Abstract
The recent sequencing of the virulence plasmid of Campylobacter jejuni 81-176 revealed the presence of genes homologous to type IV secretion systems (TFSS) that have subsequently been found in Helicobacter pylori and Wolinella succinogenes. Mutational analyses of some of these genes have implicated their involvement in intestinal epithelial cell invasion and natural competence. In this report, we demonstrate that one of these type IV secretion homologs, Cjp3/VirB10, is a glycoprotein. Treatment with various glycosidases and binding to soybean agglutinin indicated that the structure of the glycan present on VirB10 contains a terminal GalNAc, consistent with previous reports of N-linked glycans in C. jejuni. Site-directed mutagenesis of five putative N-linked glycosylation sites indicated that VirB10 is glycosylated at two sites, N32 and N97. Mutants in the N-linked general protein glycosylation (pgl) system of C. jejuni are significantly reduced in natural transformation, which is likely due, in part, to lack of glycosylation of VirB10. The natural transformation defect in a virB10 mutant can be complemented in trans by using a plasmid expressing wild-type VirB10 or an N32A substitution but not by using a mutant expressing VirB10 with an N97A substitution. Taken together, these results suggest that glycosylation of VirB10 specifically at N97 is required for the function of the TFSS and for full competence in C. jejuni 81-176.
Campylobacter jejuni 81-176 is unique for a prokaryotic organism in that it has a general system of N-linked protein glycosylation (pgl), affecting a substantial number of periplasmic and surface proteins (29, 38). The structure of the N-linked glycan present on C. jejuni NCTC 11168 glycoproteins was recently found to be a heptasaccharide with a mass of 1,406 Da composed of GalNAc-α1,4-GalNAc-α1,4-(Glc-β1,3-)GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-bacillosamine (38). There are several genes in the C. jejuni pgl locus that have been shown to be involved in distinct steps in N-linked glycosylation (38). The addition of the glycan is dependent on the activity of the PglB protein, which is predicted to function as an oligosaccharide transferase (29, 32, 38) based on its homology to an oligosaccharide transferase subunit (STT3) of Saccharomyces cerevisiae (39). Other pgl genes (pglF, pglE, and pglD) have been proposed to be involved in synthesis of bacillosamine, a sugar that appears to be specific to this N-linked glycan (32, 38). Mutation of either pglB or pglE diminished the ability of 81-176 to invade INT407 cells and colonize the intestinal tracts of mice (28), reinforcing the importance of protein glycosylation to the pathogenesis of C. jejuni. However, the precise functional contribution of N-linked glycosylation to the pathogenesis of C. jejuni remains unclear.
C. jejuni strain 81-176 possesses two plasmids, one of which, pVir, is nonconjugative and affects both virulence and natural competence (2). Sequence analysis of this plasmid revealed the presence of eight genes with greatest homology to a type IV secretion system (TFSS) subsequently shown to be present in the ruminant commensal Wolinella succinogenes (1, 3). There is also significant homology to two TFSS found in Helicobacter pylori. These are the com system, which is responsible for natural transformation in H. pylori, and a more recently described TFSS of unknown function found in the H. pylori J99 plasticity zone (15, 16, 19). In contrast, the pVir TFSS shows much less homology to the well-characterized TFSS found on the cag pathogenicity island of H. pylori (7, 31). TFSS, which are present in a variety of plant and mammalian pathogens, are involved in the transfer of DNA, protein, or nucleoprotein complexes across bacterial membranes (8). The TFSS genes present on pVir have been proposed to encode proteins that form a functional secretion channel that appears to affect both intestinal epithelial cell invasion and natural competence (2, 3).
Herein we report that a putative structural component of the pVir TFSS, VirB10 (Cjp3) (3), is glycosylated by the pgl system at two asparagines residues and that lack of glycosylation at one site results in a competence defect comparable to that of the virB10 mutant. Further, we demonstrate that pgl mutants exhibit a major defect in natural competence, suggesting that N-linked glycosylation is required for full competence in C. jejuni 81-176.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni DB179 (81-176 cured of the second plasmid, pTet) has been described previously (2). All C. jejuni mutants used were constructed by insertional inactivation of the target gene by either an aph3A cassette (Kmr) (2) or insertion of a cat cassette using an EZ::TN transposon containing a campylobacter cat gene (Cmr) (3). The 81-176 pgl mutants were Kmr insertions into pglE or pglB and have been described previously (29). The virB8::Cm and virB9::Cm mutations in pVir have been previously described (3). The pVir virB10::Km mutation (2) was reconstructed in DB179 to avoid any potential interaction with the conjugative TFSS system encoded by the pTet plasmid also resident in 81-176. All mutants were tested by PCR using primers that bracket the insertion point of the drug resistance gene to confirm a double crossover (2, 3). C. jejuni was grown at 37°C under microaerobic conditions on Mueller-Hinton (MH) agar (Difco). Escherichia coli DH5α transformed with the C. jejuni pgl genes present on pACYC184 (pBTLPS) has been described previously (32). E. coli strains were grown on Luria agar. E. coli DH5α was used as the host strain for cloning experiments, and DH5α containing pRK212.1 was used as the donor in conjugation experiments (10). E. coli ER2566 (New England Biolabs, Beverly, Mass.) was used as the host strain for protein expression experiments. Antibiotics were added when appropriate to the following concentrations: 100 μg of ampicillin per ml, 20 μg of chloramphenicol per ml, 25 μg of kanamycin per ml, 20 μg of streptomycin per ml, 20 μg of tetracycline per ml, and 10 μg of trimethoprim per ml. Plasmids used are listed in Table 1.
TABLE 1.
Plasmids used
| Plasmid | Description | Reference or source |
|---|---|---|
| pBTLPS | Entire pgl operon cloned in pACYC184 (Cmr Tcs) | 32 |
| pCE107/70 | C. jejuni/E. coli expression vector; derivative of pRY107, a Kmr campylobacter shuttle vector, with σ70 promoter from Cj1291 | 37 and this work |
| pCE111/28 | C. jejuni expression vector; derivative of pRY111, a Cmr campylobacter shuttle vector, with σ28 promoter from flaA | 37 and this work |
| pJL101 | virB10 cloned into pCE107/70 | This work |
| pJL102 | virB10 cloned into pCE111/28 | This work |
| pJL102/N32A | N32A substitution in virB10 coding sequence in pJL102 | This work |
| pJL102/N42A | N42A substitution in virB10 coding sequence in pJL102 | This work |
| pJL102/N97A | N97A substitution in virB10 coding sequence in pJL102 | This work |
| pJL102/N32A, N97A | N32A and N97A substitutions in virB10 coding sequence in pJL102 | This work |
| pCS101 | pRY111, Cmr campylobacter shuttle vector carrying pglE under control of pglE promoter | 29 |
Construction of campylobacter expression vectors.
The region upstream of Cj1291, designated accB, a putative biotin carboxyl carrier protein of acetyl-coenzyme A carboxylase (11, 27) containing a putative σ70 promoter, was PCR amplified with HF2 DNA polymerase (Clontech, Palo Alto, Calif.) with the following primers: 5′-CGGGATCCCGAAAATTCTCCTACAAAATTTAAGAAC-3′ and 5′-GCTCTAGAGCTTTTAACCTTTTAATATTAGTAATTTTTT-3′. These primers introduced BamHI and XbaI sites bracketing the promoter region of Cj1291. The PCR product was digested with BamHI and XbaI and was cloned into BamHI-XbaI digested pRY107, a kanamycin-resistant shuttle vector (37), to generate pCE107/70. The region upstream of the flaA gene containing the σ28 promoter was PCR amplified from 81-176 by using HF2 DNA polymerase (Clontech) with the following primers: 5′-GCTCTAGAGCGTAAAATTGAAGATGAAAGAGAG-3′ and 5′-CGGGATCCCGTTTTAAATCCTTTTAAATAATTTC-3′. These primers introduced XbaI and BamHI sites, respectively. The PCR product was digested with XbaI and BamHI (New England Biolabs) and cloned into XbaI-BamHI-digested pRY111, a chloramphenicol-resistant campylobacter shuttle plasmid (37), to generate pCE111/28.
Complementation in trans of the virB10 mutation.
PCR amplification was used to amplify virB10 (cjp3) from the pVir plasmid by using HF2 DNA polymerase (Clontech). The primers to amplify cjp3/virB10 were JCL 075 (5′-CGCGGATCCATGAAAAAATCCTTTTTAAGCC-3′) and JCL 076 (5′-GGCTGCAGTTAATTATCTTGGAAATATTGG-3′), which introduced BamHI and PstI sites (5′ and 3′, respectively) flanking the virB10 coding sequence. The amplicon was digested with BamHI and PstI and was cloned into the BamHI and PstI sites of pCE107/70 or pCE111/28 to create pJL101 or pJL102, respectively. The pJL102 construct and mutant derivatives were mobilized from E. coli DH5α containing pRK212.1 into C. jejuni DB179 virB10::Km cells. Transconjugants were selected on MH agar containing kanamycin, chloramphenicol, and trimethoprim. Plasmid pJL101 was transformed into E. coli DH5α with or without pBTLPS (32).
Site-directed mutagenesis of virB10.
Mutation of five of the six predicted N-linked glycosylation sites of VirB10 was carried out using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Mutations were confirmed by sequencing with a Big Dye Terminator sequencing kit (Applied Biosystems, Foster City, Calif.) on an ABI Prism 3100 genetic analyzer (Applied Biosystems). Oligonucleotides used are listed in Table 2. The construction of the single mutants was carried out with pJL102 as a template, and pJL102/N32A was used as the template to construct the double mutant, pJL102/N32, N97A.
TABLE 2.
Oligonucleotides used for the site-directed mutagenesis of C. jejuni virB10
| Name | Sequence |
|---|---|
| 5′ virB10 N32A | 5′ GCAGAAGATATATTTGATCAAACAAGCGAAGAAGCTGTATCTAAAAATATATCTAAAAAAGACAATCAAAGC 3′ |
| 3′ virB10 N32A | 5′ GCTTTGATTGTCTTTTTTAGATATATTTTTAGATACAGCTTCTTCGCTTGTTTGATCAAATATATCTTCTGC 3′ |
| 5′ virB10 N42A | 5′ CGAAGAAAATGTATCTAAAAATATATCTAAAAAAGACGCTCAAAGCCAAAATTTGCTTAACAAAGATTTAG 3′ |
| 3′ virB10 N42A | 5′ CTAAATCTTTGTTAAGCAAATTTTGGCTTTGAGCGTCTTTTTTAGATATATTTTTAGATACATTTTCTTCG 3′ |
| 5′ virB10 N97A | 5′ CATAGTGAAGAAAAACCTAAAAAAGAAGAAGATAATGCTATTACTAAGTTAGCAAAAATTGAAGAAAAAAAGCAAGAAC 3′ |
| 3′ virB10 N97A | 5′ GTTCTTGCTTTTTTTCTTCAATTTTTGCTAACTTAGTAATAGCATTATCTTCTTCTTTTTTAGGTTTTTCTTCACTATG 3′ |
| 5′ virB10 N126A | 5′ CAGCAAATTGCAAAAGAAATTCATCAAGATGCTATTAGTTCTCAAGAAAGAAAAATC 3′ |
| 3′ virB10 N126A | 5′ GATTTTTCTTTCTTGAGAACTAATAGCATCTTGATGAATTTCTTTTGCAATTTGCTG 3′ |
| 5′ virB10 N156A | 5′ CAACACGCAAATTTATTTTCAAGAAGCTTCAAAATACGGCGTTGATGGTTTTTC 3′ |
| 3′ virB10 N156A | 5′ GAAAAACCATCAACGCCGTATTTTGAAGCTTCTTGAAAATAAATTTGCGTGTTG 3′ |
Cloning and expression of VirB8, VirB9, and VirB10.
The genes encoding VirB8 (Cjp1), VirB9 (Cjp2), and VirB10 (Cjp3) were fused to intein in the expression vector pTYB12 (New England Biolabs) by the following procedure. First, the DNA fragments were generated by PCR amplification using HF2 DNA polymerase (Clontech). The primers, which introduced SpeI and EcoRI sites, were cjp1-F (5′-GACTAGTGGAATGAGTAATAATACTATTGT-3′), cjp1-R (5′-GCGAATTCGTTACTTCGCTCCTTTCGTTTG-3′), cjp2-F (5′-GACTAGTGGAGACAACATACAAATTCAAGATGTTCC-3′), cjp2-R (5′-GCGAATTCGTCATTTCTTAGCCTT-3′), cjp3-F (5′-GACTAGTGGACAAACAAGCGAAGAAAATGTATC-3′), and cjp3-R (5′-GGAATTCTTAATTATCTTGGAAATATTGGATCAATA-3′). The PCR products were digested with SpeI and EcoRI and were cloned into pTYB12 (New England Biolabs). The pTYB12-Cjp1 clone contains an N-terminal intein fusion containing residues 52 to 225 of Cjp1. The pTYB12-Cjp2 and pTYB12-Cjp3 clones contain residues 22 to 356 and 28 to 378, respectively. E. coli ER2566 containing these constructs was grown overnight in Luria-Bertani medium at 37°C. The cells were diluted 1:100 in fresh medium containing ampicillin, and 1-liter cultures were grown to an optical density at 600 nm of approximately 0.5. Isopropyl-β-d-galactoside was added to a final concentration of 0.5 mM, and cultures were grown overnight at 16°C. Bacteria were pelleted by centrifugation at 5,000 × g for 10 min. Cell pellets were stored frozen at −20°C. Affinity chromatography was carried out using the IMPACT-CN protein purification system according to the manufacturer's recommendations (New England Biolabs).
Generation of polyclonal antisera.
Protein samples were sent for injection into New Zealand White Rabbits at Harlan Bioproducts (Indianapolis, Ind.). Following the manufacturer's immunization protocol, polyclonal antisera were obtained and used at the indicated dilutions.
Electrophoresis and immunoblotting.
Campylobacter spp. and E. coli whole cells were resuspended in 1× sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) sample buffer to a final protein concentration of 10 μg/μl. Protein samples were aliquoted and resuspended in an equal volume of 2× SDS-PAGE sample buffer. Samples were boiled and loaded onto 10% acrylamide gels. Proteins were separated by SDS-PAGE (21) and were detected by staining with Coomassie brilliant blue G250 or, after transfer to nitrocellulose, Western blot analysis using the indicated rabbit antisera. The secondary antibody was goat anti-rabbit antiserum conjugated to alkaline phosphatase (Caltag, Burlingame, Calif.) used at a 1:5,000 dilution.
SBA affinity columns.
Glycine extracts were prepared by resuspending a loopful of campylobacter organisms in 0.2 M glycine-HCl, pH 2.2, and placing it on ice for 10 min. Samples were centrifuged at 16,000 × g and suspended in an equal volume of 2× SDS-PAGE sample buffer. Large-scale glycine extracts were prepared with 100 ml of C. jejuni grown in biphasic culture as previously described (24). Prepared glycine extracts were incubated with 2 ml of soybean agglutinin (SBA) agarose (Vector Labs, Burlingame, Calif.) at 4°C. Following the collection of flowthrough fractions, the column was washed in 20 ml of column buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1 mM CaCl2, and 0.01 mM MnCl2) and wash fractions were collected. Proteins were eluted from the column by washing with 3 bed volumes of column buffer containing 0.2 M galactose.
Enzymatic deglycosylation.
Neutralized glycine extracts of C. jejuni were treated with α-N-acetyl-galactosaminidase or β-N-acetylhexosaminidase (New England Biolabs) according to the manufacturer's recommendations. After the addition of an equal volume of 2× SDS-PAGE loading buffer, samples were boiled and loaded onto a 10% acrylamide gel.
Natural transformation of C. jejuni.
The biphasic natural transformation procedure was used as previously described (33). C. jejuni strains were grown overnight on plates and were resuspended in MH broth to an optical density at 600 nm of 1.0. Aliquots of 250 μl of each strain were grown for an additional 2 h at 37°C in biphasic culture tubes (13). DNA (500 ng) from a streptomycin-resistant mutant of 81-176 (13) was added to cultures, and incubation continued for 4 h at 37°C. Cultures were serially diluted and plated in duplicate to MH agar containing streptomycin. The results were expressed as the number of transformants per microgram of Strr DNA. Negative controls were treated identically without the addition of DNA.
RESULTS
Discrepancy between predicted and observed mass of VirB10.
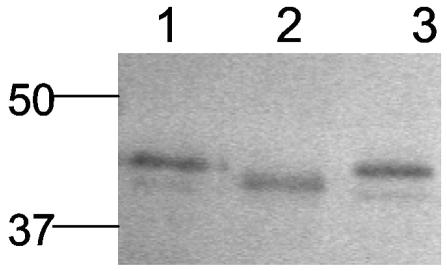
As shown in Fig. 1, lane 1, VirB10 expressed in C. jejuni DB179 presents two bands: a major band with an apparent molecular mass of approximately 43 kDa and a minor band with an apparent molecular mass of 41.5 kDa. The major 43-kDa band is substantially larger than that of the recombinant form of VirB10 expressed in E. coli (40.1 kDa) (lane 3) or the predicted mass of the mature VirB10 lacking its signal peptide (40.5 kDa). The significant discrepancy in mass between the recombinant protein expressed in E. coli and native forms of VirB10 suggested that VirB10 may be glycosylated in C. jejuni.
FIG. 1.

Discrepancy in mass of VirB10 expressed in C. jejuni and E. coli. Immunoblot of recombinant VirB10 isolated from E. coli and glycine extracts from C. jejuni DB179 and isogenic virB10 mutant. Protein samples were separated on a 10% acrylamide gel. Blots were incubated with an anti-VirB10 antiserum at 1:50,000 dilution. Lane 1, DB179; lane 2, DB179 (pVir/virB10::Km); lane 3, recombinant VirB10.
VirB10 possesses affinity for SBA.
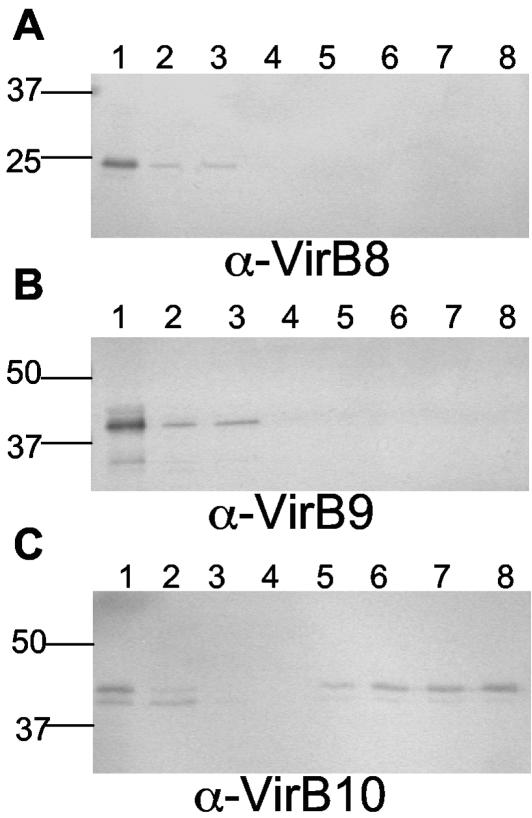
Glycine extracts of DB179 contained VirB8, -9, and -10, suggesting a periplasmic or surface localization for these proteins (Fig. 2A to C, lanes 1). Glycine extracts were passed through an SBA-agarose column to bind N-linked glycoproteins containing a terminal GalNAc (23). Western blot analysis of elution fractions using antiserum against whole cells of 81-176 revealed the presence of multiple proteins, indicating that enrichment of glycoproteins had occurred (data not shown), consistent with Linton et al. (23). When blotted with anti-VirB10 antiserum, a major band corresponding to the native molecular mass (43 kDa) of VirB10 was detected in the elution fractions (Fig. 2C, lanes 5 to 8). The lower 41.5-kDa form of VirB10 also bound to and eluted from the column, but it did so in smaller amounts than the 43-kDa form. In contrast, neither VirB8 nor VirB9 was detected in the elution fractions that used antisera against recombinant forms of these proteins (Fig. 2A and B).
FIG. 2.
VirB10 possesses affinity for SBA. Neutralized glycine extracts of DB179 were subjected to column chromatography with a SBA-agarose column. Extract, flowthrough, wash, and elution fractions were subjected to SDS-PAGE, blotted onto membranes, and incubated with either anti-VirB8 (A), anti-VirB9 (B), or anti-VirB10 antisera (C) at a 1:50,000 dilution. Lane 1, glycine extract from DB179; lane 2, column flowthrough fraction; lane 3, wash fraction; lanes 4 to 8, elution fractions.
VirB10 susceptibility to glycosidases.
Glycine-extracted proteins of C. jejuni DB179 were digested with glycosidases specific for HexNAc. After treatment with α-N-acetylgalactosaminidase, which cleaves the internal α1,3-linked N-acetylgalactosamine residue from the rest of the glycan, VirB10 mobility decreased on SDS-PAGE gels (Fig. 3, lane 2). Treatment with β-N-acetylhexosaminidase, which cleaves terminal β1-, β2-, β3-, β4-, and β6-linked GalNAc and N-acetylglucosamine residues, resulted in no discernible difference in mass (Fig. 3, lane 3). These data indicate that the glycan present on VirB10 contains an α1,3-linked GalNAc, consistent with the structure of the campylobacter N-linked glycan previously reported (38).
FIG. 3.
VirB10 is susceptible to treatment with N-acetylgalactosaminidase. Neutralized glycine extracts of C. jejuni DB179 were untreated (lane 1), treated with N-acetylgalactosaminidase (lane 2), or treated with β-N-acetylhexosaminidase (lane 3). Samples were separated on a 10% acrylamide gel, blotted onto membranes, and incubated with anti-VirB10 antiserum at a 1:50,000 dilution.
Restoration of wild-type mobility of VirB10 in E. coli expressing the pgl system.
Recently it was demonstrated that the general protein glycosylation system of C. jejuni could be functionally reconstituted in E. coli (32). We took advantage of this information to provide genetic evidence that VirB10 is a glycoprotein. When VirB10 was expressed in trans from pJL101 (a C. jejuni/E. coli Kmr expression vector; see Table 1 and Material and Methods) in E. coli DH5α in the absence of the pgl system, two bands were visible (Fig. 4, lane 3). The major band corresponded to the mass of recombinant, unglycosylated VirB10 (Fig. 4, lane 1) lacking a signal peptide (40.5 kDa). The minor band in these whole-cell extracts, of approximately 41.5 kDa, likely represents VirB10 without its leader sequence removed (see below). In DH5α containing both pBTLPS, carrying the intact pgl operon (32), and pJL101, bands of similar apparent mass were observed (Fig. 4, lane 5) as well as an additional band that corresponded to the mass of the glycosylated form of VirB10 expressed in DB179 (Fig. 4, lane 6). When a clarified whole-cell extract from DH5α(pBTLPS, pJL101) was subjected to column chromatography using the SBA-agarose column, the major band detected in the elution fractions (Fig. 4, lane 7) corresponded in mass to that of glycosylated VirB10 expressed in DB179 (lane 6); there was also a minor band that had the same apparent mass as the middle band shown in lane 5 (see below). When a lysate from DH5α containing pJL101 but not pBTLPS was passed over SBA agarose, no VirB10 could be detected in the elution fractions by immunoblot (data not shown).
FIG. 4.
Analysis of VirB10 expressed in E. coli DH5α in the presence or absence of the C. jejuni pgl system. Whole-cell extracts of E. coli were prepared and separated on a 10% acrylamide gel, blotted, and immunodetected with VirB10 antisera at 1:100,000 dilution. Lane 1, purified recombinant VirB10 from E. coli; lane 2, E. coli DH5α(pCE107/70), the vector-only control; lane 3, E. coli DH5α(pJL101), expressing virB10 in the absence of the pgl system; lane 4, E. coli DH5α(pBTLPS) containing the pgl genes cloned into pACYC184 (32); lane 5, E. coli DH5α(pBTLPS, pJL101), expressing virB10 in the presence of the pgl system; lane 6, control of a glycine extract from C. jejuni DB179; lane 7, elution fraction from SBA column of lysates from E. coli DH5α(pBTLPS, pJL101).
VirB10 is not detected in pglB or pglE mutants.
Western blot analysis of glycine extracts of the pglB::Km mutant (data not shown) or the pglE::Km mutant (29) failed to detect any VirB10 (Fig. 5C, lane 3). When the pglE mutation was complemented in trans with plasmid pCS101 (29), VirB10 was detected in the glycine extract (Fig. 5C, lane 4). Some expression of what appeared to be unglycosylated VirB10 was detected in whole cells of pgl mutants (data not shown). No detectable differences in expression patterns of VirB8 and VirB9 were observed between the wild-type and the pglE mutant (Fig. 5A and B).
FIG. 5.
VirB10 is absent in the periplasm of mutants defective in the general protein glycosylation pathway of C. jejuni. Glycine extracts of C. jejuni DB179 and mutants were separated on a 10% acrylamide gel and were blotted, and membranes were probed with either VirB8 (A), VirB9 (B), or VirB10 (C) antisera at a 1:50,000 dilution. Lanes 1, C. jejuni DB179; lanes 2, 81-176 virB8::Cm (A), 81-176 virB9::Cm (B), or DB179 virB10::Km (C); lanes 3, 81-176 pglE::Km; lane 4 of panel C, 81-176 pglE::Km (pCS101) (29).
Site-directed mutagenesis.
VirB10 is predicted to contain six potential N-linked glycosylation sites (Asn-X-Ser/Thr). A series of site-directed mutagenesis experiments was done on plasmid pJL102, expressing virB10 under control of the flaA σ28 promoter, to generate asparagine-to-alanine substitutions in five of these six possible glycosylation sites. Figure 6 shows a Western blot of glycine-extracted proteins of the DB179 virB10::Km mutant complemented in trans with selected mutated derivatives of pJL102. Alanine substitution of N32 of VirB10 (Fig. 6, lane 4) resulted in a decrease in mass such that the VirB10 band migrated at a position similar to that of the minor 41.5-kDa band seen in glycine extracts of DB179 (Fig. 6, lane 1). Mutation of N97 of VirB10 resulted in the presence of two equally intense bands (lane 5). The first band corresponded to the minor 41.5-kDa form of VirB10 detected in glycine extracts of DB179 (lane 1) and to that seen in N32A mutant (lane 4). The lower band migrated in parallel to the recombinant unglycosylated form of the protein expressed in E. coli (Fig. 6, lane 7). When both N32 and N97 were mutated in the same plasmid (lane 6), only one band was detected that was of the same apparent mass as the recombinant, nonglycosylated form of VirB10 (lane 7). Mutation of three other putative N-linked glycosylation sites of VirB10 (N42, N126, and N156) resulted in no discernible difference in mass; representative results for N42A are shown in lane 8. These results suggest the presence of two glycosylation sites within VirB10: N32 and N97.
FIG. 6.
VirB10 contains two N-linked glycosylation sites. Immunoblot of glycine extracted proteins of C. jejuni DB179 and mutants. Lane 1, DB179; lane 2, DB179 virB10::Km; lane3, DB179 virB10::Km (pJL102); lane 4, DB179 virB10::Km (pJL102N32A); lane 5, DB179 virB10::Km (pJL102N97A); lane 6, DB179 virB10::Km (pJL102N32A,N97A); lane 7, recombinant VirB10; lane 8, DB179 virB10::Km (pJL102N42A). Anti-VirB10 antiserum was used at a 1:50,000 dilution.
Effects of glycosylation on natural competence.
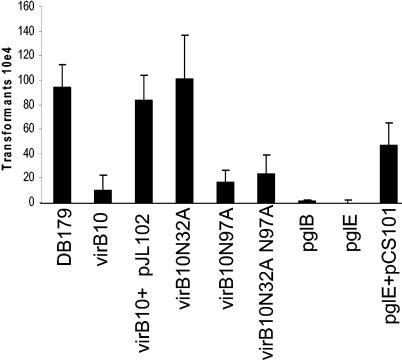
It has previously been reported that mutation of virB10 resulted in modest reductions in natural competence (2, 35). Because VirB10 is a glycoprotein, pgl mutants of 81-176 were tested for natural competence by using a chromosomal Strr marker (13). Figure 7 demonstrates that a mutation in either pglB or pglE resulted in a significant decrease in the number of transformants. C. jejuni DB179 produced an average of 9.3 × 104 transformants per μg of Strr DNA, which translates into efficiencies of 2.35 × 10−4 per μg/cell, within previously reported ranges (33, 35, 36). The pglB and pglE mutants transformed at frequencies of approximately 1.0 × 10−8, an efficiency that is 10,000-fold lower than that of the wild type. When the pglE mutation was complemented in trans with pCS101, an increase in the level of competence was observed. The lack of complete complementation likely reflects instability of the plasmid, a phenomenon that has been observed with some genes in trans and with the complementation of other competence genes in C. jejuni (35 and P. Guerry, unpublished data). The virB10 mutant was transformed as previously reported (2, 35) at a frequency of 2.3 × 10−5, or approximately 10-fold lower than the frequency of the wild-type strain. The defect in transformation frequency observed in the virB10 mutant was complemented in trans with pJL102 and restored it to wild-type levels. Wild-type levels of competence were exhibited in the mutant containing pJL102/N32A. However, pJL102/N97A failed to complement the virB10 mutant, as did pJL102/N32A, N97A, which encodes a double mutation. These results suggest that glycosylation of VirB10 at N97, but not at N32, is essential for wild-type levels of competence in C. jejuni 81-176.
FIG. 7.
Contribution of glycosylation to natural transformation. DNA (500 ng) from a streptomycin-resistant mutant of 81-176 was used to transform C. jejuni strains. Results are expressed as the total number of transformants per microgram of DNA and represent the means and standard deviations of at least three independent experiments. There was no difference in transformation ability of 81-176 and DB179 (data not shown).
DISCUSSION
Evidence suggests that the glycosylation of bacterial proteins may contribute to a variety of cellular and pathogenic processes. Reduction in bacterial adherence and invasion was demonstrated in several organisms when glycosylation was prevented (5, 12, 20, 22, 26, 29). Other work has implicated protein glycosylation in antigenic variation, protection from proteolytic cleavage, and solubility (14, 17, 18, 25). The flagella of C. jejuni 81-176 are extensively modified with O-linked pseudaminic acid residues and derivatives (30). In the absence of any of these modifications, flagella filaments do not assemble, rendering the bacteria nonmotile and thus nonvirulent (11). Additionally, mutants in the C. jejuni pgl system have a reduced capacity to invade INT407 cells and a deficiency in their ability to colonize the intestinal tracts of mice (28).
The findings reported here demonstrate that the C. jejuni 81-176 pVir TFSS protein, VirB10, is glycosylated at two sites, N32 and N97. Thus, the two forms of VirB10 observed in wild-type DB179, both of which bound to the SBA-lectin column, represent mono- and diglycosylated forms. The minor band seen in E. coli whole cells containing pJL101 (Fig. 4, lane 3) most likely represents unprocessed VirB10. The small difference in mass (906 Da) between the VirB10 signal peptide and the N-linked glycan was not resolved in SDS-PAGE. Glycosylation of VirB10 at N97, but not N32, was essential for wild-type levels of competence. The predominant modification site appears to be N97, because the unmodified form of VirB10 (40.5 kDa) was present in glycine extracts of the VirB10 N97A mutant but not the VirB10 N32A mutant (Fig. 6). This suggests that the N97 site may be in a more favorable context for glycosylation than the N32A site, perhaps due to increased surface exposure.
The original phenotype described for pgl mutants was loss of immunoreactivity with a variety of antisera made against C. jejuni (29). This was interpreted as being due to the immunodominance of the glycan on proteins that were expressed at low levels. However, here we have reported that a glycosylated protein appeared to lose reactivity in a pgl mutant background with antiserum generated against a recombinant, unglycosylated form of the same protein. This would suggest that in the absence of glycosylation VirB10 either was not transported to the periplasm or, upon transport, was unable to interact with the other components of the TFS apparatus and was rapidly degraded. However, the VirB10 N32A, N97A mutant protein was detected in glycine extracts when overexpressed in trans from the flaA σ28 promoter, which is approximately 10-fold stronger than the native virB10 promoter (P. Guerry, unpublished). This would suggest that the lack of detection of VirB10 in the pgl mutants reflects instability of the nonglycosylated protein, perhaps a result of an inability to interact with other TFSS proteins.
The six sites of potential glycosylation of the VirB10 homolog encoded by pVir are in contrast to the one or two putative glycosylation sites present in the H. pylori and Agrobacterium tumefaciens homologs, respectively. The A. tumefaciens homolog of VirB10 has been previously shown to be an inner membrane protein that spans the periplasm and interacts with other TFSS components to form a functional secretion channel (9). It is proposed for A. tumefaciens that VirB10 spans the periplasm in an oligomeric state and stabilizes interactions with other VirB proteins (4, 9, 34). In the absence of VirB10, substrates were not secreted, suggesting that the secretion channel was not formed, which underscores its role in the functionality of the system (6). From computer prediction analysis, it is believed that the C. jejuni VirB10 is structurally similar to the A. tumefaciens VirB10 and is localized and functions similarly. Mutational analyses of the genes in the ComB system of H. pylori, which share homology with the pVir TFSS, resulted in severe reductions in natural competence, suggesting that these ComB proteins form a TFSS that is involved in DNA uptake (15, 16). In C. jejuni 81-176, mutation of virB10 resulted in a modest effect on natural competence and a lesser effect on intestinal cell invasion (2, 3, 35). The pgl mutants were previously shown to have a decreased capacity to adhere and invade INT407 cells (28), and in this study we have demonstrated that pglB and pglE mutants are severely reduced in natural competence, likely due, to a limited degree, to lack of VirB10 glycosylation. The greater competence defect exhibited in the pgl mutants compared to that of the virB10 mutant suggests that additional glycoproteins are required for other steps in natural transformation. This notion is also consistent with the recent description of a putative type II secretion system involved in natural competence in C. jejuni (35). It is interesting that 8 of the 10 proteins described by Wiesner et al. (35) contain putative N-linked glycosylation sites by computer prediction. It remains to be determined if any of these proteins are glycosylated and if their function will be affected in the absence of glycosylation. Additionally, the observation that C. jejuni strains that lack pVir are competent also reinforces the notion that the pVir TFSS, while modulating competence levels, does not function as the primary DNA uptake system. Nevertheless, the modest effect of virB10 mutation on natural competence has been demonstrated by two independent groups and was able to be complemented in trans, suggesting the defect is genuine (2, 35, and this study). Speculatively, mutation of virB10 may have an indirect effect on natural competence by destabilizing other proteins that exist in the periplasm or membrane in the absence of a functional TFSS channel.
The identification of a TFSS structural protein that is glycosylated is significant on a number of levels. This is the first example, to our knowledge, of glycosylation of any TFSS protein, as well as the first function ascribed to an N-linked glycan in C. jejuni. Secondly, the plasmid-encoded pVir TFSS was presumably acquired through horizontal transfer from an unknown donor. Interestingly, the closest homolog of C. jejuni VirB10 is found in W. succinogenes, which is also the only other bacterium known to contain a putative N-linked glycosylation system homologous to the C. jejuni pgl system (1). Although the biochemical advantage of this general protein glycosylation system remains unknown, it would appear that the gene products of horizontally acquired DNA may be subject to functional restraints from the pgl glycosylation system and may need to be further modified to acclimate them to life within the C. jejuni host.
Acknowledgments
We thank Cheryl Ewing for construction of the expression plasmids, Scarlett Goon for helpful discussions, and Michael Wacker, Saba Amber, and Marcus Aebi for the gift of the E. coli BL21 isolate containing the pgl genes.
This work was supported by the Military Infectious Diseases Research Program.
REFERENCES
- 1.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaupre, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 6.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A., and Y. H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 12.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 13.Guerry, P., P. M. Pope, D. H. Burr, J. Leifer, S. W. Joseph, and A. L. Bourgeois. 1994. Development and characterization of recA mutants of Campylobacter jejuni for inclusion in attenuated vaccines. Infect. Immun. 62:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, L. A., S. M. Logan, P. Guerry, and T. J. Trust. 1987. Antigenic variation of Campylobacter flagella. J. Bacteriol. 169:5066-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 16.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 17.Jennings, M. P., M. Virji, D. Evans, V. Foster, Y. N. Srikhanta, L. Steeghs, P. van der Ley, and E. R. Moxon. 1998. Identification of a novel gene involved in pilin glycosylation in Neisseria meningitidis. Mol. Microbiol. 29:975-984. [DOI] [PubMed] [Google Scholar]
- 18.Kahler, C. M., L. E. Martin, Y. L. Tzeng, Y. K. Miller, K. Sharkey, D. S. Stephens, and J. K. Davies. 2001. Polymorphisms in pilin glycosylation locus of Neisseria meningitidis expressing class II pili. Infect. Immun. 69:3597-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayme, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo, C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S. Hakomori. 1996. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J. Clin. Investig. 98:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linton, D., E. Allan, A. V. Karlyshev, A. D. Cronshaw, and B. W. Wren. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497-508. [DOI] [PubMed] [Google Scholar]
- 24.Logan, S. M., and T. J. Trust. 1983. Molecular identification of surface protein antigens of Campylobacter jejuni. Infect. Immun. 42:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marceau, M., K. Forest, J. L. Beretti, J. Tainer, and X. Nassif. 1998. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol. Microbiol. 27:705-715. [DOI] [PubMed] [Google Scholar]
- 26.Moormann, C., I. Benz, and M. A. Schmidt. 2002. Functional substitution of the TibC protein of enterotoxigenic Escherichia coli strains for the autotransporter adhesin heptosyltransferase of the AIDA system. Infect. Immun. 70:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The complete genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 28.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 30.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 31.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 32.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, J. E., Jr., E. M. Dale, E. W. Nester, and A. N. Binns. 1990. Identification of a virB10 protein aggregate in the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 172:5200-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, D. L., J. A. Bell, V. B. Young, S. R. Wilder, L. S. Mansfield, and J. E. Linz. 2003. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149:3603-3615. [DOI] [PubMed] [Google Scholar]
- 37.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 38.Young, N. M., J. R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St. Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]
- 39.Zufferey, R., R. Knauer, P. Burda, I. Stagljar, S. te Heesen, L. Lehle, and M. Aebi. 1995. STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14:4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]