Abstract
Mounting evidence has called into question our understanding of the role that the central dogma of molecular biology plays in human pathology. The conventional view that elucidating the mechanisms for translating genes into proteins can account for a panoply of diseases has proven incomplete. Landmark studies point to epigenetics as a missing piece of the puzzle. However, technological limitations have hindered the study of specific roles for histone post-translational modifications, DNA modifications, and non-coding RNAs in regulation of the epigenome and chromatin structure. This feature highlights CRISPR systems, including CRISPR-Cas9, as novel tools for targeted epigenome editing. It summarizes recent developments in the field, including integration of optogenetic and functional genomic approaches to explore new therapeutic opportunities, and underscores the importance of mitigating current limitations in the field. This comprehensive, analytical assessment identifies current research gaps, forecasts future research opportunities, and argues that as epigenome editing technologies mature, overcoming critical challenges in delivery, specificity, and fidelity should clear the path to bring these technologies into the clinic.
Keywords: Epigenome editing, epigenome engineering, CRISPR, CRISPR-Cas9, optogenetics, epigenetics, histone and DNA epigenetic modifications, transcription activation, transcription repression, CRISPR systems for epigenome editing
Introduction
The realization that phenotypic inheritance is not merely a consequence of genetic processes, but implicates responses to environmental stimuli is one of the great discoveries of the twentieth century. Conrad Waddington's seminal Drosophila melanogaster experiments establishing phenotypic plasticity induced by chemical or temperature stimuli [1] cemented the notion of "epigenetic landscapes" [2] as the supreme phenomenon driving cellular differentiation, and the molecular basis bridging the gap between genotype and phenotype. Today, epigenetics is a dynamic and prolific field broadly aimed at studying fundamental processes related to mitotic and meiotic stable and heritable changes—in gene expression or cellular phenotype—that occur without alteration of DNA sequences [3].
An explosion of research spanning the last two decades has revealed remarkable insights into epigenetic mechanisms, including the discovery of myriad phenotypic outcomes associated with histone post-translational modifications (PTMs), DNA modifications, and non-coding RNAs (ncRNAs). Together, these concomitant processes orchestrate a highly complex system that mediates organization of chromatin structure and the epigenetic regulation of two meters of genomic DNA tightly packed into a 5µm [4] three-dimensional region. Despite the progress made, the precise functional roles for these epigenetic processes in development, cellular programming, biology, disease, and medicine still remain poorly understood. A pressing need exists for widely accessible methods to overcome current technological limitations, which have hindered our ability to study epigenetic regulation and gene expression at local and genome-wide levels.
In recent years, the CRISPR-Cas9 (clustered, regularly interspaced, short palindromic repeats (CRISPR)-CRISPR-associated protein 9) system has been adopted as a robust and versatile genome editing tool [5] for bacterial [6] and eukaryotic [7] organisms. The CRISPR-Cas9 complex is an adaptive immunity, type II CRISPR-Cas system in bacteria and archaea that uses antisense RNAs to recognize and cleave foreign DNA [8]. CRISPR-based technologies have become popular for scientific research and the CRISPR-Cas9 system has recently been repurposed for epigenome editing by engineering a nuclease-null or “dead” Cas9 (dCas9) (Figure 1), which permits targeting specific DNA loci without cleavage [9]. CRISPR-based epigenome editing technologies are poised to become powerful tools to further our understanding of the roles that epigenetic marks and effectors play in cancer and other human diseases [10,11]. More importantly, this new technology holds great promise for the future of medicine.
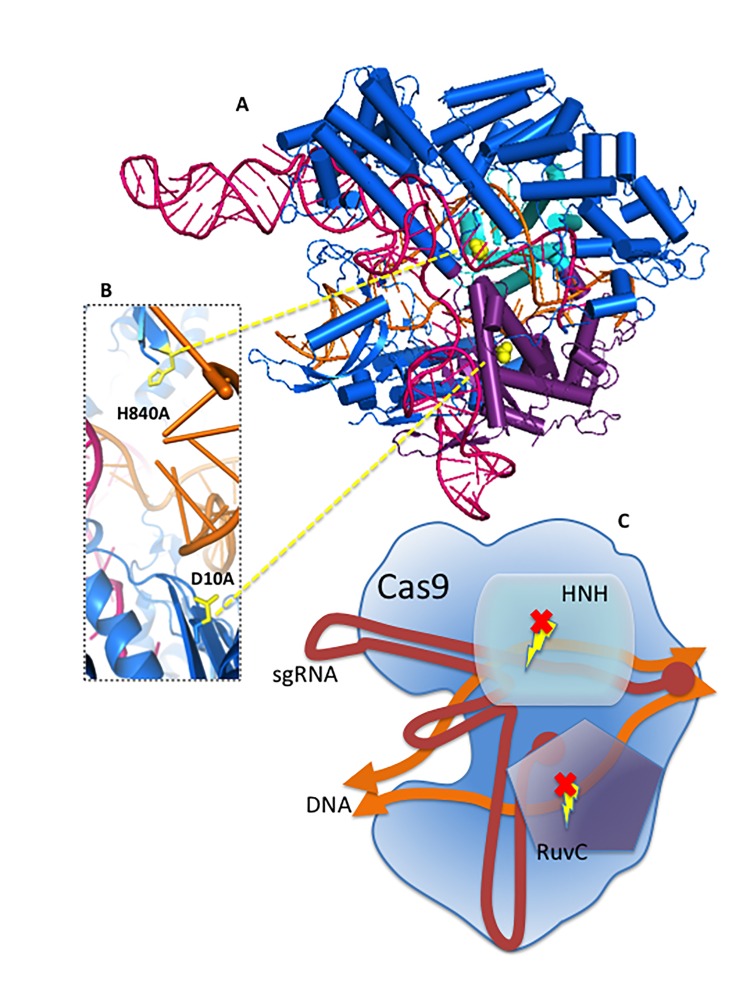
Figure 1.
Structural representation of the nuclease-null Cas9 (dCas9) from S. pyogenes. A. Crystal structure of Cas9 in complex with a single guide RNA (red) and its target DNA (orange). The HNH (cyan) and RuvC (purple) catalytic domains are shown. The non-catalytic regions of Cas9 are colored in blue. Mutation of the catalytic residues (D10A and H840A) that render Cas9 inactive (dCas9) are colored in yellow. B. Close-up view of the active site and position of the catalytic residues shown in A. C. Schematic representation of nuclease-null Cas9 in complex with sgRNA and target DNA; colors as shown in A. [PDB 4OO8].
This feature highlights CRISPR systems, including CRISPR-Cas9, as novel tools for targeted epigenome editing. The history behind the concept of epigenome editing, which emerged with the advent of Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs), has been the subject of two recent reviews [12,13]. As a result, I will not rehash what has already been discussed in great detail. Instead, this paper identifies current research gaps and critically examines shortcomings in the literature that have not been previously analyzed. Furthermore, it emphasizes recent discoveries that expand the catalog of DNA modifications and ncRNAs that can be interrogated using epigenome editing technologies. A summary of recent developments in the field, including integration of chemical, optogenetic, and functional genomic approaches to explore new therapeutic opportunities is provided briefly. Overall, this piece underscores the importance of mitigating current limitations in the field, forecasts future research opportunities beyond CRISPR-Cas9 systems, and argues that as epigenome editing tools mature, overcoming critical challenges in delivery, specificity, and fidelity should clear the path to bring these technologies into the clinic.
Targeting Histone Epigenetic Modifications
Epigenome editing technologies have demonstrated the feasibility of targeting histone epigenetic modifications. Two seminal papers published in 2015 provide important insights into the capabilities of repurposing the CRISPR-Cas9 system as an epigenome editing platform to trigger changes in transcriptional activation or repression by targeting histone PTMs known to play important roles in chromatin structure and accessibility. One involved the activation of gene expression using a nuclease-null dCas9 tethered to an acetyltransferase (HAT) enzyme that catalyzes direct covalent modification of ε-lysine residues on histone tails [14]. In contrast, the other demonstrated gene silencing using a nuclease-null dCas9 fused to the Krüpel-associated box (KRAB), a naturally-occurring transcriptional repression domain involved in recruiting a heterochromatin-forming complex that mediates histone methylation and deacetylation [15].
Both approaches fuse a catalytically inactive dCas9 to an epigenetic effector. However, activation is achieved by direct catalysis to transfer an acetyl moiety onto histone lysine residues, while repression is mediated by indirect recruitment of enzymes that catalyze the transfer of methyl or removal of acetyl groups from histone residues. I discuss these epigenome editing approaches below to highlight the progress and shortcomings of the concept of targeting histone marks to study epigenetic regulation.
Transcriptional Activation
Manipulation of transcription and gene expression without directly modifying DNA sequences has been a long-standing goal in chromatin biology. Previous efforts to control transcriptional activation led to the creation of programmable, viral-based, transactivation effector domain—e.g., VP16 and its VP64 tetrameric form—fusions to TALEs [16] and Zinc Finger [17] proteins. These tools were limited by their physical and biological properties, namely, the fact that VP16 and VP64 are not chromatin modifying enzymes, but rather recruit chromatin remodeling factors that facilitate access to specific chromatin loci [18]. In the last few years, however, the first reports of TALE fusions to chromatin modifiers were published as proof-of-concept for site-specific epigenome editing [18-20], thereby opening the path for adoption of these principles to CRISPR-Cas systems.
In 2015, a study reported the use of a dCas9 fusion to the catalytic histone acetyltransferase (HAT) core of p300 [14] (Figure 2). dCas9-p300 Core fusions were shown to be more potent at achieving transactivation of genes from proximal and distal enhancers than engineered transcription factors made with activation domains [14]. Moreover, the fusion established robust gene activation by a single guide RNA (sgRNA) at promoters and characterized enhancers [14]. The study provides an important contribution to the scientific literature and marks an important step toward programmable, facile, epigenome editing of histone PTMs. However, a few significant shortcomings not discussed in the article or subsequent literature can be identified following closer scrutiny of the findings and conclusions averred by the authors. I highlight some of the shortcomings later in this section.
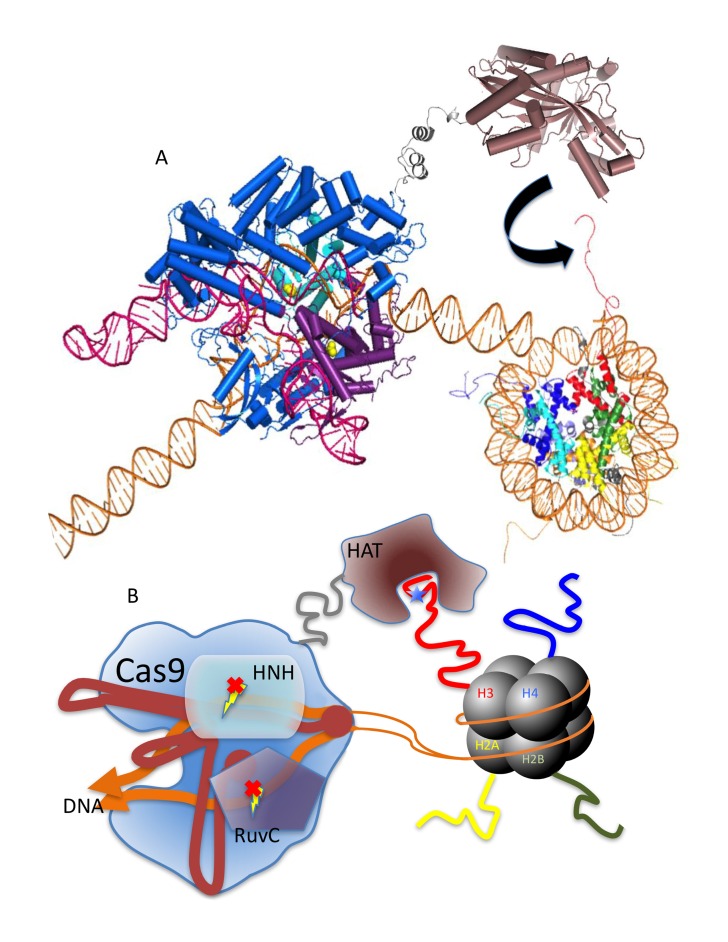
Figure 2.
Schematic representation of epigenome editing using dCas9 fused to an epigenetic effector. A. dCas9 (colors are the same as in Figure 1) binds to target DNA without triggering double-stranded DNA cleavage. An epigenetic effector—p300 histone acetyltransferase (HAT) in this drawing (brown)—is fused to dCas9 by a linker region (gray). Once positioned near the target epigenetic loci, the effector catalyzes covalent modification of the target histone tail on nearby nucleosomes (the reaction is represented by the black arrow). B. Cartoon illustration of A. Covalent modification illustrated by a blue star on the substrate histone tail. [PDB 4OO8, 3BIY, 1KX5].
Transcriptional Repression
Targeting of promoter regions or other cis-regulatory elements using the DNA binding domains of Zinc Finger proteins [21,22] and TALEs [19,23] has been reported in the past as capable of achieving effective gene silencing. More recently, evidence of effector-mediated gene repression using CRISPR-dCas9 fusions has surfaced to demonstrate proof-of-concept for CRISPR-mediated epigenome editing to silence proximal and distal regulatory elements [15,24].
Two studies published in 2015 offer differing CRISPR-based approaches to repress transcription. The first utilized a nuclease-null dCas9 fused to a KRAB repressor domain [15], a motif commonly found in eukaryotic Zinc Finger proteins [25]. This type of silencing is achieved by KRAB-mediated recruitment of complexes that trigger formation of heterochromatin at specified loci, primarily through protein-protein interactions between the KRAB-associated Protein 1 (KAP1) corepressor and other factors [26-28]. The study sought to interrogate the genome-wide specificity and chromatin remodeling activity catalyzed by dCas9-KRAB at the HS2 enhancer, a 400 bp distal regulatory element comprising several enhancer regions that orchestrate expression of hemoglobin subunit genes in erythroid cells throughout development [15]. Targeting of the HS2 enhancer by dCas9-KRAB fusions was shown to silence the expression of globin genes with relatively little off-target effects in expression of non-targeted genes [15]. Additionally, the study revealed decreased levels of chromatin accessibility at targeted promoters and enhancers, as well as increased deposition of H3K9me3 repressive epigenetic marks within the selected HS2 region [15].
The second study sought to compare the repressive properties of dCas9-KRAB and a fusion of dCas9 to the Lysine-specific demethylase 1 (LSD1) histone demethylase, which is responsible for catalyzing the elimination of methyl marks on H3K4 [29] and H3K9 [30] residues, and has been associated with repression of enhancer regions [24]. The study demonstrated that dCas9-LSD1 triggers repression through decommissioning of target enhancers in an effector-dependent manner without significant disturbance of local chromatin architecture, whereas dCas9-KRAB repression is likely the result of promoter silencing or heterochromatin spreading [24]. Interestingly, these results stand in contradiction to findings reported in the dCas9-KRAB HS2 enhancer study [15], which concluded that the dCas9-KRAB system can trigger deposition of H3K9me3 marks at endogenous enhancers, suggesting that epigenome editing via dCas9-KRAB-mediated histone methylation may be context and cell-type dependent.
Current Limitations
Epigenome editing directed by CRISPR-dCas9-p300 Core, dCas9-KRAB, and dCas9-LSD1 fusions detailed in the studies above provides tangible evidence of the feasibility of developing CRISPR-based technologies for targeted manipulation of histone epigenetic modifications. However, this proof-of-concept underscores the fact that epigenome editing has yet to come full-term. Whereas genome editing research is currently in its infancy, at this point in time, epigenome editing can be considered to be at an embryonic stage. Therefore, to propel epigenome editing forward into the clinical stage, CRISPR researchers must heed research gaps and deficiencies hidden in these foundational studies. Awareness of early technological shortcomings will permit protocol optimization and lead to data reproducibility that will prove essential in building the next generation of epigenome editing technologies.
With respect to the claim that highly specific epigenome editing was achieved across promoters and enhancers genome-wide, the data presented by the CRISPR-dCas9-p300 Core study merely corroborates targeting of desired DNA loci, not the deposition of unique epigenetic marks on histone tails. Although the authors correctly point out that acetylation of histone H3 at lysine 27 (H3K27ac) is catalyzed by p300 [14], they did not mention or consider the inherent promiscuity of the enzyme. The HAT domain of the p300 transcriptional coactivator is all but highly specific and has been shown to catalyze the acetylation of a laundry list of residues on all four—H2A, H2B, H3, and H4—core histones [31-33], transcription factors [34], and other proteins [35] to regulate a number of key biological processes. Thus, the allegation that unique targeted acetylation of H3K27 occurred at enhancers and promoters is only as good as the concession that H3K27ac was the one and only PTM the authors sought to identify. Given the poor specificity of the p300 HAT toward histone substrates, it is likely that other putative acetylation events would have been detected had chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) been performed using other histone PTM antibodies.
Despite evidence for robust transactivation at target genes using dCas9-p300 Core [14], it can hardly be said that highly specific epigenome editing took place in the transfected cell lines. The touchstone principle of epigenome editing concerns uncovering direct functional roles for unique PTMs in specific contexts without encumbrance from correlative approaches or the potential secondary effects driven by unintended factors. Besides transactivation of the target genes, non-H3K27 acetylation could likely have unspecified functional roles including recruitment of specific readers of other acetylation marks, structural effects on higher order chromatin structure, or PTM-dependent signaling cascades. This highlights a key obstacle for current CRISPR-directed epigenome editing technologies: contrary to the prevailing wisdom in genome editing, mitigation of off-target effects in epigenome editing is not exclusively related to the DNA-binding activity of dCas9 at specific loci, but also off-target effects related to deposition of PTM marks at specified nucleosomes with spatio-temporal precision.
The potential for non-specific hyperacetylation of nucleosomal substrates by dCas9-p300 Core also raises concerns about antibody specificity during epigenetic experiments. It has been reported recently that some commercially available histone PTM antibodies cannot frequently detect their intended targets [36]. Although the issue of histone antibody specificity arguably applies to many epigenetic experiments, the problem is of particular importance in dCas9 fusions to catalytic domains. For instance, evidence shows that many site-specific H4-acetyl antibodies preferentially bind to poly-acetylated histone peptides [36]. Based on these observations, hyperacetylation of proximal nucleosomes by a tethered dCas9-p300 Core HAT might confound ChIP-qPCR experiments using anti-H3K27ac antibodies. Thus, potential non-H3K27 acetylation events may have played a role in detecting highly enriched regions of H3K27ac at the enhancers and promoters surveyed.
The underlying principles applicable to transcriptional activation also apply to epigenome editing involving transcriptional repression and regulation of gene expression. Similar to HATs, other enzymes including histone deacetylases, methyltransferases, demethylases, kinases, phosphatases, ubiquitin and SUMO ligases, etc. have varying degrees of substrate specificity, and involve the use of commercial antibodies to detect PTM marks deposited on histones.
Although the effectiveness of transcriptional repression directed by epigenome editing via the dCas9-KRAB system has been aptly demonstrated, the use of KRAB domains as scaffolding platforms for recruitment of heterochromatin-forming complexes raises numerous questions, particularly concerning the dCas9-KRAB system's future applicability in clinical settings. Compared to the relatively straight forward characterization of functional roles for individual epigenetic effectors—e.g. LSD1—that catalyze either deposition or removal of specific histone epigenetic marks responsible for transcriptional suppression, the manipulation and control of functional outcomes associated with KRAB-recruited protein complexes working in tandem to trigger suppression is inherently more intricate.
Consider the KAP1 corepressor, a multi-domain protein that features a putative catalytic RING finger domain—a common E3 ubiquitin ligase motif that mediates the transfer of ubiquitin from an E2 ubiquitin-conjugating enzyme to substrate [37]—and two epigenetic reader domains [38]—a PHD domain and a bromodomain—capable of reading methyl- [39] and acetyl- lysine [40] residues. Recently, the KAP1 RING domain was characterized as a specific small ubiquitin-like modifier (SUMO) E3 ligase involved in the regulation of innate immunity [41].
Although SUMOylation has been linked to transcriptional repression [42,43] and, thus, fits within the overall purpose of the dCas9-KRAS system, new SUMOylation functional roles beyond repression have been reported, such as activation of DNA damage signaling cascades [44]. Like ubiquitination, it is likely that SUMOylation is involved in a widespread range of context-dependent, substrate functional roles including proteasome-dependent proteolysis, signaling, assembly, cellular localization, and others [37].
Consequently, dCas9-KRAB epigenome editing aimed at repression could be confounded by physical and biochemical events that obfuscate the specific contributions of each component of the system's repressing machinery. This could have serious repercussions for clinical therapies, in which CRISPR-dCas9-KRAB-mediated epigenome editing is capable of eliciting a desired silencing event, but triggers a deleterious secondary effect caused by unknown or unforeseen processes. Indeed, on this point, KRAB's corepressor, KAP1, is known to serve as a scaffold to recruit a myriad of factors and complexes associated with repressive epigenetic states including heterochromatin protein 1 (HP1) [45], SETDB1—a methyltransferase that catalyzes deposition of methyl histone marks on H3K9 [46] and other substrates [47], NuRD—a corepressor complex that catalyzes histone deacetylation and ATP-dependent chromatin remodeling [48], and N-COR1—a histone deacetylase complex [49]. KAP1 has also been linked to functional roles in DNA double-stranded repair [50], restriction of retrovirus replication [51], and regulation of self-renewal in embryonic stem cells [52].
Given the vast range of KRAB- and KAP1-mediated interactions, it is not far-fetched to foresee hurdles associated with epigenome editing, particularly when repression is orchestrated by multiple enzymes of varying degrees of substrate specificity, and proteins involved in long-range interactions. This is a significant concern—even if recruitment of repressive complexes is only targeted to a specific locus—mainly because of potential effects on chromatin architecture, and looping of discrete distant regulatory regions. Thus, individual characterization of functional roles for repressive epigenetic effectors using CRISPR-based methods will be an important first step in the path toward targeting of concomitant repressive processes.
Overcoming Current Limitations
As pointed out above, major limitations currently exist in the development and future translational applications of epigenome editing technologies. Nevertheless, the scientific community is working hard to try to tackle these obstacles head-on. As new research and technologies improve, it is likely that current impediments to efficient epigenome editing will gradually be surmounted.
One promising and appealing technique to ameliorate the problem of antibody specificity and reproducibility that often plagues epigenetic experiments was recently published and highlights the feasibility of using recombinant epigenetic readers as reliable substitutes for histone PTM antibodies [53]. The study validated the use of recombinant TAF3 PHD domain, a motif that specifically reads H3K4me3 histone marks [54], as a robust anti-H3K4me3 affinity reagent in techniques ranging from western blotting to ChIP experiments coupled with qPCR and deep sequencing [53]. This study illustrates a key methodology to circumvent current problems associated with commercial antibodies. In the last decade and a half, the epigenetics research community has characterized numerous epigenetic readers—many of which exhibit high substrate specificity and can be easily produced recombinantly—that could be repurposed as anti-histone PTM reagents to abolish the need for substandard commercial antibodies.
Other existing limitations concerning inadequate antibody specificity for epigenome editing experiments are likely to become less prominent with increased collaborations and partnerships between industry and academia that give rise to common interests for developing highly specific antibodies. The research community ought to speak in unison to exercise its commercial power and demand improved standardized protocols and internal quality controls to minimize batch-to-batch variations commonly seen among commercial suppliers of antibodies, especially polyclonal ones. Given the great significance of using reliable and adequate antibodies that facilitate reproducibility of results by independent researchers, it is also imperative that the research community avail itself of open source resources aimed at characterizing and validating antibody performance in epigenetic experiments such as the Antibody Validation Database and the Antibody Specificity Database [36,55].
The limitations surrounding substrate specificity of HAT and other epigenetic effector enzymes present a greater obstacle to the development of efficient and programmable CRISPR-mediated epigenome editing. Detailed structural information and cumbersome protein engineering is likely necessary to boost efforts to reduce substrate promiscuity and increase cognate target selectivity among certain epigenetic effectors. Structural characterization of readers, writers, and erasers of the epigenome—in their isolated states as well as within the complexes they form—is also critical to guide structure-based drug design efforts aimed at developing chemical probes to modulate epigenetic regulatory proteins. Unfortunately, many of these endeavors are time-consuming, labor-intensive, and require expenditure of considerable resources, which may not always be available to researchers. As CRISPR-mediated epigenome editing technologies mature, it will also be important to incorporate functional genomic, pharmacogenomic, and high-throughput approaches to ensure studies properly identify predictable events, as well as non-specific epigenome editing of target loci.
Interrogation of DNA Epigenetic Modifications
Following propitious reports describing the viability of CRISPR-mediated epigenome editing of histone PTMs to trigger transcriptional activation or repression in the last year, CRISPR researchers have swiftly begun to probe DNA epigenetic modifications at select regulatory elements. In 2016, several reports from independent research teams established—almost simultaneously—proof-of-concept for up- or down-regulation of target genes elicited by dCas9 fusions to catalytic domains, directing either methylation or demethylation of CpG islands that span promoter regions [58,59,66,67].
DNA Methylation
Programmable DNA methylation at specific CpG islands has been reported via fusion of dCas9 to the catalytic domain of the de novo DNA methyltransferase 3A (DNMT3A), a cytosine-5 methyltransferase [56,57], at the human CDKN2A and ARF promoters, and the mouse Cdkn1a promoter [58]. This dCas9-DNMT3A study showed that the induction of DNA methylation was confined within a region spanning approximately 50 bp of the sgRNA target site, and is strongest between adjacent and inwardly directed sgRNA binding sites [58]. The induced methylation at the promoter sites was sufficient to lower expression of all three genes targeted and generated little off-target activity [58].
Similarly, a separate published study using a dCas9-DNMT3A fusion, featuring a Gly4Ser flexible linker, showed efficient and specific CpG island methylation of the BACH2 and IL6ST promoters [59]. Notably, the study provides evidence that targeting multiple promoter sites by co-expression of diverse sgRNAs produces synergistic effects that lead to increased methylation of larger regions of the promoter [59]. Despite previous attempts to alter methylation patterns using older technologies such as Zinc Finger proteins and TALEs [60-62], which suffered from low induction levels of methylation and off-targeting effects, CRISPR-mediated approaches appear to be more robust and reliable at inducing methylation across promoters. Moreover, CRISPR-based methods can help overcome the DNA binding domain sensitivity to CpG methylation that have been shown to occur with TALEs [63].
DNA Demethylation
Promoter-specific demethylation of CpG islands has been shown to increase the expression of endogenous genes using TALE-demethylase fusions [20,64]. However, due to technological shortcomings of TALEs—e.g., the need for intricate protein engineering, high-throughput epigenome editing to regulate demethylation of promoter regions has been an unreachable goal. Two reports recently described alternative approaches using CRISPR-dCas9 fusions to the catalytic domain of Ten-Eleven translocation dioxygenase 1 (TET1), a maintenance DNA demethylase that prevents abnormal hypermethylation of CpG islands [65].
The first study involved using dCas9-TET1 alongside various sgRNAs targeting the promoter region of BRCA1, a tumor suppressor gene [66]. Experiments showed that demethlyase-directed epigenome editing selectively targeted the BRCA1 promoter and induced robust gene expression [66]. The second study used dCas9-TET1 to direct promoter-specific DNA demethylation as well [67]. However, this report also disclosed a method to exploit sgRNA plasticity by inserting bacteriophage MS2 RNA elements—which bind the MS2 coat proteins—into the scaffolding of the sgRNAs [67]. Guided by these engineered sgRNAs, dual and simultaneous recruitment of dCas9-TET1 and MS2-TET1 fusions to targeted loci was shown to occur [67]. This unusual CRISPR-based demethylase system was able to significantly upregulate RANKL, MAGEB2, and MMP2 gene expression through demethylation of their respective promoters with little off-target activity [67]. Engineering modular elements into sgRNAs could offer many advantages for high-throughput screening of genome-wide targets for epigenome editing.
Using CRISPR-mediated Approaches to Probe New DNA Epigenetic Modifications
As the above studies demonstrate, it is evident that epigenome editing can also be used as a tool to tease out the function and mechanisms of epigenetic marks deposited directly on DNA [58,59,66,67]. Early efforts have currently focused on studying epigenetic effectors responsible for the regulation of patterns commonly associated with the well-established methylation of cytosine deoxynucleotides at the C-5 position to create 5-methyldeoxycytosine (m5dC) [68]. However, CRISPR-based approaches could also be used to study functional roles for other marks. To this effect, recent findings have expanded our knowledge of the range of DNA modifications, which had previously been confined to m5dC. Catalytically inert Cas9 fusions and future editing technologies will likely facilitate ripe opportunities to further our understanding of underlying mechanisms inherent in a diverse repertoire of newly discovered DNA epigenetic marks.
Oxidation of m5dC Derivatives
The m5dC epigenetic mark is covalently attached to DNA at CpG and non-CpG sites by a class of enzymes known as DNA Methyltransferases (DNMTs) [69], and has been shown to mediate heritable transcriptional repression in the cell [70]. For nearly six decades, m5dC was thought to be the only DNA modification in higher eukaryotes until the recent discovery that Ten-Eleven Translocation (TET) “writer” proteins are capable of catalyzing the sequential oxidation of m5dC to form 5-hydroxymethylcytosine (hm5C) [71], 5-formylcytosine (f5C) [72], and 5-carboxylcytosine (ca5C) [72,73].
Although very little is known about the function of these m5dC oxidation derivatives, studies report the accumulation of such epigenetic marks at major satellite repeats [74], within exons [75], and near transcriptional start sites enriched with dual histone H3 lysine 27 trimethylation (H3K27me3) and histone H3 lysine 4 trimethylation (H3K4me3) marks [75]. Following the identification and characterization of the m5dC derivatives and their “writers,” scientists have discovered sets of “reader” proteins that exhibit preferential and specific binding to hm5C, f5C, or ca5C [76,77]. Moreover, the marks appear to be involved in recruitment of distinct transcription regulators and DNA repair proteins in mouse embryonic stem cells [76]. These findings suggest that m5dC-oxidized intermediates play important roles in regulation of transcription, control of gene expression, and chromatin dynamics.
Given the recent reported findings using CRISPR-mediated epigenome editing in DNA methylation, it is quite likely that future studies probing functional roles for m5dC derivatives will shed insights into the molecular and mechanistic underpinnings of these DNA epigenetic modifications.
Discovery of N6-methyldeoxyadenosine
Within the last year, a trio of papers marked the debut of methylation of adenine deoxynucleotides at the N-6 position of the purine ring as the latest epigenetic mark decorating the DNA of higher eukaryotes [78-80]. N6-methyladenosine had previously been known as a ubiquitous RNA modification found in viruses [81] and eukaryotes [82,83], including mammals [84,85]. However, although N6-methyldeoxyadenosine (m6dA) had been detected as an epigenetic feature of bacteria, it had not been identified in higher eukaryotes. Thanks in part to technological advances in methods of detection to overcome low sensitivity thresholds, m6dA has now been identified in the genomes of Caenorhabditis elegans [78], Drosophila [79], and Chlamydomonas [80]. Interestingly, unlike m5dC, which is associated with gene repression, m6dA appears to localize at ApT di-nucleotides near transcription start sites and is linked to gene activation [80]. The identification of m6dA in C. elegans also dispels the prevailing view that the nematode's genome lacks any DNA methylation and suggests possible mechanisms of m6dA-mediated epigenetic inheritance in eukaryotes [78].
During the first half of 2016, two additional studies established that the m6dA mark is widely distributed across the genomes of frogs, mice, mouse embryonic stem cells, and humans [86,87]. Remarkably, the published reports on Drosophila [79] and Chlamydomonas [80] squarely contradict findings in vertebrates and mammals [86,87]. Whereas m6dA appears to be enriched near transcription start sites and is associated with gene activation in simpler eukaryotes and invertebrates, m6dA seems to play opposite roles in vertebrates and mammals in which the mark triggers transcriptional suppression [87]. Future work will likely reveal a rationale for this peculiar contradiction. A hypothesis proposes that m6dA acquired new functions during evolution [87]. But there is also the possibility that m6dA activation or repression could depend on the type of tissue or species assayed.
CRISPR-mediated epigenome technologies can now be used by researchers to tease out answers to many important questions posed by these latest studies focusing on previously uncharacterized DNA modifications. Now that putative “writers,” “readers,” and “erasers” of m6dA marks have been reported, chromatin biologists and structural biochemists will lead efforts to further characterize the molecular and structural underpinnings of the repertoire of factors involved in this novel mode of epigenetic regulation as well as its potential unique roles in simpler and more complex eukaryotes.
Uncovering New DNA Epigenetic Modifications
The discovery of hm5C, f5C, ca5C, and m6dA marks, alongside the ensuing development of modern high-throughput technologies and highly sensitive methods to detect transient DNA modifications, brings exciting prospects for future research. It may be the case that other types of DNA epigenetic modifications await characterization in higher eukaryotic organisms. For instance, hypermodification to produce the DNA base β-d-glucosyl-hydroxymethyluracil, commonly known as base J [88], has been documented in unicellular eukaryotes as an epigenetic factor regulating Polymerase II (Pol II) transcription initiation [89]. Phosphothioration, which involves the incorporation of sulfur directly into the DNA phosphate backbone, has been shown to occur pervasively in bacterial genomes [90]. Earlier in 2016, insertion of 7-deazaguanine derivatives into DNA was shown to be a previously unrecognized bacterial and phage defense system targeting foreign DNA [91]. Although neither of these modifications have been identified in higher eukaryotes to date, it is possible that their future characterization will be facilitated by upcoming breakthroughs in detection methods. After all, hm5C was first discovered in phage DNA nearly sixty-five years ago [92], but only recently documented in Purkinje neurons and the human brain [93]. Similarly, m6dA was known to occur in bacterial DNA for decades before evidence of m6dA in higher eukaryotes surfaced within the last year [78-80,86,87].
Because DNA modifications likely occur in distinct contexts, probing functional roles of specific modifications in different tissues, settings, and genomic regions will be pivotal to linking sets of epigenomic marks to phenotypic outcomes. Furthermore, dynamic and reversible DNA epigenetic modifications are likely to not only affect global processes such as replication, transcription, and gene regulation, but also chromatin dynamics and the three-dimensional structure of the DNA biopolymer. This could be significant in ways we do not yet understand.
For example, earlier in 2016, the first reported crystal structure of a DNA deoxyribozyme revealed an intricate network of tertiary interactions stabilizing the DNA catalyst [94]. It is not difficult to imagine the structural consequences triggered by DNA-modified steric interference, which may ultimately promote or inhibit unknown DNA catalytic mechanisms. Thus, the new generation of epigenome editing biotechnologies aimed at interrogating the functional roles for known and yet-unknown DNA epigenetic effectors (i.e., writers, readers, and erasers) will prove invaluable to elucidate biological processes that have remained out of our technological reach.
Probing Functional Epigenetic Roles for Non-Coding RNAs
Epigenome editing can now be used to probe functional roles for non-coding RNAs (ncRNAs) involved in gene regulation and epigenetic chromatin dynamics. Although we have long known that RNA is an integral part of chromatin, our understanding and appreciation of the role RNA plays in chromatin regulation has only begun to form within the last decade [95,96]. Few recent studies demonstrate the feasibility of using CRISPR-systems to probe ncRNA function by repurposing the sgRNAs that bind dCas9 as scaffolding molecules to trigger loci- specific regulation [97,98]. Orthogonal incorporation of engineered sgRNA molecules featuring protein-binding RNA aptamers—e.g., MS2, PP7—has been established as an efficient platform for parallel gene regulation in mammalian and yeast cells [97]. Protein-binding cassettes, RNA aptamers, and long ncRNAs at least 4.8 kb long have been used to build CRISPR-Cas9 complexes that may enable precise ectopic targeting of functional RNAs and ribonucleoprotein complexes to specific genomic loci [99]. Incorporation of protein-binding motifs and ncRNAs directly into the stem-loop structures of sgRNAs—at the 5' or 3' positions—is likely to usher in a long-awaited era of ncRNA functional characterization relevant to chromatin regulation.
Chemical- and Photo-Inducible Epigenome Editing
Controlling gene activation or repression in a spatio-temporal fashion to mimic the natural chromatin dynamics of gene expression is the next frontier of epigenome editing-based biotechnologies. To date, several publications detail the use of CRISPR-Cas9 systems for chemical-[100] and photo-inducible [101-103] genome editing and transcriptional regulation. Chemically inducible epigenome editing comprises the use of small molecules that act as chemical inducers. For example, rapamycin-sensitive dimerization domains have been shown to chemically induce CRISPR-Cas9-based genome editing and transcription regulation when Cas9 is split into two fragments that, upon chemical induction, reconstitute a full-length, catalytic nuclease [100]. This approach could also be applied with other chemically inducible dimerization domains, such as abscisic acid or gibberalin sensing domains [100]. A myriad of small molecules have been established as chemical inducers such as anhydrotetracycline [9], doxycycline [104], rapamycin [100], and steroid hormone receptor ligands [105], all of which could be exploited in epigenome editing platforms.
Many photo- or light-inducible “optogenetic” approaches typically rely on a light-dependent dimerization system comprising the proteins cryptochrome circadian clock 2 (CRY2) and its binding partner CIB1 [106]. Using this approach, groups have tested the feasibility of fusing a split dCas9 together with the CRY2-CIB1 system for photoactivation of genome editing [101], or demonstrated that CRY2 and CIB1 can be separately fused to dCas9 and a transcriptional activator respectively for subsequent photo-inducible transcriptional regulation [102,103]. Interestingly, a new approach using engineered protein photoswitches (“magnets”) to enhance photo-induced heterodimerization was recently reported in the literature [107]. These optogenetic “magnets” overcome some of the obstacles inherent with the CRY2-CIB1 system and have already been shown to be a more efficient and powerful tool for optogenetic manipulation of transcription regulation [101].
Chemical and photo-induced epigenome editing to interrogate histone PTMs and modulate endogenous gene expression has been demonstrated in the past using older technologies such as TALE and Zinc Finger proteins [108,109]. It will be interesting to witness the repurposing of chemistry- and photo-based platforms into CRISPR-directed epigenome editing as a means to interrogate unexplored areas of chromatin biology and dynamics. Few significant limitations such as optimization of light delivery methods, availability of effectors capable of generating endogenous light sources with spatiotemporal precision, need for expensive equipment, and the use of favorable intensities and wavelengths for the activation of select photosensitive proteins and chemicals will need to be addressed as the technologies continue to develop. But promising signs of progress offer a glimpse into the bright future ahead for these chemical- and photo-inducible systems [110].
Given that optogenetic induction systems have already been used to study neural processing, behavior, and epigenetic chromatin modifications in mammalian cells [108,111,112], there is no doubt that the use of these spatio-temporal technologies coupled to epigenome editing will elucidate fundamental biological processes and causal relationships underlying human disease.
Linking Epigenome Editing to Pathology and Therapeutic Opportunities
Targeted epigenome editing can be an extremely useful tool for basic scientific research. However, for many researchers in the field, one important goal is to develop technological advances that may, in the near future, be used to combat disease and form the foundation of epigenetic therapies. We are still some time away from taking the leap into the realm of translational medicine. Until that time arrives, it is imperative that the scientific community continues to document and expand the knowledge related to the extensive catalog of pathologies associated with misregulation of the epigenome, which arise as a result of aberrant patterns of DNA and histone epigenetic modifications, as well as defects in regulation by ncRNAs.
Recent studies in animal models of human disease and human cell lines have demonstrated the promise of genome editing technologies to clinically address human pathologies brought on by viruses, such as HIV [113,114], hereditary diseases like Cystic Fibrosis and hereditary tyrosinemia type I [115,116], neurological disorders [117], and various monogenic conditions [118]. Likewise, genome editing has facilitated testing disease-associated gene disruption in large animals [119]. Despite the fact that many diseases have genetic components, we have only begun to uncover the epigenetic mechanisms underlying some of these conditions. CRISPR systems appear to be particularly well poised to make substantial contributions to shed light on epigenetic studies just as they have done in the field of genome editing. Indeed, many reports are already providing important insights into the functional roles played by epigenetic factors in a plethora of pathologies.
For example, recent publications reveal crucial roles for DNA methylation and histone PTM patterns for synaptic plasticity, learning, and memory, which have been useful to study behavioral abnormalities associated with fetal alcohol spectrum disorder [120]. Complex patterns of DNA methylation at CpG islands and other di-nucleotides within promoters have uncovered key roles for epigenetic processes in dementia and neurodegenerative diseases [121]. Studying putative functional roles for methylation patterns other than the canonical CpG methylation is important given that methyltransferases and demethylases have various degrees of specificity and have been shown to catalyze reactions at CpA and CpT di-nucleotides—both of which are poorly understood in the field [122].
Targeting the tumor suppressor BRCA1 promoter to decrease levels of DNA methylation has been shown to cause reactivation and restoration of BRCA1 functional activity in cervical and breast cancers [66]. Epigenome editing to achieve epigenetic silencing or activation could be modulated to treat active or latent viruses such as HSV1 and HIV [123]. Induction of H3K9 methylation and reduced histone H3 acetylation has shown that the human epidermal growth factor receptor-2 (HER2/neu/ERBB2), which is over expressed in many types of cancer, is amenable to down regulation [124]. Diabetic retinopathy has been associated with several epigenetic markers, including DNA methylation and demethylation of H3K4 and H3K9 [125]. Epigenetic silencing of genes that over express factors linked to protein aggregation diseases could open avenues for treatment, and slowing down the progression, of devastating neurodegenerative disorders [126].
Another key area of research where epigenome editing technologies could have a lasting impact is the study of therapeutic avenues to ameliorate imprinting disorders. Genomic imprinting is a unique type of epigenetic mechanism where differential patterns of allele-specific DNA methylation result in discrete expression of “imprinted” genes, depending on whether the paternal or maternal allele is inherited [127]. For instance, differential methylation patterns of the same locus in chromosome 15q form the underlying molecular basis for the neurogenetic disorders Prader-Willi syndrome—paternally-linked—and Angelman syndrome—maternally- linked [128]. Imprinting has been associated with DNA methylation [129,130], as well as histone PTMs [131], and ncRNAs [132]. CRISPR-mediated, allele-specific, epigenome editing offers remarkable possibilities to alter specific epimutations and epigenetic modifications at precise targeted genomic regions, thereby opening a path for rational therapeutic opportunities to correct aberrations associated with genomic imprinting disorders.
Of course, it would be impossible to list every example of the potential therapeutic benefits of targeted epigenome editing for mitigating human diseases in the context of this review. However, taken together, these findings and research paths present a promising and hopeful picture of the future of epigenetic therapies.
Beyond CRISPR-Cas9
CRISPR-Cas9 has been widely adapted in laboratories across the globe at an exponential rate for genome editing and, more recently as detailed in this piece, a tool for programmable epigenome editing. However, the future of the field will likely witness introduction of orthogonal and other repurposed systems. The bulk of published literature presently describes use of Streptococcus pyogenes nuclease-null Cas9, but other orthogonal Cas9 proteins such as those from Neisseria meningitides [24], Streptococcus thermophilus [133], Staphylococcus aureus [134], and Treponema denticola [135] have been used to demonstrate genome editing and transcriptional regulation. Researchers could avail themselves of these and other yet- uncharacterized orthogonal Cas9 proteins to interrogate the function of epigenetic marks genome-wide or at specific loci. The untapped diversity of Cas9 proteins is likely to prove invaluable as distinct Cas9s target unique, longer, and more discriminatory PAM sites. Ultimately, the advantage of targeting PAM sites other than the NGG motif characteristic of S. pyogenes Cas9 (Figure 3D) may offer alternatives to ameliorate some of the off-target effects reported in this system [136].
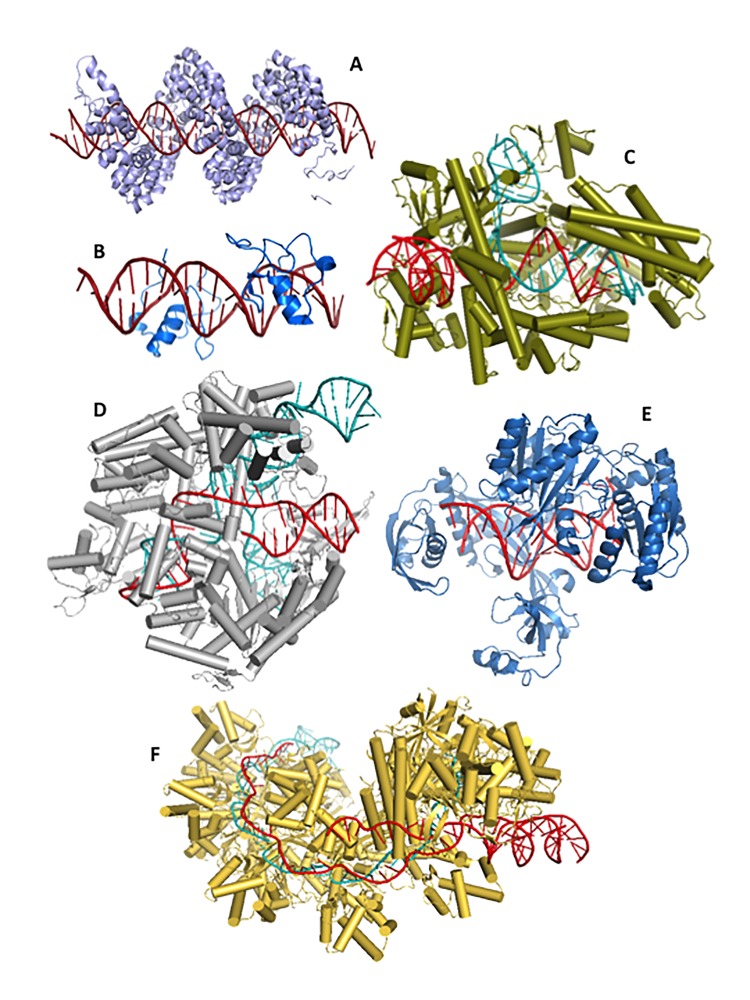
Figure 3.
Repurposing CRISPR- and non-CRISPR-based tools for epigenome editing. A. Structure of a TALE bound to its target DNA. B. Zinc Finger proteins in complex with DNA. C. Structure of Cpf1 in complex with crRNA and target DNA. D. Cas9 bound to sgRNA and PAM-containing target DNA. E. Structure of an Argonaute protein in complex with guide and target DNA fragments. F. E. coli Cascade bound to crRNA and PAM-containing dsDNA. All structures feature DNA in red and RNA (where applicable) in teal. [PDB 3UGM, 3DFX, 5B43, 4UN3, 4NCB, 5H9F].
In addition to exploiting orthogonal CRISPR-Cas9 systems for epigenome editing purposes, other CRISPR and non-CRISPR-based systems are also poised to contribute important insights to the advancement of the field. Older technologies such as Zinc Finger Nucleases (Figure 3B) and TALENs (Figure 3A) have proven valuable, and may continue to be used, as epigenome editing tools [24,137]. Newly-discovered systems, however, are likely to accelerate the rate of epigenome editing-related biotechnologies. For instance, the Cpf1 enzyme from Francisella novicida (Figure 3C) was shown in 2015 to mediate robust DNA interference distinguishable from Cas9 [138]. Orthogonal Cpf1 enzymes from Acidaminococcus sp. BV3L6 and Lachnospiraceae bacterium ND2006 are capable of mediating genome editing in human cells, suggesting that Cpf1-family proteins from diverse bacteria could also be repurposed for epigenome editing applications [138].
Moreover, the type I-E CRISPR-Cas system in Escherichia coli can be programmed to efficiently suppress gene expression by the DNA-binding Cascade complex (Figure 3F) [139,140] and could also be co-opted for epigenome editing uses. Surprisingly, the most recent addition to the list of genome editing systems in the literature was recently reported for the Natronobacterium gregoryi Argonaute endonuclease (Figure 3E—Thermus thermophilus Argonaute), which was shown to mediate DNA-guided genome editing in human cells and does not require a PAM for DNA targeting [141]. Collectively, these Cas- and non-Cas-effector enzymes, along with orthogonal and still-uncharacterized systems, may usher a new era of discoveries in basic research that could lay the foundation to propel epigenome editing into the clinical setting.
Conclusion and Future Perspectives
CRISPR systems are revolutionizing genome editing and are well-positioned to exert a similar effect in the nascent field of epigenome editing. The next generation of tools to investigate the functional and biological relevance of DNA and chromatin modifications as well as regulatory RNAs in vivo will provide scientists with a unique opportunity to study long awaited fundamental questions in chromatin biology. As the field continues to develop, it is imperative that researchers focus not merely on performing experiments to study particular epigenetic modifications with a myopic lens, but also on identifying critical obstacles that must be overcome in order to uncover the specific contributions—in local and genome-wide contexts—of writer, reader, and eraser mono- and multivalent epigenetic effectors. Likewise, as chemical- and photo-inducible epigenome editing systems become more accessible, spatio-temporal optogenetic manipulation of chromatin remodeling and epigenetic landscapes will likely provide invaluable insights into precise mechanisms for numerous biological processes including development, cellular reprogramming, and the epigenetic basis of human pathophysiology.
It is incumbent upon the scientific community to galvanize enthusiasm for using CRISPR-based technologies for designing complementary experiments that will potentially provide answers to a myriad of important questions in the field. For example, now that CRISPR systems have been established as useful tools for multiplex genome editing [142], it will be important to study the structure-function relationships of specific histone residues directly—by epigenome editing—and indirectly—by probing the epigenome with mutations of all copies of histones at the genomic level. While some progress has been made toward interrogating the function of metazoan histone residues without the use of CRISPR systems [143], it has been a difficult challenge to overcome. Although the latter approach concerns genome editing, insights from these experiments can offer valuable complementary information to those gathered through epigenome editing experiments aimed at studying the same marks from multi-faceted approaches.
Consider acetylation of the H3K14 mark. Mutating the mark to a residue that cannot be acetylated may result in a phenotype that is distinct from either rationally manipulating the acetyltransferase that deposits the mark, or purposefully preventing the locus-specific deposition of the mark. Similarly, the resulting phenotype from mutating all histones at H3K14 loci may not necessarily correlate to H3K14 acetylation alone. This is particularly true given that the histone-modifying acetyltransferase responsible for catalyzing covalent attachment of the acetyl moiety is likely to have many non-histone targets; the H3K14 site may be a cognate target for other types of modifications—e.g. methylation, butyrylation, crotonylation, ubiquitination, etc.; and there may be various readers and erasers of the mark unable to signal downstream factors and effectors. All of these scenarios—individually—are likely to mask or confound interpretation of results from a single experiment. But the collective study of these functional relationships could be used to methodically elucidate complementary roles for epigenetic marks, effectors, and other signaling factors targeting the same epigenetic locus. In short, epigenetic regulation is far more complex than its genetic counterpart and the only way to truly understand epigenetic mechanisms is through careful investigation of epigenetic processes from multiple perspectives.
Other problems that will need to be addressed in coming years relate to issues of chromatin accessibility, specificity, fidelity, and delivery of CRISPR systems repurposed for epigenome editing applications. It has been reported that nucleosomes inhibit Cas9 activity in vitro, suggesting that higher order chromatin structures are more inhibitory to Cas9 activity unless targeting stimulates chromatin remodeling at the targeted site [144]. The issues of specificity and fidelity to mitigate off-targeting effects of CRISPR-Cas systems are currently a critical area of study [145-147]. Similarly, research into optimizing delivery methods of CRISPR-based systems into target cells and tissues has paved the way for development of modern adeno-associated virus technologies and promising nanoparticle delivery approaches [148]. All these factors will play indispensable roles in determining whether CRISPR-based epigenome editing biotechnologies will successfully reach the clinical realm.
The development of new bioinformatics tools for Cas9 orthologs and other CRISPR systems is also an area that deserves attention. Currently, genome-wide off-target effects prediction is available for S. pyogenes Cas9, but other CRISPR systems are lagging behind in this regard [149]. Lastly, a combination of genome and epigenome technologies coupled to high-throughput functional genomics [150,151] to modulate transcriptional activity is likely to yield identification of much needed novel therapeutic targets for many diseases.
It will be up to the scientific community to ensure that this nascent scientific field develops into an established discipline. Already, exciting signs of progress are illustrating how epigenome editing can be used as an efficient, programmable tool to uncover biological roles of epigenomic marks and effectors in their native contexts. Undoubtedly, as detailed in this analytical assessment, epigenome editing technologies hold great promise for the future of chromatin biology and personalized medicine. But more importantly, pushing beyond the scientific frontiers of the epigenome may one day, in the near future, establish a solid foundation for the development of powerful tools to combat the suffering inherent in human disease.
Acknowledgments
I thank Robert B. Rose for comments on an earlier version of the manuscript and discussion of the figures.
Glossary
- PTMs
post-translational modifications
- ncRNAs
non-coding RNAs
- CRISPR
clustered, regularly interspaced, short palindromic repeats
- Cas9
CRISPR-associated protein 9
- ZFNs
zinc finger nucleases
- TALENs
transcription activator-like effector nucleases
- TALE
transcription activator-like effector
- HAT
histone acetyltransferase
- KRAB
Krüpel-associated box
- KAP1
KRAB-associated Protein 1
- LSD1
lysine-specific demethylase 1
- dCas9
nuclease-null, 'dead,' or 'deactivated' Cas9
- VP16
virion protein 16
- VP64
tetrameric VP16
- sgRNA
single guide RNA
- PCR
polymerase chain reaction
- ChIP-qPCR
chromatin immunoprecipitation coupled with quantitative PCR
- SUMO
small ubiquitin-like modifier
- RING
really interesting new gene
- E3
ubiquitin ligase
- E2
ubiquitin-conjugating enzyme
- PHD
plant homeodomain
- HP1
heterochromatin protein 1
- SETDB1
SET domain bifurcated 1
- NuRD
nucleosome remodeling and deacetylase complex
- ATP
adenosine triphosphate
- NCOR1
nuclear receptor corepressor 1
- TAF3
TBP-associated factor 3
- DNMT
DNA methyltransferase
- TET
ten-eleven translocation
- TET1
TET dioxygenase 1
- m5dC
5-methyldeoxycytosine
- hm5C
5-hydroxymethylcytosine
- f5C
5-formylcytosine
- ca5C
5-carboxylcytosine
- m6dA
N6-methyldeoxyadenosine
- H3K14ac
histone H3 lysine 14 acetylation
- H3K27me3
histone H3 lysine 27 trimethylation
- CRY2
cryptochrome circadian clock 2
- CIB1
cryptochrome-interaction basic helix–loop–helix 1
- HIV
human immunodeficiency virus
- HSV1
herpes simplex virus 1
- PAM
protospacer adjacent motif
- Cpf1
CRISPR from Prevotella and Francisella 1
References
- Waddington CH. Genetic Assimilation of the Bithorax Phenotype. Evolution. 1956;10(1):1–13. [Google Scholar]
- Waddington CH. The Strategy of the Genes. London: George Allen & Unwin; 1957. [Google Scholar]
- Goldberg AD. et al. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Young IT. et al. Characterization of chromatin distribution in cell nuclei. Cytometry. 1986;7(5):467–474. doi: 10.1002/cyto.990070513. [DOI] [PubMed] [Google Scholar]
- Hsu P. et al. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Qi LS. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Lassmann S. Epigenetics: development, dynamics and disease. Cell Tissue Res. 2014;356(3):451–455. doi: 10.1007/s00441-014-1916-7. [DOI] [PubMed] [Google Scholar]
- Kanwal R. et al. Cancer epigenetics: an introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- Kungulovski G, Jeltsch A. Epigenome editing: state of the art, concepts, and perspectives. Trends Genet. 2016;32(2):101–113. doi: 10.1016/j.tig.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Thakore PI. et al. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13(2):127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI. et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR. et al. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97(4):1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S. et al. Genome-scale transcriptional activation by an engineered CRISPR- Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML. et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden AW. et al. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- Kungulovski G. et al. Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenetics Chromatin. 2015;8:12. doi: 10.1186/s13072-015-0002-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 2014;42:e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA. et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12(5):401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF. et al. Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci U S A. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR. et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Groner AC. et al. KRAB-zinc finger proteins and KAP1 mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genetics. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani S. et al. Krüppel-Associated Box-Associated Protein 1 Negatively Regulates TNF-α-Induced NF-κB Transcriptional Activity by Influencing the Interactions among STAT3, p300, and NF-κB/p65. J Immunol. 2011;187:2476–2483. doi: 10.4049/jimmunol.1003243. [DOI] [PubMed] [Google Scholar]
- Shi Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Whyte WA. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;82:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz RL. et al. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274(3):1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Henry RA. et al. Differences in Specificity and Selectivity Between CBP and p300 Acetylation of Histone H3 and H3/H4. Biochemistry. 2013;52(34):5746–5759. doi: 10.1021/bi400684q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebben WR. et al. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci U S A. 2010;107(45):19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A. et al. CREB-binding Protein/p300 Activates MyoD by Acetylation. J Biol Chem. 2000;75:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- Kiernan RE. et al. HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB. et al. An Interactive Database for the Assessment of Histone Antibody Specificity. Mol Cell. 2015;59(3):502–511. doi: 10.1016/j.molcel.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. et al. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Schultz DC. et al. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Liang Q. et al. Tripartite Motif-Containing Protein 28 Is a Small Ubiquitin-Related Modifier E3 Ligase and Negative Regulator of IFN Regulatory Factor 7. J Immunol. 2011;187:4754–4763. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyret-Kahn H. et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decque A. et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016;17:140–149. doi: 10.1038/ni.3342. [DOI] [PubMed] [Google Scholar]
- Wu C-S. et al. SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev. 2014;28:1472–1484. doi: 10.1101/gad.238535.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y. et al. The Krüppel-associated box repressor domain induces reversible and irreversible regulation of endogenous mouse genes by mediating different chromatin states. Nucleic Acids Res. 2015;43:1549–1561. doi: 10.1093/nar/gkv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC. et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002 doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q. et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651. doi: 10.1038/ncomms9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y. et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Underhill C. et al. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Noon AT. et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Rowe HM. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Hu G. et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kungulovski G. et al. Application of recombinant TAF3 PHD domain instead of anti‑H3K4me3 antibody. Epigenetics Chromatin. 2016;9:11. doi: 10.1186/s13072-016-0061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM. et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA. et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H. et al. Mechanism of Stimulation of Catalytic Activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- Aoki A. et al. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JI. et al. Reprogrammable CRISPR/Cas9-based system for inducing site specific DNA methylation. Biology Open. 2016;5:866–874. doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta A. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AN. et al. Targeted Methylation and Gene Silencing of VEGF-A in Human Cells by Using a Designed Dnmt3a-Dnmt3L Single-Chain Fusion Protein with Increased DNA Methylation Activity. J Mol Biol. 2013;425:479–491. doi: 10.1016/j.jmb.2012.11.038. [DOI] [PubMed] [Google Scholar]
- Bernstein DL. et al. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Invest. 2015;125:1998–2006. doi: 10.1172/JCI77321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–112. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton J. et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem. 2012;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014;42(3):1563–1574. doi: 10.1093/nar/gkt1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C. et al. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury SR. et al. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7:46545–46556. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discovery. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RD. et al. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- Arand J. et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genetics. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Bird A. et al. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Tahiliani M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5- carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. et al. Genome-wide analysis reveals TET- and TDG-dependent 5‑methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG. et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Iurlaro M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman E. et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868–906. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. et al. N6-Methyladenine DNA Modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- Fu Y. et al. N6-Methyldeoxyadenosine Marks Active Transcription Start Sites in Chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon K, Keith J. et al. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- Kennedy TD, Lane BG. et al. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5'-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Can J Biochem. 1979;57:927–931. doi: 10.1139/o79-112. [DOI] [PubMed] [Google Scholar]
- Clancy MJ. et al. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R. et al. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ. et al. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2016;23:24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP. et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers-Ampt JH. et al. Beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Ekanayake DK. et al. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol Cell Biol. 2011;31(8):1690–1700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci U S A. 2011;108(7):2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville JJ. et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci U S A. 2016;113(11):E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170:1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Salvatierra A. et al. Crystal structure of a DNA catalyst. Nature. 2016;529:231–233. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Rinn JL. et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG. et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S. et al. Genome-scale transcriptional activation by an engineered CRISPR- Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechner DM. et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR- Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B. et al. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y. et al. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- Nihongaki Y. et al. CRISPR-Cas9-based Photoactivatable Transcription System. Chem Biol. 2015;22(2):169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. et al. A light-inducible CRISPR/Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC. et al. Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol. 2014;3(10):723–730. doi: 10.1021/sb400114p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Kawano F. et al. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- Konermann S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land BB. et al. Optogenetic inhibition of neurons by internal light production. Front Behav Neurosci. 2014;8:108. doi: 10.3389/fnbeh.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR. et al. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. J Neurosci. 2011;31(30):10829–10835. doi: 10.1523/JNEUROSCI.2246-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES. et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Hirotaka E. et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-K. et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- Schwank G. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Yin H. et al. Genome editing with Cas9 in adult mice correlates a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P. et al. Genetic correction of a LRRK2 mutation in human iPSCs links Parkinson's neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Courtney DG. et al. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108. doi: 10.1038/gt.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y. et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Basavarajappa Bs. et al. Epigenetic Mechanisms in Developmental Alcohol-Induced Neurobehavioral Deficits. Brain Sci. 2016;6:12. doi: 10.3390/brainsci6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H-U, De Jager PL. et al. Uncovering the Role of the Methylome in Dementia and Neurodegeneration. Trends Mol Med. 2016;22:687–700. doi: 10.1016/j.molmed.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH. et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Margolis DM. et al. Emerging strategies to deplete the HIV reservoir. Curr Opin Infect Dis. 2014;27:29–35. doi: 10.1097/QCO.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahi F. et al. Towards Sustained Silencing of HER2/neu in Cancer By Epigenetic Editing. Mol Cancer Res. 2013;11:1029–1039. doi: 10.1158/1541-7786.MCR-12-0567. [DOI] [PubMed] [Google Scholar]
- Kowluru RA. et al. Epigenetic Modifications and Diabetic Retinopathy. BioMed Res Int. 2013:1–9. doi: 10.1155/2013/635284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele YS. et al. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discov. 2015;14:759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JL. et al. Parental Legacy Determines Methylation and Expression of an Autosomal Transgene: A Molecular Mechanism for Parental Imprinting. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- Buiting K. et al. Sporadic imprinting defects in Prader-Willi syndrome and Angelman syndrome: Implications for imprint-switch models, genetic counseling, and prenatal diagnosis. Am J Human Genetics. 1998;63(1):170–180. doi: 10.1086/301935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS. et al. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- DeChiara TM. et al. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Ciccone DN. et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Sleutels F. et al. Imprinted silencing of Slc22a2 and Slc22a3 does not need transcriptional overlap between Igf2r and Air. EMBO J. 2003;22(14):3696–3704. doi: 10.1093/emboj/cdg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24(3):636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. et al. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Rep. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B. et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D. et al. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43(1):237–246. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML. et al. Repurposing endogenous type I CRISPR-Cas systems for programmable gene expression. Nucleic Acids Res. 2015;43(1):674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. et al. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016 doi: 10.1038/nbt.3547. [DOI] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ. et al. Interrogating the Function of Metazoan Histones using Engineered Gene Clusters. Dev Cell. 2015;32:373–386. doi: 10.1016/j.devcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM. et al. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome- wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM. et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9- induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Zuris JA. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. et al. Cas9 chromatin binding information enables more accurate CRISPR off- target prediction. Nucleic Acids Res. 2015;43:e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A. et al. Adapting CRISPR/Cas9 for functional genomics screens. Methods Enzymol. 2014;546:193–213. doi: 10.1016/B978-0-12-801185-0.00010-6. [DOI] [PubMed] [Google Scholar]
- Shalem O. et al. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Gen. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]