Abstract
Robustness, a relative insensitivity to perturbations, is a key characteristic of living cells. However, the specific structural characteristics that are responsible for robust performance are not clear, even in genetic circuits of moderate complexity. Formal sensitivity analysis allows the investigation of robustness and fragility properties of mathematical models representing regulatory networks, but it yields only local properties with respect to a particular choice of parameter values. Here, we show that by systematically investigating the parameter space, more global properties linked to network structure can be derived. Our analysis focuses on the genetic oscillator responsible for generating circadian rhythms in Drosophila as a prototypic dynamical cellular system. Analysis of two mathematical models of moderate complexity shows that the tradeoff between robustness and fragility is largely determined by the regulatory structure. Rank-ordered sensitivities, for instance, allow the correct identification of protein phosphorylation as an influential process determining the oscillator's period. Furthermore, sensitivity analysis confirms the theoretical insight that hierarchical control might be important for achieving robustness. The complex feedback structures encountered in vivo, however, do not seem to enhance robustness per se but confer robust precision and adjustability of the clock while avoiding catastrophic failure.
Cells must establish robust functions to be able to adapt to external changes, to tolerate an uncertain internal environment in terms of stochastic phenomena and variability in the concentrations of cellular components, and to cope with mutations. This requirement likely accounts for the complexity of cellular control. Redundant components or pathways and feedback control, as well as modular and hierarchical organization of intracellular networks, are means for achieving robustness, but they also increase the complexity of a system (1, 2). A high level of complexity implies a large number of components and interactions as targets for potentially deleterious attacks, requiring additional mechanisms for stabilization, which leads to “spiraling complexity” (1). As a consequence, investigations into the causes for the robustness of specific biological systems prove especially demanding.
Currently, it is largely unclear which structural characteristics of specific cellular networks are responsible for particular types of robust performance. Structural determinants of robustness have been elucidated only for steady-state (3, 4) or relatively simple dynamic (5–7) descriptions of cellular subsystems. In principle, formal sensitivity analysis of mathematical models describing more complex networks could allow for linking robustness properties to network structure. It is, however, rarely used for robustness analysis in biological systems. Moreover, this type of analysis sheds light only on local (i.e., in the neighborhood of a particular choice of parameter values) characteristics of a system. We propose a systematic analysis of sensitivities for many plausible sets of parameters to ultimately reveal robustness properties rooted in the structure of a control circuit. The central challenge addressed here is to what extent can (computationally) feasible sensitivity analyses yield results that comply with the biological knowledge and/or lead to novel hypotheses on the function of cellular networks of realistic complexity.
As a prototypic example system, we chose the core architecture of the genetic oscillator responsible for generating circadian rhythms in Drosophila. Circadian clocks provide endogenously controlled oscillations at the cellular level with a period of ≈24 h, allowing the organism to adapt to the day–night rhythms imposed by the environment. They generate complex behavior, are relatively well understood, and have been shown to be robust (8–11).
In Drosophila, a negative autoregulatory feedback loop established by the period (per) and timeless (tim) genes is at the heart of the circadian oscillator (Fig. 1). A current view is that, after their expression, these proteins are phosphorylated at multiple residues. This leads to a time delay between the rise of mRNAs and of the PER/TIM heterodimer acting as transcriptional repressor for both genes. Alternating protein production, gene repression, and protein degradation may, thus, lead to self-sustained oscillations. The network is further complicated by an interconnected positive feedback loop via dclock (dclk) (12) and by the influence of the kinase doubletime (dbt) on degradation and transport of the PER/TIM complex. Moreover, the rate of TIM degradation is (indirectly) controlled by light, which enables synchronization (entrainment) with the environment (9, 10).
Fig. 1.
Molecular interactions governing circadian rhythms in Drosophila. Genes (names in lowercase) and protein products (shaded ellipses) are connected by regulatory interactions that are either direct (solid lines) or indirect (dashed lines). Arrows and bar heads indicate positive and negative regulation, respectively. The box delimits the nucleus. Time delays are thought to result from protein phosphorylation cascades; cyc was omitted because it seems to be unregulated (10).
Methods
Mathematical Models. Over the past years, a number of deterministic models for the highly conserved circadian clock in Neurospora (13–15), in Drosophila (15–19), and in mammals (20, 21) have been proposed. They differ largely in the detail of the specific oscillator and, consequently, in their complexity. Our attention in this work is on ordinary differential equation models; however, there are lower-dimensional models that can be derived when the feedback delays that arise from, for example, phosphorylation are replaced by explicit delay terms, yielding differential-delay models (22, 23). The cited models track between 2 and 73 components, by using 8–55 kinetic parameters. Here, we searched for suitable test cases for inferring robustness properties mainly from the knowledge of the system's structural characteristics. Our initial approach of systematically investigating the parameter space, in particular, placed fundamental limitations on the dimensionality and, hence, complexity of the models. In this regard, it has been suggested that the circadian oscillator could be dissected into simpler units having characteristic functional roles (11, 24). We therefore focused on the core negative feedback loop established by per and tim as a starting point for the analysis.
Two deterministic mathematical models of moderate complexity (16, 17) seemed to be most appropriate, particularly because they are very closely related. Common characteristics are to be expected and can be used to validate the methods; systematic differences would point to a discriminative power of the analysis. The simpler model captures the clock by a single negative feedback loop via per. It encompasses five state variables for the concentrations of the components and 18 model parameters reflecting the kinetic constants of the molecular interactions (16). The more complex model describes both branches of negative feedback, comprising 10 states and 38 parameters (17). Hereafter, they are referred to as the single- and dual-feedback model, respectively.
Previous studies showed that, in principle, a feedback structure relying on a single gene (per) should be sufficient for robust oscillations (16, 25). The function of the additional branch via tim is not intuitively clear. The PER/TIM complex exerts transcriptional repression, and, hence, the branches are not redundant. We were therefore additionally interested in whether a second branch of feedback would enhance system robustness under conditions of constant darkness and, if so, in what manner.
Parameter Sensitivities. Parameter sensitivity analysis is a method frequently used in systems theory but rarely applied to dynamical biological systems (26, 27). Parameter sensitivities yield a quantitative measure of the deviations in characteristic system properties resulting from perturbation of system parameters. A higher (absolute) sensitivity of a parameter implies a lower robustness of the corresponding element of a model. It should be noted that, for the models we considered, single parameters may characterize more complex layers of regulation. Model parameters, for instance, in gene expression do not correspond to kinetic constants at the level of elementary reactions. In those cases, sensitivity of the behavior to changes in a coefficient reflects properties of underlying regulatory control structures.
We employed three types of parameter sensitivities that cover different aspects of the system: (overall) state sensitivities, period sensitivities, and amplitude sensitivities. Formally, the selected circadian clock models are autonomous dynamical systems described by ordinary differential equations of the form dx/dt = f(x(t), p, t) with time t ≥ t0, the nS × 1 vector of state variables x, the nP × 1 vector of model parameters p, and initial conditions x(t0) = x0. Parameter sensitivities with respect to the system's states along a specific trajectory S(t) (the nS × nP matrix of state sensitivities) are defined by
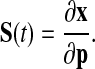 |
[1] |
To obtain a global indicator of robustness that captures aspects of the behavior including shape, phase, period, and amplitude of oscillations, overall state sensitivities So(t) were determined by integration over discrete time t0... tnT and normalization to relative sensitivity (log-gain sensitivity) as follows
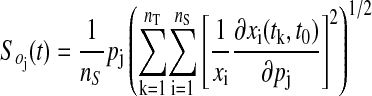 |
[2] |
to calculate the vector's element for parameter pj. The overall state sensitivities are normalized with respect to the number of states, the parameters, and the states to allow for model comparison. Moreover, they apply to all operating regimes such as steady state, oscillations, birhythmicity, or chaos.
Period and amplitude sensitivities are the quantities of primary interest in oscillating systems. Period sensitivities Sτ capture the change of period length, τ, upon changes in parameters (28):
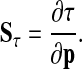 |
[3] |
Accordingly, variations in the amplitude Ai of the ith state (the absolute value of half the difference between minima and maxima of the oscillations) are described by the amplitude sensitivities
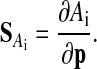 |
[4] |
It is important to note that all parameter sensitivities are only locally valid with respect to parameter space, that is, in a neighborhood of a specific parameter set. They provide information on the robustness of a particular (parametrization of a) model.
Results
Global Indicators of Robustness. Our initial observations were that period sensitivities obtained for two different published parameter sets of the single-feedback model (16, 25) showed considerable agreement (28). Hence, network structure, rather than specific parameter values, could determine system behavior. We therefore determined parameter sensitivities for both models by analyzing large sections in parameter space centered on the reference parameter values given in refs. 16 and 17. In these regions, the models exhibited stable steady state, birhythmicity, and chaotic behavior in addition to regular oscillations. We thus used overall state sensitivities that capture changes in the behavior of all components described by the models (see Supporting Text, Figs. 6–9, and Tables 1–4, which are published as supporting information on the PNAS web site, for details).
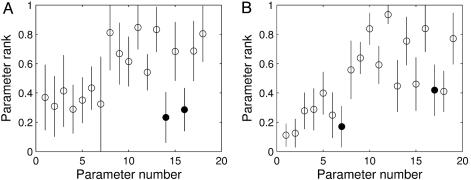
Within the parameter regions for single parameters, as well as between model parameters, the overall state sensitivities varied over several orders of magnitude (data not shown). Being local measures of parameter influence, the variations in absolute parameter sensitivities come as no surprise. Because we were interested in the relative importance of model components for robustness of the system, we ranked the parameters in order from greatest to least sensitivity (29). Importantly, these parameter ranks point to a certain conservation of robustness properties, because they show a relatively low variation (Fig. 2). To facilitate model comparison, we grouped parameters in the symmetric dual-feedback model such that the parameter numbers correspond to the single-feedback model (see Supporting Text). When one compares the error bars, the invariance is more pronounced for the dual-feedback structure (Fig. 2B) than for the simpler network (Fig. 2 A). It also is largely independent of the operating regime, namely oscillations, chaos, or stable steady states. Moreover, random sampling of a limited number of sensitivities yielded results that agreed very well with those obtained by the much more computationally intensive systematic approach (see Supporting Text). Rank-ordered sensitivities can, thus, be interpreted as global indicators of relative robustness and fragility for a given model structure and also be computed efficiently.
Fig. 2.
Conservation of robustness properties. (A) Mean values and standard errors for rank-ordered sensitivities of the single-feedback model. Parameters were assigned arbitrary numbers. Sensitivities were determined by ±1% variation of parameter values for all two-dimensional sections in parameter space (n = 1.8 × 104 parameter sets). Parameters describing maximal phosphorylation rates are emphasized (filled circles). (B) Rank-ordered parameter sensitivities are as in A for the dual-feedback model (n = 8.5 × 104). Here, parameters in the symmetric feedback loops were grouped pairwise to facilitate comparison with A; the last two parameter groups do not occur in the simpler model. Filled circles indicate constants for inhibition of gene expression.
Influence of Regulatory Processes. Parameters in the mathematical models are used to describe regulatory processes such as transcriptional control or phosphorylation/dephosphorylation of proteins. Analysis of parameter sensitivities can, hence, provide clues on the importance of individual regulatory processes on the function of the clock. For instance, the single-feedback model shows high sensitivity toward perturbations affecting protein phosphorylation (Fig. 2 A), whereas the regulation of per and tim expression constitutes a fragile part of the dual-feedback model (Fig. 2B). This indicates that different regulatory mechanisms are of different importance for the robustness of the two network structures.
To systematically investigate potential biological implications of these results, we introduced a functional classification scheme that groups model parameters according to the biochemical processes with which they are associated. We distinguish between (i) transcriptional and translational control, (ii) degradation of mRNA and protein, (iii) transport reactions, (iv) protein phosphorylation, and (v) protein dephosphorylation (see Supporting Text). Period and amplitude of the oscillations (in addition to shape and phase) are two characteristics with physiological significance. Here, we employ rank-ordered period and amplitude sensitivities, respectively, to characterize the influence of the regulatory processes.
Plotting these measures of relative sensitivity against each other enables the assessment of the properties of the oscillations that are most affected by perturbations in individual parameters (Fig. 3). Parameters situated below the diagonal primarily impact the amplitude of oscillations; those above the diagonal are biased toward changing the period length. Functionally related parameters exhibited similar sensitivity properties, but, between groups, larger variations occurred. For both clock architectures, protein phosphorylation predominantly influences the period length. This intuitive finding complies with experimental evidence linking human disorders of the sleep–wake cycle to the phosphorylation status of PER protein (30, 31). Interestingly, phosphorylation systematically is more important for the oscillator's period than dephosphorylation, contrary to the situation in cellular signaling pathways (32, 33). The relative insensitivity of the period to changes in phosphatase activities may be one reason for the fact that specific kinases for PER and TIM were discovered early, but only recently was a phosphatase for PER identified (9, 10, 34). Processes of gene regulation, transcription, and translation turned out to influence predominantly the amplitude of circadian rhythms. This corresponds to experimental observations, for instance, focusing on the transcription factor dclk (35).
Fig. 3.
Influence of biochemical processes on oscillator function. Rank-ordered period and amplitude (based on the concentration of the transcriptional repressor) sensitivities were determined for the single-feedback model (A) and the dual-feedback model (B) (see Supporting Text for details). Parameters associated with transcriptional/translational regulation (filled circles), degradation processes (open circles), transport (triangles), phosphorylation (filled squares), and dephosphorylation (open squares) are distinguished. The diagonal indicates the positions at which both measures of relative sensitivity are identical.
The above-mentioned difference in the impact of regulatory mechanisms in the two models, protein phosphorylation for single-feedback vs. gene regulation for dual-feedback (Fig. 2), reappears when considering period and amplitude sensitivities (Fig. 3), as well as in overall state sensitivities grouped by the regulatory processes (see Supporting Text). Although the models are structurally similar, sensitivity analysis provides a consistent discriminant for robustness properties. Biologically, these differences are difficult to interpret in detail. However, it is interesting to note that circadian clocks in simpler prokaryotes seem to be centered on the control of protein phosphorylation (36). We conclude that the global indicators used lead to consistent characterizations of robustness properties that can be interpreted meaningfully in biological terms.
Control Hierarchies. Based on the overall state sensitivities (Fig. 2), the parameters appear to segregate into two broad groups with high sensitivity (low rank) and with low sensitivity (high rank). In particular, for both models, a group of highly sensitive parameters with low ordinal numbers emerges. Closer inspection of this group revealed that the parameters mirror, for instance, maximal rates of transcription, translation, and mRNA or protein degradation (see Supporting Text). These kinetic parameters can be related to cellular properties that affect many regulatory processes beyond the clock function.
For example, the properties of the cell's transcriptional apparatus, and not of a single gene, determine maximal transcription rates. Apparently, the general cellular machinery, RNA polymerase complexes or the proteasome, needs recognition factors to operate on a specific target. The adapter's functions, however, are not confined to only the circadian clock. For instance, the F box recognition factor Slimb that directs phosphorylated PER toward ubiquitin-dependent proteolysis also controls Wnt signaling in development (37, 38). The more general components are not explicitly included in the circadian clock models, but a modified model that incorporates them shows qualitatively similar sensitivity characteristics (see Supporting Text). This reasoning led us to subdivide the model parameters into three classes: “global” parameters reflecting characteristics of well regulated core cellular machineries, “local” parameters primarily confined to the circadian oscillator, and an intermediary category of “mixed” character (see Supporting Text for a detailed discussion of this classification).
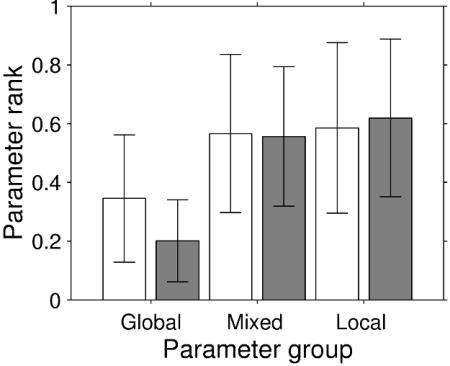
Reevaluation of the global indicators of robustness in terms of the three categories reveals that fragile parts of both clock architectures tend to be associated with global parameters (Fig. 4). Differences among parameter classes are much clearer for the dual-feedback model than for the simpler model. Thus, the additional branch of negative feedback incorporated in the more complex model contributes to a high degree of separation of robustness and fragilities. In the extreme case, such a design combines a strategy of local risk-aversion (39) with a concentration of unavoidable fragilities in few points that when severely affected lead to breakdown of the entire system.
Fig. 4.
“Global” and “local” parameter groups. Average ranks for parameters groups in the single-feedback model (open bars) and the dual-feedback model (filled bars) according to the degree of local character of the parameters. Analysis used the raw data on rank-ordered state sensitivities underlying Fig. 2.
Impact of Perturbations. The parameter sensitivity analyses thus far have the disadvantage of yielding only linear approximations of a system's reactions to perturbations. Moreover, they do not allow for a direct comparison of the quantitative impact that realistic perturbations have on the two circadian clock models. To circumvent these limitations and to validate the sensitivity results by using an independent method, we performed direct perturbation studies. In brief, for both models, we first generated random parameter sets that led to physiological rhythms of 24 ± 1 h period length to diminish the potential influences of particular choices of parameter values. These reference parameter sets were then subjected to random perturbations (up to 2-fold variation of parameter values; see Supporting Text). As an indicator for the physiological function, we used a normalized period deviation
 |
[5] |
with τ(p*) and τ(p) being the period of the perturbed and unperturbed system, respectively, to characterize the relative precision of the circadian clock. An oscillator deviation of 10–2 corresponds to a 1% change in period length relative to the 24 ± 1 h period for the reference parameter sets.
Systems analysis requires characterization of robustness in terms of both the affected system properties and the types of disturbances (40). Here, we consider both scalar perturbations, involving large changes in individual parameters, and vector perturbations, involving smaller, simultaneous changes in all parameters. Whereas the former regime simulates, for instance, single mutations, the latter one reflects changes on evolutionary timescales or in ubiquitously influential factors such as temperature. For identical magnitudes of perturbations, vector perturbations may have a larger impact on oscillator function than disturbances in single parameters (28). Hence, it is crucial to cover both types of perturbations for systematic model analysis.
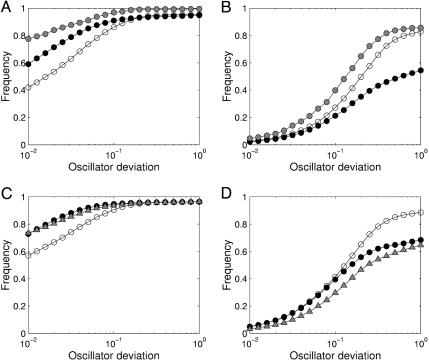
We first emulated local disturbances by random variation of single parameters, which involved identical perturbation strength for both models. We used the frequency of obtaining a given or lower clock deviation after perturbation as a measure of robustness. As shown in Fig. 5A, an additional branch of feedback proves advantageous for the system's robustness in terms of the clock's precision under these conditions. When we assumed that all parameters might be mutated simultaneously, increased model complexity led to higher absolute parameter variation for the dual-feedback model. As would be expected, the dual-feedback structure turned out to be more fragile than the simpler structure (Fig. 5B). The more complex network structure, thus, does not confer higher robustness per se, but it may support the physiologically important fine-tuning of the circadian clock (11) in the case of single perturbations.
Fig. 5.
Impact of perturbations on oscillator function. (A) Effects of scalar perturbations (up to 2-fold variation; see Supporting Text) for the single-feedback model (white circles), the dual-feedback model (black circles), and the redundant-feedback architecture (gray circles). Frequencies of obtaining a given or higher precision of the clock (lower deviation of period length) were averaged over all reference parameter sets. (B) Reaction to vector perturbations, where all parameters were varied simultaneously but independently in the given range. (C and D) Effects of selective perturbations. Frequency distributions for reduced (1%) variations in global parameters for the single-feedback model (open circles) and the dual-feedback model (filled circles), respectively. Variability of local parameters in the dual-feedback model was diminished accordingly (triangles).
Sensitivity analysis highlighted global parameters as important points of fragility for the dual-feedback system. Hence, one would expect the more complex architecture of the investigated circadian clocks to lead to improvements in robustness (compared with the single-feedback architecture) from the elaborate control for hierarchically superimposed regulatory mechanisms. To test this hypothesis, we repeated the previous perturbation studies with reduced variability of global parameters. Hierarchical control increased the robustness of both models similarly for scalar perturbations (Fig. 5C). The dual-feedback model, however, approaches the precision of the simpler model for vector perturbations, toward which it was initially more sensitive (Fig. 5D). Stabilization of local parameters leads to a less pronounced effect, despite a higher number of local vs. global model parameters. Hierarchical control modes can, thus, specifically enhance the robust precision of a dual-feedback loop architecture by avoiding catastrophic failure.
Alternative Circuit Design. One objection that could be raised against our comparison of robustness properties of the circadian clock models is that these models differ significantly in structural complexity, which influences the properties analyzed. Hence, we studied an alternative circuit design with two truly redundant branches of negative feedback. In brief, minor modifications of the dual-feedback model were introduced that allow PER and TIM individually to act as transcriptional repressors. The core model structure and the numbers of system states and model parameters, as well as most of the reference parameter values, remained unaffected (see Supporting Text).
Perturbation studies of the redundant-feedback model elucidated interesting robustness properties. In terms of preserving the oscillator's precision, the new model performs better than either of the other two models for scalar as well as for vector perturbations (Fig. 5 A and B). This result further confirms that system structure is the major determining factor for robustness properties. It also raises the question as to why a nonredundant feedback structure might be incorporated into the Drosophila circadian clock rather than a redundant architecture. Most obviously, maintenance of the period length under conditions of constant darkness may not be the function toward which the clock has been optimized during evolution. Entrainment by light is another capability of primary physiological importance. It corresponds to adaptability in cellular signaling, for which sensitive points processing the inputs have to be provided (41). We speculate that, in this regard, nonredundant feedback is advantageous because it will tend to prevent annihilation of input signals. Future studies should, thus, analyze robustness and sensitivity in more complex representations of the circadian clock, for instance, in mammalian systems with redundant components (9, 10), and include considerations of entrainment by light.
Discussion
We investigated the applicability of sensitivity methods for the analysis of mathematical models that describe the behavior of genetic circuits in terms of relating robustness properties to structural features of the networks. As test cases, we focused on the comparative analysis of moderately complex circadian clock models for Drosophila. Systematic sensitivity analyses showed that network structure largely determines robustness and fragility properties of the very similar models. In particular, rank-ordered sensitivities proved to be consistent global measures for elucidating common network properties as well as differences that seem biologically plausible. Compared with previous studies relying on sensitivities at only one particular location in parameter space (15, 21), this approach provides a comprehensive picture of more general robustness properties. Computation of the ranges of parameter values in which oscillations occur (17, 20, 21), in contrast, does not elucidate how parameter changes influence the oscillator's characteristics in this domain. Additionally, efficient determination of sensitivity properties via random sampling allows the extension of this approach to more complex systems.
More generally, our findings relate to the influential concept of “highly optimized tolerance,” which states that complexity, in biological and engineered systems alike, is primarily a consequence of design aimed at achieving robustness of desired functionalities in case of anticipated perturbations. The inherent drawback of complexity is catastrophic failure when unexpected errors occur (1, 42). The comparison of single- and dual-feedback architectures under conditions of continuous darkness supports this view. We propose that the dual-feedback structure contributes to robust fine-tuning of the clock in the case of single perturbations. Additionally, parallels to an engineering design of deliberate concentration of fragilities (and, conversely, of “export” of a specialized control circuit's points of fragility to more general, well controlled systems) point to connections between control hierarchies and robustness in biology that warrant further research. As the analysis of the redundant-feedback architecture showed, however, appropriate specifications of “complexity” and “desired functionalities” of cellular networks are critical.
For the field of circadian rhythms, we consider this study to provide a starting point for a more formal treatment of the important issue of how the clock's individual parts contribute to the overall functionality (11, 24). The models analyzed capture the real oscillator's complexity in a partial way. It will be intriguing to apply the general approach developed herein, for instance, to conditions of light entrainment. Control theoretic approaches are relevant in this context, because the circadian oscillator displays integral action (i.e., offset free tracking of a 24-h cycle) for appropriate parameter values and properties of the entraining signal. Based on studies in other biological systems (6), it is plausible that the circadian oscillator also contains an internal model leading to a robust regulatory structure (43). The method proposed herein will be applicable to complex models describing interlocked feedback loops. As for artificial genetic circuits (44), however, starting to ask simple questions on simple systems might be an appropriate strategy for ultimately uncovering the “logic” of living systems.
Supplementary Material
Acknowledgments
We thank Uwe Sauer and Daniel E. Zak for comments on this work. F.J.D. gratefully acknowledges financial support from The Alexander von Humboldt Foundation and from the Institute for Collaborative Biotechnologies through U.S. Army Research Office Grant DAAD19-03-D-0004.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Csete, M. E. & Doyle, J. C. (2002) Science 295 1664–1669. [DOI] [PubMed] [Google Scholar]
- 2.Lauffenburger, D. (2000) Proc. Natl. Acad. Sci. USA 97 5031–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eldar, A., Dorfman, R., Weiss, D., Ashe, H., Shilo, B. Z. & Barkai, N. (2002) Nature 419 304–308. [DOI] [PubMed] [Google Scholar]
- 4.Stelling, J., Klamt, S., Schuster, S. & Gilles, E. D. (2002) Nature 420 190–193. [DOI] [PubMed] [Google Scholar]
- 5.Alon, U. M., Surette, G., Barkai, N. & Leibler, S. (1999) Nature 397 168–171. [DOI] [PubMed] [Google Scholar]
- 6.Yi, T. M., Huang, Y., Simon, M. & Doyle, J. (2000) Proc. Natl. Acad. Sci. USA 97 4649–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma, L. & Iglesias, P. A. (December 13, 2002) BMC Bioinformatics, 10.1186/1471-2105-3-38.
- 8.Goldbeter, A. (2002) Nature 420 238–245. [DOI] [PubMed] [Google Scholar]
- 9.Young, M. & Kay, S. (2001) Nat. Rev. Genet. 2 702–725. [DOI] [PubMed] [Google Scholar]
- 10.Panda, S., Hogenesch, J. & Kay, S. (2002) Nature 417 329–335. [DOI] [PubMed] [Google Scholar]
- 11.Roenneberg, T. & Merrow, M. (2003) Curr. Biol. 13 R196–R207. [DOI] [PubMed] [Google Scholar]
- 12.Cyran, S. A., Buchsbaum, A. M., Reddy, K. L., Lin, M. C., Glossop, N. R., Hardin, P. E., Young, M. W., Storti, R. V. & Blau, J. (2003) Cell 112 329–341. [DOI] [PubMed] [Google Scholar]
- 13.Leloup, J.-C., Gonze, D. & Goldbeter, A. (1999) J. Biol. Rhythms 14 433–448. [DOI] [PubMed] [Google Scholar]
- 14.Ruoff, P., Vinsjevik, M., Monnerjahn, C. & Rensing, L. (2001) J. Theor. Biol. 209 29–42. [DOI] [PubMed] [Google Scholar]
- 15.Smolen, P., Baxter, D. A. & Byrne, J. H. (2001) J. Neurosci. 21 6644–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldbeter, A. (1995) Proc. R. Soc. London Ser. B 261 319–324. [Google Scholar]
- 17.Leloup, J.-C. & Goldbeter, A. (1998) J. Biol. Rhythms 13 70–87. [DOI] [PubMed] [Google Scholar]
- 18.Tyson, J., Hong, C., Thron, D. & Novak, B. (1999) Biophys. J. 77 2411–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda, H. R., Hagiwara, M. & Kitano, H. (2001) J. Theor. Biol. 210 401–406. [DOI] [PubMed] [Google Scholar]
- 20.Leloup, J.-C. & Goldbeter, A. (2003) Proc. Natl. Acad. Sci. USA 100 7051–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forger, D. B. & Peskin, C. S. (2003) Proc. Natl. Acad. Sci. USA 100 14806–14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smolen, P., Baxter, D. A. & Byrne, J. H. (2002) Biophys. J. 83 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angeli, D. & Sontag, E. D. (2004) in Optimal Control, Stabilization, and Nonsmooth Analysis, eds. de Queiroz, M., Malisoff, M. & Wolenski, P. (Springer, Heidelberg), pp. 135–154.
- 24.Hastings, M. H. (2000) Nat. Rev. Neurosci. 1 143–146. [DOI] [PubMed] [Google Scholar]
- 25.Gonze, D., Halloy, J. & Goldbeter, A. (2002) Proc. Natl. Acad. Sci. USA 99 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savageau, M. (1971) Nature 229 542–544. [DOI] [PubMed] [Google Scholar]
- 27.Ingalls, B. P. & Sauro, H. M. (2003) J. Theor. Biol. 222 23–36. [DOI] [PubMed] [Google Scholar]
- 28.Zak, D., Stelling, J. & Doyle, F. J., III (2004) Comp. Chem. Eng., in press.
- 29.Helton, J. & Davis, F. (2000) in Sensitivity Analysis, eds. Saltelli, A., Chan, K. & Scott, E. (Wiley, Chichester, U.K.), pp. 101–153.
- 30.Toh, K. L., Jones, C. R., He, Y., Eide, E. J., Hinz, W. A., Virshup, D. M., Ptacek, L. J. & Fu, Y. H. (2001) Science 291 1040–1043. [DOI] [PubMed] [Google Scholar]
- 31.Ebisawa, T., Uchiyama, M., Kajimura, N., Mishima, K., Kamei, Y., Katoh, M., Watanabe, T., Sekimoto, M., Shibui, K., Kim, K., et al. (2001) EMBO Rep. 2 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich, R., Neel, B. G. & Rapoport, T. A. (2002) Mol. Cell 9 957–970. [DOI] [PubMed] [Google Scholar]
- 33.Bhalla, U. S., Ram, P. T. & Iyengar, R. (2002) Science 297 1018–1023. [DOI] [PubMed] [Google Scholar]
- 34.Santhyanarayanan, S., Zheng, R., Xiao, R. & Seghal, A. (2004) Cell 116 603–615. [DOI] [PubMed] [Google Scholar]
- 35.Allada, R., Kadener, S., Nadakumar, N. & Rosbash, M. (2003) EMBO J. 22 3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golden, S. S. (2003) Curr. Opin. Microbiol. 6 535–540. [DOI] [PubMed] [Google Scholar]
- 37.Grima, B., Lamouroux, A., Chelot, F., Papin, C., Limbourg-Bouchon, B. & Rouyer, F. (2002) Nature 420 178–182. [DOI] [PubMed] [Google Scholar]
- 38.Ko, H. W., Jiang, J. & Edery, I. (2002) Nature 420 673–678. [DOI] [PubMed] [Google Scholar]
- 39.Newman, M., Girvan, M. & Farmer, J. (2002) Phys. Rev. Lett. 89 28301. [DOI] [PubMed] [Google Scholar]
- 40.Morohashi, M., Winn, A. E., Borisuk, M. T., Bolouri, H., Doyle, J. & Kitano, H. (2002) J. Theor. Biol. 216 19–30. [DOI] [PubMed] [Google Scholar]
- 41.Lee, E., Salic, A., Krüger, R., Heinrich, R. & Kirschner, M. W. (October 13, 2003) PLoS Biol., 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed]
- 42.Carlson, J. & Doyle, J. (2002) Proc. Natl. Acad. Sci. USA 99 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sontag, E. D. (2003) Syst. Control Lett. 50 119–126. [Google Scholar]
- 44.Hasty, J., McMillen, D. & Collins, J. J. (2002) Nature 420 224–230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.