Abstract
Type II platelet-activating factor-acetylhydrolase [PAF-AH (II)] is an N-myristoylated enzyme that contains a lipase/esterase catalytic motif and selectively hydrolyzes the sn-2 acetyl ester of PAF and other short-chain acyl groups attached to phosphoglycerides. However, the physiological role of this enzyme remains to be elucidated. PAF-AH (II) is conserved in a variety of species ranging from a simple multicellular organism, Caenorhabditis elegans, to mammals. C. elegans possesses two homologous PAF-AH (II) genes, named paf-1 and paf-2. In this study, we generated these two loss-of-function mutants to elucidate the in vivo PAF-AH (II) function. Surprisingly, mutants of paf-2, a major isoform of C. elegans PAF-AH (II)s, exhibits gross defects in epithelial sheet formation, resulting in unsuccessful subsequent morphogenesis with complete penetrance. Moreover, paf-2 RNA interference worms show a variable abnormal morphology, including ectopic protrusions and a lumpy shape at the late embryonic and early larval stages due to epithelial organization defects. Consistent with these phenotypes, PAF-AH (II) is predominantly expressed in epithelial cells of C. elegans. This study demonstrates that PAF-AH (II) is essential for epithelial morphogenesis.
Epithelial cells play essential roles in multicellular animals in which changes in epithelial cell shape orchestrate morphogenesis of the embryo and individual organs. Caenorhabditis elegans has been used as a genetic model organism for the analysis of epithelial morphogenesis. The worm-like shape of C. elegans is a result of the morphogenesis of the embryonic epidermis, the epithelium that encloses the embryo. The epidermis originates as six rows of epidermal cells positioned on the dorsal surface (1) that subsequently develop mature cell junctions to form an epidermal sheet. The two dorsal-most rows of epidermal cells interdigitate to form a single row in a movement known as dorsal intercalation (2). During dorsal intercalation, the epidermal sheet spreads to enclose the embryo, sealing at the ventral midline (3). After ventral enclosure is complete, circumferential constriction in the epidermis squeezes the embryo longitudinally in a process known as elongation (4). A number of molecules required for epidermal morphogenesis have already been identified, including axon guidance molecules, cell-junction molecules, and transcription factors (5–7).
Platelet-activating factor (PAF; systematic name 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine) is a potent signaling phospholipid involved in diverse physiological and pathological events such as inflammation, anaphylaxis, reproduction, and fetal development (8). PAF-acetylhydrolase (PAF-AH) was originally identified as an enzyme that hydrolyses the acetyl group attached at the sn-2 position of PAF (9). At least three types of PAF-AH exist in mammals, namely the intracellular types I and II and a plasma type (10). Intracellular type I PAF-AH is an oligomeric enzyme consisting of two homologous catalytic subunits and a noncatalytic subunit (11, 12). The noncatalytic subunit is identical to a product of the LIS1 gene, mutations of which cause Miller–Dieker lissencephaly, a disease characterized by a severe malformation of the brain cortex (13). In addition, targeted disruption of the intracellular type I PAF-AH catalytic subunits causes severe impairment in murine spermatogenesis (14).
Intracellular type II PAF-AH [PAF-AH (II)] is a monomeric 40-kDa enzyme. The amino acid sequence of PAF-AH (II) does not show any significant similarity to any subunit of intracellular type I PAF-AH but shows ≈42% identity with that of the plasma-type PAF-AH (15). PAF-AH (II), as well as plasma PAF-AH, hydrolyze not only acetyl groups but also short-length acyl chains such as propionyl and butyloyl groups attached to phosphoglyceride (16). Both enzymes also hydrolyze truncated acyl chains derived from oxidative cleavage of long-chain polyunsaturated fatty acyl groups, suggesting that they have a role in scavenging selectively oxidized phospholipid species without acting on structural membrane phospholipids (17). PAF-AH (II) is N-myristoylated at the N terminus and is distributed in both the cytosol and membranes like other N-myristoylated proteins (17). However, the physiological function of PAF-AH (II) is still obscure due to the lack of a genetic deficiency state in human subjects and the lack of appropriate animal models deficient in this enzyme.
A search of the database for sequences similar to human PAF-AH (II) found homologues in various animals, including the simple, multicellular C. elegans, suggesting that PAF-AH (II) plays a fundamental role in animals. To elucidate the function of PAF-AH (II) in vivo, we isolated PAF-AH (II) loss-of-function mutants of C. elegans. In the PAF-AH (II) mutant embryo, organization of the epidermal sheet was grossly defective.
Materials and Methods
General Methods and Strains. Maintenance and genetic manipulation of C. elegans were carried out as described (18). The Bristol strain N2 was used as the standard wild-type strain. For phenotypic analysis of paf-2(tj12), eff-1(oj55)II, jcIs1(ajm-1::gfp)IV, wIs51(scm::gfp), mcIs28(let-413::gfp), and zuEx24(hmp-1::gfp) were used. An allele used for a balancer chromosome of paf-2(tj12) was lon-2(e678)X.
Cloning of C. elegans PAF-AH (II)s paf-1 and paf-2. A paf-1 cDNA clone, yk92g8, was kindly supplied by Y. Kohara (National Institute of Genetics, Shizuoka, Japan). Full-length paf-2 cDNA was amplified from a C. elegans cDNA library with the following primers: GTT TAG GAT CCA TGG GTA GCT ATA TCT CGT CGC CAC AAG TTC TA; and GGA TAC TCG AGT TAA AGT TTG TAT TTT TCA CGT CC. The PCR fragment of paf-2 cDNA was cloned into pBluescript SK(–) vector by using the BamHI and XhoI sites. paf-1 and paf-2 sequences have been submitted to the GenBank database, accession nos. AF386744 and AF386745, respectively.
Isolation of paf-1 and paf-2 Deletion Mutants. A worm library that was mutagenized with a combination of trimethylpsoralen and UV irradiation (19) was screened for a deletion in the paf-1 and paf-2 locus by using their specific PCR primers. The primers for paf-1 deletion screen were: ACA GAA GGC ATT GAG AGA AGA G; GAA GTT GAA GGA AGC GGA TGT T; CCC GAT TTT CAA CAT TTC AGC C; and TTG CTG CTT ATT GTT TTT CCG G; and the primers for paf-2 deletion screen were CTC TTC TGC TTT CCG ATA GTG G; GGC TCA TCC ATT ATC CCA TCC A; CTC TTC CGT TTG TTC CAG TCT C; and CGC GAA CAA GGC AAT ACC AGA G. Both paf-1 and paf-2 deletion mutants were isolated and backcrosses were performed seven times.
Antibody Production. A recombinant PAF-1 protein that was expressed and purified by an E. coli pET expression system (Novagen) was injected into the hind foot pads of WYK rats by using Freund's complete adjuvant. The enlarged medial iliac lymph nodes were used for cell fusion (20) with mouse myeloma PAI cells. In the present investigation, SY8, which recognizes PAF-1 and PAF-2 equally, was used for Western blotting at 1:500 dilution.
Immunofluorescence and Microscopy. Embryos were prepared for antibody staining and were staged as described (21). The antibodies MH27, anti-LIN-26, and MH33 were used at dilutions of 1:1,500, 1:2,000, and 1:50, respectively. Three-dimensional reconstruction of digital images stained by MH27 was performed by using a Zeiss LSM510 confocal microscope. Four-dimensional microscopy images were taken with the 4d grabber ppc software on a Zeiss Axiophot with a focus controller (ASI), and characterized with the 4d viewer ppc software (kindly provided by C. Thomas, University of Wisconsin, Madison). Scanning electron microscopy analysis was carried out as described (3), except that the samples were coated with 5 nm of platinum/palladium and observed with a Hitachi-650 scanning electron microscope.
Preparation of Transgenic Worms. DNA injection into the C. elegans germ line was carried out as described (22). The array qaEx3101 contains the plasmid pIT3 (paf-2::GFP) and pRF4 [rol-6(su1006)] for analysis of paf-2 expression. pIT3 was generated by inserting a sequence containing the paf-2 promoter region (a 3.5-kb region upstream of the paf-2 initiation codon) and the adjacent first exon into the PstI–BamHI sites of pPD96.04. qaIs3102 (hsp::paf-2 RNAi), which is a transgenic line expressing both strands of paf-2 RNA, was engineered by using a mixture of six plasmids: pIT6 (hsp16–2::paf-2), pIT7 (hsp16–41::paf-2), pIT8 (hsp16–2::paf-2 antisense), pIT9 (hsp16–41::paf-2 antisense), pEF-1α::GFP (kindly provided by M. Koga, Kyushu University, Fukuoka-shi, Japan), and pRF4. Plasmids pIT6, 7, 8, and 9 were generated by cloning paf-2 sense or antisense cDNAs into the heat-shock vectors pPD49.78 and pPD49.83 of the NheI and KpnI sites (23). pPD49.78, pPD49.83, and pPD96.04 were kindly provided by A. Fire (Carnegie Institution of Washington, Washington, DC). The array qaEx3102 was produced by injection of the plasmid pXH01, which carries a GFP transcriptional reporter for the lbp-1 gene, and pRF4. The lbp-1p::gfp reporter is expressed at high levels in dorsal epidermis (24, 25). pXH01 was kindly provided by J. Plenefisch (University of Toledo, Toledo, OH).
RNA Interference (RNAi). For paf-2 RNAi during embryogenesis, paf-1(tj11); qaIs3102 (hsp::paf-2 RNAi) were allowed to lay eggs for 2–3 h, and 4–5 h later, the eggs were heat-shocked at 33°C for 20–25 min. Unhatched eggs were examined 16–18 h after being laid, and hatched but arrested L1 larvae were examined 48 h after being laid. We confirmed that the control strain N2 did not show any defects under the same heat treatment. For Western blotting and PAF-AH assays, mixed-stage animals were heat-shocked at 33°C for 2 h and were collected from plates 24 h after heat treatment.
Assays. The hydrolase assays using PAF, phosphatidylcholine, and oxidized phosphatidylcholine as substrates were performed as described (10). Recombinant PAF-1 and PAF-2 proteins that were expressed and purified by an E. coli pET expression system (Novagen) were used as the enzymes. C. elegans were lysed by sonication and were centrifuged at 20,000 × g for 20 min. The supernatant was used as the enzyme source.
Results
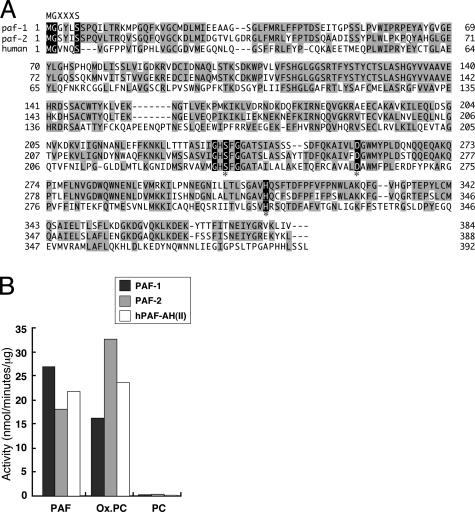
Identification of C. elegans PAF-AH (II)s paf-1 and paf-2. A database search of the complete C. elegans genome sequence revealed the presence of two PAF-AH (II) homologues on chromosomes I and X (W03G9.6 and C52B9.7, respectively). We named these W03G9.6 and C52B9.7 homologues paf-1 and paf-2, respectively. The paf-1 and paf-2 gene products consist of 384- and 388-aa residues, respectively, and are 69% identical at the amino acid level (Fig. 1A). They also show a 35% identity with the mammalian PAF-AH (II)s. Both PAF-1 and PAF-2 have a myristoylation signal at the N terminus as in mammalian PAF-AH(II)s. The lipase/esterase catalytic center (Gly-X-Ser-X-Gly motif) was also conserved in PAF-1 and PAF-2 with Ser as the attacking nucleophile, with His and Asp likely forming the other two elements of the catalytic-triad at the regions corresponding to the mammalian PAF-AH(II)s. In addition, PAF-1 and PAF-2 show limited amino acid sequence similarities to bacterial lipases (Fig. 5, which is published as supporting information on the PNAS web site, and ref. 26). Next, we examined the substrate specificities by using PAF-1 and PAF-2 recombinant proteins. Like the mammalian PAF-AH (II)s, both PAF-1 and PAF-2 efficiently hydrolyzed the acetyl moieties at the sn-2 position of PAF, but did not hydrolyze long acyl chains attached to phosphatidylcholine to any extent (Fig. 1B). They also showed hydrolytic activity toward oxidized fragments of unsaturated fatty acids at the sn-2 position of phosphoglycerides similar to mammalian PAF-AH (II)s (Fig. 1B). These observations indicate that the two PAF-AH (II)s in C. elegans, paf-1 and paf-2, are orthologues of mammalian PAF-AH (II)s with respect to their primary structure and catalytic activity.
Fig. 1.
Predicted amino acid sequences and enzyme activities of PAF-1 and PAF-2. (A) Amino acid sequences of C. elegans PAF-AH (II)s (PAF-1 and PAF-2) and human PAF-AH (II). Residues identical to those of PAF-AH (II)s are shaded in gray. Ser, Asp, and His residues that are indicated by asterisks are the predicted amino acid residues that form catalytic triads in the PAF-AH (II)s. The GXSXG motifs, which are highly conserved in lipases and esterases, and the N-myristoylation signal (MGXXXS) are highlighted in black. (B) Substrate specificities of PAF-1 and PAF-2. Hydrolytic activities of PAF-1, PAF-2, and human PAF-AH (II) were measured by using PAF, phosphatidylcholine (PC), and oxidized PC (Ox.PC) as substrates.
paf-2 Deletion Mutants Show Embryonic Lethality. To address the function of PAF-AH (II)s, we generated paf-1 and paf-2 mutants by using a PCR-based deletion screen (19). One deletion allele each for the paf-1 and paf-2 genes was isolated and named tj11 and tj12, respectively (Fig. 6, which is published as supporting information on the PNAS web site). Both deletions lack the exons containing the ATG translational initiation codon and catalytic triad, indicating that these alleles eliminate gene function completely (Fig. 6). The following observations indicate that loss of paf-2 caused a zygotic embryonic lethal phenotype with complete penetrance (n > 2,000), whereas the paf-1 homozygous mutant grew normally and exhibited no aberrant phenotype.
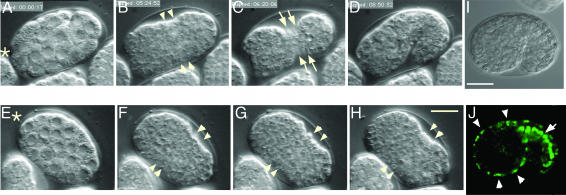
paf-2 Is Expressed in Epithelial Cells and Is Essential for Epithelial Sheet Formation. We first analyzed the paf-2 mutant embryos [paf-2(tj12)] by using four-dimensional time-lapse microscopy (Fig. 2). No significant alterations during the cell proliferation stage were noticed in the paf-2(tj12) embryos (Fig. 2 E and F). After the cell-proliferation stage in wild-type embryos, the epidermal sheet encloses the embryo and circumferential constriction within the epidermis squeezes the embryo into a long, thin worm (refs. 3, 4, and 27 and Fig. 2 B–D). Although paf-2(tj12) embryos started narrowing on the ventral side (Fig. 2 F and G, arrowheads), exhibiting the characteristic lima bean shape, all paf-2(tj12) embryos were arrested at the bean stage and failed to elongate (Fig. 2H and Movie 1, which is published as supporting information on the PNAS web site). Consistent with these phenotypes, paf-2::GFP expression was observed in epidermal cells at the beginning of elongation (Fig. 2 I and J, arrowheads). Strong paf-2::GFP was also visible in the intestine, another epithelial organ (Fig. 2 J, arrow).
Fig. 2.
paf-2(tj12) mutant fails to elongate. (A–H) Nomarski micrographs of wild-type (A–D) and paf-2(tj12) (E–H) by four-dimensional microscopy. (A–D) A wild-type embryo is enclosed by an epidermal sheet (B and C, ventral view) and subsequently elongates (D, lateral view, dorsal up). The ventral epidermal cells are visible on the ventral surface (C, arrows). (E–H) A paf-2(tj12) embryo appears to be normal during the cell proliferation stage (E and F), but does not elongate (G and H, see text). The anterior of the worm is indicated by asterisks. (I and J) paf-2::GFP expression in embryos (lateral view, dorsal is up). Nomarski micrographs (I) and corresponding paf-2::GFP expression (J). paf-2 reporter gene constructs are expressed in epidermal cells (J, arrowheads) and intestinal cells (J, arrow) at the beginning of elongation. Anterior is to the left. (Bar, 10 μm.)
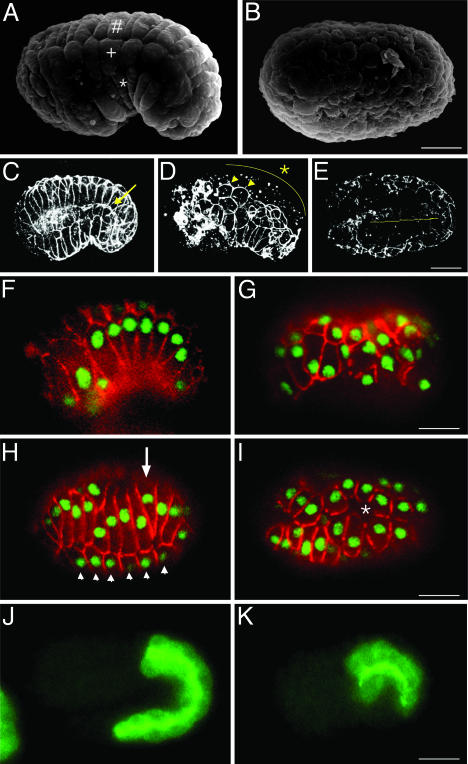
Next, we observed paf-2(tj12) embryos at the terminal stage by scanning electron microscopy to examine the epidermal cells surrounding the embryo. A lateral view of a wild-type embryo that has completed the enclosure process is shown in Fig. 3A. In contrast to wild-type embryos, paf-2(tj12) embryos at the terminal stage showed a drastically abnormal surface structure in which no rectangular cells characteristic of epidermal cells can be seen (Fig. 3B). To visualize epidermal cells, embryos were stained with mAb MH27, which recognizes AJM-1, which is present at the junctions between the epidermal, pharyngeal, and intestinal cells (28, 29). MH27 staining of a wild-type embryo shows a rectilinear pattern around the epidermal cells (Fig. 3C, and Fig. 7A, which is published as supporting information on the PNAS web site). In paf-2(tj12) terminal embryos, however, MH27 staining was discontinuous and irregular or absent throughout the embryonic surface (Figs. 3 D and E and 7 B and C). MH27 staining was punctate or was faint in the dorsal regions (Fig. 3D, asterisk), and the lateral row of epidermal cells on each side of the embryos still seemed to be expressing MH27 antigen (Figs. 3D and 7B, arrowheads, and Fig. 8D, which is published as supporting information on the PNAS web site). Epidermal membranes appeared to aggregate and were stained intensely in the anterior part of paf-2(tj12) embryos. The paf-1;paf-2 double mutants also showed the same phenotype as the paf-2 mutant (data not shown). Furthermore, injection of paf-2 dsRNA into jcIs1(ajm-1::GFP) animals (30) resulted in epidermal defects similar to those of paf-2(tj12) mutants (data not shown). These observations demonstrate that epidermal sheet formation in the paf-2(tj12) embryo is severely impaired.
Fig. 3.
paf-2(tj12) mutants display epidermal abnormalities. (A and B) Scanning electron micrographs of wild-type (A) and paf-2(tj12)(B) embryos at the terminal stage. In the wild-type embryo, dorsal (#), lateral (+), and ventral (asterisk) rows of epidermal cells are visible (A, lateral view), whereas paf-2(tj12) embryos show aberrant surface structure (B, presumably the dorsal view). (C–E) Comparison of three-dimensional reconstructions of MH27 immunostaining in wild-type (C, lateral view) and terminal stage paf-2(tj12) embryos (D, lateral view; E, dorsal view). In terminal paf-2(tj12) embryos, the MH27 staining is punctate and irregular or is absent in many parts of the epidermis, especially on the dorsal side (D, asterisk; see text). Note that the MH27 staining of intestinal cells is also discontinuous in the paf-2(tj12) embryos (E, yellow line), whereas the staining is continuous in the wild-type (C, arrow). (F–I) Embryos were costained with MH27 (red) and LIN-26 antiserum (green). (F and G) Ventral enclosure. In the wild-type embryo, the epidermis fully enclosed the embryo (F, ventrolateral external focal plane showing the ventral epidermal cells). In paf-2(tj12) embryos, the shapes and patterning of epidermal cells are aberrant and the incomplete epidermal sheet fails to fully enclose the embryo (G, lateral external focal plane). (H and I) Dorsal intercalation (dorsal external focal planes showing the dorsal epidermal cells). In wild-type embryos, two rows of dorsal epidermal cells have intercalated to form a single row (H, arrow), whereas the dorsal epidermal cells in the paf-2(tj12) embryo were mislocalized and MH27 staining soon diminished at the cell junction (I, asterisk). Arrowheads in H indicate the lateral epidermal cells. (J and K) Embryos were stained with MH33, which specifically recognizes intestinal cells (internal focal planes, lateral view). Comparison of wild-type embryos (J, twofold stage) and paf-2(tj12) embryos (K, terminal lima bean stage). The intestinal cells of the paf-2(tj12) embryo appear to be differentiated at least partially (K). Anterior is to the left in all images. (Bar, 10 μm.)
To understand the basis of the severe epidermal defect, embryos at various stages were stained by using MH27 and anti-LIN-26 antibody, the latter of which is used as a nuclear marker of nonneuronal ectodermal cells, including epidermal cells (31). During ventral enclosure in the paf-2(tj12) embryo, the positions and alignment of epidermal cells were abnormal (Fig. 3G). The anterior epidermal cells appeared to enclose the embryo, whereas the posterior ventral cells failed to form junctions and the incomplete epidermal sheet retracted to the dorsal surface (data not shown). In the dorsal regions, the alignment of dorsal epidermal cells was also abnormal, and junctional MH27 staining soon diminished on the dorsal surface (Fig. 3I, asterisk). In an eff-1(oj55) background in which epidermal cell fusion is suppressed (32), the dorsal epidermal cells became wedge-shaped and initiated dorsal intercalation, but could not complete intercalation in the paf-2(tj12) embryo (Fig. 9 C and D, and Fig. 10 C and D, which are published as supporting information on the PNAS web site). The AJM-1::GFP localization was discontinuous and punctate in the dorsal regions of eff-1;paf-2 embryos (Fig. 9D), indicating that AJM-1::GFP cannot localize at cell junctions in the dorsal epidermis of the paf-2(tj12) embryo, even under the condition where the dorsal epidermal cells contact, but do not fuse with each other. We also analyzed the expression pattern of LET-413, a leucine-rich repeats and PDZ domains family protein, which is essential for AJM-1 localization to apical junctions (33), and found that the LET-413::GFP localization was also affected in the paf-2(tj12) embryos like AJM-1 (Fig. 8F).
The epidermal cell junction in C. elegans contains at least two distinct complexes, an AJM-1/DLG-1 complex (29, 33–36), and an HMR-1 (E-cadherin)/HMP-1 (α-catenin)/HMP-2 (β-catenin) complex (27). To address whether the localization of the cadherin/catenin complex is affected in paf-2(tj12) mutant embryos, we generated paf-2(tj12) mutant expressing HMP-1::GFP (37). In a wild-type embryo, HMP-1::GFP was expressed in the cytoplasm and plasma membrane in epidermal cells (Fig. 11 A and B, which is published as supporting information on the PNAS web site). In a paf-2(tj12) embryo, however, HMP-1::GFP was not detected in the plasma membrane in many parts of the epidermis (Fig. 11 C and D). Some of the lateral epidermal cells appeared to express the continuous HMP-1::GFP at cell junctions like AJM-1 (Figs. 8D and 11 C and D, arrows).
These data indicate that the localization of AJM-1 and HMP-1 are severely impaired in paf-2(tj12) mutant embryos and that paf-2 is essential for early epidermal formation, including epidermal cell alignment, rearrangement, and/or adhesion.
Because paf-2 expression was observed in intestinal cells during embryogenesis (Fig. 2 J), we next stained paf-2(tj12) embryos with mAb MH33, which is a specific marker of intestinal cells (28). In the paf-2(tj12) embryos, the MH33 staining appeared to be normal (Fig. 3K), whereas MH27 staining of the paf-2(tj12) intestine was discontinuous and faint (Fig. 3E, yellow line). These results indicate that intestinal cells of the paf-2(tj12) are differentiated, but their organization is impaired at some levels.
Taken together, these results indicate that disruption of paf-2 results in embryonic lethality due to severe defects in epidermal sheet organization.
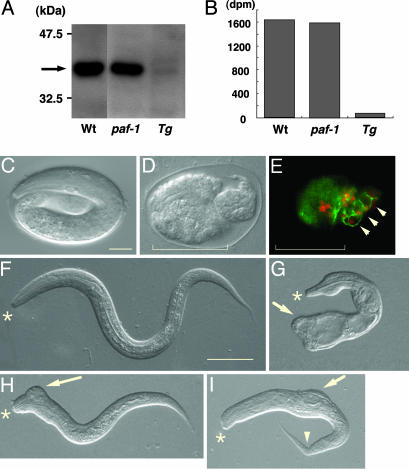
Heat-Shock-Inducible paf-2 RNAi Causes Defects in Epidermal Organization During Morphogenesis. After ventral enclosure, the epidermis begins circumferential constrictions that elongate the embryo into a worm. To analyze the function of paf-2 during the elongation process, we generated transgenic animals in which heat shock induces paf-2 RNAi, producing both senses of paf-2 RNA in vivo [qaIs3102 (hsp::paf-2 RNAi); see Materials and Methods]. We first examined the effect on PAF-AH (II) protein expression of removal of paf-1 and paf-2 function singly and in combination by using mutants and RNAi. Western blotting analysis by using mAb SY8, which recognizes both PAF-1 and PAF-2 to a similar extent, revealed a similar immunoreactive band in the paf-1(tj11) mutants (Fig. 4A). Consistent with this observation, the PAF-AH activity of the paf-1(tj11) homogenates was not decreased appreciably (Fig. 4B), indicating that PAF-1 makes a negligible contribution to PAF-AH activity in C. elegans. Upon heat-shock treatment, Tg(paf-2 RNAi), which a is paf-1(tj11) mutant carrying a qaIs3102(hsp::paf-2 RNAi) array, showed significantly reduced paf-2 expression (Fig. 4A), and exhibited negligible enzyme activity (Fig. 4B). These data demonstrate that heat-shock-inducible paf-2 RNAi by the qaIs3102 array was achieved, and that PAF-2 is the major isoform conferring PAF-AH enzyme activity in C. elegans. In ≈11% of the heat-shocked Tg(paf-2 RNAi) animals, development was arrested during embryogenesis, and in ≈5% of these animals it was arrested at the early larval stages (n = 573). The arrested Tg(paf-2 RNAi) showed variable abnormal morphology phenotypes (Fig. 4 C–I). Most embryos that were arrested during embryogenesis had a very lumpy morphology or were ruptured, with internal cells oozing through the holes in the epidermis (data not shown). In some embryos, no MH27 staining was seen in the anterior part of the embryo, whereas the posterior part was enclosed by epidermal cells and elongated partially, which is indicative of anterior rupture during enclosure (Fig. 4 D and E). Hatched but arrested Tg(paf-2 RNAi) also exhibited morphology defects, including ectopic protuberances (Fig. 4 G–I, arrows) and a kinked tail (Fig. 4I, arrowhead). These observations suggest that inhibition of paf-2 expression during late embryogenesis also affected epidermal organization in the elongation process.
Fig. 4.
Phenotypes of transgenic worms in which heat shock induces paf-2 RNAi. (A) Western blotting of total protein extract from heat-shocked wild-type (Wt), paf-1(tj11) (paf-1), and paf-1(tj11); qaIs3102 (hsp::paf-2 RNAi) (Tg) with SY8, which recognizes both PAF-1 and PAF-2 equally. The arrow indicates an immunoreactive band against PAF-1 and PAF-2. (B) PAF acetylhydrolase activity of total protein extract from heat-shocked Wt, paf-1, and Tg(paf-2 RNAi). (C–I) Wild-type and examples of the range of defects seen in heat-shocked Tg(paf-2 RNAi) animals. (C and F) Wild type. (D, E, and G–I) Tg(paf-2 RNAi). Nomarski micrographs of wild-type embryos (C, elongation stage) and unhatched Tg(paf-2 RNAi) embryos (D, terminal stage) are compared. The corresponding Tg embryo was costained with MH27 (green) and with LIN-26 antiserum (red) (E). The posterior of the Tg(paf-2 RNAi) is enclosed and is partially elongated, whereas the head has ruptured. (F–I) Hatched L1 animals. Comparison of wild-type (F) and Tg(paf-2 RNAi) (G–I). Some of the hatched Tg(paf-2 RNAi) show variable abnormal morphologies and development is arrested at the early larval stage. Ectopic protrusions in the tail (G, arrow), in the head (H, arrow), in the body (I, arrow), and kinked tail (I, arrowhead). Asterisks indicate the anterior of the larvae. (Bars, 10 μmin C–E; 20 μmin F–I.)
Discussion
PAF-AH (II) is conserved from a simple multicellular organism, C. elegans, to higher vertebrates. We show here that this enzyme is expressed predominantly in epithelial cells and is essential for epithelial sheet formation during C. elegans embryogenesis. Although the epidermal cells could be specified in the paf-2(tj12) mutant embryo, as judged from the expression of epidermal cell markers, the following phenotypes were observed in the epidermis: (i) abnormal positioning and alignment of epidermal cells, (ii) incomplete rearrangement of the dorsal epidermal cells, and (iii) irregular and punctate expression pattern of junctional molecules such as AJM-1, LET-413, and HMP-1. These observations suggest that late-stage epidermal differentiation, such as cell alignment, rearrangement and adhesion, is severely affected in the paf-2(tj12) mutant embryo.
Recent genetic analyses have identified a number of genes required for epidermal organization. The epidermal cell junction in C. elegans contains at least two distinct domains; one is adherens junction, which contains an HMR-1/HMP-1/HMP-2 complex (27), and the other is a domain basal to the adherens junction defined by an AJM-1/DLG-1 complex (29, 33–36). It has been reported that the localization of the AJM-1/DLG-1 complex is controlled by LET-413, and that dlg-1 or let-413 mutants show aberrant AJM-1-distribution like paf-2(tj12) mutants (33–36). However, the phenotypes of these mutants are different from those of paf-2(tj12) mutants in such a way that the dlg-1 or let-413 mutant exhibits junctional HMP-1 distribution and can elongate, whereas paf-2(tj12) mutants show aberrant HMP-1 localization and are arrested at the bean stage. On the other hand, mutants of the HMR-1/HMP-1/HMP-2 complex show normal AJM-1 localization at cell junctions, but exhibit a rupture phenotype similar to that of paf-2 RNAi worms. Comparing the phenotypes between the junctional molecule mutants and the paf-2(tj12) mutant, PAF-2 may play a role in epidermal sheet formation by controlling as-yet-unknown fundamental processes common to epidermal morphogenesis rather than by controlling one of these adhesion complexes.
PAF-AH (II) was originally identified as an enzyme that hydrolyzes the acetyl group attached at the sn-2 position of PAF. However, PAF is not detected in C. elegans (38). PAF-AH (II) also hydrolyzes oxidized phospholipids that possess truncated acyl chains derived from oxidative cleavage of long-chain poly-unsaturated fatty acids (PUFAs) as efficiently as PAF (16). It has been reported that C. elegans contains large amounts of PUFAs (38, 39). Membrane phospholipids containing PUFAs are oxidized readily by molecular oxygen because of the high susceptibility of bis-allylic hydrogens to oxidation. Such an oxidation, called autoxidation, proceeds by a free radical-mediated chain oxidation, resulting in oxidation of a number of lipids. Thus, lipids containing PUFAs are not only very susceptible to oxidation but also can propagate oxidative stress in the cell. It is interesting to note here that oxidative stress increases the paracellular permeability in epithelial cell monolayers by inducing the dissociation of the E-cadherin/β-catenin and occludin/ZO1 complexes from intracellular junctions without affecting cell viability (40, 41). PAF-AH (II) may function as an antioxidant phospholipase in C. elegans by hydrolyzing oxidized phospholipids and thereby reducing oxidative stress, thus maintaining the proper epithelial organization.
Recently, we found that mammalian PAF-AH (II) possesses transacetylase activity as well as hydrolase activity under certain conditions (42, 43). In an in vitro assay, the enzyme can transfer an acetyl group from PAF to acceptor lipids such as lysophospholipids and sphingosine. This finding raises the possibility that PAF-AH (II) functions as a synthetic enzyme by producing physiologically active lipids such as acetylated phospholipids and N-acetylsphingosine (C2-ceramide; ref. 44). PAF-AH (II) may alternatively play a role in epidermal morphogenesis by generating specific lipids that form a microdomain in the lipid bilayer at cell junctions or produce a certain lipid mediator involved in epidermal morphogenesis. Interestingly, in preliminary observations, we found that in mammals the enzyme is predominantly expressed in epithelial cells, such as kidney proximal and distal tubules, intestinal column epithelium, and lung tracheal epithelium. This result suggests that PAF-AH (II) has an essentially similar role in higher organisms.
Most of the molecules affecting epidermal morphogenesis that have been identified so far are localized in the plasma membrane where they form cell junctions, or in the nucleus where they transcribe target molecules involved in epidermal organization. In this respect, PAF-AH (II) is a unique protein that is localized in the endoplasmic reticulum membrane and cytosol, and possesses hydrolyzing activity against glycerolipids (17). Identification of the physiological substrate or metabolite of PAF-AH (II) and delineation of its function should therefore provide new insights into epithelial morphogenesis and organization.
Supplementary Material
Acknowledgments
We thank Y. Tomita and T. Jike (Nihon University, Tokyo) for the scanning electron microscopy analysis; Y. Kohara for cDNA clones; A. Fire for convenient vectors; J. Plenefisch for the vector lbp-1p::GFP; M. Koga for the vector pEF-1α::GFP; R. Waterston (Washington University, St. Louis) for MH27 and MH33; M. Labouesse (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Paris) for anti-Lin-26 antibody; H. Qadota and K. Kaibuchi (Nagoya University, Nagoya, Japan) for supplying antibodies; R. Legouis (Centre de Génétique Moléculaire, Centre National de la Recherche Scientifique, Paris) for let-413::GFP worms; and C. Thomas for 4d viewer ppc software. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Abbreviations: PAF, platelet-activating factor; PAF-AH, PAF-acetylhydrolase; PAF-AH (II), intracellular type II PAF-AH; RNAi, RNA interference.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF386744 and AF386745).
References
- 1.Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. (1983) Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 2.Williams-Masson, E. M., Heid, P. J., Lavin, C. A. & Hardin, J. (1998) Dev. Biol. 204, 263–276. [DOI] [PubMed] [Google Scholar]
- 3.Williams-Masson, E. M., Malik, A. N. & Hardin, J. (1997) Development (Cambridge, U.K.) 124, 2889–2901. [DOI] [PubMed] [Google Scholar]
- 4.Priess, J. R. & Hirsh, D. I. (1986) Dev. Biol. 117, 156–173. [DOI] [PubMed] [Google Scholar]
- 5.Ching-Sang, I. D. & Chisholm, A. D. (2000) Trends Genet. 16, 544–551. [DOI] [PubMed] [Google Scholar]
- 6.Simske, J. S. & Hardin, J. (2001) BioEssays 23, 12–23. [DOI] [PubMed] [Google Scholar]
- 7.Michaux, G., Legouis, R. & Labouesse, M. (2001) Gene 277, 83–100. [DOI] [PubMed] [Google Scholar]
- 8.Prescott, S. M., Zimmerman, G. A., Stafforini, D. M. & McIntyre, T. M. (2000) Annu. Rev. Biochem. 69, 419–445. [DOI] [PubMed] [Google Scholar]
- 9.Stafforini, D. M., Prescott, S. M., Zimmerman, G. A. & McIntyre, T. M. (1996) Biochim. Biophys. Acta 1301, 161–173. [DOI] [PubMed] [Google Scholar]
- 10.Hattori, M., Arai, H. & Inoue, K. (1993) J. Biol. Chem. 268, 18748–18753. [PubMed] [Google Scholar]
- 11.Hattori, M., Adachi, H., Tsujimoto, M., Arai, H. & Inoue, K. (1994) J. Biol. Chem. 269, 23150–23155. [PubMed] [Google Scholar]
- 12.Hattori, M., Adachi, H., Aoki, J., Tsujimoto, M., Arai, H. & Inoue, K. (1995) J. Biol. Chem. 270, 31345–31352. [DOI] [PubMed] [Google Scholar]
- 13.Hattori, M., Adachi, H., Tsujimoto, M., Arai, H. & Inoue, K. (1994) Nature 370, 216–218. [DOI] [PubMed] [Google Scholar]
- 14.Koizumi, H., Yamaguchi, N., Hattori, M., Ishikawa, T. O., Aoki, J., Taketo, M. M., Inoue, K. & Arai, H. (2003) J. Biol. Chem. 278, 12489–12494. [DOI] [PubMed] [Google Scholar]
- 15.Hattori, K., Adachi, H., Matsuzawa, A., Yamamoto, K., Tsujimoto, M., Aoki, J., Hattori, M., Arai, H. & Inoue, K. (1996) J. Biol. Chem. 271, 33032–33038. [DOI] [PubMed] [Google Scholar]
- 16.Hattori, K., Hattori, M., Adachi, H., Tsujimoto, M., Arai, H. & Inoue, K. (1995) J. Biol. Chem. 270, 22308–22313. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzawa, A., Hattori, K., Aoki, J., Arai, H. & Inoue, K. (1997) J. Biol. Chem. 272, 32315–32320. [DOI] [PubMed] [Google Scholar]
- 18.Brenner, S. (1974) Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yandell, M. D., Edgar, L. G. & Wood, W. B. (1994) Proc. Natl. Acad. Sci. USA 91, 1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishiro, Y., Kagawa, M., Naito, I. & Sado, Y. (1995) Cell Struct. Funct. 20, 151–156. [DOI] [PubMed] [Google Scholar]
- 21.Miller D. M. & Shakes, D. C. (1995) Caenorhabditis elegans: Modern Biological Analysis of an Organism, eds. Epstein, H. F. & Shakes, D. C. (Academic, San Diego), pp. 365–393.
- 22.Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, D., Russnak, R. H., Kay, R. J. & Candido, E. P. (1986) J. Biol. Chem. 261, 12006–12015. [PubMed] [Google Scholar]
- 24.Plenefisch, J., Xiao, H., Mei, B., Geng, J., Komuniecki, P. R. & Komuniecki, R. (2000) Mol. Biochem. Parasitol. 105, 223–236. [DOI] [PubMed] [Google Scholar]
- 25.Heid, P. J., Raich, W. B., Smith, R., Mohler, W. A., Simokat, K., Gendreau, S. B., Rothman, J. H. & Hardin, J. (2001) Dev. Biol. 236, 165–180. [DOI] [PubMed] [Google Scholar]
- 26.Wei, Y., Swenson, L., Castro, C., Derewenda, U., Minor, W., Arai, H., Aoki, J., Inoue, K., Servin, G. L. & Derewenda, Z. S. (1998) Structure (London) 6, 511–519. [DOI] [PubMed] [Google Scholar]
- 27.Costa, M., Raich, W., Agbunag, C., Leung, B., Hardin, J. & Priess, J. R. (1998) J. Cell Biol. 141, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis, G. R. & Waterston, R. H. (1985) J. Cell Biol. 101, 1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koppen, M., Simske, J. S., Sims, P. A., Firestein, B. L., Hall, D. H., Radice, A. D., Rongo, C. & Hardin, J. D. (2001) Nat. Cell Biol. 3, 983–991. [DOI] [PubMed] [Google Scholar]
- 30.Mohler, W. A., Simske, J. S., Williams-Masson, E. M., Hardin, J. D. & White, J. G. (1998) Curr. Biol. 8, 1087–1090. [DOI] [PubMed] [Google Scholar]
- 31.Labouesse, M., Hartwieg, E. & Horvitz, H. R. (1996) Development (Cambridge, U.K.) 122, 2579–2588. [DOI] [PubMed] [Google Scholar]
- 32.Mohler, W. A., Shemer, G., del, C. J., Valansi, C., Opoku, S. E., Scranton, V., Assaf, N., White, J. G. & Podbilewicz, B. (2002) Dev. Cell 2, 355–162. [DOI] [PubMed] [Google Scholar]
- 33.Legouis, R., Gansmuller, A., Sookhareea, S., Bosher, J. M., Baillie, D. L. & Labouesse, M. (2000) Nat. Cell Biol. 2, 415–422. [DOI] [PubMed] [Google Scholar]
- 34.Bossinger, O., Klebes, A., Segbert, C., Theres, C. & Knust, E. (2001) Dev. Biol. 230, 29–42. [DOI] [PubMed] [Google Scholar]
- 35.McMahon, L., Legouis, R., Vonesch, J. L. & Labouesse, M. (2001) J. Cell Sci. 2265–2277. [DOI] [PubMed]
- 36.Firestein, B. L. & Rongo, C. (2001) Mol. Biol. Cell 12, 3465–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raich, W. B., Agbunag, C. & Hardin, J. (1999) Curr. Biol. 9, 1139–1146. [DOI] [PubMed] [Google Scholar]
- 38.Satouchi, K., Hirano, K., Sakaguchi, M., Takehara, H. & Matsuura, F. (1993) Lipids 28, 837–840. [DOI] [PubMed] [Google Scholar]
- 39.Watts, J. L. & Browse, J. (2002) Proc. Natl. Acad. Sci. USA 99, 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao, R. K., Baker, R. D., Baker, S. S., Gupta, A. & Holycross, M. (1997) Am. J. Physiol. 273, G812–G823. [DOI] [PubMed] [Google Scholar]
- 41.Rao, R. K., Basuroy, S., Rao, V. U., Karnaky, J. K. & Gupta, A. (2002) Biochem. J. 368, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karasawa, K., Qiu, X. & Lee, T. (1999) J. Biol. Chem. 274, 8655–8661. [DOI] [PubMed] [Google Scholar]
- 43.Bae, K., Longobardi, L., Karasawa, K., Malone, B., Inoue, T., Aoki, J., Arai, H., Inoue, K. & Lee, T. (2000) J. Biol. Chem. 275, 26704–26709. [DOI] [PubMed] [Google Scholar]
- 44.van Blitterswijk, W. J., van der Luit, A. H., Veldman, R. J., Verheij, M. & Borst, J. (2003) Biochem. J. 369, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.