Abstract
All jawed vertebrates have highly diverse lymphocyte receptors, which allow discrimination between self and nonself antigens as well as the recognition of potential pathogens. Key elements of the anticipatory recombinatorial immune system in jawed vertebrates are the TCR, Ig, and MHC genes, but their ancestral genes have not been found in more basal vertebrates. In this study, we extended our analysis of the transcriptome of lymphocyte-like cells in the lamprey to identify the TCR-like and CD4-like genes. The structural features of these genes and their preferential expression in lymphocytes make them attractive candidates for ancestral TCR and CD4 genes. The TCR-like gene contains both V (variable) and J (joining) sequences in its first exon and exists as a single-copy gene that is invariant. Thus, the TCR-like gene cannot account for the receptor diversity that is required for the immune responses reported for lamprey, but it could have been easily modified to serve as an evolutionary precursor of modern TCR and Ig genes.
Specific adaptive immunity emerged during vertebrate evolution as a new strategy for coping with an antigenic world of infinite variety. This anticipatory immune system (AIS) in jawed vertebrates is based on diverse lymphocyte receptors, which are assembled by means of combinatorial rearrangement of Ig V (variable), D (diversity), and J (joining) gene segments. The recombination-activating genes, RAG1 and RAG2, catalyze the joining of V(D)J gene segments to generate unique antigen-binding regions of the B cell receptors and T cell receptors (TCRs). The complementary arm of the AIS is provided by the polymorphic MHC genes for class I and II molecules that present antigenic peptides to TCR-bearing lymphocytes. The TCR, Ig, and MHC genes are conserved in all jawed vertebrates that have been studied, including representatives of the oldest extant group of cartilaginous fish: the sharks, skates, and rays (1-3).
The evolutionary origin of the TCR, Ig, and MHC genes is still unknown, but precursors of these key AIS elements would be anticipated in ancestors of the jawed vertebrates. The chordate phylum derived from urochordate ancestors, some of which evolved into the cephalochordates. However, these basal chordates lack identifiable orthologs of pivotal AIS genes. No AIS genes were identified in genome of the tunicate Ciona intestinalis, a urochordate representative (4, 5). In Amphioxus, which is a cephalochordate representative, secreted VCBP proteins consisting of two diverse V-type Igs and a chitin-binding domain were found (6), as well as molecules containing one V domain and a domain spanning the membrane five times (7), but these molecules are unlikely to be direct precursors of the vertebrate antigen-specific receptors. The first jawless fish appeared ≈550 million years ago (mya), and these early craniates diverged into two lineages. One lineage included the ancestors of hagfish and lamprey, and the other lineage included armored shell-skinned fish (Ostracoderms), which were ancestors of the jawed vertebrates that became extinct ≈400 mya (8-10). Hence, contemporary jawless fish are the closest surviving relatives of the early jawed vertebrates and the best available candidates in which to search for the molecular and cellular precursors of the combinatorial AIS.
The lamprey hematopoietic tissues and blood stream abound with cells resembling mammalian lymphocytes (11-15). These cells can undergo lymphoblastic transformation and proliferation in response to stimulation with antigens and plant mitogens (16), and they have been implicated as key cellular elements in adaptive immune responses that have been reported for these jawless vertebrates (17), including rejection of second-set skin allografts at an accelerated pace (17, 18) and delayed-type hypersensitivity (19). Humoral immune responses include the production of specific agglutinins in response to lamprey immunization with particulate antigens (1, 17, 20, 21).
A previous survey of ≈8,000 ESTs from larval sea lamprey (Petromyzon marinus) lymphocyte-like cells isolated from the hematopoietic typhlosole yielded homologs of various genes involved in vertebrate lymphocyte development, differentiation, proliferation, signal transduction, activation, chemotaxis, and innate immune responses (10, 22-27). However, the cardinal elements of the combinatorial AIS, TCR, Ig, and MHC were not identified (24). In this study, we analyzed 10,230 additional ESTs of lymphocyte-like cells isolated from the hematopoietic tissues typhlosole and anterior kidney and from blood samples of lamprey larvae to identify other ancestral components of the vertebrate AIS.
Materials and Methods
Animals and Lymphocyte-Like Cells. Sea lamprey larvae from Lake Michigan tributaries (Lamprey Services, Ludington, MI) were cultured in 80-liter tanks, with 10 cm of bottom sand (Play Sand, W. R. Bonsai, Charlotte, NC), 100-150 animals per tank at 16°C and fed Brewer's Yeast tablets (Walgreen, Deerfield, IL). Dissection of tissues from anesthetized larvae (0.5% MS222; Sigma) was performed as described (15). Blood was collected from tail-severed larvae, diluted 1:1 with 0.57× PBS (1× PBS per liter, containing 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4, pH 7.4) and 30 mM EDTA. Buffy-coat peripheral blood lymphocytes were collected after a 5-min centrifugation at 50 × g and sorted by using a MoFlo cytometer (Cytomation, Fort Collins, CO) as described (15). Larvae sedated in 0.01% MS222 for 10 min were injected i.p. with 75 μl of an antigen mixture containing 107 live Escherichia coli BL21(DE3), 107 sheep erythrocytes (50% Alsever's mixture, Colorado Serum, Denver), 50 μg of phytohemagglutinin, and 25 μg of pokeweed mitogen (Sigma) four times at weekly intervals, and blood cells were collected 3-4 days after the last immunization.
Lymphocyte cDNA Library and Subtracted Libraries. A lymphocyte cDNA library was constructed (Lambda ZAP Express cDNA synthesis kit; Staratagene) from 4 × 108 cells isolated by fluorescence-activated cell sorting from typhlosole (67%) and pronephros (33%) of 150 unstimulated larvae. From the primary phage library, 3 × 106 plaque-forming units were used for in vivo excision of the phagemid pBK-CMV, and 10,368 colonies were sequenced commercially with the T3 primer (SeqWright, Houston), resulting in 8,723 sequences.
The Super SMART PCR cDNA synthesis kit (BD Biosciences) was used for cDNA synthesis from mRNA of large blood lymphocytes, myeloid cells, and erythrocytes sorted from blood of immunostimulated larvae. Activated blood lymphocyte cDNA was subtracted in two separate reactions against cDNA of myeloid cells or erythrocytes (PCR-Select cDNA subtraction kit; BD Biosciences). Subtracted products were cloned in pGEM-T Easy (Promega, Madison, WI). We sequenced 864 clones from the subtraction against activated myeloid cells and 864 clones from the subtraction against erythrocytes with the T7 primer (SeqWright), resulting in 1,507 sequences.
Genomic Clones. A lamprey genomic DNA library in lambda FIXII was screened as described (24) by using as probes the cDNA molecules encoding the lamprey TCR-like and CD4-like. Inserts from overlapping clones were restriction enzyme-digested and blotted. Positive fragments were subcloned and sequenced.
Virtual Northern and Southern Blot Analyses. Virtual Northern blot analysis was prepared according to the manufacturer's recommendations (BD Biosciences). cDNA was synthesized from mRNA of larval tail, liver, and sorted lymphocytes from blood, typhlosole, and kidneys of unstimulated animals, or small and large blood lymphocytes, myeloid cells, and erythrocytes sorted from blood of immunostimulated larvae.
Genomic DNA was isolated from whole-larval carcass. DNA from three larvae (10 μg per lane) was digested with BamHI, EcoRI, or HindIII (Roche), electrophoresed in 0.7% agarose, and blotted onto GeneScreen Plus hybridization membrane (NEN).
The following 32P-labled probes were used. The IgV domain in PmTCR-like was PCR amplified by using the following primers: TCR-IgV.F, 5-TGGTGTAACAAATGAAGAGGC (nucleotides 445-465 in AY686861); and TCR-IgV.R, 5-TTCCACTGTA AGT TGTGT TCC (nucleotides 719-739 in AY686861). The IgV domain in PmCD4-like was PCR amplified by using the following primers: CD4-IgV.F, 5-ACAGTGTCTGGCTACCGTG (nucleotides 114-132 in AY686862); and CD4-IgV.R, 5-CAAGTATATCTCCCCGCATC (nucleotides 326-345 in AY686862). GAPDH was amplified from clone PmGAPDH (AY578058) with the following primers: GAPDH.F, 5-GAACATCGGCATCAATGGGT (nucleotides 71-90); and GAPDH.R, 5-GAGGCCTTATCGATGGTGGT (nucleotides 366-385).
Sequence Analysis. Stand-alone blast was downloaded from (ftp.ncbi.nih.gov/blast/executables). Homologs of the PmTCR-like were identified by using blastp with the IgV domain as query. The highest score was with porcine TCRα (E = 0.003; BAC66581). blastp with the entire PmCD4-like scored highest with rabbit CD4 (E = 0.001; P46630). For detection of EST homology to known GenBank sequences, a blast cutoff was set at E < 10-5. clustalx was used for multiple sequence alignments (28). Neighbor-joining trees with pairwise gap deletions were drawn by using mega 2.1 phylogenetic and molecular evolutionary analyses (29). Structural modeling was performed by using the 3d-pssm server (30).
Results
Transcripts from Lymphocyte-Like Cells in the Typhlosole and Kidney. Typhlosole and kidney lymphocyte-like cells were isolated on the basis of light-scatter characteristics, and their transcripts were sampled by analysis of 8,723 ESTs. Search for homologs in the GenBank database identified 339 (3.9%) candidate lamprey immune-related molecules (see Table 1 and Supporting Text, which are published as supporting information on the PNAS web site, for categorized sequence analysis and a list of putative immune system molecules). One group of the ESTs encoded 51 homologs of vertebrate lymphocyte surface receptors. These receptors included one TCR-like molecule, one CD4-like molecule, and five additional transcripts encoding other Ig superfamily members.
Interestingly, 35 different homologs of genes that are characteristically expressed by vertebrate hematopoietic stem cells and progenitor cells (31) were identified among these ESTs. This finding suggested that the lymphocyte-like population in the lamprey typhlosole and kidney is enriched in hematopoietic stem cells (32). Given this cell-lineage ambiguity, we turned to activated lymphoblastoid cells in blood, reasoning that these would be more likely to express the genes that are responsible for AIS responses. To activate their lymphocytes, lamprey larvae were stimulated by weekly i.p. injections of a mixture of antigens and mitogens (33). Blood cells from the immunostimulated larvae were sorted into fractions of small and large activated lymphocytes and myeloid cells. Libraries enriched in messages of activated lymphocytes were then constructed by subtraction against cDNA from lamprey-activated myeloid cells and erythrocytes, and 1,507 sequences were analyzed.
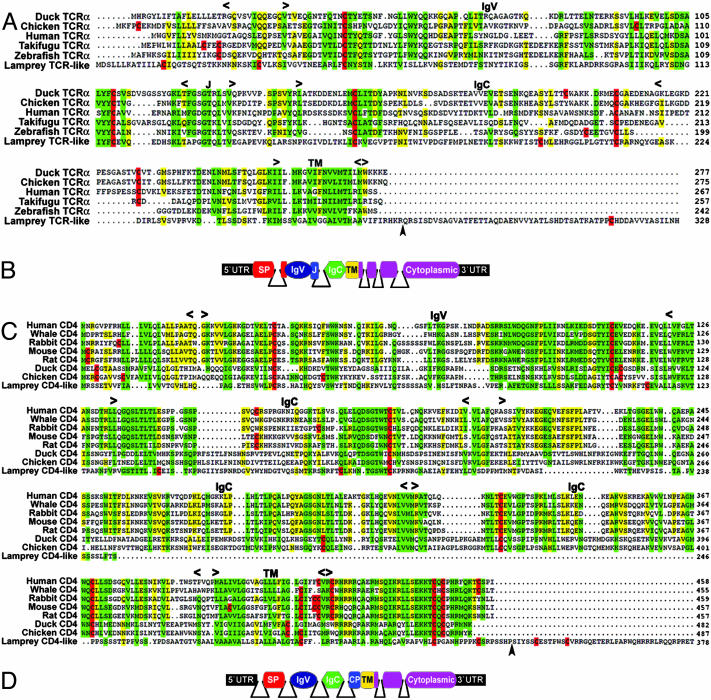
Lamprey TCR-Like and CD4-Like Molecules. A TCR-like sequence was identified in three ESTs derived from unstimulated lamprey and 10 additional sequences from the subtracted libraries, all of which encoded the same polypeptide. The longest clone had an insert of ≈3.5 kb, and the two shorter inserts of ≈2 kb were missing 140 or 281 nucleotides from the 5′ UTR. These sequences were assembled into a contig of 1,750 bp. The predicted polypeptide of 328 residues (35.6 kDa) includes a signal peptide of 20 residues, followed by an IgV region spanning residues 37-136, an IgC2 region spanning residues 149-222, a transmembrane region spanning residues 248-270, and a 57-residue cytoplasmic tail (Figs. 1A and 4A, which is published as supporting information on the PNAS web site). The C-terminal part of the IgV encodes a peptide, GGTQLTV, with similarity to the goat TCRα J-segment sequence GGTRLSV (GenBank accession no. AAK09314). Both of these sequences are slightly divergent forms of the consensus TCR J sequence GXGTXLXV (34). Two Tyr residues are present in the cytoplasmic domain. The first residue, Tyr-299, is included in a VXYXXL motif that conforms to the immunoreceptor Tyr-based inhibitory consensus (I/V/L/S)XYXX(L/V). The second Tyr, Tyr-322, is found in a degenerate immunoreceptor Tyr-based inhibitory motif with a noncanonical terminal isoleucine residue, VXYXXI. Six exons encode the TCR-like polypeptide, with the IgV domain being encoded by exon 2 that also includes the J segment. Exon 3 encodes the IgC2 domain, a membrane proximal serine-rich domain, a transmembrane region, and the first four residues of the cytoplasmic tail (Figs. 1B and 4A). This membrane proximal portion of the cytoplasmic tail consists of basic residues like those found in the cytoplasmic tail of avian TCRs (Fig. 1 A).
Fig. 1.
Multiple alignment of the lamprey TCR-like and CD4-like molecules with related vertebrate lymphocyte receptors. (A and C) Jawed vertebrate TCRα and CD4 molecules. Approximate location of Ig and transmembrane (TM) domains as well as the location of the J region in TCRs are indicated. Arrowheads indicate the positions of intron 4 in the lamprey TCR-like and intron 6 in the CD4-like. Cysteine residues are highlighted in red. Green, ≥80% identity and similarity; yellow, 60-79% identity/similarity. Amino acid similarity groups are as follows: acidic, D and E; aromatic, F, W, and Y; basic, H, K, and R; hydrophobic, A, I, L, M, and V; polar, N, Q, S, and T; and ungrouped, G and P. (B and D) Stick models of exons and introns in the TCR-like and CD4-like genes. The position of introns was determined by sequencing genomic clones and are indicated by solid lines. SP, signal peptide; CP, connecting peptide. GenBank accession nos. for the sequences shown in A are as follows: duck, AN3311A9; chicken, AAC98541; human, AAC14929; takifugu (Takifugu rubripes), AAF97794; zebrafish (Danio rerio), AAL23935; and lamprey TCR-like, AY686861. GenBank accession nos. for the sequences shown in C are as follows: human, NP_000607; white whale (Delphinapterus leucas), AAD23738; rabbit, P46630; mouse, NP_038516; rat, NP_036837; duck, AAK59279; chicken, NP_989980; and lamprey CD4-like, AY686862.
The lamprey CD4-like molecule was identified in two ESTs, one clone of 1,444 bp and the other missing 29 nucleotides from the 5′ UTR. The predicted polypeptide of 378 residues (42.2 kDa) consists of a signal peptide of 20 residues, followed by an IgV region spanning residues 21-108, an Ig-like region spanning residues 131-208, a transmembrane domain spanning residues 274-296, and a 77-residue cytoplasmic tail (Figs. 1C and 4B). The ectodomain in the sea lamprey CD4-like molecule consists of only two Ig domains, whereas CD4 molecules of mammals feature three Ig domains and avian CD4 molecules consist of four Ig domains. Seven exons encode the CD4-like polypeptide, with each Ig domain being encoded by a separate exon (Figs. 1D and 4B). As in the case of the lamprey TCR-like gene, the last exon of the CD4-like gene encodes the C-terminal portion of the cytoplasmic tail, which is unrelated to the sequence of CD4 in jawed vertebrates.
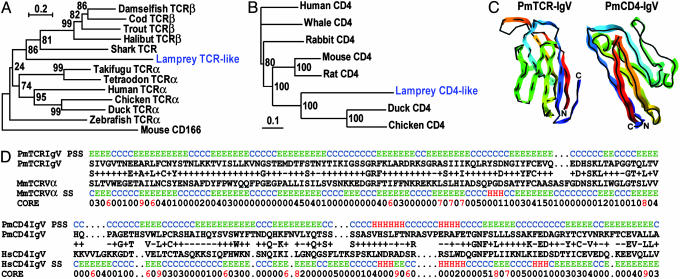
Phylogenetic analysis supports the classification of these lamprey transcripts as prototypic TCR and CD4. A neighbor-joining tree of TCR ectodomains places the lamprey TCR-like molecule in a cluster with fish TCR, separated from the mouse CD166, which is an Ig superfamily member cell-adhesion molecule (Fig. 2A). The lamprey CD4-like ectodomain clusters with the two N-terminal Ig domains of avian CD4 (Fig. 2B). Because the initial blast scores used to identify the lamprey TCR-like and CD4-like molecules were marginal (see Materials and Methods), we analyzed these sequences by using the 3d-pssm program, which is a bioinformatic tool that is sensitive to structural protein similarities (30). The IgV domain from the lamprey TCR-like was threaded on the crystal-structure coordinates of mouse TCRVα, and the IgV domain from the CD4-like molecule was threaded on the crystal-structure coordinates of the N-terminal IgV in human CD4 (Fig. 2C). Alignment of the lamprey TCR-IgV domain with mouse TCRVα (Fig. 2D) revealed a statistically significant threading value (E = 9e - 7), based on 25% amino acid identity and only a single three-residue gap within the complementarity-determining region 3 (CDR3). More gaps were required to align the lamprey CD4-IgV with the corresponding domain from human CD4 (27% identity, E = 3e - 6), suggesting that it may be more closely related to the avian CD4 molecules (Fig. 2B), the structures of which have not been resolved.
Fig. 2.
Phylogenetic and structural analysis of the lamprey TCR-like and CD4-like molecules. (A) Neighbor-joining trees of the two N-terminal Ig domains of the TCRα and TCRβ chains are shown for the following: takifugu, residues 1-234; tetraodon (Tetraodon nigroviridis), residues 1-232 (GenBank accession no. CAC86241); zebrafish, residues 1-226; damselfish (Stegastes partitus), residues 1-256 (GenBank accession no. AAG46047); Atlantic cod (Gadus morhua), residues 1-239 (GenBank accession no. CAB39549); Rainbow trout (Oncorhynchus mykiss), residures 1-238 (GenBank accession no. AAK63017); Bastard halibut (Paralichthys olivaceus), residues 1-254 (GenBank accession no. BAB82597); Horn shark (Heterodontus francisci), residues 1-230 (GenBank accession no. AAA61563); human, residues 1-228; chicken, residues 1-235; duck, residues 1-237; lamprey, residues 1-243; mouse CD166, residues 1-239 (GenBank accession no. Q61490). (B) Neighbor-joining trees of the two N-terminal Ig domains of the CD4 coreceptors are shown for the following: human, residues 1-201; whale, residues 1-201; rabbit, residues 1-206; mouse, residues 1-205; rat, residues 1-204; duck, residues 1-213; chicken, residues 1-217; and lamprey, residues 1-208. (C) Structural 3D model of the N-terminal IgV domains from the lamprey TCR-like and CD4-like molecules (color) threaded on the crystal-structure coordinates (black) of the mouse TCRVα (PDB ID code 1B88) and human CD4 N-terminal IgV (PDB ID code 1CDY) created by using the 3d-pssm program. (D) Sequence and structural alignments of the lamprey TCR-like and CD4-like Ig domains used for the 3D-structure modeling. SS, secondary structure (helix, red; strand, green; coil, blue); PSS, predicted structure; CORE, measure of burial and contacts (0, unburied).
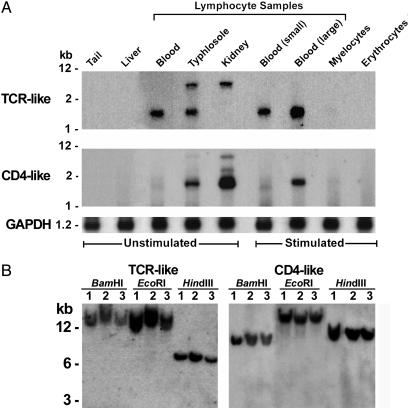
Selective Expression of TCR-Like and CD4-Like Transcripts by Lymphocyte-Like Cells. Like their relatives in jawed vertebrates, the TCR- and CD4-like transcripts were found exclusively in lymphocyte-rich populations from the lamprey typhlosole, kidney, and blood (Fig. 3A). Elevated levels of TCR-like messages were noted for the large activated blood lymphocytes. The 3.5-kb band observed among lymphocyte transcripts from the hematopoietic tissues may represent alternatively spliced forms or differential utilization of polyadenylation signals in the lamprey TCR-like messages, indicated by the two insert sizes found in our cDNA clones. Interestingly, the longer variant was identified only in lymphocytes from hematopoietic tissues, which could suggest a developmental role for the extended 3′ UTR. Expression levels of the lamprey CD4-like messages were highest in lymphocyte-like cells derived from the kidney. Southern blot hybridization with lamprey TCR-like and CD4-like probes revealed a single band for genomic DNA isolated from three individuals and digested with three restriction enzymes (Fig. 3B). These results indicate that each of these genes occurs in a single copy and that the TCR-like gene does not undergo rearrangement.
Fig. 3.
Lamprey TCR-like and CD4-like expression patterns and genomic loci. (A) Virtual Northern blots using cDNA amplified from tissues and cell populations purified from hematopoietic organs and blood of unstimulated and immunostimulated larvae. GAPDH hybridization served as a control. (B) Genome blots of three lampreys. Genomic DNA was digested with the indicated restriction enzymes.
Discussion
Whereas Ig superfamily members constitute approximately one-third of the lymphocyte cell-surface receptors in jawed vertebrates (35), they accounted for only 14% (7/51) of the putative surface receptors that were identified in this analysis of lamprey lymphocyte-like transcripts. Nevertheless, two of these molecules have sequence and structural homology to the jawed vertebrate TCR and CD4 coreceptor, and both molecules are expressed exclusively by lamprey lymphocytes. The features of the lamprey TCR-like gene fit the model proposed for an ancestral TCR gene (36, 37). It is made up of one IgV domain that includes a J motif, one IgC-type domain that is connected to a membrane-spanning domain, and a cytoplasmic tail that encodes one or two immunoreceptor Tyr-based inhibitory signaling motifs. Furthermore, the transmembrane region contains conserved antigen receptor transmembrane (CART) residues (38). The cytoplasmic domain of the lamprey TCR-like protein is much longer than the corresponding region of the TCR chains in gnathostomes, yet evolutionary modification of this cytoplasmic tail by a single mutation could introduce a stop codon at the end of exon 3 of the lamprey TCR-like gene to convert its C terminus into a short cytoplasmic tail that characterizes the TCR chains in jawed vertebrates.
Speculation about the function of the lamprey TCR-like molecule is constrained by a lack of information about its ligand(s), its ability to associate with itself or other cell surface proteins, and its signaling capability. However, the available data indicate that it cannot provide the lymphocyte receptor diversity that would be required to explain the immune responses reported for lamprey. Only a single TCR-like gene could be identified in the sea-lamprey genome, and this gene did not have V(D)J gene segments with flanking recombinatorial signal sequences. Also, only limited single-nucleotide variations were found in the 13 TCR-like cDNA sequences. Therefore, the function of the lamprey TCR-like molecule may depend on its interaction with a nonvariant ligand, either of conserved pathogen epitopes or a self-antigen. Importantly, the TCR-like ligand is unlikely to be an MHC molecule, given that none of our >18,000 ESTs of lamprey lymphocyte-like cells correspond to MHC class I or II molecules.
The question of whether the TCR or Ig genes came first has been a topic of discussion since their discovery. Most immunologists favor the idea that both genes were derived from a primordial gene that encoded a TCR-like protein in the primitive lymphocyte (36), and our data are consistent with this hypothesis. Although the fact that we failed to find a counterpart antibody-like molecule in our analysis of the lamprey lymphocyte-like transcriptome is noteworthy in this regard, the sequencing of the lamprey genome will be required for final resolution of this issue.
The identification of a lamprey CD4-like cDNA provides an ancestral candidate for another important T cell molecule that serves an important role as coreceptor for the TCR in modern vertebrates. Likewise, the CD4-like gene is preferentially expressed in lamprey lymphocyte-like cells. The predicted lamprey CD4-like molecule has only two extracellular Ig domains, an IgV and an Ig-like domain, whereas mammals and birds have three or four (39, 40). This difference suggests duplication and diversification of these Ig exons, the evolutionary timing of which may become apparent when CD4 genes are characterized in fish, amphibians, and reptiles. The predicted lamprey CD4-like molecule has an exceptionally long cytoplasmic tail comprised of a C-terminal portion encoded in the last exon that is unrelated to jawed vertebrate CD4, but a single mutation introducing a stop codon at the end of exon 5 would generate a short cytoplasmic tail with residues characteristic of avian and mammalian CD4 molecules. One potentially important evolutionary pressure to modify this CD4-like gene would have been the appearance of MHC class II genes because their protein products serve as CD4 ligands.
Although this extended survey of the lamprey lymphocyte-like transcriptome reveals many of the ancestral genetic components of the lymphocyte-based AIS of jawed vertebrates, our findings reinforce the conclusion that this agnathan representative lacks a recombinatorial immune system based on Ig gene segments. However, we have very recently identified lamprey variable lymphocyte receptors (VLRs) that are composed of diverse leucine-rich repeat (LRR) motifs. Mature VLR genes are assembled by somatic rearrangement of LRR cassettes that flank an incomplete germline VLR gene. The resultant highly diverse mature VLR genes are expressed in lymphocytes in monoallelic fashion. The VLR proteins may exist either as glycosylphosphatidylinositol-anchored surface receptors or as circulating agglutinins after glycosylphosphatidylinositol-specific phospholipase release from the cell surface. In this way, the lamprey VLRs could potentially mediate both cellular and humoral immune responses (33). Ancient components of innate defense proteins from both animals and plants, the LRR motifs are logical building blocks for the construction of a combinatorial immune system. A more difficult question is where the immediate evolutionary precursors of the jawed vertebrate rearranging Ig receptors are. The answer to this question could lie in the agnathan ostracoderm species, which unfortunately, we can know only by their paleontological remains.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Robert A. Good. We thank Roger Bergstedt and William Swink (Hammond Bay Biological Station, Millersburg, MI) for assistance with lamprey biology and supply; Gene Hines, Jim Crawford, and the University of Alabama at Birmingham Animal Resources Program staff for lamprey care; and Ann Brookshire (University of Alabama at Birmingham) for help in preparing the manuscript. This work was supported by National Science Foundation Grants MCB-0317460 and IBN-0321461, National Institutes of Health Grant HG02526-01, and the Max Planck Institute. M.D.C. is an Investigator of the Howard Hughes Medical Institute. Z.P. was supported by a Frederick Gardner Cottrell Postdoctoral Enhancement Award.
Abbreviations: AIS, anticipatory immune system; TCR, T cell receptor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY686861, AY686862, and CO542795-CO553159).
References
- 1.Litman, G. W., Frommel, D., Finstad, F. J., Howell, J., Pollara, B. W. & Good, R. A. (1970) J. Immunol. 105, 1278-1285. [PubMed] [Google Scholar]
- 2.Flajnik, M. F. & Kasahara, M. (2001) Immunity 15, 351-362. [DOI] [PubMed] [Google Scholar]
- 3.Flajnik, M. F. (2002) Nat. Rev. Immunol. 2, 688-698. [DOI] [PubMed] [Google Scholar]
- 4.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- 5.Azumi, K., De Santis, R., De Tomaso, A., Rigoutsos, I., Yoshizaki, F., Pinto, M. R., Marino, R., Shida, K., Ikeda, M., Ikeda, M., et al. (2003) Immunogenetics 55, 570-581. [DOI] [PubMed] [Google Scholar]
- 6.Cannon, J. P., Haire, R. N. & Litman, G. W. (2002) Nat. Immunol. 3, 1200-1207. [DOI] [PubMed] [Google Scholar]
- 7.Sato, A. Mayer, W. E. & Klein, J. (2003) Immunogenetics 55, 423-427. [DOI] [PubMed] [Google Scholar]
- 8.Forey, P. L. & Janvier, P. (1993) Nature 361, 129-134. [Google Scholar]
- 9.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917-920. [DOI] [PubMed] [Google Scholar]
- 10.Takezaki, N., Figueroa, F., Zaleska-Rutczynska, Z. & Klein, J. (2003) Mol. Biol. Evol. 20, 287-292. [DOI] [PubMed] [Google Scholar]
- 11.Finstad, J., Papermaster, B. W. & Good, R. A. (1964) Lab. Invest. 13, 490-512. [PubMed] [Google Scholar]
- 12.Piavis, G. W. & Hiatt, J. L. (1971) Copeia 4, 722-728. [Google Scholar]
- 13.Fujii, T. (1982) J. Morphol. 173, 87-100. [DOI] [PubMed] [Google Scholar]
- 14.Ardavin, C. F. & Zapata, A. (1987) Dev. Comp. Immunol. 11, 79-93. [DOI] [PubMed] [Google Scholar]
- 15.Mayer, W. E., Uinuk-Ool, T., Tichy, H., Gartland, L. A., Klein, J. & Cooper, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 14350-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper, A. J. (1971) in 4th Leukocyte Culture Conference, ed. McIntyre, O. R. (Appleton Century-Crofts, New York), pp. 137-147.
- 17.Good, R. A., Finstad, J. & Litman, G. W. (1972) in The Biology of Lampreys II: Immunology, eds. Hardisty, M. V. & Potter, I. C. (Academic, London), pp. 405-432.
- 18.Perey, D. Y., Finstad, J., Pollara, B. & Good, R. A. (1968) Lab. Invest. 19, 591-597. [PubMed] [Google Scholar]
- 19.Finstad, J. & Good, R. A. (1964) J. Exp. Med. 120, 1151-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollara, B., Litman, G. W., Finstad, J., Howell, J. & Good, R. A. (1970) J. Immunol. 105, 738-745. [PubMed] [Google Scholar]
- 21.Marchalonis, J. J. & Edelman, G. M. (1968) J. Exp. Med. 127, 891-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shintani, S., Terzic, J., Sato, A., Saraga-Babic, M., O'hUigin, C., Tichy, H. & Klein, J. (2000) Proc. Natl. Acad. Sci. USA 97, 7417-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer, W. E., O'Huigin, C., Tichy, H., Terzic, J. & Saraga-Babic, M. (2002) Scand. J. Immunol. 55, 162-170. [DOI] [PubMed] [Google Scholar]
- 24.Uinuk-Ool, T., Mayer, W. E., Sato, A., Dongak, R., Cooper, M. D. & Klein, J. (2002) Proc. Natl. Acad. Sci. USA 99, 14356-14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, N., Uinuk-ool, T. S., Sato, A., Samonte, I. E., Figueroa, F., Mayer, W. E. & Klein, J. (2003) Immunogenetics 54, 884-895. [DOI] [PubMed] [Google Scholar]
- 26.Sato, A., Uinuk-ool, T. S., Kuroda, N., Mayer, W. E., Takezaki, N., Dongak, R., Figueroa, F., Cooper, M. D. & Klein, J. (2003) Dev. Comp. Immunol. 27, 401-412. [DOI] [PubMed] [Google Scholar]
- 27.Uinuk-Ool, T. S., Mayer, W. E., Sato, A., Takezaki, N., Benyon, L., Cooper, M. D. & Klein, J. (2003) Immunogenetics 55, 38-48. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 30.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. E. (2000) J. Mol. Biol. 299, 499-520. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Q. H., Ye, M., Wu, X. Y., Ren, S. X., Zhao, M., Zhao, C. J., Fu, G., Shen, Y., Fan, H. Y., Lu, G., et al. (2000) Genome Res. 10, 1546-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potter, I. C., Perey, L. R., Barber, D. L. & Macey, D. J. (1982) in The Biology of Lampreys (Academic, London), pp. 233-292.
- 33.Pancer, Z., Amemiya, C. T., Ehrhardt, G. R. A., Ceitlin, J., Gartland, G. L. & Cooper, M. D. (2004) Nature 430, 174-180. [DOI] [PubMed] [Google Scholar]
- 34.Strong, S. J., Mueller, M. G., Litman, R. T., Hawke, N. A., Haire, R. N., Miracle, A. L., Rast, J. P., Amemiya, C. T. & Litman, G. W. (1999) Proc. Natl. Acad. Sci. USA 96, 15080-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van den Berg, T. K., Jeffrey, A., Yoder, J. A. & Litman, G. W. (2004) Trends Immunol. 25, 11-16. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, C. B. (1995) Immunity 3, 531-539. [DOI] [PubMed] [Google Scholar]
- 37.Marchalonis, J. J., Schluter, S. F. & Bernstein, R. M. (1998) Adv. Immunol. 70, 417-506. [DOI] [PubMed] [Google Scholar]
- 38.Campbell, K. S., Backstrom, B. T., Tiefenthaler, G. & Palmer, E. (1994) Semin. Immunol. 6, 393-410. [DOI] [PubMed] [Google Scholar]
- 39.Parnes, J. (1989) Adv. Immunol. 44, 265-311. [DOI] [PubMed] [Google Scholar]
- 40.Koskinen, R., Lamminmaki, U., Tregaskes, C. A., Salomonsen, J., Young, J. R. & Vainio, O. (1999) J. Immunol. 162, 4115-4121. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.