Abstract
The leukemic fusion gene AML1-MDS1-EVI1 (AME) encodes a chimeric transcription factor that results from the t(3,21)(q26;q22) translocation seen in patients with acute myeloid leukemia, with therapy-related myelodysplastic syndrome, or with chronic myeloid leukemia in blast crisis. The myeloid transcription factor CEBPA is crucial for normal granulopoiesis. Here, we found that conditional expression of AME suppresses CEBPA protein by 90.8% and DNA-binding activity by 93.9%. In contrast, CEBPA mRNA levels remained unchanged. In addition, we detected no differences in CEBPA mRNA levels in leukemic blasts of patients carrying the AME translocation (n = 8) compared to acute myeloid leukemia patients with a normal karyotype (n = 9). CEBPA protein and binding activity, however, were reduced significantly (100% and 92.1%, respectively) in AME patient samples. Furthermore, we observed that calreticulin (CRT), a putative inhibitor of CEBPA translation, was strongly activated after induction of AME in the cell-line system (14.8-fold) and in AME patient samples (12.2-fold). Moreover, inhibition of CRT by small interfering RNA powerfully restored CEBPA levels. These results identify CEBPA as a key target of the leukemic fusion protein AME and suggest that modulation of CEBPA by CRT may represent a mechanism involved in the differentiation block in AME leukemias.
Acute myeloid leukemia (AML) is a clonal malignant disease characterized by a block in normal myeloid differentiation leading to the accumulation of immature hematopoietic cells in the bone marrow and peripheral blood (1). AML is characterized further by the presence of specific balanced chromosome rearrangements that create novel fusion genes (2). However, little is known about the mechanisms of how such fusion genes contribute to the differentiation block.
AML1-MDS1-EVI1 (AME) is a chimeric fusion gene observed in patients with de novo or therapy-related AML, with therapy-related myelodysplastic syndrome (MDS), or with chronic myeloid leukemia in blast crisis (CML-BC) (3-5). AME is an in-frame fusion of the AML1 and MDS1/EVI1 genes (6). AML1 (also known as RUNX1) is one of the most frequently translocated or mutated genes in human cancer (7-12). EVI1 is abnormally expressed in human MDS, AML, and CML-BC that are associated with the t(3,3)(q21q26) or inv(3)(q21q26) (3, 13, 14). MDS1 is a gene of largely unknown function located upstream of EVI1. Mice transplanted with syngeneic bone marrow cells expressing the AME fusion gene develop a disease similar to human acute myelomonocytic leukemia (15). In addition, AME has been shown to induce proliferation and inhibit differentiation in myeloid cells (16, 17).
CEBPA plays distinct roles in the differentiation process of various cell types (18-25). In the hematopoietic system, CEBPA is expressed exclusively in myelomonocytic cells (18, 25). Conditional expression of CEBPA is sufficient to trigger terminal neutrophil differentiation (25-28) and block the monocytic differentiation program (25, 27). In addition, no mature granulocytes are observed in cebpa knock-out mice, whereas all other blood-cell types are present in normal numbers (24).
We showed previously that dominant-negative mutations of the CEBPA gene are found in a significant proportion of patients with myeloblastic subtypes (M1 and M2) of AML (29-31). Furthermore, we demonstrated that the AML1-ETO fusion protein suppresses CEBPA expression (32). Here, we found that AME suppresses CEBPA protein; in contrast to the AML1-ETO fusion, it fails to suppress CEBPA mRNA expression. We identified translational inhibition of CEBPA mediated by induction of calreticulin (CRT), a ubiquitous protein with calcium storage and chaperone function, as a mechanism involved in leukemia.
Materials and Methods
Patient Samples. Ficoll-separated, fresh, mononucleated peripheral blood or bone marrow cells of AML patients were collected at the time of diagnosis before initiation of treatment. Conventional cytogenetic analysis was performed in each patient (Table 1).
Table 1. Clinical presentation of patients.
| No. | Sex | Age, y | FAB | Karyotype | WBC, G/I | % blasts in PBLs | LDH, units/ml |
|---|---|---|---|---|---|---|---|
| 1 | M | 52 | M1 | 46;XY;t(3;21) | 22.2 | 78 | 954 |
| 2 | F | 60 | M1 | 45;XX;t(3;21);−7 | 14.3 | 54 | 752 |
| 3 | M | 68 | M2 | 46;XY;t(3;21) | 13.2 | 58 | 674 |
| 4 | M | 48 | M1 | 46;XY;t(3;21) | 38.4 | 95 | 1,455 |
| 5 | F | 58 | M1 | 46;XX;t(3;21) | 55.8 | 98 | 1,025 |
| 6 | F | 64 | M2 | 46;XX;t(3;21) | 18.2 | 45 | 770 |
| 7 | M | 56 | M4 | 46;XY;t(3;21);inv(1)(q25q44) | 8.2 | 25 | 482 |
| 8 | M | 59 | M2 | 46;XY;t(3;21);+8 | 15.3 | 38 | 920 |
| 9 | F | 72 | M1 | 46;XX;t(3;21) | 32.8 | 90 | 1,285 |
| 10 | M | 44 | M4 | 46;XY | 6.6 | 30 | 380 |
| 11 | M | 54 | M2 | 46;XY | 24.5 | 85 | 842 |
| 12 | M | 58 | M4 | 46;XY | 14.4 | 55 | 710 |
| 13 | M | 58 | M4 | 46;XY | 98.8 | 98 | 1,662 |
| 14 | F | 74 | M4 | 46;XX | 30.5 | 72 | 908 |
| 15 | M | 62 | M4 | 46;XY | 84.5 | 98 | 1,585 |
| 16 | F | 61 | M4 | 46;XX | 18.8 | 65 | 880 |
| 17 | M | 56 | M2 | 46;XY | 21.5 | 55 | 560 |
| 18 | F | 70 | M1 | 46;XX | 19.5 | 72 | 980 |
| 19 | F | 52 | M1 | 46;XX | 52.2 | 92 | 1,440 |
M, male; F, female; FAB, French-American-British classification; WBC, white blood cell count; PBL, peripheral blood leukocytes; LDH, lactate dehydrogenase; G/l: 109 per liter.
Generation of Cell Line with Conditional AME Expression. The U937T cell line with the tetracycline transactivator under the control of a tetracycline-responsive element was obtained from Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN). A 4.0-kb ScaI/XbaI fragment of the pcDNA3 vector, containing the neomycin resistance gene, was ligated with the 0.95-kb ScaI/XbaI fragment of the tetracycline-off response plasmid pTRE. A 5.2-kb EcoR1/XbaI fragment encoding for the entire AME cDNA was introduced into the pTRE-neo plasmid. The plasmid was transfected into U937T cells by electroporation. Eighteen single-cell clones were tested for AME induction. The clone with the maximum increase of AME mRNA transcripts was selected for additional experiments.
Real-Time PCR and Sequencing. For isolation of total RNA, the RNeasy minikit (Qiagen, Hilden, Germany) was used. Real-time PCR was performed on the ABI PRISM 7700 sequence-detection system by using TaqMan Universal PCR Master Mix. For CEBPA and CRT mRNA quantitation by Assays-on-Demand gene-expression probes (Applied Biosystems) were used. Primers for AME detection were targeting the AML1/MDS1 transition (Assays-by-Design gene-expression probes, Applied Biosystems). The primers were 5′-AACCACTCCACTGCCT-3′ and 5′-ATACCGTTGATGGGACTTTATGGAAA-3′, and the probe was 5′-FAM-CAGTCTACGTCTTACT-TAMRA-3′ (FAM, 6-carboxyfluorescein; TAMRA, N,N,N′,N′-tetramethyl-6-carboxyrhodamine). 7S was used as reference gene. N-fold changes were calculated as: n-fold = (Ct1 - Ct2)2 × PCR efficiency (Ct, cycle threshold). The PCR efficiency was calculated based on a standard curve. Sequencing of the CEBPA gene was done as described (29).
Western Blot Analysis. CEBPA, CEBPB, CEBPE, granulocyte colony-stimulating factor (G-CSF) receptor, AME, and CRT proteins were detected with rabbit polyclonal antibody against CEBPA (1:500; Santa Cruz Biotechnology), a rabbit polyclonal antibody against CEBPB (1:1,000; Santa Cruz Biotechnology), a rabbit polyclonal antibody against CEBPE (1:1,000; Santa Cruz Biotechnology), a rabbit polyclonal antibody against G-CSF receptor (1:500; Santa Cruz Biotechnology), a rabbit polyclonal antibody against AML1B (1:500; Oncogene Science), and a rabbit polyclonal antibody against CRT (1:200,000; Sigma) followed by an IgG-horseradish peroxidase-conjugated secondary antibody against rabbit (Amersham Pharmacia Biosciences). A monoclonal anti-rabbit β-actin antibody served as a loading control (Sigma).
Electrophoretic Mobility Shift Assays. The G-CSF receptor promotor oligonucleotide (bp -57 to -38) had the sequence 5′-AAGGTGTTGCAATCCCCAGC-3′ (the CEBP-binding site is underlined). An electrophoretic mobility shift assay was performed as described (25, 29, 32, 33). Quantitative CEBPA- and CEBPB-binding activity was assessed further by using an ELISA-based assay (TransAM, Active Motif, Carlsbad, CA). Briefly, a 96-well plate was coated with the immobilized oligo 5′-CTTGCGCAATCTATA-3′ (the CEBP consensus binding site is underlined). Nuclear extracts were added together with a CEBPA antibody. Addition of a secondary antibody conjugated to horseradish peroxidase provided colorimetric quantitation by spectrophotometry.
UV Cross-Link Assay for CRT. A double-stranded RNA oligomer covering a CRT-binding site within the CEBPA mRNA was generated as follows: oligomer A (5′-CCCCACGGGCGGCGGCGGCGGCGGCGACUU-3, containing CGG repeats) and oligomer B(5′-UAACCAGCCGCCGCCGCCGCCGCCGCCGCCGCCC-3′, containing CCG repeats) were annealed. The double-stranded oligomers were separated from single-stranded oligomers by gel electrophoresis and subsequent extraction. The double-stranded oligomers were labeled by using [γ-32P]ATP, and equal amounts were incubated with whole-cell protein extracts for 30 min at room temperature and subjected to UV treatment for 5 min at 125 mJ (34). After electrophoresis, the proteins were transferred to the membrane and autoradiographed.
RNA Interference. CRT small interfering RNA (siRNA) (Ambion, Austin, TX) had the sequences 5′-GGAGCAGUUUCUGGACGGATT-3′ and 5′-UCCGUCCAGAAACUGCUCCTT-3′. Ascontrol, the Silencer negative control no. 2 siRNA (Ambion) was used. U937 AME cells were set to a density of 1.4 × 106 in 100 μl of Amaxa solution V (Nucleofector kit V, Amaxa, Cologne, Germany) and mixed with 800 ng of siRNA. Cells were transfected by electroporation applying nucleofector technology (software version 2.1, Amaxa, Gaithersburg, MD).
Statistical Analysis. Mean and SD were calculated. Statistical analysis was performed by using the Mann-Whitney rank sum test (sigmastat 3.0).
Results
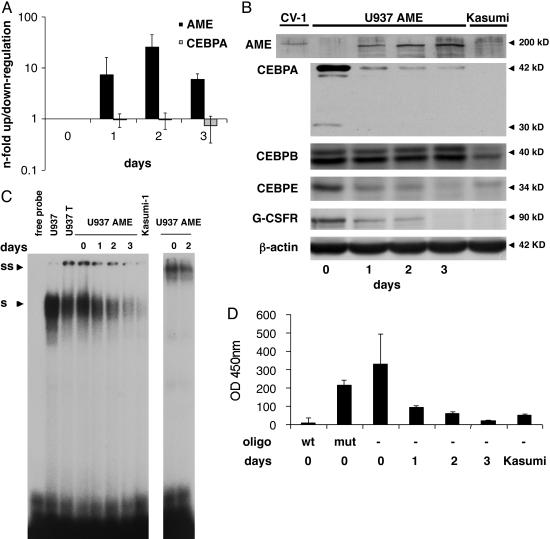
Conditional Expression of AME Suppresses CEBPA Protein in U937 Leukemic Cells. We established single-cell clones of the myeloid leukemic cell line (U937) that conditionally express the AME protein after withdrawal of tetracycline. Real-time PCR analysis showed in 4 of 18 clones a >10-fold increase of AME mRNA transcripts 48 h after withdrawal of tetracycline. No morphological changes were observed after the induction of AME (data not shown). The clone selected for the experiments in this study showed an increase in AME mRNA expression of 25-fold on day 2 (Fig. 1A). However, no CEBPA mRNA changes were observed (n-fold range, 1.03-1.37; three independent experiments) (Fig. 1A). On day 2, the median absolute cycle threshold values for AME and CEBPA were 21.7 (SD, 0.1) and 18.4 (SD, 0.3), respectively. We thus concluded that forced expression of AME had no effect on CEBPA mRNA levels.
Fig. 1.
Conditional expression of AME in U937 leukemic cells. (A) U937 cells were analyzed before (day 0) and 1, 2, and 3 days after withdrawal of tetracycline by real-time PCR analyses for AME and CEBPA expression. Mean values and SD (error bars) are depicted. (B) Western blot analyses at the same time points as in A. CV1 cells transiently transfected with an AME expression construct served as positive control (left lane). The membrane was incubated further with antibodies against CEBPA, CEBPB, CEBPE, G-CSF receptor, and β-actin. (C) CEBPA-binding activity to a CEBP site as present in the G-CSF receptor promoter was assessed by electrophoretic mobility shift assays at the time points indicated after withdrawal of tetracycline. Kasumi-1 cells served as a CEBPA negative control. S, shifted CEBPA protein; SS, supershifted CEBPA-protein complex. (D) CEBPA-binding activity measured with the TransAM assay. A CEBP wild-type (wt) or a CEBP mutated oligonucleotide (mut) were added to nuclear extracts from day 0 as a control. Mean values and SD (error bars) are depicted. Nuclear extracts from Kasumi-1 cells served as a negative control.
Western blot analysis verified the induction of the 200-kDa AME protein (Fig. 1B). In contrast to CEBPA mRNA levels, CEBPA protein was rapidly suppressed after AME induction (Fig. 1B). The blot was subsequently incubated with antibodies against other CEBP family members and the G-CSF receptor (Fig. 1B). CEBPB protein remained unchanged after AME induction, consistent with findings after AML1-ETO induction (32). CEBPE is reported to be a downstream target of CEBPA (29, 32). As expected, we observed a marked decrease in CEBPE protein after AME induction. In addition, the G-CSF receptor protein as another direct target of CEBPA (24, 32, 33) was similarly suppressed after AME induction. To ensure that the induction of the tetracycline system itself had no effect on CEBPA mRNA and protein levels, parental U937 cells with the tetracycline-transactivator constructs but lacking the AME cDNA were analyzed. Indeed, no changes on CEBPA mRNA and protein levels were detectable after withdrawal of tetracycline (data not shown).
To investigate CEBPA DNA-binding activity, we performed gel-shift analyses. In unstimulated U937 cells, almost the entire binding activity to a CEBP site in a downstream target such as the G-CSF receptor promoter is contributed by CEBPA (33). Starting 24 h after AME induction, we observed a consistent decrease of CEBPA binding to this site. At day 3, binding activity was hardly detectable (Fig. 1C), which is equivalent to a 93.6% reduction of binding activity as further verified by the TransAM assay (Fig. 1D). In contrast, we observed no changes in DNA-binding activity in the parental U937T cells after withdrawal of tetracycline (data not shown). In conclusion, these experiments confirm that AME indeed suppresses the CEBPA-protein production and function.
CEBPA Protein Is Specifically Suppressed in AML Patients Carrying the AME Translocation. We aimed to verify the results obtained in induced U937 cells in malignant cells from eight patients carrying the AME translocation. We compared them to nine AML patients with a normal karyotype. As assessed by direct sequencing, none of the patients had CEBPA mutations. Real-time PCR analysis demonstrated similar CEBPA mRNA levels (P = 0.779) in AML patients with the AME translocation and with a normal karyotype (Fig. 2A). Again, no CEBPA protein was detectable by Western blot in any of the samples with the AME translocation. In contrast, significant amounts of CEBPA protein were seen in AML patients with a normal karyotype (Fig. 2B).
Fig. 2.
CEBPA protein is specifically suppressed in AME patients. Eight patient samples carrying the AME translocation and nine AML patients with a normal karyotype were analyzed. (A) Real-time PCR analysis of CEBPA levels from AME patients and AML patients with a normal karyotype. Mean and SD (error bars) are shown. (B) Western blot analyses from lysates of three AME patient samples and of three representative AML patients with a normal karyotype. The same membrane was incubated with an antibody against β-actin for control (Lower). (C) CEBPA-binding activity was measured by using the TransAM assay. Mean and SD (error bars) are shown.
We analyzed patient samples with and without the AME translocation for their binding activity to a CEBP consensus binding site by using the TransAM assay. We found a dramatically reduced CEBPA-binding activity (92.1% reduction) in the eight samples with the AME translocation as compared to AML patients with a normal karyotype (Fig. 2C). In contrast, no difference in binding activity was observed for CEBPB (P = 0.29) (data not shown).
These results suggest that the leukemic fusion protein AME suppresses CEBPA protein and DNA-binding activity. In contrast to our previous findings with the AML1-ETO fusion (32), no changes on CEBPA mRNA levels were detected after induction of AME. We therefore conclude that a posttranscriptional mechanism must be involved in the regulation of CEBPA in AML with AME.
CRT Levels and Activity Are Increased After Conditional Expression of AME in U937 Cells and in AML Patients with AME. There are only a few reports about mechanisms involved in posttranscriptional regulation of CEBPA (34-36). It has been shown that the poly(rC)-binding protein hnRNP E2 inhibits CEBPA expression at the translational level in patients with CML-BC but not in those in the chronic phase of CML (CML-CP) (36). In contrast to patients with CML-BC, we observed no changes of hnRNP E2 mRNA and protein expression as assessed by real-time PCR and by Western blot analysis in U937 cells following AME induction (data not shown).
CRT has been reported to interact with CEBPA mRNA and thereby to repress translation of the CEBPA protein (34). We therefore hypothesized that the posttranscriptional down-regulation of CEBPA after conditional expression of AME might be caused by an increase of CRT expression and/or activity. Real-time PCR measurements of CRT mRNA transcripts showed a 4.4-fold increase on day 2 (Fig. 3A). Western blot analysis of whole-cell lysates further demonstrated an increase of CRT protein after induction of AME (Fig. 3B).
Fig. 3.
CRT expression and activity are induced after conditional expression of AME in U937 cells and AME patient samples. (A) Measurement of CRT mRNA by real-time PCR. Mean and SD (error bars) are shown. (B) CRT proteins were assessed by Western blot analysis. HeLa cells served as positive control. (C) CRT activity of U937 AME cells after withdrawal of tetracycline was assessed by UV cross-linking. The assay demonstrates the direct interaction of CRT protein to a CRT-binding site within the CEBPA mRNA. Coomassie blue (C. blue) staining is given as a loading control. (D) CRT activity by UV cross-linking. Two representative AME patient samples (lanes 1 and 2) are compared to two AML patient samples with a normal karyotype (lanes 3 and 4). Coomassie blue staining is depicted as a control. (E) U937 cells were transfected with GFP expression plasmid or GFP and CRT expression plasmids. Forty-eight hours after transfection, the expression of CEBPA was examined by immunostaining for CEBPA (Top). Levels of CEBPA expression were determined in 100 cells transfected with CRT and GFP and in 100 cells transfected with GFP alone (Bottom). DAPI, 4′,6-diamidino-2-phenylindole.
CRT activity can be measured by UV cross-linking, thereby enabling visualization of the direct interaction of CRT protein to a CRT-binding site within the CEBPA mRNA (34). Fig. 3C gives evidence of a dramatic increase in CRT activity starting early on day 1 after induction of AME (14.8-fold up-regulation). Finally, we detected increased CRT activity in patient samples carrying the AME translocation (12.2-fold) compared to AML patients with a normal karyotype (Fig. 3D). In conclusion, results obtained from patient samples and cell lines indicate that CEBPA-protein and -binding activity seem to be regulated on a translational level by modulation of CRT protein and activity.
We also tested in a single-cell assay whether overexpression of CRT in U937 cells inhibits CEBPA translation in these cells. To visualize cells expressing CRT, U937 cells were cotransfected with CRT and with a vector expressing GFP at a 10:1 ratio. Under these conditions, each green cell containing GFP is assumed to express CRT (Fig. 3E). We observed that expression of CRT in U937 cells inhibits translation of CEBPA. This CRT-dependent inhibition of CEBPA translation is specific, because GFP alone does not affect CEBPA expression. We examined the levels of CEBPA in 100 cells transfected with CRT, and we found that CEBPA expression was inhibited in 85 of the cells. Similar analysis of CEBPA protein in 100 cells transfected with GFP alone showed only nine cells with reduced levels of CEBPA (Fig. 3E). Interestingly, no differences in CEBPA mRNA levels using real-time PCR were detected between U937 cells transfected with or without CRT (data not shown). Thus, these studies indicate that overexpression of CRT blocks translation of CEBPA in U937 cells.
Inhibition of CRT by siRNA Restores CEBPA Protein Levels in AME U937 Cells After Induction of AME. The experiments described above suggest that CEBPA suppression is mediated by modulation of CRT. We therefore hypothesized that functional knock-down of CRT by siRNA might be able to restore efficient CEBPA translation. We thus induced the AME protein and transfected siRNA designed to target CRT. We observed an 87% knock-down of CRT mRNA levels 48 h after transfection (Fig. 4A). Moreover, CRT-protein suppression was also evident 48 h after siRNA transfection (Fig. 4B). The block of CRT-protein expression was equally observed in U937 cells after AME induction as well as in the parental U937T cells.
Fig. 4.
Inhibition of CRT expression by siRNA restores CEBPA-protein expression. AME protein was induced in U937 cells by withdrawal of tetracycline, and siRNA designed to knock-down CRT or mock siRNA was transfected by electroporation. As further control, U937T cells were transfected with siRNA. (A) Real-time PCR analysis of CRT levels 48 h after transfection. Mean and SD (error bars) are shown. (B) Western blot analysis of CRT, CEBPA, CEBPB, and β-actin.
Most interestingly, transfection of CRT siRNA prevented suppression of CEBPA protein after AME induction (Fig. 4B). CEBPA-protein levels after inhibition of CRT by siRNA even exceeded the levels of U937 cells before AME induction (Fig. 4B). In addition, the block of CRT in U937T cells (thus in the absence of AME) also resulted in a significant increase of CEBPA protein. However, and in contrast to CEBPA, no significant changes in CEBPB expression were observed. We therefore conclude that CRT in myeloid cells indeed is a potent inhibitor of CEBPA translation.
Discussion
Here we show that the myeloid transcription factor CEBPA is specifically suppressed in AML patients carrying the AME translocation. We also demonstrate that this suppression is mediated on a translational level and is caused by CRT, a putative inhibitor of CEBPA translation (34).
Previous reports by us and others have pointed to a crucial role of CEBPA in the pathogenesis of AML (29, 32, 36). In particular, the CBF complex seems to target CEBPA (32). Chromosomal abnormalities affecting AML1B/RUNX1, one of the two subunits of the transcription factor CBF, have been shown to suppress CEBPA transcription (32). Here we focused on the AME translocation. Because the AME fusion equally affects the AML1 gene (the DNA-binding subunit of the CBF complex), our findings further support the hypothesis that CBF leukemias target the myeloid key transcription factor CEBPA and that this pathway may contribute to the differentiation block seen in these particular subsets of AML.
The mechanisms of how the AME fusion contributes to leukemogenesis are largely unknown. Expression of AME has been reported to increase proliferation and abnormal differentiation in 32D cells and in murine bone marrow progenitors (17, 37). Furthermore, it has been shown that AME inhibits the antiproliferative effect of transforming growth factor β. It also blocks the granulocytic differentiation of IL-3-dependent 32D cells when stimulated with G-CSF (16, 17). In addition, AME seems to require functions and/or functional cooperation of both aml1 and evi1 to induce AML in mice (38). However, the target genes involved in the differentiation block seen in AME leukemias remain unknown.
In murine transplant models, AME can induce a disease similar to human acute myelomonocytic leukemia (15). Coexpression of bcr-abl and ame fusion genes in mice rapidly induces AML, suggesting that a cooperation between mutations that dysregulate tyrosine kinase signaling (bcr-abl) and those that disrupt differentiation (ame) is necessary (39). Interestingly, in CML-CP, CEBPA-protein levels are normal, whereas patients with CML-BC have suppressed CEBPA protein, thereby possibly contributing to the differentiation block seen in CML-BC but not in CML-CP (36). Because AME does occur in patients with CML-BC, but not in CML-CP, one could speculate that the block in differentiation observed in patients with AME in CML-BC might be caused by CEBPA suppression. Moreover, a recent report suggests that restoration of CEBPA in a BCR-ABL-positive cell line rapidly induces terminal granulocytic differentiation (28).
Various mechanisms have been reported thus far to account for a disruption of CEBPA function in AML patients. We and others have reported that wild-type CEBPA function is abrogated in some AML patients by dominant-negative mutations in the CEBPA gene (29-31). We have further shown that CEBPA expression can be inhibited by AML1-ETO on the transcriptional level by suppressing its autoregulatory loop (32). Furthermore, a recent report indicates that CEBPA expression in AML is abolished at the RNA level by the tyrosine kinase receptor FLT3 (40). In addition, CEBPA function seems to be inhibited by phosphorylation at Ser-21 mediated by overexpression of FLT3 or by FLT3 mutants (41). Posttranscriptional modulation of CEBPA is involved in patients with CML-BC through the inhibitory action of the poly(rC)-binding protein hnRNP E2 by direct interaction with the upstream ORF of CEBPA (36). A novel posttranscriptional mechanism for the modulation of CEBPA and CEBPB expression in HeLa cells was reported recently, involving the chaperone CRT (34). CRT protein binds to GCN repeats in the CEBPA mRNA and thereby impedes translation of CEBPA mRNA. Our data suggest that in AME leukemias, this mechanism is involved in the suppression of CEBPA in vitro and in vivo, which highlights a role of RNA-binding proteins for modulation of CEBPA expression in AML. It also suggests the design of additional studies investigating the role of CRT in other subsets of AML.
The myeloid key transcription factor CEBPA is believed to suppress the leukemic phenotype through combined induction of direct transcriptional targets crucial for normal myeloid differentiation and inhibition of cell-cycle progression. We and others have shown in leukemic cells that restoring CEBPA expression is sufficient to induce neutrophil differentiation (25-29, 32, 40, 41), thereby pointing to potential therapeutic implications. Here we report that the block of CRT expression by siRNA powerfully restores CEBPA expression. Therefore, modulation of CRT expression might be a potent target for subsets of AML in which CEBPA protein is suppressed.
Acknowledgments
The AML1-MDS1-EVI1 cDNA was kindly provided by Giuseppina Nucifora (Department of Pathology, University of Illinois, Chicago), and the U937T cell line was obtained from Gerard Grosveld. This work was supported by Swiss National Science Foundation Grants SF 31-666899.01 (to T.P.), SF 3100-67213 (to M.F.F.), and SF 3100A0-100445 (to B.U.M.).
Abbreviations: AML, acute myeloid leukemia; AME, AML1-MDS1-EVI1; MDS, myelodysplastic syndrome; CML, chronic myeloid leukemia; CML-BC, CML in blast crisis; CRT, calreticulin; G-CSF, granulocyte colony-stimulating factor; siRNA, small interfering RNA; CMLCP, chronic phase of CML.
References
- 1.Tenen, D. G. (2003) Nat. Rev. Cancer 3, 89-101. [DOI] [PubMed] [Google Scholar]
- 2.Rowley, J. D. (2001) Nat. Rev. Cancer 1, 245-250. [DOI] [PubMed] [Google Scholar]
- 3.Nucifora, G. (1997) Leukemia 11, 2022-2031. [DOI] [PubMed] [Google Scholar]
- 4.Rubin, C. M., Larson, R. A., Anastasi, J., Winter, J. N., Thangavelu, M., Vardiman, J. W., Rowley, J. D. & Le Beau, M. M. (1990) Blood 76, 2594-2598. [PubMed] [Google Scholar]
- 5.Rubin, C. M., Larson, R. A., Bitter, M. A., Carrino, J. J., Le Beau, M. M., Diaz, M. O. & Rowley, J. D. (1987) Blood 70, 1338-1342. [PubMed] [Google Scholar]
- 6.Nucifora, G., Begy, C. R., Kobayashi, H., Roulston, D., Claxton, D., Pedersen-Bjergaard, J., Parganas, E., Ihle, J. N. & Rowley, J. D. (1994) Proc. Natl. Acad. Sci. USA 91, 4004-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osato, M., Asou, N., Abdalla, E., Hoshino, K., Yamasaki, H., Okubo, T., Suzushima, H., Takatsuki, K., Kanno, T., Shigesada, K. & Ito, Y. (1999) Blood 93, 1817-1824. [PubMed] [Google Scholar]
- 8.Nucifora, G., Birn, D. J., Erickson, P., Gao, J., LeBeau, M. M., Drabkin, H. A. & Rowley, J. D. (1993) Blood 81, 883-888. [PubMed] [Google Scholar]
- 9.Song, W. J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I. C., Haworth, C., Hock, R., et al. (1999) Nat. Genet. 23, 166-175. [DOI] [PubMed] [Google Scholar]
- 10.Roulston, D., Espinosa, R., 3rd, Nucifora, G., Larson, R. A., Le Beau, M. M. & Rowley, J. D. (1998) Blood 92, 2879-2885. [PubMed] [Google Scholar]
- 11.Mikhail, F. M., Serry, K. A., Hatem, N., Mourad, Z. I., Farawela, H. M., El Kaffash, D. M., Coignet, L. & Nucifora, G. (2002) Cancer Genet. Cytogenet. 135, 96-100. [DOI] [PubMed] [Google Scholar]
- 12.Zent, C., Rowley, J. D. & Nucifora, G. (1997) Leukemia 11, Suppl. 3, 273-278. [PubMed] [Google Scholar]
- 13.Mucenski, M. L., Taylor, B. A., Ihle, J. N., Hartley, J. W., Morse, H. C., 3rd, Jenkins, N. A. & Copeland, N. G. (1988) Mol. Cell. Biol. 8, 301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morishita, K., Parker, D. S., Mucenski, M. L., Jenkins, N. A., Copeland, N. G. & Ihle, J. N. (1988) Cell 54, 831-840. [DOI] [PubMed] [Google Scholar]
- 15.Cuenco, G. M., Nucifora, G. & Ren, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sood, R., Talwar-Trikha, A., Chakrabarti, S. R. & Nucifora, G. (1999) Leukemia 13, 348-357. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka, T., Mitani, K., Kurokawa, M., Ogawa, S., Tanaka, K., Nishida, J., Yazaki, Y., Shibata, Y. & Hirai, H. (1995) Mol. Cell. Biol. 15, 2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott, L. M., Civin, C. I., Rorth, P. & Friedman, A. D. (1992) Blood 80, 1725-1735. [PubMed] [Google Scholar]
- 19.Cao, Z., Umek, R. M. & McKnight, S. L. (1991) Genes Dev. 5, 1538-1552. [DOI] [PubMed] [Google Scholar]
- 20.Birkenmeier, E. H., Gwynn, B., Howard, S., Jerry, J., Gordon, J. I., Landschulz, W. H. & McKnight, S. L. (1989) Genes Dev. 3, 1146-1156. [DOI] [PubMed] [Google Scholar]
- 21.Flodby, P., Barlow, C., Kylefjord, H., Ahrlund-Richter, L. & Xanthopoulos, K. G. (1996) J. Biol. Chem. 271, 24753-24760. [DOI] [PubMed] [Google Scholar]
- 22.Chandrasekaran, C. & Gordon, J. I. (1993) Proc. Natl. Acad. Sci. USA 90, 8871-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swart, G. W., van Groningen, J. J., van Ruissen, F., Bergers, M. & Schalkwijk, J. (1997) Biol. Chem. 378, 373-379. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, D. E., Zhang, P., Wang, N. D., Hetherington, C. J., Darlington, G. J. & Tenen, D. G. (1997) Proc. Natl. Acad. Sci. USA 94, 569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radomska, H. S., Huettner, C. S., Zhang, P., Cheng, T., Scadden, D. T. & Tenen, D. G. (1998) Mol. Cell. Biol. 18, 4301-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, X., Scott, E., Sawyers, C. L. & Friedman, A. D. (1999) Blood 94, 560-571. [PubMed] [Google Scholar]
- 27.Miyamoto, T., Iwasaki, H., Reizis, B., Ye, M., Graf, T., Weissman, I. L. & Akashi, K. (2002) Dev. Cell 3, 137-147. [DOI] [PubMed] [Google Scholar]
- 28.Tavor, S., Park, D. J., Gery, S., Vuong, P. T., Gombart, A. F. & Koeffler, H. P. (2003) J. Biol. Chem. 278, 52651-52659. [DOI] [PubMed] [Google Scholar]
- 29.Pabst, T., Mueller, B. U., Zhang, P., Radomska, H. S., Narravula, S., Schnittger, S., Behre, G., Hiddemann, W. & Tenen, D. G. (2001) Nat. Genet. 27, 263-270. [DOI] [PubMed] [Google Scholar]
- 30.Preudhomme, C., Sagot, C., Boissel, N., Cayuela, J. M., Tigaud, I., de Botton, S., Thomas, X., Raffoux, E., Lamandin, C., Castaigne, S., Fenaux, P. & Dombret, H. (2002) Blood 100, 2717-2723. [DOI] [PubMed] [Google Scholar]
- 31.Gombart, A. F., Hofmann, W. K., Kawano, S., Takeuchi, S., Krug, U., Kwok, S. H., Larsen, R. J., Asou, H., Miller, C. W., Hoelzer, D. & Koeffler, H. P. (2002) Blood 99, 1332-1340. [DOI] [PubMed] [Google Scholar]
- 32.Pabst, T., Mueller, B. U., Harakawa, N., Schoch, C., Haferlach, T., Behre, G., Hiddemann, W., Zhang, D. E. & Tenen, D. G. (2001) Nat. Med. 7, 444-451. [DOI] [PubMed] [Google Scholar]
- 33.Smith, L. T., Hohaus, S., Gonzalez, D. A., Dziennis, S. E. & Tenen, D. G. (1996) Blood 88, 1234-1247. [PubMed] [Google Scholar]
- 34.Timchenko, L. T., Iakova, P., Welm, A. L., Cai, Z. J. & Timchenko, N. A. (2002) Mol. Cell. Biol. 22, 7242-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lincoln, A. J., Monczak, Y., Williams, S. C. & Johnson, P. F. (1998) J. Biol. Chem. 273, 9552-9560. [DOI] [PubMed] [Google Scholar]
- 36.Perrotti, D., Cesi, V., Trotta, R., Guerzoni, C., Santilli, G., Campbell, K., Iervolino, A., Condorelli, F., Gambacorti-Passerini, C., Caligiuri, M. A., et al. (2002) Nat. Genet. 30, 48-58. [DOI] [PubMed] [Google Scholar]
- 37.Senyuk, V., Chakraborty, S., Mikhail, F. M., Zhao, R., Chi, Y. & Nucifora, G. (2002) Oncogene 21, 3232-3240. [DOI] [PubMed] [Google Scholar]
- 38.Cuenco, G. M. & Ren, R. (2004) Oncogene 23, 569-579. [DOI] [PubMed] [Google Scholar]
- 39.Cuenco, G. M. & Ren, R. (2001) Oncogene 20, 8236-8248. [DOI] [PubMed] [Google Scholar]
- 40.Zheng, R., Friedman, A. D., Levis, M., Li, L., Weir, E. G. & Small, D. (2004) Blood 103, 1883-1890. [DOI] [PubMed] [Google Scholar]
- 41.Ross, S. E., Radomska, H. S., Wu, B., Zhang, P., Winnay, J. N., Bajnok, L., Wright, W. S., Schaufele, F., Tenen, D. G., & MacDougald, O. A. (2004) Mol. Cell. Biol. 24, 675-686. [DOI] [PMC free article] [PubMed] [Google Scholar]