Abstract
Proper centrosome duplication and spindle formation are crucial for prevention of chromosomal instability, and BRCA1 plays a role in this process. In this study, transient inhibition of BRCA1 function in cell lines derived from mammary tissue caused rapid amplification and fragmentation of centrosomes. Cell lines tested that were derived from nonmammary tissues did not amplify the centrosome number in this transient assay. We tested whether BRCA1 and its binding partner, BARD1, ubiquitinate centrosome proteins. Results showed that centrosome components, including γ-tubulin, are ubiquitinated by BRCA1/BARD1 in vitro. The in vitro ubiquitination of γ-tubulin was specific, and function of the carboxy terminus was necessary for this reaction; truncated BRCA1 did not ubiquitinate γ-tubulin. BRCA1/BARD1 ubiquitinated lysines 48 and 344 of γ-tubulin in vitro, and expression in cells of γ-tubulin K48R caused a marked amplification of centrosomes. This result supports the notion that the modification of these lysines in living cells is critical in the maintenance of centrosome number. One of the key problems in understanding the biology of BRCA1 has been the identification of a specific target of BRCA1/BARD1 ubiquitination and its effect on mammary cell biology. The results of this study identify a ubiquitination target and suggest a biological impact important in the etiology of breast cancer.
Cancer cells frequently have unstable numbers of chromosomes (reviewed in reference 20). One mechanism for chromosomal instability is improper centrosome duplication, since the centrosome is the organelle that organizes the spindle for separation of chromosomes during mitosis. The presence of more than two centrosomes in a cell can result in lost or fragmented chromosomes after cell division. Human tumors derived from breast and other tissues have abnormal centrosome numbers in early-stage lesions. As an example, abnormal centrosome numbers have been detected in ductal carcinoma in situ, the first stage of breast cancer (21, 33), and BRCA1 has been shown to have a role in regulating centrosome number (reviewed in reference 9).
BRCA1 is a tumor suppressor that is mutated in inherited breast and ovarian cancer cases, and it is also epigenetically down-regulated in sporadic breast cancers. Strikingly, BRCA1 function is required for nearly all cell types to grow; it has many roles in the cell. These functions include the regulation of DNA damage repair, transcription, and X-chromosome inactivation (reviewed in references 37 and 41). All of these processes could be important in protecting mammary cells from uncontrolled growth, but it is not clear why loss of BRCA1 specifically results in breast and ovarian cancer.
There is growing evidence that BRCA1 functions as a regulator of centrosome number. First, BRCA1 is localized to the centrosome in mitotic cells (17, 23). Second, interference with BRCA1 function by various methods can cause an increased centrosome number. For example, mouse fibroblasts derived from BRCA1 exon 11 knockouts have amplified centrosomes (46). Amino acid residues 504 to 803 of BRCA1 bind γ-tubulin, a major component of centrosomes, and stable overexpression of the γ-tubulin binding domain of BRCA1 causes centrosome amplification in tissue culture cells (16). BRCA1 function can be inhibited by a peptide that binds to its carboxy terminus. Acute expression of this peptide causes a rapid rise in centrosome number in a mammary cell line (36).
BRCA1 is an 1,863-amino-acid protein with BRCT repeats at its carboxy terminus; at its amino terminus is a RING domain, which is a powerful ubiquitin ligase when this domain of BRCA1 is associated with BARD1 (15, 45). Candidate substrates identified in vitro include autoubiquitination of BRCA1/BARD1 and monoubiquitination of histone monomers and of the p53 protein (8, 10, 25). The cellular consequences of the BRCA1/BARD1-mediated ubiquitination of these substrates remain unclear.
In this study, we inhibited BRCA1 function in several tissue culture cell lines and found that the mammary tissue-derived cell lines rapidly accumulated extra centrosomes after BRCA1 inhibition. Intriguingly, BRCA1 inhibition did not result in centrosome amplification in the nonmammary cell lines that we tested. Since BRCA1 and BARD1 together constitute a ubiquitin ligase, we tested whether BRCA1/BARD1 ubiquitinates centrosome-associated proteins, and we found that several centrosome proteins, including γ-tubulin, are specifically ubiquitinated by full-length BRCA1/BARD1. The lysines of γ-tubulin modified by BRCA1-dependent ubiquitination were identified, mutated to arginine, and expressed in cells. Expression of these mutant γ-tubulin proteins caused centrosome amplification, indicating that the ubiquitination of γ-tubulin by BRCA1 is an important step in the regulation of centrosomes.
MATERIALS AND METHODS
Cell culture.
MCF10A, T47D, ZR75-1, HS578T, U2OS, HeLa, PC3, and DLD-1 tissue culture cells were cultured according to American Type Culture Collection recommendations. MTSV1-7 cells were a gift from J. Taylor-Papadimitriou (Cancer Research UK, Guy’s Hospital, London, United Kingdom) and were cultured according to established protocols (4). Cells were infected with adenovirus as described previously (36) at a multiplicity of infection of 200 to 400 PFU/cell. The virus expressing BRCA1 interfering peptide fragment (BIF) was compared to viruses expressing either green fluorescent protein (GFP) or LacZ that were infected at an equal multiplicity of infection. In comparisons of different cell lines to MCF10A, the level of expression of the BIF peptide was equal. Small interfering RNA (siRNA) transfection was performed by using Oligofectamine (Invitrogen), and cells were harvested 2 days posttransfection. The control oligonucleotide targets luciferase mRNA (5′ to 3′, AAC GUA CGC GGA AUA CUU CGA), the BRCA1-a oligonucleotides have been published elsewhere (14), and the BRCA1-b oligonucleotide target is BRCA1 mRNA (5′ to 3′, AAG GUU UCA AAG CGC CAG UCA) (Dharmacon). Lipofectamine 2000 (Invitrogen) was utilized for transfection of γ-tubulin and GFP-centrin plasmids.
Immunofluorescence.
Cells were fixed and stained with anti-γ-tubulin (Sigma) as described elsewhere (36). Centrin staining is described in reference 22. Images were viewed at an original magnification of ×400 or ×1,000 with a Zeiss Axioscope and were captured by using a model 2.3.1 SPOT digital camera.
Plasmids and viruses.
BRCA1-FLAG and BARD1 baculoviruses were constructed according to the FastBac system (Invitrogen). Full-length BRCA1 and BRCA1(1-1852) have one amino-terminal FLAG tag. BRCA1(1-1527), BRCA1(1-1000), and BRCA1(1-500) each have one carboxy-terminal FLAG tag. The BARD1 gene (43) was kindly provided by J. Chen and D. M. Livingston. Hemagglutinin (HA) ubiquitin was as described previously (40). The human γ-tubulin gene (IMAGE clone 3345973; Invitrogen) and the human centrin-1 gene (32) were subcloned into pET21A for expression in bacteria with a six-His tag. PCR-based point mutagenesis (QuikChange XL kit; Stratagene) was used to mutate the human γ-tubulin gene in the pET21A vector. The wild-type and mutant γ-tubulin genes were then cloned into the pOZ-N vector (29), which contains a FLAG and an HA epitope tag.
Protein purification.
SF9 cells were coinfected with the BRCA1 and BARD1 baculoviruses by using standard protocols. The BRCA1/BARD1 complex was affinity purified by using M2-agarose (Sigma) according to a protocol described previously (18). Briefly, cells were collected, washed once in phosphate-buffered saline, and then lysed in 8 ml of lysis buffer consisting of 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 4 mM MgCl2, 0.4 mM EDTA, 2 mM dithiothreitol, 20 mM β-glycerophosphate, 20% glycerol, 0.4 mM phenylmethylsulfonyl fluoride, 4 μg of leupeptin/ml, and 2 μg of aprotinin/ml on ice. The lysate was Dounce homogenized and diluted twofold with dilution buffer, consisting of 20 mM Tris-HCl (pH 7.9), 10% glycerol, and 0.02% Nonidet P-40. The cell debris was removed by centrifugation. The supernatant was incubated with 200 μl of prewashed M2 agarose (Sigma) for 5 h and then washed four times in 10 ml of ice-cold wash buffer (20 mM Tris-HCl [pH 7.9], 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 15% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, 1 μg of aprotinin/ml, and 0.01% Nonidet P-40). The BRCA1/BARD1 complexes were then eluted by adding 0.2 mg of FLAG peptide/ml and 0.1 mg of insulin or lysozyme/ml to wash buffer and adding 4 × 100 μl of elution buffer to M2 agarose beads for 5 min on ice and collecting the supernatant. Protein content was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie brilliant blue.
Centrosome fractions were prepared according to the protocol described previously (5). Both of the recombinant centrosomal proteins, γ-tubulin and centrin-1, were hexahistidine tagged, separately expressed in bacteria, and purified with a nickel-nitrilotriacetic acid matrix by using standard protocols. Each was further purified by ion-exchange chromatography using a Bioscale Q column.
The BIF fragment of RNA helicase A (amino acids 89 to 344) and the control fragment of RNA helicase A (amino acids 1 to 250) were cloned into pGEX-2TK vectors, expressed in Escherichia coli, and purified by using standard procedures for glutathione S-transferase fusion proteins.
In vitro ubiquitination.
Reaction mixtures contained 10 mM HEPES (pH 7.9), 0.5 mM EDTA, 5 mM MgCl2, 2 mM NaF, 2 mM ATP, 60 mM KCl, 1 μM ubiquitin, 200 nM E1-His (a kind gift from P. Howley), 5 μM UbcH5c-His (a kind gift from R. Baer), and 200 to 400 ng of BRCA1-Flag/BARD1. Several forms of modified ubiquitin were used: biotin-tagged (28), histidine-tagged (Boston Biochem), histidine-tagged with a lysine 48 mutation to arginine (Boston Biochem), or purified bovine ubiquitin (Sigma). The centrosome fraction (400 ng) with the largest amount of γ-tubulin was added to the reaction mixtures as indicated. In reaction mixtures containing purified recombinant γ-tubulin or centrin, the individual proteins were added at 700 ng each. Reaction mixtures were incubated at 37°C for 30 min, and then proteins were resolved on protein gels and blotted to polyvinylidene difluoride membranes. Total ubiquitinated proteins were visualized by using streptavidin fused to horseradish peroxidase (HRP) and chemiluminescence. Ubiquitinated γ-tubulin was visualized by Western blotting with mouse anti-γ-tubulin (Sigma) followed by chemiluminescence. Antibodies specific for α- and ɛ-tubulin were purchased from Sigma. Streptavidin affinity purification was performed according to standard protocols (Dynal).
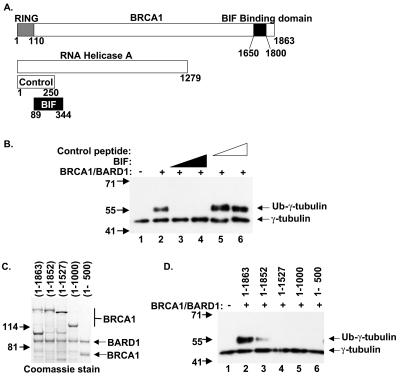
The BRCA1 full-length and carboxy-terminal truncation proteins were balanced by BARD1 content. The BARD1 concentration in each reaction mixture was 15 nM, making the deduced BRCA1 concentration 15 nM in each ubiquitination reaction. In the experiment for which results are shown in Fig. 6B, BIF or control peptide was added at two concentrations for each protein fragment, 900 nM and 3.5 μM.
FIG. 6.
The function of the carboxy terminus of BRCA1 is necessary for ubiquitination of γ-tubulin in vitro. (A) Schematic of BRCA1, RNA helicase A, BIF, and control peptide. (B) Increasing amounts of purified BIF (lanes 3 and 4) or the same amounts of control peptide (lanes 5 and 6) were added to ubiquitination reaction mixtures. Reaction products were analyzed by Western blotting with anti-γ-tubulin. (C) BRCA1(1-1863)/BARD1, BRCA1(1-1852)/BARD1, BRCA1(1-1527)/BARD1, BRCA1(1-1000)/BARD1, and BRCA1(1-500)/BARD1 were analyzed by protein gel and visualized by staining with Coomassie brilliant blue. (D) The BRCA1/BARD1 complexes containing 15 nM (each) BRCA1 and BARD1, with BRCA1 of varying lengths, shown in panel C, were added to ubiquitination reaction mixtures as indicated and analyzed by Western blotting with anti-γ-tubulin.
Mass spectroscopy.
Excised gel bands were subjected to trypsin proteolysis; MS/MS spectra of resulting peptides were acquired by nanoscale microcapillary LC-MS/MS as described previously (31) on an LTQ-DECA mass spectrometer (ThermoElectron). Spectra were searched with the SEQUEST (11) algorithm against human entries from the Nonredundant Protein sequence database (NRP), available from the National Center for Biotechnology Information.
RESULTS
BRCA1 function is necessary for maintenance of centrosome number in mammary cell lines.
Evidence from many tumors reveals that cancer-causing mutations can occur in the carboxy terminus of BRCA1. For example, truncation of the last 11 amino acids can render BRCA1 nonfunctional as a tumor suppressor (38), suggesting that this domain of BRCA1 is vital for its tumor suppressor function. In a previous study, we had developed a strategy to inhibit the function of the BRCA1 carboxy terminus by expressing a peptide that specifically binds to amino acids 1650 to 1800 of BRCA1. That peptide, termed BIF in this study, is a fragment of RNA helicase A (RHA), a protein that mediates the interaction of BRCA1 and RNA polymerase II holoenzyme (1). BIF contains amino acid residues 89 to 344 of RHA and an epitope tag; it lacks other functional domains of RHA important for transcription. Expression of BIF in the mammary cell line MCF10A and in the osteosarcoma cell line U2OS was shown to interfere with BRCA1 localization to nuclear foci after DNA damage (36). This observation supports the notion that BIF expression inhibits a known function of BRCA1. Interestingly, in the previous study, when the two cell lines analyzed were compared, expression of BIF caused a centrosome amplification phenotype in the mammary cell line but not in the osteosarcoma cell line. Since centrosome amplification was also observed in the BRCA1-mutant breast cancer cell line HCC1937 (36), it was suggested that this centrosome amplification was in fact caused by disruption of BRCA1 function. In this study we tested whether other cell lines derived from breast tissue also show centrosome amplification after inhibition of BRCA1.
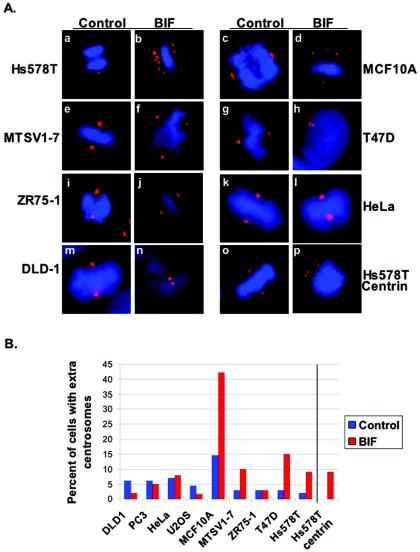
In order to express BIF in multiple cell lines, we utilized a recombinant adenovirus (adBIF). Cells were infected with adBIF and compared to cells that were infected at the same multiplicity of infection with recombinant adenoviruses expressing either GFP or LacZ. All nine cell lines derived from both breast and other cell types that were chosen had a low baseline of centrosome anomalies. Each cell line was infected with adBIF, and differences in BIF expression between cell types were normalized by assessing protein levels of the BIF polypeptide by Western blot analysis (data not shown). After 24 to 27 h, each infected cell line was fixed and stained with antibodies specific for the centrosome protein γ-tubulin. The number of cells with abnormal numbers of centrosomes (n > 2) was scored by fluorescence microscopy. The results in Fig. 1Aa to n and B show that only the mammary tissue-derived cell lines expressing BIF exhibited a threefold increase in cells with amplified centrosome numbers (four out of five tested). Cell lines derived from other cell types—prostate (PC3), cervix (HeLa), colon (DLD-1), and osteosarcoma (U2OS)-did not have amplified centrosomes caused by expression of BIF (none of four tested). These results reveal a striking correlation: a BRCA1 function disrupted by BIF is important for maintenance of centrosome number in breast cells, but this BRCA1 function is not important in other cell types.
FIG. 1.
Expression of BIF caused centrosome amplification in several tissue culture cell lines derived from mammary tissue. (A) The indicated cell lines either were infected with a control adenovirus expressing either GFP or LacZ (a, c, e, g, i, k, m, and o) or were infected with an adenovirus expressing BIF at an equal multiplicity of infection (b, d, f, h, j, l, n, and p). Twenty-four to 27 h after infection, cells were fixed and stained for DNA content (DAPI; blue) and with an antibody specific to γ-tubulin (a to n) (red). Centrosomes were also stained with an antibody specific to centrin (o and p) (red). (B) The cells from the experiments for which results are shown in panel A were counted, and the percentage of cells with amplified (more than two) centrosomes were scored. Results of one representative experiment are shown. The number of cells with more than two centrosomes was determined at least twice for each cell line, and at least 100 cells were counted in each experiment. The data were collected from γ-tubulin-stained cells for the first nine experimental pairs and from centrin-stained cells for the last pair on the right (Hs578T, centrin).
The cell lines used represented a spectrum of phenotypes regarding cancer and p53 status. The MCF10A and MTSV1-7 lines are nontumorigenic and are thought to reflect normal mammary epithelium. The ZR75 and T47D cells are from breast cancer lines, and they are estrogen receptor positive. The Hs578T breast cancer cell line is estrogen-receptor negative. Thus, the effects of BIF expression on centrosome number were obtained in all of these breast-derived cell lines, except for ZR75, regardless of cancer status. Interestingly, p53 status does not correlate with BIF-induced centrosome amplification, either. Among the mammary tissue-derived cell lines, only MCF10A is wild type for p53. The other cell lines, MTSV1-7, T47D, Hs578T, and ZR75, are all negative for p53. MTSV1-7 cells express simian virus 40 T antigen (3), and the others harbor p53 mutations (4, 19, 24, 30). Among the nonbreast cell lines, U2OS cells have wild-type p53, whereas HeLa, PC3, and DLD-1 cells either express proteins that degrade p53 or have mutant p53 (2, 7, 26, 34, 35). Determining whether the p53 status of these cell lines correlates with the centrosome amplification phenotype is important, since p53 plays a role in centrosome number regulation, and p53 is known to interact with BRCA1 pathways (13, 47). It should be noted that several cell lines with mutant p53 could not be used in this study because they had centrosome amplification independent of BIF expression.
In analysis of the centrosome number in cells by use of an antibody specific to γ-tubulin, it is possible that fragmentation, rather than replication, of centrosomes was scored as extra centrosomes. In order to test this possibility, the experiment for which results are shown in Fig. 1Aa and b was repeated, but the cells were stained with an antibody specific to centrin. Centrin is located at each centriole, and by immunofluorescence each centrosome should have two pinpoints of centrin. Normal cells with one or two centrosomes would have two or four points of centrin in pairs. It was found that Hs578T cells expressing the control protein had no examples of cells with more than four centrin-positive spots in two pairs. By contrast, expression of BIF in these cells resulted in a rapid increase in the number of cells with more than four centrin-positive spots (9%), and these were frequently found not to be in pairs (Fig. 1Ao-p). These results suggest that both the proper regulation of centrosome replication and the proper maintenance of the centrosome structure require BRCA1 function in this cell line.
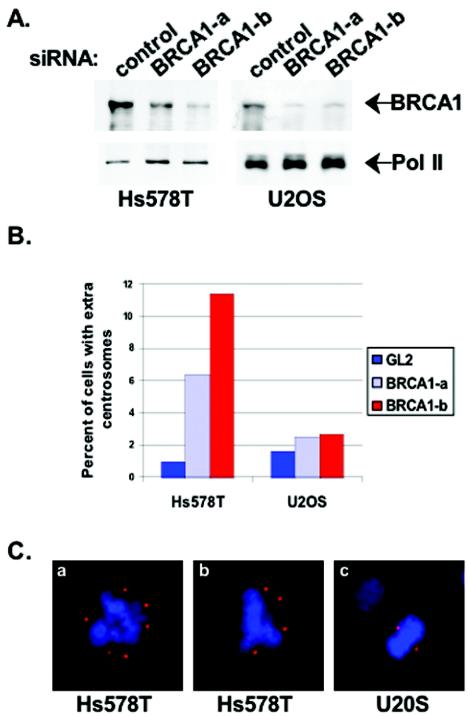
There is supporting evidence that the effects of BIF expression on centrosome number reflected an inhibition of BRCA1. BIF interferes with BRCA1 localization after DNA damage, and cells that lack wild-type BRCA1 (HCC1937) show a similar centrosome amplification phenotype (36). Nevertheless, it is possible that BIF alone, and not its interaction with BRCA1, caused centrosome amplification. To use a third method to test whether BRCA1 is indeed necessary for proper centrosome maintenance in mammary cells, siRNA oligonucleotides were designed to decrease the expression of BRCA1. The Western blot in Fig. 2A shows that the two different siRNA oligonucleotide pairs directed against BRCA1, but not a control siRNA, significantly reduced the amount of BRCA1 protein in transfected Hs578T and U2OS cells. To determine if a reduction in BRCA1 levels by siRNA caused amplified centrosomes, the number of cells with abnormal centrosome numbers was scored by immunofluorescence using a γ-tubulin-specific antibody. The results summarized in Fig. 2B show that Hs578T mammary cells that have reduced BRCA1 levels have a 5- to 10-fold increase in the fraction of cells with more than two centrosomes. Figure 2C shows examples of mitotic Hs578T cells with amplified centrosomes. While the expression of BRCA1 was reduced to the same level in U2OS cells, centrosomes from this cell line were not amplified. This suggests that the centrosome amplification phenotype caused by siRNA knockdown of BRCA1 followed the same mammary cell-specific trend observed for expression of BIF. This also suggests that the function of BRCA1 at the centrosome is inhibited by BIF binding.
FIG. 2.
Reduction in BRCA1 protein levels by siRNA leads to amplified centrosomes in Hs578T cells. (A) Western blots for BRCA1 protein 48 h after transfection with BRCA1-directed or control siRNA. The same filter was restained with an antibody specific for the large subunit of RNA polymerase II (Pol II) in order to evaluate equal loading of samples. (B) Cells with more than two centrosomes were scored by fluorescence microscopy as for Fig. 1B. The histogram shows the percentage of cells with amplified centrosomes. (C) Cells were fixed and stained with an antibody specific to γ-tubulin (as for Fig. 1A) 48 h after transfection with BRCA1-directed siRNA (BRCA1-b oligonucleotide). Examples of mitotic Hs578T cells (a and b) and U2OS cells (c) are shown.
HU abolishes centrosome amplification in cells expressing BIF.
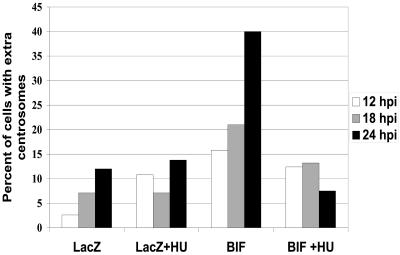
As an initial characterization of the centrosome amplification in these mammary epithelial cell lines expressing BIF, we asked whether passage through the cell cycle is important for the increase in centrosome number. It is known that centrosome duplication in mammalian cells is dependent on the activity of cyclin A (27). Thus, passage through S phase of the cell cycle would be critical to duplicate the centrosomes if the BIF-dependent amplification was utilizing the normal cellular machinery for this duplication process. This was tested directly by treating cells with hydroxyurea (HU), which blocks cells early in S phase, and assaying centrosome amplification. MCF10A cells were infected at time zero with adBIF or a control, and HU was added after 3 h. The centrosome number was scored by immunofluorescence for γ-tubulin at 12, 18, and 24 h postinfection (hpi). In the absence of HU, amplification of centrosomes in these cells was apparent after 18 h. HU blocked the amplification of centrosomes when added to the medium after adBIF infection (Fig. 3). These results suggest that treatment of cells with HU halted the cell cycle upstream of the point at which BRCA1 affected centrosome replication.
FIG. 3.
HU abolishes BIF-associated centrosome amplification in MCF10A cells. MCF10A cells were treated with 2 mM HU 3 h after infection with adBIF or adLacZ. Cells were fixed and stained for γ-tubulin at 12, 18, and 24 h postinfection (hpi). The histogram shows the percentage of cells with abnormal centrosome numbers.
The BRCA1/BARD1 heterodimer ubiquitinates centrosome proteins in vitro.
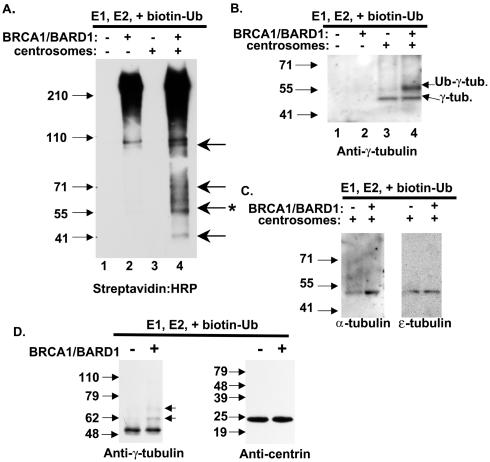
Our data clearly indicate that BRCA1 executes a major role in the maintenance of centrosome number in mammary cells (Fig. 1 and 2). BRCA1 has also been shown to localize to centrosomes during mitosis and to bind to γ-tubulin by coimmunoprecipitation (16). BRCA1, together with BARD1, forms a heterodimer associating via the respective RING domains and adjacent α-helices of each protein, and the BRCA1/BARD1 heterodimer is an active ubiquitin ligase (6, 15). We hypothesized that ubiquitination of centrosome-associated proteins by BRCA1 plays a role in centrosome regulation. To this end, centrosome fractions were prepared from MCF10A cells in order to identify targets of BRCA1/BARD1 ubiquitination in vitro. We tested whether BRCA1/BARD1 ubiquitinated any of the centrosome proteins by carrying out in vitro reactions that included copurified, full-length BRCA1/BARD1, purified E1, purified E2 UbcH5c, and biotinylated ubiquitin. Reaction products were resolved on protein gels and transferred to polyvinylidene difluoride membranes, and ubiquitinated species were identified by using streptavidin conjugated to HRP. In Fig. 4A, lane 2, autoubiquitination of BRCA1 (>220 kDa) is apparent along with ubiquitination of a 110-kDa polypeptide, which may be BARD1. In the presence of centrosomes, E1, UbcH5c, and ubiquitin but no BRCA1/BARD1, no ubiquitinated species were apparent (lane 3). Addition of BRCA1/BARD1 to these reaction mixtures resulted in four or more new ubiquitinated products (lane 4). Comparison with the total polypeptide composition of the centrosome preparation (data not shown) reveals that only a subset of the centrosome-associated proteins was modified by ubiquitin.
FIG. 4.
BRCA1/BARD1-dependent ubiquitination of centrosome proteins. (A) Partially purified centrosomes were tested as a substrate for the BRCA1/BARD1 ubiquitin ligase activity. Ubiquitination reaction mixtures contained the centrosome fraction, E1, UbcH5c, biotinylated ubiquitin, full-length BRCA1/BARD1, and centrosomes as indicated. Ubiquitinated protein products were visualized by HRP-conjugated streptavidin and chemiluminescence. (B) Reaction products as in panel A were analyzed by Western blotting with anti-γ-tubulin. (C) In vitro ubiquitination reaction products, as above, were analyzed by Western blotting with anti-α-tubulin and anti-ɛ-tubulin as indicated. (D) In vitro ubiquitination reaction products containing either γ-tubulin or centrin-1, each expressed and purified from bacteria, were analyzed by Western blotting with anti-γ-tubulin or anti-centrin as indicated.
One of the ubiquitinated polypeptides had a migration consistent with monoubiquitinated γ-tubulin (48 kDa unmodified plus 8 kDa of ubiquitin). To determine whether the ubiquitinated species at 56 kDa (Fig. 4A, lane 4) was γ-tubulin, the same ubiquitination reaction, including the centrosome fraction, BRCA1/BARD1, E1, UbcH5c, and biotinylated ubiquitin, was analyzed for modification of γ-tubulin. Monoclonal antibodies to γ-tubulin were used to detect unmodified γ-tubulin at 48 kDa in lanes containing centrosomes (Fig. 4B, lanes 3 and 4), and in the presence of BRCA1/BARD1, a shifted polypeptide consistent with monoubiquitinated γ-tubulin was apparent (lane 4). To test whether the reaction was specific for γ-tubulin, we also analyzed reaction products for ubiquitination of the related α- and ɛ-tubulin polypeptides, and they were negative for BRCA1/BARD1-dependent ubiquitination under these conditions (Fig. 4C). These results suggest that γ-tubulin is a specific target of BRCA1/BARD1 for ubiquitination.
In order to define the specificity of the BRCA1/BARD1 ubiquitination reactions for γ-tubulin, we tested as substrates γ-tubulin and centrin-1, which had been expressed and purified from bacteria. Equal amounts of recombinant protein were included in reaction mixtures containing BRCA1/BARD1, E1, UbcH5c, and ubiquitin. Dependent on the presence of BRCA1/BARD1, the γ-tubulin band was shifted by 8 and 16 kDa, consistent with ubiquitination (Fig. 4D). By contrast, inclusion of BRCA1/BARD1 along with the other ubiquitination factors showed no modification of the centrin protein (Fig. 4D). This result implies that BRCA1/BARD1 specifically modifies γ-tubulin, either as a purified protein or in association with the centrosome. Interestingly, purified γ-tubulin was modified by one or two ubiquitin moieties, whereas centrosome-associated γ-tubulin was only modified by one ubiquitin molecule. It is possible that the recombinant protein has a second site that is monoubiquitinated, whereas the γ-tubulin in the centrosome would have the second site blocked.
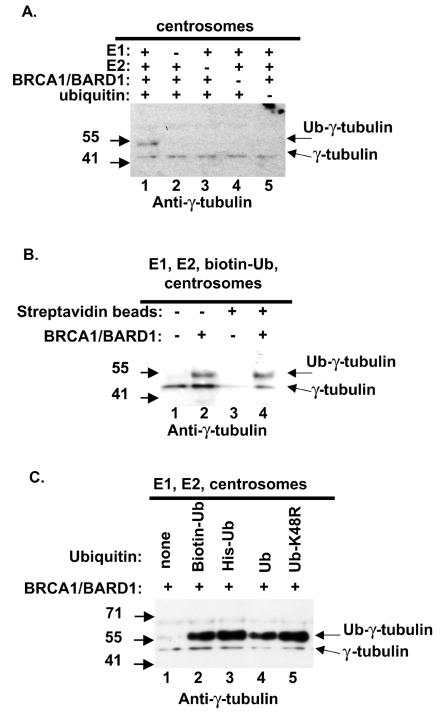
In order to test whether the slower-migrating band in reactions using centrosome fractions was in fact a ubiquitin modification, the requirement for ubiquitination factors was tested. In the complete reaction, a band at 56 kDa was detected by using an antibody specific for γ-tubulin (Fig. 5A, lane 1). Omission of any single ubiquitination protein from the reaction resulted in a loss of the 56-kDa γ-tubulin species (Fig. 5A, lanes 2 to 5). Thus, the modification of γ-tubulin by BRCA1/BARD1 was also dependent on the presence of E1, UbcH5c, and ubiquitin.
FIG. 5.
The in vitro modification of γ-tubulin by BRCA1/BARD1 is monoubiquitination. (A) Individual components of the ubiquitination reaction mixtures in Fig. 4B were singly omitted as indicated. Reaction products were analyzed by Western blotting with anti-γ-tubulin. (B) Ubiquitination reactions were carried out as for Fig. 4B; reaction products were affinity purified by magnetic streptavidin beads (lanes 3 and 4) and analyzed by Western blotting with anti-γ-tubulin. (C) Biotinylated ubiquitin, six-His-tagged ubiquitin, purified bovine ubiquitin, and six-His-tagged ubiquitin K48R were added to the ubiquitination reaction mixtures as indicated. Reaction products were analyzed by Western blotting with anti-γ-tubulin.
To ensure that the 56-kDa band detected by Western blotting with anti-γ-tubulin was indeed γ-tubulin plus biotinylated ubiquitin, we utilized magnetic streptavidin beads to purify ubiquitinated proteins. Lanes 1 and 2 of Fig. 5B show the products of the standard ubiquitination reaction before affinity purification by streptavidin beads. Following purification using streptavidin beads, the 48-kDa γ-tubulin did not bind to the matrix (lane 3). The 56-kDa band, dependent on BRCA1/BARD1 in the reaction, was purified via the biotin-ubiquitin modification (lane 4). Unmodified γ-tubulin (48 kDa) was also purified on the streptavidin beads in this reaction, although at a reduced level (lane 4). The presence of the 48-kDa γ-tubulin was anticipated because γ-tubulin is present as a multimer in large multisubunit complexes in centrosomes (γ-tubulin ring complex [γ-TuRC]). An inference from the presence of the unmodified γ-tubulin associated with the monoubiquitinated γ-tubulin is that the modification does not disrupt the γ-TuRC complex.
BRCA1/BARD1 monoubiquitinates γ-tubulin from centrosomes in vitro.
Polymerization of ubiquitin into chains requires isopeptide linkages between the activated carboxy terminus of an incoming ubiquitin molecule and a lysine side chain of the protein substrate or the previously conjugated ubiquitin. Isopeptide linkages via lysine 48 of four or more ubiquitin molecules constitute a signal for proteasome-mediated degradation of the substrate protein. In order to facilitate easy detection of ubiquitinated proteins, the ubiquitin used in Fig. 4A to C and Fig. 5A and B was biotinylated via the amine group in the lysine side chains of ubiquitin. The ubiquitin, biotinylated via lysines, may inefficiently polymerize into multiubiquitin chains. In order to address this issue, we compared reactions that contained various ubiquitin molecules, including purified bovine ubiquitin, histidine-tagged-ubiquitin, and histidine-tagged-ubiquitin containing a mutation at lysine 48. In reactions in which any of the purified ubiquitin proteins was included, a 56-kDa band was observed, but ubiquitin polymers were not detected on γ-tubulin regardless of the type of ubiquitin used (Fig. 5C). This suggested that the detection of monoubiquitinated γ-tubulin was not due solely to a modification of ubiquitin. Increasing the amount of E2 factor or ubiquitin also resulted in monoubiquitination (data not shown), suggesting that the lack of a multiubiquitin chain was not due to limiting components in the reaction. It is possible that other factors not present in the reaction would cause the multiubiquitination of γ-tubulin. Since the proteasome recognizes ubiquitin chains of four units or longer, linked through lysine 48, these data suggest that γ-tubulin ubiquitination by BRCA1/BARD1 is not a signal for proteasome-mediated degradation. The pathway triggered by BRCA1 ubiquitination is currently unclear. In this study, centrosomal γ-tubulin is modified with a single ubiquitin molecule, and in other studies BRCA1/BARD1 polymerizes ubiquitin through a novel lysine 6 linkage, which has unknown consequences for the cell (44). It is possible that proteins missing from the in vitro assays described here could facilitate polyubiquitination via lysine 6 of BRCA1-dependent target proteins such as γ-tubulin.
Function of the carboxy terminus of BRCA1 is required for ubiquitination of γ-tubulin.
Although the amino-terminal 110 amino acid residues of BRCA1 contain the RING domain and are sufficient for ubiquitination (15, 45), and although BRCA1 amino acid residues 504 to 803 bind γ-tubulin (16), we noted that expression of the BIF peptide that binds to the BRCA1 carboxy terminus inhibited BRCA1-dependent regulation of centrosomes in cells. We tested whether BIF would inhibit the in vitro BRCA1/BARD1-dependent ubiquitination of γ-tubulin. The BIF peptide and a control peptide fragment from RHA (diagrammed in Fig. 6A) were included in the complete BRCA1/BARD1 ubiquitination reaction described above. While the control peptide had no effect on BRCA1/BARD1 ubiquitination of γ-tubulin, the BIF peptide powerfully inhibited the BRCA1/BARD1-dependent ubiquitination of γ-tubulin (Fig. 6B). This result shows that an unbound carboxy-terminal domain of BRCA1 is required for the ubiquitination reaction.
To test whether the carboxy terminus of BRCA1 is indeed necessary for ubiquitination of γ-tubulin in vitro, we purified mutants of BRCA1 truncated at the carboxy terminus as heterodimers associated with full-length BARD1. Protein preparations included full-length BRCA1 (amino acids 1 to 1863), a cancer-associated truncation of BRCA1, BRCA1(1-1852), and three other truncations, BRCA1(1-1527), BRCA1(1-1000), and BRCA1(1-500), each coexpressed and purified with full-length BARD1 protein. The BARD1 protein was untagged and was purified via association with the tagged BRCA1. These five BRCA1/BARD1 preparations were balanced for BARD1 content (Fig. 6C), and each was enzymatically active in a nonspecific ubiquitin polymerization assay (data not shown). Each of these heterodimers was included in a ubiquitination reaction with E1, UbcH5c, biotinylated ubiquitin, and the centrosome fraction. Only the full-length BRCA1/BARD1 complex robustly ubiquitinated γ-tubulin (Fig. 6D, lane 2). Ubiquitination of γ-tubulin by the cancer-associated mutant that truncated the protein by only 11 amino acids, BRCA1(1-1852)/BARD1, was severely compromised (Fig. 6D, lane 3). Trimming 336 or more amino acid residues from the carboxy terminus of BRCA1 completely abolished the ubiquitination of γ-tubulin by BRCA1/BARD1 (Fig. 6D, lanes 4 to 6). The requirement for full-length BRCA1 protein was striking, since the domains required for nonspecific ubiquitination and for γ-tubulin binding reside in the amino-terminal 803 amino acids of BRCA1.
Expression in cells of γ-tubulin with a mutant lysine 48 resulted in centrosome amplification.
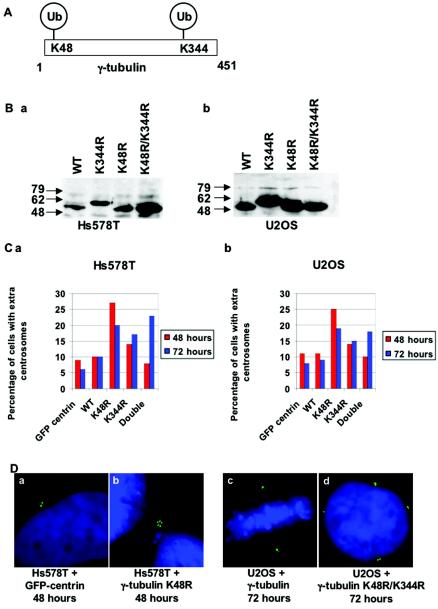
The γ-tubulin residues modified by BRCA1-dependent ubiquitination were determined by using the bacterially expressed γ-tubulin (as in Fig. 4D) and mass spectroscopy of the ubiquitinated products. Lysines 48 and 344 of γ-tubulin were found to be modified (Fig. 7A). We mutated these lysines to arginine, singly and together, and expressed these γ-tubulin mutants by transient transfection in Hs578T mammary cells and in U2OS osteosarcoma cells. Western blotting of the epitope-tagged γ-tubulin mutants revealed that they were all expressed well (Fig. 7B). The γ-tubulin K344R mutant was found to have a slowed migration, even though the DNA sequence revealed only the lysine-to-arginine mutation. We tested whether transient transfection of these γ-tubulin mutants affected the centrosome number by cotransfecting GFP-centrin in order to analyze only cells that had been transfected. Remarkably, expression of these mutated γ-tubulin proteins caused centrosome amplification (Fig. 7C and D). The γ-tubulin K48R mutant had the most pronounced effect on centrosome number after 48 h, causing a 2.5-fold increase in the number of cells with more than two centrosomes. On the other hand, expression of the γ-tubulin K344R mutant caused only a modest increase in the number of cells with amplified centrosomes after 48 and 72 h. Expression of a mutant γ-tubulin containing both modified lysines caused no effect after 48 h but a greater-than-twofold increase in centrosome number at the 72-h time point. We interpret the slowed centrosome amplification by the γ-tubulin double mutant as a deficiency in localizing to the centrosome.
FIG. 7.
BRCA1/BARD1 ubiquitinates lysines of γ-tubulin required for regulation of centrosome duplication. (A) Schematic of the ubiquitinated lysines of γ-tubulin found by mass spectroscopy. (B) Expression levels of Hs578T (a) or U2OS (b) cells transfected with epitope-tagged wild-type γ-tubulin (WT), γ-tubulin K344R, γ-tubulin K48R, or the double mutant γ-tubulin K48R K344R, as indicated, were analyzed by Western blotting with anti-HA. (C) Hs578T (a) or U2OS (b) cells were transfected with plasmids expressing the wild-type or mutant γ-tubulin and GFP-centrin. At 42 and 72 h posttransfection, cells were fixed and stained with DAPI. The centrosome content of cells was determined by counting the pinpoint GFP-centrin spots, and the percentage of cells with more than two centrosomes was determined. Each experiment was performed twice, and at least 100 cells were counted for each transfection. The histograms show the percentage of cells with extra centrosomes. (D) Examples of cells expressing GFP-centrin and the indicated γ-tubulin protein are shown with normal (a and c) or abnormal (b and d) centrosome content.
Expression of the mutant γ-tubulin in both Hs578T and U2OS cell lines resulted in centrosome amplification. In fact, the results from experiments with the two cell lines were nearly identical. This shows that the ubiquitination of γ-tubulin is important in both cell types but that the enzyme that mediates the ubiquitination may differ from cell type to cell type. Taking these findings together with the data from Fig. 1 and 2, we conclude that the mammary cell lines tested depend on BRCA1 for this important ubiquitination event and that other cell lines, such as U2OS, which are unaffected by BRCA1 inhibition, rely on another factor to modify lysine 48 of γ-tubulin.
DISCUSSION
Ever since it was discovered that BRCA1/BARD1 constitutes a highly active E3 ubiquitin ligase (15), the key question has been the identification of the specific target of the BRCA1/BARD1 ubiquitin ligase activity and the importance of this enzymatic reaction to the biology of the mammary cell. In this study, we found that BRCA1/BARD1 ubiquitinates centrosome proteins, including γ-tubulin. The ubiquitination of γ-tubulin is important for the maintenance of an appropriate centrosome number in the cell. Clearly, disruption of the regulation of centrosome duplication by the loss of BRCA1 function, as in the mammary cells we tested, would lead to harmful sequelae, such as aneuploidy and chromosomal instability.
The ubiquitination of γ-tubulin was remarkably specific. Though other centrosome proteins were ubiquitinated by BRCA1/BARD1, the α-tubulin and ɛ-tubulin present in the centrosome preparations were not. Ubiquitination of γ-tubulin by BRCA1/BARD1 required the full-length 1,863-amino-acid protein, even though the amino-terminal 110 residues containing the RING domain of BRCA1 are sufficient for ubiquitination activity (45), and even though amino acids 504 to 803 of BRCA1 are sufficient for binding to γ-tubulin (16). This reaction was markedly defective in a cancer-associated 11-amino-acid truncation from the carboxy terminus, and larger truncations from the carboxy terminus resulted in a complete loss of γ-tubulin ubiquitination activity, suggesting that ubiquitination of γ-tubulin may be important to protect mammary cells from cancerous transformation. Consistent with the truncation data, binding of the BRCA1 carboxy terminus by BIF blocked the γ-tubulin ubiquitination activity as well. In this case, the extreme carboxy terminus of BRCA1 is necessary for regulation of its amino-terminal RING domain.
We observed a profound effect on centrosome number by blocking the BRCA1-dependent ubiquitination of γ-tubulin by expressing proteins containing the mutated target lysine residues. Since the expression of the γ-tubulin K48R mutation exactly mimicked the phenotype of inhibiting BRCA1 in Hs578T cells, we conclude that γ-tubulin is a valid in vivo target of the BRCA1 ubiquitin ligase. The modification of γ-tubulin must be transient, since we did not detect ubiquitinated γ-tubulin in cells (data not shown). These results with the mutant γ-tubulin also support our hypothesis that the ubiquitination of γ-tubulin and other centrosome targets is necessary in order to protect mammary cells from aneuploidy.
Is the apparent mammary specificity in centrosome amplification the key to the breast-specific phenotype of BRCA1-associated cancers? Such a conclusion cannot be drawn from the results of this study. However, we do note that the regulation of centrosome duplication depends in part on modification of lysine 48 of γ-tubulin and that, at least among some breast cell lines, this modification requires BRCA1. Published findings report counterexamples to the apparent breast cell specificity observed in this study. In one case, murine embryonic fibroblasts that lack exon 11 of BRCA1 have been made, and these murine embryonic fibroblasts accumulate extra centrosomes in culture (46). Also, COS cells, which stably overexpress a portion of BRCA1 that binds to γ-tubulin, accumulate extra centrosomes (16). The differences in cell type specificity in centrosome amplification secondary to inhibition of BRCA1 function may be due to the natures of the assays used in each study, or, alternatively, due to species specificity. In the experiments in this study using adBIF, the centrosome phenotype in human mammary cells occurred rapidly, within 24 h. The data in Fig. 1 characterize acute loss of BRCA1, not cells that have overcome loss of BRCA1 function after multiple divisions as in previously published studies.
BRCA1 function has been implicated as important in DNA damage repair pathways. There are multiple ways in which proteins involved in genome maintenance regulate centrosome replication. As an example, the normal function of the p53 protein protects the cell from DNA damage, and mutant p53 can cause centrosome amplification (13). In a different system, the fruit fly embryo, DNA damage caused depletion of γ-tubulin from the spindle to block mitosis in a given nucleus (39). Ubiquitination is known to play a vital role in the maturation and replication of centrosomes (12, 42), and BRCA1 is in a position to link genome maintenance to centrosome regulation via ubiquitination.
Acknowledgments
We are grateful for the advice of A. Dutta, J. Ruderman, D. Finley, and Parvin lab members during the course of this project. We thank A. Goldberg, T. Ohta, R. Baer, P. Howley, Q. Pan, J. Chen, D. Livingston, J. Taylor-Papadimitriou, K. Munger, and M. Bornens for the gift of plasmids and cells critical to this project. We thank J. L. Salisbury for the gift of centrin-specific antibodies.
This project was supported by a Department of Defense Breast Cancer Research Program fellowship (L.M.S.), a Komen Foundation fellowship (S.S.), and research grant CA90281 from the NCI (J.D.P.).
REFERENCES
- 1.Anderson, S. F., B. P. Schlegel, T. Nakajima, E. S. Wolpin, and J. D. Parvin. 1998. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 19:254-256. [DOI] [PubMed] [Google Scholar]
- 2.Arita, D., M. Kambe, C. Ishioka, and R. Kanamaru. 1997. Induction of p53-independent apoptosis associated with G2M arrest following DNA damage in human colon cancer cell lines. Jpn. J. Cancer Res. 88:39-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartek, J., J. Bartkova, N. Kyprianou, E. N. Lalani, Z. Staskova, M. Shearer, S. Chang, and J. Taylor-Papadimitriou. 1991. Efficient immortalization of luminal epithelial cells from human mammary gland by introduction of simian virus 40 large tumor antigen with a recombinant retrovirus. Proc. Natl. Acad. Sci. USA 88:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berdichevsky, F., and J. Taylor-Papadimitriou. 1991. Morphological differentiation of hybrids of human mammary epithelial cell lines is dominant and correlates with the pattern of expression of intermediate filaments. Exp. Cell Res. 194:267-274. [DOI] [PubMed] [Google Scholar]
- 5.Bornens, M., M. Paintrand, J. Berges, M. C. Marty, and E. Karsenti. 1987. Structural and chemical characterization of isolated centrosomes. Cell. Motil. Cytoskeleton 8:238-249. [DOI] [PubMed] [Google Scholar]
- 6.Brzovic, P. S., P. Rajagopal, D. W. Hoyt, M. C. King, and R. E. Klevit. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8:833-837. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, A. G., H. J. Voeller, L. Sugars, and E. P. Gelmann. 1993. p53 oncogene mutations in three human prostate cancer cell lines. Prostate 23:123-134. [DOI] [PubMed] [Google Scholar]
- 8.Chen, A., F. E. Kleiman, J. L. Manley, T. Ouchi, and Z. Q. Pan. 2002. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 277:22085-22092. [DOI] [PubMed] [Google Scholar]
- 9.Deng, C. X. 2002. Roles of BRCA1 in centrosome duplication. Oncogene 21:6222-6227. [DOI] [PubMed] [Google Scholar]
- 10.Dong, Y., M. A. Hakimi, X. Chen, E. Kumaraswamy, N. S. Cooch, A. K. Godwin, and R. Shiekhattar. 2003. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12:1087-1099. [DOI] [PubMed] [Google Scholar]
- 11.Ducret, A., I. Van Oostveen, J. K. Eng, J. R. Yates III, and R. Aebersold. 1998. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 7:706-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E., K. R. Lacey, P. Huie, S. A. Lyapina, R. J. Deshaies, T. Stearns, and P. K. Jackson. 1999. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13:2242-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 14.Ganesan, S., D. P. Silver, R. A. Greenberg, D. Avni, R. Drapkin, A. Miron, S. C. Mok, V. Randrianarison, S. Brodie, J. Salstrom, T. P. Rasmussen, A. Klimke, C. Marrese, Y. Marahrens, C. X. Deng, J. Feunteun, and D. M. Livingston. 2002. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111:393-405. [DOI] [PubMed] [Google Scholar]
- 15.Hashizume, R., M. Fukuda, I. Maeda, H. Nishikawa, D. Oyake, Y. Yabuki, H. Ogata, and T. Ohta. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276:14537-14540. [DOI] [PubMed] [Google Scholar]
- 16.Hsu, L. C., T. P. Doan, and R. L. White. 2001. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 61:7713-7718. [PubMed] [Google Scholar]
- 17.Hsu, L. C., and R. L. White. 1998. BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. USA 95:12983-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffy, B. D., R. B. Chirnomas, E. J. Chen, J. M. Gudas, and D. F. Romagnolo. 2002. Activation of the aromatic hydrocarbon receptor pathway is not sufficient for transcriptional repression of BRCA-1: requirements for metabolism of benzo[a]pyrene to 7r,8t-dihydroxy-9t,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. Cancer Res. 62:113-121. [PubMed] [Google Scholar]
- 20.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 21.Lingle, W. L., S. L. Barrett, V. C. Negron, A. B. D'Assoro, K. Boeneman, W. Liu, C. M. Whitehead, C. Reynolds, and J. L. Salisbury. 2002. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA 99:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingle, W. L., and J. L. Salisbury. 2001. Methods for the analysis of centrosome reproduction in cancer cells. Methods Cell Biol. 67:325-336. [DOI] [PubMed] [Google Scholar]
- 23.Lotti, L. V., L. Ottini, C. D'Amico, R. Gradini, A. Cama, F. Belleudi, L. Frati, M. R. Torrisi, and R. Mariani-Costantini. 2002. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes Chromosomes Cancer 35:193-203. [DOI] [PubMed] [Google Scholar]
- 24.Lyakhovich, A., and M. P. Shekhar. 2003. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol. Cell. Biol. 23:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallery, D. L., C. J. Vandenberg, and K. Hiom. 2002. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21:6755-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matlashewski, G., L. Banks, D. Pim, and L. Crawford. 1986. Analysis of human p53 proteins and mRNA levels in normal and transformed cells. Eur. J. Biochem. 154:665-672. [DOI] [PubMed] [Google Scholar]
- 27.Meraldi, P., J. Lukas, A. M. Fry, J. Bartek, and E. A. Nigg. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88-93. [DOI] [PubMed] [Google Scholar]
- 28.Mitsui, A., and P. A. Sharp. 1999. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA 96:6054-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 30.Nieves-Neira, W., and Y. Pommier. 1999. Apoptotic response to camptothecin and 7-hydroxystaurosporine (UCN-01) in the 8 human breast cancer cell lines of the NCI Anticancer Drug Screen: multifactorial relationships with topoisomerase I, protein kinase C, Bcl-2, p53, MDM-2 and caspase pathways. Int. J. Cancer 82:396-404. [DOI] [PubMed] [Google Scholar]
- 31.Peng, J., and S. P. Gygi. 2001. Proteomics: the move to mixtures. J. Mass Spectrom. 36:1083-1091. [DOI] [PubMed] [Google Scholar]
- 32.Piel, M., J. Nordberg, U. Euteneuer, and M. Bornens. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science 291:1550-1553. [DOI] [PubMed] [Google Scholar]
- 33.Pihan, G. A., J. Wallace, Y. Zhou, and S. J. Doxsey. 2003. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63:1398-1404. [PubMed] [Google Scholar]
- 34.Rothmann, T., A. Hengstermann, N. J. Whitaker, M. Scheffner, and H. zur Hausen. 1998. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J. Virol. 72:9470-9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 36.Schlegel, B. P., L. M. Starita, and J. D. Parvin. 2003. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene 22:983-991. [DOI] [PubMed] [Google Scholar]
- 37.Starita, L. M., and J. D. Parvin. 2003. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr. Opin. Cell Biol. 15:345-350. [DOI] [PubMed] [Google Scholar]
- 38.Szabo, C. I., and M. C. King. 1995. Inherited breast and ovarian cancer. Hum. Mol. Genet. 4:1811-1817. [DOI] [PubMed] [Google Scholar]
- 39.Takada, S., A. Kelkar, and W. E. Theurkauf. 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113:87-99. [DOI] [PubMed] [Google Scholar]
- 40.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 41.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 42.Wojcik, E. J., D. M. Glover, and T. S. Hays. 2000. The SCF ubiquitin ligase protein Slimb regulates centrosome duplication in Drosophila. Curr. Biol. 10:1131-1134. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L. C., Z. W. Wang, J. T. Tsan, M. A. Spillman, A. Phung, X. L. Xu, M. C. Yang, L. Y. Hwang, A. M. Bowcock, and R. Baer. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14:430-440. [DOI] [PubMed] [Google Scholar]
- 44.Wu-Baer, F., K. Lagrazon, W. Yuan, and R. Baer. 2003. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278:34743-34746. [DOI] [PubMed] [Google Scholar]
- 45.Xia, Y., G. M. Pao, H. W. Chen, I. M. Verma, and T. Hunter. 2003. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278:5255-5263. [DOI] [PubMed] [Google Scholar]
- 46.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, H., K. Somasundaram, Y. Peng, H. Tian, D. Bi, B. L. Weber, and W. S. El-Deiry. 1998. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene 16:1713-1721. [DOI] [PubMed] [Google Scholar]