Abstract
Promyelocytic leukemia (PML) and Cajal bodies are mobile subnuclear organelles, which are involved in activities like RNA processing, transcriptional regulation, and antiviral defense. A key parameter in understanding their biological functions is their mobility. The diffusion properties of PML and Cajal bodies were compared with a biochemically inactive body formed by aggregates of murine Mx1 by using single-particle tracking methods. The artificial Mx1-yellow fluorescent protein body showed a very similar mobility compared with PML and Cajal bodies. The data are described quantitatively by a mechanism of nuclear body movement consisting of two components: diffusion of the body within a chromatin corral and its translocation resulting from chromatin diffusion. This finding suggests that the body mobility reflects the dynamics and accessibility of the chromatin environment, which might target bodies to specific nuclear subcompartments where they exert their biological function.
Cajal and promyelocytic leukemia (PML) bodies are essential components of the nucleus that are thought to contain activities for RNA processing, transcriptional regulation, and antiviral defense. For understanding their biological functions, parameters have to be identified that characterize their mobility and lead to a localization of Cajal and PML bodies at their target sites in the nucleus. Whereas Cajal bodies are often found associated to several different small nuclear RNA and small nucleolar RNA gene loci as well as histone gene loci (1–5), PML bodies are frequently located in the MHC gene region (6). For both nuclear bodies, different types of movements were described and assigned to distinct subgroups of Cajal and PML bodies (7–12). The analysis of a baby hamster kidney cell line revealed that a fraction of PML bodies moved over longer distances and in an energy-dependent manner. These faster moving bodies were not observed in HeLa cells (8). In a detailed study of Cajal bodies in HeLa cells, an anomalous diffusion behavior and an ATP- and transcription-dependent association with chromatin was reported (7).
Here, the mobility of PML and Cajal bodies has been compared with the nuclear body-like structures formed by Mx1 protein fused to yellow fluorescent protein (YFP). Mx proteins are IFN-induced GTPases of the dynamin super family involved in defense mechanisms against RNA viruses (13). The murine Mx1 protein, 72 kDa in size, has a natural nuclear localization sequence sequence and displays a nuclear body-like distribution (14, 15). In contrast to the Mx1 WT protein, the Mx1-YFP construct studied has no antiviral activity as tested with influenza and Thogoto virus infection and formed crystals (S. Stertz and O. Haller, personal communication). The diffusion properties of this bona fide biologically inert body were used as a reference to identify principles determining the mobility of bodies within the nucleus.
Materials and Methods
Expression Plasmids. The cDNA coding for Mx1 in the plasmid pBS-T7 (kindly provided by O. Haller, University of Freiburg, Freiburg, Germany) was cut out and inserted into the pEYFP-C1 vector (Clontech) by using ApaI and BamH I. Expression plasmids coilin-GFP and GFP-PML-III (16) were kindly provided by C. Cardoso (Max Delbrück Center, Berlin) and A. Möller (University of Bern, Bern, Switzerland), respectively. A SW13 cell line stably expressing H2A-cyan fluorescent protein (CFP) (SW13H2A-CFP) was provided by M. Reichenzeller and T. A. Knoch (Deutsches Krebforschungszentrum, Heidelberg).
Cell Culture and Transfection. HeLa cells (ATCC CCL-2) were cultured on 12-mm glass coverslips for immunofluorescence or 35-mm glass coverslips (Helmut Sauer, Reutlingen, Germany) and in Lab-Tek chambers (Nunc) for in vivo studies in DMEM for 1 day after plating. Cells were transiently transfected in duplicate or triplicate by using FuGENE 6 (Roche Diagnostics), as it was not possible to obtain a stable cell line with the Mx1-YFP crystals. The experiments were repeated with different batches of transfected cells, and the same results for all three types of nuclear bodies were obtained within the accuracy of the measurements. This finding indicates that when using transiently transfected cells reproducible expression levels were achieved, and that these cells were appropriate for studying nuclear mobility. For energy depletion sodium azide was added either to the medium to a final concentration of 10 mM or in a depletion buffer according to ref. 17. For in vivo staining of the DNA, the cells were incubated with 1 μg/ml Hoechst 33342 (Molecular Probes) for 20 min.
Fixation and Indirect Immunofluorescence. After transfection, cells were incubated for 1–2 days and then fixed in 4% formaldehyde containing 2% sucrose on ice. For indirect immunofluorescence, cells were permeabilized with 0.2% Triton X-100 in PBS. To identify nuclear bodies the following mouse mAbs were used: anti-p80 coilin (kind gift of A. Lamond, University of Dundee, Dundee, U.K.), anti-fibrillarin (kind gift of D. Olins, Bowdoin College, New Brunswick, ME), and anti-PML PG-M3 (Santa Cruz Biotechnology). Secondary Texas red-conjugated antibodies (Dianova, Hamburg, Germany) were diluted 1:200. All coverslips were mounted by using VECTASHIELD with 4′,6-diamidino-2-phenylindole (Vector Laboratories).
Confocal Laser Scanning and Correlative Electron Microscopy. 3D image stacks were acquired by using the Leica TCS SP2 confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany). A diode laser as well as argon and helium/neon lasers were used for 4′,6-diamidino-2-phenylindole/Hoechst (λ = 405 nm), GFP (λ = 488 nm), YFP (λ = 514 nm), and Texas red (λ = 543 nm) excitation. For all images a PlanApochromat ×63/1.32 numerical aperture oil objective was used. For 4D imaging, cells were cultured by using DMEM without phenol red in a humid incubation chamber (Helmut Sauer) with temperature and CO2 control (PeCon, Erbach, Germany). Either 45 or 98 stacks of 9–12 sections with 0.3-μm z interval were collected every 10 s. Laser intensities were adjusted such that the power in the sample did not exceed 10 μW, resulting in an energy deposition of ≈20 μJ per stack for the selected scan speed (800 Hz) and image size (256 × 256 pixels) similar to the illumination energy deposition in previous studies (7, 8). Single Mx1-YFP bodies were bleached by using maximum laser intensity, and recovery was monitored every 5–10 min. For 4D imaging of cells with Mx1-YFP and H2A-CFP a Zeiss LSM 510 equipped with argon and krypton lasers for CFP (λ = 413 nm) and YFP (λ = 514 nm) as well as a PlanApochromat ×63/1.4 numerical aperture oil differential interference contrast objective (Zeiss) was used. For all images 45 stacks with 0.5-μm z interval were recorded every 10 s. Maximum intensity projections were made with Leica confocal software and a Zeiss image browser. The method for correlative confocal laser scanning microscopy and electron microscopy investigation of Mx1-YFP was essentially as described (18).
Image Processing and Movement Analysis. Confocal images were processed and merged by using photoshop (Adobe Systems, San Jose, CA). The analysis on colocalization of Mx1-YFP and nuclear bodies was determined in >350 nuclei. The statistical evaluation of the vicinity was carried out by Wilcoxon rank sum tests. The cell movement was corrected if necessary by using rigid registration (19). For single-particle tracking body positions were either defined by the brightest pixel or the center of mass by using imagej software (National Institutes of Health, Bethesda). Both methods gave the same results. The pair-wise motion correlation coefficients ρ were determined for all possible combinations. For body size calculation particle pixels were extracted by using tikal software (principles described in ref. 20). The velocity was determined for various time intervals ranging from 10 to 100 s. The mean square displacement (MSD) was calculated for each body and time point of the data set with
 |
[1] |
and plotted as <d2> (μm2) versus Δt (s) by using kaleidagraph (Synergy Software, Reading, PA). The value for the MSD at Δt = 0 is 0 with an uncertainty of 2.5 × 10–3 μm2 as evaluated for 280 time points at 10-s intervals with fixed cells transfected with Mx1-YFP. The fitting of the MSD versus time was carried out by using Eqs. 3 and 4. For the average MSD plots 100 Mx1-YFP, 100 PML bodies, and 55 Cajal bodies were evaluated. For comparison of two fits, a normalized χ2 was determined according to Eq. 2 with N being the number of time points, P the number of fit parameters, yi the measured data values, σi the corresponding errors, and fi the fit function values at the data points i with 1 ≤ I ≤ N.
 |
[2] |
Bodies were further tested for anomalous diffusion by using log-log plots (21). In the case of free diffusion, the result is a horizontal line. For anomalous diffusion, this plot has a slope of α =–1. The slope was determined by using regression curve fitting, and the anomalous diffusion coefficient α was calculated. α > 0 is considered as confined diffusion, α = 1 indicates free diffusion, α < 1 is interpreted as obstructed diffusion (21), and α > 1 represents directed motion. Trajectories of random walks either measured or simulated were generated (for simulated data) and plotted on a square lattice, each site of which represents an image pixel, by using the software obstacles written in our laboratory. To define the scale for simulated free diffusion the MSD of Δt = 10 s as observed for Mx1-YFP was extrapolated linearly. The trajectory coordinates for confined diffusion were taken from the measurements, whereas the values for free diffusion were generated by allowing a particle to jump from site to site with random direction. The degree of chromatin condensation during sodium azide treatment was determined by using correlation length analysis (22).
Results
Characterization of Mx1-YFP, PML, and Cajal Bodies. Upon low expression, Mx1-YFP formed bodies, which are ≈1 μm in size (Fig. 1A). On electron micrographs Mx1-YFP was found to have a crystalline organization. The bodies are built by a regular packing of filament-like structures (Fig. 1D). Interestingly, ultrastructural analysis revealed that the chromatin around the bodies has a distinct structure, which appeared to be less dense than the bulk chromatin (Fig. 1D). This was also observed for PML and Cajal bodies (data not shown). Although it cannot be excluded that this effect is caused by shrinkage during fixation, some fine texture in this region is visible on the original electron micrographs, arguing against an artifact of sample preparation. Fusions of Mx1-YFP bodies were not observed during time intervals up to 2.5 h probably because of their crystalline nature. In contrast, fusions of Cajal and PML bodies were observed within 16 min. Cajal body fusion had been demonstrated previously (23, 24), whereas PML body fusion has been controversially discussed (8, 25, 26). Fluorescence recovery after photobleaching experiments of Mx1-YFP bodies showed no recovery within 20 min, indicating the absence of a free nuclear Mx1-YFP pool that could exchange with the Mx1-YFP bodies (data not shown). In contrast, an exchange with a 50% recovery within few minutes was reported for some proteins of PML and Cajal bodies (25, 27–29). The missing antiviral activity and the crystalline structure make Mx1-YFP bodies a suitable reference system without bona fide biological activity. In our analysis, PML bodies were frequently located next to Mx1-YFP bodies (34% of all analyzed bodies, Fig. 1B), and Cajal bodies were also found preferentially next to them (50% of all bodies, Fig. 1C). This result is in agreement with the previously reported vicinity of Mx1 and PML bodies (30, 31). In contrast, the spatial relation of Mx1-YFP with nucleoli was significantly different because only 12% of Mx1-YFP bodies were found next to nucleoli.
Fig. 1.
Mx1-YFP colocalizes with nuclear bodies. Confocal images of Mx1-YFP (A), Mx1-YFP with PML bodies (B), and Mx1-YFP with Cajal bodies (C) are shown. Mx1-YFP is shown in green, PML and Cajal bodies in red, and 4′,6-diamidino-2-phenylindole staining in blue. (D) Electron micrograph showing a Mx1-YFP body. (Scale bars: 10 μm, A–C; 1 μm, D.)
General Features of Nuclear Body Movement. We expressed Mx1-YFP (Fig. 2A), coilin-GFP (Fig. 2B), or GFP-PML-III (Fig. 2C) in HeLa cells. The diffusion properties of the corresponding bodies were quantified by single-particle tracking. Analyzing data from 3D image stacks and 2D projections of these stacks led to the same diffusion properties, implying isotropic motion in all three directions. In such a case, the components in x, y, and z directions of the body movement can be treated as independent (see Supporting Text, which is published as supporting information on the PNAS web site), and the analysis of the diffusion behavior based on 2D image projections is correctly described by models for 2D motion (see Supporting Text).
Fig. 2.
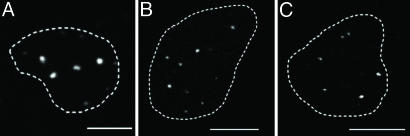
In vivo images. Confocal sections of Mx1-YFP (A), PML bodies (B), and Cajal bodies (C) are shown. The nuclear shape is outlined by a dotted line. (Scale bars: 10 μm.)
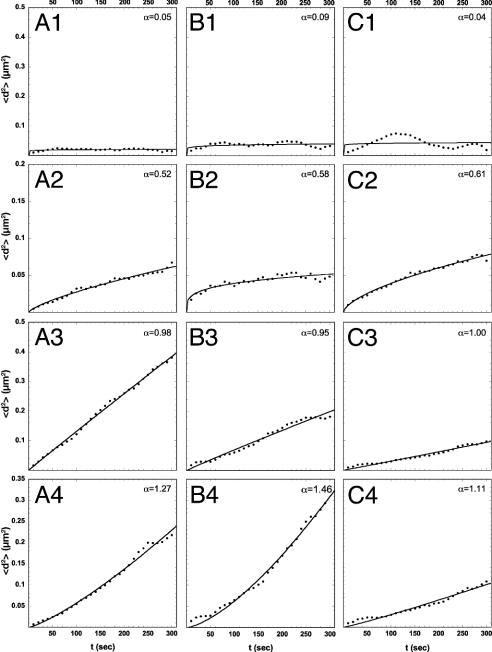
All three types of bodies displayed an independent mobility as determined by pair-wise correlation analysis. Correlation coefficients of 0.07 ± 0.01 (PML bodies), 0.04 ± 0.02 (Cajal bodies), and 0.1 ± 0.02 (Mx1-YFP) on a scale from –1 (perfect anticorrelation) to 1 (perfect correlation) were determined. These values indicate that there was no correlation of body mobility. The average body size was similar for all body types (Mx1-YFP, 1.3 ± 0.06 μm; PML bodies, 1.2 ± 0.07 μm; and Cajal bodies, 1.0 ± 0.04 μm). On an average, 4 ± 1 Mx1-YFP particles, 14 ± 2 PML bodies, and 8 ± 3 Cajal bodies were found per nucleus. These sizes and numbers for PML and Cajal bodies are similar to those described in the literature (32, 33). The body velocity was found to be similar for the three body types and displayed time dependence as expected for a nondirected movement. For a time interval of 10 s the velocity was 1.26 μm·min–1 for Mx1-YFP, 1.56 μm·min–1 for PML bodies, and 1.32 μm·min–1 for Cajal bodies. To compare these values with the literature, the different time intervals of the measurements had to be considered, and values similar to those of previous studies were obtained: 0.1 μm·min–1 versus 0.48–0.65 μm·min–1 for Cajal bodies (Δt = 180 s) (23) and 2.8 μm·min–1 versus 5.2 μm·min–1 for PML bodies (Δt = 5 s) (8). A dependence of velocity on particle size was not observed for Cajal bodies with a correlation coefficient of 0.07, whereas Mx1-YFP (ρ = 0.16) and PML bodies (ρ = 0.39) showed a weak correlation. From a plot of the MSD versus time, four different types of mobility had been distinguished previously: confined diffusion, obstructed diffusion, free diffusion, and directed motion (34, 35). Our analysis of prolonged 4D data sets recorded for 16 min showed that a single Mx1-YFP (Fig. 3 A1–A4), PML (Fig. 3 B1–B4), or Cajal body (Fig. 3 C1–C4) displayed periods of confined, obstructed, and free diffusion as well as directed motion during the observation time with a finite overall average step size. This behavior is consistent with statistical variations expected to occur for random Brownian motion. Accordingly, making use of the ergodic principle, the body displacement measured for a given time interval was averaged for all bodies of a given type to obtain the MSD plots shown in Fig. 4 A–C.
Fig. 3.
A single nuclear body displays different types of motion over time. Confined, obstructed, and free diffusion as well as directed motion for individual Mx1-YFP (A1–A4), PML (B1–B4), and Cajal bodies (C1–C4) are shown. The respective α values for the classification are noted.
Fig. 4.
Single-particle tracking analysis of nuclear bodies. Shown are average MSD plots for all Mx1-YFP (A), PML bodies (B), and Cajal bodies (C) examined. The fit for obstructed diffusion is shown in red, and the fit for diffusion of particles in corrals is shown in blue, both with weighted errors. The χ2 values for the red and the blue fits are 0.34 and 0.27 for Mx1-YFP, 0.59 and 0.31 for PML bodies, and 0.47 and 0.28 for Cajal bodies. Average log-log plots for Mx1-YFP (D), PML bodies (E), and Cajal bodies (F) are shown. The normal log-log plots are shown in red with the α values 0.34, 0.27, and 0.33 for Mx1-YFP, PML, and Cajal bodies, respectively. The log-log plots with exclusion of Db are shown in blue. The fitis a horizontal line obtained from the mean value of the curve points. The α values are accordingly 1.
The Moving Corral Model. The average mobility of PML and Cajal bodies as well as Mx1-YFP was different from free diffusion because no linear dependence of the averaged MSD versus time was observed (Fig. 4 A–C). The MSD plots showed a high initial increase followed by a shallow apparently linear increase. It should be noted that this finding is in contrast to pure confined diffusion where the MSD reaches a plateau and to pure obstructed diffusion, which displays a nonlinear increase (21). Trajectories of Mx1-YFP were indistinguishable from those of PML and Cajal bodies (representative examples are shown in Fig. 8A, which is published as supporting information on the PNAS web site). The comparison with simulated free trajectories (Fig. 8B) clearly shows that the body movement is spatially restricted. Extending the theoretical framework from previous studies (35–37), we developed the following model (Fig. 5): the bodies are diffusing within a “corral” of accessible chromatin accounting for the high initial increase. These chromatin corrals can translocate within the nucleus, leading to the apparently linear dependence at later times. Thus, the measured diffusion of a body is composed of the body diffusion with constant diffusion coefficient (D)b and chromatin motion characterized by constant Dc. This finding would be consistent with the electron micrographs, where the chromatin surrounding the bodies appeared to be less dense (see Fig. 1D for Mx1-YFP), allowing the bodies to move relatively fast within an area of a few hundred nanometers that is defined by the chromatin density distribution. An example for the restricted movement of an Mx1-YFP body in a putative chromatin corral is shown in Movie 1, which is published as supporting information on the PNAS web site. Using this model the average MSD plots fitted to an equation for diffusion of particles extending the theoretical framework developed by Saxton (36) and Saxton and Jacobson (38) (Fig. 4 A–C, blue curves):
 |
[3] |
The derivation of Eq. 3 is described in Supporting Text and yields the accessible chromatin space around the body rc, the body Db, and the chromatin Dc. The numbers for the Db values represent lower limits because it is possible that a steeper initial increase of the MSD values would be observed if a time resolution shorter than Δt = 10 s is used. The Db values (Table 1) obtained for PML (Db ≥ 6.2 × 10–3 ± 8.5 × 10–4 μm2·s–1) and Cajal bodies (Db ≥ 7.0 × 10–3 ± 1.7 × 10–3 μm2·s–1) were slightly larger than the Mx1-YFP value (Db ≥ 2.9 × 10–3 ± 4.0 × 10–4 μm2·s–1). The rc values are similar for all body types: Mx1-YFP, 0.28 ± 0.03 μm; PML bodies, 0.31 ± 0.02 μm; and Cajal bodies, 0.26 ± 0.03 μm. This finding supports the view that there is an accessible chromatin space of an average of 300 nm surrounding the bodies. The Dc values (Table 1) for the chromatin corrals were very similar for all three body types ranging from 1.1 × 10–4 ± 2.9 × 10–5 μm2·s–1 to 1.8 × 10–4 ± 9.2 × 10–5 μm2·s–1, which is consistent with the view that this component of the movement reflects rearrangements of chromatin domains on a larger scale.
Fig. 5.
Schematic illustration of nuclear body diffusion. The nuclear body is enclosed in a chromatin corral, defined by the chromatin accessibility in which they diffuse with Db as demonstrated by the deformation of the chromatin. The chromatin corrals translocate within the nucleus with the Dc indicated by the red arrow.
Table 1. Ds for nuclear bodies.
| Body | Db, × 10-3 μm2·s-1 | Dc, × 10-4 μm2·s-1 | rc, μm |
|---|---|---|---|
| Mx1-YFP | ≥ 2.9 ± 0.4 | 1.8 ± 0.92 | 0.28 ± 0.03 |
| PML | ≥ 6.2 ± 0.85 | 1.2 ± 0.31 | 0.31 ± 0.02 |
| Cajal | ≥ 7.0 ± 1.7 | 1.1 ± 0.29 | 0.26 ± 0.03 |
Db, body D; Dc, chromatin D; rc, accessible chromatin space around the body.
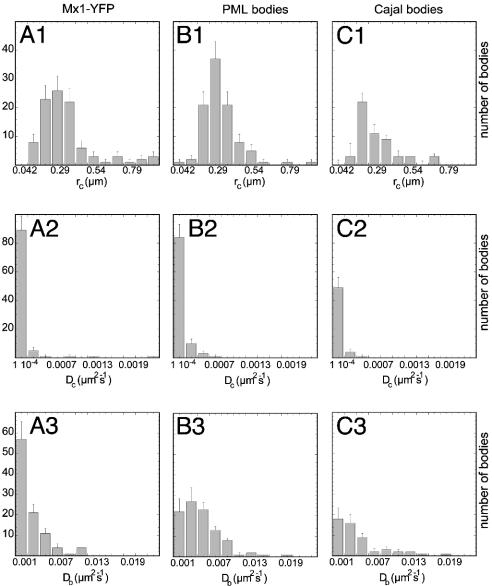
To examine whether bodies of different types were present with mobility significantly different from the average movement as depicted in Fig. 4, the rc, Dc, and Db values were calculated for all analyzed bodies individually. The resulting distributions for Mx1-YFP, Cajal, and PML bodies showed single peaks at almost identical positions (Fig. 6). No statistically relevant deviations that would be indicative of the presence of a species with higher or lower mobility were detected. This finding demonstrates that for all three nuclear body types only one population of bodies with respect to their diffusional properties was present. Within each class of bodies the values for Dc and Db were uncorrelated with correlation coefficients between –0.1 and 0.1.
Fig. 6.
Analysis of single nuclear bodies. For all bodies studied the rc, Dc, and Db values according to Eq. 3 were calculated individually. The resulting distribution of rc, Dc, and Db is plotted for Mx1-YFP (A1–A3), PML (B1–B3), and Cajal bodies (C1–C3). All histograms show essentially a distribution with a single peak indicative of only one species of each nuclear body type.
Cajal bodies had been described to diffuse anomalously (7). For anomalous diffusion the MSD is no longer linear with time, but proportional to tα with α being the anomalous diffusion coefficient and Γ a generalized transport coefficient according to Eq. 4:
 |
[4] |
Eq. 4 was used to test whether our data can also be described by an anomalous diffusion model. The anomalous diffusion model did not result in a satisfactory fit of the data (Fig. 4 A–C, red graphs). Systematic deviations were observed at short times (Δt <50 s) and longer times (Δt >250 s). This qualitative difference between the two models was confirmed by a difference in the normalized χ2 values for the two fits that is more than one order of magnitude larger for Eq. 4 and indicates that the fit to Eq. 3 is significantly better. A frequently used test for the anomalous diffusion behavior of a particle is the log-log plot (21), which yields a negative slope for anomalous diffusion (Fig. 4 D–F, red graphs). However, this type of analysis will not allow separating the two components for a biphasic diffusion behavior as proposed here. A separate plot of the slow component describing the corral motion can be obtained by subtracting the extrapolated value for Δt = 0 obtained from a linear fit of the data at Δt ≥40 s. The resulting log-log plots demonstrate apparent free diffusion behavior for the slower component during the observation period (blue lines in Fig. 4 D–F). Thus, the overall anomalous diffusion behavior can be mostly assigned to the additional fast but restricted movements of the particle within the corral.
Previously, the effect of energy depletion on the diffusion of nuclear bodies and chromatin had been investigated (7, 8, 39). We observed that ATP depletion leads to reversible chromatin condensation, which leads to a reduction of the average chromatin domain size from ≈1.66 to 0.85 μm as inferred from the change of the correlation length (Fig. 9, which is published as supporting information on the PNAS web site). This condensation was found by using either an energy depletion buffer (Fig. 9A) or adding sodium azide directly to the medium (Fig. 9B) with HeLa and SW13 cells. It is expected that this change in the chromatin organization has an intricate impact on body mobility.
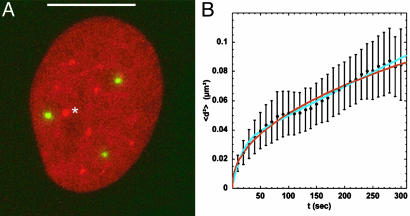
To compare the obtained results on nuclear body diffusion with other nuclear components, well defined speckle-like regions of dense chromatin (40) were analyzed by using a cell line stably expressing H2A-CFP (* in Fig. 7A). Interestingly, Mx1-YFP was never found to be localized next to a dense chromatin region (Fig. 7A). An analysis of a limited data set yielded essentially the same results with respect to the fit parameters rc, Db, and Dc for the movement of Mx1-YFP bodies in the SW13H2A-CFP, indicating that HeLa and SW13 cells were similar with respect to nuclear body movement. As shown in Fig. 7, the moving corral model (Eq. 3) did not fit the data for the dense chromatin regions significantly better than the anomalous diffusion model (Eq. 4), as shown by the χ2 values (Fig. 7B). The transport coefficient is 0.47 ± 0.07 μm2·s–α. The corresponding parameters for the moving corral model would be rc = 0.18 ± 0.08 μm, Db ≥ 5.0 × 10–4 ± 2.8 × 10–4 μm2·s–1, and Dc = 4.8 × 10–5 ± 2.1 × 10–5 μm2·s–1, i.e., all below the values for the bodies, as expected for a structure by definition embedded into and linked to chromatin regions denser than the environment of the nuclear bodies. These values are only given for comparison, because there is no need to assume a biphasic dependence of the MSD with time to describe the mobility of dense chromatin regions.
Fig. 7.
Single-particle tracking analysis of dense chromatin regions. (A) Mx1-YFP particles in a cell line stably expressing H2A-CFP (SW13H2A-CFP). An example for an evaluated dense chromatin block is labeled by *. (Scale bar: 10 μm.) (B) Average MSD plot for 15 evaluated regions. The fit for obstructed diffusion is shown in red (χ2 = 0.032), and the fit for diffusion of particles in corrals is shown in blue (χ2 = 0.018).
Discussion
The analysis of PML, Cajal, and a bona fide inert nuclear body composed of Mx1-YFP by single-particle tracking presented here demonstrates that the diffusion of these bodies can be described by the same principles: the nuclear bodies diffuse within an accessible chromatin corral, whereas these chromatin corrals translocate within the nucleus. The movement of all Mx1-YFP, PML, and Cajal bodies was not correlated as quantified by pair-wise correlation analysis. Although it has been reported that in some cases PML bodies appeared to move in groups of two or three (8), this behavior of a subfraction appears not to be the characteristic for the majority of PML bodies analyzed here. It was demonstrated that Cajal and PML bodies are frequently associated with each other (41). This result is supported by our finding that Mx1-YFP and Cajal bodies or PML bodies colocalize to 50% and 34%, respectively. The observed colocalization indicates that the nucleus has subspaces, which are preferably accessible for different kinds of bodies independent of their functionality. The analysis of longer trajectories conducted here suggests that in principle every single body can display all four different types of movement, namely confined, obstructed, and free diffusion as well as directed motion (Fig. 3). In these experiments the data acquisition was limited to ≈16 min to be able to track the body movement with high temporal resolution by confocal fluorescence microscopy and without too much bleaching of the fluorescence signal. For an ergodic system it is equivalent to measuring many bodies for a short time as opposed to analyzing a few bodies on a longer time scale. Thus, it is not expected that an increase of the observation time would reveal the existence of a special class of bodies not detected here. Furthermore, it was confirmed by determining the mobility parameters for all trajectories individually that no distinct mobility classes were present. In addition, averaging the individual parameters resulted in virtually the same values as those derived from the averaged trajectories. However, it should be noted that the finite dimensions of the cell nucleus will pose a limit to the maximum value of the MSD, which is not apparent on the time scale studied here. Our data can be described by a simple model, in which the bodies diffuse in and with a chromatin corral in the nucleus. The size of the corral is defined by the area of lower chromatin density around the bodies. Such a chromatin distribution is expected from its polymeric nature. In contrast to a liquid, polymers will not fill the available space near boundaries homogeneously unless a strong compressing force is present (42).
For Cajal bodies an association to small nuclear RNA, small nucleolar RNA, and histone genes has been reported (1–5), which suggests that Cajal bodies bind to specific chromatin loci. The binding of nuclear bodies to chromatin could contribute to the corralling, but it would have no effect on the long-range movements of corrals because of large-scale chromatin rearrangements. However, as Cajal bodies and Mx1-YFP bodies displayed essentially the same mobility and there is no evidence for a specific binding of Mx1-YFP bodies, any Cajal body interaction with chromatin is not detected in the analysis described here. This finding suggests that Cajal body binding to chromatin is transient, because otherwise the mobility should match with that of dense chromatin regions illustrated in Fig. 7. The model proposed here implies that any chromatin remodeling has a direct impact on body mobility. ATP depletion is likely to have complex effects on chromatin organization and dynamics. A marked decrease of chromatin mobility in yeast was reported after ATP depletion (39). We have also observed that ATP depletion can lead to chromatin condensation, which could increase the body mobility because of an extension of regions with lower chromatin density. On the other hand, nuclear bodies might become trapped in the condensed chromatin regions. Thus, it is difficult to assign the previously observed increased Cajal body mobility in HeLa (7) and decreased PML body mobility in baby hamster kidney cells (8) after ATP depletion simply to a direct energy-dependent binding affinity or transport mechanism. In addition, the response of Cajal and PML bodies to ATP depletion is likely to be cell line dependent because the fraction of PML bodies that moved over longer distances in an energy-dependent manner was not detected in HeLa cells (8). We therefore suggest that in energy depletion experiments of nuclear body mobility the effect of ATP depletion on chromatin structure and dynamics has to be considered.
Using different GFP-lac-repressor systems, rapid but locally restricted movements in mammalian cells as well as in yeast and Drosophila were shown to occur on a length scale of 0.4–0.5 μm (39, 43–46). This finding is in good agreement with the observed corral size (rc) of ≈0.3 μm for all nuclear bodies determined here. Interestingly, in a recent paper very similar results were obtained from particle nanotracking (47). From an analysis of the dynamics of 100-nm diameter nanospheres nuclear microdomains with a mean size of 0.29 ± 0.05 μm were identified. Thus, several lines of evidence support our finding that a chromatin space ≈0.3 μm is accessible for fast movements of nuclear bodies. Furthermore, the mean D value for the chromatin surrounding the nuclear bodies (Dc) of 1.36 × 10–4 μm2·s–1 is identical to D = 1.4 × 10–4 μm2·s–1 for the movement of chromatin loci determined in the lac repressor system (45) and similar to values from 1.8 to 5.8 × 10–4 μm2·s–1 measured for telomeres by using peptide nucleic acid probes (48). In summary, our analysis describes the movement of nuclear bodies as mostly determined by fluctuations of the chromatin density and large-scale movements of chromatin corrals within the nucleus. The artificial Mx1-YFP body introduced here showed mobility very similar to PML and Cajal bodies. This finding suggests that in our experimental system specific transport mechanisms for a certain type of body or body-specific chromatin interactions do not determine the movement of these bodies. The observed colocalization of bodies avoiding chromatin regions around the nucleolus indicates that only a subspace is accessible, consistent with the observation that the chromatin around the nucleolus is frequently more strongly condensed. These highly compacted chromatin loci had been frequently associated with silencing of biological activity. Other characteristic features of chromatin that lead to a preferred localization of nuclear bodies remain to be identified. Thus, concentrating certain transcription-related functions into nuclear bodies, which because of their physical nature are enriched in a more accessible chromatin environment, might be an effective mechanism to colocalize specific proteins with their target sites.
Supplementary Material
Acknowledgments
We thank K. Richter for the electron microscopy study; D. Spector and A. Lamond for critical reading of the manuscript; and D. Gerlich, S. Sterz, J. Fieres, J. Mattes, O. Haller, C. Cardoso, A. Möller, M. Reichenzeller, T. Knoch, A. Lamond, and D. Olins for reagents and support. This work was supported by Deutsche Forschungsgemeinschaft Grant Li406/5-3 and the Volkswagen Stiftung in the program Junior Research Groups at German Universities.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: D, diffusion coefficient; MSD, mean square displacement; PML, promyelocytic leukemia; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein.
References
- 1.Frey, M. R. & Matera, A. G. (1995) Proc. Natl. Acad. Sci. USA 92, 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schul, W., van Driel, R. & de Jong, L. (1998) Mol. Biol. Cell 9, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs, E. Y., Frey, M. R., Wu, W., Ingledue, T. C., Gebuhr, T. C., Gao, L., Marzluff, W. F. & Matera, A. G. (1999) Mol. Biol. Cell 10, 1653–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gall, J. G., Stephenson, E. C., Erba, H. P., Diaz, M. O. & Barsacchi-Pilone, G. (1981) Chromosoma 84, 159–171. [DOI] [PubMed] [Google Scholar]
- 5.Callan, H. G., Gall, J. G. & Murphy, C. (1991) Chromosoma 101, 245–251. [DOI] [PubMed] [Google Scholar]
- 6.Shiels, C., Islam, S. A., Vatcheva, R., Sasieni, P., Sternberg, M. J., Freemont, P. S. & Sheer, D. (2001) J. Cell Sci. 114, 3705–3716. [DOI] [PubMed] [Google Scholar]
- 7.Platani, M., Goldberg, I., Lamond, A. I. & Swedlow, J. R. (2002) Nat. Cell Biol. 4, 502–508. [DOI] [PubMed] [Google Scholar]
- 8.Muratani, M., Gerlich, D., Janicki, S. M., Gebhard, M., Eils, R. & Spector, D. L. (2002) Nat. Cell Biol. 4, 106–110. [DOI] [PubMed] [Google Scholar]
- 9.Gall, J. G. (2000) Annu. Rev. Cell Dev. Biol. 16, 273–300. [DOI] [PubMed] [Google Scholar]
- 10.Negorev, D. & Maul, G. G. (2001) Oncogene 20, 7234–7242. [DOI] [PubMed] [Google Scholar]
- 11.Lamond, A. I. & Sleeman, J. E. (2003) Curr. Biol. 13, R825–R828. [DOI] [PubMed] [Google Scholar]
- 12.Spector, D. L. (2003) Annu. Rev. Biochem. 72, 573–608. [DOI] [PubMed] [Google Scholar]
- 13.Haller, O. & Kochs, G. (2002) Traffic 3, 710–717. [DOI] [PubMed] [Google Scholar]
- 14.Horisberger, M. A., Staeheli, P. & Haller, O. (1983) Proc. Natl. Acad. Sci. USA 80, 1910–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreiding, P., Staeheli, P. & Haller, O. (1985) Virology 140, 192–196. [DOI] [PubMed] [Google Scholar]
- 16.Möller, A., Sirma, H., Hofmann, T. G., Rueffer, S., Klimczak, E., Droge, W., Will, H. & Schmitz, M. L. (2003) Cancer Res. 63, 4310–4314. [PubMed] [Google Scholar]
- 17.Svitkina, T. M. & Borisy, G. G. (1999) J. Cell Biol. 145, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görisch, S. M., Richter, K., Scheuermann, M. O., Herrmann, H. & Lichter, P. (2003) Exp. Cell Res. 289, 282–294. [DOI] [PubMed] [Google Scholar]
- 19.Fieres, J., Mattes, M. & Eils, R. (2001) in Pattern Recognition DAGM 2001, eds. Radig, B. & Florczyk, S. (Springer, Berlin), Vol. 2191, pp. 76–83. [Google Scholar]
- 20.Eils, R. & Athale, C. (2003) J. Cell Biol. 161, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxton, M. J. (1994) Biophys. J. 66, 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fejes Tóth, K., Knoch, T. A., Wachsmuth, M., Frank-Stöhr, M., Stöhr, M., Bacher, C. P., Müller, G. & Rippe, K. (2004) J. Cell Sci. 117, 4277–4287. [DOI] [PubMed] [Google Scholar]
- 23.Platani, M., Goldberg, I., Swedlow, J. R. & Lamond, A. I. (2000) J. Cell Biol. 151, 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudonck, K., Dolan, L. & Shaw, P. J. (1999) Mol. Biol. Cell 10, 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiesmeijer, K., Molenaar, C., Bekeer, I. M., Tanke, H. J. & Dirks, R. W. (2002) J. Struct. Biol. 140, 180–188. [DOI] [PubMed] [Google Scholar]
- 26.Eskiw, C. H., Dellaire, G., Mymryk, J. S. & Bazett-Jones, D. P. (2003) J. Cell Sci. 116, 4455–4466. [DOI] [PubMed] [Google Scholar]
- 27.Sleeman, J. E., Trinkle-Mulcahy, L., Prescott, A. R., Ogg, S. C. & Lamond, A. I. (2003) J. Cell Sci. 116, 2039–2050. [DOI] [PubMed] [Google Scholar]
- 28.Deryusheva, S. & Gall, J. G. (2004) Proc. Natl. Acad. Sci. USA 101, 4810–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handwerger, K. E., Murphy, C. & Gall, J. G. (2003) J. Cell Biol. 160, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chelbi-Alix, M. K., Pelicano, L., Quignon, F., Koken, M. H., Venturini, L., Stadler, M., Pavlovic, J., Degos, L. & de The, H. (1995) Leukemia 9, 2027–2033. [PubMed] [Google Scholar]
- 31.Engelhardt, O. G., Ullrich, E., Kochs, G. & Haller, O. (2001) Exp. Cell Res. 271, 286–295. [DOI] [PubMed] [Google Scholar]
- 32.Monneron, A. & Bernhard, W. (1969) J. Ultrastruct. Res. 27, 266–288. [DOI] [PubMed] [Google Scholar]
- 33.Zhong, S., Salomoni, P. & Pandolfi, P. P. (2000) Nat. Cell Biol. 2, E85–E90. [DOI] [PubMed] [Google Scholar]
- 34.Simson, R., Yang, B., Moore, S. E., Doherty, P., Walsh, F. S. & Jacobson, K. A. (1998) Biophys. J. 74, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sako, Y. & Kusumi, A. (1994) J. Cell Biol. 125, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxton, M. J. (1993) Biophys. J. 64, 1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxton, M. J. (1995) Biophys. J. 69, 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxton, M. J. & Jacobson, K. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 373–399. [DOI] [PubMed] [Google Scholar]
- 39.Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. (2001) Science 294, 2181–2186. [DOI] [PubMed] [Google Scholar]
- 40.Kanda, T., Sullivan, K. F. & Wahl, G. M. (1998) Curr. Biol. 8, 377–385. [DOI] [PubMed] [Google Scholar]
- 41.Schul, W., Groenhout, B., Koberna, K., Takagaki, Y., Jenny, A., Manders, E. M., Raska, I., van Driel, R. & de Jong, L. (1996) EMBO J. 15, 2883–2892. [PMC free article] [PubMed] [Google Scholar]
- 42.Grosberg, A. Y. & Khokhlov, A. R. (1994) Statistical Physics of Macromolecules (AIP Press, New York).
- 43.Marshall, W. F., Straight, A., Marko, J. F., Swedlow, J., Dernburg, A., Belmont, A., Murray, A. W., Agard, D. A. & Sedat, J. W. (1997) Curr. Biol. 7, 930–939. [DOI] [PubMed] [Google Scholar]
- 44.Vazquez, J., Belmont, A. S. & Sedat, J. W. (2001) Curr. Biol. 11, 1227–1239. [DOI] [PubMed] [Google Scholar]
- 45.Chubb, J. R., Boyle, S., Perry, P. & Bickmore, W. A. (2002) Curr. Biol. 12, 439–445. [DOI] [PubMed] [Google Scholar]
- 46.Abney, J. R., Cutler, B., Fillbach, M. L., Axelrod, D. & Scalettar, B. A. (1997) J. Cell Biol. 137, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng, Y., Lee, J. S., Kole, T. P., Jiang, I. & Wirtz, D. (2004) J. Cell Sci. 117, 2159–2167. [DOI] [PubMed] [Google Scholar]
- 48.Molenaar, C., Wiesmeijer, K., Verwoerd, N. P., Khazen, S., Eils, R., Tanke, H. J. & Dirks, R. W. (2003) EMBO J. 22, 6631–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.