Abstract
Lactobacillus plantarum is a flexible and versatile microorganism that inhabits a variety of environmental niches, including the human gastrointestinal (GI) tract. Moreover, this lactic acid bacterium can survive passage through the human or mouse stomach in an active form. To investigate the genetic background of this persistence, resolvase-based in vivo expression technology (R-IVET) was performed in L. plantarum WCFS1 by using the mouse GI tract as a model system. This approach identified 72 L. plantarum genes whose expression was induced during passage through the GI tract as compared to laboratory media. Nine of these genes encode sugar-related functions, including ribose, cellobiose, sucrose, and sorbitol transporter genes. Another nine genes encode functions involved in acquisition and synthesis of amino acids, nucleotides, cofactors, and vitamins, indicating their limited availability in the GI tract. Four genes involved in stress-related functions were identified, reflecting the harsh conditions that L. plantarum encounters in the GI tract. The four extracellular protein encoding genes identified could potentially be involved in interaction with host specific factors. The rest of the genes are part of several functionally unrelated pathways or encode (conserved) hypothetical proteins. Remarkably, a large number of the functions or pathways identified here have previously been identified in pathogens as being important in vivo during infection, strongly suggesting that survival rather than virulence is the explanation for the importance of these genes during host residence.
Traditionally, lactic acid bacteria (LAB) are applied extensively in the production of a wide variety of fermented food and feed products. In addition, certain LAB species, in particular from the genus Lactobacillus, are natural inhabitants of the gastrointestinal (GI) tract and may have probiotic effects in humans and animals (2). Moreover, LAB have great potential to serve as delivery vehicles of health-promoting or therapeutic compounds to the human GI tract (20, 48). Among the different species encompassed by this genus, Lactobacillus plantarum is encountered in many environmental niches, including some dairy, meat, and a variety of vegetable fermentations (28). Because of the long tradition of utilization of L. plantarum in industrial and artisanal fermentations, this microbe is generally regarded as safe. In addition to the occurrence of L. plantarum in our diets, this microbe is frequently encountered as a natural inhabitant of the human GI tract (2). The complete 3.3-Mbp genome sequence of L. plantarum WCFS1 has been determined (28). This strain is a single-colony isolate of strain NCIMB8826, which effectively survives passage of the human stomach in an active form, reaches the ileum in high numbers compared to other strains, and is detectable in the colon (51). Intriguingly, genome sequence comparison revealed that the closest relatives of L. plantarum include Listeria innocua and L. monocytogenes, which also naturally inhabit and persist in the human GI tract (19). The availability of its genome sequence allows effective investigation of the genes and regulation mechanisms underlying the observed persistence of L. plantarum in the GI tract.
Three main strategies have been developed for the identification of genes that are highly expressed in vivo as compared to expression in laboratory conditions, namely, selective capture of transcribed sequences (SCOTS) (11, 15), signature-tagged mutagenesis (STM) (for reviews, see references 42 and 53), and in vivo expression technology (IVET). The original IVET strategy involves a tandem set of promoterless reporter genes that were used to identify promoters that are specifically switched on in Salmonella enterica serovar Typhimurium during infection (37). Subsequently, IVET variations in analyses utilizing different auxotrophic markers, dual reporters, and antibiotic resistance genes have been used to trap promoters specifically activated during infection of several other pathogens (for reviews, see references 3 and 36). The major drawback of the aforementioned IVET variations is that the experimental setup is designed in such a way that gene activity is required throughout the residence of the bacteria in the host. Hence, genes that are weakly expressed in the laboratory or transiently expressed only in a specific compartment of the host's GI tract slip through the selection procedure without being noticed. The fourth IVET variation circumvents this disadvantage by using the irreversible enzymatic activity of resolvases as reporter gene. Recombination-based IVET (R-IVET) is the only IVET approach that functions as a genetic screen. An antibiotic resistance marker flanked by two resolvase-recognition sites is integrated into the chromosome of the bacterium of interest. Subsequently, a promoterless copy of a resolvase-encoding gene is introduced on a plasmid and used to trap transcriptional activation by monitoring changes in the antibiotic resistance phenotype. Importantly, this approach does not rely on selective pressure during the animal experiments, since promoter activations are irreversibly trapped by the excision of the antibiotic resistance marker and can be identified after recovery of the bacterium under investigation from the host (3, 36).
Although many studies have described the in vivo behavior of pathogens during infection in various animal models, the bacterial factors that allow survival and persistence of food-associated microorganisms remain largely unknown. Only a single study describes an in vivo approach in L. reuteri, resulting in the identification of three genes that are induced during colonization of Lactobacillus-free mice on an antibiotic-containing diet (52). Unfortunately, the administration of antibiotics dramatically disturbs the intestinal microflora present in these mice, probably leading to GI tract conditions that differ significantly from those in a conventional mouse. This disadvantage does not apply to R-IVET strategies (3). Here we describe the exploitation of such a R-IVET approach in the food-grade organism L. plantarum WCFS1, leading to the identification of 72 genes of which the expression is induced in this LAB during passage of the GI tract of conventional mice compared to laboratory media. Homologues of many of the genes identified here have previously been identified in pathogenic bacteria by using a variety of SCOTS, STM, and (R-)IVET approaches. In addition, several novel in vivo-induced genes of L. plantarum have been identified that potentially contribute to specific host-microbe interactions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids that were used in the present study are listed in Table 1. Escherichia coli strains MC1061 (9) was used as cloning host during construction of pNZ7109 (see below) and was grown aerobically in TY medium (45). Lactococcus lactis MG1363 (18) was used as a cloning host during construction of pNZ7125, pNZ7126, and the L. plantarum promoter library (see below). Lactococcus lactis was grown without aeration at 30°C in M17 medium (Merck, Darmstadt, Germany), supplemented with 0.5% (wt/vol) glucose (GM17). L. plantarum WCFS1 (28), its rifampin-resistant derivative WCFS1-R (see below), and its loxP-ery-loxP derivative NZ7109 (see below) were grown at 37°C in MRS medium (Difco, Surrey, United Kingdom) without aeration. L. plantarum WCFS1 was subcultured in the presence of increasing concentrations of rifampin (up to 50 μg/ml) to obtain a rifampin-resistant derivative of this strain that was designated L. plantarum WCFS1-R (Table 1), which was used for the effective and selective recovery of this bacterium from fecal samples. When appropriate, antibiotics were added to the media as follows: for E. coli, ampicillin (50 μg/ml); for Lactococcus lactis, chloramphenicol (5 μg/ml); and for L. plantarum, chloramphenicol (5 μg/ml), erythromycin (5 or 30 μg/ml, for selection after transformation or replica plating, respectively), lincomycin (10 μg/ml), and rifampin (50 μg/ml).
TABLE 1.
Strains, plasmids, and primers used in this study and their relevant characteristics
| Material | Relevant features or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli MC1061 | Cloning host | 9 |
| Lactococcus lactis MG1363 | Cloning host | 18 |
| Lactobacillus plantarum | ||
| WCFS1 | Wild-type for which the genome sequence is available | 28 |
| WCFS1-R | Rifr | This work |
| NZ7109 | Rifr Emr; L. plantarum WCFS1 derivative containing a chromosomally located loxP-ery-loxP-cassette | This work |
| Plasmids | ||
| pUC18 | Apr | 54 |
| pUC19 | Apr | 54 |
| pNZ7105 | Apr; pUC18 derivative containing a 5′-truncated fragment of lp_3504 | This work |
| pNZ7106 | Apr; pUC19 derivative containing a 5′-truncated fragment of lp_3503 | This work |
| pNZ7107 | Apr; pNZ7105 derivative containing lp_3503 originating from pNZ7106 | This work |
| pUC19lox1 | Apr; pUC18 derivative containing a loxP site | This work |
| pUC19lox2 | Apr; pUC18 derivative containing 2 loxP sites in tandem repeat | This work |
| pGhost8 | Tetr | 35 |
| pNZ7103 | Apr Tetr; pUClox2 derivative containing tetR originating from pGhost8 | This work |
| pNZ7108 | Apr Tetr; pNZ7107 derivative containing loxP-tetR-loxP cassette originating from pNZ7103 | This work |
| pUC18ery | Apr Emr | 50 |
| pNZ7109 | Apr Emr; contains ery originating from pUC18ery flanked by two synthetic loxP sites and 5′-truncated fragments of lp_3503 and lp_3504 originating from pNZ7106 and pNZ7105, respectively (Fig. 1) | This work |
| pNZ7110 | Apr, pUC18 derivative containing cre and TpepN | 6 |
| pJIM2246 | Low-copy-cloning vector; Cmr | 44 |
| pNZ7124 | Cmr; pJIM2246 derivative containing cre and TpepN originating from pNZ7110 | This work |
| pNZ7125 | Cmr; pNZ7124 derivative containing Tlox upstream of cre | This work |
| pNZ7126 | Cmr; pNZ7125 derivative containing PldhL1 upstream of cre | This work |
| Primers | ||
| cre-R2 | 5′-GTCCATCAGGTTCTTGCG-3′ | |
| BglII-cre | 5′-ATAGTTTACCCCGTCAGC-3′ | |
| lp_3503F | 5′-TGCTTTCCAAGAGCAAGCTG-3′ | |
| lp_3503R | 5′-ACGTCTGCAGTCAGGTGTGAAGTTGGCACT-3′ | |
| lp_3504F | 5′-ACGTCTGCAGCCACTCACCGACTAACACTC-3′ | |
| lp_3504R | 5′-ACGTAAGCTTGACCCACGAGTTACCAACACG-5′ | |
| flox1 | 5′-AGCTCTGCAGATAACTTCGTATAGCATACATTATACGAAGTTATTTGCA-3′ | |
| flox2 | 5′-AATAACTTCGTATAATGTATGCTATACGTAGTTATCTGCAG-3′ | |
| flox3 | 5′-CATAACTTCGTATAGCATACATTATACGAAGTTATCTGCAG-3′ | |
| flox4 | 5′-AATTCTGCAGATAACTTCGTATAATGTATGCTATACGAAGTTATGAGCT-3′ | |
| hrec3503 | 5′-GCATCGCTGCACTCTTTATG-3′ | |
| hrec3504 | 5′-GAATATGGTCCATGGTCCAC-3′ | |
| eryF | 5′-AAGCAATGAAACACGCC-3 | |
| eryR | 5′-TTAGCCAGTTTCGTCGTT-3′ | |
| lasF2 | 5′-ACGTGGATCCGGACAATATGGGGTAAGCG-3′ | |
| lasR | 5′-AAGAAGATCTCTAAAGCTGACGGGGTAAAC-3′ | |
| PldhL-F | 5′-GAAGATCTTCAATCTTCTCACCGTCTTG-3′ | |
| PldhL-R | 5′-GAAGATCTTCAATAAGTCATCCTCTCGT-3′ |
Underlined sequences indicate restriction sites subsequently used in cloning procedures. Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Rifr, rifampicin resistant; Tetr, tetracycline resistant.
DNA techniques and sequence analysis.
Plasmid DNA was isolated from E. coli on a small scale by using the alkaline lysis method (4, 45). Large-scale plasmid DNA isolations were performed by using Jetstar columns according to the manufacturer's instructions (Genomed GmbH, Bad Oberhausen, Germany). DNA isolation and transformation in Lactococcus lactis and L. plantarum were performed as described previously (12, 16, 23). Standard procedures were applied for DNA manipulations in E. coli (45). Restriction endonucleases, Taq and Pwo polymerase, T4 DNA ligase, and calf intestinal alkaline phosphatase (CIAP) were used following the recommendations of the manufacturer (Promega, Leiden, The Netherlands and Boehringer, Mannheim, Germany). Primers were purchased from Pharmacia Biotech (Roosendaal, The Netherlands). The sequences of the inserts present in the pNZ7125 derivatives (see below), were amplified by PCR with the primers cre-R2 and BglII-cre (Table 1), followed by high-throughput amplicon purification with Sephadex-G50 and multiscreen HV 96-well plates (Millipore, Amsterdam, The Netherlands). Partial insert sequences were determined with primer cre-R2 or BglII-cre, ca. 100 ng of the purified amplicons and the ABI Prism BigDye terminator cycle sequencing ready reaction kit protocol (Applied Biosystems, Nieuwekerk a/d IJssel, The Netherlands). Sequence reaction products were analyzed by using an ABI Prism 3700 DNA analyzer. The determined insert sequences were assigned to L. plantarum WCFS1 chromosomal loci by using BLAST-N (28).
Construction of L. plantarum NZ7109.
The locus flanked by lp_3503 and lp_3504 encoding a transport protein and a putative integral membrane protein, respectively, was chosen to integrate a loxP-ery-loxP cassette into the chromosome of L. plantarum (Fig. 1). First, genomic DNA of L. plantarum WCFS1 was used as a template to amplify 5′-truncated fragments of lp_3503 and lp_3504 with Pwo polymerase and the primer combinations lp_3503F and lp_3503R or lp_3504F and lp_3504R, respectively. The amplicons were digested with EcoRI-PstI or PstI-HindIII (all restriction sites introduced with the primers) and cloned into similarly digested pUC19 and pUC18 (54), respectively. The identity and sequences of the fragments cloned were verified by automatic double strand sequence analysis. The resulting plasmids were designated pNZ7106 and pNZ7105, respectively. The pNZ7106 insert, harboring lp_3503, was recovered as an EcoRI-PstI fragment and subcloned into similarly digested pNZ7105, yielding vector pNZ7107. Two tandem loxP sites were introduced into pUC19 by synthetic-oligonucleotide linker insertion. The oligonucleotides flox1 and flox2 were annealed and ligated into HindIII-PstI digested pUC19, yielding pUC19lox1. Subsequently, flox3 and flox4 oligonucleotides were annealed and ligated into SacI-EcoRI-digested pUC19lox1, resulting in pUC19lox2. Initially, and for other research purposes, a tetracycline resistance gene cassette (tetR) obtained as an Ecl136II-SmaI fragment of pGhost8 (35) was cloned into the blunted AccI-SmaI-digested pUC19lox2, yielding pNZ7103. The generated loxP-tetR-loxP cassette was subcloned as a PstI fragment from pNZ7103 into similarly digested pNZ7107, resulting in pNZ7108. Finally, the erythromycin resistance gene cassette was obtained as a BamHI-HindIII fragment from pUC18ery (50) and subcloned into NheI-SwaI-digested pNZ7108, after the filling of all sticky ends with Klenow polymerase. The resulting vector was designated pNZ7109 and harbors a loxP-ery-loxP cassette flanked by two regions of the L. plantarum WCFS1 genome, allowing homologous recombination via a double-crossover event.
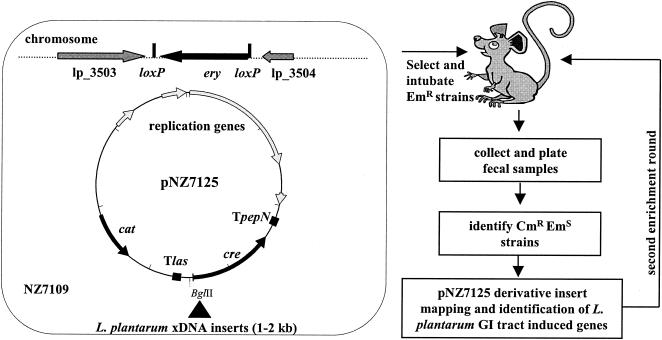
FIG. 1.
Basic principles of R-IVET. L. plantarum strain NZ7109 harbors a chromosomally located loxP-ery-loxP cassette and pNZ7125. Chromosomal loci of the L. plantarum genome (▴) are cloned upstream of cre in pNZ7125 and promoter activities in the resulting library can be trapped by monitoring the erythromycin phenotype, since cre expression will lead to excision of the erythromycin marker from the chromosome by a homologous recombination event between the two loxP sites. The colonies in the library appearing as erythromycin-resistant in the laboratory (no active promoters under laboratory conditions) were administered to mice, and changes in the erythromycin phenotype (promoter activations) were monitored in fecal samples.
The integration vector pNZ7109 was introduced into L. plantarum WCFS1-R and primary single-cross over integrants were selected on MRS plates containing 5 μg of erythromycin/ml plus 10 μg of lincomycin/ml. The integration of pNZ7109 in the anticipated chromosomal locus was verified for one of the colonies obtained, which was subsequently subcultured without antibiotic selection pressure to obtain the desired double-crossover loxP-ery-loxP cassette integrant. After ca. 150 generations, candidate double-crossover integrants were identified among individual colonies by PCR with the primer combinations hrec3503-eryF and hrec3504-eryR. Of the 150 colonies analyzed in this way, 2 appeared to contain the desired chromosomal organization, which was further confirmed by Southern blotting. One of the obtained mutants of strain WCFS1-R was designated NZ7109 and harbors a chromosomally located loxP-ery-loxP cassette integrated in the intergenic region present between lp_3503 and lp_3504.
pNZ7125, pNZ7126, and R-IVET library construction.
To implement R-IVET in L. plantarum the low-copy vector pNZ7125 was constructed. First, pNZ7110 (6) was digested with PstI, and the resulting fragment harboring the resolvase encoding gene cre (1) was cloned into the low-copy variant of similarly digested pJIM2246 (44), resulting in pNZ7124. Genomic DNA of L. lactis MG1363 was used as a template to amplify the terminator of the las operon (33), with the primers lasF2 and lasR. The resulting 0.35-kb amplicon was digested with BamHI and BglII and cloned into BglII-digested pNZ7124. A plasmid containing the las terminator properly oriented upstream of the cre gene to prevent readthrough transcription was designated pNZ7125.
To confirm the functionality of pNZ7125 as a R-IVET vector in combination with R-IVET strain L. plantarum NZ7109, the promoter region of the ldhL1 gene of L. plantarum WCFS1 was cloned in pNZ7125. To this end, the ldhL1 promoter was amplified by using the primers PldhL-F and PldhL-R and chromosomal DNA as a template. The resulting 0.5-kb amplicon was digested with BglII and cloned into similarly digested pNZ7125. The resulting plasmid that contains the cre gene under control of the ldhL1 promoter obtained was designated pNZ7126.
An L. plantarum WCFS1 chromosomal R-IVET library was constructed in pNZ7125. Chromosomal DNA was partially digested with Sau3AI and size fractionated on 1% agarose gels. Fragments of 1 to 2 kb were purified by using Sephaglas Bandprep (Pharmacia Biotech). These purified fragments were cloned into BglII-digested and calf intestinal alkaline phosphatase-dephosphorylated pNZ7125 (Fig. 1). Ligation mixtures were transformed to Lactococcus lactis MG1363 (18), and ca. 50,000 of the obtained colonies were collectively resuspended in GM17. Plasmid DNA was isolated from these cells and introduced into NZ7109. The approximate 37,000 colonies obtained were collectively resuspended in MRS containing 15% glycerol and stored in aliquots at −80°C.
R-IVET animal experiments.
To counterselect against clones in the R-IVET library that harbor pNZ7125 derivatives containing a promoter element that is active under the laboratory conditions applied, the collective library was subcultured for ca. 20 generations in MRS containing 5 μg of chloramphenicol/ml, 30 μg of erythromycin/ml, and 50 μg of rifampin/ml. Subsequent animal experiments were performed in an accredited establishment (no. A59107) according to guidelines N°86/609/CEE of the French government. Seven-week-old female BALB/c mice were purchased from Iffa Credo (St. Germain sur l'Arbresle, France) and had free access to tap water and standard mouse chow during the experiments. After overnight culture, bacterial cells were pelleted by centrifugation and resuspended at 1010 CFU per ml in MRS. The four mice received a 100 μl oral dose of these freshly prepared bacterial suspensions by intragastric administration on 2 consecutive days, and 24 h after the last administration individual fecal samples were collected and resuspended in MRS medium. After extensive homogenization, the complete R-IVET library was recovered from the fecal samples by plating appropriate dilutions of the suspensions on MRS plates containing 5 μg of chloramphenicol/ml and 50 μg of rifampin/ml. After 72 h full-grown colonies were replica plated onto plates containing 5 μg of chloramphenicol/ml and 50 μg of rifampin/ml, with or without 30 μg of erythromycin/ml. Another 24 h later, the plates were compared, leading to the identification of cells displaying an erythromycin-sensitive phenotype. The inserts present in the corresponding pNZ7125 derivatives harbored by these resolved clones were amplified by PCR, and from the resulting amplicons the DNA sequence was analyzed.
RESULTS
Implementation and functionality of R-IVET in L. plantarum.
Previously, R-IVET was used exclusively for the identification of genes induced in pathogenic microorganisms during infection of host tissues (for reviews, see references 3 and 36). Since it has been reported that the Cre resolvase (1), encoded by the cre gene, could be functionally implemented in the LAB Lactococcus lactis (8), the suitability of this resolvase for a R-IVET approach in L. plantarum was evaluated. Therefore, an L. plantarum WCFS1 derivative strain harboring a chromosomally located loxP-ery-loxP cassette was constructed and designated NZ7109 (Fig. 1). The growth rate of NZ7109 did not differ from that observed for wild-type L. plantarum WCFS1 and, after 50 generations of growth without antibiotic selection pressure, replica-plating revealed that all NZ7109 cells were erythromycin resistant. Moreover, PCR analysis revealed that the chromosomal loxP-ery-loxP insertion could be amplified from all colonies (data not shown), indicating that the chromosomal insertion is stable. Both pNZ7125, a low-copy vector encoding a promoterless copy of cre, and a derivative containing the cre gene under control of the L. plantarum WCFS1 ldhL1 promoter (pNZ7126) were introduced in NZ7109 cells and plated on MRS medium with chloramphenicol. Replica plating of the transformants revealed that all NZ7109 colonies harboring pNZ7125 were erythromycin resistant, whereas all NZ7109 colonies harboring pNZ7126 were erythromycin sensitive. In addition, PCR analysis confirmed that the loxP-ery-loxP locus could only be amplified from erythromycin-resistant colonies (data not shown). These results establish the functional implementation of the cre-loxP resolution system in L. plantarum WCFS1 and confirm the suitability of pNZ7125 as an R-IVET vector.
Construction of an L. plantarum R-IVET library in pNZ7125.
A genomic library of L. plantarum WCFS1 was constructed in pNZ7125, by using Lactococcus lactis MG1363 as an intermediate cloning host. Approximately 37,000 colonies were obtained in L. plantarum NZ7109, and the quality of this library was assessed in several ways. The pNZ7125 derivatives of 100 randomly picked colonies were used as a template for insert amplification by PCR, demonstrating that >95% of the investigated clones contained an insert with an estimated average size of 1.3 kb (data not shown). To assess insert redundancy, all amplicons were digested with Sau3AI, and the resulting fragments were separated by 2% agarose gel electrophoresis. No common restriction profiles were detected, indicating that redundancy in the R-IVET library is low. Moreover, 28 of these amplicons were used for partial sequence analysis, which revealed no apparent over- or underrepresentation of a specific region of the L. plantarum genome. These results support the randomness of the library and genome coverage was calculated to be ca. 98% (data not shown). The R-IVET library was replica plated to plates with or without erythromycin, which indicated that 10% of the R-IVET clones contain a pNZ7125 derivative harboring a properly oriented promoter element that is active under the laboratory conditions applied and drives cre expression at a sufficient level to excise the loxP-ery-loxP cassette from the chromosome of NZ7109.
R-IVET screen in mice.
To counterselect against clones displaying cre expression under laboratory conditions, the R-IVET library was cultured for 20 generations in the presence of erythromycin. Subsequently, full-grown cultures were used for gastric administration to four BALB/c mice. After recovery from fecal samples 6,000 of the R-IVET clones were analyzed for their erythromycin phenotype by replica plating, revealing 198 (3.3%) clones that displayed an erythromycin-sensitive phenotype. The partial sequence of 132 of the chromosomal inserts present in the pNZ7125 derivatives originating from these clones was determined and corresponded to 119 unique loci of the L. plantarum genome, since 1 locus was found three times, while 11 loci were found twice. According to the current genome annotation database of L. plantarum WCFS1 (28), these loci harbor 72 unique genes, and their upstream sequences in the proper orientation to explain the observed induction of cre expression (Table 2). Notably, nine loci contained more than one putative 5′ end of an annotated open reading frame (ORF) and their potential promoter. Remarkably, two independent Sau3AI clones corresponding to lp_0291 and its upstream sequence were found that differ in size (1.0 and 1.5 kb). Hence, the in vivo induction of this gene was independently confirmed twice during the R-IVET procedure. The identified genes appeared to be randomly located within the L. plantarum genome. Moreover, the genes appeared to be randomly distributed among the main functional categories. The ORFs identified in this R-IVET screen were functionally grouped in genes involved in nutrient acquisition and synthesis (18 ORFs), stress response and adaptation (4 ORFs), extracellular proteins (4 ORFs), regulation (3 ORFs), and others (19 ORFs). The remaining 24 genes encoded (conserved) hypothetical proteins of unknown function (Table 2).
TABLE 2.
L. plantarum genes that are induced during passage through the mouse GI tract as compared to laboratory conditions
| Class | ORF (gene)a | Product or function | Reference(s) |
|---|---|---|---|
| Nutrient acquisition and synthesis | |||
| Sugar | lp_0185a (ptsIBCA) | Sucrose PTS EIIBCA | 27b |
| lp_1164 (pts14C) | Cellobiose PTS, EIIC | 17, 27 | |
| lp_2647c (pts19A) | N-Acetylglucosamine/galactosamine PTS, EIIA | ||
| lp_3473 (ram2) | Alpha-l-rhamnosidase | ||
| lp_3522c (pts32BC) | Sucrose PTS, EIIBC | 27b | |
| lp_3526a (pbg10) | 6-Phospho-beta-glucosidase | ||
| lp_36182,c (pts37A) | Sorbitol PTS EIIA | 22 | |
| lp_36593,d (rbsD) | Ribose transport protein | 11, 31 | |
| lp_36603,d (rbsK3) | Ribokinase | 11, 31 | |
| Nonsugar | lp_0017c (proA) | Glutamate-5-semialdehyde dehydrogenase | 25, 47 |
| lp_0228e (pepD1) | Dipeptidase | ||
| lp_0696 | Cytosine/adenosine deaminase | 43 | |
| lp_0775 (argG) | Argininosuccinate synthase | 7 | |
| lp_0854a,c (birA2) | Biotin (acetyl-coenzyme A-carboxylase) ligase and biotin operon repressor | 10, 26 | |
| lp_1058 (adk) | Adenylate kinase | ||
| lp_1319d (rsuA) | Pseudouridylate synthase | ||
| lp_16027 | Geranyltranstransferase/famesyl di-P-transferase | ||
| lp_2031 (ribC2) | Bifunctional protein: riboflavin kinase and FMN adenylyl- transferase | 34 | |
| Stress | lp_1019 (clpC) | ATP-dependent Clp protease | 24, 32 |
| lp_30554,c (copA) | Copper transporting ATPase | 21, 36 | |
| lp_3288c | Cation efflux protein | ||
| lp_33035 | Putative multidrug transport protein | 25 | |
| Extracellular | |||
| LPQTNE containing | lp_0800 | Cell surface protein precursor | |
| lp_2940c | Cell surface protein precursor | ||
| Other | lp_0141e | Extracellular protein | |
| lp_1403a,d | Cell surface protein | ||
| Regulators | lp_3176c (pkn2) | Serine/threonine protein kinase | |
| lp_3514c (bglG4) | Transcription antitermination | 38 | |
| lp_3646a,d | Transcription regulator | ||
| Other | lp_0237 | Integral membrane protein | |
| lp_0299c | ABC transporter, ATP-binding protein | ||
| lp_0305d (gcsH1) | Glycine cleavage system, H protein | ||
| lp_03931,d (thgA1) | Galactoside O-acetyltransferase | ||
| lp_03941,d | Transport protein | ||
| lp_0419a (plnI) | Immunity protein | ||
| lp_0489 (tktI-N) | Transketolase | ||
| lp_0698a,d (dnaX) | DNA-directed DNA polymerase III, gamma/tau subunit | 27 | |
| lp_07406 (prfB-N) | Peptide chain release factor 2, N-terminal fragment | ||
| lp_07416 (prfB-C) | Peptide chain release factor 2, C-terminal fragment | ||
| lp_16037 (ispA) | Hemolysin homologue | ||
| lp_1847 (codV) | Integrase/recombinase | ||
| lp_3051 (dhaT) | 1,3-Propanediol dehydrogenase | 39 | |
| lp_3082d | Transport protein | ||
| lp_3281d | Transport protein | ||
| lp_3500 | Short-chain dehydrogenase/oxidoreductase | 39 | |
| lp_3505 (est2) | Acetylesterase | ||
| lp_36172,c (tal3) | Transaldolase | ||
| lp_3662c (adhE) | Bifunctional protein: alcohol and acetaldehyde dehydrogenase | 40 | |
| Hypothetical | |||
| Conserved | lp_0026c, lp_0139, lp_0190d, lp_0291a,c, lp_0476∞, lp_0477∞, lp_0907d, lp_1969d, lp_2337c, lp_25079,d, lp_25089,d, lp_2713, lp_2718, lp_30574,c, lp_30584,c and lp_3312c | 52 (for lp_2718, see text) | |
| Not conserved | lp_0118a,d, lp_0292, lp_1788, lp_1872, lp_2112c, lp_2584, lp_3246c, lp_33055 |
Genes with the same superscript number originate from one R-IVET clone. Genes denoted by a superscript “a” were identified twice during the first-round R-IVET screen. After the second-round passage and subsequent sequence analysis, some genes were identified exclusively in the erythromycin sensitive (superscript c) or resistant (superscript e) group. Genes indicated by a superscript “d” were identified in both the erythromycin-resistant and the erythromycin-sensitive groups.
In the original reference, 13 in vivo-induced loci were identified in Streptococcus gordonii. However, these loci were only partially sequenced, and for six loci this was not sufficient to identify a promoter element. At present, with more detailed sequence analysis and the partially available genome sequence, these loci have been assigned a promoter element (Lin Tao, unpublished data).
To verify the primary R-IVET results, the 132 partially sequenced clones were divided into four groups of 33 clones that were used for collective plasmid DNA isolation. These mixtures of pNZ7125 derivatives were reintroduced into L. plantarum NZ7109, and the resulting colonies were collectively stored. Replica plating of the transformants revealed that all tested colonies displayed an erythromycin-resistant phenotype, confirming the absence of cre expression in these clones under laboratory conditions. These four groups were separately subjected to a second-round passage through eight mice (two animals per group) using the same procedure applied in the first-round passage, including recovery from fecal samples and analysis of the erythromycin phenotype. This second-round passage revealed a dramatic increase in the percentage of erythromycin-sensitive colonies that was recovered from the fecal samples of all individual mice (Table 3), which is clearly apparent from the average of 38.1% erythromycin-sensitive clones compared to 3.3% in the experimentally identical first-round passage.
TABLE 3.
A second-round passage of R-IVET positives through mice results in an increased percentage of erythromycin-sensitive NZ7109 colonies compared to the first-round passage (3.3%)
| Mouse | Promoter group | Erys (%)a |
|---|---|---|
| 1 | 1 | 58.0 |
| 2 | 1 | 71.7 |
| 3 | 2 | 52.9 |
| 4 | 2 | 28.8 |
| 5 | 3 | 34.2 |
| 6 | 3 | 33.3 |
| 7 | 4 | 31.5 |
| 8 | 4 | 37.5 |
The average of the erythromycin sensitivity (Erys) values for mice 1 to 8 was 38.1%.
To obtain more detailed insight in the results of this second-round screening procedure, an exemplary group of the recovered colonies was selected. For each of the four groups of clones, both erythromycin-sensitive (64 colonies per group) and erythromycin-resistant (32 colonies per group) clones were randomly picked from the library-recovery plates and subjected to clone identification by partial sequence analysis of the insert present in the pNZ7125 derivative harbored by these clones. This analysis revealed that 84 of the original 132 R-IVET-positive clones were represented within this random sample of second-round recovery clones. Of these 84 clones, 37 were found to only be erythromycin sensitive (44%), whereas 4 were identified exclusively in the erythromycin-resistant group (5%). Notably, the residual 43 clones were found both as erythromycin sensitive and resistant (51%). The latter finding probably reflects the single-cell response selection that is an intrinsic characteristic of an R-IVET screen, which implies that the specific condition leading to promoter activation for a specific R-IVET-library clone is not encountered by each individual cell representing that clone. By analogy, for only 4 of the 84 clones in this second-round analysis the possibility that they represent false-positives obtained in the first round of R-IVET selection could not be excluded. On the other hand, it also cannot be excluded that the in vivo regulation of these clones has remained undetected in this second round due to the relatively small size of the random sample analyzed and that more extensive analyses of the clones recovered in the second round would validate their in vivo induction. The results from this second-round R-IVET analysis have been incorporated in Table 2 for individual genes. Overall, these results both confirm the in vivo induction of the vast majority of the clones and corresponding genes selected in the first round and exclude the possibility that a relatively large proportion of the clones identified represent false positives.
DISCUSSION
Three major strategies have been utilized for the in vivo identification of promoters. The major disadvantages of SCOTS are the instability of bacterial mRNA for the construction of cDNA libraries, the low abundance of mRNA from transiently expressed genes, and the difficulty in isolation of sufficient high-quality mRNA from small populations of bacteria in vivo (36). Using STM only limited numbers of mutants can be screened per animal model. Moreover, mutants that are slow-growing, nonviable, contain mutations in genes encoding redundant functions or that can be complemented in a mixed population may be underrepresented. The disadvantages mentioned above for SCOTS and STM do not apply to IVET and R-IVET strategies (36). For reasons mentioned in the introduction, we preferred a R-IVET screening in L. plantarum. To our knowledge, this is the first R-IVET approach in a food-grade bacterium, which resulted in the identification of 72 genes that are induced in situ in L. plantarum WCFS1 during passage through the mouse GI tract. The identified genes appeared to be randomly distributed over the chromosome. Moreover, genes from many functional classes were identified and grouped into six functional domains (Table 2).
Nine of the genes identified by using R-IVET are involved in sugar transport and utilization (Table 2), including five (components of) phosphotransferase systems (PTS), specific for N-acetylglucosamine, sorbitol, sucrose (twice), and cellobiose, a ribose permease and a ribose kinase, and two di- and polysaccharide hydrolyzing enzymes. A diverse carbohydrate potential has been associated with several gram-positive microbes inhabiting the GI tract, including L. plantarum (28), Listeria innocua and Listeria monocytogenes (19), and Bifidobacterium longum (46). The finding that several of these functions are induced in situ in the GI tract supports their importance for survival and persistence under GI tract conditions. Moreover, genes involved in the metabolism of the same sugars have been identified as being important for pathogenesis in various bacteria (11, 17, 22, 27, 31; Lin Tao, unpublished data). Remarkably, the IIC transport component of the cellobiose PTS system found (lp_1164) is not located in a typical PTS-operon structure. In Listeria monocytogenes, similar “orphan” cellobiose-PTS-IIC components have been shown to play a role in host-specific signaling, leading to modulation of virulence gene expression (30), suggesting host-factor mediated gene regulation in bacteria, possibly including L. plantarum.
Nine genes were identified that are involved in the acquisition and biosynthesis of nonsugar compounds, including amino acids, nucleotides, cofactors, and vitamins (Table 2). These results suggest that limiting amounts of these compounds are readily available in the GI tract, leading to activation or derepression of these L. plantarum genes. Accordingly, in vitro studies in different bacteria confirm the induction of several of these metabolic pathways under limiting conditions of the corresponding end products (10, 34, 43). Moreover, in vivo approaches have demonstrated that genes involved in arginine, biotin, and proline metabolism in Vibrio cholerae (7), E. coli (26) and Helicobacter pylori (25), respectively, are induced during mouse infection. In one of the closest relatives of L. plantarum, Listeria monocytogenes, proline metabolism is induced under high-osmolarity conditions (47). Such conditions could potentially be found in the colon and suggest differential colonic expression of lp_0017 (Table 2). In analogy, other experiments in our laboratory have identified lp_3473, encoding an α-rhamnosidase, as induced by high osmolarity (unpublished observation), suggesting its colonic induction.
The copper-transporting ATPase identified here (lp_3055, Table 2) could be involved in copper acquisition. Alternatively, this transporter could act as an exporter, thereby preventing accumulation of copper in the cytoplasm. Arbitrarily, this gene was categorized as a stress-related protein involved in copper detoxification. Three other genes were categorized as stress related, namely, clpC, a multidrug transporter, and a cation efflux protein (Table 2). The fact that three possible exporters were identified suggests that the efficient transport of toxic compounds is important for the GI tract persistence of L. plantarum. Genes important in the transport of metals have been identified in many IVET screens (36). Moreover, in H. pylori an STM strategy revealed two genes encoding multidrug transporters to be essential for gastric colonization of mice (25). In several gram-positive microorganisms, clp genes were demonstrated to be involved in stress responses (13). In Streptococcus mutans the clpC operon is induced at low pH (32), which could suggest that the expression of this gene might be induced in L. plantarum during passage through the mouse stomach. Moreover, mutations in the ctsR gene, encoding the clpC regulator, in Listeria monocytogenes displayed reduced survival during the initial stages of murine infection in mice (24).
Four genes encoding extracellular proteins, including two proteins (lp_0800 and lp_2940) that contain an LPXTG-like motif (LPQTNE) involved in anchoring them to the bacterial cell wall (41), were identified in the R-IVET screen (Table 2). lp_0141 contains a high number of positive charges that could be involved in the interaction of the encoded protein with the cell wall. No putative binding domains were found in the protein encoded by lp_1403, suggesting secretion of this protein. The bacterial surface is the primary site of interaction with the host, and numerous surface-exposed adhesion factors have been described (29). Therefore, the surface-anchored proteins identified here might represent factors that mediate interaction with host cells in the GI tract or with components excreted in the GI tract lumen of the host. lp_0800 encodes a protein that is extremely rich in serine and threonine. For a serine-rich surface protein encoded by Streptococcus pneumoniae it has been suggested that the serine residues might be glycosylated by glycosyltransferases that are encoded by genes flanking the surface protein encoding gene (49). These glycosylated serine residues could resemble mucin-like structures that coat the bacterial surface or interact with host cell mucins (49). Although no glycosyltransferase encoding genes appear to be genetically linked to lp_0800, a similar role might be fulfilled in L. plantarum by the lp_0800 encoded protein.
Three regulators of different families were found to be induced in vivo (Table 2). The bgl operon in L. plantarum was previously shown to be downregulated in the presence of glucose (38). Therefore, the BglG transcription antiterminator (lp_3514) might be involved in the regulation of the response to the different sugars L. plantarum ferments during passage of the GI tract. Remarkably, among the best homologues of lp_3514 in the Listeria monocytogenes genome is the bvrA gene (33% identity), which encodes a BglG-family antiterminator involved in the regulation of virulence gene expression (5).
Nineteen of the in vivo-induced genes that were identified here are involved in diverse pathways, including DNA and energy metabolism, protein fate and synthesis, and fermentation (Table 2). Several genes in these pathways have previously been described as being important for pathogenesis in various bacteria, including the 1,3-propanediol regulator and a short-chain dehydrogenase of Klebsiella pneumonia (39), a bifunctional protein possessing alcohol and acetaldehyde dehydrogenase activity in E. coli K1 (40), and a DNA polymerase in Streptococcus gordonii (27). Another interesting observation is the apparent induction of PlnI, a plantaricin immunity protein (14), suggesting that the production of this bacteriocin is important for L. plantarum in the highly competitive environment in the GI tract.
Finally, 24 hypothetical proteins (16 conserved and 8 unique) apparently play a role during the passage of L. plantarum through the GI tract (Table 2). Strikingly, the protein encoded by lp_2718 is a homologue (32% identity) of the only conserved hypothetical protein that was identified with IVET in L. reuteri (52; Christian Hertel, unpublished data). Although the other putative genes were subjected to extensive analysis by using the available (R-)IVET literature and BLAST searches, no significant homologies could be found between these L. plantarum genes encoding hypothetical proteins and in vivo-induced genes found in other species. One reason for this could be that a large amount of the nucleotide sequence data obtained for the hypothetical genes found in in vivo screens in pathogens is not publicly available. The role of these hypothetical genes in L. plantarum GI tract persistence remains to be determined.
Overall, a striking number of parallels can be drawn between the pathogenic and nonpathogenic in vivo response, strongly suggesting that survival rather than virulence is the explanation for the importance of these genes during host residence. This suggestion is further corroborated by the fact that the gene encoding the peptide methionine sulfoxide reductase has previously been identified by using IVET in the food-associated microbe L. reuteri during passage through the GI tract (52) and IVET in the non-food-associated Streptococcus gordonii during endocarditis (27). The similarities found for the L. plantarum R-IVET screen presented here are most prominent with in vivo screens that are performed in the same host (mice) with pathogens that infect the same host organ (GI tract), suggesting that organ- and host-specific factors play a key role in the determination of the microbial response. In addition, a number of functions that are induced in vivo in L. plantarum during passage of the mouse GI tract, which (thus far) have not been identified in similar in vivo studies in other bacteria, might contribute to specific interactions between this bacterium and factors encountered in this niche. Moreover, parallels between the in vivo study presented here and in vitro studies performed in the same bacterium (osmolarity induction [see above]) or closely related species (osmolarity induction or low pH [see above]) might hint at a spatial differentiation of L. plantarum gene expression during passage of the mouse GI tract, i.e., specific induction in the stomach, small intestine, or colon. The recent observation in our laboratory that the expression of lp_0237 and lp_0775 is induced by bile, suggesting in vivo induction in the duodenum, is interesting in this respect since this is the site of bile release by the host. In conclusion, the R-IVET screen performed in L. plantarum is an important step in understanding the behavior of this food-associated microbe related to stress, persistence, and host-microbe and microbe-microbe interactions in the complex GI tract environment.
Acknowledgments
This study was supported in part by the EU project LABDEL (EU-QLRT-2000-00340).
We thank Iris van Swam for construction of pUC19lox1, pUC19lox2, and pNZ7103 and Thamara Hesselink, René Klein Lankhorst, and Michiel Wels for sequence and BLAST analyses. We gratefully acknowledge Catherine Daniel and Sally Hoffer for fruitful discussions and Lin Tao, Cormac Gahan, and Christian Hertel for sharing unpublished sequence information on their IVET clones.
REFERENCES
- 1.Abremski, K., R. Hoess, and N. Sternberg. 1983. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32:1301-1311. [DOI] [PubMed] [Google Scholar]
- 2.Ahrne, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. [DOI] [PubMed] [Google Scholar]
- 3.Angelichio, M. J., and A. Camilli. 2002. In vivo expression technology. Infect. Immun. 70:6518-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bron, P. A., M. G. Benchimol, J. Lambert, E. Palumbo, M. Deghorain, J. Delcour, W. M. De Vos, M. Kleerebezem, and P. Hols. 2002. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 68:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campo, N., M. L. Daveran-Mingot, K. Leenhouts, P. Ritzenthaler, and P. Le Bourgeois. 2002. Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl. Environ. Microbiol. 68:2359-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 10.Cronan, J. E., Jr. 1988. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 263:10332-10336. [PubMed] [Google Scholar]
- 11.Daigle, F., J. E. Graham, and R. Curtiss III. 2001. Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS). Mol. Microbiol. 41:1211-1222. [DOI] [PubMed] [Google Scholar]
- 12.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 13.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 14.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gahan, C. G., and C. Hill. 2000. The use of listeriolysin to identify in vivo induced genes in the gram-positive intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 36:498-507. [DOI] [PubMed] [Google Scholar]
- 18.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 20.Grangette, C., H. Muller-Alouf, M. Geoffroy, D. Goudercourt, M. Turneer, and A. Mercenier. 2002. Protection against tetanus toxin after intragastric administration of two recombinant lactic acid bacteria: impact of strain viability and in vivo persistence. Vaccine 20:3304-3309. [DOI] [PubMed] [Google Scholar]
- 21.Heithoff, D. M., C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel, and M. J. Mahan. 1997. Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. USA 94:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, M. L., D. J. Boucher, J. D. Boyce, and B. Adler. 2001. In vivo-expressed genes of Pasteurella multocida. Infect. Immun. 69:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 24.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 25.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan, M. A., and R. E. Isaacson. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 70:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiliç, A. O., M. C. Herzberg, M. W. Meyer, X. Zhao, and L. Tao. 1999. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid 42:67-72. [DOI] [PubMed] [Google Scholar]
- 28.Kleerebezem, M., J. Boekhorst, R. Van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. De Vries, B. Ursing, W. M. De Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 30.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 31.Lai, Y. C., H. L. Peng, and H. Y. Chang. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llanos, R. M., C. J. Harris, A. J. Hillier, and B. E. Davidson. 1993. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J. Bacteriol. 175:2541-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack, M., A. P. van Loon, and H. P. Hohmann. 1998. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 180:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahan, M. J., D. M. Heithoff, R. L. Sinsheimer, and D. A. Low. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139-164. [DOI] [PubMed] [Google Scholar]
- 37.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 38.Marasco, R., I. Salatiello, M. De Felice, and M. Sacco. 2000. A physical and functional analysis of the newly identified bglGPT operon of Lactobacillus plantarum. FEMS Microbiol. Lett. 186:269-273. [DOI] [PubMed] [Google Scholar]
- 39.Maroncle, N., D. Balestrino, C. Rich, and C. Forestier. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 70:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 41.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 42.Mecsas, J. 2002. Use of signature-tagged mutagenesis in pathogenesis studies. Curr. Opin. Microbiol. 5:33-37. [DOI] [PubMed] [Google Scholar]
- 43.Muse, W. B., C. J. Rosario, and R. A. Bender. 2003. Nitrogen regulation of the codBA (cytosine deaminase) operon from Escherichia coli by the nitrogen assimilation control protein, NAC. J. Bacteriol. 185:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renault, P., G. Corthier, N. Goupil, C. Delorme, and S. D. Ehrlich. 1996. Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene 183:175-182. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sleator, R. D., C. G. Gahan, and C. Hill. 2001. Identification and disruption of the proBA locus in Listeria monocytogenes: role of proline biosynthesis in salt tolerance and murine infection. Appl. Environ. Microbiol. 67:2571-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 49.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 50.van Kranenburg, R., J. D. Marugg, S. van, I. I., N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 51.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]
- 52.Walter, J., N. C. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West, N. P., P. J. Sansonetti, G. Frankel, and C. M. Tang. 2003. Finding your niche: what has been learned from STM studies on GI colonization. Trends Microbiol. 11:338-344. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved m13 phage cloning vectors and host strains; nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]