Abstract
In the period since LRRK2 (Leucine-Rich Repeat Kinase 2) was identified as a causal gene for late-onset autosomal-dominant parkinsonism, a great deal of work has been aimed at understanding whether the LRRK2 protein might be a druggable target for Parkinson’s disease (PD). As part of this effort, animal models have been developed to explore both the normal and the pathophysiological roles of LRRK2. However, LRRK2 is part of a wider family of proteins whose functions in different organisms remain poorly understood. In this review, we compare the information available on biochemical properties of LRRK2 homologues and orthologues from different species from invertebrates (e.g., Caenorhabditis elegans and Drosophila melanogaster) to mammals. We particularly discuss the mammalian LRRK2 homologue, LRRK1, and those species where there is only a single lrrk homologue, discussing examples where each of the LRRK family of proteins has distinct properties as well as those cases where there appear to be functional redundancy. We conclude that uncovering the function of LRRK2 orthologues will help elucidate the key properties of human LRRK2 as well as to improve understanding of the suitability of different animal models for investigation of LRRK2-related PD.
Keywords: LRRK2 homologue, animal models, kinase, GTPase, dopaminergic neurons, neurodegeneration, Preclinical research
INTRODUCTION
Parkinson’s disease (PD) is characterized clinically by a range of motor symptoms, including postural instability, resting tremor, bradykinesia, and rigidity as well as non-motor features such as hyposmia [1,2], constipation [3], and mood disturbances [4]. Pathologically, the disease is associated with progressive loss of dopaminergic (DA) neurons in the substantia nigra and the formation of fibrillar aggregates called Lewy bodies in surviving neurons throughout the brain [5].
The cause of most cases of PD is not known, leading to the description as a sporadic or idiopathic disease. However, in recent years it has been increasingly appreciated that monogenic forms of PD exist and that cloning the underlying gene mutations can lead to important insights into pathobiology [6,7]. A locus on chromosome 12 (historically termed PARK8) was first linked to familial parkinsonism in 2002 through analysis of a large Japanese family in which affected members exhibited clinical phenotypes resembling those of idiopathic PD [8]. In 2004, late-onset autosomal-dominant parkinsonism was linked to the same locus in multiple families, and a number of pathogenic mutations in the LRRK2 gene were identified [9–11]. Importantly, subsequent investigations of the genetic contribution to sporadic PD using genome-wide association studies have nominated variation around the wild-type LRRK2 gene as a risk factor for the common form of this disease [12,13]. Thus, the LRRK2 gene likely links sporadic and inherited PD.
The LRRK2 gene encodes a large multi-domain protein, LRRK2. The N-terminal region of LRRK2 comprises a series of repeats predicted to adopt armadillo-like and ankyrin repeat folds as well as a leucine-rich repeat (LRR) domain for which LRRK2 and its homologues are named. The C-terminal region also contains a set of repeats that form a predicted β-sheet rich WD40 domain. These protein interaction motifs flank a catalytic tridomain: a Roc (Ras of complex proteins) GTPase domain in tandem with its associated C-terminal of Roc (COR) domain, followed by a kinase domain [14]. Notably, the COR domain always follows the Roc domain within proteins belonging to the Roco protein family [15] and may play an important role in intramolecular interactions between the two catalytic domains of LRRK2 [16].
As well as LRRK2, there are three other Roco proteins that have been identified in the human proteome, with the closest homologue being leucine-rich repeat kinase 1 (LRRK1), death-associated protein kinase 1 (DAPK1) and malignant fibrous histiocytoma amplified sequences with leucine-rich tandem repeats 1 (MASL1). All four have been found capable of binding guanosine nucleotides via the Roc domain and LRRK2 and DAPK1 are active kinases [17–22], suggesting that there are functional similarities between the human Roco proteins.
These human proteins are part of an evolutionarily older superfamily of Roco proteins [23,24] that include representatives from the slime mold Dictyostelium discoideum through most of the animal kingdom, with a few representatives in plants such as the gene TORNADO1 in Arabidopsis. The evolutionary history of the modern LRRK proteins can be traced to the cnidarian Nematostella vectensis, which has four LRRK genes. Two of these genes are orthologues of the LRRK1 and LRRK2 genes present in deuterostomes (including humans and rodents), while a third is an orthologue of the LRRK gene in protostomes such as C. elegans and Drosophila. The fourth appears to be cnidarian-specific [24,25]. Thus, the C. elegans (lrk-1) and Drosophila (Lrrk) genes have an ancient origin distinct from that of LRRK2.
Here, we will focus on LRRK proteins across the animal kingdom, discussing the functional redundancy between these different proteins but also those examples where different proteins seem to have different functions within or between different species. This will also help to frame later discussions of the suitability of different animal models for the study of Parkinson’s disease and for preclinical trials of novel therapeutic treatments. For further discussion, including an overview of two studies of zebrafish LRRK2 models, the interested reader may refer to another recent review article about LRRK2 animal models [26]. We will start with two well-characterized invertebrate model organisms, C. elegans and Drosophila melanogaster.
The Caenorhabditis elegans protein, Lrk-1
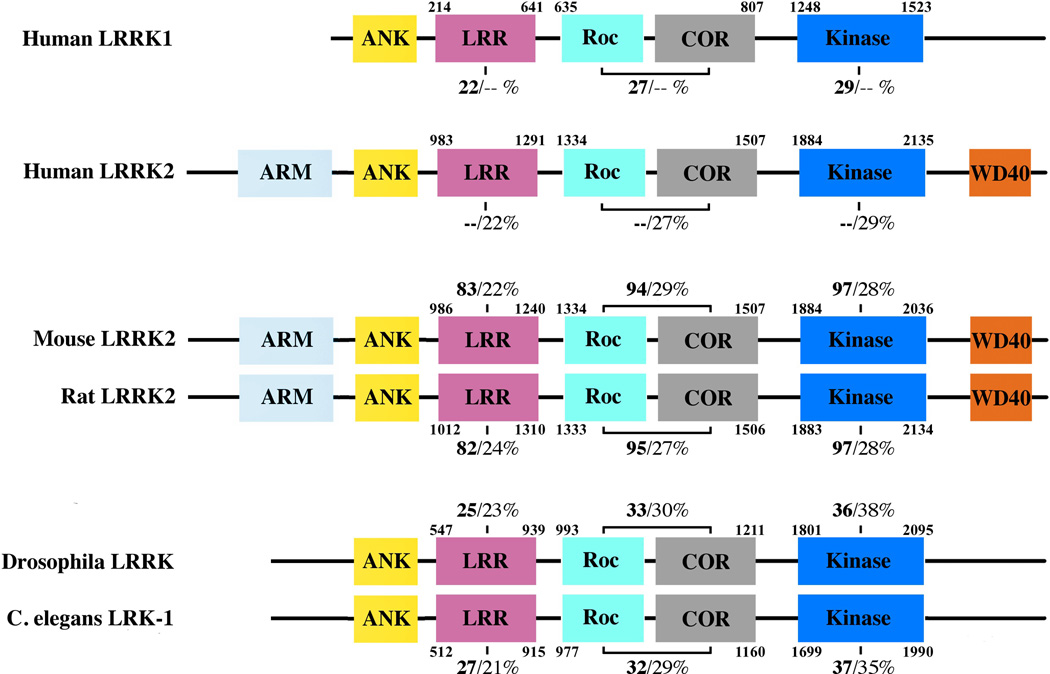
The transparent nematode Caenorhabditis elegans offers a unique model system for neurodegenerative disease as the 302 neurons that constitute the nervous system of an adult C. elegans hermaphrodite can be examined in vivo and, of particular importance to PD research, the eight dopaminergic neurons can be easily visualized [27–29]. C. elegans has a single orthologue of mammalian LRRK genes, lrk-1. The lrk-1 protein is slightly shorter than LRRK2 (2,393 amino acids) but similarly organized with clear LRR, Roc, COR, and kinase domains [28] (Figure 1). Sequence homology between LRRK2 and lrk-1 is modest, with 24/40% identity/similarity over a region of ~1,500 amino acids [30,31]. As discussed above, because lrk-1 has a distinct evolutionary history from LRRK1 and LRRK2, this relatively low level of similarity is not surprising and suggests that lrk-1 knockouts should be considered distinct from knockouts of LRRK2, which we will discuss later in this article.
Figure 1.
Schematic diagrams of LRRK proteins in different species, including human LRRK2 and its human homologue LRRK1, as well as orthologues from different species Predicted domains are indicated by labelled boxes. Percentage identities based on a Clustal 2.1 multiple sequence alignment are given relative to human LRRK2 (left, bolded) and relative to human LRRK1 (right) for LRR, Roc-COR and kinase domains.
Worms carrying deletion mutations in lrk-1 exhibit a range of phenotypes, including subtle motor defects [28], aberrant localization of synaptic vesicle proteins [28], and increased sensitivity to a variety of pharmacological stressors [30–33]. Along with regulation of polarized sorting of synaptic vesicle proteins in neurons [28], lrk-1 has also been functionally linked to regulation of neurite outgrowth [31]. Evidence that LRRK2 can substitute for lrk-1 in reducing neurotoxicity [32] and that knockdown of endogenous lrk-1 and expression of human LRRK2 have reciprocal effects [29,30,33] suggests that lrk-1 and LRRK2 can interact with the same pathways in C. elegans. However, overexpression of either lrk-1 wild-type or lrk-1 G1876S (corresponding to the human LRRK2 G2019S mutation) caused embryonic lethality [31], which might imply that the native protein possesses an intrinsic toxicity to nematodes not conserved in the human LRRK2 protein.
Transgenic expression of wild-type human LRRK2 in nematodes was associated with increased lifespan and protection against whole organism toxicity caused by rotenone or paraquat [30,33]. Transgenic worms expressing mutant human LRRK2 were protected against rotenone and paraquat toxicity to a lesser extent than observed in LRRK2 wild-type animals [30,33] and displayed adult-onset dopaminergic neuron loss sometimes accompanied by motor deficits [29,30,32,34,35]. R1441C and G2019S mutants caused a more severe phenotype than wild-type LRRK2, but expression of the GTPase binding defective mutant K1347A did not induce the parkinsonian phenotype [29]. Similarly, there is evidence to support the contribution of kinase activity of LRRK2 mutants in PD pathology [29,35]. Some phenotypes in these animals could be rescued by administration of L-DOPA [32,34]. However, when examining dopamine neurons directly, G2019S-LRRK2 transgenic nematodes displayed significant loss of dopaminergic markers on adult days 2 and 3 relative to wild-type, both basally and in response to mitochondrial toxins [30]. Interestingly, knockdown of lrk-1 was found to attenuate the pathogenic effects of LRRK2 expression [29,32], suggesting that overexpression of mutant LRRK2 may work, in part, by a dominant negative effect on the endogenous protein. However, why LRRK2 is protective in some assays but toxic in others is not yet resolved.
Collectively, these results suggest that there may be some shared functions between human LRRK2 and C. elegans lrk-1. An important caveat, particularly in relation to the transgenic approaches, is that levels of proteins are not usually assessed. Thus, whether the two proteins can be directly compared or whether there are non-specific events related to overexpression is difficult to determine. Furthermore, the observation that there are effects seen with lrk-1 not shared with LRRK2 [31] suggests that the two proteins have at least some distinct properties. It would be important in the future to understand the similarities and differences between human LRRK2 and worm lrk-1 with more mechanistic approaches, particularly at the level of protein interactions or substrates.
The Drosophila melanogaster protein, Lrrk
Like C. elegans, Drosophila melanogaster has a single orthologue of LRRK1/2 named Lrrk, an approximately 2,400 amino acid long protein containing LRR, ROC-COR, and kinase motifs [36,37]. However, Lrrk does not appear to contain the LRRK2-like N-terminal ankyrin repeats or a C-terminal WD40 domain. Many of the residues involved in LRRK2 PD pathology are conserved between human LRRK2 and Drosophila Lrrk, which share 24% identity and 38% similarity overall [36–38] (Figure 1). Lrrk is ubiquitously expressed in Drosophila tissues, including in fly brain [36,39,40]. Within cells, Lrrk protein is largely cytoplasmic with some association with membranous structures including endosomes, lysosomes, and synaptic vesicles [36,41], thus being broadly similar to human LRRK2.
The Lrrk protein is an active kinase capable of autophosphorylation [36] and of phosphorylation of various proposed substrates, including 4E-BP, Futsch, Endophilin A, and ribosomal protein s15 [39,42,43]. Lrrk has been shown to physically interact with Rab5, Rab7, and Rab9 in Drosophila follicle cells [41,44] and to associate with Drosophila Argonaute-1 (dAgo1) of the RNA-induced silencing complex in fly head extracts [45]. As human LRRK2 is known to interact with multiple Rab proteins [46–49], and is claimed to phosphorylate some of the same substrates [39,42,43] and to associate with Ago1 [45], there may be at least partial conservation of function between Lrrk and LRRK2. Supporting this hypothesis, several experiments have indicated that Lrrk knockdown and human LRRK2 expression have reciprocal effects [42,43,50] or that overexpression of Lrrk and overexpression of human LRRK2 have the same effect [51]. However, not all phenotypes related to loss of Lrrk have been reproducible. For example, locomotor dysfunction and abnormal morphology of dopaminergic neurons in Lrrk deficient flies from one study [37] were not reproduced in independently generated Lrrk deficient lines [36,38]. Therefore, some aspects of Lrrk function remain to be clarified.
Transgenic expression of mutant Lrrk in Drosophila dopaminergic neurons has consistently resulted in age-dependent dopamine cell death. Imai et al. reported that 60-day-old but not 10-day-old transgenic flies expressing Drosophila Lrrk Y1383C (corresponding to human LRRK2 Y1699C) or I1915T (corresponding to human I2020T mutant) driven by a TH-Gal4 showed a significant reduction in both number of dopaminergic neurons and dopamine content compared to those expressing wild-type Lrrk [36]. Gehrke et al. also observed DA neuron loss as well as climbing defects in TH-Gal4-directed pathogenic Drosophila Lrrk I1915T and human LRRK2 G2019S transgenic animals at 65 days of age [45].
Similarly, mutant human LRRK2 is toxic in flies. Aged flies expressing ddc-GAL4-driven UAS-G2019S-LRRK2 exhibit selective degeneration of DA neurons, motor dysfunction, and reduced lifespan [35,40,43,51–53]. Linhart et al. expressed ddc-GAL4-driven mutant human LRRK2 I2020T in transgenic Drosophila lines, which also resulted in the loss of dopaminergic neurons, locomotor deficits, and decreased lifespan [54]. Ng et al. generated mutant flies with ddc-Gal4-directed expression of human LRRK2 Y1699C. These flies were phenotypically similar to flies expressing human LRRK2 G2019S. Interestingly, human LRRK2 G2385R mutant flies did not show significant degeneration of DA neurons or motor deficits, but did have a significantly reduced lifespan [52]. Flies overexpressing mutant Lrrk/LRRK2 were also found to be more vulnerable to hydrogen peroxide (H2O2) compared to those overexpressing wild-type protein [36,55], along with decreased sensitivity to the H2O2 treatment in Drosophila Lrrk deletion mutant flies [36]. However, Wang et al. found Lrrk deletion mutant flies to be more sensitive to H2O2 than wild-type flies [38]. Expression of wild-type human LRRK2 did not induce a phenotype significantly different from that of control in some studies [35,51–53,56], but a mild phenotype has been reported in others [40,57]. Hindle et al. did not see reduction in number of DA neurons in 28-day-old flies expressing human LRRK2 G2019S directed by TH- or HL9- dopaminergic GAL4 drivers, but did find retinal degeneration [56].
Despite some discrepancies between different transgenic lines, these models have led to several important insights into the pathogenesis of LRRK2-mediated disease. For example, both Drosophila Lrrk and human LRRK2 modulate neurite growth and synaptic morphogenesis via phosphorylation of Futsch, the Drosophila homolog of microtubule-associated protein 1B (MAP1B), at the pre-synapse and of eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP) at the post-synapse [39]. While it remains unclear how 4E-BP, a negative regulator of protein translation, mediates the synaptic effects of LRRK2, this hypothesis would be consistent with prior claims that 4E-BP is a substrate of Drosophila Lrrk [36], although data from human cells suggests that the phosphorylation of 4E-BP might be regulated by other stress-induced kinases such as p38 [58]. LRRK2/Lrrk have also been claimed to phosphorylate FoxO, a transcription factor that regulates 4E-BP transcription, potentially suggesting a mechanism other than direct phosphorylation of 4E-BP by LRRK2/Lrrk to control 4E-BP [59]. Additional experiments linked deregulated protein synthesis with increased LRRK2 kinase activity in human LRRK2 G2019S transgenic flies through increased phosphorylation of ribosomal protein s15, an activator of protein synthesis [43]. As yet another potential mechanism, Gehrke et al. showed that pathogenic Drosophila Lrrk I1915T and human LRRK2 G2019S negatively regulate translational repression via the microRNA pathway [45], although subsequent studies looking at miRNA abundance in LRRK2 knockout or transgenic mice showed a very modest effect on transcription [60], leaving open the question as to whether the observed effects on translation are a direct consequence of LRRK2 or subsequent to the accumulation of age-related pathology in Drosophila Lrrk/LRRK2 models.
Another concept that has been derived largely from fly models is that bioenergetics might be important in Lrrk/LRRK2 pathogenesis. For example, retinal degeneration in flies expressing human LRRK2 G2019S can be prompted by increased activity/energy demand in the visual system [56]. Mitochondria, which are clearly important in energy generation, show pathology in the flight muscles of G2019S–LRRK2 transgenic Drosophila [53]. These effects are modulated by both parkin and LRRK2 via the AMPK signaling pathway, further implying that compromised neuronal energy homeostasis could contribute to neurodegeneration [53]. Multiple studies of transgenic LRRK2 Drosophila models reported a genetic linkage with parkin [52,53,57], PINK1 [57], or DJ-1 [57], genes that are known to be important in mitochondrial function [61].
An apparently distinct set of interactions between Drosophila Lrrk and the Rab protein family in Drosophila follicle cells suggest roles for Lrrk in Rab7-dependent lysosomal positioning and in endosomal recycling via Rab9 [41,44]. Matta et al. suggested an involvement of Drosophila Lrrk in synaptic vesicle endocytosis at the Drosophila neuromuscular junction through phosphorylation of Endophilin A, a result later supported by experiments in rodent cells [42,62]. Linhart et al. provided evidence of interaction between human LRRK2 and vacuolar protein sorting 35 (Vps35) using a genetic modifier screen with transgenic human LRRK2 I2020T mutant flies [54]. Mutation of the Vps35 gene, which encodes a core component of the retromer complex critically involved in protein sorting and recycling and endosome-trans-Golgi trafficking, has been identified as causative of PD [63,64]. These results would then suggest that one or more aspects of vesicular transport, particularly related to turnover of endosomes, might be impacted by loss of Drosophila Lrrk.
Finally, Drosophila Lrrk may also have effects on regulation of the cytoskeleton. It has been claimed that human LRRK2 can phosphorylate the microtubule binding protein Futsch, leading to altered synaptic microtubule organization and, hence, synaptic growth [39]. Additionally, human LRRK2 is thought to bind to the Drosophila glycogen synthase kinase (GSK3b) homologue, Shaggy, that can then phosphorylate tau [51], which would then also be expected to affect microtubule dynamics.
How do we interpret claims that a single protein is involved in at least four distinct cellular processes, namely protein translation, mitochondrial bioenergetics, vesicular transport and cytoskeletal regulation? Two disparate answers to this question are either that LRRK2 genuinely participates in all of these processes, perhaps playing a coordinating role, or that some effects are secondary to others. For example, autophagy is at least one homeostatic mechanism by which dicer and argonaute are degraded [65], raising the possibility that the effects of LRRK on miRNA generation [45] are secondary to its effects on lysosomal positioning [41]. Similar arguments may be made for the relationship between autophagy and energetics as these two processes are tightly co-regulated and selective autophagy is one way in which mitochondria are degraded [66]. For these reasons, mechanistic details such as whether claimed Lrrk/LRRK2 substrates can be shown to be reliable are very important as they might help resolve which events are closest to LRRK. A related question is the extent to which we can extrapolate from those organisms that have a single homologue to those that have distinct LRRKs. We will therefore discuss next the relationship between LRRK1 and LRRK2, the two paralogous genes found in mammals.
The mammalian paralogues LRRK1 and LRRK2
LRRK1 has 1,981 amino acids [25], sharing a similar overall domain structure [20,25,67,68] and an overall sequence homology of 26/45% identity/similarity [69] to the larger (2,527 amino acid) LRRK2. Both LRRK1 and LRRK2 bind GDP/GTP with a similar affinity via the GTPase-like Roc domain [20,67,69]. Although the regulatory interplay between the Roc domain and the kinase domain is not yet well understood, LRRK2 kinase function does require an intact Roc domain [16,70,71] whereas GTP-binding was shown to stimulate LRRK1 kinase activity [20]. However, differences between the two proteins exist in both the N- and C- terminal regions as LRRK2 has a larger N-terminal region containing several unique repeats that are absent in the equivalent section of LRRK1 [23,25,67,72,73] (Figure 1).
A reasonable question, therefore, is whether LRRK1 and LRRK2 act in the same pathway or have functionally distinct roles. Both LRRK1 and LRRK2 are widely expressed in the mammalian brain including in the frontal cortex, striatum, and hippocampus [69,74–76] with the overall level of LRRK2 mRNA expression in brain higher than that of LRRK1 [77]. However, studies using in situ hybridization detected very low if any LRRK1 expression in striatum and also showed that, unlike LRRK2, LRRK1 is not expressed in all neuronal layers [78,79]. Regarding subcellular distribution, LRRK1 is predominantly cytoplasmic as is LRRK2 [18,20,69]. Because they are co-expressed and co-localized in some areas of the brain, a heterodimeric interaction between LRRK1 and LRRK2 could potentially occur in vivo as in vitro [73,74,80]. It is therefore possible that LRRK1 could play a modulatory role in the LRRK2 signaling network and hence change risk of PD [73,74,80]. A study by Dachsel et al. identified a LRRK1 variant, L416M, that was associated with a 6-year earlier age of onset of PD when carried alongside the LRRK2 G2019S mutation, supporting the idea of LRRK1 as a potential disease modifier [74]. However, attempts to directly link variants in LRRK1 to PD pathogenesis have so far been unsuccessful [68,76,81].
A further development of the concept that LRRK1 and LRRK2 might be in the same pathway is that the two proteins might be functionally redundant [77,82], i.e., that LRRK1 could compensate for lack of LRRK2. Potentially arguing against this possibility are the observations that (i) disturbance of LRRK2 does not alter expression of LRRK1 [83,84], (ii) mutations in LRRK1 do not mimic the pathological effects of the equivalent mutations in LRRK2 [69], and (iii) LRRK2 possesses an inherent toxicity not matched by LRRK1, possibly due to the higher basal level of LRRK2 kinase activity [67,69]. Kinase assays of LRRK2 demonstrate robust autophosphorylation as well as phosphorylation of model substrates including LRRKtide, Nictide, myelin basic protein (MBP), and moesin in vitro [67,85]. LRRK1 also binds ATP, but autophosphorylation activity is much more modest [86] and is inactive against LRRK2 model peptides [67,85]. Therefore, although both LRRK1 and LRRK2 are at least recognizable as kinases they do not have precisely the same enzymatic function.
Though like many other kinases both proteins are basally phosphorylated [69], Reyniers et al. ascertained that different residues are phosphorylated in each protein by mapping LRRK1 and LRRK2 phosphosites in parallel [73]. One example is the LRRK2-specific binding of 14-3-3 mediated by phosphoresidues S910/S935 that are not found in LRRK1 [73,87,88]. Each protein also has distinct interaction partners. As well as 14-3-3 proteins, LRRK2 has interactions with a number of pre-synaptic proteins mediated by its WD40 domain [89] that do not appear to be shared with LRRK1. In contrast, LRRK1 specifically binds the adaptor protein Grb2 that has been shown to mediate complex formation between LRRK1 and BCR-ABL1, a fusion tyrosine kinase that causes leukemia [90]. Additionally, LRRK1 plays a role in cell division downstream of Polo-like kinase 1 (PLK1) as mitotic spindle orientation depends on centrosome maturation, which requires LRRK1-mediated phosphorylation of CDK5RAP2 [91]. LRRK1 also has a specific interaction with epidermal growth factor receptor (EGFR) [92] that is not shared with LRRK2 [73]. Finally, a study of Lrrk1 knockout mice presented evidence that severe osteopetrosis is observed in these mice because Lrrk1 regulates bone mass through interaction with the cellular Rous sarcoma oncogene (c-Src) signaling pathway, whereas Lrrk2 knockout mice do not display a bone phenotype [93]. These results are substantiated by a high-throughput screening of mouse gene knockouts that identified Lrrk1 as a gene affecting bone mass [94]. Again, these data support some distinctions between LRRK1 and LRRK2.
At the cellular level, both proteins are cytosolic or associated with membranous and vesicular structures [20,69,92,95–97]. LRRK1 has been proposed to regulate EGFR trafficking in endosomes [73,92] via phosphorylation of its substrate CLIP-170, a microtubule plus-end protein [98]. LRRK2 has also been linked to vesicular trafficking, but to date it appears to modulate distinct vesicular transport events related to vesicular sorting and/or the autophagy/lysosome system [46,54,73], likely mediated by protein interactions at the late endosome or the trans-Golgi network (TGN). So while LRRK1 and LRRK2 are both involved in vesicular events that fundamentally alter protein trafficking, it would initially appear that LRRK1 is directed more to the plasma membrane and early recycling endosomes while LRRK2 is involved in later endosomal events and potentially in retromer function. With this in mind, we will next discuss the attempts to model loss of function and PD-related alleles of LRRK2 in rodents.
Rodent models of LRRK2
The LRRK2 protein is highly conserved between humans, mice and rats, with over 88% identity in the amino acid sequences of each species [99] (Figure 1). Lrrk2 mRNA and protein are expressed throughout both mouse and rat brain with highest expression levels in cortex and striatum, and an overall distribution pattern similar to that of LRRK2 in the human brain [75,95,99–104]. Much like human LRRK2, mouse and rat Lrrk2 have been shown to localize in the cytoplasm and also to associate with membranous and vesicular structures within cells [95,97,101,105]. It has recently been confirmed that mouse Lrrk2 and human LRRK2 can form a cross-species heterodimer by co-transfection and immunoprecipitation, further supporting that they are functionally similar proteins [106].
There has been some debate as to whether or not Lrrk2 is present in the rodent substantia nigra as some studies did find nigral expression [75,95,101,103,104] and some did not [100,102], an issue which may be due to variation in antibody specificity and sensitivity. West et al. saw Lrrk2 expression in the substantia nigra in mice but not in rats and also pointed out that although the amino acid sequence of human and mouse LRRK2 is highly conserved, there are marked differences in the regulatory regions across species. These species-specific elements may lead to important distinctions in how the LRRK2 gene is expressed in different animals [99]. Because of these potentially important distinctions between how LRRK2 behaves in even relatively closely related species, we will discuss mouse and rat models separately, starting first with knockout models.
Loss of function models
Lrrk2 knockout mice
Because LRRK2 mutations are associated with PD, an important question is whether deletion of the LRRK2 gene influences neurodegeneration and any attendant behavioral outcomes. Several Lrrk2 knockout mouse models yielded animals that did not show loss of dopaminergic neurons (evaluated from 18–24 months of age) or abnormal locomotive behavior [82,107–110]. Another knockout model also showed no neuronal loss, but had a subtly enhanced motor performance in the rotarod test at 7 months and increased thigmotaxic behavior in the open field test at 7 and 16 months [83]. Collectively, these results suggest that expression of LRRK2 is not required for dopamine neuron survival in the adult brain.
In contrast to the modest effects in the brain, several studies have shown a reproducible kidney pathology in Lrrk2 knockout mice. Dramatic morphological changes in color, size, weight, and texture are reported in conjunction with a range of other observations including altered levels of autophagy markers [82,83,107,110], increased inflammatory response [82,83], indications of oxidative damage [82,110], and evidence of interplay between LRRK2 and α-synuclein [82,110] in kidney. The reason that kidneys are affected is proposed to be that LRRK2 expression is high, and LRRK1 expression is low, in this tissue. The relative expression of the two genes might therefore indicate some partial redundancy in other tissues.
The mechanism by which LRRK2 deficiency results in kidney pathology may relate to the autophagy pathway [111,112]. Observations using light and electron microscopy suggest the accumulation of vesicle-derived structures that might be related to autophagosomes or lysosomes. At the protein level, Tong et al. noted increased levels of p62, a ubiquitin-binding protein involved in autophagy, and significantly decreased levels of LC3-II in Lrrk2 knockout mouse kidneys at 20 months of age [82]. In a later study, it was noted that there are age-dependent alterations of these markers, with increased levels of LC3-II and decreased levels of p62 in kidneys of 7-month-old Lrrk2 knockout mice but decreased LC3-II and increased p62 in kidneys of 20-month old Lrrk2 knockout mice [110]. In an independent knockout line, increased levels of p62 and of mTOR (mammalian target of rapamycin), a key regulator of autophagy, were seen but the autophagy marker LC3-II was similar to wild-type in kidneys of Lrrk2 knockout mice aged 14 months [107]. Finally, Hinkle et al. found no evidence of biphasic changes, but instead observed consistently increased levels of LC3-II and p62 in the kidneys of 3-, 12-, and 18–20-month-old Lrrk2 knockout mice, suggesting a progressive increase in autophagic activity in the kidneys [83] (Table 1). Therefore, the majority of available evidence suggests some change in autophagy and/or lysosomal markers but the data is still inconsistent as to which changes are seen and the role of aging.
Table 1.
Rodent models organized by group and characterized by genetic construction, expression level of LRRK2 protein, and the presence, +, or absence, —, of neurodegeneration and/or behavioral phenotypes.
| REFERENCE | GENETIC CONSTRUCT |
LRRK2 PROTEIN EPXRESSION LEVEL |
PHENOTYPE | ||
|---|---|---|---|---|---|
| NEURODEGENERATION | MOTOR ACTIVITY | ||||
| MOUSE MODELS | Li 2010 | BAC LRRK2 WT | x 6 | — | ↑↑ |
| BAC LRRK2 G2019S | x 6 | — | — | ||
| Herzig 2011 | Lrrk2 KO | — | — | — | |
| Lrrk2 KI G2019S❖ | x 1 | — | — | ||
| Longo 2014 | Lrrk2 KI G2019S❖ | x 1 | — | ↑↑ | |
| Melrose 2010 | BAC LRRK2 WT | x 3.5 | — | — (with increased thigmotaxis) |
|
| BAC LRRK2 G2019S■ | x 2.5 | — | — | ||
| Winner 2010 | BAC LRRK2 G2019S■ | NR | NR | NR | |
| Lee 2010 | HSV-LRRK2 G2019S | NR | +++ | NR | |
| Lin 2009 |
tetO-LRRK2 G2019S/CamKII-tTA |
x 8-16 | — | ↑↑ | |
| Lrrk2 KO | — | — | — | ||
| Chen 2012 | CMVE-PDGFβ-LRRK2 G2019S |
NR | +++ | ↓↓ (+) | |
| Ramonet 2011 | CMVE-PDGFβ-LRRK2 G2019S |
x 3-5 | + | — | |
| CMVE-PDGFβ-LRRK2 R1441C |
x 3-5 | — | ↓ | ||
| Li 2009 | BAC LRRK2 R1441G′ | x 5-10 | — | ↓↓↓ (+) | |
| Bichler 2013 | BAC LRRK2 R1441G′ | NR | NR | ↓ | |
| Dranka 2013 | BAC LRRK2 R1441G′ | NR | — | ↓ | |
| Dranka 2014 | BAC LRRK2 R1441G′ | NR | NR | ↓ | |
| Tong 2009 | Lrrk2 KI R1441C | x 1 | — | — | |
| Maekawa 2012 | CMV-LRRK2 I2020T | x 1.3 | — | ↓↑ | |
| Andres-Mateos 2009 | Lrrk2 KO | — | — | — | |
| Tong 2010 | Lrrk2 KO* | — | — | NR | |
| Tong 2012 | Lrrk2 KO* | — | NR | NR | |
| Hinkle 2012 | Lrrk2 KO | — | — | ↑ (with increased thigmotaxis) |
|
| RAT MODELS | Ness 2013 | Lrrk2 KO† | — | NR | NR |
| Baptista 2013 | Lrrk2 KO† | — | NR | NR | |
| Dusonchet 2011 | rAd-LRRK2 WT | x 2 | — | NR | |
| rAd-LRRK2 G2019S | x 2 | + | NR | ||
| Zhou 2011 | tTA-LRRK2 G2019S; constitutive expression |
NR | — | — | |
| tTA-LRRK2 G2019S; temporal expression |
NR | — | ↑ | ||
| Lee 2015 | BAC LRRK2 G2019S | x 5-8 | — | ↓↑ | |
| Daher 2014 | Lrrk2 KO +rAAv α-synuclein |
— | — | NR | |
| wild-type +rAAv α-synuclein |
x 1 | + | NR | ||
| Daher 2015 | wild-type +rAAv α-synuclein |
x 1 | + | NR | |
| BAC LRRK2 G2019S +rAAv α-synuclein |
x 20-30 | ++ | NR | ||
Superscript symbols (❖, ■, ○, * and †) are used to denote rodents from the same line. Neurodegeneration is defined here as loss of TH+ cells. Up or down arrows are used to indicate an observed increase or decrease, respectively, in motor activity. (+) = phenotype reversed by administration of L-DOPA. NR = not reported.
Lrrk2 knockout mice have also been used to highlight a potential role for LRRK2 in the immune system. For example, Lrrk2 knockout mice are more sensitive than their wild type counterparts to inflammatory colitis induced by dextran sulfate sodium [113]. In contrast, the same animals are less sensitive to another inflammatory condition, experimental autoimmune uveitis [114]. It has been suggested [114] that the difference between these two studies relates to the relative contribution of the adaptive immune system compared to innate immunity, the latter being more important in experimental colitis. Supporting this idea, there is evidence from human systems that LRRK2 is responsive to signaling pathways relevant to innate immunity [88,115]. It is likely that the involvement of LRRK2 in both autophagy and responses to pathogens are related to each other as a form of autophagy, xenophagy, is important in the removal of pathogens. By extension, this may mean that inflammation plays a role in the pathology of human LRRK2-mediated PD (reviewed in [116]). Overall, these studies show that important insights into the normal biological function of Lrrk2 have been obtained from studies of knockout mice. We will next examine which aspects of function of this protein are conserved in rats.
Lrrk2 knockout rats
Similarly to Lrrk2 knockout mouse models, robust pathophysiological changes were detected in the kidneys of Lrrk2 knockout rats [117–119]. Gross morphological and histological alterations that appear to be progressive with age [118] were observed, implicating a range of pathogenic mechanisms including impairment of the autophagy-lysosomal pathway [118], disrupted immune function [117,119], and deregulated metabolism [117,118]. Biochemical profiling of Lrrk2 knockout rat kidneys indicated that their abnormally dark pigmentation is due to accumulation of hemoglobin and lipofuscin [119]. Pathophysiological alterations were also seen in rat lungs [118], liver [118], and spleen [117]. Therefore, although the underlying function of LRRK2 is not known, the function is likely to be highly conserved between rats and mice.
Interestingly, overexpression of human α-synuclein via adeno-associated virus-mediated transduction elicited dopaminergic neurodegeneration in wild-type rats but not in Lrrk2 knockout rats, suggesting that LRRK2 inhibition could have neuroprotective effects [120] (Table 1). This is consistent with results using inducible expression of of human α-synuclein A53T generated in the Lrrk2 knockout background that showed that LRRK2 expression is required for the toxic effects of α-ynuclein [108]. Diminished recruitment of proinflammatory myeloid cells was also observed in the Lrrk2 knockout rats, which would be consistent with a role of Lrrk2 in innate immunity leading to neurodegeneration as discussed above..
Models of dominant mutations
LRRK2 R1441C/G mouse models
The most dramatic behavioral phenotype in mice using pathogenic mutations in LRRK2 was generated using a BAC transgenic approach to express human R1441G LRRK2. These mice were hypokinetic beginning at 6 months of age, with motor activity deficits progressing to a visually apparent immobility by 10–13 months [121]. Additionally, the motor deficits could be rescued by treatment with the dopamine precursor L-DOPA (L-3,4-dihydroxyphenylalanine) or apomorphine, a non-selective D1 and D2 dopamine agonist. There was no loss of dopaminergic neurons, though the DA neurons had abnormally small cell bodies on average and reduced dendritic density in mice aged 9–10 months [121]. This line can be obtained from The Jackson Laboratory, but subsequent reports were not able to reproduce the originally observed motor dysfunction. Bichler et al. saw only subtle motor deficits after the age of 16 months in R1441G mice subjected to open field, rotarod, cylinder, and grip strength tests [122]. Dranka et al. measured defects in coordinated motor function by rotarod and pole tests in the mice by 15 months of age, but observed unaltered gross motor function in open field and gait tests. Re-examination of dopaminergic neurons revealed normal tyrosine hydroxylase staining in both substantia nigra and striatum of R1441G mice at 16 months of age [123,124] (Table 1). The reasons for this loss of phenotype are not clear, but could potentially relate either to genetic modifiers in the background of the original animals or a loss of copies of the integrated BAC that would then lead to lower expression levels of human LRRK2. It is likely that lower expression of mutant protein would not result in dramatic phenotypes, as a murine Lrrk2 R1441C knockin model also did not display dopaminergic neurodegeneration or abnormal spontaneous motor activity in mice up to 2 years of age [125].
LRRK2 G2019S mouse models
Only one mouse model has shown both dopaminergic neurodegeneration and motor dysfunction. Using neuronal-specific expression of human G2019S LRRK2, progressive deterioration of SNpc dopaminergic neurons was observed in mice. Reduced locomotor activity was evident via measurement of spontaneous ambulatory activity in an activity cage and performance of the pole test at 12 months, with a slightly more severe phenotype at 16 months. Furthermore, administration of L-DOPA reversed the hypoactive phenotype in 12-month-old G2019S mice [105]. A second group also generated a transgenic mouse line with expression of human LRRK2 G2019S driven by the same promoter. In this line, loss of DA neurons and reduced neurite density of remaining DA neurons were found in mice aged to 19–21 months, but the mice displayed normal locomotor activity at 6 and 15 months [126]. Another mouse model using viral-based expression of human LRRK2 G2019S showed a similar loss of dopaminergic neurons and reduced neurite density but no atypical motor phenotype [86].
However, a series of other G2019S mouse models reported normal dopaminergic neurons but with a range of behavioral abnormalities. One knock-in Lrrk2 G2019S mouse model exhibited a hyperkinetic phenotype and enhanced motor performance that could be reversed with administration of LRRK2 kinase inhibitors [127]. Expression of human G2019S LRRK2 in the forebrain results in normal performance in the rotarod test but significantly increased ambulatory activity in the open field test in mice [108]. Neither wild-type nor G2019S LRRK2 BAC transgenic mice aged to 7–8 months performed significantly differently from control non-transgenic animals in beam-crossing, gait, or negative geotaxis tests, but G2019S LRRK2 BAC transgenic mice did show reduced exploratory behavior (increased thigmotaxis) in the open field test [84]. BAC transgenic mice overexpressing murine Lrrk2 G2019S demonstrated normal locomotor activity in the open field, beam and gait tests at 6 and 12 months, but mice overexpressing murine Lrrk2 wild-type displayed hyperactivity and enhanced performance in these tests [128] (Table 1).
Although these studies have generally focused on the dopamine system, there are some reported abnormalities that could represent symptoms of disease preceding overt neuropathology. Several groups identified signs of altered dopamine neurotransmission [84,105,121,125,128,129], irregular immune response [86,123,124], and/or tau alterations [84,105,121,128]. Gastrointestinal dysfunctions beginning at 6 months of age [122] and olfactory impairment at 15 months of age [123,124] were observed in LRRK2 mice, although not all groups that assessed olfaction ability in LRRK2 mice saw a deficit [122,129]. How these findings may be relevant to human Parkinson’s disease patients is not yet clear; regardless, they could indicate promising new strategies for understanding and targeting disease mechanisms.
LRRK2 I2020T mouse model
In the only I2020T mouse model reported in the literature, human LRRK2 I2020T is expressed specifically in neurons [129]. No loss of dopaminergic neurons was observed in either young (10 weeks of age) or aged (18 months of age) mice, but the mice had an abnormal locomotor phenotype. A higher frequency of rearing was seen in 22-week-old transgenic human LRRK2 I2020T mice compared to non-transgenic controls. Additionally, transgenic mice subjected to the beam test (at 23 and 43 weeks of age) and the rotarod test (at 34 weeks of age) exhibited impaired locomotor ability. However, the phenotype did not persist in older animals as no significant difference was observed between transgenic and non-transgenic mice in the performance of the beam test at 73 weeks of age, or the rotarod test at 42 and 59 weeks of age [129] (Table 1).
LRRK2 G2019S rat models
Several models involving overexpression of human LRRK2 G2019S in rats have been described in the literature. Dusonchet et al. (2011) instigated neuron-specific expression of human wild-type or G2019S LRRK2 via adenoviral vector injection in the striatum of adult rats. Expression of wild-type LRRK2 did not induce significant neuronal loss in the rats, but animals overexpressing the G2019S LRRK2 mutant displayed progressive degeneration of nigral dopaminergic neurons up to 42 days after injection [130]. Temporal overexpression of human G2019S LRRK2 in 5-month-old inducible transgenic rats did not cause loss of DA neurons, though it did promote enhanced locomotor activity in the open field test in rats at 18 months of age. In contrast, animals constitutively overexpressing human LRRK2 G2019S mutant did not have an atypical behavioral phenotype, which may be indicative of an as yet undefined developmental compensatory mechanism [131]. In a human BAC LRRK2 G2019S transgenic rat model, there was no loss of DA neurons but the neuron cell bodies were significantly flatter than those in wild-type rats at 12 months of age. In a test of postural instability, these human LRRK2 G2019S transgenic rats showed increased instability compared to non-transgenic littermate controls at 8 months of age, but not at 4 or 12 months, and an increased number of rearings in a cylinder test at 12 months of age, but not at 4 or 8 months [132]. When coupled with adeno-associated viral-mediated overexpression of human α-synuclein, transgenic overexpression of human LRRK2 G2019S in rats induced exacerbated DA neurodegeneration as well as increased microglial activation and myeloid cell recruitment compared with wild-type rats [133]. LRRK2 kinase inhibition mitigated both neurodegeneration and neuroinflammation, supporting the proposal of LRRK2 as a modulator of neuroinflammatory response. Overall, these results show that in rats as in mice, the chronic expression of LRRK2 has relatively modest effects. However, the acute expression from viral vectors in rats, which might be accompanied by modest disruption of the brain parenchyma and transient inflammation, does result in a stronger phenotype.
LRRK2 animal models as a bridge from bench to bedside
The ultimate goal of the extensive research into LRRK2 biology is to develop targeted therapy for people living with PD. To date, most efforts in drug development for LRRK2-associated PD have related to the design of specific, potent, and brain permeable LRRK2 kinase inhibitors. Like any kinase inhibitor, no LRRK2 inhibitor has perfect selectivity and many are only poorly permeable across the blood-brain barrier [134]. Furthermore, recent results have suggested that LRRK2 kinase inhibitors can have effects on other organ systems that may produce detrimental effects [135]. Assuming that these technical concerns can be overcome, kinase inhibitors might be an appropriate treatment option for patients with the G2019S mutation as it is well established that G2019S LRRK2 has an increased kinase activity in vitro. However, other LRRK2 mutations apparently have kinase activity similar to wild type protein or even diminished phosphorylation activity [136,137] and therefore may require alternative pharmacological approaches [134]. For example, mutations in the ROC domain (R1441C/G/H) may be targeted with compounds that interfere with GTP binding [138,139]. Importantly for the current discussion, the optimal choice of animal models for each treatment modality stratified by pathogenic mutation would allow estimating in vivo efficacy of the drugs as well as revealing potential side effects more effectively during preclinical research prior to human trials.
It is important to be clear what one might expect from an optimal animal for LRRK2. In humans, it is clear that the consistent phenotype associated with mutations is progressive dopaminergic neuronal cell loss in the substantia nigra pars compacta with more variable α-synuclein pathology [140]. Therefore, an ideal animal model would replicate the biochemical effect of each mutation and have dopamine cell loss so that the mutations can be tied to a pathology that has face validity for the human condition. Unfortunately, this crucial phenotype has generally been lacking in many animal models of genetic forms of PD (see [141] for review). One way to provide a stronger effect on dopamine cell loss would be by addition of stressors such as a-synuclein as reported recently in rats [133]. Alternatively, it might be possible to examine intermediate phenotypes, likely autophagy or inflammation, that while not sufficient to induce toxicity in the lifetime of rodents might faithfully report the activity of LRRK2 in vivo.
CONCLUSIONS
This survey of the currently available literature has covered concepts related to distinctness and commonality for LRRK2 and its homologues within and between species. Our overall impression at this stage is that while LRRK1 and LRRK2 have importantly different properties they are likely involved in related processes in the cell. An analogy would be the larger family of Rab proteins – all have a similar biochemical function but due to targeting to different subcellular (and in some cases sub-organellar) compartments they can each have different effects in cells. In the case of LRRK1 and LRRK2, it is likely that each might impact similar pathways but in different mechanistic ways. We would therefore predict that knocking out both homologues in tissues would have additive effects rather than demonstrating epistasis and might result in phenotypes that overlap more with those seen in the invertebrate lrk-1/Lrrk knockouts than with single LRRK1 or LRRK2 knockouts. The orthologues from invertebrate species may subsume functions of both LRRK1 and LRRK2 in mammals.
Collectively these models have therefore produced substantial and consistent insights into the normal physiological functions of LRRK2 and its homologues across species. A major current focus is on the effects of LRRK2 on the autophagy-lysosome system, but these proteins are also likely involved in cytoskeletal regulation, mitochondrial function and protein translation. It is our view that each of these processes are likely related to each other either directly or indirectly.
All mutations in LRRK2 that cause PD in humans have the ability to produce toxic effects, which has been consistently shown in Drosophila models in particular. It is very likely that the toxic effects of mutant LRRK2 relate to its normal biochemical and cellular function(s), particularly to kinase activity. However, mutations do not appear to be simple loss of function as the knockout and mutation phenotypes differ dramatically, especially in vertebrate models. This leads to the most likely current interpretation that mutations are gain of function in some way that remains to be fully defined. Overall, the utility of animal models to dissect mechanisms related to human mutations is easily demonstrated by studies on LRRK2 and it is hoped that this will be useful in the development of therapeutic approaches in the near future.
Abbreviations
- BAC
bacterial artificial chromosome
- BBB
blood-brain barrier
- COR
C-terminus of ROC
- DA
dopaminergic
- EGFR
epidermal growth factor receptor
- L-DOPA
L-3,4-dihydroxyphenylalanine
- LRR
leucine rich repeat
- LRRK
leucine rich repeat kinase
- lrk-1
leucine-rich repeats-1, Ras-like domain, kinase
- PD
Parkinson’s disease
- ROC
Ras of complex proteins
- SNpc
substantia nigra pars compacta
Contributor Information
Rebekah G. Langston, Email: rebekah.langston@nih.gov.
Iakov N. Rudenko, Email: iakov.rudenko@stonybrookmedicine.edu.
Mark R. Cookson, Email: cookson@mail.nih.gov.
References
- 1.Ponsen MM, Stoffers D, Booij J, van Eck-Smit BL, Wolters Ec, Berendse HW. Idiopathic hyposmia as a preclinical sign of Parkinson’s disease. Ann. Neurol. 2004;56:173–181. doi: 10.1002/ana.20160. [DOI] [PubMed] [Google Scholar]
- 2.Xiao Q, Chen S, Le W. Hyposmia: a possible biomarker of Parkinson’s disease. Neurosci. Bull. 2014;30:134–140. doi: 10.1007/s12264-013-1390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blonder LX, Slevin JT. Emotional dysfunction in Parkinson’s disease. Behav. Neurol. 2011;24:201–217. doi: 10.3233/BEN-2011-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 6.Kumaran R, Cookson MR. Pathways to Parkinsonism Redux: convergent pathobiological mechanisms in genetics of Parkinson’s disease. Hum. Mol. Genet. 2015 doi: 10.1093/hmg/ddv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cookson MR. LRRK2 Pathways Leading to Neurodegeneration. Curr. Neurol. Neurosci. Rep. 2015;15:42. doi: 10.1007/s11910-015-0564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 9.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Zimprich A, Muller-Myhsok B, Farrer M, Leitner P, Sharma M, Hulihan M, Lockhart P, Strongosky A, Kachergus J, Calne DB, et al. The PARK8 locus in autosomal dominant parkinsonism: confirmation of linkage and further delineation of the disease-containing interval. Am. J. Hum. Genet. 2004;74:11–19. doi: 10.1086/380647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 14.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Taymans JM. The GTPase function of LRRK2. Biochem. Soc. Trans. 2012;40:1063–1069. doi: 10.1042/BST20120133. [DOI] [PubMed] [Google Scholar]
- 17.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 18.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp. Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korr D, Toschi L, Donner P, Pohlenz HD, Kreft B, Weiss B. LRRK1 protein kinase activity is stimulated upon binding of GTP to its Roc domain. Cell. Signal. 2006;18:910–920. doi: 10.1016/j.cellsig.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Carlessi R, Levin-Salomon V, Ciprut S, Bialik S, Berissi H, Albeck S, Peleg Y, Kimchi A. GTP binding to the ROC domain of DAP-kinase regulates its function through intramolecular signalling. EMBO Rep. 2011;12:917–923. doi: 10.1038/embor.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dihanich S, Civiero L, Manzoni C, Mamais A, Bandopadhyay R, Greggio E, Lewis PA. GTP binding controls complex formation by the human ROCO protein MASL1. FEBS J. 2014;281:261–274. doi: 10.1111/febs.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marin I. The Parkinson disease gene LRRK2: evolutionary and structural insights. Mol. Biol. Evol. 2006;23:2423–2433. doi: 10.1093/molbev/msl114. [DOI] [PubMed] [Google Scholar]
- 24.Marin I, van Egmond WN, van Haastert PJ. The Roco protein family: a functional perspective. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008;22:3103–3110. doi: 10.1096/fj.08-111310. [DOI] [PubMed] [Google Scholar]
- 25.Marin I. Ancient origin of the Parkinson disease gene LRRK2. J. Mol. Evol. 2008;67:41–50. doi: 10.1007/s00239-008-9122-4. [DOI] [PubMed] [Google Scholar]
- 26.Daniel G, Moore DJ. Modeling LRRK2 Pathobiology in Parkinson’s Disease: From Yeast to Rodents. Curr. Top. Behav. Neurosci. 2015;22:331–368. doi: 10.1007/7854_2014_311. [DOI] [PubMed] [Google Scholar]
- 27.Dimitriadi M, Hart AC. Neurodegenerative disorders: insights from the nematode Caenorhabditis elegans. Neurobiol. Dis. 2010;40:4–11. doi: 10.1016/j.nbd.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr. Biol. CB. 2007;17:592–598. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- 29.Yao C, Khoury R El, Wang W, Byrd TA, Pehek EA, Thacker C, Zhu X, Smith MA, Wilson-Delfosse AL, Chen SG. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson’s disease. Neurobiol. Dis. 2010;40:73–81. doi: 10.1016/j.nbd.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha S, Guillily MD, Ferree A, Lanceta J, Chan D, Ghosh J, Hsu CH, Segal L, Raghavan K, Matsumoto K, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan Y, Cao P, Smith MA, Kramp K, Huang Y, Hisamoto N, Matsumoto K, Hatzoglou M, Jin H, Feng Z. Dysregulated LRRK2 signaling in response to endoplasmic reticulum stress leads to dopaminergic neuron degeneration in C. elegans. PloS One. 2011;6:e22354. doi: 10.1371/journal.pone.0022354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolozin B, Saha S, Guillily M, Ferree A, Riley M. Investigating convergent actions of genes linked to familial Parkinson’s disease. Neurodegener. Dis. 2008;5:182–185. doi: 10.1159/000113697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng M, Gorelenkova O, Yang J, Feng Z. A liquid phase based C. elegans behavioral analysis system identifies motor activity loss in a nematode Parkinson’s disease model. J. Neurosci. Methods. 2012;204:234–237. doi: 10.1016/j.jneumeth.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z, Hamamichi S, Lee BD, Yang D, Ray A, Caldwell GA, Caldwell KA, Dawson TM, Smith WW, Dawson VL. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum. Mol. Genet. 2011;20:3933–3942. doi: 10.1093/hmg/ddr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SB, Kim W, Lee S, Chung J. Loss of LRRK2/PARK8 induces degeneration of dopaminergic neurons in Drosophila. Biochem. Biophys. Res. Commun. 2007;358:534–539. doi: 10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, Tang B, Zhao G, Pan Q, Xia K, Bodmer R, Zhang Z. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Mol. Neurodegener. 2008;3:3. doi: 10.1186/1750-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, Liu HP, Lin WY, Guo H, Lu B. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. J. Neurosci. Off. J. Soc. Neurosci. 2010;30:16959–16969. doi: 10.1523/JNEUROSCI.1807-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, Ren Q, Jiao Y, Sawa A, Moran T, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, et al. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 43.Martin I, Kim JW, Lee BD, Kang HC, Xu JC, Jia H, Stankowski J, Kim MS, Zhong J, Kumar M, et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell. 2014;157:472–485. doi: 10.1016/j.cell.2014.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dodson MW, Leung LK, Lone M, Lizzio MA, Guo M. Novel ethyl methanesulfonate (EMS)-induced null alleles of the Drosophila homolog of LRRK2 reveal a crucial role in endolysosomal functions and autophagy in vivo. Dis. Model. Mech. 2014;7:1351–1363. doi: 10.1242/dmm.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beilina A, Rudenko IN, Kaganovich A, Civiero L, Chau H, Kalia SK, Kalia LV, Lobbestael E, Chia R, Ndukwe K, et al. Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2626–2631. doi: 10.1073/pnas.1318306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, Marder KS, Honig LS, Clark LN, Small SA, et al. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77:425–439. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 49.Waschbusch D, Michels H, Strassheim S, Ossendorf E, Kessler D, Gloeckner CJ, Barnekow A. LRRK2 transport is regulated by its novel interacting partner Rab32. PloS One. 2014;9:e111632. doi: 10.1371/journal.pone.0111632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang CL, Lu YN, Wang HC, Chang HY. Genetic dissection reveals that Akt is the critical kinase downstream of LRRK2 to phosphorylate and inhibit FOXO1, and promotes neuron survival. Hum. Mol. Genet. 2014;23:5649–5658. doi: 10.1093/hmg/ddu281. [DOI] [PubMed] [Google Scholar]
- 51.Lin CH, Tsai PI, Wu RM, Chien CT. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ss. J. Neurosci. Off. J. Soc. Neurosci. 2010;30:13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J. Neurosci. Off. J. Soc. Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng CH, Guan MS, Koh C, Ouyang X, Yu F, Tan EK, O’Neill SP, Zhang X, Chung J, Lim KL. AMP kinase activation mitigates dopaminergic dysfunction and mitochondrial abnormalities in Drosophila models of Parkinson’s disease. J. Neurosci. Off. J. Soc. Neurosci. 2012;32:14311–14317. doi: 10.1523/JNEUROSCI.0499-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Linhart R, Wong SA, Cao J, Tran M, Huynh A, Ardrey C, Park JM, Hsu C, Taha S, Peterson R, et al. Vacuolar protein sorting 35 (Vps35) rescues locomotor deficits and shortened lifespan in Drosophila expressing a Parkinson’s disease mutant of Leucine-Rich Repeat Kinase 2 (LRRK2) Mol. Neurodegener. 2014;9:23. doi: 10.1186/1750-1326-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang D, Li T, Liu Z, Arbez N, Yan J, Moran TH, Ross CA, Smith WW. LRRK2 kinase activity mediates toxic interactions between genetic mutation and oxidative stress in a Drosophila model: suppression by curcumin. Neurobiol. Dis. 2012;47:385–392. doi: 10.1016/j.nbd.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 56.Hindle S, Afsari F, Stark M, Middleton CA, Evans GJ, Sweeney ST, Elliott CJ. Dopaminergic expression of the Parkinsonian gene LRRK2-G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Hum. Mol. Genet. 2013;22:2129–2140. doi: 10.1093/hmg/ddt061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venderova K, Kabbach G, Abdel-Messih E, Zhang Y, Parks RJ, Imai Y, Gehrke S, Ngsee J, Lavoie MJ, Slack RS, et al. Leucine-Rich Repeat Kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson’s disease. Hum. Mol. Genet. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- 58.Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, Wolozin B, Cookson MR. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PloS One. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanao T, Venderova K, Park DS, Unterman T, Lu B, Imai Y. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Hum. Mol. Genet. 2010;19:3747–3758. doi: 10.1093/hmg/ddq289. [DOI] [PubMed] [Google Scholar]
- 60.Dorval V, Mandemakers W, Jolivette F, Coudert L, Mazroui R, De Strooper B, Hebert SS. Gene and MicroRNA transcriptome analysis of Parkinson’s related LRRK2 mouse models. PloS One. 2014;9:e85510. doi: 10.1371/journal.pone.0085510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beilina A, Cookson MR. Genes associated with Parkinson’s disease: regulation of autophagy and beyond. J. Neurochem. 2015 doi: 10.1111/jnc.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arranz AM, Delbroek L, Van Kolen K, Guimaraes MR, Mandemakers W, Daneels G, Matta S, Calafate S, Shaban H, Baatsen P, et al. LRRK2 functions in synaptic vesicle endocytosis through a kinase-dependent mechanism. J. Cell Sci. 2014 doi: 10.1242/jcs.158196. [DOI] [PubMed] [Google Scholar]
- 63.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melser S, Lavie J, Benard G. Mitochondrial degradation and energy metabolism. Biochim. Biophys. Acta. 2015 doi: 10.1016/j.bbamcr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 67.Civiero L, Vancraenenbroeck R, Belluzzi E, Beilina A, Lobbestael E, Reyniers L, Gao F, Micetic I, De Maeyer M, Bubacco L, et al. Biochemical characterization of highly purified leucine-rich repeat kinases 1 and 2 demonstrates formation of homodimers. PloS One. 2012;7:e43472. doi: 10.1371/journal.pone.0043472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulte EC, Ellwanger DC, Dihanich S, Manzoni C, Stangl K, Schormair B, Graf E, Eck S, Mollenhauer B, Haubenberger D, et al. Rare variants in LRRK1 and Parkinson’s disease. Neurogenetics. 2014;15:49–57. doi: 10.1007/s10048-013-0383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greggio E, Lewis PA, van der Brug MP, Ahmad R, Kaganovich A, Ding J, Beilina A, Baker AK, Cookson MR. Mutations in LRRK2/dardarin associated with Parkinson disease are more toxic than equivalent mutations in the homologous kinase LRRK1. J. Neurochem. 2007;102:93–102. doi: 10.1111/j.1471-4159.2007.04523.x. [DOI] [PubMed] [Google Scholar]
- 70.Biosa A, Trancikova A, Civiero L, Glauser L, Bubacco L, Greggio E, Moore DJ. GTPase activity regulates kinase activity and cellular phenotypes of Parkinson’s disease-associated LRRK2. Hum. Mol. Genet. 2013;22:1140–1156. doi: 10.1093/hmg/dds522. [DOI] [PubMed] [Google Scholar]
- 71.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochem. J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jorgensen ND, Peng Y, Ho CC, Rideout HJ, Petrey D, Liu P, Dauer WT. The WD40 domain is required for LRRK2 neurotoxicity. PloS One. 2009;4:e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reyniers L, Del Giudice MG, Civiero L, Belluzzi E, Lobbestael E, Beilina A, Arrigoni G, Derua R, Waelkens E, Li Y, et al. Differential protein-protein interactions of LRRK1 and LRRK2 indicate roles in distinct cellular signaling pathways. J. Neurochem. 2014 doi: 10.1111/jnc.12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dachsel JC, Nishioka K, Vilarino-Guell C, Lincoln SJ, Soto-Ortolaza AI, Kachergus J, Hinkle KM, Heckman MG, Jasinska-Myga B, Taylor JP, et al. Heterodimerization of Lrrk1-Lrrk2: Implications for LRRK2-associated Parkinson disease. Mech. Ageing Dev. 2010;131:210–214. doi: 10.1016/j.mad.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taymans JM, Van den Haute C, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J. Neurochem. 2006;98:951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- 76.Taylor JP, Hulihan MM, Kachergus JM, Melrose HL, Lincoln SJ, Hinkle KM, Stone JT, Ross OA, Hauser R, Aasly J, et al. Leucine-rich repeat kinase 1: a paralog of LRRK2 and a candidate gene for Parkinson’s disease. Neurogenetics. 2007;8:95–102. doi: 10.1007/s10048-006-0075-8. [DOI] [PubMed] [Google Scholar]
- 77.Biskup S, Moore DJ, Rea A, Lorenz-Deperieux B, Coombes CE, Dawson VL, Dawson TM, West AB. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci. 2007;8:102. doi: 10.1186/1471-2202-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westerlund M, Belin AC, Anvret A, Bickford P, Olson L, Galter D. Developmental regulation of leucine-rich repeat kinase 1 and 2 expression in the brain and other rodent and human organs: Implications for Parkinson’s disease. Neuroscience. 2008;152:429–436. doi: 10.1016/j.neuroscience.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 79.Giesert F, Hofmann A, Burger A, Zerle J, Kloos K, Hafen U, Ernst L, Zhang J, Vogt-Weisenhorn DM, Wurst W. Expression analysis of Lrrk1, Lrrk2 and Lrrk2 splice variants in mice. PloS One. 2013;8:e63778. doi: 10.1371/journal.pone.0063778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J. Neurochem. 2009;111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 81.Haugarvoll K, Toft M, Ross OA, White LR, Aasly JO, Farrer MJ. Variants in the LRRK1 gene and susceptibility to Parkinson’s disease in Norway. Neurosci. Lett. 2007;416:299–301. doi: 10.1016/j.neulet.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 82.Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ, Shen J., 3rd Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hinkle KM, Yue M, Behrouz B, Dachsel JC, Lincoln SJ, Bowles EE, Beevers JE, Dugger B, Winner B, Prots I, et al. LRRK2 knockout mice have an intact dopaminergic system but display alterations in exploratory and motor co-ordination behaviors. Mol. Neurodegener. 2012;7:25. doi: 10.1186/1750-1326-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melrose HL, Dachsel JC, Behrouz B, Lincoln SJ, Yue M, Hinkle KM, Kent CB, Korvatska E, Taylor JP, Witten L, et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol. Dis. 2010;40:503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat. Chem. Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat. Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dzamko N, Inesta-Vaquera F, Zhang J, Xie C, Cai H, Arthur S, Tan L, Choi H, Gray N, Cohen P, et al. The IkappaB kinase family phosphorylates the Parkinson’s disease kinase LRRK2 at Ser935 and Ser910 during Toll-like receptor signaling. PloS One. 2012;7:e39132. doi: 10.1371/journal.pone.0039132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piccoli G, Onofri F, Cirnaru MD, Kaiser CJ, Jagtap P, Kastenmuller A, Pischedda F, Marte A, von Zweydorf F, Vogt A, et al. Leucine-rich repeat kinase 2 binds to neuronal vesicles through protein interactions mediated by its C-terminal WD40 domain. Mol. Cell. Biol. 2014;34:2147–2161. doi: 10.1128/MCB.00914-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Titz B, Low T, Komisopoulou E, Chen SS, Rubbi L, Graeber TG. The proximal signaling network of the BCR-ABL1 oncogene shows a modular organization. Oncogene. 2010;29:5895–5910. doi: 10.1038/onc.2010.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanafusa H, Kedashiro S, Tezuka M, Funatsu M, Usami S, Toyoshima F, Matsumoto K. PLK1-dependent activation of LRRK1 regulates spindle orientation by phosphorylating CDK5RAP2. Nat. Cell Biol. 2015;17:1024–1035. doi: 10.1038/ncb3204. [DOI] [PubMed] [Google Scholar]
- 92.Hanafusa H, Ishikawa K, Kedashiro S, Saigo T, Iemura S, Natsume T, Komada M, Shibuya H, Nara A, Matsumoto K. Leucine-rich repeat kinase LRRK1 regulates endosomal trafficking of the EGF receptor. Nat. Commun. 2011;2:158. doi: 10.1038/ncomms1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xing W, Liu J, Cheng S, Vogel P, Mohan S, Brommage R. Targeted disruption of leucine-rich repeat kinase 1 but not leucine-rich repeat kinase 2 in mice causes severe osteopetrosis. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2013;28:1962–1974. doi: 10.1002/jbmr.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brommage R, Liu J, Hansen GM, Kirkpatrick LL, Potter DG, Sands AT, Zambrowicz B, Powell DR, Vogel P. High-throughput screening of mouse gene knockouts identifies established and novel skeletal phenotypes. Bone Res. 2014;2:14034. doi: 10.1038/boneres.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 96.Higashi S, Biskup S, West AB, Trinkaus D, Dawson VL, Faull RL, Waldvogel HJ, Arai H, Dawson TM, Moore DJ, et al. Localization of Parkinson’s disease-associated LRRK2 in normal and pathological human brain. Brain Res. 2007;1155:208–219. doi: 10.1016/j.brainres.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 97.Rudenko IN, Kaganovich A, Hauser DN, Beylina A, Chia R, Ding J, Maric D, Jaffe H, Cookson MR. The G2385R variant of leucine-rich repeat kinase 2 associated with Parkinson’s disease is a partial loss-of-function mutation. Biochem. J. 2012;446:99–111. doi: 10.1042/BJ20120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kedashiro S, Pastuhov SI, Nishioka T, Watanabe T, Kaibuchi K, Matsumoto K, Hanafusa H. LRRK1-phosphorylated CLIP-170 regulates EGFR trafficking by recruiting p150Glued to microtubule plus ends. J. Cell Sci. 2015;128:385–396. doi: 10.1242/jcs.161547. [DOI] [PubMed] [Google Scholar]
- 99.West AB, Cowell RM, Daher JP, Moehle MS, Hinkle KM, Melrose HL, Standaert DG, Volpicelli-Daley LA. Differential LRRK2 expression in the cortex, striatum, and substantia nigra in transgenic and nontransgenic rodents. J. Comp. Neurol. 2014;522:2465–2480. doi: 10.1002/cne.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Melrose H, Lincoln S, Tyndall G, Dickson D, Farrer M. Anatomical localization of leucine-rich repeat kinase 2 in mouse brain. Neuroscience. 2006;139:791–794. doi: 10.1016/j.neuroscience.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 101.Higashi S, Moore DJ, Colebrooke RE, Biskup S, Dawson VL, Arai H, Dawson TM, Emson PC. Expression and localization of Parkinson’s disease-associated leucine-rich repeat kinase 2 in the mouse brain. J. Neurochem. 2007;100:368–381. doi: 10.1111/j.1471-4159.2006.04246.x. [DOI] [PubMed] [Google Scholar]
- 102.Galter D, Westerlund M, Carmine A, Lindqvist E, Sydow O, Olson L. LRRK2 expression linked to dopamine-innervated areas. Ann. Neurol. 2006;59:714–719. doi: 10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- 103.Simon-Sanchez J, Herranz-Perez V, Olucha-Bordonau F, Perez-Tur J. LRRK2 is expressed in areas affected by Parkinson’s disease in the adult mouse brain. Eur. J. Neurosci. 2006;23:659–666. doi: 10.1111/j.1460-9568.2006.04616.x. [DOI] [PubMed] [Google Scholar]
- 104.Melrose HL, Kent CB, Taylor JP, Dachsel JC, Hinkle KM, Lincoln SJ, Mok SS, Culvenor JG, Masters CL, Tyndall GM, et al. A comparative analysis of leucine-rich repeat kinase 2 (Lrrk2) expression in mouse brain and Lewy body disease. Neuroscience. 2007;147:1047–1058. doi: 10.1016/j.neuroscience.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 105.Chen CY, Weng YH, Chien KY, Lin KJ, Yeh TH, Cheng YP, Lu CS, Wang HL. (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ. 2012;19:1623–1633. doi: 10.1038/cdd.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miyajima T, Ohta E, Kawada H, Maekawa T, Obata F. The mouse/human cross-species heterodimer of leucine-rich repeat kinase 2: Possible significance in the transgenic model mouse of Parkinson’s disease. Neurosci. Lett. 2015 doi: 10.1016/j.neulet.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 107.Herzig MC, Kolly C, Persohn E, Theil D, Schweizer T, Hafner T, Stemmelen C, Troxler TJ, Schmid P, Danner S, et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet. 2011;20:4209–4223. doi: 10.1093/hmg/ddr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) J. Neurosci. Off. J. Soc. Neurosci. 2009;29:15846–15850. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol. Neurodegener. 2012;7:2. doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bravo-San Pedro JM, Gomez-Sanchez R, Niso-Santano M, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, Climent V, Lopez Maturanade Maturana R, Sanchez-Pernaute R, Lopez de Munain A, et al. The MAPK1/3 pathway is essential for the deregulation of autophagy observed in G2019S LRRK2 mutant fibroblasts. Autophagy. 2012;8:1537–1539. doi: 10.4161/auto.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yakhine-Diop SM, Bravo-San Pedro JM, Gomez-Sanchez R, Pizarro-Estrella E, Rodriguez-Arribas M, Climent V, Aiastui A, Lopez de deMunain A, Fuentes JM, Gonzalez-Polo RA. G2019S LRRK2 mutant fibroblasts from Parkinson’s disease patients show increased sensitivity to neurotoxin 1-methyl-4-phenylpyridinium dependent of autophagy. Toxicology. 2014;324:1–9. doi: 10.1016/j.tox.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 113.Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011;12:1063–1070. doi: 10.1038/ni.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wandu WS, Tan C, Ogbeifun O, Vistica BP, Shi G, Hinshaw SJH, Xie C, Chen X, Klinman DM, Cai H, et al. Leucine-Rich Repeat Kinase 2 (Lrrk2) Deficiency Diminishes the Development of Experimental Autoimmune Uveitis (EAU) and the Adaptive Immune Response. PloS One. 2015;10:e0128906. doi: 10.1371/journal.pone.0128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ, et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J. Immunol. Baltim. Md 1950. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dzamko N, Geczy CL, Halliday GM. Inflammation is genetically implicated in Parkinson’s disease. Neuroscience. 2015;302:89–102. doi: 10.1016/j.neuroscience.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 117.Ness D, Ren Z, Gardai S, Sharpnack D, Johnson VJ, Brennan RJ, Brigham EF, Olaharski AJ. Leucine-rich repeat kinase 2 (LRRK2)-deficient rats exhibit renal tubule injury and perturbations in metabolic and immunological homeostasis. PloS One. 2013;8:e66164. doi: 10.1371/journal.pone.0066164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baptista MA, Dave KD, Frasier MA, Sherer TB, Greeley M, Beck MJ, Varsho JS, Parker GA, Moore C, Churchill MJ, et al. Loss of leucine-rich repeat kinase 2 (LRRK2) in rats leads to progressive abnormal phenotypes in peripheral organs. PloS One. 2013;8:e80705. doi: 10.1371/journal.pone.0080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boddu R, Hull TD, Bolisetty S, Hu X, Moehle MS, Daher JPL, Kamal AI, Joseph R, George JF, Agarwal A, et al. Leucine-rich repeat kinase 2 deficiency is protective in rhabdomyolysis-induced kidney injury. Hum. Mol. Genet. 2015;24:4078–4093. doi: 10.1093/hmg/ddv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Daher JP, Volpicelli-Daley LA, Blackburn JP, Moehle MS, West AB. Abrogation of alpha-synuclein-mediated dopaminergic neurodegeneration in LRRK2-deficient rats. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9289–9294. doi: 10.1073/pnas.1403215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bichler Z, Lim HC, Zeng L, Tan EK. Non-motor and motor features in LRRK2 transgenic mice. PloS One. 2013;8:e70249. doi: 10.1371/journal.pone.0070249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dranka BP, Gifford A, Ghosh A, Zielonka J, Joseph J, Kanthasamy AG, Kalyanaraman B. Diapocynin prevents early Parkinson’s disease symptoms in the leucine-rich repeat kinase 2 (LRRK2R(1)(4)(4)(1)G) transgenic mouse. Neurosci. Lett. 2013;549:57–62. doi: 10.1016/j.neulet.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dranka BP, Gifford A, McAllister D, Zielonka J, Joseph J, O’Hara CL, Stucky CL, Kanthasamy AG, Kalyanaraman B. A novel mitochondrially-targeted apocynin derivative prevents hyposmia and loss of motor function in the leucine-rich repeat kinase 2 (LRRK2(R1441G)) transgenic mouse model of Parkinson’s disease. Neurosci. Lett. 2014;583:159–164. doi: 10.1016/j.neulet.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tong Y, Pisani A, Martella G, Karouani M, Yamaguchi H, Pothos EN, Shen J. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ramonet D, Daher JP, Lin BM, Stafa K, Kim J, Banerjee R, Westerlund M, Pletnikova O, Glauser L, Yang L, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PloS One. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Longo F, Russo I, Shimshek DR, Greggio E, Morari M. Genetic and pharmacological evidence that G2019S LRRK2 confers a hyperkinetic phenotype, resistant to motor decline associated with aging. Neurobiol. Dis. 2014;71:62–73. doi: 10.1016/j.nbd.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li X, Patel JC, Wang J, Avshalumov MV, Nicholson C, Buxbaum JD, Elder GA, Rice ME, Yue Z. Enhanced striatal dopamine transmission and motor performance with LRRK2 overexpression in mice is eliminated by familial Parkinson’s disease mutation G2019S. J. Neurosci. Off. J. Soc. Neurosci. 2010;30:1788–1797. doi: 10.1523/JNEUROSCI.5604-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maekawa T, Mori S, Sasaki Y, Miyajima T, Azuma S, Ohta E, Obata F. The I2020T Leucine-rich repeat kinase 2 transgenic mouse exhibits impaired locomotive ability accompanied by dopaminergic neuron abnormalities. Mol. Neurodegener. 2012;7:15. doi: 10.1186/1750-1326-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dusonchet J, Kochubey O, Stafa K, Young SM, Jr, Zufferey R, Moore DJ, Schneider BL, Aebischer P. A rat model of progressive nigral neurodegeneration induced by the Parkinson’s disease-associated G2019S mutation in LRRK2. J. Neurosci. Off. J. Soc. Neurosci. 2011;31:907–912. doi: 10.1523/JNEUROSCI.5092-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou H, Huang C, Tong J, Hong WC, Liu YJ, Xia XG. Temporal expression of mutant LRRK2 in adult rats impairs dopamine reuptake. Int. J. Biol. Sci. 2011;7:753–761. doi: 10.7150/ijbs.7.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee JW, Tapias V, Di Maio R, Greenamyre JT, Cannon JR. Behavioral, neurochemical, and pathologic alterations in bacterial artificial chromosome transgenic G2019S leucine-rich repeated kinase 2 rats. Neurobiol. Aging. 2015;36:505–518. doi: 10.1016/j.neurobiolaging.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Daher JP, Abdelmotilib HA, Hu X, Volpicelli-Daley LA, Moehle MS, Fraser KB, Needle E, Chen Y, Steyn SJ, Galatsis P, et al. Leucine-rich Repeat Kinase 2 (LRRK2) Pharmacological Inhibition Abates alpha-Synuclein Gene-induced Neurodegeneration. J. Biol. Chem. 2015;290:19433–19444. doi: 10.1074/jbc.M115.660001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rudenko IN, Chia R, Cookson MR. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson’s disease? BMC Med. 2012;10:20. doi: 10.1186/1741-7015-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fuji RN, Flagella M, Baca M, Baptista MA, Brodbeck J, Chan BK, Fiske BK, Honigberg L, Jubb AM, Katavolos P, et al. Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci Transl Med. 2015;7:273ra15. doi: 10.1126/scitranslmed.aaa3634. [DOI] [PubMed] [Google Scholar]
- 136.Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro. 2009:1. doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rudenko IN, Cookson MR. Heterogeneity of leucine-rich repeat kinase 2 mutations: genetics, mechanisms and therapeutic implications. Neurotherapeutics. 2014;11:738–750. doi: 10.1007/s13311-014-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li T, He X, Thomas JM, Yang D, Zhong S, Xue F, Smith WW. A novel GTP-binding inhibitor, FX2149, attenuates LRRK2 toxicity in Parkinson’s disease models. PLoS One. 2015;10:e0122461. doi: 10.1371/journal.pone.0122461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li T, Yang D, Zhong S, Thomas JM, Xue F, Liu J, Kong L, Voulalas P, Hassan HE, Park JS, et al. Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson’s disease cell and mouse models. Hum Mol Genet. 2014;23:6212–6222. doi: 10.1093/hmg/ddu341. [DOI] [PubMed] [Google Scholar]
- 140.Cookson MR, Hardy J, Lewis PA. Genetic neuropathology of Parkinson’s disease. Int. J. Clin. Exp. Pathol. 2008;1:217–231. [PMC free article] [PubMed] [Google Scholar]
- 141.Blesa J, Przedborski S. Parkinson’s disease: animal models and dopaminergic cell vulnerability. Front. Neuroanat. 2014;8:155. doi: 10.3389/fnana.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Winner B, Melrose HL, Zhao C, Hinkle KM, Yue M, Kent C, Braithwaite AT, Ogholikhan S, Aigner R, Winkler J, et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiol. Dis. 2011;41:706–716. doi: 10.1016/j.nbd.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]