Abstract
We report here that the bzd genes for anaerobic benzoate degradation in Azoarcus sp. strain CIB are organized as two transcriptional units, i.e., a benzoate-inducible catabolic operon, bzdNOPQMSTUVWXYZA, and a gene, bzdR, encoding a putative transcriptional regulator. The last gene of the catabolic operon, bzdA, has been expressed in Escherichia coli and encodes the benzoate-coenzyme A (CoA) ligase that catalyzes the first step in the benzoate degradation pathway. The BzdA enzyme is able to activate a wider range of aromatic compounds than that reported for other previously characterized benzoate-CoA ligases. The reduction of benzoyl-CoA to a nonaromatic cyclic intermediate is carried out by a benzoyl-CoA reductase (bzdNOPQ gene products) detected in Azoarcus sp. strain CIB extracts. The bzdW, bzdX, and bzdY gene products show significant similarity to the hydratase, dehydrogenase, and ring-cleavage hydrolase that act sequentially on the product of the benzoyl-CoA reductase in the benzoate catabolic pathway of Thauera aromatica. Benzoate-CoA ligase assays and transcriptional analyses based on lacZ-reporter fusions revealed that benzoate degradation in Azoarcus sp. strain CIB is subject to carbon catabolite repression by some organic acids, indicating the existence of a physiological control that connects the expression of the bzd genes to the metabolic status of the cell.
After carbohydrates, aromatic compounds are the most widely distributed class of organic compounds and hence constitute a common carbon source for many microorganisms (32). The thermodynamic stability of the benzene ring increases its persistence in the environment; this fact, together with the massive release of aromatics into the field, has caused many such compounds, e.g., benzene, toluene, xylenes, ethylbenzene, phenol, etc., to be major environmental pollutants. Since many ecosystems are often anoxic, e.g., aquifers, aquatic sediments, and submerged soils, the anaerobic catabolism of aromatic compounds by microorganisms becomes crucial in biogeochemical cycles and in the sustainable development of the biosphere (44, 58). In contrast to the aerobic degradation of aromatic compounds, which has been extensively studied in many microorganisms, the anaerobic processes leading to mineralization of the aromatic ring have been studied in some detail in only a few microorganisms, i.e., in the denitrifying bacteria Thauera aromatica and Azoarcus evansii, in the photosynthetic bacterium Rhodopseudomonas palustris, and to a lesser extent in some Fe(III)-reducing, sulfate-reducing, and fermenting bacteria (30, 31).
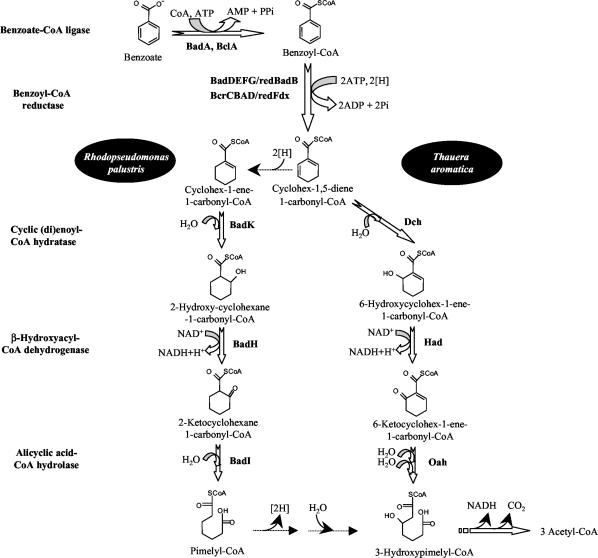
Benzoate has been used as a model compound for the study of the anaerobic catabolism of aromatic compounds (30, 31). Benzoate degradation starts with its activation to benzoyl-coenzymeA (CoA) via an ATP-dependent benzoate-CoA ligase (1, 28, 54) (Fig. 1). The subsequent ring reduction to a nonaromatic compound is an oxygen-sensitive process that is carried out by a four-subunit benzoyl-CoA reductase which requires the low-potential electron donor ferredoxin and ATP hydrolysis. Whereas benzoyl-CoA is reduced to a cyclohexadienecarbonyl-CoA intermediate in T. aromatica, a probable four-electron reduction to give cyclohex-1-ene-carbonyl-CoA was reported for R. palustris (Fig. 1) (10). Next in the pathway, two slightly different β-oxidation-like sets involving the introduction of a hydroxyl group, a dehydrogenation reaction, and hydrolytic ring fission lead to the formation of pimelyl-CoA in R. palustris and 3-hydroxypimelyl-CoA in T. aromatica (Fig. 1). Further degradation of the dicarboxylic acid via β-oxidation and decarboxylation steps yields three molecules of acetyl-CoA which are channeled into the central metabolism of the cell (Fig. 1) (31).
FIG. 1.
Scheme of anaerobic benzoate degradation pathway in R. palustris and T. aromatica. The enzymes and pathway intermediates are indicated. Bad proteins are from R. palustris; the Bcl, Bcr, Fdx, Dch, Had, and Oah proteins are from T. aromatica. Modified from reference 31.
Benzoyl-CoA reductase activity has been detected in cell extracts of T. aromatica (8, 38), R. palustris (38), and A. evansii (23). In T. aromatica, the reductase is an αβγδ heterotetramer that contains three cysteine-ligated [4Fe-4S]1+/2+ clusters; a ferredoxin containing two [4Fe-4S]1+/2+ clusters serves as the electron donor (10). A two-subunit ferredoxin-reducing enzyme, the 2-oxoglutarate:ferredoxin oxidoreductase that replaces the 2-oxoglutarate dehydrogenase complex of the Krebs cycle during anaerobic growth on aromatic compounds, directly regenerates reduced ferredoxin in T. aromatica (10, 22). Recently, a three-subunit NADP+-dependent 2-oxoglutarate:ferredoxin oxidoreductase was shown to regenerate, in combination with an inducible NADPH:ferredoxin oxidoreductase, reduced ferredoxin in A. evansii (23).
The genes responsible for the anaerobic catabolism of benzoate have been described for R. palustris, which is a member of the α-Proteobacteria, and T. aromatica, a member of the β-Proteobacteria (14, 25, 54). An initial analysis of the genes involved in the anaerobic catabolism of benzoate in the β-proteobacterium A. evansii was also reported (31). In both R. palustris and T. aromatica, the genes encoding the anaerobic pathway for the catabolism of benzoate are arranged as a single cluster, with the only exception being the gene encoding the benzoate-CoA ligase that is located within the cluster predicted for aerobic benzoate degradation in T. aromatica (54). In this work, we provide experimental evidence for the first time of the existence of a set of genes (bzd cluster) involved in anaerobic benzoate degradation in a strain of the genus Azoarcus, Azoarcus sp. strain CIB, as well as for its transcriptional organization. Transcriptional analyses and enzymatic assays revealed that benzoate degradation in Azoarcus sp. strain CIB is subject to carbon catabolite repression by some organic acids, indicating the existence of a physiological control that connects expression of the bzd genes to the metabolic status of the cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Azoarcus sp. strain CIB was deposited in the Spanish Type Culture Collection (CECT #5669). The plasmids used for this study are indicated in Table 1, and the oligonucleotides employed for PCR amplification of the cloned fragments are summarized in Table 2.
TABLE 1.
Plasmids used for this work
| Plasmid | Relevant features | Reference or source |
|---|---|---|
| pUC18/19 | AproriColE1 lacZα | 51 |
| pK18mob | KmroriColE1 Mob+lacZα, used for directed insertional disruption | 53 |
| pSJ3 | AproriColE1 ′lacZ promoter probe vector, lacZ fusion flanked by NotI sites | 27 |
| pUCBZDA | Apr, pUC18 containing the bzdA gene under control of the Plac promoter | This work |
| pECOR2 | Apr, pUC19 containing a 6.6-kb EcoRI DNA fragment from λBzd1 | This work |
| pECOR7 | Apr, pUC19 containing a 7.1-kb EcoRI DNA fragment from λBzd1 | This work |
| pECOR8 | Apr, pUC19 harboring a 5.4-kb EcoRI DNA fragment from Azoarcus sp. strain CIB that contains the right end of the bzd cluster | This work |
| pSJ3PR | Apr, 675-bp KpnI/XbaI bzdR fragment (PR promoter) cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pSJ3PN | Apr, 598-bp KpnI/XbaI bzdRN fragment (PN promoter) cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pSJ3QM | Apr, 444-bp KpnI/XbaI bzdQ-bzdM intergenic fragment cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pSJ3VW | Apr, 313-bp KpnI/XbaI bzdV-bzdW intergenic fragment cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pSJ3WX | Apr, 602-bp KpnI/XbaI bzdW-bzdX intergenic fragment cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pSJ3YZ | Apr, 277-bp KpnI/XbaI bzdY-bzdZ intergenic fragment cloned into KpnI/XbaI double-digested pSJ3 vector | This work |
| pK18mobPNlacZ | Kmr, EcoRI/HindIII PN::lacZ fragment from pSJ3PN cloned into EcoRI/HindIII double-digested pK18mob vector | This work |
| pK18mobbzdN | Kmr, 523-bp EcoRI bzdN internal fragment cloned into EcoRI-digested pK18mob | This work |
| pK18mobbzdY | Kmr, 366-bp EcoRI bzdY internal fragment cloned into EcoRI-digested pK18mob | This work |
TABLE 2.
List of primers used in this work for PCR and RT-PCR reactions
| Primer no. | Primer name | Sequence (5′-3′)a | Amplified fragmentb |
|---|---|---|---|
| 1 | NIN35 | GGAGTCGGTGGTCGCAAGC | 313-bp bzdR internal fragment |
| 2 | REG | GTGCCAGGACTTCGAGGATGTC | |
| 3 | BzdN5′ | CGAATTCTGATCGAGGCCGGCATGCTG (EcoRI) | 523-bp bzdN internal fragment cloned in pK18mobbzdN |
| 4 | BzdN3′ | CGAATTCAGCATCTCGTTGTGCTC (EcoRI) | |
| 5 | BzdP5′ | CCAAGGAGCCGAGCTTCGC | 399-bp bzdP internal fragment |
| 6 | BzdP3′ | CCGAATTCCTTGAGCGACAGC | |
| 7 | δBcr5′ | GCTACGGCCGGGTGAACGTGC | 487-bp bzdQ internal fragment, bzdQ probe |
| 8 | δBcr3′ | CTTCACGACGCCCGGGTTCTTC | |
| 9 | BzdQM5′ | GCGGCGCTGTTCGGCTAC | 217-bp bzdQM intergenic region |
| 10 | BzdQM3′ | GATGCTGATGTAATACTCGCCGG | |
| 11 | EcoR8 | GGCGTCGATCCGCGCTTCC | 503-bp bzdTU intergenic region |
| 12 | BzdU3′ | CATCGGGAGGTAGTTGTCGCG | |
| 13 | EcoR11 | CTGATCGAATCCGGCTGTAC | 523-bp bzdV internal fragment |
| 14 | BzdV4-11 | GGAACGCGGTAGGACGGG | |
| 15 | BzdVW5′ | GGAACAGCTCGAACGTGGCGG | 515-bp bzdVW intergenic region |
| 16 | BzdVW3′ | GGGCGACGTTCATCTCCTCCATC | |
| 17 | BzdWXII5 | GCACCCGCAAGACGATCCG | 544-bp bzdWX intergenic region |
| 18 | BzdWXII3 | GCCGATCACTGTGCCGGAG | |
| 19 | BzdY5′ | CGAATTCAGTACAACTCCTACACCACC (EcoRI) | 366-bp bzdY internal fragment cloned in pK18mobbzdY, bzdY probe |
| 20 | BzdY3′ | GGAATTCTGCCGTGCTTCGGGCCGGC (EcoRI) | |
| 21 | YZ5′ | CGGCAAGGACGTCATCGACTTC | 322-bp bzdYZ intergenic region |
| 22 | YZ3′ | CCGCCGCGCCGATGCCCTG | |
| 23 | BzdZ-15′ | GTTGAAGGACAGGGTCGCAATCG | 447-bp bzdZ internal fragment |
| 24 | 8.3.4. | CTGCCCGATCGTGCCGAGC | |
| 25 | BzdA-25′ | GGGCACGCGCGGTGGACATC | 577-bp bzdA internal fragment |
| 26 | 8.3.2. | CCCCTGCCCTGTTCGAGAG | |
| 27 | 3REG | GGGGTACCCGTGCATCAATGATCCGGCAAG (KpnI) | 490-bp bzdRN intergenic region |
| 28 | N-INV-2 | GCAGTGCAGGCGATGTTGAT | |
| 29 | 5PBM | GGGGTACCGAAGTCCGAAGCGCTGGGTCTGC (KpnI) | 444-bp bzdQM fragment cloned in pSJ3QM |
| 30 | 3PBM | GCTCTAGACCCATTTTTTCCCTCCTCGGGCACTTAGTAGG (XbaI) | |
| 31 | 5PBZDX | GGGGTACCACATGCTCGTCGCTGCCTCCAAC (KpnI) | 602-bp bzdWX fragment cloned in pSJ3WX |
| 32 | 3PBZDX | GCTCTAGACCCACGCTTCGTCTCCCTTTAGCTTTCGG (XbaI) | |
| 33 | 5PBZDYZ | GGGGTACCGGCAAGGACGTCATCGACTTC (KpnI) | 277-bp bzdYZ fragment cloned in pSJ3YZ |
| 34 | 3PBZDYZ | GCTCTAGACCCATGCAAGTCCCCTTTAACCGC (XbaI) | |
| 35 | RIVW5′ | GCGGTACCTGCAGCAGTACGGAAAGATG (KpnI) | 313-bp bzdVW fragment cloned in pSJ3VW |
| 36 | RIVW3′ | GCTCTAGACCCATGGTGTCTCCCCTGGTTC (XbaI) | |
| 37 | 3REG | GGGGTACCCGTGCATCAATGATCCGGCAAG (KpnI) | 598-bp bzdRN fragment (PN) cloned in pSJ3PN |
| 38 | 5BZN | GCTCTAGACCCATCGAACTATCTCCTCTGATG (XbaI) | |
| 39 | 5REG | GGGGTACCGGTTTCGTCGCAGGTGCTGTCTGGC (KpnI) | 675-bp bzdR fragment (PR) cloned in pSJ3PR |
| 40 | 3PREG | GCTCTAGACCCATCAGCAGGTAGTTGTTCTCTTAACG (XbaI) | |
| 41 | AINI | CCGAATTCTGAATAGATAAGGAGAGGAGGAGCAAATGGCAGAAC (EcoRI) | 1,913-bp bzdA gene cloned in pUCBZDA |
| 42 | BZDB′3′ | CGCCGAACGAGTATTTCCAGCTC | |
| 43 | 1600R | AAGGAGGTGATCCAGCC | 1,525-bp fragment of the 16S rDNA genes |
| 44 | 16SF1 | GAGASTTTGATCHTGGCTCAG |
Engineered restriction sites are underlined, and the corresponding restriction enzyme is shown in parentheses.
PCR reactions were carried out with the sets of two primers indicated to the left.
Escherichia coli strains W (17), XL1-Blue MRA (P2) [Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 gyrA96 supE44 relA1 thi-1 lac, P2 lysogen] (Stratagene), MC4100 [araD319 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR] (15), DH10B [F′, mcrA Δ(mrr hsdRMS-mcrBC) φ80dlaZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK λ rpsL endA1 nupG] (Life Technologies), and S17-1λpir (Tpr Smr recA thi hsdRM+ RP4::2-Tc::Mu::Km Tn7 λpir phage lysogen) (18) were grown at 37°C in Luria-Bertani (LB) medium (51). Where appropriate, antibiotics were added to the LB medium at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml.
Azoarcus sp. strain CIB was grown anaerobically on MA basal medium (composed of the following, per liter of distilled water: 0.33 g of KH2PO4, 1.2 g of Na2HPO4, 0.11 g of NH4Cl, 0.1 g of MgSO4 · 7H2O, 0.04 g of CaCl2 [pH 7.5]) supplemented with trace elements [stock solution, 100×; 1.5 g of nitrilotriacetic acid, 3 g of MgSO4 · 7H2O, 0.5 g of MnSO4 · 2H2O, 1 g of NaCl, 0.1 g of FeSO4 · 7H2O, 0.18 g of CoSO4 · 7H2O, 0.1 g of CaCl2 · 2H2O, 0.18 g of ZnSO4 · 7H2O, 0.01 g of CuSO4 · 5H2O, 0.02 g of KAl(SO4)2 · 12H2O, 0.01 g of H3BO3, 0.01 g of Na2MoO · 2H2O, 0.025 g of NiCl2 · 6H2O, and 0.3 mg of Na2SeO3 · 5H2O (pH 6.5) per liter of deionized water], vitamin solution (stock, 1,000×; 20 mg of biotin, 20 mg of folic acid, 10 mg of pyridoxine-HCl, 50 mg of thiamine-HCl · 2H2O, 50 mg of riboflavin, 50 mg of nicotinic acid, 50 mg of calcium d-pantothenic acid, 50 mg of vitamin B12, and 50 mg of p-aminobenzoic acid per liter of distilled water), and 10 mM potassium nitrate. Fifteen milliliters of MC medium (MA basal medium plus trace elements, vitamins, and nitrate) was flushed with N2, and the bottles were sealed with rubber stoppers and aluminum crimp seals before being autoclaved. The carbon sources and the bacterial inoculum were injected through the stopper with a syringe. The cultures were incubated at 30°C with static growth. Azoarcus sp. strain CIB was also grown aerobically with slow shaking at 30°C on MC medium without nitrate. When they were used as carbon sources, aromatic compounds were added to 3 mM, except for toluene and m-xylene, which were added to 2 mM. Bacterial growth was monitored by measuring the absorbance at 600 nm (A600). Denitrification was monitored by measuring the levels of NO3 and NO2 by a nitrate test (Merck). The concentration of benzoate in the culture medium was monitored spectrophotometrically at 273 nm (41).
[ring-14C]benzoate mineralization.
To investigate the mineralization of benzoate by Azoarcus sp. strain CIB, we grew cells anaerobically in benzoate-containing MC medium (10 ml) supplemented with 5 μCi of [ring-14C]benzoate (55 mCi/mmol; Sigma-Aldrich). A culture of E. coli W grown anaerobically on MC minimal medium containing 0.2% succinate plus [ring-14C]benzoate (5 μCi) was used as a control strain that does not mineralize benzoate. Two culture samples (1 ml) were collected after incubation for 16 h (complete consumption of benzoate). One sample was passed through a filter to quantify the total amount of radioactivity incorporated into the cells. The filter was washed twice with saline solution, mixed with 3 ml of scintillation liquid, and analyzed in a liquid scintillation counter. The second sample was used to detect the 14C incorporated into the macromolecules of the cell. To this end, the cells were centrifuged, resuspended in 1 ml of saline solution, frozen, and thawed, and the resulting cell extract was precipitated with trichloroacetic acid for 2 h at 4°C. After centrifugation for 15 min at 15,000 × g, the pellet was washed three times with saline solution containing trichloroacetic acid and was finally resuspended in 200 μl of saline solution for 14C quantification in a liquid scintillation counter. For detection of the 14CO2 produced during the mineralization of [ring-14C]benzoate, a trap containing 0.1 N NaOH was used.
Molecular biology techniques.
Standard molecular biology techniques were performed as previously described (51). DNA fragments were purified with Gene-Clean Turbo (BIO101 Systems). PCR products were purified with a High Pure plasmid isolation kit (Roche). Nucleotide sequences were determined directly from plasmids and the λBzd1 phage by the use of fluorescently labeled dideoxynucleotide terminators (52) and AmpliTaq FS DNA polymerase (Applied Biosystems Inc.). The sequencing reactions were analyzed with an ABI Prism 377 automated DNA sequencer (Applied Biosystems Inc.). Nucleotide sequence analyses were done at the Infobiogen server (http://www.infobiogen.fr/services/menuserv.html). The amino acid sequences of open reading frames were compared with those present in finished and unfinished microbial genome databases by use of the TBLASTN algorithm (2) at the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Nucleotide and protein sequence similarity searches were also performed with the BLAST programs at the BLAST server of NCBI (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi). Pairwise and multiple protein sequence alignments were made with the ALIGN (59) and CLUSTALW (57) programs, respectively, at the Infobiogen server. Phylogenetic analysis of the 16S ribosomal DNA (rDNA) genes was carried out according to Kimura's two-parameter method (37) and a tree was reconstructed by use of the neighbor-joining method (50) of the PHYLIP program (26).
For reverse transcription-PCR (RT-PCR) experiments, cultures of Azoarcus sp. strain CIB grown anaerobically in either benzoate or succinate were collected at an A600 of 0.3. Total mRNAs were obtained with a High Pure RNA isolation kit (Roche). Any contamination by DNA was eliminated by the use of a DNase treatment and removal kit (Ambion). One microgram of purified total RNA was used to prepare cDNA by the use of 3 U of avian myeloblastosis virus reverse transcriptase (Promega). PCRs were carried out with 2.5 U of AmpliTaq DNA polymerase (Roche). Control reactions in which reverse transcriptase was omitted from the reaction mixture ensured that DNA products resulted from the amplification of cDNA rather than from DNA contamination.
DNA preparation for pulsed-field gel electrophoresis of Azoarcus sp. strain CIB cells was performed as previously described (49). Samples were electrophoresed at 170 V with a 45-s pulse setting for 24 h at 15°C. DNA size standards (225 to 2,200 kb) from Saccharomyces cerevisiae (Bio-Rad) were used. Rhodococcus sp. strain IGTS8 was used as a control for cells containing megaplasmids of 50, 120, and 400 kb (19). The DNA fragments were then blotted onto a nylon membrane as previously described (51) and were probed with a digoxigenin-labeled bzdQ gene. Hybridization of the probe was observed with the chromosomal DNA band.
Construction of Azoarcus sp. strain CIBdbzdN, Azoarcus sp. strain CIBdbzdY, and Azoarcus sp. strain CIBlacZ.
For gene disruption through single homologous recombination, internal fragments of the bzdN and bzdY genes to be disrupted were cloned into the polylinker of pK18mob (a mobilizable plasmid which does not replicate in Azoarcus) (Table 1), and the resulting constructs, pK18mobbzdN and pK18mobbzdY (Table 1), were introduced into Azoarcus sp. strain CIB. By means of RP4-mediated mobilization, plasmids pK18mobbzdN and pK18mobbzdY were transferred from E. coli S17-1λpir (donor strain) into Azoarcus sp. strain CIB (recipient strain). Biparental filter mating was performed aerobically at 30°C for 16 h as described previously (18), but with a ratio of 109 donor cells to 1010 recipient cells, on MC medium plates containing 0.4% succinate. Exconjugants harboring the disrupted gene by insertion of the suicide plasmid, namely Azoarcus sp. strain CIBdbzdN and Azoarcus sp. strain CIBdbzdY, were isolated aerobically on kanamycin (50 μg/ml)-containing MC medium lacking nitrate and containing 0.4% citrate as the sole carbon source for counterselection of donor cells. The mutants were analyzed by PCR to confirm the disruption of the target gene.
For the construction of Azoarcus sp. strain CIBlacZ, plasmid pK18mobPNlacZ, a derivative of the suicide plasmid pK18mob harboring a PN::lacZ translational fusion (Table 1), was transferred from E. coli S17-1λpir into Azoarcus sp. strain CIB. Biparental filter mating was performed as described above. Exconjugants containing the PN::lacZ translational fusion stably inserted into the chromosomal PN promoter were merodiploids with an intact copy of the bzd catabolic genes, and they were selected for the plasmid marker, kanamycin, on aerobic MC medium lacking nitrate and containing 0.4% citrate as the sole carbon source for counterselection of donor cells. One of the exconjugants, Azoarcus sp. strain CIBlacZ, was confirmed by PCR and selected for further studies.
Cloning and expression of bzdA from Azoarcus sp. strain CIB in E. coli.
The bzdA gene was PCR amplified from the chromosomal DNA of Azoarcus sp. strain CIB by using primers AINI and BZDB′3′ (Table 2). The resulting 2.2-kb DNA fragment, which contained the structural bzdA gene with its own Shine-Dalgarno region, was digested with EcoRI and ScaI and cloned into an EcoRI- and SmaI-digested pUC18 vector to produce plasmid pUCBZDA (Table 1). E. coli DH10B cells harboring pUCBZDA express the bzdA gene under the control of the Plac promoter. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with 12.5% polyacrylamide slab gels as described by Laemmli (40).
Construction of genome libraries of Azoarcus sp. strain CIB.
Thirty micrograms of genomic DNA from Azoarcus sp. strain CIB was partially digested with Sau3AI, and fragments of 15 to 25 kb were ligated to a BamHI-digested λ DASH II vector (Stratagene Cloning Systems). Gigapack III XL packaging extract (Stratagene) was used to package the recombinant λ phage, and phage particles were obtained by infecting E. coli XL1-Blue MRA(P2) as described previously (51). Screening of the library was carried out by hybridization with a digoxigenin-labeled bzdQ probe. For preparation of the DNA probe, a 487-bp bzdQ internal fragment was PCR amplified with primers δBcr5′ and δBcr3′ (Table 2) and randomly labeled with a DIG DNA labeling kit (Roche). Positive plaques (4 of 2,000 plaques) were detected with a DIG luminescence detection kit (Roche), and phage DNA was purified as previously reported (51).
An EcoRI DNA library of Azoarcus sp. strain CIB was also constructed in E. coli DH10B by use of an EcoRI-digested pUC19 vector. Screening of the library by colony hybridization with a digoxigenin-labeled bzdY probe (Table 2) identified a plasmid, pECOR8, containing the right end of the bzd cluster (Table 1; also see Fig. 3A).
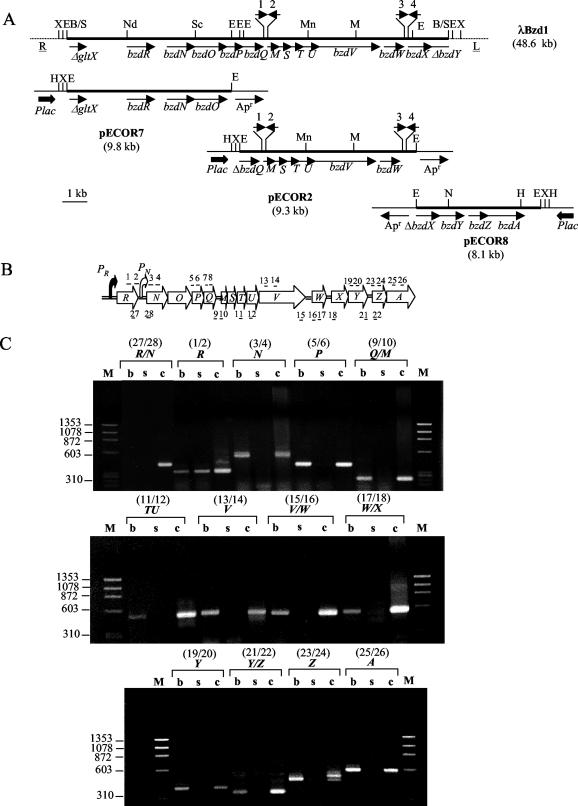
FIG. 3.
Physical, genetic, and transcriptional analysis of region encoding anaerobic benzoate catabolism in Azoarcus sp. strain CIB. (A) Azoarcus sp. strain CIB fragments (thick lines) cloned in the recombinant λ DASH II phage (λBzd1) and in three pUC19 derivatives (pECOR7, pECOR2, and pECOR8) are indicated. R and L represent the right and left arms, respectively, of the λ phage. Arrows indicate the direction of transcription of the genes. Thick arrows show the Plac promoter from the pUC19 vector. Apr, gene that confers resistance to ampicillin. Δ, indicates a truncated gene. 1, 2, 3, and 4, corresponding REP elements. Restriction sites: E, EcoRI; H, HindIII; M, MluI; Mn, MunI; N, NcoI; Nd, NdeI; Sc, ScaI; X, XbaI; B/S, a ligation of compatible ends generated by BamHI and Sau3AI cleavage. A truncated gene encoding a putative glutamate tRNA synthetase (gltX) is shown at the left end of the bzd cluster. (B) Schematic representation of bzd gene cluster. Arrows indicate the direction of transcription of the genes. Filled and open bent arrows show the PR and PN promoters, respectively. Relevant intergenic regions are represented by a double line. The primer pairs used for the RT-PCRs shown in panel C are indicated above (structural regions) and below (intergenic regions) the corresponding genes. The odd and even numbers refer to primers that hybridize with the lower and upper strands of the DNA, respectively (Table 2). (C) Agarose gel electrophoresis of RT-PCR products. RT-PCRs with Azoarcus sp. strain CIB cells grown under denitrifying conditions on benzoate (lanes b) or succinate (lanes s) were performed as described in Materials and Methods with the primer pairs indicated at the top (Table 2). PCRs performed with the primer pairs indicated at the top and with genomic DNA as a positive control are also indicated (lanes c). Lanes M, molecular size markers (HaeIII-digested ΦX174 DNA). Numbers on the left represent the sizes of the markers (in base pairs).
Benzoate-CoA ligase assay.
E. coli cells containing the bzdA gene were grown in LB medium to an A600 of 2. Azoarcus sp. strain CIB cells were grown anaerobically in MC medium with the appropriate carbon source to reach an A600 of 0.3. The cells were collected and washed with 100 mM Tris-HCl buffer, pH 8.5. The cells were broken by sonication and the cell debris was removed by centrifugation. The clear supernatant fluid was decanted and used as a crude cell extract. The protein concentration was determined by the method of Bradford (13), with bovine serum albumin as a standard. Benzoate-CoA ligase activity was determined at 30°C with crude cell extracts through a direct spectrophotometric assay or via a coupled enzyme assay. For the direct assay, product formation (CoA derivatives) was monitored spectrophotometrically as described previously (48). The assay mixture (600 μl) contained 100 mM Tris-HCl (pH 8.5), 2 mM dithiothreitol, 5 mM MgCl2, 1 mM ATP, 0.4 mM CoA, 1 mM aromatic acid, and 6 μl of crude cell extract. Benzoyl-CoA was measured at 290 nm (ɛ = 3.9 mM−1 cm−1), 2-hydroxybenzoyl-CoA was measured at 320 nm (ɛ = 5.9 mM−1 cm−1), 3-hydroxybenzoyl-CoA was measured at 310 nm (ɛ = 3.3 mM−1 cm−1), 4-hydroxybenzoyl-CoA was measured at 300 nm (ɛ = 10.55 mM−1 cm−1), and 2-aminobenzoyl-CoA was measured at 365 nm (ɛ = 5.5 mM−1 cm−1) (48, 60). For the coupled enzyme assay, AMP formation was monitored by coupling the CoA ligase reaction to the myokinase, pyruvate kinase, and lactate dehydrogenase system and by spectrophotometrically measuring the rate of NADH oxidation at 365 nm (60). The reaction mixture (600 μl) contained 100 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 2 mM ATP, 1 mM CoA, 2 mM phosphoenolpyruvate, 0.5 mM aromatic compound, 0.5 mM NADH, 1.2 μl of a mixture of pyruvate kinase (1 U) and lactate dehydrogenase (1 U) from rabbit muscle (Roche), 0.6 μl (0.6 U) of myokinase from chicken muscle (Sigma-Aldrich), and 6 μl of crude extract.
Benzoyl-CoA reductase assay.
For the preparation of crude cell extracts, 2 g (wet mass) of Azoarcus sp. strain CIB cells grown on benzoate-containing MC medium was anaerobically suspended in 2 ml of 100 mM Tris-HCl (pH 7.8), 20% (wt/vol) glycerol, 2 mM dithiothreitol, and 1 to 2 mg of DNase I. A French press treatment (137 MPa) and ultracentrifugation (100,000 × g) were performed under anaerobic conditions as described previously (12). The clear supernatant fluid was used as a crude cell extract. Benzoyl-CoA reductase activity was tested in crude cell extracts by a radioactive assay as described previously (8, 23, 39); each step was performed under strictly anaerobic conditions. The test was carried out in gas-tight-sealed glass tubes at 30°C. The standard assay mixture (0.35 ml) contained 100 mM morpholinepropanesulfonic acid-KOH buffer (pH 7.2), 10 mM MgCl2, 0.5 mM CoA, 7 mM ATP, 200 μM [ring-14C]benzoate (4,810 kBq/μmol; Amersham Biosciences), 5 mM sodium dithionite (as an electron donor), and 50 μl of cell extract with a protein concentration of 70 mg ml−1. When the assay was performed with benzoyl-CoA reductase from T. aromatica, dithionite was replaced with Ti(III)-citrate (5 mM). For the formation of benzoyl-CoA by benzoate-CoA ligase, the mixture was preincubated for 10 min at 30°C with 10 μl of a partially purified fraction containing the benzoate-CoA ligase activity of T. aromatica (6 mg ml−1; 0.3 μmol min−1 mg−1). The reaction was started by the addition of crude cell extracts of Azoarcus sp. strain CIB. Residual [14C]benzoyl-CoA and derived 14C-labeled CoA ester products were hydrolyzed under alkaline conditions. Samples (50 μl) taken between 0.1 and 15 min were added to 5 μl of 4 M KOH and hydrolyzed by heating at 80°C for 10 min. After acidification to pH 2 by the addition of 2 μl of 50% H2SO4 and centrifugation, the released carboxylic acids (5 μl of each sample) were analyzed by thin-layer chromatography (TLC) on silica gel (Kieselgel 60, F254; Merck) (the solvent system contained 25% 1-butanol and 75% diisopropyl ether, by volume). For testing of the formation of thiol esters, control samples were not hydrolyzed. For this purpose, 50-μl samples were added to 5 μl of 10% formic acid. After centrifugation, 5 μl of the supernatant was applied to the TLC plate. To test whether the aerobic benzoate-CoA ligase preparation from T. aromatica contained any residual benzoyl-CoA reductase activity, we performed an additional control experiment in which the Azoarcus sp. strain CIB extract was omitted from the reaction. For comparison, the reductase assay was also performed with partially purified benzoyl-CoA reductase from T. aromatica (0.4 mg ml−1; 0.25 U mg−1). Radioactive bands were visualized by autoradiography with a phosphorimager plate (Fuji Photo Film). Semiquantitative analysis was performed with Quantity One software (Bio-Rad).
For further analysis of the products formed by the benzoyl-CoA reductase assay, samples were subjected to C18 reversed-phase high-performance liquid chromatography (HPLC) analysis as described previously (38, 39). Sample preparation was performed as described above for TLC, including the hydrolysis of the thiol esters to the free carboxylic acids by KOH (pH ∼12) at 80°C. After acidification and subsequent centrifugation (see above), 25 μl of the supernatant was applied to an HPLC column, with 40 mM formic acid (pH 2.5) and methanol as the mobile phase. Detection of the 14C-labeled products was done with a UV spectroscopic (260 nm) cell and a radioactivity monitoring analyzer (Raytest, Straubenhart, Germany).
β-Galactosidase assay.
β-Galactosidase activities were measured with permeabilized cells as described by Miller (47).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rDNA and the bzd cluster have been submitted to GenBank under accession numbers AF515816 and AF521665, respectively.
RESULTS AND DISCUSSION
Aromatic growth substrates and morphological and phylogenetic analyses of Azoarcus sp. strain CIB.
Azoarcus sp. strain CIB was isolated from a culture (DSMZ 12184) which was supposed to be Azoarcus sp. strain M3 (33) by growing the cells in benzoate-containing MC medium under denitrifying conditions. Surprisingly, the sequence of the 16S rDNA from the DSMZ 12184 culture did not match with the reported sequence of Azoarcus sp. strain M3 (accession no. Y11040), and on the contrary, it was shown to correspond to a new strain that was named Azoarcus sp. strain CIB. Azoarcus sp. strain CIB is able to use several aromatic compounds as a sole carbon and energy source under anaerobic (denitrifying) conditions. Thus, the strain grows on both nonhydroxylated (benzoate, phenylacetate, phenylpropionate, and tropic acid), and hydroxylated (3-hydroxybenzoate, 4-hydroxybenzoate, 4-hydroxyphenylacetate, and 3-hydroxyphenylpropionate) aromatic acids, aromatic amines (benzylamine and phenylethylamine), aromatic aldehydes (benzaldehyde and phenylacetaldehyde), aromatic alcohols (phenylethanol and benzyl alcohol), aromatic amino acids (tyrosine and phenylalanine), and aromatic hydrocarbons (toluene and m-xylene). The doubling time under batch conditions was about 6 to 8 h when benzoate (3 mM) was used as a carbon source and nitrate (10 mM) was used as the sole electron acceptor. Benzoate-grown cells were motile and rod-shaped, as are most Azoarcus strains reported so far (46, 56). Azoarcus sp. strain CIB was also able to use several aromatic compounds, e.g., benzoate, 3-hydroxybenzoate, benzylamine, phenylacetate, 3-hydroxyphenylpropionate, and toluene, as sole carbon and energy sources under aerobic conditions.
A comparative 16S rDNA analysis of Azoarcus sp. strain CIB revealed a close phylogenetic relationship to the Azoarcus toluvorans group, which includes organisms that are able to degrade aromatic hydrocarbons under anaerobic conditions and whose G+C content is about 67.8 mol% (56). Nevertheless, a more detailed characterization is needed for a definite taxonomic classification of strain CIB within the genus Azoarcus.
Anaerobic degradation of benzoate by Azoarcus sp. strain CIB.
To demonstrate that Azoarcus sp. strain CIB is able to mineralize benzoate in an oxygen-free medium supplemented with nitrate as the sole electron acceptor, we used [ring-14C]benzoate as a substrate. Whereas 36,000 cpm (about 11% of the applied radioactivity) was incorporated into the cell material from Azoarcus sp. strain CIB, only 195 cpm was measured with the control E. coli extracts. Moreover, we detected the formation of 14CO2 by Azoarcus sp. strain CIB extracts (see Materials and Methods). These data indicate that benzoate is indeed mineralized in Azoarcus sp. strain CIB.
As shown for other benzoate degraders, the anaerobic catabolism of benzoate begins with the activation of the aromatic substrate to benzoyl-CoA by the action of a benzoate-CoA ligase (AMP forming) (1, 27, 55) (Fig. 1). Extracts of Azoarcus sp. strain CIB cells grown anaerobically on benzoate rendered about 0.17 μmol of benzoyl-CoA formed min−1 mg of protein−1 when benzoate was used as the substrate. Benzoate-CoA ligase activity was not observed when the CIB strain was grown anaerobically on succinate, indicating that the benzoate degradation pathway is inducible. The benzoate-CoA ligase activity was absolutely dependent on the aromatic acid, ATP, Mg2+, and CoA, as previously described for other aromatic-CoA ligases (1, 28, 54). Although similar levels of CoA ligase activity were reported for benzoate-grown A. evansii cells and although this activity was ascribed to a single benzoate-CoA ligase enzyme (1), we cannot discard the existence of different enzymes catalyzing the formation of CoA thioesters of benzoate in extracts from Azoarcus sp. strain CIB, as already reported for benzoate-grown R. palustris cells (24).
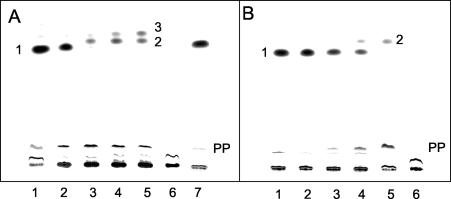
The only step in the anaerobic degradation of benzoate that requires strict anoxic conditions is that catalyzed by the iron-sulfur enzyme benzoyl-CoA reductase (10) (Fig. 1). We therefore tested whether extracts from benzoate-grown Azoarcus sp. strain CIB cells contained benzoyl-CoA reductase activity. For this purpose, we used an anaerobic radioactive assay for benzoyl-CoA reductase in cell extracts (see Materials and Methods). This assay monitors the formation of labeled products derived from [ring-14C]benzoate after hydrolysis of the CoA thiol esters to the free acids and analysis of the products formed by TLC and autoradiography. Figure 2A shows the time-dependent consumption of benzoyl-CoA and the formation of products by an Azoarcus sp. strain CIB cell extract. After 5 min, most of the formed [ring-14C]benzoyl-CoA (compound 1) was converted to two less polar products (compounds 2 and 3) and to more polar products (compound PP and other products). In control samples, the alkaline hydrolysis step was omitted and the radioactively labeled compounds remained in the starting line (Fig. 2A, lane 6), which confirms that the 14C-labeled compounds are highly polar thiol esters that do not migrate in the TLC system used. When the Azoarcus sp. strain CIB extract was omitted from the assay, no product was formed from [ring-14C]benzoyl-CoA after a 10-min incubation (Fig. 2A, lane 7), which rules out the presence of benzoyl-CoA reductase contamination in the partially purified benzoate-CoA ligase preparation from T. aromatica. Moreover, it should be noted that the preparation of benzoate-CoA ligase from T. aromatica was carried out under aerobic conditions that irreversibly inactivate the benzoyl-CoA reductase within a few seconds (8). For a comparison of the product patterns, the radioactive benzoyl-CoA reductase assay was also performed with partially purified benzoyl-CoA reductase from T. aromatica (Fig. 2B). The product of benzoyl-CoA reductase from T. aromatica is the cyclohexa-1,5-diene-1-carbonyl-CoA (11), and compound 2 represents the free acid of this dienoyl-CoA compound. Compound PP represents the hydrated next product in the pathway, the corresponding 6-hydroxycylohex-1-ene-1-carbonyl-CoA. The latter compound is formed by dienoyl-CoA hydratase, a frequent impurity of benzoyl-CoA reductase preparation (11). As shown in Fig. 2, the compound 2 formed from benzoyl-CoA by cell extracts of Azoarcus sp. strain CIB migrated by TLC similarly to the compound 2 obtained with the benzoyl-CoA reductase preparation from T. aromatica (8, 23, 39). Moreover, both compounds also comigrated in HPLC analysis (data not shown), which strongly suggests that benzoyl-CoA reductase from Azoarcus sp. strain CIB behaves similarly to the enzyme from T. aromatica. Most possibly, compound 3 represents a cyclohex-1-ene-1-carbonyl-CoA, which would be formed by a further two-electron reduction of the formed diene, as demonstrated earlier for purified benzoyl-CoA reductase from T. aromatica (11). The product pattern of benzoyl-CoA conversion by cell extracts of Azoarcus sp. strain CIB was therefore similar to that obtained with extracts from T. aromatica (8, 39). The specific activity of benzoyl-CoA conversion in Azoarcus sp. strain CIB, 5 nmol min−1 mg of protein−1, was approximately 25 to 30% of the values reported for cell extracts of A. evansii (23) and T. aromatica (39). Note that in T. aromatica the benzoyl-CoA reductase activity was found to be threefold higher with the natural electron donor ferredoxin than with artificial electron donors (9). Since sodium dithionite was used as an electron donor in the reductase assay, the in vivo activity of benzoyl-CoA reductase from Azoarcus sp. strain CIB might also be higher than that reported here.
FIG. 2.
Conversion of [ring-14C]benzoyl-CoA to radioactively labeled products by cell extracts of Azoarcus sp. strain CIB or partially purified benzoyl-CoA reductase from T. aromatica. The anaerobic assay mixture contained CoA, ATP, and [ring-14C] benzoate. Benzoyl-CoA was formed enzymatically by preincubation with the partially purified benzoate-CoA ligase of T. aromatica (see Materials and Methods). Samples were taken at the indicated times and analyzed by TLC, and autoradiographs are shown. (A) Assay with cell extracts from Azoarcus sp. strain CIB. Lanes 1 to 5, samples taken after 5 s, 1 min, 3 min, 5 min, and 10 min of incubation, respectively; lane 6, the same as lane 5 but without alkaline hydrolysis; lane 7, assay without a cell extract from Azoarcus sp. strain CIB after 10 min of incubation. 1, [14C]benzoate; 2 and 3, free acids of the putative diene and monoene products; PP, free acids of more-polar products. (B) Assay with partially purified benzoyl-CoA reductase from T. aromatica. Lanes 1 to 5, samples taken after 5 s, 1 min, 3 min, 5 min, and 10 min of incubation, respectively; lane 6, the same as lane 5 but without alkaline hydrolysis. 1, [14C]benzoate; 2, free acid of the diene product; PP, free acids of more-polar products.
The enzyme assays performed with the Azoarcus sp. strain CIB extracts suggest a reaction scheme for the initial benzoate degradation based on a benzoate-CoA ligase and a benzoyl-CoA reductase similar to those reported for other anaerobic benzoate degraders such as T. aromatica (31) (Fig. 1).
Cloning, sequencing, and sequence analysis of the bzd cluster for benzoate degradation in Azoarcus sp. strain CIB.
When the total DNA from Azoarcus sp. strain CIB was analyzed by pulsed-field gel electrophoresis, no plasmid was observed, suggesting that the genetic determinants for anaerobic benzoate degradation are chromosome encoded. At the beginning of this research, the only sequence available for the genes responsible for anaerobic benzoate degradation in Azoarcus was that of an internal fragment of the gene encoding the α subunit of the benzoyl-CoA reductase from A. evansii (7). This sequence was used to design two primers for PCR amplification of the equivalent gene in Azoarcus sp. strain CIB. The resulting 487-bp PCR fragment was then used as a probe to screen a BamHI DNA library of Azoarcus sp. strain CIB constructed in E. coli by use of the λ phage (see Materials and Methods), and a positive clone, λBzd1, was selected. The recombinant λBzd1 phage contains a 16-kb DNA insert that was digested with EcoRI and partially subcloned into the pUC19 vector, producing plasmids pECOR7 and pECOR2 (Fig. 3A; Table 1). An EcoRI DNA library of Azoarcus sp. strain CIB was also constructed in E. coli by use of an EcoRI-digested pUC19 vector, and a clone (plasmid pECOR8) (Table 1) that hybridized with the downstream end of the 16-kb DNA fragment (bzdY probe) (Table 2) was selected. An overall analysis of the 19.6-kb DNA cloned from Azoarcus sp. strain CIB revealed 15 open reading frames transcribed in the same direction that might constitute the anaerobic benzoate degradation cluster, the bzd cluster (benzoate degradation) (Fig. 3A and 4). Although the results of the sequence comparison analyses (Table 3) strongly suggest that the bzd gene cluster codes for the anaerobic degradation of benzoate in Azoarcus sp. strain CIB, to confirm this assumption we constructed two mutant strains, Azoarcus sp. strain CIBdbzdN and Azoarcus sp. strain CIBdbzdY, with insertional disruptions within genes bzdN and bzdY, respectively. Both mutant strains were unable to grow in MC medium containing benzoate as the sole carbon and energy source, thus demonstrating that the bzd genes are indeed involved in the anaerobic catabolism of benzoate. Moreover, the two mutant strains were also defective in growth on other aromatic compounds such as phenylacetate, 4-hydroxybenzoate, 4-hydroxyphenylacetate, and 3-hydroxybenzoate, which suggests that either the catabolism of these compounds leads to benzoate or it shares some enzymatic steps with that of benzoate in Azoarcus sp. strain CIB.
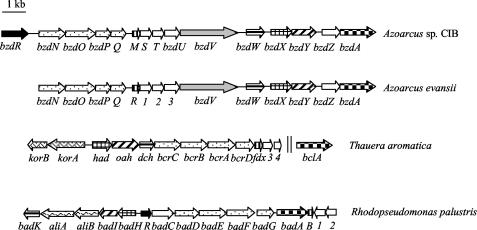
FIG. 4.
Organization of the gene clusters involved in the anaerobic catabolism of benzoate in Azoarcus sp. strain CIB, A. evansii (accession no. AJ428529) (31), T. aromatica (accession no. AJ224959 and AF373594), and R. palustris (accession no. U75363). Genes are represented by arrows as follows: black, regulatory genes; white, genes of unknown function; checkerboard pattern, genes encoding the benzoate-CoA ligases; stippling, genes encoding the four subunits of the benzoyl-CoA reductase; vertical stripes, ferredoxins; horizontal stripes, genes encoding enoyl-CoA hydratases; cross hatching, genes encoding NAD-dependent dehydrogenases; hatching, genes encoding ring-cleavage hydrolases; gray, gene encoding a putative ferredoxin oxidoreductase; wavy, genes encoding enzymes of alicyclic acid degradation; intertwined lines, genes encoding the 2-oxoglutarate:ferredoxin oxidoreductase. Two vertical lines indicate that the genes are not adjacent in the genome.
TABLE 3.
The bzd genes, their products, and related gene products
| Gene | G+C content (%) | Distance to next gene (bp)a | Gene product (aa/kDa) | Putative function of gene product | Related gene products
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Name/size (aa) | Function | Organismb | % Identity | Accession no. | |||||
| bzdR | 63.6 | 582 | 298/33.5 | Transcriptional regulator | Reut0152/316 | Unknown | R. metallidurans | 53 | ZP_00021253 |
| ORF1/303 | Putative regulator | T. aromatica | 51 | AAN32624 | |||||
| ORF10/300 | Putative regulator | A. evansii | 50 | AAN39374 | |||||
| bzdN | 61.1 | 21 | 379/43.4 | γsubunit of BCRc | BadD/394 | γ subunit of BCRc | R. palustris | 28 | AAC23925 |
| FldC/374 | R-phenyllactate dehydratase small subunit | C. sporogenes | 28 | AAL18811 | |||||
| BcrC/386 | γ subunit of BCRc | T. aromatica | 25 | CAA12247 | |||||
| bzdO | 62.1 | 28 | 437/49.9 | β subunit of BCRc | HgdA/391 | 2-hydroxyglutaryl-CoA dehydratase α subunit | A. fulgidus | 27 | NP_070782 |
| BcrB/432 | β subunit of BCRc | T. aromatica | 23 | CAA12248 | |||||
| BadE/436 | β subunit of BCRc | R. palustris | 22 | AAC23926 | |||||
| bzdP | 65.2 | 38 | 269/27.9 | δ subunit of BCRc | HgdC/251 | 2-hydroxyglutaryl-CoA dehydratase activator | A. fulgidus | 36 | NP_070783 |
| BcrD/282 | δ subunit of BCRc | T. aromatica | 32 | CAA12250 | |||||
| BadG/277 | δ subunit of BCRc | R. palustris | 30 | AAC23928 | |||||
| bzdQ | 62.7 | 138 | 301/32.5 | α subunit of BCRc | BcrA/438 | α subunitof BCRc | T. aromatica | 43 | CAA12249 |
| BadF/437 | α subunit of BCRc | R. palustris | 40 | AAC23927 | |||||
| FldI/264 | R-phenyllactate dehydratase activator | C. sporogenes | 40 | AAL18809 | |||||
| bzdM | 61 | 10 | 94/10.2 | Ferredoxin | Desu3036/240 | Ferredoxin | D. hafniense | 39 | ZP_00099885 |
| CPE1065/268 | Ferredoxin-like | C. perfringens | 34 | NP_561981 | |||||
| BadB/81 | Ferredoxin | R. palustris | 21 | AAC13360 | |||||
| bzdS | 61.5 | 51 | 108/12.2 | Unknown | |||||
| bzdT | 57.6 | 30 | 150/17.1 | Unknown | ORF3/156 | Unknown | T. aromatica | 33 | CAA12252 |
| ORF2/159 | Unknown | R. palustris | 23 | ACC13362 | |||||
| bzdU | 63.1 | 57 | 212/23 | Unknown | Meth3594/224 | Putative phosphoesterase | M. barkeri | 29 | ZP_00078942 |
| bzdV | 66.8 | 238 | 849/90.8 | NADPH:acceptor oxidoreductase | Desul139/844 | NADPH-dependent glutamate synthase | D. hafniense | 42 | ZP_00098034 |
| bzdW | 65.5 | 161 | 259/27.6 | Enoyl-CoA hydratase | Dch/258 | Cyclohex-1,5-diene 1-carbonyl-CoA hydratase | T. aromatica | 38 | CAA12246 |
| Crt/259 | Crotonase | T. thermosaccharolyticum | 37 | CAB07495 | |||||
| bzdX | 63.8 | 35 | 355/38.4 | β-Hydroxyacyl-CoA dehydrogenase | Magn5225/388 | Zn-dependent alcohol dehydrogenase | M. magnetotacticum | 46 | ZP_00052492 |
| Had/368 | 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase | T. aromatica | 42 | CAA12244 | |||||
| bzdY | 62.6 | 156 | 376/41.8 | Alicyclic acid CoA hydrolase | Oah/377 | 6-ketocyclohex-1-ene 1-carbonyl-CoA hydrolase | T. aromatica | 51 | CAA12245 |
| Gmet2899/374 | Dihydroxynaphthoic acid synthetase | G. metallireducens | 51 | ZP_00082132 | |||||
| BadI/260 | 2-ketocyclohexane 1-carbonyl-CoA hydrolase | R. palustris | 35 | AAC23921 | |||||
| bzdZ | 64.5 | 48 | 248/26.2 | Unknown | FabG/246 | 3-ketoacyl carrier protein reductase | B. subtilis | 46 | AAC44307 |
| bzdA | 65 | 61 | 533/57.4 | Benzoate-CoA ligase | ORF7d/529 | Benzoate-CoA ligase | A. evansii | 69 | AAN39371 |
| BclAe/532 | Benzoate-CoA ligase | T. aromatica | 68 | CAD21683 | |||||
| BclA/527 | Benzoate-CoA ligase | T. aromatica | 68 | AAN32623 | |||||
| BadA/523 | Benzoate-CoA ligase | R. palustris | 54 | AAK39104 | |||||
Length of intergenic region to next gene.
The full names of the organisms are Ralstonia metallidurans, Thauera aromatica, Azoarcus evansii, Rhodopseudomonas palustris, Clostridium sporogenes, Archaeoglobus fulgidus, Desulfitobacterium hafniense, Clostridium perfringens, Methanosarcina barkeri, Thermoanaerobacterium thermosaccharolyticum, Magnetospirillum magnetotacticum, Geobacter metallireducens, and Bacillus subtilis.
BCR, benzoyl-CoA reductase.
ORF7, aerobic benzoate-CoA ligase from A. evansii.
BclA (532 aa), putative benzoate-CoA ligase from T. aromatica.
According to the pathway that has been described for anaerobic benzoate degradation in T. aromatica and R. palustris (31), the role of most of the bzd genes from Azoarcus sp. strain CIB can be foreseen (Fig. 4; Table 3). Genes bzdS, bzdU, and bzdZ encode proteins of unknown function and do not have homologues in other benzoate degradation clusters (Fig. 4). The bzdR gene encodes a putative transcriptional regulatory protein (Table 3) whose N-terminal and C-terminal halves show similarity to the DNA-binding domain of the SinR regulator of Bacillus subtilis (45) and to the shikimate kinase I of E. coli (43), respectively. At the downstream end of the bzd cluster there is a partial sequence that shows similarity to genes encoding the substrate-binding subunit of an ABC transporter (34). Although genes encoding ABC transporters have been found in the vicinity of aromatic degradation clusters (25, 29, 55), the role of such transporters in the uptake of the cognate aromatic compounds has not yet been demonstrated.
It is worth noting that during the course of this research a homologous bzd gene cluster from A. evansii was submitted to GenBank (accession no. AJ428529). The level of identity between each couple of bzd homologues ranged from 91 to 98%, thus suggesting that A. evansii and Azoarcus sp. strain CIB (closely related to the A. toluvorans species; see above) might share a similar catabolic pathway for anaerobic benzoate degradation. Although the gene arrangements within the bzd gene clusters from Azoarcus sp. strain CIB and A. evansii were similar (Fig. 4), the CIB strain contains four repeats (REP elements) of a 38-bp DNA sequence (5′-CCNTCCCCTTCAAGGGGANGGNNNGGNTGGGGATGGGT-3′; N indicates that the nucleotide at that position is not conserved and underlining indicates a palindromic region) that are lacking in the bzd cluster from A. evansii. The REP sequences are arranged as pairs of convergent elements separated by 17 and 23 bp in the bzdQ-bzdM and bzdW-bzdX intergenic regions, respectively (Fig. 3A). Since it was shown previously that some repetitive extragenic elements (BOX-like sequences) are present in the genome of Azoarcus (35), the repeats reported here may represent a new repetitive DNA sequence that is present in the Azoarcus genus. A similar extragenic sequence is present in the genome of Vibrio cholerae O1, although in this bacterium such repetitive elements are not associated as pairs. Although repetitive extragenic palindromic elements have been shown to be involved in several functions (5, 36), the physiological role of the REP elements within the bzd cluster of Azoarcus sp. strain CIB is still unknown.
The amino acid sequences of the four subunits of benzoyl-CoA reductase from T. aromatica and R. palustris have higher similarities (64 to 76% identity) to each other than to the corresponding subunits of the putative enzyme from Azoarcus sp. strain CIB (22 to 43% identity). Notably, whereas the two subunits carrying a putative ATP-binding motif, the bzdP and bzdQ gene products, have a similar size in the benzoyl-CoA reductase from Azoarcus, the corresponding subunits in T. aromatica and R. palustris differ largely in size (Table 3). Nevertheless, the benzoyl-CoA reductases from Thauera and Azoarcus appear to form the same product when acting on benzoyl-CoA in vitro (described above). In contrast, the deduced amino acid sequences of the putative ligase, hydratase, dehydrogenase, and hydrolase from Azoarcus are more similar to those of the corresponding enzymes from T. aromatica than to those of the enzymes from R. palustris (Table 3; Fig. 4). This observation might indicate that generally the benzoate degradation pathway in Azoarcus is more similar to that reported for T. aromatica than to that reported for R. palustris (Fig. 1), which is consistent with the fact that both Thauera and Azoarcus are denitrifying β-Proteobacteria, whereas Rhodopseudomonas is a phototrophic α-Proteobacteria. Interestingly, the korA and korB genes encoding the two subunits of the 2-oxoglutarate:ferredoxin oxidoreductase (KGOR) that reduces the oxidized ferredoxin (Fdx) in T. aromatica (22) are not present in the bzd and bad clusters (Fig. 4). In this sense, it has been recently demonstrated in A. evansii that oxidized ferredoxin is reduced by the combined action of a three-subunit 2-oxoglutarate:acceptor oxidoreductase (KGOR) and an NADPH:acceptor (ferredoxin) oxidoreductase, with the latter likely encoded by the bzdV gene (23) (Fig. 4).
The G+C contents of the bzd gene cluster (Table 3) and of the bcr cluster from T. aromatica averaged 63.1 and 64.7%, respectively, which are slightly lower than the mean G+C content (about 67%) of Azoarcus and Thauera species (4, 56). In contrast, the bad cluster presents a G+C content (64.7%) that fits the mean G+C content of R. palustris (65%) (42). Whether the bzd and bcr clusters represent catabolic islands within their respective genomes with distinct G+C contents and evolutionary histories requires further research.
The bzdA gene from Azoarcus sp. strain CIB encodes the benzoate-CoA ligase.
Sequence comparison analyses (Table 3) suggested that the bzdA gene encodes the benzoate-CoA ligase that activates benzoate to benzoyl-CoA as the first step in the anaerobic catabolism of benzoate in Azoarcus sp. strain CIB. To demonstrate this, we PCR amplified a 2.2-kb EcoRI-ScaI fragment containing the bzdA gene from Azoarcus sp. strain CIB and cloned it under control of the Plac promoter into the double-digested EcoRI-SmaI pUC18 vector, producing plasmid pUCBZDA (Table 1). Cell extracts prepared from an E. coli DH10B strain harboring plasmid pUCBZDA showed a high benzoate-CoA ligase activity (8.9 μmol min−1 mg of protein−1), indicating that the overexpressed bzdA gene indeed encodes a benzoate-CoA ligase. Moreover, sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of crude lysates from E. coli DH10B cells containing plasmid pUCBZDA showed the presence of an intense band corresponding to a protein with an apparent molecular mass of 54 kDa (data not shown), in good agreement with the predicted molecular mass for the BzdA protein (57.4 kDa).
As shown previously with equivalent benzoate-CoA ligases from different microorganisms (28, 54), the optimum pH of BzdA was near pH 8.5 and the enzyme was specific for ATP. No activity was observed when ATP was replaced with GTP. The substrate specificity of BzdA (Table 4) revealed that this enzyme is able to activate a wider range of aromatic compounds than other benzoate-CoA ligases described thus far (54). In addition to the three monofluorobenzoate isomers, the BzdA enzyme also shows significant activity with 2-aminobenzoate and two benzoate derivatives, 4-hydroxybenzoate and 3-chlorobenzoate, that are not substrates of other previously reported bacterial benzoate-CoA ligases (54). Remarkably, aromatic heterocycles, such as isonicotinate and nicotinate, showed a reaction rate similar to those of benzoate and 2-aminobenzoate, respectively, a behavior that has not been observed yet for other benzoate-CoA ligases. As reported previously with other benzoate-CoA ligases (1), the thiol-modifying reagent N-ethylmaleimide (1 mM) inhibited the BzdA enzyme, which suggests that the presence of SH groups in this protein is essential for its catalytic activity.
TABLE 4.
Substrate specificity of benzoate-CoA ligase (BzdA)
| Substrate | Relative activity (%)a |
|---|---|
| Benzoate | 100 |
| 2-Fluorobenzoate | 91 |
| 3-Fluorobenzoate | 92 |
| 4-Fluorobenzoate | 84 |
| 2-Chlorobenzoate | 5 |
| 3-Chlorobenzoate | 13 |
| 4-Chlorobenzoate | 3 |
| 2-Aminobenzoate | 35 |
| 3-Hydroxybenzoate | 8 |
| 4-Hydroxybenzoate | 16 |
| Nicotinate | 32 |
| Isonicotinate | 95 |
Relative activities were determined with a coupled assay, except for those for 2-aminobenzoate and the three hydroxybenzoates, which were assayed with a direct assay (see Materials and Methods). The specific activities with benzoate as the substrate were 8.9 μmol min−1 mg of protein−1 for the direct assay and 1.6 μmol min−1 mg of protein−1 for the coupled assay. The values given are averages of at least two measurements with <10% deviation. The following compounds were not substrates of BzdA (<1% relative activity): protocatechuate, 2-hydroxybenzoate, 3-methylbenzoate, 4-aminobenzoate, phenylacetate, 4-hydroxyphenylacetate, 6-hydroxynicotinate, vanillate, phenylglyoxylate, phenylpropionate, phthalate, and pimelate.
As far as we know, this is the first time that a gene encoding a benzoate-CoA ligase from a denitrifying bacterium has been expressed in heterologous hosts. E. coli biocatalysts that overexpress the bzdA gene may constitute an interesting strategy for the enzymatic synthesis of different aromatic-CoA esters from the corresponding aromatic acids (6).
Transcriptional organization and induction of the bzd cluster.
The organization of the genes within the bzd cluster in Azoarcus sp. strain CIB differs from that observed in the equivalent clusters from T. aromatica and R. palustris (Fig. 4). The bzd catabolic genes are arranged in the same orientation and most of them are separated by short distances (Table 3), suggesting that they constitute a single operon. Nevertheless, the lengths of the intergenic regions bzdR-bzdN (582 bp), bzdQ-bzdM (138 bp), bzdV-bzdW (238 bp), bzdW-bzdX (161 bp), and bzdY-bzdZ (156 bp) (Table 3; Fig. 3B) do not allow us to discard the possibility of the existence of promoter activity in such regions. To check the existence of transcriptional promoters in such intergenic regions as well as upstream of the bzdR gene, we PCR amplified them and ligated them to the lacZ reporter gene of the pSJ3 promoter probe vector (Table 1). Whereas pSJ3 derivatives containing the upstream region of bzdR (plasmid pSJ3PR) and the bzdR-bzdN intergenic region (plasmid pSJ3PN) conferred to the host strain E. coli MC4100 the ability to produce blue colonies on media containing the β-galactosidase indicator 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, plasmids harboring other intergenic regions (pSJ3QM, pSJ3VW, pSJ3WX, and pSJ3YZ) did not confer the blue phenotype to E. coli MC4100 (data not shown). These data suggest the presence of a functional promoter upstream of bzdR (PR promoter) and bzdN (PN promoter), which is consistent with the existence of two different transcriptional units, the bzdR regulatory gene and the bzdNOPQMSTUVWXYZA catabolic operon. To further study the transcriptional organization of the bzd cluster, we performed RT-PCR experiments using total RNAs harvested from benzoate-grown cells of Azoarcus sp. strain CIB and primer pairs that were complementary to neighboring genes for amplification of the five intergenic regions mentioned above. Whereas no transcript between genes bzdR and bzdN was observed, RT-PCR fragments of the expected sizes were obtained for genes bzdQ-bzdM, bzdV-bzdW, bzdW-bzdX, and bzdY-bzdZ (Fig. 3C). These results strongly suggest that the bzd catabolic genes are cotranscribed, whereas the bzdR gene is transcribed separately.
The organization of the bzdNOPQMSTUVWXYZA genes as a catabolic operon was also confirmed by checking whether there are polar effects derived from the insertional disruption of the bzdN and bzdY genes in Azoarcus sp. strain CIBdbzdN and Azoarcus sp. strain CIBdbzdY. To this end, we assayed the benzoate-CoA ligase activities in the wild type and two mutant strains growing anaerobically in Casamino Acid-containing MC medium in the presence of benzoate. As expected, crude extracts from the wild-type strain showed benzoate-CoA ligase activity (0.06 μmol of benzoyl-CoA formed min−1 mg of protein−1), whereas no benzoate-CoA ligase activity was detected in extracts from both mutant strains. These data indicate that transposon insertions within the catabolic bzd genes cause polar effects, thus suggesting that these genes constitute an operon.
An analysis of the benzoate-CoA ligase levels in Azoarcus sp. strain CIB cells suggested that the benzoate degradation pathway was inducible (described above). To confirm the induction of the bzd genes, we performed RT-PCR experiments with mRNAs from Azoarcus sp. strain CIB cells grown anaerobically on benzoate and compared the results with those obtained with cells grown on succinate. Whereas there was a clear induction of the bzd catabolic genes in the presence of benzoate, the putative bzdR regulatory gene was expressed constitutively (Fig. 3C). The differential expression of catabolic and regulatory genes is a common feature in the catabolism of aromatic compounds and reflects the complexity of the regulatory mechanisms controlling such catabolic pathways (21).
Carbon catabolite repression of bzd genes.
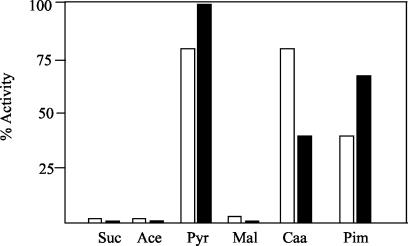
To study the existence of carbon catabolite repression of the anaerobic benzoate degradation pathway in Azoarcus sp. strain CIB, we monitored both the consumption of benzoate and the benzoate-CoA ligase activity in cells grown on benzoate and on mixtures of benzoate plus an additional carbon source. As shown in Fig. 5, whereas malate, succinate, and acetate caused a severe repression of the benzoate-CoA ligase activity, pimelate and Casamino Acids caused a moderate repression and pyruvate was a nonrepressing carbon source. The levels of benzoate-CoA ligase activity matched the rate of benzoate consumption (data not shown). To confirm that the repressive effect of some carbon sources was carried out at the level of transcription from the PN promoter, we constructed the strain Azoarcus sp. strain CIBlacZ by inserting the PN::lacZ translational fusion into the chromosome of Azoarcus sp. strain CIB (see Materials and Methods). β-Galactosidase assays were performed after 48 h of anaerobic growth, and they revealed a repression pattern similar to that obtained with the benzoate-CoA ligase assays (Fig. 5), thus suggesting that PN is the primary target of the catabolite repression effect. As expected, compounds that cannot be used as sole carbon sources under anaerobic conditions, e.g., citrate, glycerol, fructose, and maltose, did not decrease the β-galactosidase levels of Azoarcus sp. strain CIBlacZ cells growing on benzoate (data not shown). All of these data taken together indicate that carbon catabolite repression of aromatic catabolic pathways, previously reported for aerobic catabolism (3, 16, 20), is also a typical feature of organisms under anaerobic growth conditions, although understanding the molecular mechanisms controlling such a repression effect will require further research.
FIG. 5.
Carbon catabolite repression of benzoate degradation in Azoarcus sp. strain CIB. Azoarcus sp. strain CIB and Azoarcus sp. strain CIBlacZ cells were grown anaerobically for 48 h in MC medium containing 1 mM benzoate and 0.4% (wt/vol) of an additional carbon source, i.e., succinate (Suc), acetate (Ace), pyruvate (Pyr), malate (Mal), Casamino Acids (Caa), and pimelate (Pim). Benzoate-CoA ligase (filled blocks) and β-galactosidase (open blocks) activities were measured in Azoarcus sp. strain CIB and Azoarcus sp. strain CIBlacZ cells as described in Materials and Methods, and they are represented as percentages of the activity in cells growing in 3 mM benzoate as the sole carbon source. One hundred percent benzoate-CoA ligase and β-galactosidase correspond to 0.17 μmol min−1 mg of protein−1 and 2,500 Miller units, respectively. The results of one experiment are shown, and values were reproducible in three separate experiments, with standard deviations of <10%.
In summary, in this work we have described for the first time a gene cluster involved in the anaerobic degradation of benzoate in the genus Azoarcus. Moreover, the bzd genes have become an interesting model system for unraveling both the specific regulation and the superimposed control that governs the expression of genes involved in the bacterial catabolism of aromatic compounds under anaerobic conditions.
Acknowledgments
The technical work of I. Alonso, E. Cano, and F. Morante is greatly appreciated. We thank E. García and E. Aporta for their help with pulsed-field gel electrophoresis and oligonucleotide synthesis, respectively. The help of A. Díaz, S. Carbajo, M. Cayuela, and G. Porras with sequencing is gratefully acknowledged. We are indebted to J. Heider for providing us the partial sequence of the bzdQ gene from A. evansii.
This work was supported by grants 07 M/0076/2002 and 07 M/0127/2000 from the Comunidad Autónoma de Madrid and by grants BIO2000-1076, BIO2003-01482, and VEM2003-20075-CO2-02 from the Comisión Interministerial de Ciencia y Tecnología. M. J. L. Barragán was a recipient of a predoctoral fellowship from the Plan Nacional de Formación de Personal Investigador-MCYT; M. T. Zamarro was a recipient of a postdoctoral fellowship from the Comunidad Autónoma de Madrid; and M. Carmona is a holder of the Ramón y Cajal Program of the Spanish Ministerio de Ciencia y Tecnología.
REFERENCES
- 1.Altenschmidt, U., B. Oswald, and G. Fuchs. 1991. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J. Bacteriol. 173:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ampe, F., D. Léonard, and N. D. Lindley. 1998. Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl. Environ. Microbiol. 64:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying Pseudomonad strains K172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 5.Bachellier, S., J.-M. Clément, and M. Hofnung. 1999. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 150:627-639. [DOI] [PubMed] [Google Scholar]
- 6.Beuerle, T., and E. Pichersky. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal. Biochem. 302:305-312. [DOI] [PubMed] [Google Scholar]
- 7.Blum, C. 2000. M.S. thesis. University of Freiburg, Freiburg, Germany.
- 8.Boll, M., and G. Fuchs. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 9.Boll, M., and G. Fuchs. 1998. Identification and characterization of the natural electron donor ferredoxin and of FAD as a possible prosthetic group of benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 251:946-954. [DOI] [PubMed] [Google Scholar]
- 10.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 11.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, and G. Fuchs. 2000. Nonaromatic products from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 275:21889-21895. [DOI] [PubMed] [Google Scholar]
- 12.Brackmann, R., and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:563-571. [DOI] [PubMed] [Google Scholar]
- 13.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 14.Breese, K., M. Boll, J. Alt-Mörbe, H. Schägger, and G. Fuchs. 1998. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur. J. Biochem. 256:148-154. [DOI] [PubMed] [Google Scholar]
- 15.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 16.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5 and Tn10-derived mini-transposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 19.Denome, S. A., C. Oldfield, L. J. Nash, and K. D. Young. 1994. Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. J. Bacteriol. 176:6707-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Díaz, E., A. Ferrández, M. A. Prieto, and J. L. García. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 22.Dörner, E., and M. Boll. 2002. Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J. Bacteriol. 184:3975-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebenau-Jehle, C., M. Boll, and G. Fuchs. 2003. 2-Oxoglutarate:NADP+ oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J. Bacteriol. 185:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egland, P. G., J. Gibson, and C. S. Harwood. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177:6545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein, J. 1993. PHYLIP (phylogenetic inference package), version 3.5.1. Department of Genetics, University of Washington, Seattle.
- 27.Ferrández, A., B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 28.Geissler, J. F., C. S. Harwood, and J. Gibson. 1998. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J. Bacteriol. 170:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gescher, J., A. Zaar, M. Mohamed, H. Schägger, and G. Fuchs. 2002. Genes coding for a new pathway of aerobic benzoate metabolism in Azoarcus evansii. J. Bacteriol. 184:6301-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 31.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 32.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 33.Hess, A., B. Zarda, D. Hahn, A. Häner, D. Stax, P. Höhener, and J. Zeyer. 1997. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 63:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 35.Hurek, T., B. Wagner, and B. Reinhold-Hurek. 1997. Identification of N2-fixing plant- and fungus-associated Azoarcus species by PCR-based genomic fingerprints. Appl. Environ. Microbiol. 63:4331-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khemici, V., and A. J. Carpousis. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51:777-790. [DOI] [PubMed] [Google Scholar]
- 37.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 38.Koch, J., W. Eisenreich, A. Bacher, and G. Fuchs. 1993. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 211:649-661. [DOI] [PubMed] [Google Scholar]
- 39.Koch, J., and G. Fuchs. 1992. Enzymatic reduction of benzoyl-CoA to alicyclic compounds, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 205:195-202. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 41.Laempe, D., M. Jahn, K. Breese, H. Schägger, and G. Fuchs. 2001. Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica. J. Bacteriol. 183:968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 43.Lobner-Olesen, A., and M. G. Marinus. 1992. Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J. Bacteriol. 174:525-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 45.Mandic-Mullec, I., L. Douckhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 177:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mechichi, T., E. Stackebrandt, N. Gad'on, and G. Fuchs. 2002. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 178:26-35. [DOI] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Niemetz, R., U. Altenschmidt, S. Brucker, and G. Fuchs. 1995. Benzoyl-coenzyme-A 3-monooxygenase, a flavin-dependent hydroxylase. Purification, some properties and its role in aerobic benzoate oxidation via gentisate in a denitrifying bacterium. Eur. J. Biochem. 227:161-168. [DOI] [PubMed] [Google Scholar]
- 49.Rainey, P. B., I. P. Thompson, and N. J. Palleroni. 1994. Genome and fatty acid analysis of Pseudomonas stutzeri. Int. J. Syst. Bacteriol. 44:54-61. [DOI] [PubMed] [Google Scholar]
- 50.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 52.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 54.Schühle, K., J. Gescher, U. Feil, M. Paul, M. Jahn, H. Schägger, and G. Fuchs. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 185:4920-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schühle, K., M. Jahn, S. Ghisla, and G. Fuchs. 2001. Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 183:5268-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song, B., M. M. Häggblom, J. Zhou, J. M. Tiedje, and N. J. Palleroni. 1999. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of A. toluvonrans sp. nov. and A. toluclasticus sp. nov. Int. J. Syst. Bacteriol. 49:1129-1140. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 59.Wilbur, W. J., and D. J. Lipman. 1983. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. USA 80:726-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziegler, K., R. Buder, J. Winter, and G. Fuchs. 1989. Activation of aromatic acids and aerobic 2-aminobenzoate metabolism in a denitrifying Pseudomonas strain. Arch. Microbiol. 151:171-176. [Google Scholar]