Abstract
Deregulated expression of MYC is a driver of colorectal carcinogenesis, suggesting that inhibiting MYC may have significant therapeutic value. The PI3-kinase and mTOR pathways control MYC turnover and translation, respectively, providing a rationale to target both pathways to inhibit MYC. Surprisingly, inhibition of PI3-kinase does not promote MYC turnover in colon carcinoma cells, but enhances MYC expression since it promotes FOXO-dependent expression of growth factor receptors and MAPkinase-dependent transcription of MYC. Inhibition of mTOR fails to inhibit translation of MYC, since levels of 4E-BPs are insufficient to fully sequester eIF4E and since an IRES-element in the 5’-UTR of the MYC permits translation independent of eIF4E. A small molecule inhibitor of the translation factor, eIF4A, silvestrol, bypasses the signaling feedbacks, reduces MYC translation and inhibits tumor growth in a mouse model of colorectal tumorigenesis. We propose that targeting translation initiation is a promising strategy to limit MYC expression in colorectal tumors.
Keywords: MYC, eIF4A, FBXW7, silvestrol, colorectal tumors
Introduction
With over 1.2 million newly diagnosed cases per year, colorectal cancer (CRC) is the most common gastrointestinal malignancy (1). Sequence analysis shows that each tumor genome carries multiple mutations that deregulate major signaling pathways that control growth and survival of colon epithelial cells (2). Despite their genomic heterogeneity, enhanced expression of MYC proteins is universally observed in colon cancers and gene expression analyses show that a signature of activated and repressed MYC-target genes is present in a vast majority of CRCs (2). Deletion of the MYC gene ablates tumorigenesis in mouse models that faithfully mimic the human disease (3). Collectively, these data argue that targeting MYC might achieve significant therapeutic efficacy in CRCs.
MYC is a transcription factor that binds broadly to thousands of promoters and enhancers and activates or represses its target genes as part of several DNA binding protein complexes (4). Both direct and indirect strategies have been proposed to inhibit MYC function and expression (5–7). The MYC protein is highly unstable in non-tumor cells and is constantly degraded by the proteasome system (8). Several ubiquitin ligases are known that ubiquitinate MYC and ubiquitination by FBXW7 targets MYC for proteasomal degradation (8). FBXW7 is frequently mutated in human CRC enhancing the stability of MYC (9). Furthermore, CRCs express high levels of USP28, an ubiquitin protease that binds to FBXW7 and antagonizes its function; deletion of USP28 reduces MYC levels and extends life span in colon tumor models (10). Enhancing MYC turnover therefore may be a valid strategy to inhibit MYC function in CRC.
Degradation of MYC by FBXW7 is initiated by phosphorylation at S62, which primes subsequent phosphorylation at T58 by GSK3 (8). Subsequent de-phosphorylation at S62 by PP2A allows recognition and ubiquitination of T58-phosphorylated MYC by FBXW7 (8). GSK3 itself is inhibited by PI3K/AKT-dependent phosphorylation at S9 and inhibitors of PI3K or dual mTOR/PI3K-inhibitors enhance N-MYC turnover in pediatric tumors (11). Conversely, ectopic expression of MYC confers resistance of mammary tumor cells to PI3-kinase inhibition (12). A second rationale to target the PI3-kinase/mTOR pathway is provided by its ability to enhance CAP-dependent translation initiation. mTORC1 and the downstream S6 kinase promote translation since they phosphorylate and thereby inactivate the 4E-BP and PDCD4 proteins that inhibit the eIF4F translation initiation complex (13, 14). As consequence, inhibition of mTORC1 blocks MYC expression in myeloma cells and targeting protein translation limits the growth of MYC-driven hematopoietic tumors (15).
Here we have explored whether targeting signaling pathways that control MYC turnover and translation can be used to eliminate MYC expression in CRC, using the dual mTOR/PI3-kinase inhibitor BEZ235 and the eIF4A helicase inhibitor, silvestrol, as tools (16, 17). We show that targeting PI3K and mTOR fails to increase MYC turnover and instead enhances MYC expression and functionality. In contrast, directly targeting translation initiation bypasses the feedback mechanisms that cause this surprising response, reduces MYC expression and inhibits tumor growth in mouse models of colorectal carcinoma.
Results
FBXW7-pathway is active in colon carcinoma cells
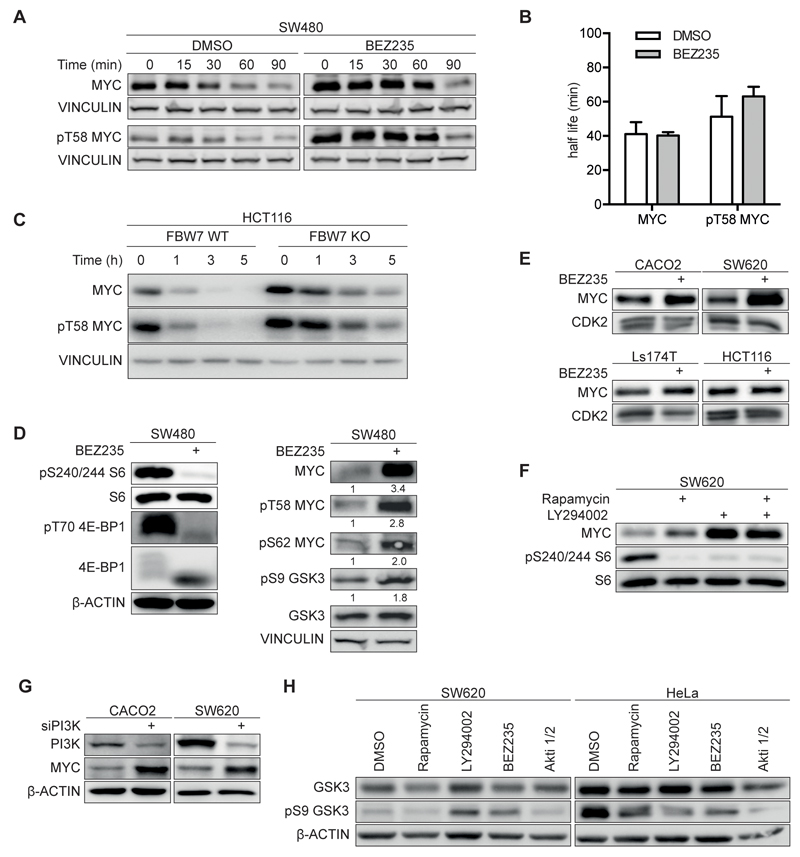
In many cells, MYC proteins turn over with a half-life of approximately 20 minutes (8). To determine the stability of MYC in CRC, we added cycloheximide to block new protein synthesis and determined the amount of MYC by immunoblotting at several time points afterwards (Figure 1A,B,C and Supplementary Figure 1A,B; see Supplementary Table 1 for all antibodies and primer sequences). MYC turned over with a half-life that was between 41 to approximately 60 minutes in SW480, SW620 and HCT116 cells, respectively (Figure 1 A,B,C and Supplementary Figure 1 A,B). MYC that is phosphorylated at T58, the recognition site for FBXW7, turned over with a slightly longer half-life in all three cell lines. This is consistent with the notion that FBXW7 is actively degrading a large fraction, but not all of MYC in these cells. In support of this notion, both MYC and pT58MYC turned over with a greatly extended half-life in HCT116 cells, in which the endogenous FBXW7 gene has been disrupted (Figure 1C and Supplementary Figure 1A) (9).
Figure 1. Effect of PI3K/mTORC inhibition on MYC expression and stability in colorectal cancer cells.
A. Immunoblots documenting MYC and pT58 MYC stability. SW480 cells were treated with 200nM BEZ235 or solvent control for 24h and cycloheximide (50µg/ml) and harvested at the indicated time points. Vinculin was used as loading control. Exposures of MYC and pT58 MYC blots were adjusted to equalize exposure at 0 min (n=3; unless otherwise indicated, n indicates the number of independent biological repeat experiments in the following legends).
B. Calculated half-life of total MYC and pT58 MYC. Immunoblots shown in panel A.
C. Immunoblots show MYC and pT58 MYC stability in wild type (WT) and FBXW7 deficient (KO) HCT116 cells (n=1).
D. SW480 cells were incubated with 200nM BEZ235 for 24h. Left panel document effect on mTOR targets S6 and 4E-BP1, right panel on MYC and GSK3 (n=2).
E. Immunoblots of four colorectal cell lines upon treatment with BEZ235 (500nM; 24h) or solvent control (n=3).
F. SW620 cells were treated for 24h with Rapamycin (100nM), LY294002 (50µM) or both and analyzed by immunoblotting.
G. The indicated cell lines were transfected with siRNA targeting the p110alpha subunit of PI3K or control siRNA. 72h after transfection protein levels were determined by immunoblotting (n=2).
H. Immunoblot of cells treated for 24h with indicated inhibitors or solvent control. (Rapamycin 100nM, LY294002 50µM, BEZ235 500nM, Akti 1/2 1µM).
The critical kinase that phosphorylates T58 and promotes degradation of MYC and N-MYC proteins is GSK3 (8). Since GSK3 in turn is inhibited by PI3-kinase/AKT-dependent phosphorylation, we tested the effect of BEZ235, a dual mTOR/PI3-kinase inhibitor, which destabilizes N-MYC in neuroblastoma cells (11). BEZ235 was used at a concentration of 200nM, which is sufficient to inhibit both PI3K and mTOR activity (16). Immunoblotting showed an altered migration of 4E-BP1 and confirmed de-phosphorylation of 4E-BP1 at T70 and of S6 at S240/244, downstream targets of mTOR in response to BEZ235 (Figure 1D and Supplementary Figure 1C). Furthermore, BEZ235 inhibited the AKT-dependent phosphorylation of FOXO3A, demonstrating that PI3-kinase is also inhibited (see below). Consistent with previous observations, exposure of several human colon cancer cell lines to 200nM BEZ235 suppressed proliferation and led to a moderate accumulation in the G1 phase of the cell cycle, but did not induce apoptosis (Supplementary Figure 1D,E,F) (16). Cells resumed proliferation after withdrawal of BEZ235, arguing that the BEZ235-induced cell cycle arrest is reversible (Supplementary Figure 1F).
Surprisingly, the analysis also showed that exposure to BEZ235 increased rather than decreased MYC levels in SW480 cells (Figure 1A,D). Similarly, exposure to BEZ235 led to a robust increase in MYC levels in SW620 and CACO2 cells and a weaker increase in Ls174T and HCT116 cells (Figure 1E). Titration of BEZ235 revealed an IC50 value around 20nM for this increase, consistent with an on-target effect for either mTOR or PI3-kinase (see below). Using rapamycin, a specific inhibitor of mTORC1 and LY294002, an inhibitor of PI3-kinase, showed that inhibition of PI3-kinase was sufficient to induce expression of MYC (Figure 1F). Consistent with this interpretation, siRNA-mediated depletion of the catalytic subunit of PI3-kinase alpha (p110alpha) enhanced MYC levels in both SW620 and CACO2 cells (Figure 1G).
Cycloheximide treatment revealed that treatment with BEZ235 did not accelerate MYC turnover (Figure 1A,B and Supplementary Figure 1A,B). To understand this result, we analyzed phosphorylation of serine 9 of GSK3. Inhibition of either PI3-kinase or AKT, using specific inhibitors reduced phosphorylation at this site in HeLa cells, which were used as positive control (Figure 1H). In contrast, inhibition of neither AKT nor PI3-kinase decreased phosphorylation of GSK3 S9 in colorectal tumor cells, arguing that AKT activity is not rate limiting for phosphorylation of this site in colorectal tumor cells (see Discussion). Consistently, exposure of colon cancer cells to BEZ235 did not strongly alter the relative fraction of MYC that is phosphorylated at T58 (Figure 1D). We concluded that PI3/AKT-kinase activity is not critical for stabilizing MYC proteins in colon carcinoma cells.
MYC protein is functional after inhibition of PI3-kinase and mTOR
To test whether MYC is functional in cells exposed to BEZ235, we performed microarray analyses of SW620 cells exposed to 200nM BEZ235 for 24hrs relative to control cells. To ascertain which changes depend on MYC, we compared control siRNA-treated cells to cells, in which endogenous MYC had been depleted by a specific siRNA (Supplementary Figure 2A). Consistent with the arrest in proliferation observed upon exposure to BEZ235, multiple gene sets encoding proteins involved in cell proliferation were robustly downregulated upon exposure of BEZ235 (Supplementary Figure 2B,C,D). In contrast, exposure of cells to BEZ235 led to a robust increase in expression of genes encoding proteins involved in ribosome function and translation (Supplementary Figure 2B,C,D). Many of the genes encoding proteins involved in translation are direct target genes of MYC (18). Consistent with this observation, siRNA-mediated depletion of MYC reduced both their basal and BEZ235-increased expression (Supplementary Figure 2C). Several well-characterized sets of MYC target genes contain both genes involved in proliferation and in ribosome function and translation: consistent with these data, depletion of MYC reduced expression of such gene sets even in the presence of BEZ235 (Supplementary Figure 2C, left panel). We concluded that the MYC protein present in BEZ235-treated cells is capable of activating MYC target genes and that genes involved in cell cycle progression are downregulated in a MYC-independent manner upon BEZ235 treatment.
FOXO-dependent MAP-kinase signaling increases MYC levels upon PI3-kinase inhibition
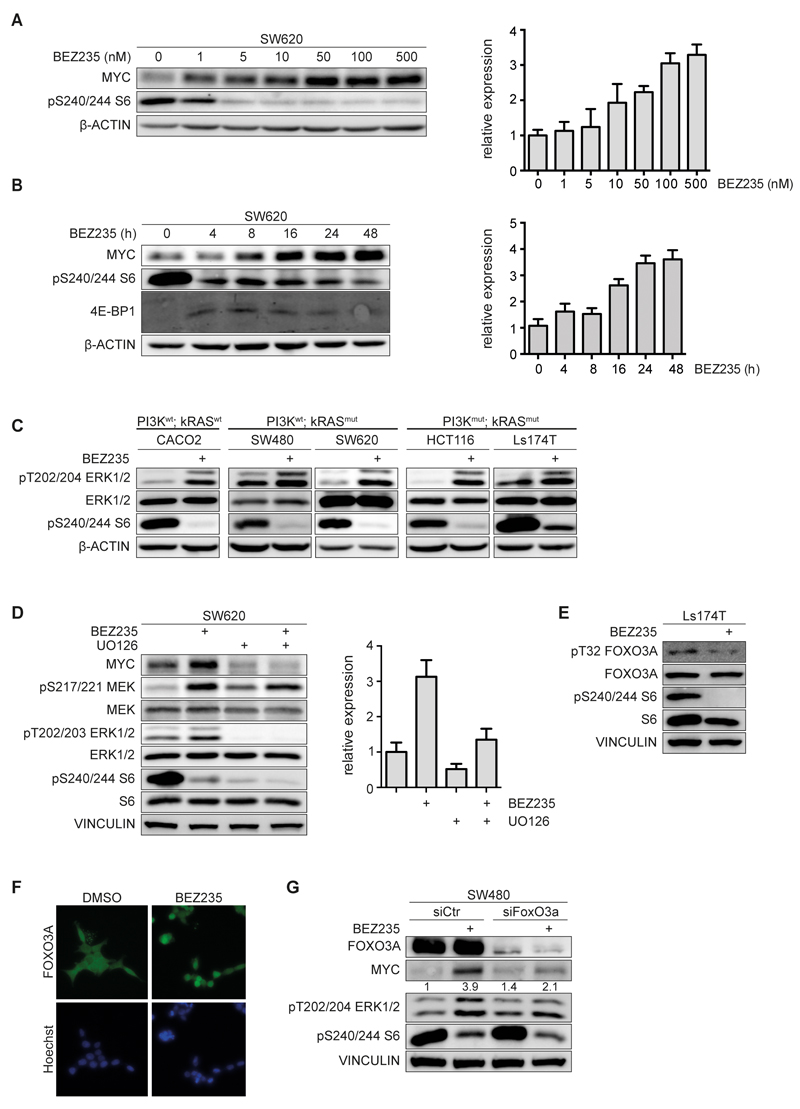
Incubation of SW620 cells with BEZ235 induced a dose- and time-dependent increase in MYC mRNA levels that paralleled the increase in MYC protein (Figure 2A,B). Transcription of MYC is under control of the MAP-kinase pathway via a joint ETS/E2F-site in the MYC promoter (19). Consistent with a role for MAP-kinase activation, incubation with BEZ235 enhanced phosphorylation of ERK, in several CRC cell lines tested (Figure 2C). Surprisingly, this increase was observed in both KRASwt and KRASmut cell lines. In line with the phosphorylation of ERK, inhibition of PI3-kinase induced phosphorylation of the upstream kinases c-RAF and MEK at growth factor-dependent phosphorylation sites (Supplementary Figure 3A). Depletion of PI3K using a specific siRNA activated the MAP- kinase pathway, consistent with previous observations (Supplementary Figure 3B) (20). Blockade of MAP-kinase activity using the MEK inhibitor UO126 abolished induction of MYC protein and attenuated induction of MYC mRNA, arguing that an increase in MAP- kinase activity is critical for induction of MYC upon PI3-kinase inhibition (Figure 2D).
Figure 2. BEZ235 induce MAPK-signaling in a FOXO3A dependent manner.
A. SW620 cells were treated with indicated concentrations of BEZ235 for 24h. Cell lysates were probed with indicated antibodies (left panel). MYC mRNA levels were assessed by RQ-PCR (right panel) (n=3).
B. SW620 cells were treated with 200nM BEZ235 and harvested at indicated time points. Cell lysates were probed with indicated antibodies (left panel). MYC mRNA levels were assessed by RQ-PCR (right panel) (n=2).
C. Cell lines were treated with BEZ235 (200nM, 24h) or solvent control. Immunoblots of lysates were probed with the indicated antibodies (n=3).
D. SW620 cells were incubated with BEZ235 (500nM), UO126 (20µM) or both for 24h. Protein levels were determined by immunoblotting (left panel). MYC mRNA levels were assessed by RQ-PCR analysis (right panel) (n=3).
E. Ls174T cells were treated with BEZ235 (200nM, 24h). Immunoblots of cell lysates were probed with the indicated antibodies (n=2).
F. Ls174T cells were treated with BEZ235 (200nM, 24h), fixed and subjected to immunofluorescence using a FOXO3A antibody. Nuclei were stained using Hoechst33342 (n=1).
G. SW480 cells were transfected with siRNA targeting FOXO3A or control siRNA for 48h followed by treatment with BEZ235 (200nM) or solvent control for 24h (n=2).
FOXO proteins are inhibited by PI3-kinase via AKT-dependent phosphorylation (21). Consistently, treatment of CRC cells with BEZ235 resulted in de-phosphorylation of FOXO3A at threonine 32, one of the sites phosphorylated by AKT (Figure 2E), as well as in its nuclear translocation (Figure 2F) and activation of known FOXO target genes (Supplementary Figure 3C) (22). Depletion of FOXO3A, a member of the FOXO family that is strongly expressed in colorectal tumor cells, attenuated both induction of MYC expression and activation of MAP-kinase signaling, demonstrating that activation of FOXO3A is critical for activation of MYC upon inhibition of PI3-kinase (Figure 2G). FOXO proteins are part of an evolutionary conserved feedback loop that limits expression of growth factor receptors at the cell surface in response to PI3-kinase activity (20, 23). As a result, treatment with BEZ235 strongly induced expression of HER3, of the insulin receptor (IR) and of the insulin-like growth factor receptor (IGFR) mRNAs in a FOXO3A-dependent manner (Supplementary Figure 3D). The data suggest that enhanced growth factor signaling induces MAP-kinase activity, which in turn leads to enhanced MYC expression in CRC cells upon BEZ235 treatment.
Targeting eIF4F activity restricts MYC expression in colon carcinoma cells
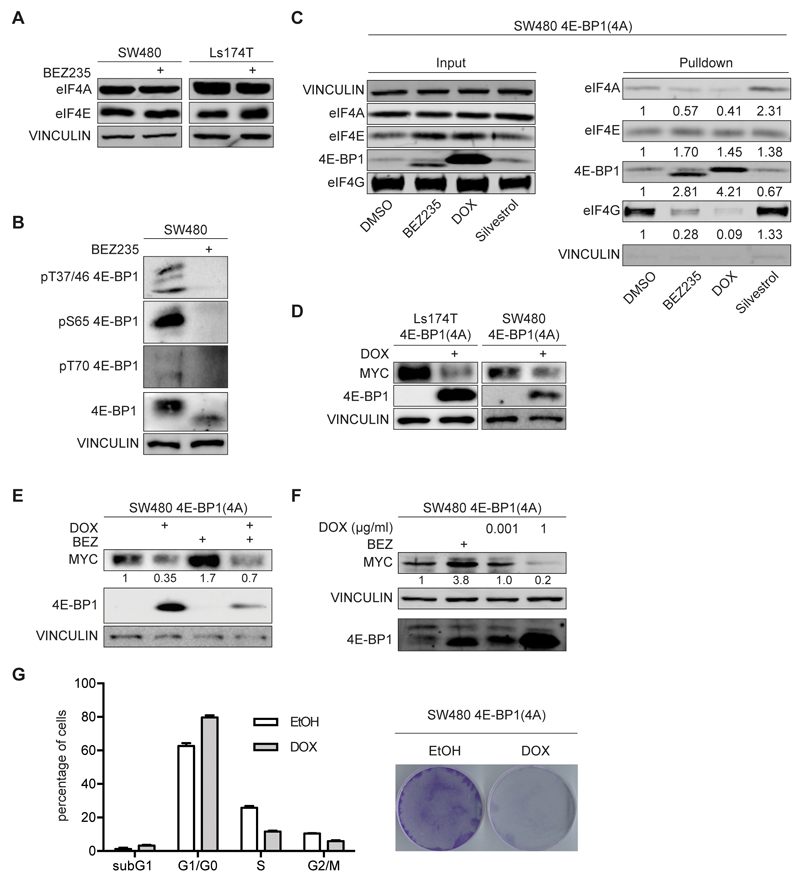
Upregulation of MYC after treatment with BEZ235 is also surprising since inhibition of mTORC1 is expected to inhibit the eIF4F translation initiation factor complex (see Introduction). Treatment with BEZ235 did not alter the expression of eIF4A or eIF4E, subunits of the eIF4F complex (Figure 3A). Antibodies directed against the four different mTORC1-dependent phosphorylation sites in 4E-BP1 (Thr37/46; Ser65; Thr70) confirmed that these sites were dephosphorylated upon exposure to BEZ235 (Figure 3B). To test whether BEZ235 inhibits translation initiation, we isolated CAP-binding complexes using m7G-affinity chromatography. Exposure of cells to BEZ235 did not interfere with binding of eIF4E, but reduced CAP-binding of eIF4A and eIF4G, which are recruited by eIF4E (Figure 3C); this is consistent with previous observations (24). The result is compatible with two interpretations: Either, translation of MYC, like that of the insulin receptor, does not depend on either eIF4A or eIF4E (25) or the amount of 4E-BPs is insufficient to fully sequester eIF4E in colon carcinoma even when de-phosphorylated.
Figure 3. Effect of eIF4F inhibition on MYC protein levels.
A. The indicated cell lines were incubated with BEZ235 (200nM, 24 h). Immunoblots of cell lysate were probed with the indicated antibodies (n=2).
B. SW480 cells were incubated with BEZ235 (200nM, 24h) and immunoblots probed for the indicated proteins (n=2).
C. m7GTP-CAP pull down assay was performed in SW480 cells after treatment with BEZ235 (200nM, 24h), DOX (24h), Silvestrol (25nM, 24h) or solvent control. Cell lysates were incubated with m7GTP beads and bound proteins immunoblotted for indicated proteins. Left panel demonstrates input of cell lysate, right panel the m7GTP bound protein fraction (n=2).
D. SW480 and Ls174T cells were infected with a lentivirus expressing 4E-BP1(4A) under the control of a doxycycline (DOX) inducible promoter. 4E-BP1(4A) harbors four mutations on mTOR phosphosites (T37A, T46A, S65A and T70A). Cells were incubated for 24h with DOX (1µg/ml) or ethanol as control. Protein levels were determined by immunoblotting (n=2).
E. SW480 cells expressing doxycycline-inducible 4E-BP1(4A) were incubated for 24h with DOX, BEZ235 (200nM) or the combination of both and cell lysates probed for the indicated proteins (n=2).
F. SW480 cells expressing doxycycline-inducible 4E-BP1(4A) were incubated with BEZ235 (200nM), low DOX (0.001µg/ml) or high DOX (1µg/ml) concentrations for 24h. Cell lysates were immunoblotted with the indicated antibodies (n=2).
G. SW480 cells described in panel C were incubated with DOX (1µg/ml). Left panel shows FACS analysis in response to DOX (24h) or solvent control. Error bars indicate SD of biological triplicates from one representative experiment (n=3). Right panel shows a colony assay stained with crystal violet after 5 days of DOX treatment.
To test whether expression of MYC depends on eIF4E, we expressed a doxycycline-inducible allele of 4E-BP1 that carries alanine substitutions at four serine/threonine residues that are targets for mTORC1-dependent phosphorylation and that acts as a dominant inhibitor of eIF4E (26) (Figure 3D). Induction of 4E-BP1(4A) by addition of doxycycline in Ls174T and SW480 cells inhibited expression of MYC, both in the absence and the presence of BEZ235 (Figure 3 D,E). Induction of 4E-BP1(4A) blocked CAP-binding of eIF4A and eIF4G; for both proteins, inhibition by 4E-BP1(4A) was stronger than observed for BEZ235 (Figure 3C). Since levels of 4E-BP1 in the presence of doxycycline exceeded those of endogenous 4E-BP, this suggested that endogenous levels of 4E-BP are insufficient to sequester eIF4E in colon carcinoma cells. Consistent with this suggestion, titration of doxycycline showed that suppression of MYC expression by 4E-BP1(4A) required protein levels that exceeded those found in BEZ235-treated cells (Figure 3F and Supplementary Figure 4A). Inhibition of MYC expression by 4E-BP1(4A) correlated with an accumulation of cells in the G1 phase of the cell cycle and suppression of cell proliferation (Figure 3G and Supplementary Figure 4B). We concluded that efficient translation of MYC requires active eIF4E, but that endogenous levels of 4E-BPs are insufficient to fully inhibit expression of MYC in colon carcinoma cells (27).
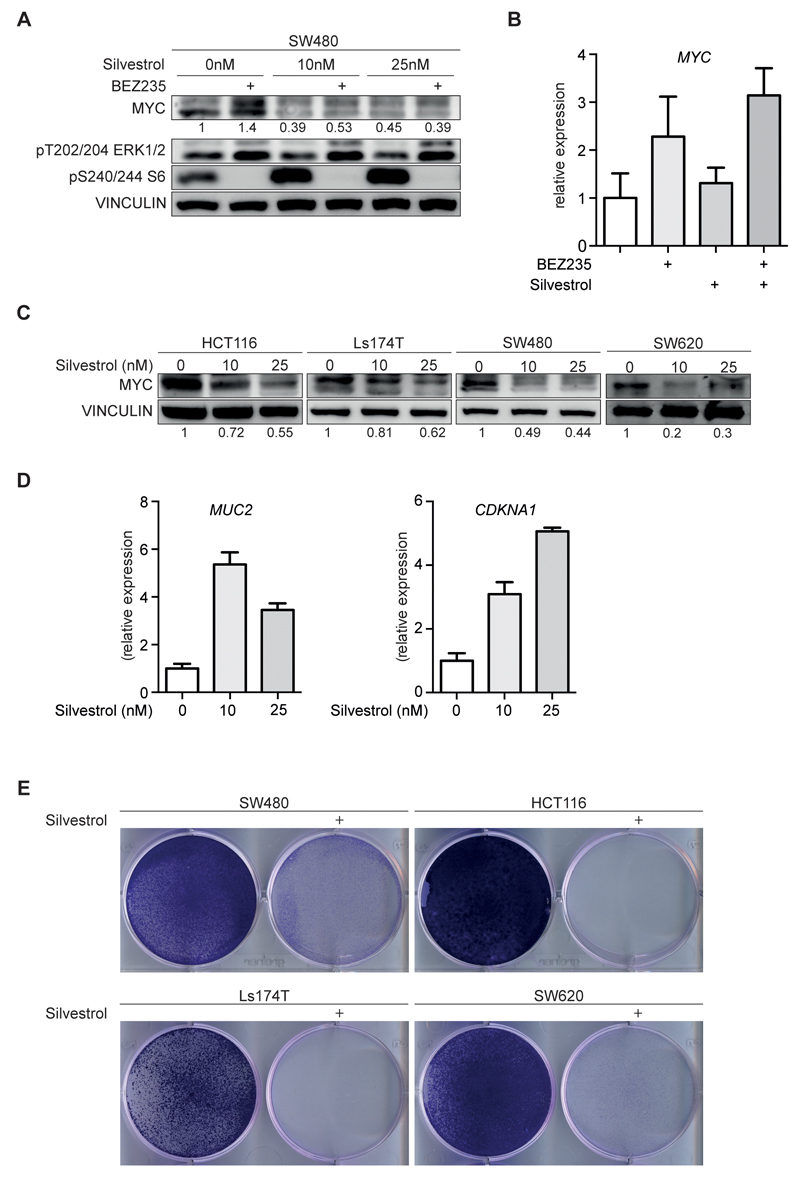
As an alternative means to inhibit eIF4F activity, we used silvestrol, a small molecule inhibitor of the eIF4A helicase (17). Incubation of SW480 cells with nanomolar concentrations of silvestrol suppressed both basal MYC protein expression and the increase in MYC levels observed in response to treatment with BEZ235 (Figure 4A). Identical results were obtained with rocaglamide, a structurally related compound that also inhibits the eIF4A helicase (28) (Supplementary Figure 4C). Neither compound decreased ERK or mTOR activity (Figure 4A and Supplementary Figure 4C). Furthermore, silvestrol did not suppress MYC mRNA levels; on the contrary, MYC mRNA levels slightly increased upon exposure to low concentrations of silvestrol possibly since high levels of MYC represses transcription from its own promoter (Figure 4B). Incubation with silvestrol also suppressed MYC protein levels in HCT116, Ls174T, SW480 and SW620 cells (Figure 4C) and led to an increase in CDKN1A (encoding p21, Cip1) and MUC2 (encoding Mucin2, which is a marker of terminal differentiation of colon cancer cells) mRNA levels, both of which are repressed by MYC (Figure 4D) (29). Incubation with silvestrol suppressed proliferation of colon carcinoma cells but did not induce a significant degree of apoptosis (Figure 4E and Supplementary Figure 4D,E). FACS analysis showed that silvestrol did not induce major changes in the cell cycle distribution, arguing that silvestrol arrests proliferation in all phases of the cell cycle (Supplementary Figure 4E). FACS analyses showed that a siRNA-mediated knockdown of MYC, in contrast to silvestrol, led to an accumulation of cells in the G1 phase; combining depletion of MYC with treatment with silvestrol further increased the accumulation of cells in G1 phase (Supplementary Figure 4F). Both observations suggest that silvestrol suppresses translation of proteins that are critical for progression through the cell cycle in addition to MYC. We concluded that inhibition of eIF4A helicase activity is a valid approach to inhibit proliferation and to suppress MYC expression in colon carcinoma cells.
Figure 4. Small molecule inhibitors of eIF4A reduce MYC protein levels and suppress cancer cell proliferation.
A. SW480 cells were treated with BEZ235 (200nM) and the indicated concentrations of silvestrol for 48h and analyzed by immunoblotting (n=3).
B. SW480 cells were incubated with BEZ235 (200nM), Silvestrol (25nM) or both. RNA was isolated after 48h and subjected to RQ-PCR analysis (n=2).
C. Immunoblots of four colorectal cell lines upon treatment with increasing concentration of silvestrol or solvent control (n=2).
D. Ls174T cells incubated with increasing concentration of silvestrol for 48h were subjected to RQ-PCR and analyzed for markers of cell cycle arrest (CDKN1A) and differentiation (MUC2) (n=2).
E. Colony formation assay stained with crystal violet. The indicated cell lines were incubated with silvestrol (25nM) for 5 days.
Silvestrol targets both CAP-and IRES-dependent translation of MYC
Measurements of 35S-methionine incorporation showed that incubation of colon carcinoma cells with BEZ235 or silvestrol or induction of 4E-BP1(4A) reduced global protein synthesis to a similar extent (Figure 5A), raising the question why MYC protein levels are differentially affected. To address this question, we performed polysome profiling from control and inhibitor-treated cells and measured the association of different mRNAs with polysomes by RQ-PCR. Consistent with their effects on CAP-binding complexes, induction of 4E-BP1(4A) strongly inhibited association of two control mRNAs, ACTB and TUBB3, with polysomes, while BEZ235 had moderate effects (Figure 5B). In contrast, induction of 4E-BP1(4A) had only moderate and BEZ235 no effects on association of MYC mRNA with polysomes, arguing that MYC mRNA remains associated with polysomes even when CAP-recognition is strongly impaired (Figure 5B).
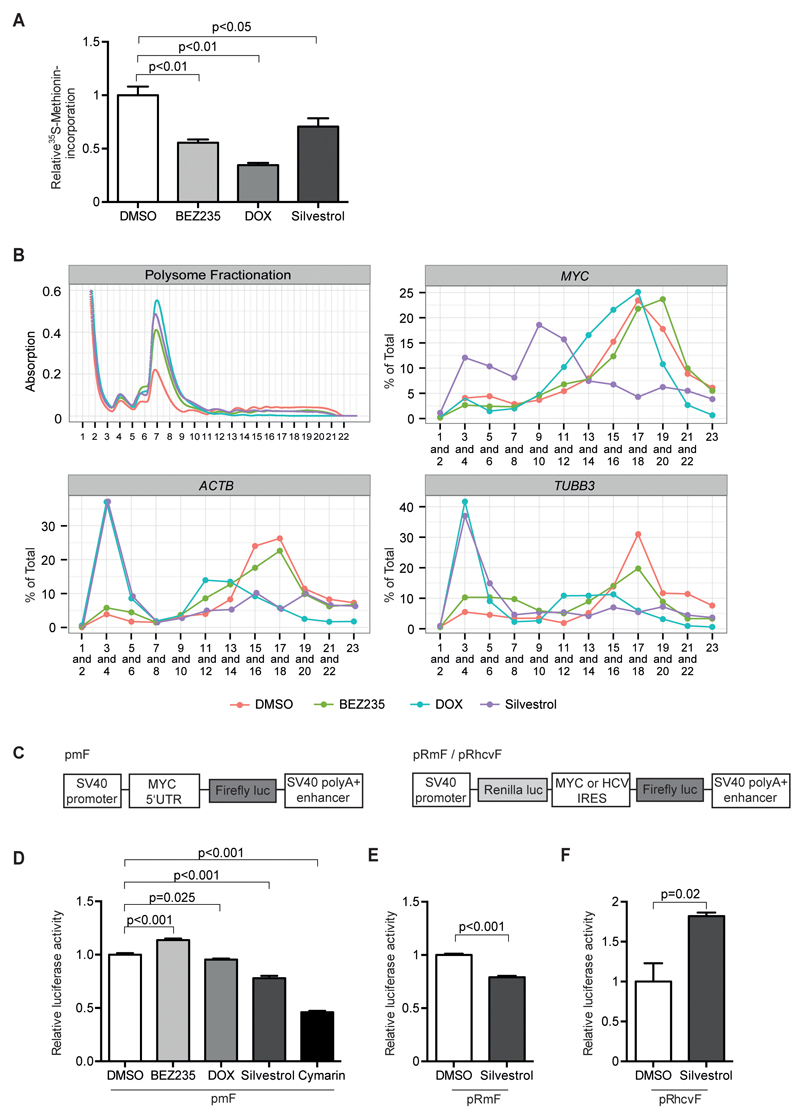
Figure 5. Effects of silvestrol and BEZ235 on translation of MYC.
A. Incorporation of 35S-labeled methionine in SW480 cells treated with BEZ235 (200nM), Silvestrol (25nM), DOX or solvent control for 24h. Shown are mean+SD (n=3).
B. Polysome fractionation of SW480 cells (Top left), treated with BEZ235 (200nM), DOX, Silvestrol (25nM) or solvent control for 24 h. RNA was isolated from the indicated fractions and relative mRNA content per fraction was measured by RQ-PCR. Top right MYC mRNA distribution, bottom left ACTB mRNA and bottom right TUBB3 mRNA distribution (n=2).
C. Schematic illustration describing the luciferase reporter systems used in (D), (E) and (F). The pmF reporter construct contains the MYC 5’UTR inserted into the control vector pGL3 (Promega) proximal to firefly luciferase coding sequence. The bicistronic pRmF and pRhcvF reporter constructs contain the MYC or the HCV (hepatitis C virus) IRES sequence distal to renilla and proximal to firefly luciferase gene.
D. SW480 cells were transfected with pmF luciferase reporter and treated with BEZ235 (200nM), DOX, silvestrol (25nM), Cymarin (100nM) or solvent control for 24h. Luciferase activity is shown relative to a co-transfected beta-Gal reporter (n=3).
E. SW480 cells were transfected with pRmF luciferase reporter and treated with silvestrol (25nM) or solvent control. Relative firefly luciferase activity was calculated using the ratio of firefly to renilla luciferase (n=3).
F. SW480 cells were transfected with pRhcvF luciferase reporter and analyzed as in panel E (n=3).
The 5'-UTR of the MYC mRNA contains an internal ribosome entry site and therefore MYC is translated both in CAP- and in an IRES-dependent manner; translation from the IRES element depends on eIF4A, but is independent of eIF4E (27). We therefore tested how inhibition of eIF4A affects polysome profiles. Similar to the induction of 4E-BP1(4A), incubation with silvestrol blocked polysome association of the ACTB and TUBB3 mRNAs. In contrast to 4E-BP1(4A), silvestrol also strongly affected association of the MYC mRNA with polysomes. The dependence on eIF4A correlates with a complex secondary structure in the 5`-UTR of a mRNA and association of some mRNAs, such as PFN2, with polysomes shows little dependence on eIF4A (30); this was also observed in SW480 cells (Supplementary Figure 5A).
The data argue that the eIF4A-dependent, but eIF4E-independent translation from the IRES maintains polysome association of MYC mRNA when CAP-dependent translation is inhibited. Relative to the CAP-dependent translation of a control mRNA, incubation with silvestrol reduced translation of a luciferase under the control of the MYC 5'-UTR (Figure 5C,D). Furthermore, silvestrol inhibited translation under the control of the MYC IRES, which depends on eIF4A (27), but did not inhibit translation under the control of an eIF4A-independent IRES element that is present in the HCV genome (Figure 5C,E,F). Collectively, the data argue that the presence of an IRES-element facilitates translation of MYC when CAP-dependent translation is inhibited.
To confirm these data, we used a second inhibitor of translation initiation, cymarin, that has been identified as an inhibitor of MYC IRES-dependent translation by high-throughput screening (31). Reporter assays showed that incubation with 100nM cymarin inhibited both translation under the control of the 5`-UTR of MYC relative to a control reporter (Figure 5D). Consistently, incubation of SW480 cells with cymarin mimicked the effect of silvestrol on MYC expression (Supplementary Figure 5B).
Silvestrol suppresses MYC expression and proliferation of colon cancers in vivo
To test whether targeting translation initiation may open a therapeutic window in colon carcinoma, we initially analyzed publically available gene expression databases. These analyses showed that expression of the mRNA encoding PDCD4 is strongly suppressed in colon carcinoma relative to normal tissue (Supplementary Figure 5C). In contrast, expression of mRNAs encoding eIF4A, eIF4E, 4E-BP1 and 4E-BP2 showed minor changes in tumor relative to normal tissue. Histopathological analysis of 10 human CRC samples confirmed the downregulation of PDCD4 in colon tumor relative to normal mucosa; this data is consistent with previous findings (Supplementary Figure 5D) (32). In contrast, we did not observe significant differences in expression of eIF4E, eIF4A and 4E-BPs proteins between normal mucosa and CRCs in the same tumor samples (Supplementary Figure 5D). This suggested that eIF4A activity may be enhanced due to silencing of its negative regulator PDCD4 in colon carcinoma and that targeting eIF4A may therefore be suitable for targeting MYC expression in vivo.
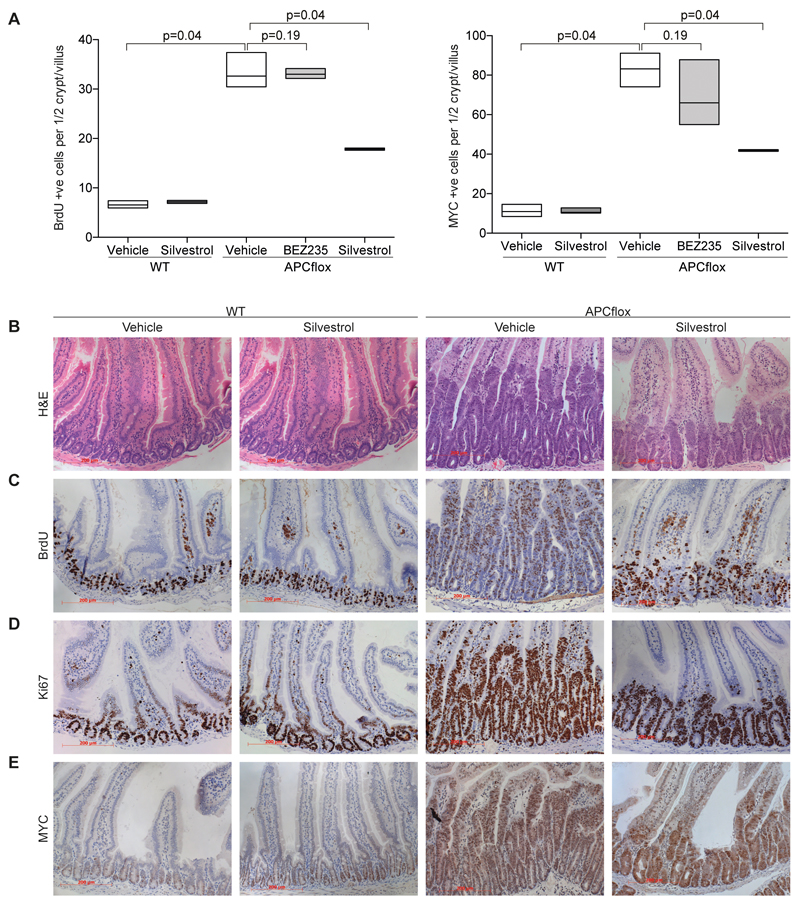
To explore this possibility, we assessed the ability of silvestrol to suppress the proliferation induced by acute APC deletion in the intestine. Our previous experiments have shown the “crypt progenitor phenotype” induced by Apc loss to be MYC dependent (3). To achieve APC deletion in the murine intestine we intercrossed mice carrying the VillinCreER transgene to mice bearing the conditional knockout Apc580s allele to generate VilCreER Apcfl/fl mice (labeled “APCflox“ in Figure 6). To achieve high penetrant deletion of the Apc tumor suppressor, mice were given a daily injection of 2 mg of tamoxifen for 2 days (33). Mice were then given either a 1 mg/kg (i/p) injection of silvestrol or vehicle on days 2 and 3 post induction and harvested on day 4. For BEZ235 treatment mice were gavaged with 45 mg/kg of BEZ235 on days 2 and 3 post induction. Four days post induction mice were euthanized and the intestinal crypt hyperproliferative phenotype was examined. Exposure to BEZ235 had no significant impact on hyperproliferation following Apc loss, with similar crypt size, BrdU incorporation, Ki67 and MYC levels (Figure 6A and Supplementary Figure 6A,B,C,D). BEZ235 was active since it led to a significant increase in expression of CDKN1A (p<0.01; Supplementary Figure 6E). In marked contrast, exposure of mice to silvestrol robustly suppressed the hyperproliferation following APC loss (Figure 6A-D). Upon treatment with silvestrol, intestinal crypts from VilCreER Apcfl/fl mice were significantly smaller and showed a marked reduction in both BrdU incorporation and Ki67 positivity. Importantly, there was a clear reduction in MYC positivity via immunohistochemistry within APC-deficient crypt (p=0.04, Figure 6 A-E). In situ hybridization showed that silvestrol did not reduce levels of Myc mRNA (Supplementary Figure 7A), demonstrating that it reduces MYC expression posttranscriptionally. Furthermore, quantitative evaluation documented that silvestrol led to a reduction in MYC protein levels in the lower half of the crypts (Supplementary Figure 7B), arguing that the reduction in MYC level is not due to an indirect effect of silvestrol on crypt/villus differentiation. No impact on proliferation or MYC levels was observed in wild type intestinal crypts (Figure 6A-E) suggesting a clear therapeutic window exists between APC-deficient and wild type intestinal enterocytes.
Figure 6. Silvestrol reduces proliferation and MYC levels in APC-deficient intestinal enterocytes but not in wildtype cells.
A. Graph documenting number of proliferating cells (shown for BrdU incorporation; left panel) and number of cells staining positive for MYC (right panel) in silvestrol, BEZ235 or vehicle-treated wild type or APC-deficient intestines. The number of BrdU or MYC positive nuclei per crypt-villus axis was scored in 30 full crypts in at least 3 mice. Data are presented as Box- and Whisker plot.
B. Representative H&E stained sections showing effects of silvestrol on wild type ("WT") and APC-deficient crypts. Remark that crypts are enlarged due to APC loss and that this is reduced following silvestrol treatment.
C. Representative BrdU staining showing that silvestrol reduced the proliferation APC-deficient and not wild type intestines.
D. Representative Ki67 stained sections showing a reduction in the proliferation in APC-deficient crypts following silvestrol treatment.
E. Representative MYC staining showing reduction by silvestrol in APC-deficient but not wild type intestines.
Discussion
Deregulated and enhanced expression of MYC is a driver of colorectal tumorigenesis, necessitating strategies to inhibit MYC function or expression for tumor therapy. Here we have explored the possibility to target protein turnover and translation initiation to inhibit MYC expression. As a tool to dissect the regulatory circuits that maintain elevated MYC expression, we used BEZ235 and silvestrol, well-characterized inhibitors of the PI3-kinase/mTOR pathway and of eIF4A helicase, respectively (16).
We expected that BEZ235 would decrease MYC expression via promoting FBXW7-dependent turnover and via inhibition of eIF4F-dependent translation of MYC. We confirmed that turnover of MYC proteins in CRC cells depends on FBXW7 (8). Inhibition of PI3-kinase or AKT can increase MYC turnover since AKT phosphorylates and inhibits GSK3 at serine 9 (34). Hence, AKT inhibition can increase phosphorylation of MYC at T58 by GSK3. Surprisingly, phosphorylation of GSK3 at serine 9 does not depend on PI3-kinase and AKT activity in colon carcinoma cells, arguing that one of several AKT-independent kinases that can phosphorylate this site (e.g. Aurora-A (35) or p90RSK (36)) maintain GSK3 phosphorylation upon inhibition of PI3-kinase or AKT.
Instead of promoting degradation, inhibition of PI3-kinase increased MYC levels in several colon cancer cell lines due to a FOXO-dependent transcriptional upregulation of growth factor receptor genes and, downstream of receptor activity, to a MAP-kinase dependent increase in MYC mRNA levels (see Figure 7). A similar crosstalk between the PI3K/AKT-pathway and MAP-kinase activity has been identified previously in breast cancer cells (20). Most likely, it reflects an evolutionary conserved regulatory circuit that couples expression of cell surface receptor genes to PI3-kinase activity (23).
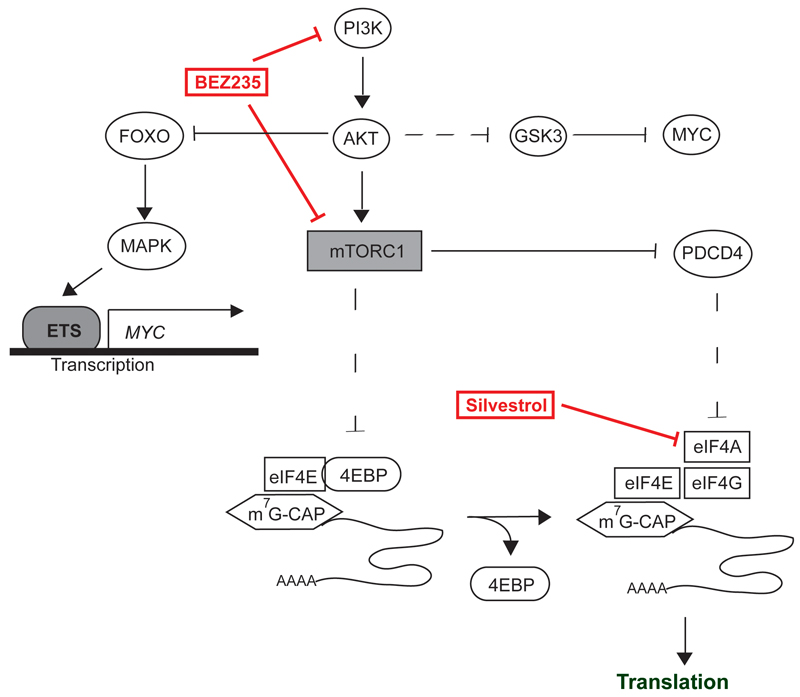
Figure 7. Model summarizing our findings.
Treatment with BEZ235 upregulates MYC via a FOXO/MAPK-dependent pathway (black blunt line, upper left part). Negative regulation of MYC levels via GSK3 or inhibitors of translation like PDCD4 and 4E-BPs is lost in CRC (dashed lines). Treatment with silvestrol reduces MYC expression by inhibition of the eIF4A (right lower part).
Inhibiting protein translation has emerged as a therapeutic strategy to target MYC-dependent tumor growth, since translation initiation is deregulated in MYC-driven lymphomas and supraphysiological protein synthesis rates are required for their growth (37). In MYC-driven lymphomas, targeting protein translation via inhibition of mTORC1 and mTORC2 has therapeutic efficacy, because two inhibitors of eIF4F-dependent translation initiation, 4E-BP and PDCD4, are inactivated via mTORC1-dependent phosphorylation and, in the case of PDCD4, subsequent ubiquitin-dependent degradation (14, 38, 39). Expression of PDCD4 is strongly downregulated in CRC. In response to BEZ235, 4E-BP1 is dephosphorylated on mTORC1-dependent sites, but this does not inhibit translation of MYC. We identify two causes for this effect: First, CAP-binding of eIF4A und eIF4G in response to BEZ235 is only partially inhibited, arguing that the amount of 4E-BPs are insufficient to fully sequester eIF4E in CRC cells. Second, MYC mRNA remains associated with polysomes even when CAP-binding is fully inhibited by a non-phosphorylatable allele of 4E-BP1. Most likely, this is due to the presence of an IRES in the 5’-UTR of MYC, which is known to be independent of eIF4E (27). Our findings are consistent with recent observations that the 4E-BP proteins are not the critical targets of the mTORC1 inhibitor, rapamycin, and that even genetic ablation of mTORC1 activity does not inhibit MYC expression in a mouse model of CRC (40).
Our data also show that dual PI3-kinase/mTOR inhibition is not an effective therapeutic strategy for CRCs since BEZ235 has only small effect on MYC levels and no effect proliferation and cellularity in a mouse model of CRC that is driven by deletion of the Apc tumor suppressor gene. In contrast, selective targeting of mTORC1 by rapamycin, while not targeting MYC, is effective in suppressing growth of colon carcinoma (40). We suggest that the BEZ235, but not rapamycin-dependent inhibition of PI3Kinase and subsequent FOXO-dependent activation of MAPkinase limits the therapeutic efficacy of BEZ235 in this model.
In contrast to BEZ235, silvestrol inhibited expression of MYC in colorectal tumor cell lines at nanomolar concentrations. At the same time, silvestrol reduced proliferation and cellularity of colon tumors in vivo, arguing that inhibition of the eIF4A helicase is effective to inhibit MYC expression in CRC and extending similar observations made in a NOTCH-driven model of T-ALL lymphomas (41). Surprisingly, concentrations of silvestrol that strongly reduce MYC levels and proliferation in colorectal tumor cells are well tolerated without apparent toxicity; this correlates with the observation that the effects of silvestrol on MYC levels, proliferation and cellularity of normal colon are small. Furthermore, translation of MYC is not affected by mTOR inhibition in murine fibroblasts, arguing that the dependence of MYC translation on eIF4A and eIF4G function is not uniformly high (26). The dependence of eIF4A is mediated by the presence of G-quadruplexes in the 5´-UTR (41). Since other RNA helicases such as RHAU (42) can target G-quadruplexes, it is possible that the dependence of colon carcinoma cells on eIF4A for translation of MYC opens a therapeutic window since other helicases carry out this function in normal colon cells.
Methods
Reagents
BEZ235 (Lclabs), Rapamycin (Lclabs), UO126 (Promega), Akti 1/2 (Sigma), Silvestrol (Med-Chemexpress), and Cymarin (Sigma), Rocaglamid (Sigma) were dissolved in DMSO. Doxycycline and cycloheximide (both Sigma) were dissolved in Ethanol.
Cell culture and transfection
CACO2, HCT116, SW480, SW620 and HeLa cells were cultured in DMEM, Ls174T cells in RPMI-1640 medium containing 10% FCS and 1% penicillin/streptomycin. Cell lines were purchased from ATCC (CACO2 in 2012, SW620 in 2012), Cell Line Services (SW480 in 2013) or the German Collection of Microorganisms and Cell Cultures (HCT116 in 2012) and were maintained according to company recommendations. Ls174T and Hela cells were kind gifts of Hans Clevers and Michael Bishop, respectively. All cell lines were authenticated via STR Analysis in 2014.
For depletion experiments, cells were transfected with siRNAs (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer´s instructions. Expression plasmid encoding 4E-BP1(4A) was provided by David Sabatini (via Addgene). Translation reporter constructs for luciferase assays were a kind gift of Anne Willis (27). FACS analysis was performed using FACS Canto II (Becton Dickinson). Antibodies are listed in Supplementary Table 1. Global translation was measured using 35S-Methionine and TCA precipitation.
Gene expression analysis
Agilent Human Genome Microarray 4x44 K v2 was used. The complete data set can be seen at Array express (E-MTAB-2882). Human PDCD4, eIF4A, eIF4E, 4E-BP1 and 4E-BP2 expression data from the Skrzypczak Colorectal 2 dataset were downloaded from Oncomine (43). Statistical evaluation was performed by 2-tailed Student’s unpaired t test or with Mann-Whitney test. Data are presented as mean ± SD.
Polysome fractionation and CAP pull-down assays
12 x 106 SW480 cells were plated on 3x15 cm cell culture dishes for 16 hours and then treated as indicated for 24 hours. Cell lysis was performed on ice in 100μl gradient buffer (1mM DTT; 100mM KCl, 20mM Tris-HCl, pH 7.5, 5mM MgCl2, 0.5% NP40, 20μl 0.1mg/ml cycloheximide containing protease and RNAse inhibitors). Lysates were cleared by centrifugation. Lysates were layered on top of 5-45% sucrose gradients and centrifuged in an SW41-Ti rotor at 34.500 rpm for 1 hour at 4°C. Profiles were fractionated using a Piston gradient fractionator (BioComp). RNA was analyzed by quantitative RQ-PCR. CAP pull-downs were performed as described (44).
Animal Experiments
All animal experiments were performed under UK Home Office guidelines using the project license: 60-4183. This undergoes local ethical review at Glasgow University. VillinCREER APCfl/fl mice have been described previously (38). Silvestrol (Medchemexpress, H-13251) was dissolved in 20%(w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107) at a concentration of 125 µg/ml and injected into mice i.p.. BEZ235 (Synkinase, SYN-1018) was dissolved in 10%(v/v) 1 methyl-2-pyrrolidinone; 90%(v/v) poly(ethylene glycol), SigmaP3265 at a concentration of 4.5mg/ml and mice gavaged. Sample size was decided using NC3Rs guidelines, the smallest number of animal to yield a significant difference.
Immunohistochemistry
Antibodies are listed in Supplementary Table 1. Intestinal scoring was performed in a blinded manner. 30 full crypt-villus axis were scored for BRDU, CDKN1A and MYC positivity. For quantification of the histoscore, 25 crypts from each MYC-stained section at 200x magnification were scored. Per crypt, each nucleus was scored either: 0= no stain; 1= weak stain; 2= moderate stain; 3= strong stain. The numbers assigned to each category were multiplied by the relevant multiplication factor for that score. The average histoscore of 25 crypts per mouse section was used. Statistical analysis was by non-parametric Mann Whitney using Minitab version 17.
RNAscope
RNAscope images were developed from paraffin embedded formalin fixed samples using Advanced Cell Diagnostics' RNAscope 2.0 HD (brown) kit (#310035), following manufacturers instructions. Probes: Mm Myc (#413451); DapB (RNA negative control (#310043)); Mm Ppib (RNA positive control (#313911)). RNA control data not shown.
Supplementary Material
Statement of significance.
Inhibiting MYC function is likely to have a significant therapeutic impact in colorectal cancers. Here we explore several strategies to target translation initiation in order to block MYC expression. We show that a small molecule inhibitor of eIF4A inhibits MYC expression and suppresses tumor growth in vivo.
Acknowledgements
This work was supported by a grant from the interdisciplinary center for clinical research (IZKF B-186) of the medical faculty of Würzburg (to A.W.). O.J.S. holds an ERC investigator award COLONCAN and T.J./O.J.S. are funded by a CRUK (C596/A17196). We thank Anne Willis for translation reporter constructs and members of the Eilers laboratory for critical reading of the manuscript.
Financial support: AW: Interdisciplinary center for clinical research (IZKF B-186); OS and TJ: CRUK (C596/A17196).
Footnotes
Authors contributions
Conception and design: A. Wiegering, S. Herold, C-T. Germer, O Sansom, M. Eilers
Acquisition of data: A. Wiegering, T. Jamieson, M. Hüttenrauch, L. Taranets, C.Nixon, M.Küspert, C. Pfann, Y. Ruoss, F. Uthe, S. Herold, S.Walz, A. Rosenwald
Analysis and interpretation of data: A. Wiegering, S. Herold, O. Sansom, S.Walz, A. Rosenwald, M. Eilers, Y. Ruoss, F. Uthe
Writing, review, and/or revision of the manuscript: A. Wiegering, S. Herold, O. Sansom, M. Eilers
Administrative, technical, or material support: C-T. Germer, A. Rosenwald, O Sansom, M. Eilers
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.CancerGenomeAtlasNetwork. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–9. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 4.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harbor perspectives in medicine. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–9. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 7.Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Muller J, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO molecular medicine. 2014;6:1525–41. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell AS, Sears RC. MYC degradation. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 10.Diefenbacher ME, Popov N, Blake SM, Schulein-Volk C, Nye E, Spencer-Dene B, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest. 2014;124:3407–18. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer research. 2006;66:8139–46. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–37. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 15.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–63. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 17.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–60. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roussel MF, Davis JN, Cleveland JL, Ghysdael J, Hiebert SW. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene. 1994;9:405–15. [PubMed] [Google Scholar]
- 20.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 22.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–9. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes & development. 2003;17:2006–20. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proud CG. The eukaryotic initiation factor 4E-binding proteins and apoptosis. Cell Death Differ. 2005;12:541–6. doi: 10.1038/sj.cdd.4401588. [DOI] [PubMed] [Google Scholar]
- 25.Olson CM, Donovan MR, Spellberg MJ, Marr MT., 2nd The insulin receptor cellular IRES confers resistance to eIF4A inhibition. eLife. 2013;2:e00542. doi: 10.7554/eLife.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spriggs KA, Cobbold LC, Jopling CL, Cooper RE, Wilson LA, Stoneley M, et al. Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Molecular and cellular biology. 2009;29:1565–74. doi: 10.1128/MCB.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem. 2012;55:558–62. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 30.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. doi: 10.1186/s13059-014-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didiot MC, Hewett J, Varin T, Freuler F, Selinger D, Nick H, et al. Identification of cardiac glycoside molecules as inhibitors of c-Myc IRES-mediated translation. J Biomol Screen. 2013;18:407–19. doi: 10.1177/1087057112466698. [DOI] [PubMed] [Google Scholar]
- 32.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 33.Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell stem cell. 2013;12:761–73. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–9. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 35.Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–75. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J. 1994;303(Pt 3):701–4. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–5. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 39.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11988–93. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–33. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.