Abstract
Previously, we described the AtGRF [Arabidopsis thaliana growth-regulating factor (GRF)] gene family, which encodes putative transcription factors that play a regulatory role in growth and development of leaves and cotyledons. We demonstrate here that the C-terminal region of GRF proteins has transactivation activity. In search of partner proteins for GRF1, we identified another gene family, GRF-interacting factor (GIF), which comprises three members. Sequence and molecular analysis showed that GIF1 is a functional homolog of the human SYT transcription coactivator. We found that the N-terminal region of GIF1 protein was involved in the interaction with GRF1. To understand the biological function of GIF1, we isolated a loss-of-function mutant of GIF1 and prepared transgenic plants subject to GIF1-specific RNA interference. Like grf mutants, the gif1 mutant and transgenic plants developed narrower leaves and petals than did wild-type plants, and combinations of gif1 and grf mutations showed a cooperative effect. The narrow leaf phenotype of gif1, as well as that of the grf triple mutant, was caused by a reduction in cell numbers along the leaf-width axis. We propose that GRF1 and GIF1 act as transcription activator and coactivator, respectively, and that they are part of a complex involved in regulating the growth and shape of leaves and petals.
Growth-regulating factor (GRF) genes occur in the genomes of all seed plants thus far examined (1-3). The deduced protein products of GRF genes contain, in their N-terminal region, the conserved QLQ and WRC domains, which define the GRF protein family. The QLQ domain shows similarity to the N-terminal part of the yeast SWI2/SNF2 protein (1-3). Because the N-terminal region of SWI2/SNF2 mediates the interaction with another component of the SWI2/SNF2 chromatin-remodeling complex in yeast (4), the QLQ domain of GRF may also function in protein-protein interactions (1-3). The WRC domain contains a functional nuclear localization signal and a DNA-binding motif consisting of the conserved spacing of three Cys and one His residues (1, 2). The C-terminal regions of GRF proteins diverge in length and amino acid sequence from each other but nevertheless have common features that are reminiscent of transcription factors. We suggested that the GRF proteins may be a previously undescribed class of transcription regulators (1-3).
The GRF proteins of Arabidopsis thaliana comprise a family of nine members and are involved in regulating the growth and development of leaves and cotyledons in a functionally redundant manner (2). Transgenic plants overexpressing GRF1 or GRF2 developed larger leaves than did wild-type plants, whereas triple null mutants of GRF1/2/3 had narrower leaves and fused cotyledons (2).
By using GRF1 as bait in a yeast two-hybrid screen, we identified an Arabidopsis transcription coactivator, GRF-interacting factor (AtGIF1). The loss-of-function gif1 mutant and transgenic plants with reduced expression of GIF1 developed small and narrow leaves and petals, which had fewer cells, indicating that GIF1 affects growth and morphology of leaves and petals by regulating cell numbers. We provide biochemical and genetic evidence that GRF1 and GIF1 act as transcription activator and coactivator, respectively, and form a functional complex.
Materials and Methods
Plant Materials and Growth Conditions. Arabidopsis plants were grown in soil in a growth chamber at 23°C in 16-h light/8-h dark cycles. The grf triple mutants were constructed in the Wassilewskija (Ws) ecotype (2), and the gif1 mutant was identified in the Columbia (Col) ecotype.
Plasmid Construction. Procedures for plasmid construction and primer sequences for PCR are available in Supporting Text and Table 1, respectively, which are published as supporting information on the PNAS web site.
Transactivation and Protein-Protein Interaction Assays in Yeast Cells. For transactivation assay, various plasmid constructs for GRF and its segments fused to the GAL4-DNA-binding domain (DBD) were introduced into yeast HF7c cells. Growth and β-galactosidase activities, which are indicators for transcriptional activation of the HIS3 and LacZ reporter genes, were determined as described in the Yeast Protocols Handbook of Clontech. For the protein-protein interaction assay, plasmid constructs for the GAL4-DBD fusion of the truncated form of GRF1 and GRF2, which lack the last 44 and 35 aa, respectively, were introduced into yeast together with an empty partner vector or a construct for the GAL4 activation domain (AD) fused to GIF1. Yeast transformants were subjected to the growth and β-galactosidase assays.
Yeast Two-Hybrid Screening. Yeast HF7c cells expressing, as bait, the truncated form of GRF1 lacking the last 44 amino acids and fused to GAL4-DBD were transformed with a cDNA expression library that was constructed to express prey proteins fused to GAL4-AD. Transformants carrying the bait-prey interaction were selected according to the MatchMaker GAL4 Two-Hybrid User Manual of Clontech, except that we screened at 25°C instead of 30°C.
RNA Gel Blot Analysis. Membrane filters previously used to determine the expression pattern of GRF genes (2) were rehybridized with GIF-specific probes.
In Vitro Binding Assay. Plasmid constructs for the GRF1-hemagglutinin (HA) epitope tag and for GIF1-GST fusion protein were introduced to Escherichia coli BL21 cells. Expression was induced with 0.1 mM isopropyl-1-thio-β-D-galactoside for 3 h at 28°C. Fifty milliliters of cultured cells expressing GST-GIF1 or GST was extracted in 10 ml of PBS buffer by sonication. After centrifugation, 3 ml of supernatant containing the GST-GIF1 fusion or GST proteins was applied to glutathione (GSH)-Sepharose 4B columns (Amersham Pharmacia Biosciences) followed by five washes with PBS. Fifty milliliters of cultured cells expressing HA-tagged protein was extracted in binding buffer [10 mM Hepes/1 mM KCl/0.4 M MgCl2/60 mM NaCl/0.8 mM EDTA/8% (vol/vol) glycerol, pH 7.9], and 3 ml of supernatant was applied to the column charged with GST-GIF1 fusion protein or GST. After being washed five times with binding buffer, HA-tagged proteins retained in the column were eluted with 600 μl of GSH solution according to the manufacturer's instructions (Bulk GST Purification Modules, Amersham Pharmacia Biosciences). Twenty microliters of the eluate was subjected to SDS/PAGE and transferred to a nitrocellulose membrane, which was probed with anti-HA antibody conjugated to peroxidase (Roche Molecular Biochemicals). X-ray film was exposed to the membrane according to the enhanced chemiluminescence protocol (Amersham Pharmacia Biosciences).
Transient Expression in Onion Epidermal Cells. A biolistic PDS-1000/He particle gun (Bio-Rad) was used to transiently transform onion epidermal cells with the GIF1-EYFP fusion gene. After bombardment, cell layers were incubated for 24 h at 28°C in the dark on Murashige and Skoog (MS) agar plates [0.5× MS salts (GIBCO/BRL); 0.05% (wt/vol) Mes-KOH (pH 5.7); 1% (wt/vol) sucrose; 0.8% (wt/vol) phytoagar (GIBCO/BRL)]. Fluorescence of cells was observed under a Zeiss Axiophot microscope.
Genotyping of a T-DNA Insertion Mutant of the AtGIF1 Gene. A T-DNA insertion line, Salk_150407, for the GIF1 gene was obtained from the Salk T-DNA collection (5). The T-DNA insertion site was determined by sequencing PCR products amplified with the T-DNA- and GIF1-specific forward primers. The homozygosity of the insertion was confirmed by genotyping with gene-specific primers.
Preparation of AtGIF1-RNA Interference (RNAi) Transgenic Plants. The GIF1-RNAi plasmid was introduced into Arabidopsis by Agrobacterium-mediated transformation (6). Seventeen independent transgenic lines were selected on MS agar plates containing 50 μg/ml kanamycin. Homozygous transgenic plants of the T3 generation were selected and studied.
Measurement of Dimensional Parameters of Leaves and Petals. Digital images of detached leaves and mature petals were acquired, and the surface area, length, and width were determined with the image-analyzing program scionimage (Scion, Frederick, MD).
Numbers and Size of Leaf-Blade and Petal Cells. Leaf tissues were cleared with chloral hydrate/glycerol/water (8:1:2), as described (7). Micrographs of the abaxial petal epidermis were obtained by scanning electron microscopy (2). The numbers and size of leaf palisade and petal cells in the maximum-width region were determined with the scionimage software.
Results
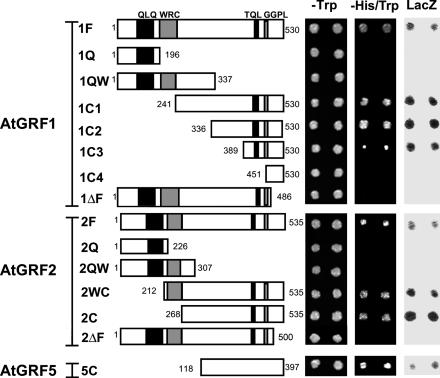
AtGRF Proteins Act As Transcriptional Activators. The full-length GRF1 and GRF2 proteins fused to GAL4-DBD activated the expression of the HIS3 and LacZ reporter genes, enabling the yeast transformants to grow on medium lacking His and to show a positive β-galactosidase reaction (Fig. 1, constructs 1F and 2F). This result indicates that GRF1 and GRF2 act as transcription activators. Further dissection of GRF1 and GRF2 proteins showed that the transactivation activity was present in the C-terminal region of both GRF1 (1C1, 1C2, and 1C3) and GRF2 (2WC and 2C) but not in the N-terminal region containing the QLQ or WRC domain (1Q, 1QW, 2Q, and 2QW). However, full-length GRF1 and GRF2 lacking the last 44- and 35-aa residues, respectively, did not show any activity (1ΔF and 2ΔF) and neither did the last 80-aa fragment of GRF1 (1C4). These results show that the terminal amino acid residues of GRF1 and GRF2 are essential although not sufficient for transactivation activity. The TQL and GGPL motifs present in the C-terminal region of GRF1-4, OsGRF1, and in some of other rice homologs (1-3) are not essential for transactivation activity, because the complete C-terminal fragment of GRF5, which does not contain any of these motifs, still possesses a significant level of transactivation activity (Fig. 1, construct 5C).
Fig. 1.
AtGRF proteins show transactivation activity in the yeast GAL4 system. DNA fragments encoding the depicted portions of GRF1 (1F-1ΔF), GRF2 (2F-2ΔF), and GRF5 (5C) were fused to the DNA sequence encoding GAL4-DBD and introduced into yeast cells. Two independent transformants for each fusion construct were tested for their ability to activate the expression of the HIS3 and LacZ reporter genes. -Trp and -His/Trp denote the colonies grown on plates lacking Trp and His/Trp, respectively; LacZ denotes β-galactosidase activities on filters after a colony-lift assay.
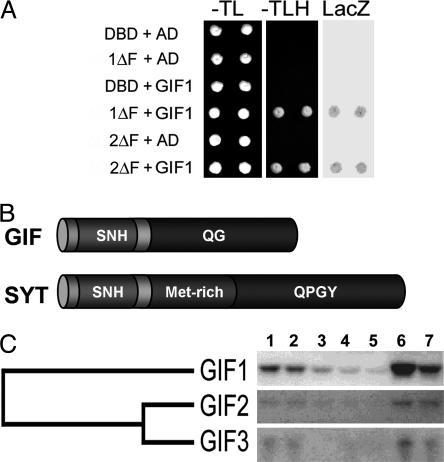
Isolation and Characterization of AtGRF1-Interacting Factors (AtGIFs). To identify the partner protein(s) that interact with GRF1, we performed the yeast two-hybrid screen by using, as bait, GRF1 lacking the last 44 amino acid residues (ΔGRF1; Fig. 1, 1ΔF fragment). The ΔGRF1 bait enabled us to select candidate yeast colonies without interference caused by autotransactivation activity of the full-length GRF1. Yeast cells harboring the bait were transformed with a cDNA library, which contains inserts for prey proteins fused to GAL4-AD. From 2.2 × 106 colonies, we selected 193 that were positive for the expression of the HIS3 and LacZ reporter genes. Among the first 12 preys, all except one contained coding sequences for two related genes, which, together with a third, form a small gene family in Arabidopsis. We annotated this gene family as GRF-Interacting Factor (GIF). Ten of these 12 clones corresponded to GIF1, one to GIF2, and one to a gene encoding a putative subunit of Gly decarboxylase. When the remaining 181 positive clones were analyzed by DNA gel blotting with a GIF1-specific probe, only 13 did not hybridize. Sequencing of the clones that did not hybridize to the GIF1 probe showed that four represented GIF2 and nine various other genes that appeared only once in this collection. Thus, of the 193 interacting proteins, 178 were encoded by GIF1 and 5 by GIF2, which is strong evidence that GIF1 and GIF2 are partner proteins of GRF1. Yeast cells expressing both ΔGRF1 and GIF1 grew on His-deficient medium and gave the color reaction for β-galactosidase activity, but none of the other combinations of ΔGRF1 or GIF1 gave a positive response, thereby confirming the screening result (Fig. 2A). We also found that ΔGRF2 interacted with GIF1.
Fig. 2.
Characterization of the AtGIF gene family. (A) Protein-protein interaction studies between GRF1 and GIF1 proteins in the yeast two-hybrid system. The 1ΔF or 2ΔF construct (see Fig. 1) was introduced into yeast cells together with the construct for GIF1 fused to GAL4-AD. As control, empty vectors containing inserts only for DBD or AD were also introduced with each of the 1ΔF, 2ΔF, and GIF1 constructs. -TL and -TLH denote the colonies grown on plates lacking Trp/Leu and His/Leu/Trp, respectively; and LacZ indicates β-galactosidase activity. (B) Schematic comparison of the domain structures of the GIF and SYT proteins. (C) Tissue-specific expression of GIF genes. Lanes: 1, root tips of seedlings; 2, uppermost stem; 3, internode; 4, fully grown leaves; 5, young leaves; 6, shoot tips containing the shoot apical meristem and early flower buds; 7, mature flowers. A phylogenetic tree derived from amino acid sequence shows the relatedness between GIF proteins.
Searching databases with the amino acid sequence of GIF1 as query, we found that GIF proteins are similar to the human synovial translocation (SYT) protein (Fig. 2B, and see Fig. 8, which is published as supporting information on the PNAS web site). The SYT protein is a transcription coactivator whose biological function has not yet been elucidated (8, 9). It has three distinct regions: the SNH domain near the N terminus, the QPGY domain in the C-terminal region, and the Met-rich region between the SNH and QPGY domains (Fig. 2B) (8). The overall amino acid identity of GIF proteins to SYT is low, ranging from 24% to 28%. However, the N-terminal region of GIF displays 53-57% amino acid identity to the SNH domain of the SYT protein. Although the C-terminal region of the GIF proteins did not align with that of SYT in a colinear manner, the amino acid composition of the C-terminal region, nevertheless, is similar enough to indicate a high level of relatedness between SYT and the GIF proteins (Fig. 8). GIF proteins and SYT share an unusually high frequency of Gln (19% in SYT and 13-17% in GIFs) and Gly (15% in SYT and 10-15% in GIFs). The GIF proteins, however, lack the repetitive occurrence of Pro and Tyr residues that are abundant in the QPGY domain of SYT (8). Therefore, we call the Gln/Gly-rich region of GIFs the QG domain (Fig. 2B). A comparison of the GIF proteins shows 61% amino acid identity between GIF2 and GIF3 but only 31% between GIF1 and the other two GIF proteins because of the divergent C-terminal regions. GIF1 was strongly expressed in roots, upper stems, shoot tips containing the shoot apical meristem, flower buds, and mature flowers but weakly in leaves and mature stems (Fig. 2C). GIF2 and GIF3 were also expressed with a pattern similar to that of GIF1 but at a much lower level. To compare the tissue-specific expression pattern of GRF1 and GIF1 in more detail, we compiled the microarray data by using the Affymetrix Arabidopsis ATH1 GeneChip (www.arabidopsis.org). The expression pattern of GRF1 matched exactly that of GIF1 in all tissues and at all developmental stages (Fig. 9, which is published as supporting information on the PNAS web site).
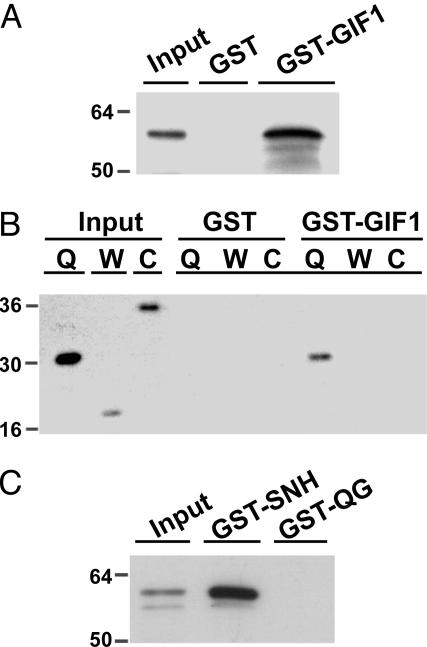
AtGRF1 and AtGIF1 Interact Directly by means of the QLQ and SNH Domains. We used an in vitro test to confirm the interaction of GRF1 and GIF1 in the yeast GAL4 system. GIF1 and fragments containing its SNH or QG domains were expressed as GST fusion proteins and were bound to glutathione (GSH)-Sepharose columns. GRF1 or fragments containing its QLQ, WRC, or C-terminal domains tagged with the HA epitope were loaded onto these affinity columns, which were then washed repeatedly to remove nonspecifically bound protein and eluted subsequently with buffer containing GSH. The eluates were analyzed by immunoblotting with antibody against the HA epitope as probe. The GRF1 protein was retained only on the GST-GIF1 column but not on a GST column (Fig. 3A), confirming direct interaction between GRF1 and GIF1. Among the fragments containing the QLQ, WRC, or C-terminal domains of GRF1, only that containing the QLQ domain was able to bind to GIF1 (Fig. 3B). Only the SNH but not the QG domain of GIF1 was able to bind GRF1 (Fig. 3C).
Fig. 3.
In vitro column-binding assay. (A and B) HA-tagged full-length GRF1 protein (A) or HA-tagged GRF1 fragments (B) were applied to a GST or GST-GIF1 column, as indicated. After extensive washing with the binding buffer to remove nonspecifically bound protein, HA-tagged GRF1 or its fragments retained on the column were eluted and subjected to immunoblot analysis with anti-HA antibody. Q, W, and C above the lanes indicate the GRF1 fragments containing the QLQ, WRC, and C-terminal regions, respectively. (C) The full-length HA-GRF1 protein was applied to a GST-SNH or GST-QG fusion column followed by immunoblot analysis with anti-HA antibody. “Input” indicates the signal elicited by 1% of applied protein as a control.
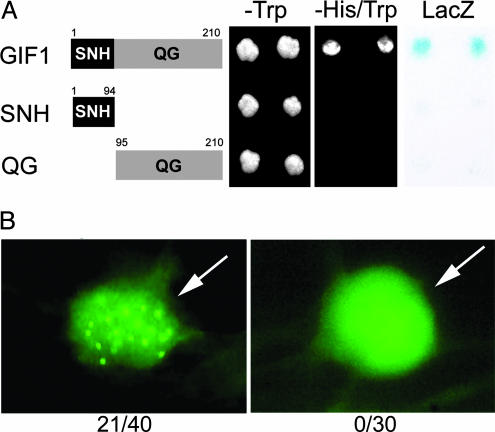
AtGIF1 Possesses Transactivation Activity and Forms Nuclear Speckles. Because the human SYT protein was shown to have transactivation activity in the QPGY domain (8), we examined whether GIF1 also has this activity by using the yeast GAL4 system. When fused to GAL4-DBD, the full-length GIF1 proteins activated the transcription of the HIS3 and LacZ reporter genes (Fig. 4A). However, neither the SNH (1-94) nor QG (95-210) domain showed transactivation activity.
Fig. 4.
Molecular activities of GIF1 proteins. (A) Transactivation assay in the yeast GAL4 system. DNA encoding full-length GIF1 or its segments, as depicted, was fused to DNA encoding GAL4-DBD and introduced into yeast cells. See Fig. 1 legend for procedures and labeling. (B) GIF1-EYFP fusion proteins form nuclear speckles. Onion epidermal cells were transformed transiently with the plasmid construct for the GIF1-EYFP fusion protein or empty vector for EYFP alone, as indicated. Fluorescence of cells was observed 24 h after incubation. Arrows indicate nucleus, and the numbers at the bottom denote the number of cells exhibiting nuclear speckles per the number of cells examined.
The SYT protein was found to form nuclear speckles, which colocalized with the human SWI2/SNF2-type protein BRAHMA (8). Onion epidermal cells transformed transiently to express GIF1 fused to enhanced yellow fluorescent protein (EYFP) also showed nuclear speckles, which were distributed throughout the nucleus in 21 of 40 transformants, but EYFP alone did not, among 30 transformants examined (Fig. 4B). We failed, for unknown reasons, to express in onion cells GRF1 proteins fused to various fluorescent proteins, such as EYFP, GFP, and cyan fluorescent protein. However, OsGRF1 fused to β-glucuronidase was shown to be targeted to the nucleus of onion epidermal cells (1).
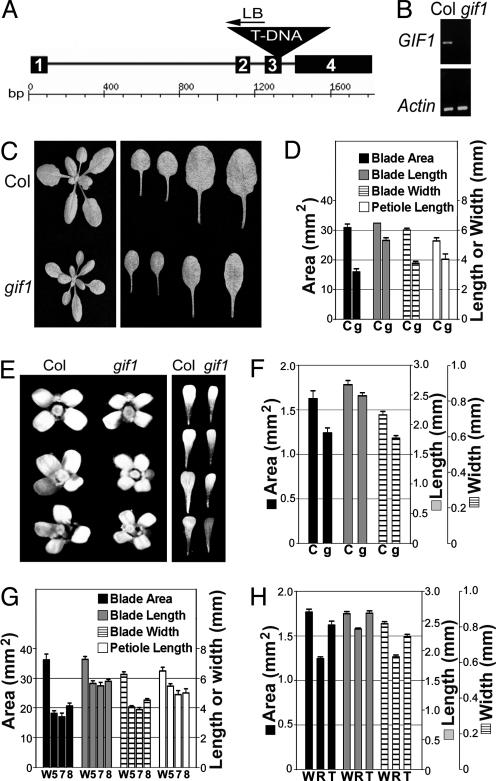
Isolation and Genetic Characterization of the Atgif1 Mutant. To investigate the biological function of the GIF1 protein, we isolated a homozygous T-DNA insertional mutant, gif1, by PCR-assisted genotyping. Sequencing of the PCR products showed that the T-DNA was inserted between the last two nucleotides of the third exon (Fig. 5A). By performing RT-PCR on RNA prepared from the gif1 mutant by using gene-specific primers flanking the T-DNA insertion site, we confirmed that there was no GIF1 mRNA in the mutant (Fig. 5B). Genetic analysis combined with genotyping of F2 progeny from a cross between the gif1 mutant and wild-type plant established that the mutant phenotype (see below) and T-DNA insertion cosegregated, and that the gif1 phenotype was inherited as a single recessive mutation (data not shown).
Fig. 5.
The phenotypes of Atgif1 mutant and AtGIF1-RNAi transgenic plants. (A) Representation of the gif1 T-DNA insertional mutation. Black boxes, exons; solid line, introns; inverse triangle, T-DNA integration site. The ruler is a scale of base pairs. (B) RT-PCR determination of GIF1 mRNA content in the mutant. Amplification of actin mRNA was used as control. (C) Col and gif1 plants (Left) and rosette leaves from the first to fourth leaf 20 d after germination (Right). (D) Dimensional parameters of the first two rosette leaves from Col and gif1 plants at 20 d. (E and F) Col and gif1 flowers and petals (E) and their dimensional parameters (F). (G and H) Dimensional parameters of the first two rosette leaves (G) and dimensional parameters of petals (H) from Ws and GIF1-RNAi plants. C, Col; g, gif1; W, Ws; R, GIF1-RNAi line 7; T, the grf1-2 grf2 grf3 triple mutant; and 5, 7, 8, GIF1-RNAi lines. Error bars indicate standard error.
The AtGIF1 Protein Is Involved in the Regulation of Leaf and Petal Shape. Compared to Col wild-type plants, gif1 mutants developed shorter and mainly narrower rosette leaves (Fig. 5C). The surface area of the first two mature rosette leaves was reduced by nearly 50% (Fig. 5D). This reduction in leaf area was primarily due to a nearly 40% decrease in leaf width. Leaf and petiole length were reduced by ≈20%. The reduction in the width of the other rosette and cauline leaves of gif1 mutants was similar to that of the first two rosette leaves, although there was only a marginal decrease in the length of the leaf blade and the petiole (Fig. 5C and data not shown). The gif1 mutant also displayed narrow petals (Fig. 5E). The area and width of the gif1 petals were decreased by 24% and 18%, respectively, and the length by 7% (Fig. 5F).
To verify the phenotype of the gif1 mutant, an RNAi construct designed to reduce the level of GIF1 mRNA was prepared and introduced into Ws wild-type plants. Fourteen of 17 independent RNAi transgenic lines showed a phenotype closely similar to that of the gif1 insertional mutant (Fig. 5 G and H). We selected the three strongest lines, 5, 7, and 8, to quantify the size of rosette leaves. These lines displayed 43-53% reduction in blade area with much greater decrease in blade width than in blade and petiole lengths. RT-PCR results showed that these lines had ≈10% of GIF1 mRNA in comparison to wild-type plants (data not shown). We used line 7 for further studies and observed a 30% decrease in petal area and a close to 20% decrease in petal width (Fig. 5H). The petal phenotype of the gif1 and RNAi plants prompted us to examine the petal size of a grf triple mutant (grf1-2 grf2 grf3) in the Ws background (2). The petals of the grf triple mutant were, indeed, slightly narrower than those of wild-type Ws, but there was no difference in petal length (Fig. 5H).
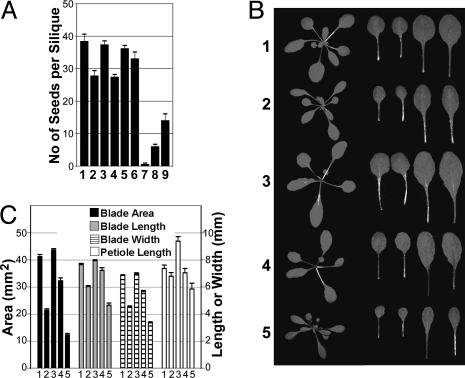
AtGIF1 and AtGRF Proteins Act in a Cooperative Manner. Because of the overlapping phenotype of the gif1 and grf triple mutants with respect to leaf size, it is conceivable that a grf1 grf2 grf3 gif1 quadruple mutant may show a stronger phenotype than did the grf1 grf2 grf3 triple mutants (2) or the gif1 single mutant (Fig. 5 C and D). To test this hypothesis, we tried to isolate a quadruple mutant from the F2 progeny of the cross between the grf1-2 grf2 grf3 triple mutant and gif1. However, we failed to get any quadruple mutant from 64 candidate plants that had narrower leaves than did the parental mutants. Instead, we were able to obtain different combinations of multiple mutants: grf1-2 grf3 gif1; grf1-2 grf2/GRF2 grf3/GRF3 gif1; grf1-2 grf2/GRF2 grf3 gif1; and grf1-2 grf2 grf3 gif1/GIF1. All these mutants were partially or completely sterile, as was the parental gif1 mutant, which produced 30% fewer seed than did GIF1 (Fig. 6A). The parental grf triple mutant, however, was as fertile as the wild-type plant. As the dosage of grf mutations in the homozygous gif1 background increased from grf1-2 grf3 gif1 through grf1-2 grf2/GRF2 grf3/GRF3 gif1 to grf1-2 grf2/GRF2 grf3 gif1, seed productivity decreased from 51% to 22% to 2% of that of the gif1 single mutant (Fig. 6A). These results indicate that the gif1 and grf mutations act synergistically to cause sterility. By contrast, a mutant heterozygous for the gif1 allele in the background of three homozygous grf mutations produced almost as many seeds as wild-type plants, indicating that the gif1 mutation plays a principal role in this synergism. We failed to get a quadruple mutant from 18 progeny of self-fertilized grf1-2 grf2 grf3 gif1/GIF1 mutants, even though the selfing should have produced homozygous quadruple mutants with a probability of 0.25. Sterility apparently was caused by lack of viable pollen (data not shown) and also by defective functioning of the female reproductive system, because sterility was not alleviated by pollination with wild-type pollen (data not shown). These results indicate that the GIF1 and GRF proteins are required for the production of viable pollen and normal functioning of the female reproductive system.
Fig. 6.
Cooperative effects of Atgif1 and Atgrf triple mutations. (A) Seed production. Bars: 1, Col; 2, gif1; 3, Ws; 4, GIF1-RNAi; 5, grf1-2 grf2 grf3; 6, grf1-2 grf2 grf3 gif1/GIF1;7, grf1-2 grf2/GRF2 grf3 gif1;8, grf1-2 grf2/GRF2 grf3/GRF3 gif1;9, grf1-2 grf3 gif1. (B) Leaf growth. (C) Bars 1, 2, and 3, are as in A. Bar 4, grf1-2 grf2 grf3; bar 5, grf1-2 grf2 gif1. Error bars indicate standard error.
To investigate the cooperative action of GRF and GIF1 proteins on leaf size, we chose the triple mutant grf1-2 grf3 gif1 because it displayed the strongest phenotype among the available multiple mutants. The triple mutant had much smaller leaves than did the parental grf triple and gif1 single mutants (Fig. 6 B and C). Its leaf area was reduced to one-half and one-third, respectively, of that of gif1 and the grf triple mutant, clearly showing an additive effect of both parental mutations.
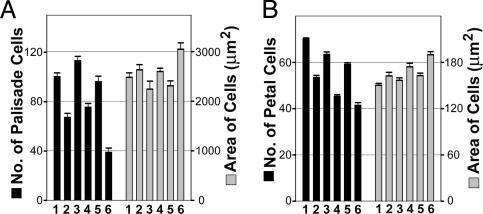
The Reduced Width of Leaves and Petals in gif1 Mutants and GIF1-RNAi Transgenic Plants Is Due to Decreased Cell Numbers. The numbers of leaf palisade and petal cells in a transverse line at the site of maximum leaf and petal width were determined. The cell numbers were reduced in both the gif1 and RNAi plants by ≈35% and 25% for the first two leaves and petal tissues (Fig. 7), respectively, which matches the reduction of width of the leaves and petals (Fig. 5). The cell size in gif1 and GIF1-RNAi leaves and petals, however, was slightly larger than that in the corresponding wild-type plants (Fig. 7), probably because of a compensatory mechanism determining cell number and size (10). Thus, the reduction in size of gif1 and RNAi leaves and petals results from a decrease in cell number and not in cell size. Furthermore, the grf1-2 grf3 gif1 triple mutant displayed an additional decrease in cell numbers compared to its parental mutants.
Fig. 7.
Number and area of cells in a transverse line at maximum width of leaves and petals. (A) Subepidermal palisade cells of the first two rosette leaves. (B) Abaxial epidermal cells of petals were analyzed. Bars: 1, Col; 2, gif1; 3, Ws; 4, GIF1-RNAi; 5, grf1-2 grf 2 grf3; 6, grf1-2 grf3 gif1.
Discussion
AtGRF1 and AtGIF1 Form a Functional Complex Involved in the Regulation of Cell Numbers in Leaves and Petals. GRF proteins function as transcription activators because Arabidopsis GRF1, GRF2, and GRF5 as well as the rice GRF1 showed transactivation activity (Fig. 1; ref. 3). This conclusion is supported by the presence of a putative DNA-binding domain and nuclear localization signal in GRF proteins (1-3).
GIF1 protein shows sequence similarity to SYT (Fig. 8), possesses transactivation activity, and forms nuclear speckles (Fig. 4), as does SYT, indicating that it is a functional homolog of human SYT and related proteins that are transcription coactivators (8, 9, 11). Therefore, GIF proteins may also act as transcription coactivators.
The yeast two-hybrid (Fig. 2) and in vitro binding assays (Fig. 3) showed that GRF1 and GIF1 are interacting partners. The biological significance of this interaction is supported by the phenotype of gif1 and grf mutants. In comparing the phenotypes of the gif1 and grf mutants, one has to keep in mind that there is a 9-fold redundancy and overlapping expression pattern of GRF genes (2). Although there are three GIF genes, it is very likely that GIF1 plays a predominant role, because the single null mutation or reduced expression of the GIF1 gene yields a stronger phenotype than do the grf triple mutations (Fig. 6), and because the expression level of GIF1 is much higher than that of the other two GIF genes (Fig. 2C). Whereas single grf mutants do not have an altered phenotype, the grf1/2/3 triple mutants and the gif1 single mutant, both form small narrow leaves and petals (Fig. 5 C-F and Fig. 6 B and C; ref. 2), which is due to reduced cell numbers (Fig. 7). There is an apparent inconsistency between our previous (2) and current results in that we could not ascertain before an effect of the grf triple mutation on cell numbers in the leaf blade. However, the cooperative effect of the grf and gif1 mutations on cell numbers along the transverse axis of the leaf blade shows clearly that GRF proteins also exert a control over cell proliferation affecting, thereby, leaf size and shape (Figs. 6 and 7). The similar expression pattern of GIF and GRF genes (Figs. 2 and 9; ref. 2) is further support for the notion that GIF and GRF interact in influencing the size and shape of leaves and petals as well as fertility.
Neither the grf1 grf3 gif1 triple mutant nor the gif1 and GIF1-RNAi plants developed fused cotyledons, which is a distinctive phenotype of the grf1/2/3 triple mutants (2). One possible explanation for this discrepancy is that the GRF-signaling pathway may diverge into two branches, one dependent on GIF for leaf and petal development and another independent of GIF for cotyledon development.
AtGIF and AtGRF Proteins Are Positive Regulators of Cell Proliferation Involved in Growth of Leaves and Petals. The angustifolia (an) mutant of Arabidopsis has narrow leaves of wild-type length, whereas the rotundifolia3 (rot3) mutant has shorter leaves of wild-type width (7). The phenotype of an and rot3 mutants was caused by reduced cell expansion along the transverse and longitudinal axes of the leaf, respectively. Thus, leaf shape is under the control of separate genetic pathways determining cell expansion in either the longitudinal or transverse direction (7, 12). The AN gene encodes a plant homolog of the animal transcription corepressor family CtBP (C-terminal binding protein) (13, 14) and the ROT3 gene a member of the cytochrome P450 family (15). Recently, Narita et al. (16) reported that a dominant Arabidopsis mutant, rot4-1D, formed short leaves caused by a reduction in cell numbers along the longitudinal axis; the ROT4 gene encodes a member of a novel peptide family. In contrast, the gif1 mutant developed narrow leaves, which was also due to a reduction of cell numbers, but along the transverse axis (Figs. 5 and 7). This result indicates that GIF1 is a component of the pathway controlling leaf growth by regulating cell proliferation in a transverse direction. Despite a weak phenotype of the grf triple mutation alone on cell numbers in leaves and petals, GRF proteins are also involved in the control of cell proliferation along the transverse axis (Fig. 7, and see above). Therefore, Arabidopsis appears to regulate the growth of leaf width by two independent mechanisms: the AN pathway is involved in controlling cell expansion and the GRF/GIF in cell proliferation.
Possible Molecular Roles of GIF. The SYT family exists in all metazoans examined, but its biological function has been elusive (8, 9). Because the SYT family proteins interact with SWI2/SNF2-chromatin remodeling proteins, it has been suggested that the interaction between them may regulate transcription by means of chromatin modification (8, 11, 17). Because GIF is a plant homolog of SYT, it may interact with plant SWI2/SNF2 to effect transcriptional activation of GRF target gene(s).
Supplementary Material
Acknowledgments
We thank Dr. Jae-Heung Ko (Michigan State University, East Lansing) for compiling the Affymetrix microarray data; Dr. Detlef Weigel (Max-Planck-Institut, Tübingen, Germany) for providing the cDNA expression library; Dr. Myeong Min Lee (University of Michigan, Ann Arbor) for the pGBT8 vector and a yeast cell line; Dr. Albrecht von Arnim (University of Tennessee, Knoxville) for a series of pAVA vectors; Dr. Peter Waterhouse (CSIRO Plant Industry, Canberra, Australia) for the RNAi vectors; and Katherine Krive and Samantha Warwick for excellent technical help. We also thank the Arabidopsis Biological Resource Center for the mutant seeds. This work was supported by U.S. Department of Energy Grant DE-FG-02-91ER20021.
Abbreviations: GRF, growth-regulating factor; GIF, GRF-interacting factor; Ws, Wassilewskija; Col, Columbia; DBD, DNA-binding domain; AD, activation domain; HA, hemagglutinin; RNAi, RNA interference; SYT, synovial translocation; EYFP, enhanced yellow fluorescent protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY102639, AY102640, and AY102641 for GIF1, GIF2, and GIF3, respectively).
References
- 1.van der Knaap, E., Kim, J. H. & Kende, H. (2000) Plant Physiol. 122, 695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim, J. H., Choi, D. & Kende, H. (2003) Plant J. 36, 94-104. [DOI] [PubMed] [Google Scholar]
- 3.Choi, D., Kim, J. H. & Kende, H. (2004) Plant Cell Physiol. 45, 897-904. [DOI] [PubMed] [Google Scholar]
- 4.Treich, I., Cairns, B. R., de los Santos, T., Brewster, E. & Carlson, M. (1995) Mol. Cell. Biol. 15, 4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 6.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 7.Tsuge, T., Tsukaya, H. & Uchiyama, H. (1996) Development (Cambridge, U.K.) 122, 1589-1600. [DOI] [PubMed] [Google Scholar]
- 8.Thaete, C., Brett, D., Monaghan, P., Whitehouse, S., Rennie, G., Rayner, E., Cooper, C. S. & Goodwin, G. (1999) Hum. Mol. Genet. 8, 585-591. [DOI] [PubMed] [Google Scholar]
- 9.Ladanyi, M. (2000) Oncogene 20, 5755-5762. [DOI] [PubMed] [Google Scholar]
- 10.Tsukaya, H. (2002) Curr. Opin. Plant Biol. 6, 57-62. [DOI] [PubMed] [Google Scholar]
- 11.Aizawa, H., Hu, S.-C., Bobb, K., Balakrishnan, K., Ince, G., Cowan, M. & Ghosh, A. (2004) Science 303, 197-202. [DOI] [PubMed] [Google Scholar]
- 12.Tsukaya, H. (2002) Plant Cell Physiol. 43, 372-378. [DOI] [PubMed] [Google Scholar]
- 13.Kim, G.-T., Shoda, K., Tsuge, T., Cho, K.-H., Uchimiya, H., Yokoyama, R., Nihitani, K. & Tsukaya, H. (2002) EMBO J. 21, 1267-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folkers, U., Kirik, V., Schöbinger, U., Falk, S., Krishnakumar, S., Pollock, M. A., Oppenheimer, D. G., Day, I., Reddy, A. R., Jürgens, G., et al. (2002) EMBO J. 21, 1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, G.-T., Tsukaya, H. & Uchimiya, H. (1998) Genes Dev. 12, 2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narita, N. N., Moore, S., Horiguchi, G., Kubo, M., Demura, T., Fukuda, H., Goodrich, J. & Tsukaya, H. (2004) Plant J. 38, 699-713. [DOI] [PubMed] [Google Scholar]
- 17.Kato, H., Tjernberg, A., Zhang, W., Krutchinsky, A. N., An, W., Takeuchi, T., Ohtsuki, Y., Sugano, S., de Bruijn, D. R., Chait, B. T., et al. (2002) J. Biol. Chem. 277, 5498-5505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.