Abstract
A reversion assay system previously implemented to demonstrate the existence of adaptive or stationary-phase-induced mutagenesis in Bacillus subtilis was utilized in this report to study the influence of the mismatch DNA repair (MMR) system on this type of mutagenesis. Results revealed that a strain deficient in MutSL showed a significant propensity to generate increased numbers of stationary-phase-induced revertants. These results suggest that absence or depression of MMR is an important factor in the mutagenesis of nongrowing B. subtilis cells because of the role of MMR in repairing DNA damage. In agreement with this suggestion, a significant decrease in the number of adaptive revertant colonies, for the three markers tested, occurred in B. subtilis cells which overexpressed a component of the MMR system. Interestingly, the single overexpression of mutS, but not of mutL, was sufficient to decrease the level of adaptive mutants in the reversion assay system of B. subtilis. The results presented in this work, as well as in our previous studies, appear to suggest that an MMR deficiency, putatively attributable to inactivation or saturation with DNA damage of MutS, may occur in a subset of B. subtilis cells that differentiate into the hypermutable state.
Adaptive or stationary-phase-induced mutagenesis occurs in nondividing cells during prolonged nonlethal selective pressure, e.g., starvation for an essential amino acid (25). While most of the research has involved Escherichia coli model systems, similar observations have been made in other prokaryotes (14, 30) as well as in eukaryotic organisms (8). The most widely studied system thus far has been the F′ lac frameshift-reversion construct of E. coli (25). In this system it has been demonstrated that generation of Lac+ stationary-phase-associated revertants is dependent on (i) a functional Rec system (10), (ii) F′ transfer functions (6), and (iii) a component(s) of the SOS system (18, 19). In addition, both DNA polymerase III and the SOS-regulated DNA polymerase IV (19) have been shown to be responsible for the synthesis of errors that lead to these mutations (for review of the SOS regulon see reference 33). More recent evidence demonstrates that the mutations generated by this lac system during stationary phase are the result of actual cell growth and amplification of the plasmid-borne gene that is followed by SOS-induced mutagenesis and selection (11, 28).
The existence of stationary-phase-induced mutagenesis was recently demonstrated in Bacillus subtilis following the utilization of a reversion assay system (30). In contrast to the F′ lac system of E. coli, this type of mutagenesis in B. subtilis is not dependent upon a functional RecA protein (i.e., recombination or the activation of type 1 SOS functions [35] was not required). Moreover, it was also demonstrated that generation of B. subtilis adaptive mutants did not require a functional σB factor (RNA polymerase σB controls the general stress response in B. subtilis [34]). However, one of the most relevant outcomes derived from studying adaptive mutagenesis in B. subtilis was the observation that transcription factors such as ComA and ComK (21) did influence the eventual production of stationary-phase-induced mutants (30). More recent evidence has demonstrated that a null mutation in yqjH, which encodes a homolog of the UmuC/DinB or Y superfamily of DNA polymerases, affected the generation of revertants of the hisC952 allele in the B. subtilis system (32). Thus, for the adaptive mutagenesis phenomenon of B. subtilis a hypothesis was advanced that basically proposed that during periods of environmental stress subpopulations are differentiated within a culture in order to generate genetic diversity (30). Furthermore, it was proposed that within some of these subpopulations mutation frequencies can be increased (hypermutability) by the suppression of DNA repair systems and/or the activation of mechanisms that would increase the introduction of DNA damage into the genome (30).
The MutS and MutL proteins of the mismatch DNA repair (MMR) system have been conserved across the domains of life and play important roles in several DNA repair pathways (20). In addition to mutation avoidance, MMR has been linked to the control of genome rearrangements (22), gene transfer among species (17), and the development of cancer (4, 23).
MMR was first linked to adaptive mutagenesis in the F′ reversion system of E. coli, following the observation that the type of mutants that gave the lac+ revertant genotype were almost all −1 deletions in a region of small mononucleotide repeats (5, 26). This was the category of growth-dependent revertants found in cells lacking a functional MMR system (16). These findings are apparently contradictory because production of adaptive lac+ mutants still occurs in wild-type cells where MMR is functional. Based on these collective observations, it was suggested that the function of the MMR system (namely, MutL) is in some way inhibited exclusively during the adaptive or stationary-phase-induced mutagenesis process (9). Further evidence showed that overproduction of MutL (but not MutS) decreased the number of mutations generated by the adaptive process(es) without affecting growth-dependent mutagenesis (9). In addition to these findings in E. coli, a homolog of the MutS protein has also been found to be involved in the generation of stationary-phase-induced mutants in Saccharomyces cerevisiae (8).
In the present communication, we took advantage of the reversion system of B. subtilis (30) to study the effects of the MMR proteins, MutS and MutL, on the production of stationary-phase-induced mutants in this bacterium. Our results demonstrate that a deficiency of the MMR system increases the number of revertants during the stationary phase of B. subtilis. On the other hand overexpression of the entire mutSL operon or just the mutS gene product (but not the mutL product) significantly diminished the production of stationary-phase-associated revertants for the three markers tested. Thus, our results support the hypothesis that at least one mechanism for the generation of adaptive mutants in B. subtilis is the loss or decrease of MMR activity during stationary phase.
MATERIALS AND METHODS
The bacterial strains used are listed in Table 1. B. subtilis YB955 is a prophage “cured” strain that contains the hisC952. metB5, and leuC427 alleles (30, 36, 37). Procedures for transformation and isolation of chromosomal and plasmid DNA were as described previously (1, 2, 27). B. subtilis strains were maintained on tryptose blood agar base medium (PB medium; Difco Laboratories, Detroit, Mich.). Liquid cultures of B. subtilis strains were routinely grown in NOB medium (nutrient broth; Oxoid) supplemented with 2% yeast extract (NOB-YE). Neomycin and kanamycin (KAN) were added as appropriate to final concentrations of 10 μg/ml. E. coli cultures were grown in Luria-Bertani medium supplemented with ampicillin to a final concentration of 100 μg/ml.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or phenotype | Source or reference |
|---|---|---|
| YB955 | hisC952 metB5 leuC427 xin-1 Sp−βSENS | 37 |
| MPRYB148 | YB955 containing pMPRYB146 | This studya |
| MPRYB149 | YB955 containing pMPRYB147 | This study |
| MPRYB150 | YB955 containing pDG148 | This study |
| MPRYB151 | YB955 carrying mutSL::neo | This study |
| MPRYB156 | YB955 containing pMPRYB155 | This study |
See Materials and Methods for description of construction.
Stationary-phase mutagenesis assays.
Where appropriate, strains (10-ml cultures) were grown in NOB-YE medium containing KAN (10 μg/ml) to an optical density, at 600 nm, of 0.5, and then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures to a final concentration of 5 mM in order to induce the transcription of the cloned genes. The stationary-phase mutagenesis assays were performed as previously described (30, 32). Essentially, cultures were grown in nephelometric flasks with aeration (250 rpm) at 37°C to 90 min after the cessation of exponential growth (designated T90). Growth was monitored with a Klett-Summerson colorimeter (no. 66 filter; Klett Manufacturing Co., Inc.; 1 Klett unit ≈ 106 CFU/ml). The cells were harvested by centrifugation at 10,000 × g for 10 min at room temperature and then resuspended in 10 ml of 1× Spizizen salt solution (29) in order to reduce the amount of trace nutrients. The cells were then plated in quintuplicate and incubated at 37°C on Spizizen minimal medium (1× Spizizen salts supplemented with 0.5% glucose and either 50 μg or 200 ng of the required amino acid/ml and 50 μg each of isoleucine and glutamic acid/ml). The selection medium was also supplemented with 2.5 mM IPTG/ml. The experiments were repeated at least three times.
In addition, each time that an isogenic derivative strain of YB955 was examined for its ability to perform stationary-phase-induced mutagenesis, a YB955 control was tested simultaneously. The concentration of the amino acid used depended on the reversion that was being selected. For instance, when His+ revertants were selected, 50 μg of methionine and leucine/ml was added to the medium and 200 ng of histidine/ml was added. Isoleucine and glutamic acid were added as described previously (31) in order to protect the viability of the cells. Where needed appropriate concentrations of antibiotics and/or IPTG were maintained throughout the experiments. The number of revertants was scored daily. The initial number of bacteria plated for each experiment was determined by serial dilution of the bacterial cultures and then by plating the cells on a minimal medium containing all three essential amino acids.
The survival rates of the bacteria plated on the minimal selective medium were determined as follows. Three agar plugs were removed from each selection plate daily. The plugs were removed with sterile Pasteur pipettes and taken from areas of the plates where no growth of revertants was observed. The plugs were suspended in 400 μl of 1× Spizizen salts mixed, diluted, and plated on Spizizen minimal medium containing all the essential amino acids (50 μg/ml). Again, the number of colonies was determined following 48 h of growth at 37°C.
Analysis of mutation rates.
The growth-dependent reversion frequencies for the His+, Met+, and Leu+ alleles were measured by fluctuation tests. In order to determine the mutation rates of various strains, the bacteria were grown to saturation at 37°C with aeration in PB medium. The saturated cultures were then used to make a 10−4-fold dilution into fresh PB medium and dispensed into 1 ml of PB medium. Thirty-eight 18-mm test tubes, each containing almost the same number of cells, were incubated overnight to saturation (about 14 to 16 h with aeration at 37°C). The saturated cultures were then pelleted and resuspended in 100 μl of 1× Spizizen salts medium. The cells were then spread onto selection medium as described previously for detecting His+, Met+, and Leu+ stationary-phase-generated mutations. The revertants were scored and recorded within 48 h after plating. The median (r) is the mean of the 19th and 20th values of r (observed number of mutants per culture) when the r values are ranked. The number of mutations per culture (m) is calculated with the Lea-Coulson formula, r/m − ln(m) = 1.24 (15). Three parallel cultures were used to determine the total number of CFU plated on each plate (Nt) by titration. The mutation rates were calculated as previously described with the formula m/2Nt (24, 30, 32).
Design of a plasmid to disrupt the mutSL operon.
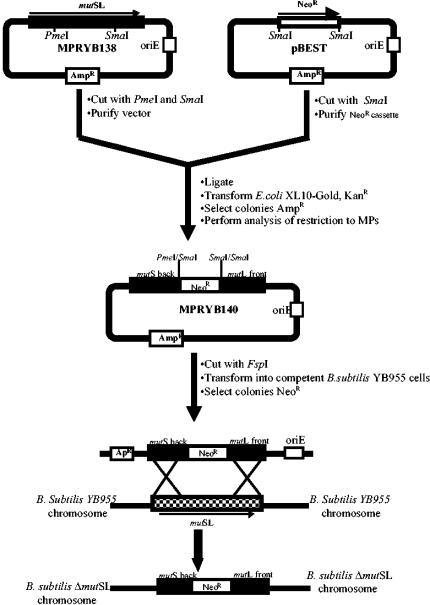
The mutSL (7) operon was amplified by PCR with use of 0.1 μg of chromosomal DNA from B. subtilis YB955 and the oligonucleotide primers 5′-GGCGGGATCCGCCGGTTATACGCCTATGATACAGC-3′ (forward) and 5′-ACGCGTCGACCACTGCTCACACGTTTACAACCG-3′. The primers were designed to insert BamHI and SalI sites, respectively (underlined). Amplification was performed with Vent DNA polymerase (New England Biolabs, Beverly, Mass.). The PCR fragment (4,777 bp) was ligated into SmaI-treated pUC19 to generate pMPRYB138, which was replicated into E. coli XL-10 Gold Kanr (Stratagene, La Jolla, Calif.). A 2.5-kbp SmaI/PmeI internal DNA fragment of the mutSL operon was released from pPERMYB138 and replaced with the neomycin cassette released from plasmid pBEST (13) with SmaI. The resulting construction (pMPRYB140) was replicated into E. coli XL-10 Gold Kanr. pMPRYB140 was linearized with FspI and then introduced by transformation into competent cells of B. subtilis YB955 to generate B. subtilis MPRYB151 with a disrupted mutSL operon.
The mutation rates of the ΔmutSL B. subtilis strain for the generation of rifampin-resistant cells were determined as follows. Essentially, the knockout mutSL strain was grown in PB medium supplemented with 10 μg of neomycin/ml for 12 h. Mutation frequencies were determined by plating aliquots on five Luria-Bertani medium plates containing rifampin to a final concentration of 5 μg/ml. The revertant colonies were counted after 1 day of incubation at 37°C to estimate mutation frequencies. The experiment was repeated at least two times.
Construction of plasmids to overexpress the mutS and mutL genes and the mutSL operon.
The open reading frames (ORFs) of mutS. mutL, and the mutSL operon were PCR amplified and cloned into the SalI/XbaI sites of pDG148, under the control of the IPTG-inducible pSPAC promoter. The plasmids were amplified in E. coli XL-10 Gold Kanr and termed pMPRYB146 (mutL), pMPRYB147 (mutS), and pMPRYB155 (mutSL) (Table 2). The three plasmids were introduced by transformation into the strain B. subtilis YB955, to generate the B. subtilis strains MPRYB148, MPRYB149, and MPRYB156, respectively. A control strain was also made by transforming the plasmid pDG148 into B. subtilis YB955 to generate B. subtilis MPRYB150.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pDG148 | spac expression vector; Ampr Kanr | W. L. Nicholson |
| pBEST | pGEM4 containing the neomycin resistance cassette under control of repU promoter of B. subtilis | 13 |
| pMPRYB146 | mutL ORF cloned into the SalI/XbaI sites of pDG148 | This studya |
| pMPRYB147 | mutS ORF cloned into the SalI/XbaI sites of pDG148 | This study |
| pMPRYB155 | mutSL ORF cloned into the SalI/XbaI sites of pDG148 | This study |
| pMPRYB138 | 4,777-bp PCR fragment of mutSL operon ligated into SmaI-treated pUC19 | This study |
| pMPRYB140 | pMPR138 with a 2.5-kbp SmaI/PmeI internal deletion of the mutSL operon containing the neomycin cassette of pBEST released with SmaI | This study |
See Materials and Methods for description of construction.
RESULTS
Generation of stationary-phase-induced mutants in an MMR-deficient B. subtilis strain.
In B. subtilis the reading frames of mutS and mutL, which encode the MMR proteins, are part of an operon (7). Therefore, we designed a plasmid termed pMPRYB140 to generate a mutSL-null mutant by eliminating part of the mutS and mutL ORFs and inserting a neomycin cassette in the middle of the operon (Fig. 1). This construction was transformed into B. subtilis YB955. A double crossover recombination event replaced the mutSL operon with the construction generating the Neor strain B. subtilis PERMYB151 (Fig. 1). Interruption of the mutSL operon in this strain was confirmed by PCR (results not shown). To better assess the mutSL-deficient genotype of the strain B. subtilis PERMYB151, the frequencies of mutation to rifampin resistance were determined for both the ΔmutSL strain and its parental strain YB955. For strain YB955, the frequencies were 0.4 × 10−8 and 0.1 × 10−8 for experiments 1 and 2, respectively; for the ΔmutSL strain, the frequencies were 52.0 × 10−8 and 11.5 × 10−8 for experiments 1 and 2, respectively. Thus, the ΔmutSL strain had a mutation frequency that was at least 2 orders of magnitude greater than that of its parental repair-proficient strain.
FIG. 1.
Construction of a plasmid to disrupt the mutSL operon of the strain B. subtilis YB955 as described in Materials and Methods.
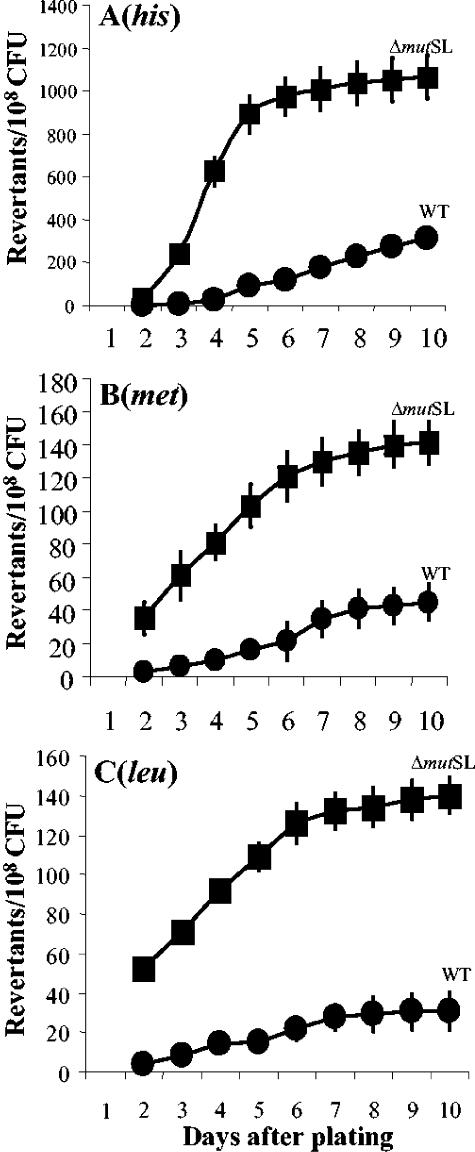
As mentioned above, a reversion system to study stationary-phase-induced mutations in B. subtilis has recently been characterized (30, 32). We utilized this system to analyze the production of His+, Met+, and Leu+ revertant colonies during the stationary phase for the repair-proficient strain B. subtilis YB955 and its isogenic mutSL mutant derivatives. The results shown in Fig. 2A demonstrated that the number of his+ revertants accumulating during days 2 to 10 after plating was around three times greater in the MMR-deficient strain than with the parental strain YB955. As shown in Fig. 2B and C the ΔmutSL strain produced between five and six times more met+ and leu+ revertant colonies during the stationary phase than did its parental strain. In general, Fig. 2 reveals that the number of adaptive mutants for the three markers in the mutSL mutant increased until day 6 after plating.
FIG. 2.
Stationary-phase-induced reversions for the his (A), met (B), and leu (C) alleles of YB955 (wild-type [WT]) (•) and MPRYB151 (ΔmutSL) (▪) B. subtilis strains as described in Materials and Methods. Results are the average numbers of accumulated revertants from five different selection plates.
Effects of MMR overproduction in the generation of B. subtilis stationary-phase-induced mutants.
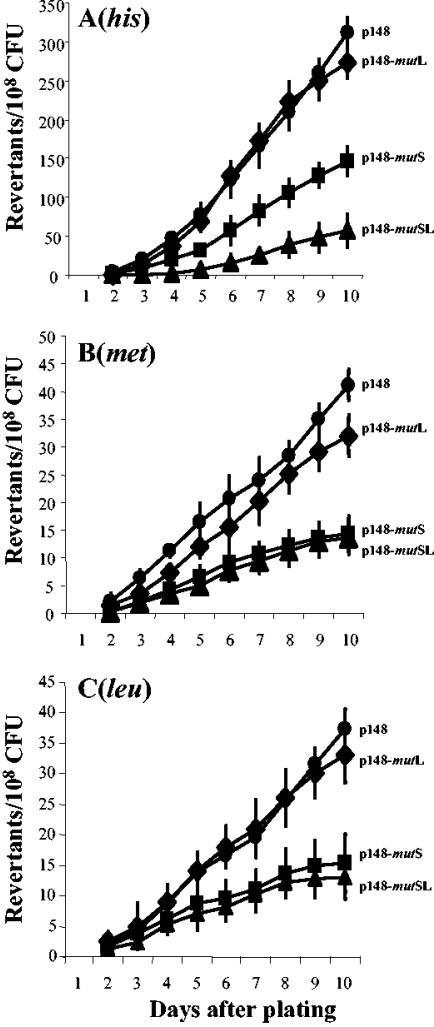
As demonstrated above, B. subtilis cells lacking a functional MMR system had a significantly increased stationary-phase-induced mutation frequency compared to that of isogenic repair-proficient bacteria. This result suggested that the MMR proteins could be reduced or become nonfunctional in those cells responsible for generating stationary-phase-induced mutagenesis. Therefore, we next investigated this possibility. To this end, the entire mutSL operon or each of its components was overexpressed in strain YB955 and the numbers of His+, Met+, and Leu+ revertant colonies were scored during the stationary phase. The results were compared with those obtained with the B. subtilis strain YB955 harboring only the vector pDG148. For all three markers, the results demonstrated that overexpression of the mutSL or mutS genes significantly decreased the number of revertant colonies that arose as a result of stationary-phase-induced mutagenesis (Fig. 3). Interestingly, in contrast to the results obtained with mutS, the overexpression of mutL did not significantly affect the production of stationary-phase-induced mutants for the three markers tested (Fig. 3).
FIG. 3.
Stationary-phase-induced reversions for his (A), met (B), and leu (C) alleles of MPRYB150 (pDG148) (•), MPRYB148 (pDG148-mutL) (♦), MPRYB149 (pDG148-mutS) (▪), and MPRYB156 (pDG148-mutSL) (▴) B. subtilis strains. As described in Materials and Methods, bacteria were grown in NOB-YE medium containing KAN (10 μg/ml) to an optical density at 600 nm of 0.5, and then IPTG to a final concentration of 5 mM was added to the cultures. At 150 min past T0 the strains were plated in minimum medium containing 2.5 mM IPTG. Results are the averages from six different selection plates. Each experiment was performed at least three times.
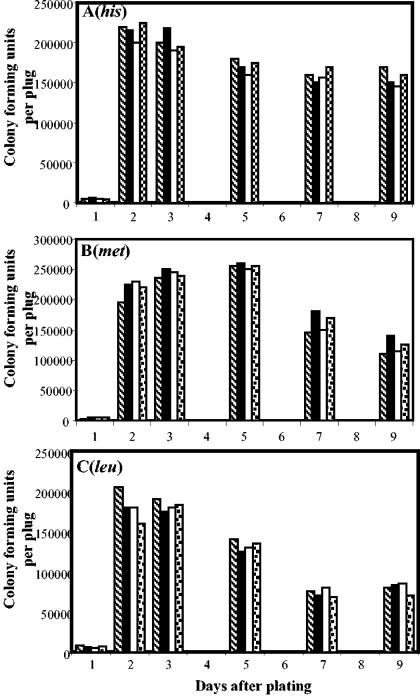
The results described in Fig. 4 revealed that the strains which overexpressed mutS. mutL, or both genes showed no differences in the rates of survival with respect to the parental strain PERMYB150, which contained only the vector pDG148. Therefore, the reduction in the number of His+, Met+, and Leu+ stationary-phase mutants generated by the strains PERMYB148 and PERMYB156, compared with those generated by the control strain PERMYB150, could not be attributed to a decreased survival of these strains on the selection medium.
FIG. 4.
Abilities of strains MPRYB150 (pDG148; hatched bars), MPRYB149 (pDG148-mutS; black bars), MPRYB156 (pDG148-mutSL; open bars), and MPRYB148 (PDG148-mutL; checkered bars) to survive histidine (A), methionine (B), and leucine (C) starvation. Three plugs of bacteria-containing agar were taken from selection plates each day for testing of the viability of bacteria on the selecting medium (see Materials and Methods for details). The experiments were repeated at least twice. The results are representative.
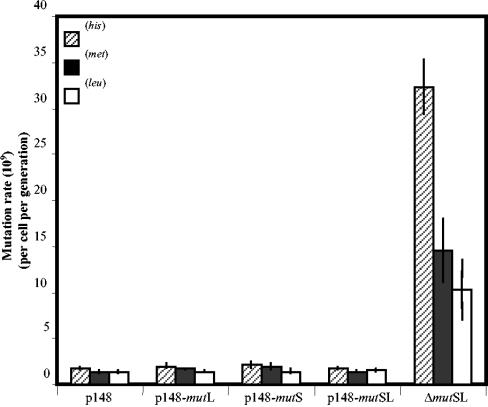
The growth-dependent mutation rates of the strains PERMYB148, PERMYB149, and PERMYB156 for the generation of His+, Met+, and Leu+ colonies were also determined and compared with those of the control strain PERMYB150. The results shown in Fig. 5 revealed that the mutation rates of the strains which overexpressed either the individual genes or both genes of the mutSL operon were indistinguishable from those obtained for the strain PERMYB150. Thus, neither the independent nor the concomitant overexpression of the genes of the mutSL operon affected the spontaneous mutation rates of these strains during active growth stages.
FIG. 5.
Analysis of mutation rates. B. subtilis strains MPRYB150, MPRYB149, MPRYB148, MPRYB156, and MPRYB151 were tested for their ability to produce His+, Met+, and Leu+ revertants during exponential growth as described in Materials and Methods. The mutation rates were calculated as previously described with the formula m/2Nt (24, 30, 32). Results presented are the average mutation rates from three individual fluctuation tests. Errors represent 1 standard error. Where appropriate, IPTG and antibiotics were added as described in Materials and Methods.
Taken together, these results strongly support the conclusion that stationary-phase-induced mutagenesis occurs as a result of a significantly reduced amount of functional MMR proteins in B. subtilis.
DISCUSSION
The previously characterized reversion system of B. subtilis is being used to study stationary-phase-induced mutagenesis in order to yield insights into some of the mechanisms which control the production of adaptive mutants in this gram-positive microorganism (30, 32). Consequently, important differences have been found between the B. subtilis model and the F′ lac reversion system of E. coli, which has been the paradigm for adaptive or stationary-phase-induced mutagenesis. For instance, while the E. coli model requires functional recombination and SOS systems, such activities have been found to be dispensable in B. subtilis (30). Moreover, the production of stationary-phase-associated mutants has been shown to be influenced by the developmentally important transcription factors ComA and ComK (21) in B. subtilis (30), while no such interaction has been shown in the E. coli model.
Because of the involvement of these regulatory genes, it was suggested that, in B. subtilis, cell subpopulations with suppressed DNA repair systems (30) might be responsible for some or all of the adaptive or stationary-phase-induced mutagenesis. This hypothesis prompted us to investigate whether the MMR system is involved in stationary-phase-induced mutagenesis and whether this system might be inactivated or down-regulated in a hypermutable subpopulation of the culture. To this end we first constructed a strain of B. subtilis which lacked a functional mutSL operon. The results demonstrated that the rates of spontaneous mutation to rifampin resistance in this repair-deficient strain were around 100 times higher than those of its parental strain (see above). Compatible with this result, the growth-dependent mutation rates for the production of His+, Met+, and Leu+ colonies were also higher in the ΔmutSL strain than in the parental strain YB955 (Fig. 5).
In addition, the B. subtilis ΔmutSL strain was tested in the adaptive mutagenesis reversion assay. Results revealed that, in the absence of a functional MMR system, the mutant strain showed a clear propensity to increase the number of His+, Met+, and Leu+ colonies during stationary phase with respect to the number of revertants produced by the parental strain. In agreement with our results, the contribution of the MMR system in the adaptive mutation phenomenon was positively demonstrated in S. cerevisiae (8). Essentially, in that report the effects of each of the mutS and mutL homologs on the production of stationary-phase-associated mutants were independently tested and the strongest effect was obtained with an S. cerevisiae strain which lacked the MutS homolog MSH2 (8).
Although our results, utilizing the B. subtilis ΔmutSL strain, suggested that the MMR system is involved in adaptive mutagenesis, we wanted to determine the individual contribution of the MutS and MutL proteins to this phenomenon. Therefore, we implemented an approach similar to that utilized in E. coli (9). Specifically, the components of the MMR system were overexpressed in B. subtilis. With use of this approach, the overproduction of the proteins encoded by the mutSL operon resulted in a significant decrease in the number of mutations generated for all three markers via stationary-phase-induced mutagenesis (Fig. 3). A similar result was obtained following overexpression of the mutS gene. On the other hand, we did not observe a significant inhibitory effect in the production of adaptive revertants for any of the markers tested in the strain which exclusively overexpressed the mutL gene. Thus, our results strongly suggest that the MMR system, in particular the MutS protein, is reduced in either amount or activity in those cells responsible for the mutation levels during the stationary phase of B. subtilis. However, overexpression of the mutSL operon did not completely suppress the production of stationary-phase-induced mutations. This result indicates either that we did not generate enough MMR proteins or that additional mechanisms are involved in the generation of stationary-phase mutations.
Our results also demonstrate that the complete elimination of the MMR system increases the production of stationary-phase-induced mutations. Thus, in B. subtilis either the overall functional level of MMR proteins is reduced in stationary-phase cells or there is a subpopulation that is lacking sufficient levels of active MMR proteins.
Although the MMR system has been shown to be involved in adaptive mutation in the F′ lac reversion system of E. coli (9), our results suggest that the mechanism of involvement could be different in B. subtilis. While the loss of function of the MutL protein was proposed to be responsible for an MMR deficiency during the stationary phase of E. coli (9), the evidence presented in this report strongly suggests that the MutS protein is the nonfunctional component of the MMR system for the production of stationary-phase-induced mutants in B. subtilis. In S. cerevisiae, the other microorganism where the MMR system has been shown to play a role in stationary-phase-induced mutagenesis, MSH2, a MutS homolog, has been implicated as the major contributor to this type of mutagenesis (8).
Taken collectively, the data tend to support the hypothesis that MMR is involved in at least a major share of the stationary-phase-induced mutagenesis process and that a completely functional MMR system might be absent or severely reduced in many if not all of those cells that mutate. Another possibility, and one that is not mutually exclusive, is that those cells responsible for producing stationary-phase mutations might have accumulated DNA damage that saturates the repair capacity of the MMR proteins. Experimental support for this latter hypothesis comes from results that demonstrate the involvement of error-prone polymerases in the generation of stationary-phase-induced mutations in the E. coli model system (6, 11, 18). In addition a recent report revealed that the generation of revertants of the hisC952 allele is significantly decreased in a B. subtilis strain that lacks the YqjH protein, a homolog of the Y superfamily of E. coli DNA polymerases (32). This result suggests that error-prone replication occurs during the stationary phase of B. subtilis, as it does in E. coli. However, unlike the E. coli system, stationary-phase-induced mutagenesis in B. subtilis does not require RecA-controlled SOS functions (30).
The evidence presented in this report clearly indicates that a deficiency of functional MMR proteins exists in cells responsible for a significant portion of stationary-phase-induced mutagenesis. The question remains whether those cells that are responsible for this type of mutagenesis represent a distinct hypermutable subpopulation of the culture. With use of reporter gene technology, a previous report indicated that the level of transcription of the mutSL operon did increase in B. subtilis as the cells approached the end of exponential growth (7). These results would seem to be contrary to the ones that we have obtained (Fig. 3) unless a small subpopulation is envisioned as being responsible for the vast majority of the revertants generated via stationary-phase-induced mutagenesis. Such a differentiated hypermutable subpopulation is also suggested by the involvement of the comA and comK eubacterial differentiation transcription factors in the regulation of stationary-phase-induced mutagenesis (30). Interestingly, in the report by Ginetti et al. (7) mutations in comA or comK did not appear to affect the transcription of the mutSL operon. However, these results were based on the use of a reporter gene and could not measure actual translation levels or activity of MMR proteins.
Although the biochemical details of DNA repair through the MMR system have not been described for B. subtilis, it has been reported that the MutS homologs are very important in DNA repair, as they bind mismatches or small insertion-deletion events which can result from DNA polymerase errors (20). Therefore, mismatches generated during error-prone repair replication, in a subpopulation of nongrowing B. subtilis cells, are possible targets which could contribute to saturating the DNA repair capacity of the MMR system. Equally plausible is the possibility that other DNA repair systems are reduced or inactivated in a subpopulation of the culture. Such activity would lead to the accumulation of mutations in these cells, especially if the MMR system had been reduced or eliminated. In this paper we believe that we have demonstrated the involvement of the MMR system in stationary-phase-induced mutagenesis of B. subtilis. Previously, we showed that one error-prone DNA polymerase was also involved in this mutagenesis process (32). We are continuing to study the genetic and developmental regulation of additional DNA repair systems in order to test the hypermutable subpopulation hypothesis. However, one problem that arises if a hypermutable subpopulation(s) actually exists is how do these cells generate a good mutation before they reach genetic load (3)? Basically, how does a hypermutable cell achieve a desired mutation (if mutations are random) before incurring a lethal mutation? Mathematical analysis of random mutagenesis in a subpopulation should argue against the existence of such a population. However, the data that we have obtained with the ComA and ComK proteins as well as with the MMR system strongly indicate that the cells responsible for stationary-phase-mutagenesis might have certain physiological differences from the general population. The answer to this conundrum may very well lie in the “retromutagenesis” hypothesis that has recently been advanced (12). Specifically, for the types of mutations that we are observing (revertants of amino acid requirements) one could imagine errors in mRNA being generated by the error-prone nature of the RNA polymerase. Such “mutant” RNA might very well yield an altered protein that would permit the transient growth of a bacterium. This transient growth could then allow for actual DNA mutations that might result in the generation of a true revertant. We are currently testing this hypothesis.
The existence of alternative explanations for the phenomenon described for the E. coli lac reversion system (28) points to the importance of examining multiple models of adaptive or stationary-phase mutagenesis. While the genetic characterizations of the B. subtilis reversion assay used in this study and the E. coli lac reversion system are basically different, it is clear that MMR proteins play important roles in both models (most likely for different reasons). Characterizing the specific involvement of the MMR proteins in these two different systems should help in our understanding of the overall importance of mismatch repair mechanisms in mutagenesis, gene transfer, species isolation, and even cancer.
Acknowledgments
R.E.Y. was supported by MCB-997514 and MCB-0317076 from the National Science Foundation. M.P.-R. was supported by a Fulbright Scholarship and by grant 31767-N from the Consejo Nacional de Ciencia y Tecnología (CONACYT) of México.
We thank Huang Mo Sung and Christian A. Ross for helpful suggestions and Juan A. Rojas and Ada A. Sandoval for technical assistance.
REFERENCES
- 1.Boylan, R. J., N. H. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, Sussex, England.
- 3.Dobzhansky, T. 1950. The genetic basis of evolution. Sci. Am. 182:32-41. [DOI] [PubMed] [Google Scholar]
- 4.Fishel, R., and R. D. Kolodner. 1995. Identification of mismatch repair genes and their role in the development of cancer. Curr. Opin. Genet. Dev. 5:382-395. [DOI] [PubMed] [Google Scholar]
- 5.Foster, P. L., and J. M. Trimarchi. 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265:407-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galitski, T., and J. R. Roth. 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268:421-423. [DOI] [PubMed] [Google Scholar]
- 7.Ginetti, F., M. Perego, A. M. Albertini, and A. Galizzi. 1996. Bacillus subtilis mutS mutL operon: identification, nucleotide sequence and mutagenesis. Microbiology 142:2021-2029. [DOI] [PubMed] [Google Scholar]
- 8.Halas, A., H. Baranowska, and Z. Policinska. 2002. The influence of the mismatch repair system on stationary phase mutagenesis in the yeast Saccharomyces cerevisiae. Curr. Genet. 42:140-146. [DOI] [PubMed] [Google Scholar]
- 9.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, M. E. Winkler, and S. M. Rosenberg. 1997. Mismatch repair protein MutL becomes limiting during stationary phase mutation. Genes Dev. 11:2426-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 11.Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Andersson, and J. R. Roth. 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmquist, G. P. 2002. Cell-selfish modes of evolution and mutations directed after transcriptional bypass. Mutat. Res. 510:141-152. [DOI] [PubMed] [Google Scholar]
- 13.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance cassette selectable in a single copy state in the B. subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasak, L., R. Horak, and M. Kivisaar. 1997. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc. Natl. Acad. Sci. USA 94:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 16.Longerich, S., A. M. Galloway, R. S. Harris, C. Wong, and S. M. Rosenberg. 1995. Adaptive mutation sequences reproduced by mismatch repair deficiency. Proc. Natl. Acad. Sci. USA 92:12017-12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic, I., C. Rayssiguler, and M. Radman. 1995. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80:507-515. [DOI] [PubMed] [Google Scholar]
- 18.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie, G. J., and S. M. Rosenberg. 2001. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr. Opin. Microbiol. 4:586-594. [DOI] [PubMed] [Google Scholar]
- 20.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination and cancer. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 21.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 7:201-207. [DOI] [PubMed] [Google Scholar]
- 22.Petit, M.-A., J. Dimpel, M. Radman, and H. Echols. 1991. Control of large chromosomal deletions in Escherichia coli by the mismatch repair system. Genetics 129:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards, B., H. Zhang, G. Phear, and M. Meuth. 1997. Conditional mutator phenotypes in hMSH2-deficient tumor cell lines. Science 277:1523-1526. [DOI] [PubMed] [Google Scholar]
- 24.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg, S. M., C. Thulin, and R. S. Harris. 1998. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics 148:1559-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg, S. M., S. Longerich, P. Gee, and R. S. Harris. 1994. Adaptive mutation by deletions in small mononucleotide repeats. Science 265:405-407. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Slechta, E. S., K. L. Bunny, E. Kugelberg, E. Kofoid, D. I. Andersson, and J. R. Roth. 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100:12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung, H.-M., and R. E. Yasbin. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J. Bacteriol. 184:5641-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung, H.-M., and R. E. Yasbin. 2000. Transient growth requirement in Bacillus subtilis following the cessation of exponential growth. Appl. Environ. Microbiol. 66:1220-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung, H.-M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2002. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 34.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasbin, R. E., D. Cheo, and D. Bol. 1993. DNA repair systems, p. 529-537. In A. L. Sonenshein, R. Losick, and J. Hoch (ed.), Bacillus subtilis and other gram-positive organisms. American Society for Microbiology, Washington, D.C.
- 36.Yasbin, R. E., P. I. Fields, and B. J. Andersen. 1980. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene 12:155-159. [DOI] [PubMed] [Google Scholar]
- 37.Yasbin, R. E., R. Miehl-Lester, and P. E. Love. 1987. Mutagenesis in Bacillus subtilis, p. 73-84. In M. Alacevic, D. Hranueli, and Z. Tomen (ed.), Genetics of industrial microorganisms. GIM-86, Split, Yugoslavia.