Abstract
Francisella tularensis is a gram-negative, facultative intracellular pathogen that causes the highly infectious zoonotic disease tularemia. We have discovered a ca. 30-kb pathogenicity island of F. tularensis (FPI) that includes four large open reading frames (ORFs) of 2.5 to 3.9 kb and 13 ORFs of 1.5 kb or smaller. Previously, two small genes located near the center of the FPI were shown to be needed for intramacrophage growth. In this work we show that two of the large ORFs, located toward the ends of the FPI, are needed for virulence. Although most genes in the FPI encode proteins with amino acid sequences that are highly conserved between high- and low-virulence strains, one of the FPI genes is present in highly virulent type A F. tularensis, absent in moderately virulent type B F. tularensis, and altered in F. tularensis subsp. novicida, which is highly virulent for mice but avirulent for humans. The G+C content of a 17.7-kb stretch of the FPI is 26.6%, which is 6.6% below the average G+C content of the F. tularensis genome. This extremely low G+C content suggests that the DNA was imported from a microbe with a very low G+C-containing chromosome.
Francisella tularensis is a highly infectious gram-negative coccobacillus that causes the zoonotic disease tularemia (11). This bacterial pathogen is known for its ability to cause a fulminating disease in humans after exposure to as few as 10 cells and has raised considerable concern as a potential bioterrorist agent (10). Because of its high infectivity and lethality, F. tularensis is one of six types of microbes classified by the U.S. Centers for Disease Control and Prevention as a category A agent, one that poses the most serious threat as a vehicle of bioterror.
There are a variety of subspecies and biotypes of F. tularensis, but they all have greater than 95% DNA sequence identity. Although the type A and type B biotype strains are highly infectious, only type A strains, which are found exclusively in North America, cause significant mortality in infected humans. An attenuated variant of a type B biotype strain formed the basis of a live vaccine strain (LVS) of F. tularensis. Understanding the molecular basis of the differences in virulence levels of F. tularensis strains may help in the development of a rationally designed LVS.
F. tularensis is a facultative intracellular pathogen. The currently available evidence suggests that F. tularensis resides inside a membrane-bound phagosome during its initial growth in a macrophage and that it may be released into the cytoplasm during a later phase of growth (3, 14). Very little is known about the bacterial virulence factors needed for infection, although it is clear that intracellular growth, especially in macrophages, is essential to the virulence of F. tularensis. A biochemical study of the LVS of F. tularensis showed that four proteins are induced after F. tularensis entry into macrophages (15). The gene encoding the most prominently induced protein, the 23-kDa IglC protein, has been molecularly cloned and sequenced. Recently, Golovliov et al. (16) deleted this gene and showed that the resulting mutant was unable to grow in macrophages and unable to cause disease in mice.
Genetic approaches have also been used to discover other F. tularensis genes needed for optimal intracellular growth. The products of mglA and mglB, thought to be global regulators, are both required for intramacrophage growth and virulence in mice (5). Random insertional mutagenesis revealed that inactivation of F. tularensis genes encoding homologues of glutamine phosphoribosylpyrophosphate amidotransferase (purine biosynthesis), alanine racemase (peptidoglycan biosynthesis), and the heat shock-inducible ClpB protease reduces the ability of F. tularensis to grow in mouse macrophages (17). Perhaps most significantly, transposon insertion into iglB and iglC, which are part of the pathogenicity island described in this work, profoundly affects intramacrophage growth (17).
The strategy of parasitizing host cells is a common theme used by both bacterial and protozoan pathogens (2). In many bacterial intracellular pathogens, a specific gene or set of genes that promotes entry into host cells has been identified (12, 13, 20). Two general types of cell entry mechanisms have been identified. One involves the tight binding of a bacterial surface protein to a host cell receptor, followed by engulfment of the bacterial cell by a zipper-like phagocytosis. A second type of uptake uses type III secretion machinery to inject effector proteins into host cells, inducing membrane ruffling and macropinocytosis. The genetic loci in Salmonella and Shigella spp. that encode the products needed for entry into mammalian cells are pathogenicity islands of common origin (18). The horizontal movement of this cluster of genes has enabled different species of bacteria to gain the ability to invade cells.
In this study we provide evidence for a Francisella pathogenicity island (FPI) that is required for intramacrophage growth and virulence in mice. The presumed effector proteins encoded by the FPI genes show no definitive similarity to known prokaryotic virulence proteins and thus represent novel factors required for virulence and intramacrophage growth. The gene encoding the PdpD protein appears to be absent in F. tularensis type B strains, and this absence may play a role in the wide difference in virulence of human infections between the type A and type B strains.
MATERIALS AND METHODS
Strains and molecular techniques.
The following F. tularensis strains were used. F. tularensis B38 (ATCC 6223) is the type strain for the highly virulent F. tularensis subsp. tularensis (type A biotype). However, the B38 strain has lost virulence through laboratory passage. The F. tularensis LVS (ATCC 29684) is the type strain for F. tularensis subsp. holarctica and represents the type B biotype. F. tularensis subsp. novicida (type strain U112; ATCC 15482) was used for all gene knockout and virulence work; the DNA sequence reported in this work is from the U112 genome. Unless otherwise stated, the F. tularensis subsp. novicida strains were grown in Trypticase soy broth supplemented with 0.1% cysteine (TSBC). The generation time for F. tularensis subsp. novicida U112 and mutant strains grown in TSBC was in the range of 80 to 90 min. F. tularensis Schu4 is a fully virulent F. tularensis subsp. tularensis strain. Initial bioinformatics analysis was performed using the DNA sequence of the genome project of F. tularensis strain Schu4 (http://artedi.ebc.uu.se/Projects/Francisella/); however, all of the analyses reported for the region covering pdpD through pdpA were done with the F. tularensis subsp. novicida strain U112 sequence. Clinical isolates of F. tularensis were from the following locations in British Columbia and were collected in the years indicated: B1, Mission, 1993; B2, Vernon, 1997; B6, Vernon, 2002; B7, Vandehoof, 2003. These clinical isolates and the Schu4 strain were manipulated under biosafety level 3 (BSL3) containment, and all other strains were handled under BSL2 conditions. Standard recombinant DNA and PCR techniques were used to manipulate or analyze DNA (29). Transposon mutagenesis (8) and transformation (4) of F. tularensis to create mutant 304-2 were performed as previously described. To construct the pdpD mutant, JL12, regions of the chromosome flanking pdpD were amplified by PCR and ligated to an erythromycin resistance cassette (see Fig. 4B). The region to the left of pdpD was apparently lethal in Escherichia coli in high-copy-number vectors. Therefore it was amplified with the proofreading Pfx polymerase (Invitrogen) and ligated to the low-copy-number vector pWSK29 (30), which had been digested with EcoRV, which generates flush ends. The primers used for this amplification were pdpDL-F, GGTACCTGGGTTATTTTTGCTGCTGA, and pdpDL-R, CTCGAGGATCCATACTTACTACTCTTACAAGTAAACC. The resulting amplicon was 1,864 bp. The right side of pdpD was amplified by standard PCR techniques and cloned into pCR2.1 (Invitrogen) with primers pdpDR-F, CTCGAGCAATGATCTGGGTTTAAATTTAGC, and pdpDR-R, GGTACCGCCATTTCTAAAGGGGTTGG. The resulting amplicon was 1,315 bp. The two recombinant clones were digested with XhoI and joined to an erythromycin resistance cassette (19) that was engineered to contain flanking XhoI sites by PCR amplification using the primers EmXhoF, CTCGAGTGAATCGTTAATAAGCAAAATTC, and EmXhoR, CTCGAGTTAAGGGATGCAGTTTATGC. The replacement of pdpD by the erythromycin cassette was verified by using PCR with three pdpD primer sets to show that pdpD was absent and combining primers for the erythromycin cassette with primers that hybridize to DNA flanking pdpD to generate amplicons.
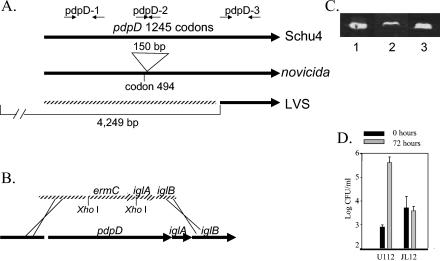
FIG. 4.
Properties of the pdpD genomic region and phenotypes of pdpD mutant. (A) Relative sizes of the pdpD genes in F. tularensis strain Schu4, F. tularensis subsp. novicida, and the LVS. (B) Recombinant constructs used to make the pdpD::Emr allelic replacement mutant JL12, as described in the Materials and Methods. (C) Immunoblot of E. coli-produced, isolated recombinant IglA (lane 1) and IglA expressed in the pdpD mutant, JL12 (lane 2), and wild-type U112 (lane 3). When normalized to the amount of total protein loaded per lane, the intensity of the IglA band is sixfold higher in the U112 lane than in the JL12 lane. (D) Relative growth of wild-type F. tularensis subsp. novicida U112 and JL12 in bone marrow-derived macrophages after 72 h. Representative data from one of three repetitions are shown.
DNA sequencing of the FPI region was performed with custom primers. Sequence assembly and analysis were done with the LaserGene (DNAStar) suite of programs. Comparison of amino acid sequences deduced from the FPI to those in protein and nucleic acid databases was done using online BLASTP (1) with the default settings. TBLASTN and BLASTN were also used. The locations of transposon insertions were determined by amplifying the area of interest and sequencing parts of the amplicon by initiating DNA sequencing reactions from the TnMax2 transposon (19) with the primers AAACATGCAGGAATTGACGA and TTCCTGAGCCGATTTCAAAG. Mutant 304-2 was genetically complemented by introducing DNA cloned into the kanamycin resistance, broad-host-range plasmid pDSK519 (23). Several primers were used to produce the amplicons shown in Fig. 2 and 3, and they are listed in Table 1; the relative positions of some of these primers are shown in Fig. 4A.
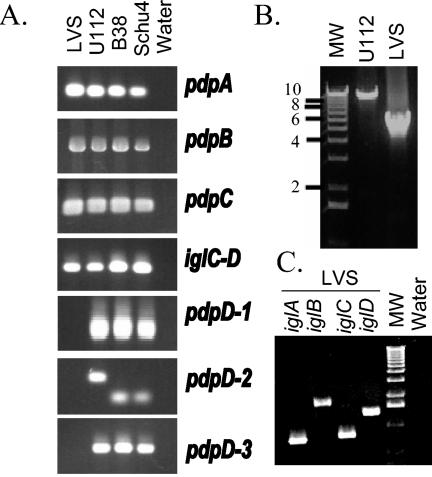
FIG. 2.
PCR amplification of FPI segments. (A) Results when primers were used to amplify internal fragments of pdpA, pdpB, pdpC, pdpD, and the junction between iglC and iglD. The resulting PCR products are shown; they provide evidence that the amplified regions in pdpA, pdpB, pdpC, and iglC-D in the F. tularensis strains that were tested are very similar. The three primer pairs used to amplify the region encompassing codons 123 to 356 (pdpD-1), codons 468 to 560 (in U112; codons 468 to 512 in Schu4; pdpD-2), and codons 906 to 1131 for pdpD-3 demonstrate that the pdpD gene is missing or is substantially different in sequence from the form found in strains U112, Schu4, and B38. The different sizes of the PCR products with the pdpD-2 set of primers with U112 DNA as the template relative to the sizes for the other template products confirm the difference in the DNA sequences. (B) Long-range PCR encompassing pdpD to iglD. Primers were used to amplify a region of approximately 10 kb in strain U112. The product in the LVS reaction is approximately 5.5 kb. Lane MW, molecular weight markers. (C) Left to right, PCR products obtained by using primers for iglA, iglB, iglC, and iglD. Molecular weight markers are as in panel B. Lane water, PCR done with primers for iglA but with no template. Similar reactions were done with the three other sets of primers with identical results (data not shown).
FIG. 3.
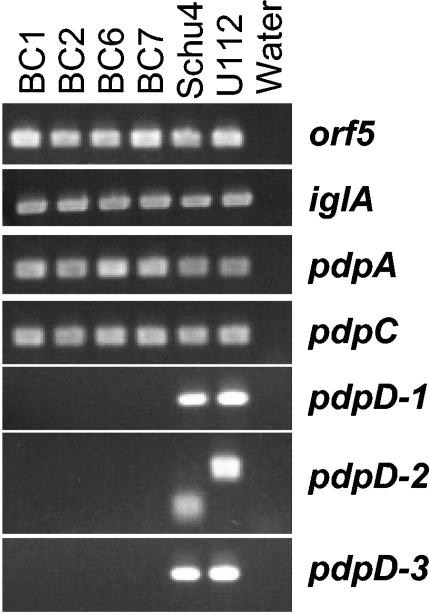
PCR analysis of clinical isolates of F. tularensis. The chromosomes of four type B isolates were amplified with primers corresponding to three regions of the pdpD gene as well as surrounding areas of the chromosome. The orf5 locus is 4 kb to the left of pdpD as shown in Fig. 1. These results show that pdpD is missing or substantially different in type B clinical isolates compared to the form found in type A strains or the F. tularensis subsp. novicida biotype. Lane water is as defined for Fig. 2.
TABLE 1.
Primers used to amplify FPI genes
| Gene or region | Gene size (bp) | Primer | Sequence (5′-) | Amplicon size(s) (bp) |
|---|---|---|---|---|
| pdpA | 2,463 | pdpA-F | TGC TTT TAG TGG GTC ATG GA | 307 |
| pdpA-R | CTA CGG CAT AAA TGG CTG GT | |||
| pdpB | 3,282 | pdpB-F | TTA GGT ACC GCC TTG CTC TTC TAA GTT GA | 1,569 |
| pdpB-R | ATA CTC GAG GCT AGC AAT GTC ATC AAA GG | |||
| pdpC | 3,657 | pdpC-F | TGC CTG AGT CAT TGC TTG AT | 305 |
| pdpC-R | TTG CCT CAA CAA CTG CTT TG | |||
| pdpD | 3,738 | pdpD-1F | CAA GTG CTT GGT GGT GGT AA | 720 |
| pdpD-1R | TGA TGT TTG ACC TGA ATT AGT GG | |||
| pdpD-2F | TGG GTT ATT CAA TGG CTC AG | 280 (U112), 136 | ||
| (Schu4) | ||||
| pdpD-2R | TCT TGC ACA GCT CCA AGA GT | |||
| pdpD-3F | TCC TGG CTT TGA TTT TGA GC | 678 | ||
| pdpD-3R | AAA TCT TGT TCA TCA AAC GCA AT | |||
| iglA-iglB | NAa | pdpDR-F | CTC GAG CAA TGA TCT GGG TTT AAA TTT AGC | 1,315 |
| pdpDR-R | GGT ACC GCC ATT TCT AAA GGG GTT GG | |||
| pdpD-iglD | NA | LA-F | TAA AAT TGC ACA GCA GAT AAG AGC | 9,932 (U112), 5,665 (LVS) |
| LA-R | CGT ATA GCT GAT GGC TGG GCC | |||
| iglA | 555 | iglA-F | CTC GAG GGC GTT GTT AAG GTA ACT TGC | 567 |
| iglA-R | CTC GAG CAA CTT CTG TAG ATC CCC CAA A | |||
| iglB | 1,545 | iglB-F | CTC GAG CTC TTG TGA TGC TGC TGA GTC T | 1,557 |
| iglB-R | CTC GAG TCG CCA CTT GTT ACC TGT TG | |||
| iglC | 636 | iglC-F | CTC GAG TTT GAA GGA ATG AAT ACT ACA ATG A | 648 |
| iglC-R | CTC GAG CCA TCT TCC CAA TAA ATC CTT | |||
| iglD | 1,197 | iglD-F | CTC GAG GCG CAG CTA GCA CAG ATA AA | 1,209 |
| iglD-R | CTC GAG GCT GGG CTA TCC CTC ATT AT | |||
| iglC-D | NA | iglCD-F | TTG CGC AGC TAG CAC AGA TA | 704 |
| iglCD-R | TCT GCG AAC TTC AAT TCT CTT TC | |||
| Region to left of pdpD | NA | pdpDL-F | GGT ACC TGG GTT ATT TTT GCT GCT GA | 1,864 |
| pdpDL-R | CTC GAG GAT CCA TAC TTA CTA CTC TTA CAA GTA AAC C | |||
| Region ca. 4 kb to left of pdpD | NA | O5-F | AGT GTA ATG GAG CCC AAC CA | 420 |
| O5-R | GGT TTG CCA AAG CAG ATG AT |
NA, not applicable.
An antibody was raised against IglA in a rabbit by injecting recombinant protein. The primers CTCGAGGGCGTTGTTAAGGTAACTTGC and CTCGAGCAACTTCTGTAGATCCCCCAAA were used to amplify the iglA gene as an XhoI-XhoI fragment. This amplicon was cloned into the SalI site of pTZ18U and transformed into BL21λDE3(pLysS) (Novagen). The hyperexpressed IglA was separated on sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gel and purified with a Bio-Rad model 491 Prep cell. The acetone-precipitated protein was emulsified in TiterMax adjuvant (Sigma) and injected subcutaneously three times at intervals of 3 weeks into a New Zealand White rabbit, in accordance with Canadian Council on Animal Care protocols. Immune serum was used in immunoblots at a dilution of 1:2,500. The antibody reactivity was visualized by reacting the blots with IRDye800-conjugated goat anti-rabbit immunoglobulin G (Rockland Immunochemicals, Gilbertsville, Pa.) and exposing the filters to excitation light in a LiCor Odyssey imaging system.
Infection of murine bone marrow-derived macrophages with bacteria.
Bone marrow macrophages were infected with F. tularensis subsp. novicida strain U112 or mutants as previously described (7). Briefly, bone marrow cells obtained from femurs of healthy BALB/cByJ male mice were plated at 2 × 106/well in 24-well plates (Costar, Corning, N.Y.) for 1 week in complete tissue culture medium containing L929 supernatants (cDMEM). Macrophages were then infected with F. tularensis strains at a multiplicity of infection of 1:20 (bacterium-to-macrophage ratio), and monolayers were incubated for 2 h in cDMEM, washed, and incubated at 37°C in 5% CO2 for the remainder of the experiment. The common practice of adding gentamicin to kill extracellular bacteria cannot be used with the F. tularensis subsp. novicida strain in this assay as it is exquisitely sensitive to this treatment; it has been demonstrated that macrophages do not support F. tularensis extracellular growth in cDMEM (3). To determine bacterial uptake and replication, infected macrophages were lysed at the time points indicated in Fig. 5 with sterile distilled water for 3 min, diluted immediately in phosphate-buffered saline, and plated on Mueller-Hinton (MH) agar plates containing the appropriate antibiotics. The plates were incubated for 1 to 2 days at 37°C in 5% CO2, and colonies were counted.
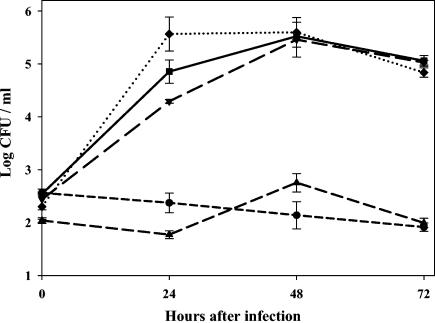
FIG. 5.
Growth of F. tularensis pdpA mutant and control strains in mouse macrophages. Bone marrow-derived macrophages were infected, and samples were lysed every 24 h to determine the number of viable bacteria (CFU) on agar medium. Squares, growth of parent strain F. tularensis (U112); inverted triangles, control strain R4 with a random transposon insertion not affecting intracellular growth; triangles, mutant 304-2; diamonds, mutant 304-2 with complementing plasmid pVIC314; circles, mutant 304-2 with plasmid pVIC311, which should not complement the genetic defect. Representative data from one of three repetitions are shown.
Bacterial stocks.
For macrophage and animal experiments, isolated colonies of bacteria were inoculated into modified MH broth (Difco Laboratories, Detroit, Mich.) supplemented with ferric pyrophosphate and IsoVitaleX (Becton Dickinson, Cockeysville, Md.) as previously described (7). Broth cultures were grown to mid-log phase as previously described (7), and 1-ml aliquots of bacteria were frozen in broth alone at −70°C. These were periodically thawed for use, and viable bacteria were quantified by plating serial dilutions on MH agar plates. The number of CFU after thawing varied less than 10% over a 12-month period.
Animals and mouse infections.
Six- to eight-week-old male specific-pathogen-free BALB/cByJ mice were purchased from the Jackson Laboratory (Bar Harbor, Maine). Animals were housed in sterile microisolator cages in a barrier environment at the Center for Biologics Evaluation and Research. Mice were fed autoclaved food and water ad libitum, and all experiments were performed under Institutional Animal Care and Use Committee guidelines. Mice were given 0.1 ml of appropriately diluted bacteria intradermally at the base of the tail; actual doses of inoculated bacteria were simultaneously determined by plate count. All materials used in animals, including bacteria, were diluted in phosphate-buffered saline (BioWhittaker, Walkersville, Md.) containing <0.01 ng of endotoxin/ml.
The sequences of the pdpD regions of two type B strains were assigned GenBank accession numbers AY626806 and AY626807.
The DNA sequence of F. tularensis subsp. novicida strain U112 has been assigned GenBank accession no. AY293579.
RESULTS
The identification of an FPI.
The sequencing of the F. tularensis genome (22, 28) and the development of simple genetic tools with F. tularensis subsp. novicida have facilitated analysis of virulence factors of Francisella. We previously isolated two mutations in the linked genes iglB and iglC that reduce the ability of F. tularensis to grow in macrophages (17). We performed bioinformatics analysis of the DNA in the region of the F. tularensis genome surrounding the location of these insertions and discovered an apparent FPI of approximately 30 kb (Fig. 1). In the left half of the FPI are eight open reading frames (ORFs), four of which, iglABCD, appear to be organized into an operon. The deduced products of iglA and iglB have about 30% identity to hypothetical proteins found in several bacterial species, most of which are animal or plant pathogens or plant symbionts. One set of similar genes, impBC of Rhizobium leguminosarum, encode proteins thought to be involved in protein secretion (6). Bladergroen and colleagues (6) noted that presumed homologues of impBC are found organized in an identical fashion in operons in a number of gram-negative bacteria; this organization is maintained in the F. tularensis iglABCD operon. The product of iglC has previously been shown (15) to be very highly induced after entry of F. tularensis into mouse macrophages and has recently been shown by us and others to be needed for growth in macrophages (16, 17). However, the deduced proteins IglC and IglD show no significant similarity to other known proteins. In the right half of the FPI are three large ORFs, named pdpABC (for pathogenicity determinant protein). The region between pdpB and pdpC has eight relatively short ORFs, seven of which are below 800 bp and one of which is 1,431 bp. None of the amino acid sequences deduced from pdpABC or the smaller ORFs show substantial similarity to those of known proteins.
FIG. 1.
Gene organization and G+C content of the FPI. Top graph, fractional G+C content of the 300-kb region of the F. tularensis subsp. tularensis (strain Schu4) chromosome that encompasses the FPI (http://artedi.ebc.uu.se/Projects/Francisella/); bottom graph, G+C content of the FPI. The ORFs (arrows) from pdpD through pdpA are derived from the DNA sequence of F. tularensis subsp. novicida strain U112, determined in this work (GenBank accession no. AY293579) and the remaining ORFs and sequence data are derived from the F. tularensis subsp. tularensis Schu4 sequence. At the left end of the FPI are ORFs tnpAB, which show exact identity to genes encoding presumed transposases previously found in F. tularensis (GenBank accession no. AAL06399 and AAL064100); at the right end is tnpA only. The small opposing arrows indicate the approximate positions of 16-bp inverted repeats that have previously been shown to be associated with tnpA (21). The ORF labeled pmcA shows 44% identity to conserved domains found in putative molecular chaperones, the most closely related of which is the vdcC gene of Bacillus anthracis (NP_656320). The short nature of seven of the eight ORFs between pdpB and pdpC suggests that the area has many sequencing errors or is composed of nonfunctional vestigial genes. However, the DNA sequence is highly conserved between F. tularensis subsp. tularensis and novicida, suggesting that these ORFs represent functioning genes. Arrows labeled 62 and 116, locations of the transposon insertions that originally indicated a cluster of virulence-associated genes; arrow labeled 304-2, location of the insertion that gave rise to the mutant described in this work, codon 401 of pdpA (821 codons). Hatched line, region of the allelic replacement in the JL12 mutant; solid thick lines, extent of the F. tularensis DNA insert in the complementing plasmid pVIC314 and the noncomplementing plasmid pVIC311. rrsH and rrlH, 16S and 23S rRNA genes, respectively; fdn, A subunit of formate dehydrogenase. The smaller ORFs are not drawn to scale; the numbers of ORFs cited in the text are based on the F. tularensis subsp. novicida DNA sequence.
Pathogenicity islands are often recognized by the aberrant G+C content in their DNA, which differs from that for the rest of the resident genome. The F. tularensis genome has an overall G+C content of 33.2% (28). The FPI has different regions that have variable G+C content. The region corresponding to pdpD through iglD has a G+C content of 31% (Fig. 1). The region from 1 kb to the left of pdpC to 204 bp to the right of the start codon of pdpA has a G+C content of 26.6% (bp 7969 to 25635 in the sequence with GenBank accession no. AY293579). Immediately to the right of the presumed promoter region of pdpA lies a 5,050-bp region that is 51% G+C and that encodes rRNA. The very different G+C content of this region is consistent with the need for conservation of the rRNA sequence, which permits changes in the G+C content primarily to adapt to the optimum growth temperature of the bacterium (27).
The recently released raw sequence data for the genome of F. tularensis LVS show that there are two copies of the FPI in this genome. (These sequence data were produced by the BBRP Sequencing Group at Lawrence Livermore National Laboratory and can be obtained from ftp://bbrp.llnl.gov/pub/cbnp/F-tularensis/F.tularensis.html.) One of the LVS forms of the FPI is essentially identical to the Schu4 form (excluding the pdpD deletion; see below) from one set of inverted repeats on the left end to the other set on the right end. A second copy of the FPI in the LVS strain is identical from the inverted repeats on the left end through the rRNA genes. Thus, the presence of two copies of this region in LVS suggests that the FPI region was capable of movement at one time and may still have the capacity to be mobile.
The pdpD gene is absent from type B strains of F. tularensis.
Upstream of iglABCD is a large ORF that we named pdpD. The deduced amino acid sequence shows no significant similarity to that of any known protein. The F. tularensis subsp. novicida form of the PdpD protein is composed of 1,245 amino acids and is predicted to be 141 kDa with a pI of 6.84. Comparison of the deduced amino acid sequence of PdpD found by us in F. tularensis subsp. novicida strain U112 to the PdpD found in F. tularensis subsp. tularensis strain Schu4 showed that the F. tularensis subsp. novicida form has 50 additional amino acids. Forty-eight of these amino acids were found in a continuous stretch between Q494 and Q543. We examined the nature of the corresponding region of the pdpD gene by amplifying this region by PCR using DNA from the three widely available type strains of F. tularensis (B38, LVS, and F. tularensis subsp. novicida U112); Schu4 DNA, which represents the sequenced genome, was also included (Fig. 2A). Several control PCRs, involving two other regions of the pdpD gene, as well as regions of iglC-D, pdpC, pdpB, and pdpA (see below), were performed. As expected, the PCR product that corresponds to codons 468 to 512 (for Schu4; codons 468 to 560 for U112) generated a product that was 136 bp when B38 or Schu4 DNA was used as the template and 280 bp when U112 DNA was used as the template. Surprisingly, DNA from the LVS showed no PCR amplicon for three different PCR primer pairs for pdpD, while showing the expected amplicons for pdpA, pdpB, pdpC, and iglC-D. To corroborate that a substantial portion of pdpD was missing from the LVS strain, we performed long-range PCR with primers that surrounded the pdpD gene (Fig. 2B). With Schu4 DNA as the template a 9.9-kp product was generated; however, the LVS template generated a 5.5-kb product. To test if the full iglABCD operon was present in LVS, we performed further PCRs. LVS template DNA generated products corresponding to the full lengths of iglA (555 bp), iglB (1,545 bp), iglC (636 bp), and iglD (1,197 bp) (Fig. 2C). DNA sequencing of this region in the LVS strain showed that a 4,249-bp region is deleted from this region. The deletion extends from 107 bp upstream of pmcA to the first base pair of codon 980 of pdpD (Schu4 form; codon 1030 of the U112 form of pdpD; Fig. 1; see Fig. 4A). The genome sequence of the LVS strain confirms this deletion and shows that it occurs in both copies of the FPI in the LVS strain.
Since the LVS strain is an attenuated variant of a type B strain, the question arose as to whether the absence of pdpD represents a feature specific to the LVS or is a feature of type B strains in general. To address this question, we examined the DNA of four type B clinical isolates for the presence of pdpD. As shown in Fig. 3, PCR amplification of three regions of pdpD indicates that this gene is missing from the clinical isolates or is significantly different in its nucleotide sequence. PCR amplification of other regions of the chromosome close to pdpD suggests that the clinical isolate forms are very similar to those found in the type strains, B38, LVS, and U112, of F. tularensis.
Disruption of pdpD results in a F. tularensis mutant defective for intramacrophage growth and virulence in mice.
The region corresponding to the pdpD gene was replaced with an erythromycin cassette by a recently described technique (25) (Fig. 4B). The resulting pdpD mutant, named JL12, still produced IglA, albeit at a lower level than the wild type (Fig. 4C). This mutant failed to grow in mouse bone marrow-derived macrophages (Fig. 4D). In two independent experiments, a total of 11 of 11 BALB/cByJ mice died within 7 days following intradermal infections with 105 cells of wild-type F. novicida U112. In contrast, 12 of 12 mice survived intradermal infection with 105 cells of JL12. Multiple attempts were made to complement the pdpD mutation. However, we were unable to recover recombinant plasmids carrying pdpD in E. coli that were not lethal in F. tularensis subsp. novicida.
Disruption of the pdpA gene renders F. tularensis unable to grow in macrophages and avirulent in mice.
We wished to test whether one or more of the genes associated with the most extreme low-G+C-content region of the FPI, pdpA through pdpC, were needed for virulence. Because of the dramatic difference between the G+C content of the rRNA genes and that of the adjacent pdpA gene, a putative promoter region for pdpA could be surmised. Hence, we inactivated pdpA of F. tularensis subsp. novicida by transposon insertion and tested the resulting mutant for virulence properties. Several insertions in pdpA were found to diminish intramacrophage growth, and one mutant, 304-2, was studied in detail. As seen in Fig. 5, the F. tularensis parent strain (U112) and a random-insertion mutant (R4) replicated exponentially in primary murine macrophages. In contrast, the mutant 304-2 failed to grow in macrophages. Efforts to complement this mutant were successful. When the pdpA gene on plasmid pVIC314 was reintroduced to F. tularensis strain 304-2, the wild-type intramacrophage growth phenotype was restored. The same pattern was seen with intradermal infections of mice. Doses of 2 × 105 CFU of F. tularensis strains U112 and R4 (about 50 times the 50% lethal dose) (24) were lethal for BALB/cByJ male mice; in contrast, mice survived doses of 2 × 107 CFU of strain 304-2, over 100-fold more bacteria (Fig. 6), demonstrating that the pdpA mutant is extremely attenuated. When pdpA on plasmid pVIC314 was reintroduced into F. tularensis strain 304-2, mice succumbed to infection with a dose of 2 × 107 CFU. Experiments in which 2 × 105 CFU of the pdpA mutant or the complement strain were used for infection gave essentially identical results as when 2 × 107 bacteria were used for infection, indicating that complementation was essentially complete. Thus, pdpA is required for growth in macrophages and virulence in mice.
FIG. 6.
Infection of mice with F. tularensis pdpA mutant and control strains. Mice were infected intradermally with doses ranging from 2 × 105 to 2 × 107 CFU of the indicated bacteria; actual infection doses were confirmed by retrospective plate count. Squares, survival of mice following infection with 2 × 105 CFU of the parent strain of F. tularensis (U112); inverted triangles, infection with 2 × 105 CFU of the control strain R4, which has a random transposon insertion not affecting intracellular growth; triangles, infection with 2 × 107 CFU of mutant 304-2; diamonds, infection with 2 × 107 CFU of mutant 304-2 with complementing plasmid pVIC314; circles, infection with 2 × 107 CFU of mutant 304-2 with plasmid pVIC311, which should not complement the genetic defect. Although lines in this graph are offset slightly for clarity, all mice survived (100% survival) infection with mutant 304-2 and mutant 304-2 with the noncomplementing plasmid pVIC311, and similarly all mice died (0% survival) following infection with the wild type and mutant 304-2 with complementing plasmid pVIC314. Representative data from one of four repetitions are shown.
DISCUSSION
There are multiple lines of evidence that the genomic region described in this work can be classified as a pathogenicity island. First, this work and others demonstrate that this region contains a cluster of genes encoding virulence factors (16, 17). Second, much of the region has DNA with a G+C content that differs significantly from that of the rest of the F. tularensis chromosome. Third, this area is surrounded by transposable elements. Although transposable elements are common in F. tularensis (21), there is good evidence that this region is actually mobile. Golovliov et al. (16) demonstrated that iglC is duplicated in the LVS of F. tularensis, and the genome sequence data on the LVS strain show that the entire FPI region is duplicated in that strain. This, in turn, suggests that this genomic area can move to and from replicons other than the F. tularensis genome. Very recent work shows that iglA, iglC, pdpA, and pdpD are all regulated by MglA, suggesting that these FPI genes are coordinately regulated to produce a virulence phenotype in F. tularensis (26). Together, these features of this genomic area justify applying the term pathogenicity island. Thus, this work represents the first description of a cluster of virulence genes, and the first description of a pathogenicity island, in F. tularensis.
There are numerous examples of large strain-to-strain differences in virulence levels among microbial pathogens. For F. tularensis, type A strains are thought to produce approximately 10% mortality in untreated human cases, and infections with the type B strains are rarely fatal (10). Until now, there was no known potential virulence factor that is present in the type A strains that is not present in type B strains. The presence or absence of pdpD may account, in part, for the strain-to-strain difference in virulence. Strangely, the pdpD mutant F. tularensis described here has a lower virulence in mice than the LVS strain, which also lacks pdpD. Conceivably, the duplication of the FPI in the LVS strain and the consequent increased dosage of the FPI-encoded genes may partially compensate for the absence of pdpD. Alternatively, the apparently polar effect on transcription of the erythromycin resistance cassette insertion may account for the lowered virulence phenotype in the pdpD mutant, JL12. Interpreting the virulence role of the altered form of pdpD in F. tularensis subsp. novicida is complicated by the different endotoxin and O antigen that this strain possesses (9). The endotoxin in particular clearly diminishes the ability of F. tularensis subsp. novicida to grow in the macrophages of some animals (9).
At present the most significant clue as to the function of IglA and IglB comes from the study of putative homologues in Rhizobium leguminosarum (6). In this species the IglAB homologues are thought to be needed for secretion of certain proteins. Hence, one possible hypothesis for the role of IglAB in F. tularensis is that they have a role in secretion of PdpABCD and perhaps other proteins.
Acknowledgments
This work was supported in part by a grant from the Natural Sciences and Engineering Council of Canada to F.E.N.
We are grateful to the members of the F. tularensis genome sequencing consortium for granting access to unpublished DNA sequence data. We thank A. Kari for use of facilities; P. Keeling, D. Burns, T. Pearson, and E. S. Stibitz for critical reading of the manuscript; and the C. Upton group and R. Roper for assistance with bioinformatics analyses.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amer, A. O., and M. S. Swanson. 2002. A phagosome of one's own: a microbial guide to life in the macrophage. Curr. Opin. Microbiol. 5:56-61. [DOI] [PubMed] [Google Scholar]
- 3.Anthony, L. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59:3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony, L. S. D., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 137:2697-2703. [DOI] [PubMed] [Google Scholar]
- 5.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29:247-259. [DOI] [PubMed] [Google Scholar]
- 6.Bladergroen, M. R., K. Badelt, and H. P. Spaink. 2003. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16:53-64. [DOI] [PubMed] [Google Scholar]
- 7.Bosio, C. M., and K. L. Elkins. 2001. Susceptibility to secondary Francisella tularensis live vaccine strain infection in B-cell-deficient mice is associated with neutrophilia but not with defects in specific T-cell-mediated immunity. Infect. Immun. 69:194-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley, S. C., C. J. Gray, and F. E. Nano. 2000. Isolation and characterization of Francisella novicida mutants defective in lipopolysaccharide biosynthesis. FEMS Microbiol. Lett. 182:63-67. [DOI] [PubMed] [Google Scholar]
- 9.Cowley, S. C., S. V. Myltseva, and F. E. Nano. 1996. Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol. Microbiol. 20:867-874. [DOI] [PubMed] [Google Scholar]
- 10.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 11.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 13.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 14.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov, I., M. Ericsson, G. Sandstrom, A. Tarnvik, and A. Sjostedt. 1997. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65:2183-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222:273-280. [DOI] [PubMed] [Google Scholar]
- 17.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215:53-56. [DOI] [PubMed] [Google Scholar]
- 18.Groisman, E. A., and H. Ochman. 1993. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 12:3779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas, R., A. F. Kahrs, D. Facius, H. Allmeier, R. Schmitt, and T. F. Meyer. 1993. TnMax-a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene 130:23-31. [DOI] [PubMed] [Google Scholar]
- 20.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 21.Johansson, A., I. Goransson, P. Larsson, and A. Sjostedt. 2001. Extensive allelic variation among Francisella tularensis strains in a short-sequence tandem repeat region. J. Clin. Microbiol. 39:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson, J., R. G. Prior, K. Williams, L. Lindler, K. A. Brown, N. Chatwell, K. Hjalmarsson, N. Loman, K. A. Mack, M. Pallen, M. Popek, G. Sandstrom, A. Sjostedt, T. Svensson, I. Tamas, S. G. Andersson, B. W. Wren, P. C. Oyston, and R. W. Titball. 2000. Sequencing of the Francisella tularensis strain Schu 4 genome reveals the shikimate and purine metabolic pathways, targets for the construction of a rationally attenuated auxotrophic vaccine. Microb. Comp. Genomics 5:25-39. [DOI] [PubMed] [Google Scholar]
- 23.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Kieffer, T. L., S. C. Cowley, F. E. Nano, and K. L. Elkins. 2003. Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 5:397-403. [DOI] [PubMed] [Google Scholar]
- 25.Lauriano, C. M., J. R. Barker, F. E. Nano, B. P. Arulanandam, and K. E. Klose. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195-202. [DOI] [PubMed] [Google Scholar]
- 26.Lauriano, C. M., J. R. Barker, S.-S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 101:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakashima, H., S. Fukuchi, and K. Nishikawa. 2003. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J. Biochem. (Tokyo) 133:507-513. [DOI] [PubMed] [Google Scholar]
- 28.Prior, R. G., L. Klasson, P. Larsson, K. Williams, L. Lindler, A. Sjostedt, T. Svensson, I. Tamas, B. W. Wren, P. C. Oyston, S. G. Andersson, and R. W. Titball. 2001. Preliminary analysis and annotation of the partial genome sequence of Francisella tularensis strain Schu 4. J. Appl. Microbiol. 91:614-620. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]