Abstract
We have developed a three-dimensional testicular co-culture system (3D-TCS) which mimics in vivo testes. In this study, transcriptomic responses to phthalate esters (PE’s) were compared in the 3D-TCS with responses in rat testes in vivo. Microarray data from the 3D-TCS and from in vivo testes were used to compare changes in gene expression patterns after exposure to developmentally toxic (DTPE) or developmentally non-toxic (DNTPE) phthalate esters. DTPE treatments clustered separately from DNTPE treatments based on principle components analysis both in vitro and in vivo. Pathway analysis using GO-Elite software showed that terms relating to steroid metabolism or reproductive development were enriched both in vitro and in vivo after DTPE exposure. Processes such as cell cycle, cell proliferation and apoptosis were enriched for DTPE treatments in vitro, but not in vivo. Based on these analyses we concluded that transcriptomic responses in the 3D-TCS reflect key aspects of in vivo phthalate toxicity.
Keywords: Phthalate esters, testes, reproductive toxicity, in vitro, testes, toxicogenomic, transcriptomic
1. Introduction
Current models of male reproductive toxicity are expensive, time consuming and require a large number of animals. Rovida and Hartung [1] estimated a cost of ~€6.9 billion and ~49 million animals would be required to implement all reproductive toxicity testing proposed under Europe’s REACH program (Registration, Evaluation, Authorisation and Restriction of Chemical substances). New in vitro methods may aid in reducing these burdens [1, 2]. However, the use of alternative models of reproductive development has been limited by difficulties in modeling the complex cellular interactions and multiple endpoints which are involved in these processes [3]. Due to these difficulties, there is currently no reliable and reproducible in vitro model for testicular toxicity screening [4]. With the aim of addressing some of these issues, we have developed an in vitro model of testes development [5]. This cellular co-culture system (3D-TCS) contains several testes cell types (Sertoli, germ and Leydig cells) grown in a three dimensional conformation facilitated by an extracellular matrix (ECM) overlay. Earlier experiments have demonstrated that addition of ECM in this co-culture model results in a more physiologically stable system and that cells form a testicular-like architectural structure representative of in vivo characteristics of seminiferous tubules [5].
Phthalate esters (PE’s) represent a class of well-known male reproductive toxicants. Male reproductive effects of exposure to certain phthalate esters (PE’s) include endpoints such as underdeveloped reproductive organs, hypospadias, cryptorchidism and decreased anogenital distance [6–8]. Previously, we demonstrated that our 3D-TCS model was able to distinguish between developmentally toxic PEs (DTPE) and developmentally non-toxic PEs (DNTPE) based on observed differences in microarray-based gene expression profiles, with significant changes occurring in the steroid biosynthetic pathway. In contrast, a general cytotoxicity assay (neutral red uptake assay) was not able to distinguish between the toxic and non-toxic phthalates at the same dose. These results suggested that the 3D-TCS system provides a sensitive tool for identifying male reproductive toxicants through alterations in important biological pathways with functional relevance to male reproductive development [9].
Previously, Liu et al. exposed pregnant rats to the same group of DTPE and DNTPE’s. This was followed by microarray-based gene expression profiling in the testes of male fetuses, which identified a number of cellular pathways disrupted by DTPE exposure. These pathways included lipid and cholesterol homeostasis, insulin signaling, oxidative stress and steroidogenesis. In addition to cellular pathway changes, DTPE exposure was associated with the phenotypic outcome of significantly decreased anogenital distance [10]. The purpose of the current study was to compare transcriptomic responses induced by PEs in the 3D-TCS model with those observed from the Liu, et al. in vivo exposure using analysis of microarray gene expression profiles and pathway based analysis.
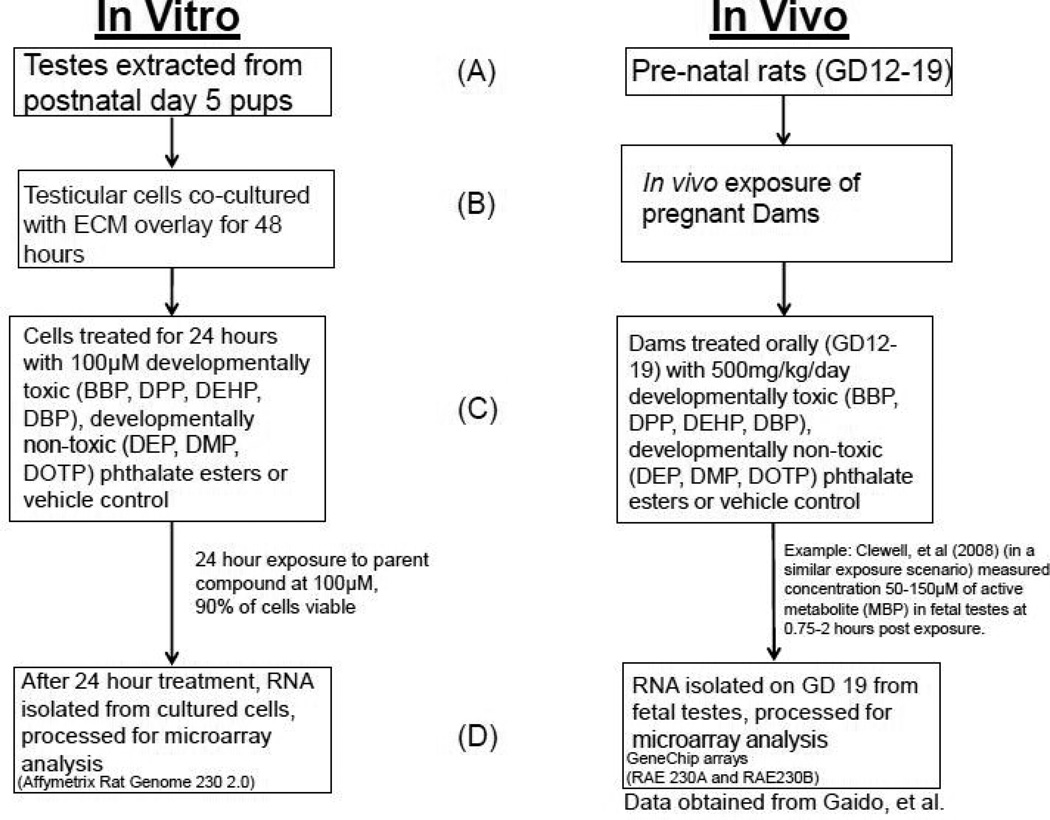
Our comparative analyses will allow us to determine the extent to which DTPE induced gene expression changes in the 3D-TCS reflect those occurring under an in vivo exposure scenario and aid in the evaluation of a proposed adverse outcome pathway (AOP) for phthalate reproductive toxicity [11, 12]. As shown in fig. 1, there were significant differences in the experimental design between the in vivo and in vitro datasets used in this study, for example the in vivo data was a repeated multiple dose exposure while the in vitro study was a 24 hours acute exposure. Bearing these differences in mind, we sought to determine whether the transcriptomic response in the 3D-TCS revealed key aspects of the mode of action (MOA) for phthalate reproductive toxicity. In order to accomplish this, we examined overall changes at the pathway level with a particular focus on the steroid biosynthesis pathway because disruption of steroidogenic gene expression has been phenotypically anchored to a number of male reproductive outcomes and is an important MOA for phthalate toxicity [6, 7, 13]. These comparisons have allowed us to link effects on gene expression observed in vitro with those observed in vivo and provide critical information in supporting in vitro models for evaluating mechanisms of male reproductive toxicity.
Fig. 1.
Experimental design for the in vitro three-dimensional testicular cell co-culture system (3D-TCS) and in vivo prenatal rats exposed to phthalate esters
2. Materials and Methods
2.1 Three dimensional co-culture model for testes (3D-TCS), treatments and gene expression profiling
Fig. 1 shows the experimental designs for both the in vitro and in vivo data used in this study. The method for the three dimensional co-culture model for testes (3D-TCS) and PE treatments was described previously [9]. Briefly, testes were dissected from 5-day-old male pups obtained from mated Sprague–Dawley rats (Charles River Laboratories, Wilmington, USA). A single cell suspension containing primarily Sertoli, germ and Leydig cells was then plated followed by an overlay of extracellular matrix medium (Matrigel™). Benzyl butyl phthalate (BBP, Sigma # 44-2503, 99% purity), dibutyl phthalate (DBP, Sigma # D2270, 99% purity), diethylhexyl phthalate (DEHP, Sigma # 4-8557, 99% purity), diethyl phthalate (DEP, Sigma #524972, 99.5% purity), dimethyl phthalate (DMP, Sigma # 525081, 99% purity), dioctyl terephthalate (DOTP, Sigma # 525189, 96% purity) or dipentyl phthalate (DPP, Sigma # 80154, 99% purity) were obtained from Sigma-Aldrich (MO, USA). Phthalate solutions (or vehicle control, DMSO, Sigma- Aldrich, MO,USA ) with a final concentration of 100 µM were then added directly to the culture medium 48 hours after the addition of ECM overlay. This dose was selected based on earlier experimental data showing changes in gene expression with a minimum effect on cell viability (90% of the cells were still viable). Total RNA was extracted from cells and used as starting material for probing of Affymetrix Rat Genome 230 2.0 microarrays (3 replicate arrays for each PE treatment and control).
2.2 Gene Expression Profiling Following In Utero Exposure to Phthalate Esters
In vivo exposure to phthalate esters was reported previously by Liu et al. [10]. Briefly, time-mated Sprague-Dawley outbred CD rats were obtained from Charles River Laboratories, Inc. (Raleigh, NC) on Gestation Day 0 (GD 0). Dams were treated by gavage daily from GD 12 to GD 19 with corn oil vehicle or BBP, DBP, DEHP, DEP, DMP, DOTP or DPP (Aldrich Chemical Company, Milwaukee, WI) in corn oil at 500 mg/kg per day. This dose was chosen based on earlier experiments showing changes in gene expression with no maternal toxicity or fetal death. DTPE exposure at this dose produced a significant decrease in anogenital distance for male fetal pups. Dams were euthanized on GD 19 by carbon dioxide asphyxiation. Right and left testes were removed from male fetuses. RNA was then isolated followed by synthesis of complementary DNA from 2 µg of total RNA. Equal amounts of purified cDNA per sample were used as the template for subsequent hybridization to GeneChip arrays (RAE230A and RAE230B, which contain the same geneset as Affymetrix Rat Genome 230 2.0 arrays on two separate chips). Three replicate arrays were used for each PE treatment except for DOTP (n=2). The resultant cell intensity (CEL) files were kindly donated by Dr. Kevin Gaido.
2.3 Statistical Analysis
2.3.1 QC/Normalization
Cell intensity files (CEL) for the in vitro 3D model and in vivo data were normalized using ArrayAnalysis software (http://www.arrayanalysis.org, accessed 11/04/14) for quality control and normalization [14]. Data normalization was performed using GC-RMA (GeneChip-Robust Multiarray Averaging). For each array, intensity values for 31,099 unique probes were normalized to one log2-transformed expression value for 12,025 EntrezGene ID’s. For in vivo arrays, A and B chips were analyzed and normalized separately and then combined for the final total of 12,025 genes. For genes which appeared on both A and B chips (1,081 genes), the normalized value from the A chip was used.
2.3.2 Principle Component Analysis
Principle Component Analysis (PCA) was performed on the normalized gene expression values for the in vitro and in vivo microarrays to visualize similarities and dissimilarities within the data using R statistical software (version 3.1.0). On the PCA plot, shorter distance between two points indicates greater similarity of gene expression profiles between two arrays [15, 16].
2.3.3 Pathway analysis for all gene expression values for each experimental condition
ToxProfiler software (http://ntc.voeding.tno.nl/toxprofiler_test/, accessed 12/11/14) was used to conduct a robust, unbiased analysis of significantly impacted biological pathways using the average log2 (fold-change/mean of controls) for all genes for on each array used in the in vitro and in vivo experiments. ToxProfiler is a web-based pathway analysis application that identifies enriched biological pathways among sets of genes without the need for statistical cutoffs. T-scores, a measure of statistical enrichment and magnitude of average fold change for genes in each biological pathway, were calculated for each enriched Gene Ontology (GO) term [17]. T-scores were input into MeV software for visualization in a heatmap [18]. Hierarchal clustering using Euclidean distance and average linkage was performed for all t-scores in order to cluster PE treatment groups based on the similarity of alterations in biological pathways.
2.3.4 Pathway analysis for genes commonly changed by toxic phthalate treatment both in vitro and in vivo
In order to identify biological pathways that were specifically impacted by the DTPE exposures, we performed t-tests using R statistical software (version 3.1.0) to find differentially expressed genes (compared to controls) for each phthalate treatment, both in vitro and in vivo (p≤0.005). Four hundred-forty (440) genes which were found to be commonly changed across all toxic phthalate treatments in the 3D-TCS; while 35 genes were commonly impacted in the rat fetal testes (4 genes were found to be common between these two lists). These sets of “toxicity signature” genes were used for two separate pathway enrichment analyses using GO-Elite software (version 1.2.5), which also calculates the average fold-change for genes within each process [19]. The average fold-changes for genes within each identified pathway were calculated for both in vitro and in vivo models. MeV software was used to visualize average expression for all enriched GO terms [18], followed by hierarchal clustering using Euclidean distance and average linkage in order to cluster PE treatment groups based on the similarity of response to PE treatments.
Following GO-Elite analysis, GO terms with particular relevance for male reproductive development (e.g. “male gonad development”, “steroid biosynthesis”, “Sertoli cell differentiation”, etc.) were identified and expression values were obtained for genes related to these terms that were appeared on either of the in vitro or in vivo “toxicity genelists”. Twenty-one (21) genes were found to be involved in one or more of these terms. Visualization and hierarchical clustering were performed for this set of genes as described above.
2.4 Pathway visualization
PathVisio software (version 3.1.3) was used to construct a visualization of the steroid biosynthetic pathway starting with fatty acid degradation or cholesterol import into the Leydig cell [20]. This pathway was constructed by combining elements from the WikiPathways steroid and cholesterol biosynthesis maps and sources from the literature [21–23]. This constructed pathway was then used to compare gene expression changes for the reproductively toxic phthalate treatments both in vitro and in vivo.
3. Results
3.1 Principle Component Analysis
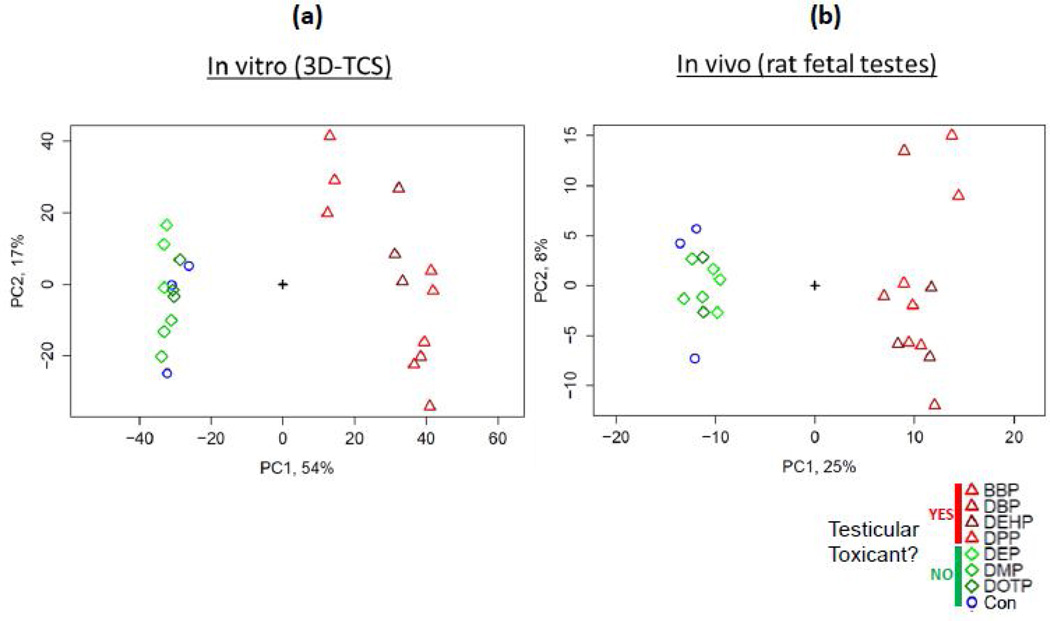
Principle Component Analysis was performed for all genes for both in vitro and in vivo datasets. For both datasets, there was a clear distinction between the two classes of phthalates. Fig. 2 shows that the non-toxic PE treatments and controls formed a distinct cluster from the toxic PE treated samples. This was observed for both the in vitro and in vivo expression data.
Fig. 2. Principle component analysis of gene expression analysis for microarrays from (a) in vitro (3D-TCS) and (b) in vivo (fetal rat testes).
Genes on each array were analyzed using principle components analysis in order to group samples by the similarity of genes expression profiles. Developmentally toxic phthalate treatments formed a distinct cluster separate from the controls and developmentally non-toxic phthalate treatments for in vitro (3D-TCS) and in vivo (fetal rat testes) model systems.
3.2 Phthalate induced gene expression changes in testicular co-culture model and in fetal rat testes
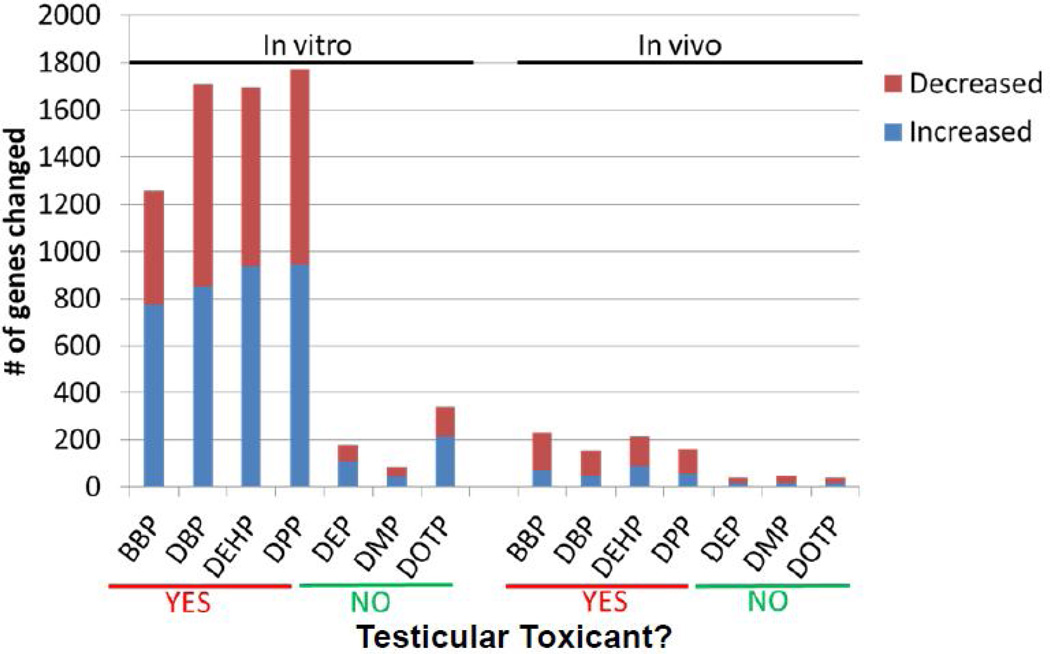
One-way ANOVA’s were used to determine the number of genes significantly changed compared to controls for each phthalate treatment. As shown in fig. 3, the number of genes significantly changed was greater after exposure to the reproductively toxic phthalates when compared to the non-reproductively toxic phthalates in both the testicular co-culture as well as the in vivo rat testes. However, the overall number of genes changes was higher in the in vitro model compared to the in vivo treatments. In general, there was greater percentage of genes with increased expression compared to controls in vitro, while a greater percentage of genes were decreased in vivo. Four genes were identified as significantly changed across all toxic PE treatments in both in vitro and in vivo models: Star (steroidogenic acute regulatory protein), Gsta1 (glutathione S-transferase alpha 1), Fdx1 (ferredoxin 1) and Pgap2 (post-GPI attachment to proteins 2).
Fig. 3. Gene expression changes following treatment with phthalate esters in vitro (3D-TCS) and in vivo (fetal rat testes).
The number of significantly changed genes was determined using a t-test for significant differences in gene expression between individual phthalate treatment and controls (p<0.005). The number of significant gene changes was higher for developmentally toxic phthalates both in vitro (3D-TCS) and in vivo (fetal rat testes).
3.2 Pathway Analysis
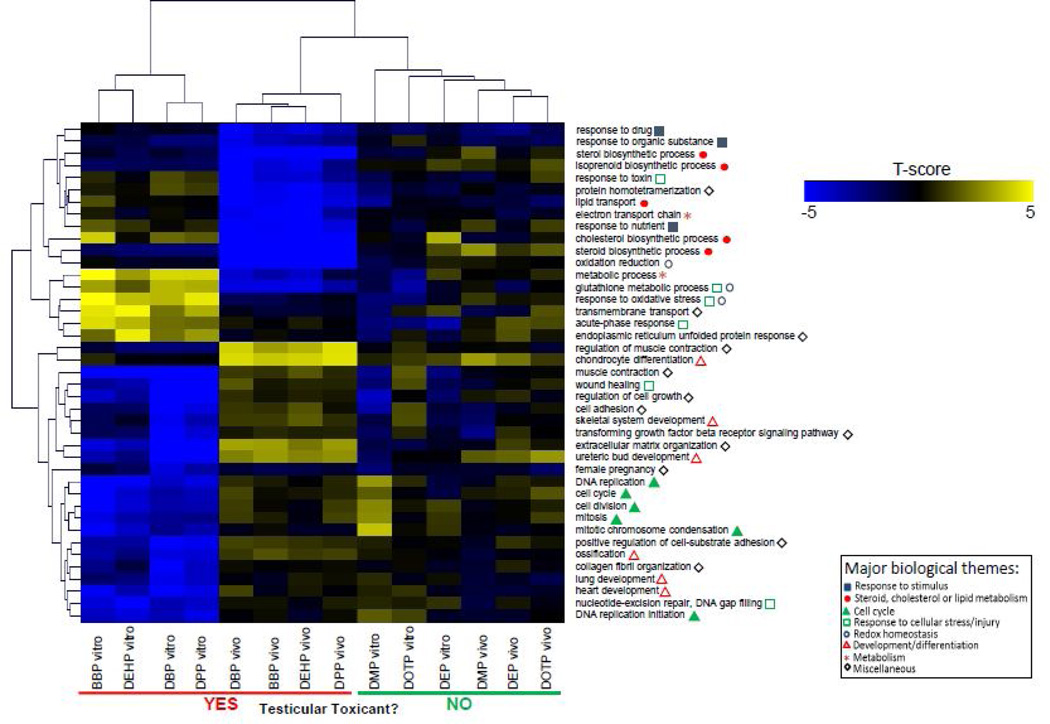
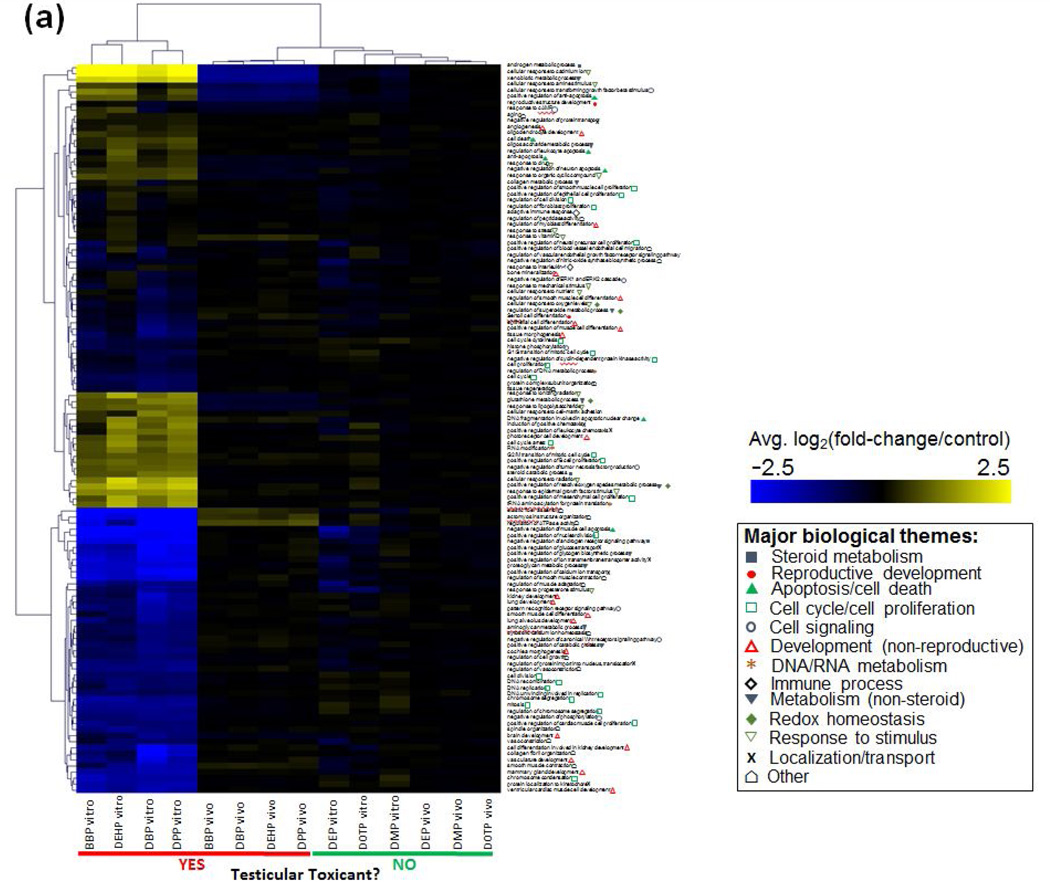
ToxProfiler was used to investigate enriched pathways for all genes on each array. Average expression values across all experimental replicates were used for pathway analysis. Significantly enriched Gene Ontology defined biological processes were given a t-score based on the significance of enrichment and the average log2(fold-change/control) in gene expression for each gene. These t-scores were then used to cluster treatment groups based on similarity of the changes in these biological processes. As shown in fig. 4, toxic and non-toxic PE treatments clustered separately from DNTPE treatments in both the testes co-culture as well as the in vivo treatments. While some processes were affected by the non-toxic phthalates, there was a clearly stronger and more consistent response after exposure to the toxic PE’s. Pathways which were altered by DTPE treatments represented several biological themes, including: response to stimuli, steroid or lipid metabolism, cell cycle, redox homeostasis and development or cell differentiation as indicated in the figure. Some of these processes were similarly regulated by DTPE treatment both in vitro and in vivo. These included the downregulation of processes related to lipid, cholesterol or steroid metabolism (GO terms: “cholesterol biosynthetic process”, “isoprenoid biosynthetic process”). By contrast, there were a number of pathways related to processes such as oxidative stress (“response to oxidative stress”, “glutathione metabolic process”, etc.) and cell cycle (“cell division”, “mitosis”, etc.), which were upregulated in vitro and downregulated in vivo.
Fig. 4. Identification of enriched Gene Ontology defined biological processes using ToxProfiler software.
Enriched processes were identified and t-scores were calculated for transcriptomic profiles from in vitro testes co-cultures and from in vivo rat testes after exposure to developmentally toxic and non-toxic phthalate esters.
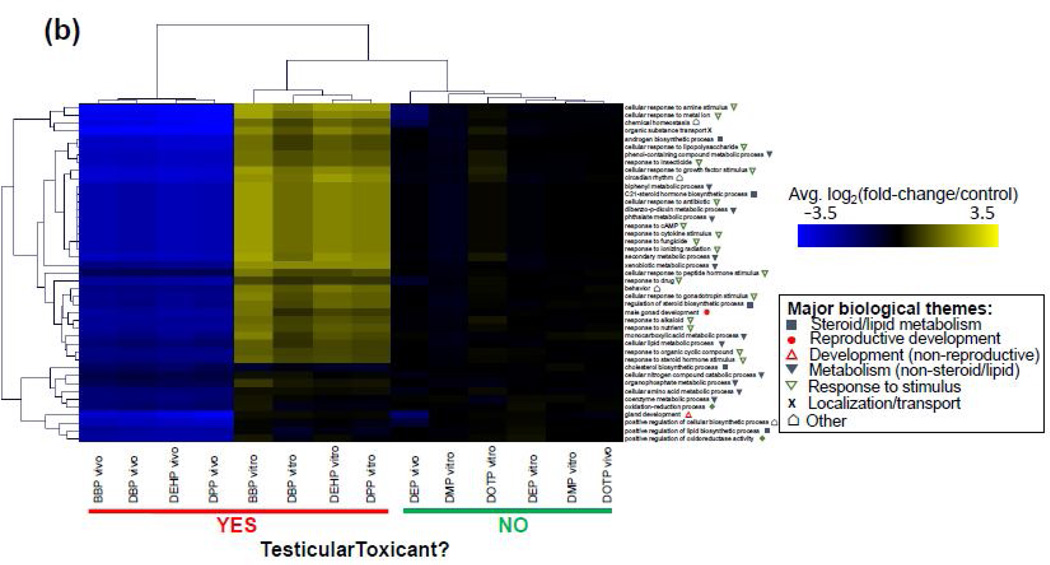
A more targeted pathway analysis using genes that were commonly impacted by all of the DTPE treatments in the 3D-TCS (in vitro) and in the fetal rat testes (in vivo) was conducted. Multiple biological processes were significantly enriched in vitro (n=120) and in vivo (n=43). Overall biological themes represented among these pathways included apoptosis, cell cycle, development, hormone metabolism, response to stimulus and cell homeostasis. Analysis of enriched GO terms showed that several cellular pathways with significant mechanistic implications for steroid metabolism or male reproductive development were significantly enriched both in vitro ("androgen metabolic process”, “steroid catabolic process”, “reproductive structure development”, see fig. 5a) and in vivo (“androgen biosynthetic process”, “male gonad development”, see fig. 5b) after exposure to developmentally toxic phthalates (see Supplementary Tables 1 and 2 for full list of enriched biological processes and z-scores). By contrast, GO terms related to cellular processes such as cell cycle, cell proliferation and apoptosis were enriched for DTPE treatments in vitro, but not in vivo. Developmentally non-toxic phthalates had comparably little impact on the expression of genes used for pathway analysis. Six processes were commonly enriched in vitro and in vivo (“cellular response to amine stimulus”, “response to cAMP”, “response to drug”, “response to ionizing radiation”, “response to organic cyclic compound” and “xenobiotic metabolic process”) that were involved with either cellular response to a stimulus (either exogenous or endogenous) or metabolism of xenobiotics. Figs 5 (a) and (b) show heatmaps for average change in gene expression for biological processes enriched in vitro and in vivo, respectively.
Fig. 5. Identification of enriched Gene Ontology defined biological processes using GO-Elite software.
Enriched processes were identified by a z-score>2 and perm. p-value<0.05) were for genes changed across toxic phthalate treatments (a) in vitro (3D-TCS) or (b) in vivo (fetal rat testes). Average log2 (fold-change/control) for genes changed in each process were calculated for all phthalate treatments.
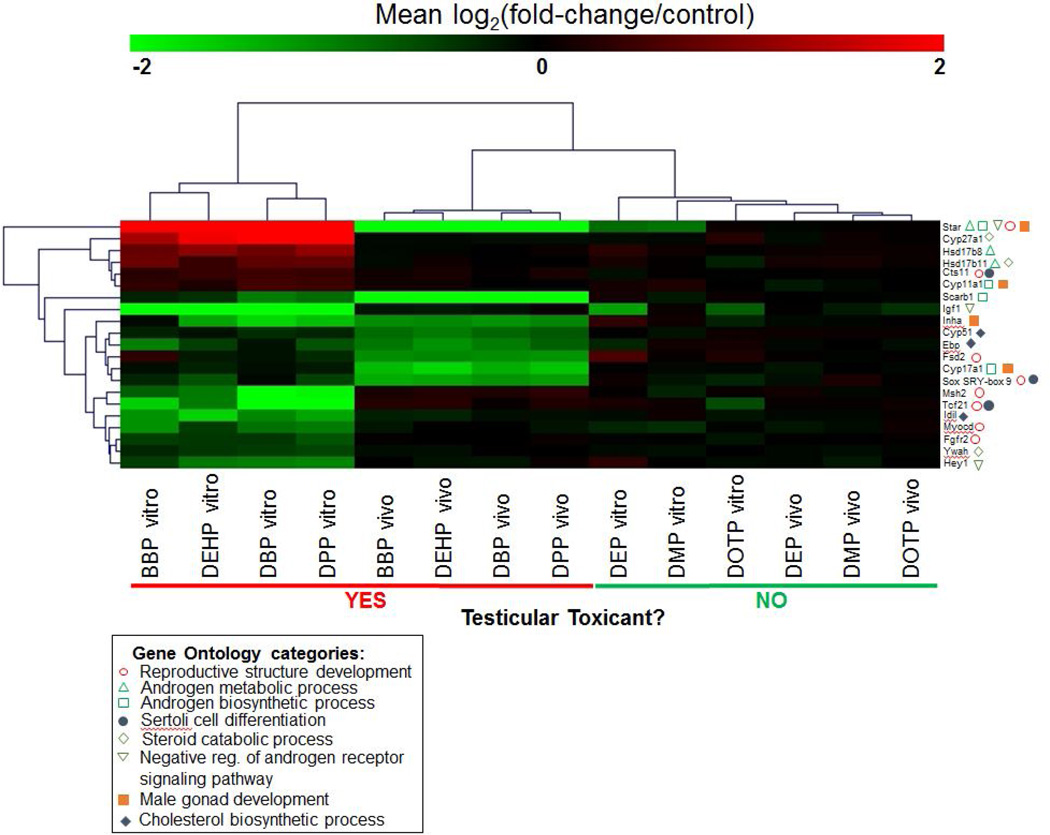
By comparing the enrichment of gene ontology terms between the in vitro and in vivo transcriptomic profiles, a list of 21 genes was compiled and which are highly relevant to the pathways of steroid biosynthesis, male reproductive development or both. Fig. 6 shows the direction and magnitude for gene changes both in vitro and in vivo for this group of genes, as well as which Gene Ontology process the genes belong to. Multiple genes belonged to >1 biological process. The pattern of gene expression changes for these individual genes was consistent with the overall patterns we had observed previously, i.e. the DTPE treatments induced gene expression changes that were clearly distinct from the DNTPE treatments. A few genes, such as Scarb1 (involved in the uptake of cholesterol for the purposes of steroid biosynthesis) and Inha, were downregulated by DTPE exposure both in vitro and in vivo. However, the majority of gene expression changes showed a distinction between the two models, either in terms of direction of regulation (e.g. upregulated in one, downregulated in the other) or genes that were changed in only one of the model systems (e.g. changed in vitro only).
Fig. 6. Individual gene expression changes for genes involved in reproductive development or steroid biosynthesis.
Based on the pathway analysis of the in vitro (3D-TCS) and in vivo (fetal rat testes) data, we identified a group of 21 genes which are involved in either steroid metabolism or reproductive development. Changes in gene expression compared to control samples are shown for both in vitro (3D-TCS) and in vivo (fetal rat testes) exposures for these 21 genes.
3.3 Steroid biosynthesis pathway
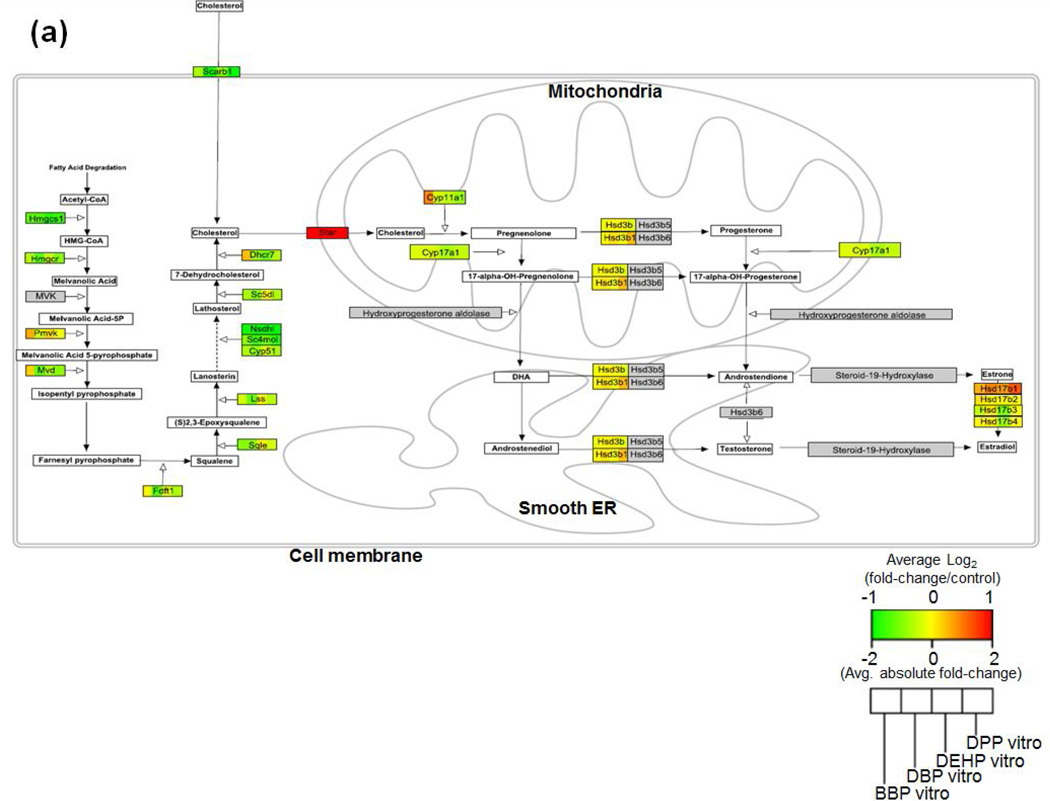
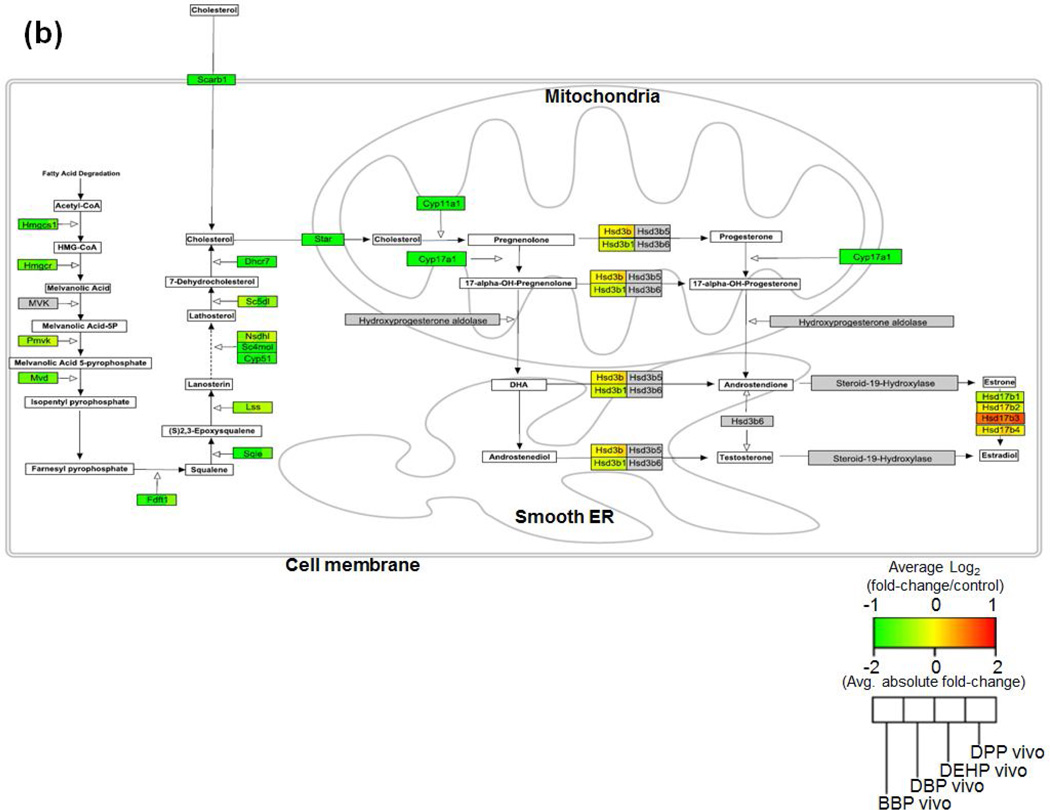
Alterations in steroid biosynthetic gene expression and associated decreases in testosterone are considered the key mode of phthalate testes toxicity [24, 25]. In addition, the current study identified that processes related to steroid metabolism were impacted by DTPE exposure in the 3D-TCS as well as in the rat fetal testes (in vitro: “androgen metabolic process”, in vivo: “androgen biosynthetic process”). Therefore, we sought to investigate effects on genes involved in the synthesis of testosterone. As shown in figs. 7 (a) and (b), multiple genes were altered after exposure to the reproductively toxic PE’s both in vitro and in vivo, respectively. In our testicular co-culture model, most of the effects appeared to be occurring upstream of cholesterol import into the mitochondria. However, Star, the key gene responsible for cholesterol import into the mitochondria (a rate limiting step in testosterone synthesis), was highly upregulated. By comparison all the genes changes in the in vivo rat testes were downregulated, including Star.
Fig. 7. Visualization of gene expression changes in the steroid biosynthesis pathway after exposure to toxic PE’s in the (a) in vitro (3D-TCS) and (b) in vivo (fetal rat testes).
Multiple references were used to construct the rat steroid biosynthesis pathway using Pathvisio software. Gene expression changes (log2fold-change/control) for genes in the pathway are shown, with red=upregulation, green=downregulation, grey=no gene data found and white=metabolite/precursors.
4. Discussion
Gene expression profiling is a powerful tool in toxicology. Changes in gene expression can be more sensitive signals than other toxicity endpoints such as cytotoxicity and can provide key insight into mechanisms of action [26]. In our previous publication, we noted that phthalate ester impacts on cell viability in the 3D-TCS were able to be used to discriminate between DTPE and DNTPE. While some DTPE could be identified using effects on cell viability as an endpoint, analysis of transcriptomic responses proved to be a much more reliable endpoint in discriminating between these two class of related compounds [9]. This comparison of the transcriptomic changes in our testes co-culture (3D-TCS) with effects observed in in vivo fetal rat testes provides important information which aids in the interpretation of data obtained from the 3D-TCS model. Despite differences in experimental design (acute exposure in vitro versus subchronic exposure in vivo) and the observed differences in expression of single genes and pathways when comparing the in vitro and in vivo testes models, we confirmed that our in vitro co-culture model was able to distinguish between developmentally toxic (DTPE) and non-toxic phthalate esters (DNTPE) based on overall gene expression changes and using a pathway based analysis. This analysis of perturbed cellular pathways showed that changes in the 3D-TCS were consistent with phthalate modes of action (MOA’s) in vivo, i.e. disruption of pathways involved in steroid biosynthesis and overall testes development [27, 28].
4.1 Patterns of gene expression changes discriminate between developmentally toxic and non-toxic phthalates in vitro and in vivo
An unsupervised principle components analysis (PCA) of all gene expression data on each array showed that the DTPE treatments formed a distinct cluster from the DNTPE treatments (and controls) both for the in vitro co-culture model and the in vivo testes. In addition, t-tests identifying genes significantly changed compared to controls showed that DTPE treatments were associated with a higher number of gene changes compared to DNTPE treatments both in vitro and in vivo. This demonstrates that for this class of structurally similar compounds (phthalates), transcriptomic profiles from the 3D-TCS are able to discriminate between those that are potent developmental toxicants from those that are weak or nondevelopmental toxicants.
4.2 Pathway based analysis shows that mechanistic aspects of DTPE toxicity in the 3D-TCS are consistent with phthalate toxicity in vivo
In order to gain insight into the mechanistic aspects of phthalate effects on transcriptomic responses, we performed an unsupervised pathway analysis using expression data for all genes represented on each array. Consistent with the PCA analysis, cluster analysis based on similarities in t-scores for the enriched Gene Ontology terms showed that the in vitro and in vivo DTPE treatments clustered separately from the DNTPE treatments. In general, DTPE impacts on pathways or processes could be said to be either: upregulated in vitro/downregulated in vivo, downregulated in vitro/upregulated in vivo or downregulated both in vitro and in vivo. Among processes that were downregulated both in vitro and in vivo, were the GO terms “cholesterol biosynthesis” and “isoprenoid biosynthesis”. Impacts on such pathways, which relate to the metabolism of cholesterol and steroid precursors, represent one of the mechanisms of phthalate reproductive toxicity. Altered expression of genes responsible for steroid, lipid and cholesterol metabolism have been shown to be associated with decreased serum testosterone, cryptorchidism, hypospadias and decreased anogenital distance (AGD) in male rats in vivo. [29, 30]. Indeed, the male rats from which gene expression data were obtained for the current study showed a decreased AGD [10]. Thus, using a pathway based approach and applying no cutoffs for significant gene changes, we showed that important mechanisms of in vivo phthalate toxicity were captured in the 3D-TCS model.
In order to further investigate in vivo toxicity responses that were reflected in the 3D-TCS transcriptomic signals, we identified genes which were commonly altered by reproductively toxic phthalates in our co-culture model (440 genes) and those commonly altered in the in vivo rat testes (35 genes). We then used GO-Elite to identify biological processes enriched in each of these genelists and quantified the average change in gene expression for these processes. GO terms enriched by DTPE treatments in vitro included a broad range of biological themes, including apoptosis, cell cycle, cell signaling, development, redox homeostasis, steroid metabolism and reproductive development. These results are consistent with previous reports discussing transcriptomic signals for male reproductive toxicity of phthalates. For example, in a recent evaluation of phthalate toxicogenomic data, Euling et al. discuss three main modes of action (MOAs) for phthalate reproductive effects in the male. One involves the decrease in fetal testosterone, which leads to perturbed development of the reproductive tract. The second is a decrease in insl3 expression, which has been demonstrated to be responsible for undescended testes. In addition, a third less characterized MOA is thought to be responsible for effects including apoptosis and multinucleation of gonocytes, altered proliferation of Sertoli cells and altered morphology of seminiferous tubules [25]. Analysis of pathway enrichment in the 3D-TCS suggests impacts on pathways that are indicative of at least two of these MOA’s. Enrichment of processes such as androgen metabolism and steroid catabolic process demonstrate a signal indicative of the first (altered testosterone production), while alteration of cell cycle pathways and apoptosis signaling demonstrate potential signals for the third, less well-characterized MOA.
Using the 35 genes which were changed across all toxic treatments in vivo, we identified enrichment for multiple pathways involved in steroid, cholesterol or lipid metabolism as well as reproductive development. By quantifying the average gene expression changes in these pathways in vitro and in vivo, we were able to confirm that impacts to on these pathways occurred in both models. However, we observed a clear distinction between the two models, with average changes in gene expression showing downregulation in vivo, while showing upregualtion in vitro.
At the level of biological processes, several similar GO terms with implications for steroid metabolism were identified for both models (in vitro: “androgen metabolic process”, “steroid catabolic process”; in vivo: “androgen biosynthetic process”, “cholesterol biosynthetic process”). Similar responses in genes related to cholesterol, lipid and steroid (CLS) metabolism have been reported in other in vitro test systems, such as the rat whole embryo culture system, which showed significant impacts on CLS pathways after exposure to the reproductive toxicant MEHP (the main metabolite of DEHP) but comparably little impact after exposure to the non-reproductively toxic MMP (the main metabolite of DMP). In the same study, apoptosis pathways were also upregulated, similar to what we observed in the 3D-TCS [31]. Thus, these results demonstrate a consistency in modes of toxicity observed in the 3D-TCS when compared to both in vitro and in vivo systems, as phthalate exposure in rats in vivo has led to disruption of CLS metabolism and apoptosis of testicular cells [32–34]. In addition to these CLS and apoptosis pathways, pathways related to general reproductive development were altered by phthalates in the 3D-TCS and in the in vivo rat testes (in vitro: “reproductive structure development”, in vivo: “male gonad development”).
Looking more specifically at the steroidogenic pathway, we observed that several genes both up and downstream of cholesterol synthesis were significantly altered by at least one DTPE under both in vitro and in vivo experimental scenarios. Notably, Star and Scarb1, which code for key proteins involved in the transport of cholesterol during the process of steroidogenesis, were significantly altered both in vitro and in vivo [35]. In addition, several important CYP450’s with important roles in the metabolism of cholesterol and testosterone (Cyp11a1, Cyp17a1 and Cyp51) were downregulated by one or more reproductively toxic phthalates both in vitro and in vivo. Furthermore, 2 of the 4 genes which were commonly altered by all DTPE treatments in vitro and in vivo have roles in steroidogenesis. Star performs the rate-limiting function of importing cholesterol into the mitochondria, while Fdx-1 is involved in transferring electrons to steroidogenic CYP450’s [36].
After performing pathway analysis, we identified a group of individual genes belonging to pathways highly relevant to male reproductive biology which were altered by DTPE treatment. After plotting gene expression for these 21 genes, we observed a response in the in vitro system in which genes were either up or downregulated after exposure to DTPE (there were few to no significant changes in the DNTPE exposure groups). In contrast, for the in vivo rat testes, almost all of the genes were downregulated. This suggests a complex and dynamic transcriptomic response to phthalates in the 3D-TCS.
4.3 Mapping changes in the steroid biosynthesis pathway
As we mentioned previously, the downregulation of steroidogenic genes and the decreased production of testosterone in the male testes is a well-studied mode of action for phthalate reproductive toxicity [34, 37]. In order to investigate the dynamics of gene expression changes in this key pathway, we used Pathvisio to map the steroid biosynthetic pathway [20]. This pathway includes major reactions from de novo cholesterol biosynthesis (or import into the cell via Scarb1) to the formation of various steroids including testosterone and estradiol. By generating this map and showing the associated changes in gene expression, we compared DTPE effects in vivo and in vitro. Fig. 7 (a+b) clearly shows that there are multiple genes upstream of cholesterol biosynthesis which were downregulated both in vitro and in vivo. However, Star, which performs the rate-limiting function of importing cholesterol into the mitochondria, was highly upregulated by DTPE in vitro, while being downregulated in vivo. This result shows that, although we can identify dysregulation of certain key pathways after toxicant exposure, it may be necessary to investigate impacts on the level of individual genes in order to fully understand the dynamics of toxicant impacts on cellular processes in the 3D-TCS.
The difference in responses that we observed could be explained by regulatory feedback mechanisms within the steroid biosynthesis pathway. Phthalate disruption of steroid metabolism may lead to decreased availability of steroid hormone precursors and associated adaptive response resulting in an increase in expression of certain steroidogenic genes. These biphasic responses to phthalate exposure have been observed in multiple studies and include reproductive or developmental endpoints such as onset of puberty, steroidogenic gene expression and testosterone production [38, 39]. Differences in response may also be due to developmental stage at which phthalate exposure occurs. For example, some studies in which prenatal animals were exposed to PE’s during the critical period of male reproductive development (approximately gestational days 12–19) resulted in damage to testicular tissue and decreased steroidogenic gene expression [23, 40]. However, other experiments in which treatments occurred outside of this window showed an increase in expression of steroidogenic genes [38]. Similarly, cells in the 3D-TCS, cultured from 5 day old rats (outside the GD 12–19 window) showed an increase in Star after PE exposure. Differences in response to phthalate exposure have also been noted for different doses administered both in vitro and in vivo. For example, Ge et al. [39] found that DEHP exposure at a dose of 10mg/kg/day for 28 days resulted in advancement of pubertal onset and increased serum testosterone for Long Evans rats while 750mg/kg/day resulted in delayed onset of puberty and decrease in serum testosterone. In the same study, MEHP (active metabolite of DEHP) treatment of cultured Leydig cells at 100µM increased testosterone production, while 10mM had the opposite effect [39].
4.4 In vitro/in vivo comparisons
Making comparisons between in vitro and in vivo transcriptomic data presents multiple challenges. Results can be strongly influenced by experimental design and a number of important factors must be taken into account when interpreting data. Comparison of in vitro and in vivo doses can be challenging. In addition, timing of exposure and kinetic factors may influence in vitro cellular responses to toxicant exposure that may make direct comparisons to in vivo exposures difficult. The doses of phthalates utilized in the 3D-TCS were chosen based on levels showing minimal impact on cell viability. For our in vivo comparison, we chose published data which a) used doses of phthalates that produced significant changes in gene expression pathways in male fetal testes without loss of the fetuses or maternal toxicity and b) was associated with a well characterized indicator of male developmental toxicity (i.e. significant decreases in anogential distance). Decreased AGD is one of several male reproductive malformations consistent with “phthalate syndrome” and the in vivo doses used in this comparison are known to generate this condition in rats. The “phthalate syndrome” observed in rats bears a striking resemblance to the testicular dysgenesis syndrome in humans and therefore a highly relevant disease state to use for linking our in vitro transcriptomic changes to in vivo outcomes relevant to humans.
One notable difference between the in vivo and in vitro responses being compared in this study were the relatively large number of gene expression changes observed in the 3D-TCS compared to the in vivo exposures. Fortunately, we were able to address these differences in the distribution of gene changes by utilizing a PCA analysis and the approach provided by the ToxProfiler software. Using these approaches we analyzed the entire set of gene expression data for principle components and pathway analysis, as opposed to applying a statistical cutoff for gene lists. This approach allowed us to demonstrate that, regardless of the differences in absolute number of gene changes, both the 3D-TCS and in vivo testes transcriptomic profiles are able to discriminate between DTPE’s and DNTPE’s.
It is possible that some differences in transcriptomic responses were due to kinetic or metabolic processes which occur in vivo but are not present in the testes co-culture model. Metabolism and kinetics of compounds in vitro present a challenge when determining how the data can be can be applied in an in vivo context [41]. For example, the monoester metabolites of PE’s are considered to be mostly responsible for male reproductive toxicity [13, 42–44]. Clewell et al. found a concentration of 100µM of MBP (the active metabolite of DBP) in fetal testes approximately 1 hour after administration of 500mg/kg DBP to pregnant Dams. This was followed by a peak of 150µM after 2 hours [45]. By comparison, the in vitro exposure to cells in the current study was to 100µM of parent compound. While the doses of phthalates used in this study are relatively high compared to typical human blood levels (often in the nanomolar range), for some scenarios such as during medical procedures (in which phthalates can leach from medical into the bloodstream) blood serum concentrations have been reported as high as 250µM for infants [46]. Furthermore, in a separate study Clewell, et al. reported levels of various phthalate metabolites appearing in rat testes under the same exposure regime (i.e. 500mg/kg of parent compounds given to pregnant Dams from postnatal days 12–19). The levels of monoester metabolites observed in fetal testes varied and ranged from as high as 400µM (for DEP, DMP exposures) to as low as 12µM (DEHP exposures) [47]. In separate experiments, we have determined that cells in the 3D-TCS metabolize phthalate parent compounds to their monoester metabolites (Harris, et al. 2015, submitted manuscript). Therefore, the 100µM levels of phthalates used in the 3D-TCS are within this range of concentrations. In the current study, in vitro responses in the 3D-TCS were compared to in vivo responses in rat testes using pathway based transcriptomic approaches. We noted distinct differences in relevant male reproductive pathways when comparing reproductively toxic and non-toxic phthalate esters exposures. Figs 4. (a+b) show various processes involved with steroid metabolism or male reproductive development that were differentially impacted by exposure to reproductively toxic phthalates both in the 3D-TCS and in the rat fetal testes (e.g. “androgen metabolic process” and “male gonad development”). Thus, using the transcriptomic approach, when we compare across the phthalates we observed that DTPE clearly impact more pathways relevant to male reproductive toxicity than DNTPE. In the supplemental data, we examined the power of detection for phthalate induced responses in terms of the number of differentially expressed genes used for analysis of critical pathways in the 3D-TCS. For example, using DAVID pathway analysis software, Gene Ontology defined Biological Processes relevant to male reproductive development were detected when ≥1000 of genes changed by phthalate treatments were included in the genelist (Suppl. Table 3). This gave us additional insight that will be critical in developing mechanism based, transcriptomic analysis of male reproductive toxicity signals in the 3D-TCS.
One of the unique strengths of the current study was the availability of transcriptomic data from seven related compounds with well-known in vivo structure activity data in regards to male reproductive effects and the ability to compare these to responses in a relevant in vitro organotypic model. It has been shown in numerous studies (as well in the case of the in vivo data used in the current study) that structurally related phthalate esters have distinct impacts on male reproductive end points, with the length of side-chain being closely correlated with testicular toxicity (side chains of four to six carbon atoms in the ortho-position are known to induce testes toxicity) and these effects are evident in transcriptomic signals [7, 29]. In the current study, we have demonstrated that the transcriptomic profiles of phthalate exposure in the 3D-TCS reflect these in vivo signals.
Additional end points with functional relevance to male reproductive development should be examined in order to compliment gene expression profiles. For example, effects on testosterone production and induction of inflammatory cytokines are currently under investigation. In addition, characterization of the metabolic capacity of the system will enhance our ability to predict and interpret responses to various toxicants in the model as well as provide crucial information of the extent to which the testes co-culture maintains the metabolic activity of in vivo testes. Proteomic analysis and assessment of enzymatic activity for key steroidogenic enzymes would also provide valuable information to compliment transcriptomic responses.
4.5 Conclusions
In conclusion, transcriptomic data showed that the 3D-TCS co-culture model was able to differentiate between developmentally toxic and non-toxic phthalate esters based on patterns of gene expression changes and through pathways analysis. Pathway analysis of in vitro and in vivo transcriptomic data showed that a number of cellular processes related to male reproductive development and hormone signaling were enriched both in vitro and in vivo. Mapping the steroidogenic pathway allowed us to compare the dynamics of changes within this important pathway between the in vitro and in vivo scenarios. Results indicate that DTPE exposure induced changes in cellular pathways in the 3D-TCS which are related to major modes of action for male reproductive toxicity of phthalates. This study facilitated the evaluation of the 3D-TCS toxicogenomic data within the context of a proposed AOP for phthalate reproductive toxicity (i.e. disruption of steroidogenesis and associated effects on male reproductive development), allowing us to link reproductive and developmental pathways impacted by phthalates in the 3D-TCS with those impacted by phthalates in other in vitro cell systems and at higher levels of biological organization (e.g. decreased anogential distance in vivo) [11, 12, 31, 39]. Despite challenges, the comparison between data obtained from this model with data generated from the in vivo study has given us valuable information which improves our ability to interpret responses to phthalate exposure in our coculture system.
Supplementary Material
Highlights.
We have developed a three-dimensional testicular co-culture system (3D-TCS)
We compared transcriptomic responses to phthalates in 3D-TCS and rat testes in vivo
Steroid metabolism and reproductive pathways were effected in vitro and in vivo
Transcriptomic responses in 3D-TCS reflect key aspects of in vivo phthalate toxicity
Acknowledgments
This work was supported by in part by the UW NIEHS Center for Ecogenetics and Environmental Health (5 P30 ES007033), UW EPA Center for Predictive Toxicology (RD-83573801), US-FDA (FDA: 1U01FD004242), and the NIH Center on Human Development and Disability (1 U54 HD083091-01) and Niels Stensen Fellowship. We gratefully acknowledge Kevin Gaido for generously providing the in vivo transcriptomic data. We would additionally like to thank Kirk Van Ness (Institute for Risk Analysis and Risk Communication, University of Washington) for his helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
References
- 1.Rovida C, Hartung T. Re-evaluation of animal numbers and costs for in vivo tests to accomplish REACH legislation requirements for chemicals - a report by the transatlantic think tank for toxicology (t(4)) ALTEX. 2009;26(3):187–208. [PubMed] [Google Scholar]
- 2.Scialli AR. The challenge of reproductive and developmental toxicology under REACH. Regul Toxicol Pharmacol. 2008;51(2):244–250. doi: 10.1016/j.yrtph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Adler S, et al. Alternative (non-animal) methods for cosmetics testing: current status and future prospects-2010. Arch Toxicol. 2011;85(5):367–485. doi: 10.1007/s00204-011-0693-2. [DOI] [PubMed] [Google Scholar]
- 4.Parks Saldutti L, et al. In vitro testicular toxicity models: opportunities for advancement via biomedical engineering techniques. ALTEX. 2013;30(3):353–377. doi: 10.14573/altex.2013.3.353. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, et al. Essential role of extracellular matrix (ECM) overlay in establishing the functional integrity of primary neonatal rat Sertoli cell/gonocyte co-cultures: an improved in vitro model for assessment of male reproductive toxicity. Toxicol Sci. 2005;84(2):378–393. doi: 10.1093/toxsci/kfi085. [DOI] [PubMed] [Google Scholar]
- 6.Mylchreest E, Cattley RC, Foster PM. Male reproductive tract malformations in rats following gestational and lactational exposure to Di(n-butyl) phthalate: an antiandrogenic mechanism? Toxicol Sci. 1998;43(1):47–60. doi: 10.1006/toxs.1998.2436. [DOI] [PubMed] [Google Scholar]
- 7.Gray LE, Jr, et al. Perinatal exposure to the phthalates DEHP, BBP, and DINP, but not DEP, DMP, or DOTP, alters sexual differentiation of the male rat. Toxicol Sci. 2000;58(2):350–365. doi: 10.1093/toxsci/58.2.350. [DOI] [PubMed] [Google Scholar]
- 8.Bajkin I, et al. Effects of phthalic acid esters on fetal health. Med Pregl. 2014;67(5–6):172–175. doi: 10.2298/mpns1406172b. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, et al. Improving in vitro Sertoli cell/gonocyte co-culture model for assessing male reproductive toxicity: Lessons learned from comparisons of cytotoxicity versus genomic responses to phthalates. Toxicol Appl Pharmacol. 2009;239(3):325–336. doi: 10.1016/j.taap.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, et al. Gene expression profiling following in utero exposure to phthalate esters reveals new gene targets in the etiology of testicular dysgenesis. Biol Reprod. 2005;73(1):180–192. doi: 10.1095/biolreprod.104.039404. [DOI] [PubMed] [Google Scholar]
- 11.Furr JR, et al. A short-term in vivo screen using fetal testosterone production, a key event in the phthalate adverse outcome pathway, to predict disruption of sexual differentiation. Toxicol Sci. 2014;140(2):403–424. doi: 10.1093/toxsci/kfu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phthalates Cumulative Risk Assessment: The Task Ahead. 2008 [Google Scholar]
- 13.Gray TJ, Gangolli SD. Aspects of the testicular toxicity of phthalate esters. Environ Health Perspect. 1986;65:229–235. doi: 10.1289/ehp.8665229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eijssen LM, et al. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Research. 2013;41(Web Server issue):W71–W76. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson K. On lines and planes of closest fit to systems of points in space. Philosophical Magazine. 1901;2:559–572. [Google Scholar]
- 16.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 17.Boorsma A, et al. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Research. 2005;33(Web Server issue):W592–W595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 19.Zambon AC, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28(16):2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Iersel MP, et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43(8):779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- 22.Willighagen E, Andrew Kwa, Daniela Digles, Manny Ramirez. Steroid Biosynthesis (Rattus norvegicus) [cited 2015 Jan. 10];2013 Available from: http://www.wikipathways.org/index.php/Pathway:WP66. [Google Scholar]
- 23.Barlow NJ, et al. Quantitative changes in gene expression in fetal rat testes following exposure to di(n-butyl) phthalate. Toxicol Sci. 2003;73(2):431–441. doi: 10.1093/toxsci/kfg087. [DOI] [PubMed] [Google Scholar]
- 24.David RM. Proposed mode of action for in utero effects of some phthalate esters on the developing male reproductive tract. Toxicol Pathol. 2006;34(3):209–219. doi: 10.1080/01926230600642625. [DOI] [PubMed] [Google Scholar]
- 25.Euling SY, et al. Use of genomic data in risk assessment case study: II. Evaluation of the dibutyl phthalate toxicogenomic data set. Toxicol Appl Pharmacol. 2011 doi: 10.1016/j.taap.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Corvi R. Genomics: an in vitro toxicology point of view. Altern Lab Anim. 2002;30(Suppl 2):129–131. doi: 10.1177/026119290203002S21. [DOI] [PubMed] [Google Scholar]
- 27.Martino-Andrade AJ, Chahoud I. Reproductive toxicity of phthalate esters. Mol Nutr Food Res. 2010;54(1):148–157. doi: 10.1002/mnfr.200800312. [DOI] [PubMed] [Google Scholar]
- 28.Parks LG, et al. The plasticizer diethylhexyl phthalate induces malformations by decreasing fetal testosterone synthesis during sexual differentiation in the male rat. Toxicol Sci. 2000;58(2):339–349. doi: 10.1093/toxsci/58.2.339. [DOI] [PubMed] [Google Scholar]
- 29.Foster PM, et al. Effects of phthalate esters on the developing reproductive tract of male rats. Hum Reprod Update. 2001;7(3):231–235. doi: 10.1093/humupd/7.3.231. [DOI] [PubMed] [Google Scholar]
- 30.Welsh M, et al. Identification in rats of a programming window for reproductive tract masculinization, disruption of which leads to hypospadias and cryptorchidism. J Clin Invest. 2008;118(4):1479–1490. doi: 10.1172/JCI34241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JF, et al. Dose-response analysis of phthalate effects on gene expression in rat whole embryo culture. Toxicol Appl Pharmacol. 2012;264(1):32–41. doi: 10.1016/j.taap.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Kasahara E, et al. Role of oxidative stress in germ cell apoptosis induced by di(2-ethylhexyl)phthalate. Biochem J. 2002;365(Pt 3):849–856. doi: 10.1042/BJ20020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, et al. The Fas system is a key regulator of germ cell apoptosis in the testis. Endocrinology. 1997;138(5):2081–2088. doi: 10.1210/endo.138.5.5110. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann KP, et al. Dose-dependent alterations in gene expression and testosterone synthesis in the fetal testes of male rats exposed to di (n-butyl) phthalate. Toxicol Sci. 2004;81(1):60–68. doi: 10.1093/toxsci/kfh169. [DOI] [PubMed] [Google Scholar]
- 35.Manna PR, Dyson MT, Stocco DM. Regulation of the steroidogenic acute regulatory protein gene expression: present and future perspectives. Mol Hum Reprod. 2009;15(6):321–333. doi: 10.1093/molehr/gap025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller WL. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology. 2005;146(6):2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 37.Foster PM. Disruption of reproductive development in male rat offspring following in utero exposure to phthalate esters. Int J Androl. 2006;29(1):140–147. doi: 10.1111/j.1365-2605.2005.00563.x. discussion 181-5. [DOI] [PubMed] [Google Scholar]
- 38.Ryu JY, et al. Time-response effects of testicular gene expression profiles in Sprague-Dawley male rats treated with di(n-butyl) phthalate. J Toxicol Environ Health A. 2008;71(23):1542–1549. doi: 10.1080/15287390802391992. [DOI] [PubMed] [Google Scholar]
- 39.Ge RS, et al. Biphasic effects of postnatal exposure to diethylhexylphthalate on the timing of puberty in male rats. J Androl. 2007;28(4):513–520. doi: 10.2164/jandrol.106.001909. [DOI] [PubMed] [Google Scholar]
- 40.Hannas BR, et al. Dipentyl phthalate dosing during sexual differentiation disrupts fetal testis function and postnatal development of the male Sprague-Dawley rat with greater relative potency than other phthalates. Toxicol Sci. 2011;120(1):184–193. doi: 10.1093/toxsci/kfq386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coecke S, et al. Metabolism: a bottleneck in in vitro toxicological test development. The report and recommendations of ECVAM workshop 54. Altern Lab Anim. 2006;34(1):49–84. doi: 10.1177/026119290603400113. [DOI] [PubMed] [Google Scholar]
- 42.Ema M, et al. Decreased anogenital distance and increased incidence of undescended testes in fetuses of rats given monobenzyl phthalate, a major metabolite of butyl benzyl phthalate. Reprod Toxicol. 2003;17(4):407–412. doi: 10.1016/s0890-6238(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 43.Howdeshell KL, et al. Cumulative effects of dibutyl phthalate and diethylhexyl phthalate on male rat reproductive tract development: altered fetal steroid hormones and genes. Toxicol Sci. 2007;99(1):190–202. doi: 10.1093/toxsci/kfm069. [DOI] [PubMed] [Google Scholar]
- 44.Creasy DM, Beech LM, Gray TJ. Effects of mono-(2-ethylhexyl) phthalate and mono-n-pentyl phthalate on the ultrastructural morphology of rat Sertoli cells in Sertoli/germ cell co-cultures: Correlation with the in vivo effects of di-n-pentyl phthalate. Toxicol In Vitro. 1988;2(2):83–95. doi: 10.1016/0887-2333(88)90018-5. [DOI] [PubMed] [Google Scholar]
- 45.Clewell RA, et al. Kinetics of selected di-n-butyl phthalate metabolites and fetal testosterone following repeated and single administration in pregnant rats. Toxicology. 2009;255(1–2):80–90. doi: 10.1016/j.tox.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Miles-Richardson S. In: Toxicological Profile for Di(2-ethylhexyl)phthalate (DEHP) U.S.D.o.H.a.H, editor. 2002. [Google Scholar]
- 47.Clewell RA, et al. Assessing the relevance of in vitro measures of phthalate inhibition of steroidogenesis for in vivo response. Toxicol In Vitro. 2010;24(1):327–334. doi: 10.1016/j.tiv.2009.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.