibitors has not yet been determined, because the number of sensitive tumors analyzed is still small.
We also demonstrate that 11 of 96 (12%) primary NSCLCs resected from untreated patients contain mutations in EGFR, all within the TK domain. None of these tumors were derived from patients of East Asian origin. Taken together with the published literature on EGFR mutations in primary lung cancers from patients in the U.S., 14 of 182 tumors (8%) are positive for EGFR mutations. These data could account for the responses seen in the phase II trials of gefitinib, in which ≈10% of European or American patients experienced radiographic regressions (12). However, other mechanisms of drug sensitivity may also apply. Remarkably, by selecting tumors for certain clinical characteristics predictive of response to TKIs, i.e., tumors from never smokers with adenocarcinoma histology, we enriched the percentage of patients with such mutations. Thus, 7 of 15 tumors from never-smoking patients with adenocarcinoma histology had EGFR mutations, whereas only 4 of 81 NSCLCs from former and current smokers contained them. Moreover, three of the four patients in the latter cohort had relatively short smoking histories. These data show that lung tumors from patients with minimal direct exposure to cigarettes and with adenocarcinoma histology, usually with features of BAC, have a distinct molecular phenotype that distinguishes them from the remainder of NSCLCs. How patients with mutation-positive NSCLC at various stages of disease should be treated is unknown, but clearly warrants prompt investigation. At the same time, critical mutations in other kinases should be sought in NSCLCs with wild-type EGFR from never, former, and current smokers.
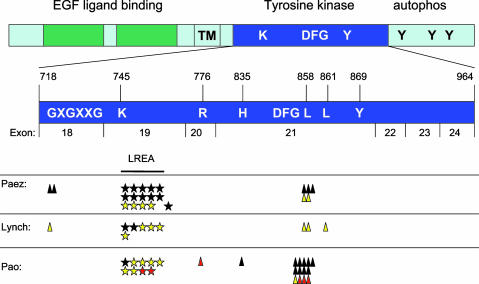
Among the NSCLC-associated EGFR mutations reported to date, 49 of 56 (88%) occur in two “hotspots” (Fig. 4). A total of 29 of these 49 (59%) are multinucleotide in-frame deletions that eliminate four amino acids (LREA) in exon 19. The other 20 of 49 “hotspot” mutations (41%) are point mutations in exon 21 that result in a specific amino acid substitution at position 858 (L858R). The remaining 7 of 56 mutations (12%) are nucleotide substitutions found in exons 18-21 outside of the common sites of mutation, including the previously unreported R776C and H835L mutations described here. In nearly all cases, only one mutation has been detected per tumor. However, in this study, we did find one tumor sample with two mutations (patient E3, R776C and L858R). The significance of the R776C mutation and whether these two mutations are on the same chromosome, different chromosomes in the same cells, or from different subclones with the same tumor specimen are all unknown.
Fig. 4.
Summary of mutations reported here and previously detected in the TK domain of EGFR in NSCLCs. Schematic view of EGFR and key domains, with an expanded view of the TK domain encoded by exons 18-24 (amino acids 718-964). Yellow, sensitive to gefitinib; red, sensitive to erlotinib; black, never treated with TKI. Data are from this paper and refs. 17 and 18. All exon 19 deletions lack amino acids LREA except for one reported by Paez et al. (18) (del S752-I759). In our series, one tumor with an L858R mutation also had an R776C mutation (see Table 1). Functional landmarks include the GXGXXG motif at position 719, the K745 critical for ATP-binding, the DFG motif at position 855, and a tyrosine at 869. The sites of described mutations are G719, L747-A750, S752, R776, H835, L858, and L861.
NSCLC-associated EGFR mutations are most frequently heterozygous. However, Paez et al. (18) reported one mutation involving exon 19 that appeared to be homozygous, and we detected two such cases. Interpretation of mutational status solely from DNA sequencing can be problematic. On the one hand, contaminating normal cells with wild-type EGFR could account for apparent heterozygosity; on the other hand, amplification of mutant EGFR, as occurs in lung cancer (23), could account for detection of only mutant sequences. Mouse models expressing mutant EGFR proteins in the lung and analysis of mutant-positive NSCLCs by fluorescence in situ hybridization and/or array-based comparative genomic hybridization may help address these issues. Interestingly, in one of these tumors (G3), we detected a heterozygous intronic polymorphism downstream of exon 19 (data not shown). In this case, it is probable that a gene conversion event occurred, encompassing the area of deletion in exon 19.
Five of 17 reported patients who experienced partial responses or marked clinical improvement while on therapy with TKIs had wild-type sequence in exons 18-24. In the series reported by Lynch et al. (17), one of nine gefitinib-sensitive patients did not have mutations within the entire coding region of EGFR. Explanations for these results include: (i) tumor fragments analyzed did not represent the tumors assessed while patients were on drug studies, (ii) mutations were present but were below the detection rate of sequencing assays, (iii) mutations lay outside the exons encoding the TK domain, and (iv) other mechanisms involving wild-type EGFR (e.g., amplification) may confer drug sensitivity. In regards to the third point, mutations outside the TK domain are unlikely, because no other mutations in EGFR were found in 240 lung tumor specimens sequenced to date [119 tumors from Paez et al. (18), 25 tumors from Lynch et al. (17), and 96 resected NSCLCs in this study]. Thus, it will be important to determine whether wild-type EGFR or other kinases play a role in tumor responses to gefitinib and erlotinib.
Why mutant EGFRs are selected for and how they confer susceptibility to TKIs are questions that require further investigation. To gain further insight, we have begun to study various EGFRs [wild type, an exon 19 deletion (del L747-S752), and an exon 21 aa substitution (L858R)] in cultured cells with low levels of endogenous EGFR by using transient transfection assays and parameters of activity, such as the ability to phosphorylate tyrosine 1092 on EGFR itself and to phosphorylate tyrosine residues on cell proteins in general. Compared to wild-type EGFR, the relative intensities of tyrosine-phosphorylated proteins induced in cells after transfection by either EGFR mutant was qualitatively and quantitatively different. The level of phosphotyrosine was especially diminished for the del L747-S752 mutant, a result that we also obtained in two other cell lines (i.e., COS1 and HPL1D, immortalized human peripheral lung epithelia, ref. 24), and in 293T cells by using another deletion mutant (del E746-A750, data not shown). Furthermore, in contrast to published data (17), we found that these two EGFR mutants had differential sensitivities to TKIs, as measured by the effect of drug on phosphorylation of EGFR at Y1092 in cells transiently transfected with various EGFRs. Whereas the L858R mutant was inhibited at ≈10-fold lower concentrations of TKI, the del L747-S752 mutant appeared to have similar sensitivities as wild-type EGFR to drug. Discrepancies with published reports could be due to by different experimental conditions. However, our data do not support the notion that EGFR mutants have enhanced activity, as has been suggested (17). Rather, our results suggest that the del L747-S752 and L858R mutants could have altered substrate specificity compared to wild-type protein. Interestingly, mutant EGFRs in which the critical K745 residue is changed to methionine or arginine also have reduced kinase activity, but can still activate the mitogen-activated protein kinase cascade, and have an “incomplete program” of cellular tyrosine phosphorylations. Such signaling is postulated to occur via heterodimerization with other ErbB family members, such as ErbB2/Neu (25-27).
The biochemical properties of the del L747-S752 mutant as evaluated above are reminiscent of certain B-RAF mutants found in human cancers (28). Although the majority of B-RAF mutants have elevated kinase activity, 3 of 22 reported B-RAF mutants were found to have reduced kinase activity toward MEK in vitro. Nevertheless, these three mutants were found to signal to ERK in cells by activating C-RAF, possibly via an allosteric or transphosphorylation mechanism.
Tumors that are sensitive to either gefitinib or erlotinib eventually progress despite continued treatment with TKIs. In patients with BCR-ABL-dependent chronic myelogenous leukemia, mutations within or amplification of BCR-ABL lead to clinical resistance (29). Whether resistance to EGFR TKIs is caused by similar mechanisms affecting EGFR and/or to other mechanisms affecting alternative molecules remains to be determined.
Supplementary Material
Acknowledgments
We thank J. Eldred, G. Fewell, T. Miner, J. Reed, C. Jeliti, H. Sun, E. Boatright, H. Bauer, and N. Hershberger from Washington University for technical assistance and data generation/analysis, and M. Watson from the Siteman Cancer Center Tissue Procurement Core for additional preparation of genomic DNA samples. We especially thank R. Bhargava and M. Ladanyi from the Memorial Sloan-Kettering Cancer Center for DNA extraction from paraffin-embedded tumors; D. Tabarini, M. Ilzarbe, and D. Wong from the Memorial Sloan-Kettering Cancer Center Sequencing Facility, and D. K. Pham for management of the surgical tumor bank. Additionally, we acknowledge M. Resh for the wild-type EGFR cDNA; E. Venkatraman for statistical calculations; P. Yurttas and N. Pavletich for discussion about EGFR structure; J. Massague for use of imagequant software; F. Cong for helpful discussions; V. Ty for peripheral blood collection; N. Shah, J. Kunkle, L. LoGuidice, R. Henry, M. Cespon, E. Ro, and J. Jones for clinical research assistance; and AstraZeneca and Genentech for supply of gefitinib and erlotinib, respectively. W.P. was supported by National Cancer Institute Grant CA009512 and the Steps for Breath Foundation. The work was also funded by an anonymous donor.
Abbreviations: TK, tyrosine kinase; TKI, TK inhibitor; EGF, epidermal growth factor; EGFR, EGF receptor; t-EGFR, total EGFR; p-EGFR, phospho-EGFR; NSCLC, non-small cell lung cancer; BAC, bronchioloalveolar carcinoma.
Footnotes
Miller, V. A., Patel, J., Shah, N., Kris, M. G., Tyson, L., Pizzo, B., Zakowski, M., Memoli, N., Sandler, A. & Johnson, D. H. (2003) Proc. Am. Soc. Clin. Oncol., abstract 2491 (abstr.).
References
- 1.Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355-365. [DOI] [PubMed] [Google Scholar]
- 2.Moody, S. E., Sarkisian, C. J., Hahn, K. T., Gunther, E. J., Pickup, S., Dugan, K. D., Innocent, N., Cardiff, R. D., Schnall, M. D. & Chodosh, L. A. (2002) Cancer Cell 2, 451-461. [DOI] [PubMed] [Google Scholar]
- 3.Huettner, C. S., Zhang, P., Van Etten, R. A. & Tenen, D. G. (2000) Nat. Genet. 24, 57-60. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein, I. (2002) Science 297, 63-64. [DOI] [PubMed] [Google Scholar]
- 5.Druker, B. (2002) Trends Mol. Med. 8, S14-S18. [DOI] [PubMed] [Google Scholar]
- 6.Demetri, G. (2001) Semin. Oncol. 28, 19-26. [PubMed] [Google Scholar]
- 7.Cools, J., DeAngelo, D. J., Gotlib, J., Stover, E. H., Legare, R. D., Cortes, J., Kutok, J., Clark, J., Galinsky, I., Griffin, J. D., et al. (2003) N. Engl. J. Med. 348, 1201-1214. [DOI] [PubMed] [Google Scholar]
- 8.Ward, W. H., Cook, P. N., Slater, A. M., Davies, D. H., Holdgate, G. A. & Green, L. R. (1994) Biochem. Pharmacol. 48, 659-666. [DOI] [PubMed] [Google Scholar]
- 9.Barker, A. J., Gibson, K. H., Grundy, W., Godfrey, A. A., Barlow, J. J., Healy, M. P., Woodburn, J. R., Ashton, S. E., Curry, B. J., Scarlett, L., et al. (2001) Bioorg. Med. Chem. Lett. 11, 1911-1914. [DOI] [PubMed] [Google Scholar]
- 10.Wakeling, A. E., Guy, S. P., Woodburn, J. R., Ashton, S. E., Curry, B. J., Barker, A. J. & Gibson, K. H. (2002) Cancer Res. 62, 5749-5754. [PubMed] [Google Scholar]
- 11.Fukuoka, M., Yano, S., Giaccone, G., Tamura, T., Nakagawa, K., Douillard, J. Y., Nishiwaki, Y., Vansteenkiste, J., Kudoh, S., Rischin, D., et al. (2003) J. Clin. Oncol. 21, 2237-2246. [DOI] [PubMed] [Google Scholar]
- 12.Kris, M. G., Natale, R. B., Herbst, R. S., Lynch, T. J., Jr., Prager, D., Belani, C. P., Schiller, J. H., Kelly, K., Spiridonidis, H., Sandler, A., et al. (2003) J. Am. Med. Assoc. 290, 2149-2158. [DOI] [PubMed] [Google Scholar]
- 13.Pao, W., Miller, V. A. & Kris, M. G. (2004) Semin. Cancer Biol. 14, 33-40. [DOI] [PubMed] [Google Scholar]
- 14.Sirotnak, F. M., Zakowski, M. F., Miller, V. A., Scher, H. I. & Kris, M. G. (2000) Clin. Cancer Res. 6, 4885-4892. [PubMed] [Google Scholar]
- 15.Baselga, J. (2004) J. Clin. Oncol. 22, 759-768. [DOI] [PubMed] [Google Scholar]
- 16.Miller, V. A., Kris, M. G., Shah, N., Patel, J., Azzoli, C., Gomez, J., Krug, L. M., Pao, W., Rizvi, N., Pizzo, B., et al. (2004) J. Clin. Oncol. 22, 1103-1109. [DOI] [PubMed] [Google Scholar]
- 17.Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., et al. (2004) N. Engl. J. Med. 350, 2129-2139. [DOI] [PubMed] [Google Scholar]
- 18.Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman, N., Boggon, T. J., et al. (2004) Science 304, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 19.Honegger, A. M., Dull, T. J., Felder, S., Van Obberghen, E., Bellot, F., Szapary, D., Schmidt, A., Ullrich, A. & Schlessinger, J. (1987) Cell 51, 199-209. [DOI] [PubMed] [Google Scholar]
- 20.Huse, M. & Kuriyan, J. (2002) Cell 109, 275-282. [DOI] [PubMed] [Google Scholar]
- 21.Doll, S. R. (2000) Am. J. Respir. Crit. Care Med. 162, 4-6. [DOI] [PubMed] [Google Scholar]
- 22.Kabat, G. C. & Wynder, E. L. (1984) Cancer 53, 1214-1221. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch, F. R., Varella-Garcia, M., Bunn, P. A., Jr., Di Maria, M. V., Veve, R., Bremmes, R. M., Baron, A. E., Zeng, C. & Franklin, W. A. (2003) J. Clin. Oncol. 21, 3798-3807. [DOI] [PubMed] [Google Scholar]
- 24.Masuda, A., Kondo, M., Saito, T., Yatabe, Y., Kobayashi, T., Okamoto, M., Suyama, M. & Takahashi, T. (1997) Cancer Res. 57, 4898-4904. [PubMed] [Google Scholar]
- 25.Ewald, J. A., Coker, K. J., Price, J. O., Staros, J. V. & Guyer, C. A. (2001) Exp. Cell Res. 268, 262-273. [DOI] [PubMed] [Google Scholar]
- 26.Selva, E., Raden, D. L. & Davis, R. J. (1993) J. Biol. Chem. 268, 2250-2254. [PubMed] [Google Scholar]
- 27.Wright, J. D., Reuter, C. W. & Weber, M. J. (1995) J. Biol. Chem. 270, 12085-12093. [DOI] [PubMed] [Google Scholar]
- 28.Wan, P. T., Garnett, M. J., Roe, S. M., Lee, S., Niculescu-Duvaz, D., Good, V. M., Jones, C. M., Marshall, C. J., Springer, C. J., Barford, D., et al. (2004) Cell 116, 855-867. [DOI] [PubMed] [Google Scholar]
- 29.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876-880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.